ABSTRACT
Small RNAs have been discovered in a wide variety of extracellular environments and are now thought to participate in communication between cells and even between different organisms and species. Helminths are parasitic worms that generally reside in extracellular niches in their hosts and can establish chronic infection through the release of immunomodulatory factors. Recent work has demonstrated that Extracellular RNA (exRNA) may be another class of immunomodulator secreted by helminths. Here we will detail what is known about small RNA pathways in helminth pathogens (focusing on nematodes) and mammalian hosts. We will then explore the computational challenges with identifying RNA-RNA interactions between 2 different species and the paradigm of RNA-RNA co-evolution that accompanies this. Finally we explore the lingering questions that require further investigation to understand the properties of exRNA that would enable it to function as an immunomodulator.
Mobile RNA: Big implications, missing mechanisms
Extracellular RNA (exRNA) is now a term used across many biologic and medical disciplines to refer to all RNA that is present outside of cells, encompassing small and large RNAs, including those that encode for proteins and those that do not. The explosion of interest in exRNA in the last decade is generally attributed to seminal papers in 2007 and 2008 detailing the movement of microRNAs and mRNAs between cells in vitro1 and the existence of microRNAs in body fluids.2-5 Since then, large initiatives have aimed to categorize “what is there” in different sample types and to try to standardize how it is assessed,6 in parallel with extensive literature touting the biomarker potential of exRNA in disease. There are fewer reports detailing exRNA functions, but a handful provide convincing evidence that RNA is functionally transported between cells in an organism. In particular, CRE/loxP tracking systems in mice demonstrated functional mRNA transfer between blood and brain7 and further proposed this as an important mechanism of cellular transformation in cancer.8,9 It is more challenging to track and test the functional transfer of non-coding RNA, but some studies using microRNA knockout mice and reporter assays make a convincing case, reviewed in.10
The implications of functional exRNA are huge: RNA moves between cells, and even between different organisms, to directly communicate information! This has been hard to fathom because we do not have a general framework for how RNA trafficking is programmed and our models for the levels of RNAs inside of cells do not account for its export and import. There are even less data to document what is required to function in the recipient cell: how do RNAs get to the right place and what concentrations have to be delivered? In the case of microRNAs (miRNAs), for example, there is not a consensus on whether these move with an Argonaute protein, or get loaded onto an Argonaute in the recipient cell10 (Fig. 1). If the latter occurs, this is hard to understand because our model for Argonaute loading involves double stranded RNA bound to Dicer: how would a single stranded small RNA get loaded? All of these missing links have kept exRNA out of the mainstream of RNA biology. Yet reports on exRNA are increasing, and RNA-binding proteins associated with exRNA are beginning to be documented, offering evidence of specificity determinants that make these processes seem more tractable.11-13
Figure 1.
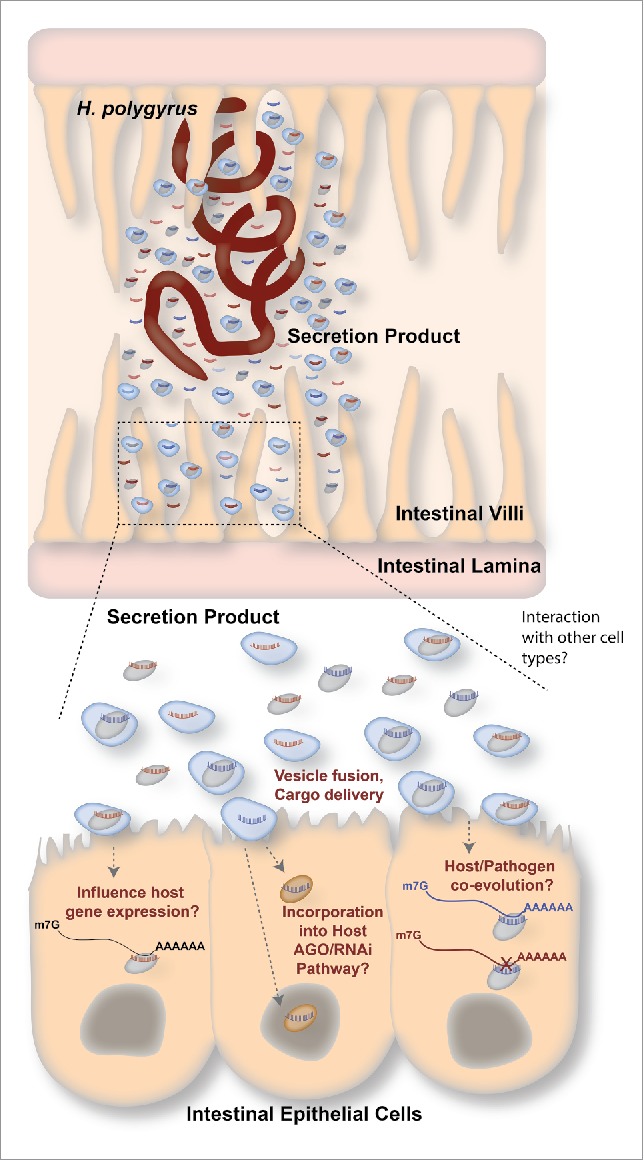
Summary of exRNA communication in a parasitic nematode model. Top: H. polygyrus is a parasitic nematode that resides in the mouse small intestine. It releases an AGO protein and various types of small RNAs in its secreted product. Some of these AGO/small RNA complexes are found within vesicles. Bottom: Extracellular vesicles and/or AGO reach the epithelial cells of the mouse intestine, or other cell types, where they may have a variety of outcomes. Left: The H. polygyrus AGO/small RNA complexes could regulate gene expression in the host (in the nucleus or cytoplasm). Middle: Small RNAs from H.polygyrus could be incorporated into host AGO/RNAi pathways, where they can be used to influence host gene expression in the nucleus or cytoplasm. Right: Over time, small RNAs and target transcripts could co-evolve, leading to optimal manipulation of host gene expression by the parasite's exRNAs. In this example, the blue transcript is capable of being recognized by the blue exRNA, but not the red exRNA because they have co-evolved.
Interestingly, the concept of mobile RNA is generally accepted in other organisms, including worms and plants, despite gaps in the knowledge of how these processes work.14 In plants mobile RNA was first described as a mechanism for spreading an antiviral response. A recent report illustrates a twist on this concept: fungal pathogens infecting plants produce mobile RNA that directly regulate plant innate immunity genes.15 Mechanistic details are lacking, but this illustrates the intriguing concept that RNA, like proteins, could be a class of immunomodulator used by extracellular pathogens. Indeed, viruses have evolved a wide range of non-coding RNA to interact with and manipulate host gene expression toward conditions that are favorable to their survival. Why wouldn't extracellular pathogens similarly use RNA for this purpose? Helminths provide an interesting system for study in this regard; these are parasitic worms (nematodes, trematodes and cestodes) that generally reside in extracellular niches in their hosts and have co-evolved with the host immune system for hundreds of millions of years. They establish chronic infections requiring parasite-encoded suppression of the immune system and this has been shown to involve active secretion of parasite factors, reviewed in.16 Research in the last 3 years has demonstrated that RNAs are present in the secretion products of diverse helminths and these are expected to be internalized by host cells through transport in extracellular vesicles17-21 (Fig. 1).
So which RNAs get secreted, in what quantities, and what do these interact with in host cells? As a framework for thinking about these questions, we detail briefly the origin and evolution of small RNAs inside of the nematode, in relation to the mammalian host, and explore the computational challenges associated with identifying RNA-RNA interactions between 2 different species.
Small RNA pathways inside nematodes and mammals
Most of what we know about the mechanisms of small RNA pathways in parasitic nematodes is inferred from extensive studies of small RNA biology in the model nematode Caenorhabditis elegans.22 Both C. elegans and mammals have the same general 3 classes of small RNA, including miRNAs, piRNAs, and endogenously produced siRNAs, but there are some striking differences in small RNA biogenesis, overall distribution, and function among these classes.23 Because piRNAs largely function in the germline and were not identified in the secretion/excretion product of the gastrointestinal nematode Heligmosomoides polygyrus17 we will not discuss them in detail here, although according to recent studies they may be present in human body fluids.24 In nematodes and mammals, miRNAs are the most abundant form of small RNA in the soma, and there is strong conservation in the miRNA pathway in terms of both biogenesis and function of the miRNAs. miRNAs are involved in cellular activities regarding development, differentiation, and disease,25 and because of their conservation and ubiquity among organisms, they are the most commonly studied for roles in extracellular communication.
Endogenous siRNAs are a broad category of small RNA that varies dramatically among organisms, both in terms of abundance and function. In mammals, siRNAs are produced by 3 predominant mechanisms: bidirectional transcription of a locus; extended hairpin molecules (distinct from miRNA); and the interaction of distinct transcripts throughout the genome that form dsRNA. Although siRNAs are not the dominant small RNA based gene regulatory pathways in mammals, they have been shown to silence protein-coding genes, pseudogenes, and transposable elements,26-28 reviewed in.29 Their general abundance is less than miRNAs across tissues, and to date, their secretion has not been reported. In nematodes, endogenous siRNAs are much more abundant than in mammals, and are found at comparable levels to miRNAs in the worm, depending on the small RNA sequencing library preparation used.30 The 22G-RNAs (named for their average length and predominant 5′ nucleotide) are synthesized by the RdRPs discretely as 22 nucleotide products, without the need for Dicer processing. Notably, while nematodes, plants and fungi generally possess orthologs of RdRPs, mammals do not. RdRP enzymes, along with the expanded and diversified set of Argonaute proteins in nematodes leads to a vast array of functional possibilities for nematode small RNA pathways and a mechanism for amplifying the silencing signal.30 These regulatory outputs include a variety of mechanisms, including transcriptional regulation via chromatin modulation, and post-transcriptional regulation via transcript degradation, translational inhibition, and transcript sequestration in RNA granules. Many of the small RNA-directed mechanisms present in nematodes are not known to exist in mammals, raising the question of whether they could operate if transferred to the host, and the minimal number of components that would be required to do so.
In terms of thinking about a host pathogen system between nematodes and mammals, which classes of small RNA might we expect to be used for extracellular communication, and how do these classes differ between organisms? The movement of RNA inside of nematodes is thought to involve endogenous small RNAs, including the 22G-RNAs. Studies of exogenously introduced double stranded RNA (dsRNA) in C. elegans clearly demonstrate that 22G-RNAs produced in the primary tissue of uptake, the gut, can be transported to other parts of the worm, where they are capable of silencing gene expression in a tissue-specific manner.31 Do we expect the classes of RNA that are involved in the spread of RNA interference-based silencing within an organism to be similar to those that are mobile between organisms? Possibly, but there is no precedent for what the targets of the 22G-RNAs would be (detailed further below).
Furthermore, even inside the nematode it is not clear how 22G-RNA production is programmed: how are target transcripts selected for the synthesis of 22G-RNAs, as some transcripts are selected to serve as templates for RdRP activity, while others are largely ignored.32 Second, how are these small RNAs (be they 22G-RNAs or miRNAs), and their cognate Argonautes, selected within parasitic cells and packaged to become exRNA-containing vesicles? Without understanding how small RNA subsets are programmed for secretion, and the quantities and mechanisms that are required for their functions inside the recipient cell, it is hard to believe that this is naturally occurring and functionally relevant. Yet identical questions exist when considering how exRNA functions in cell-to-cell communication within a single organism and mechanisms are now beginning to emerge. It seems relevant therefore to push forward similar investigations of RNA-mediated communication between different species. The added benefit, at least from a computational perspective, is that with differences at the sequence level between species, it is easier to tell which RNAs were imported. Probing RNA-RNA interactions between 2 species therefore brings with it both advantages and disadvantages compared with similar studies within just one organism.
Computational challenges of investigating RNA-mediated extracellular communication
The evidence for RNA transfer between different species ultimately comes from some form of high throughput sequencing. For example, several studies have identified nematode RNA in mammalian body fluids, reviewed in,33 and it would be expected that future studies will examine nematode RNA inside of host cells. From a computational perspective, as a starting point one needs to decide the size of RNA to be examined and the method for confidently assigning its origin. Most studies have focused on miRNAs although recent reports suggest that other classes of RNA are also there that should not be ignored: fragments from tRNAs, rRNAs, and Y RNAs in particular have been reported and postulated to be functional in some cases.17,34,35 Next, simply aligning all the sequences to the pathogen's genome to predict the exRNAs presents some major drawbacks. Many sequences are expected to be highly conserved across large phylogenetic distances, including fragments from rRNA and tRNAs. Some miRNAs, such as miR-1, miR-124 and let-7, are 100% identical between nematodes and mammals. Thus, even a perfect hit to the pathogen's genome is not sufficient proof of a foreign origin. In addition, some sequences simply by being so small, can by chance map to a genome. So, how can we confidently decide if a certain small RNA sequence was produced by one of the 2 interacting genomes? One conservative approach consists of mapping the sequences to a combination of genomes, including the 2 interactors, but also adding potential contamination sources, such as phage ϕX174 or E. coli, such that criteria can be used where only sequences that map better to one genome than the rest are kept. The next step is to think about function, and the advantage of dealing with RNA sequences is that this usually depends on RNA-RNA binding events, which in principle can be predicted.
What host molecules do pathogenic small RNAs interact with upon entering the host cells? Of all types of endogenous small RNA, more is known about how miRNAs function: in animals, they generally require perfectly complementary matches between the 6–8 nt “seed” region at their 5′ end and the 3′untranslated region (UTR) of target transcripts.36 This is likely another reason why most studies have focused on miRNAs: there are many easy-to-use tools for miRNA target prediction. But computational prediction of targets is far from perfect, and most methods show high proportions of false positives and false negatives.37 An important criterion that can improve prediction of functional miRNA targets is binding-site conservation. For endogenous miRNAs, which are important for development this makes sense, but for miRNAs coming from a pathogen it is debatable. Nevertheless, a Karposi's-sarcoma-associated herpesvirus miRNA (miR-K12–11) was shown to bind to the same sites of an endogenous miRNA (miR-155), suggesting that hijacking these sites can be a good strategy for pathogens.38 Many of the miRNAs secreted by H.polygyrus also share conserved seed sites, and in many cases share a common ancestry with those in their mammalian hosts.17 Regulating conserved endogenous sites could be effective for 2 reasons: the context of these sites already permits functional repression, and these sites cannot readily mutate to avoid pathogen regulation without disrupting the host's physiology. Nematodes could be using a similar strategy to impair host defense mechanisms.
Nevertheless, having a list of predicted targets is not equivalent to understanding the function of a miRNA. Even for a single miRNA the list can include hundreds or thousands of genes, although filtering or ranking based on which genes are expressed in the host cell-type of interest can be helpful.39 But if multiple miRNAs are considered, as is the norm when dealing with exRNA, the problem can quickly become unmanageable. The most commonly used strategy in this case is to focus on genes that are targeted by 2 (or more) miRNAs, and overlap this list with functional categories such as Gene Ontology (GO) terms, in search of functional terms that are enriched among the targets. Unfortunately, miRNA target prediction can be quite biased, due to inherent biases in the 3′UTR of genes regarding length, conservation and nucleotide composition. Thus, when overlapping targets even for randomly selected miRNAs, GO terms can appear to be significantly enriched.40 Caution should be taken before assuming that a table of enriched GO terms accurately reflects function; further validation is always desirable. In particular, biochemical approaches for identifying RNA-RNA interactions are continuing to advance,41-43 and the application of these for studying cross-species interactions is intriguing, though will likely require optimisation for low input material.
Beyond canonical miRNAs, other classes of small RNA have been identified in helminth-derived excretion-secretion products, including tRNA and Y RNA fragments,17,18 and 22G-RNAs (A. Buck, unpublished). This suggests a diverse array of pathogen-encoded molecules for which we know very little about their potential functional consequences within the host. The easiest assumption, following what we know about miRNAs and how fragments from other ncRNA can sometimes enter the miRNA pathway,44,45 is that they could be post-transcriptional silencers of gene expression. However, given the possible functions of 22G-RNAs in the nucleus in nematodes, this assumption may be false. For instance, several 22G-RNA pathways in C. elegans, including the HRDE-1, NRDE-1, and CSR-1 AGO/small RNA pathways, regulate gene expression co-transcriptionally, by influencing RNA Polymerase II activity and modulating chromatin in favor of (CSR-1) or to antagonize (HRDE-1, NRDE-1) transcription.23,46
Coevolution of small RNA/mRNA
If small RNAs are in fact part of a pathogen's arsenal, selection could act upon them, leading to detectable signals in their sequences at the population level. Although in general most mutations in a population are expected to be neutral, negative (or purifying) selection helps filter out disadvantageous mutations, while positive (or Darwinian) selection is responsible for novel advantageous mutations taking hold in the population. Thinking about pathogenic small RNAs, there are many possible scenarios, with different expectations. From the pathogen's perspective, for its own miRNAs that are important for endogenous physiology but also used as exRNA, negative selection could still be the dominant force. Positive selection could generate novel weapons for a pathogen's arsenal, and this could be more frequent among small RNAs derived from repetitive or non-coding regions in the genome. In fact, in a fungi-plant model (Botrytis cinerea and Arabidopsis thaliana) of RNA-based communication, the majority of sequences that were predicted to target host genes came from retrotransposons and intergenic regions in the fungal genome.15
Regarding the host, we have already mentioned the case of a viral miRNA potentially hijacking the sites of an important endogenous miRNA.38 If this occurs, selection is less likely to be able to act against the invading miRNA at the level of the target sites, so these would likely remain under negative selection. On the other hand, if the exRNAs bind to novel sites, hosts with otherwise neutral mutations at these sites that disrupt exRNA-binding would have an advantage over other individuals in the population, leading toward positive selection of these mutations and a potential arms-race scenario between the exRNA and its sites.
It will be intriguing to examine sequence variability within and surrounding the exRNA-producing loci in helminth populations. Selective signatures could then be calculated for different types of pathogen RNA, with a focus on those that are known to be transmitted to the host.
Outlook
Although the helminth extracellular RNA field is moving forward quickly,47 with diverse disciplines interested in its utility to understand, treat, and diagnose infection, ultimately, large gaps in our understanding remain, particularly from a basic RNA biology standpoint. In particular, there must be effective transport mechanisms to package and move sufficient quantities of RNA from the pathogen to the host and there must be mechanisms for internalization of the foreign RNA, and incorporation into a functional pathway. The discovery of exRNA-containing extracellular vesicles in helminths and the documentation of their internalization into host cells provide evidence of a transport mechanism. Yet the quantities secreted, in relation to the quantities required for function, and the mechanism(s) by which these RNAs integrate into a functional pathway in the host cell are completely unknown. Even the localization of the imported RNA has not been well studied, in any system, nor do we understand the kinetics of the exRNA signal and when it is turned over.
Although the research to date has focused largely on secreted miRNAs, the small RNA world is continually expanding. Many of the small RNA fragments that are documented to exist have not been well characterized in terms of their function inside of cells (tRNA fragments, Y RNA fragments); the meaning of their extracellular existence is therefore all the more daunting. While it has been suggested that much of the exRNA population could be degraded cellular RNA, the extracellular environment is riddled with RNases that could effectively destroy unprotected exRNA, and some of these RNases play enigmatic roles in innate immunity and defense.48 The persistence of specific stable exRNA fragments is intriguing, and the task of identifying those that might be important from those that are not is formidable. As we continue to identify the RNA-binding proteins in extracellular environments, this may advance our ability to interrogate exRNA function and understand its regulation. Finding clear answers to these and more questions related to exRNA will require multifaceted molecular and cell biology approaches along with integrated studies in both genetically tractable systems and in more natural “environmental” contexts.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors share a HFSP Young Investigator Award (RGY0069) that supports research on this topic in each laboratory. The study of H.polygyrus exRNA was also supported by WT RCDF 097394/Z/11/Z to AB.
References
- 1.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654-9; PMID:17486113; http://dx.doi.org/ 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al.. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18:997-1006; PMID:18766170; http://dx.doi.org/ 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 3.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem 2008; 54:482-90; PMID:18218722; http://dx.doi.org/ 10.1373/clinchem.2007.097972 [DOI] [PubMed] [Google Scholar]
- 4.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, et al.. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008; 141:672-5; PMID:18318758; http://dx.doi.org/ 10.1111/j.1365-2141.2008.07077.x [DOI] [PubMed] [Google Scholar]
- 5.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al.. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Nati Acad Sci U S A 2008; 105:10513-8; PMID:18663219; http://dx.doi.org/ 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill AF, Pegtel DM, Lambertz U, Leonardi T, O'Driscoll L, Pluchino S, Ter-Ovanesyan D, Nolte-'t Hoen EN. ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J Extracell Vesicles 2013; 2:22859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridder K, Keller S, Dams M, Rupp AK, Schlaudraff J, Del Turco D, Starmann J, Macas J, Karpova D, Devraj K, et al.. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol 2014; 12:e1001874; PMID:24893313; http://dx.doi.org/ 10.1371/journal.pbio.1001874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zomer A, Maynard C, Verweij FJ, Kamermans A, Schafer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SI, et al.. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015; 161:1046-57; PMID:26000481; http://dx.doi.org/ 10.1016/j.cell.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridder K, Sevko A, Heide J, Dams M, Rupp AK, Macas J, Starmann J, Tjwa M, Plate KH, Sültmann H, et al.. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology 2015; 4:e1008371; PMID:26155418; http://dx.doi.org/ 10.1080/2162402X.2015.1008371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turchinovich A, Tonevitsky AG, Burwinkel B. Extracellular miRNA: a collision of two paradigms. Trend Biochemi Sci 2016; 41:883-92; PMID:27597517; http://dx.doi.org/ 10.1016/j.tibs.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 11.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 2016; 5:e19276; PMID:27559612; http://dx.doi.org/24356509 10.7554/eLife.19276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 2013; 4:2980; PMID:24356509; http://dx.doi.org/ 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, et al.. KRAS-dependent sorting of miRNA to exosomes. eLife 2015; 4:e07197; PMID:26132860; http://dx.doi.org/ 10.7554/eLife.07197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarkies P, Miska EA. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol 2014; 15:525-35; PMID:25053358; http://dx.doi.org/ 10.1038/nrm3840 [DOI] [PubMed] [Google Scholar]
- 15.Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013; 342:118-23; PMID:24092744; http://dx.doi.org/ 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coakley G, Buck AH, Maizels RM. Host parasite communications-Messages from helminths for the immune system: parasite communication and cell-cell interactions. Mol Biochem Parasitol 2016; 208:33-40; PMID:27297184; http://dx.doi.org/ 10.1016/j.molbiopara.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, et al.. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun 2014; 5:5488; PMID:25421927; http://dx.doi.org/ 10.1038/ncomms6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowacki FC, Swain MT, Klychnikov OI, Niazi U, Ivens A, Quintana JF, Hensbergen PJ, Hokke CH, Buck AH, Hoffmann KF. Protein and small non-coding RNA-enriched extracellular vesicles are released by the pathogenic blood fluke Schistosoma mansoni. J Extracell Vesicles 2015; 4:28665; PMID:26443722; http://dx.doi.org/ 10.3402/jev.v4.28665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamanian M, Fraser LM, Agbedanu PN, Harischandra H, Moorhead AR, Day TA, Bartholomay LC, Kimber MJ. Release of small RNA-containing exosome-like vesicles from the human filarial parasite brugia malayi. PLoS Negl Trop Dis 2015; 9:e0004069; PMID:26401956; http://dx.doi.org/ 10.1371/journal.pntd.0004069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen EP, Kringel H, Williams AR, Nejsum P. Secretion of RNA-containing extracellular vesicles by the porcine whipworm, Trichuris suis. J Parasitol 2015; 101:336-40; PMID:25723295; http://dx.doi.org/ 10.1645/14-714.1 [DOI] [PubMed] [Google Scholar]
- 21.Fromm B, Trelis M, Hackenberg M, Cantalapiedra F, Bernal D, Marcilla A. The revised microRNA complement of Fasciola hepatica reveals a plethora of overlooked microRNAs and evidence for enrichment of immuno-regulatory microRNAs in extracellular vesicles. Int J Parasitol 2015; 45:697-702; PMID:26183562; http://dx.doi.org/ 10.1016/j.ijpara.2015.06.002 [DOI] [PubMed] [Google Scholar]
- 22.Zhuang JJ, Hunter CP. RNA interference in Caenorhabditis elegans: uptake, mechanism, and regulation. Parasitology 2012; 139:560-73; PMID:22075748; http://dx.doi.org/ 10.1017/S0031182011001788 [DOI] [PubMed] [Google Scholar]
- 23.Billi AC, Fischer SE, Kim JK. Endogenous RNAi pathways in C. elegans. WormBook; The C. elegans Research Community, WormBook; 2014:1-49; PMID:24816713; http://dx.doi.org/27112789 10.1895/wormbook.1.170.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R, Das S, Shah R, Danielson K, Beaulieu L, et al.. Diverse human extracellular RNAs are widely detected in human plasma. Nat Communicat 2016; 7:11106; PMID:27112789; http://dx.doi.org/ 10.1038/ncomms11106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ketting RF. microRNA biogenesis and function: An overview. Adv Exp Med Biol 2011; 700:1-14; PMID:21755468; http://dx.doi.org/ 10.1007/978-1-4419-7823-3_1 [DOI] [PubMed] [Google Scholar]
- 26.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 2006; 20:1732-43; PMID:16766679; http://dx.doi.org/ 10.1101/gad.1425706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al.. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008; 453:534-8; PMID:18404147; http://dx.doi.org/ 10.1038/nature06904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al.. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008; 453:539-43; PMID:18404146; http://dx.doi.org/ 10.1038/nature06908 [DOI] [PubMed] [Google Scholar]
- 29.Piatek MJ, Werner A. Endogenous siRNAs: regulators of internal affairs. Biochem Soc Transact 2014; 42:1174-9; PMID:25110021; http://dx.doi.org/ 10.1042/BST20140068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claycomb JM. Ancient endo-siRNA pathways reveal new tricks. Curr Biol 2014; 24:R703-15; PMID:25093565; http://dx.doi.org/ 10.1016/j.cub.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 31.Hunter CP, Winston WM, Molodowitch C, Feinberg EH, Shih J, Sutherlin M, Wright AJ, Fitzgerald MC. Systemic RNAi in Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol 2006; 71:95-100; PMID:17381285; http://dx.doi.org/ 10.1101/sqb.2006.71.060 [DOI] [PubMed] [Google Scholar]
- 32.Youngman EM, Claycomb JM. From early lessons to new frontiers: the worm as a treasure trove of small RNA biology. Front Genet 2014; 5:416; PMID:25505902; http://dx.doi.org/ 10.3389/fgene.2014.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintana JF, Babayan SA, Buck AH. Small RNAs and extracellular vesicles in filarial nematodes: from nematode development to diagnostics. Parasite Immunol 2016; PMID:27748953; http://dx.doi.org/24812611 10.1111/pim.12395 [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Silva MR, Cabrera-Cabrera F, das Neves RF, Souto-Padron T, de Souza W, Cayota A. Gene expression changes induced by Trypanosoma cruzi shed microvesicles in mammalian host cells: relevance of tRNA-derived halves. BioMed Res Internat 2014; 2014:305239; PMID:24812611; http://dx.doi.org/ 10.1155/2014/305239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambertz U, Oviedo Ovando ME, Vasconcelos EJ, Unrau PJ, Myler PJ, Reiner NE. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Gen 2015; 16:151; PMID:25764986; http://dx.doi.org/ 10.1186/s12864-015-1260-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics 2009; 25:3049-55; PMID:19789267; http://dx.doi.org/ 10.1093/bioinformatics/btp565 [DOI] [PubMed] [Google Scholar]
- 38.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al.. A viral microRNA functions as an orthologue of cellular miR-155. Nature 2007; 450:1096-9; PMID:18075594; http://dx.doi.org/ 10.1038/nature05992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ovando-Vazquez C, Lepe-Soltero D, Abreu-Goodger C. Improving microRNA target prediction with gene expression profiles. BMC Gen 2016; 17:364; PMID:27189211; http://dx.doi.org/ 10.1186/s12864-016-2695-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bleazard T, Lamb JA, Griffiths-Jones S. Bias in microRNA functional enrichment analysis. Bioinformatics 2015; 31:1592-8; PMID:25609791; http://dx.doi.org/ 10.1093/bioinformatics/btv023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Commun 2015; 6:8864; PMID:26602609; http://dx.doi.org/ 10.1038/ncomms9864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen TC, Cao X, Yu P, Xiao S, Lu J, Biase FH, Sridhar B, Huang N, Zhang K, Zhong S. Mapping RNA-RNA interactome and RNA structure in vivo by MARIO. Nat Commun 2016; 7:12023; PMID:27338251; http://dx.doi.org/ 10.1038/ncomms12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma E, Sterne-Weiler T, O'Hanlon D, Blencowe BJ. Global Mapping of Human RNA-RNA Interactions. Mol Cell 2016; 62:618-26; PMID:27184080; http://dx.doi.org/ 10.1016/j.molcel.2016.04.030 [DOI] [PubMed] [Google Scholar]
- 44.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell 2008; 32:519-28; PMID:19026782; http://dx.doi.org/ 10.1016/j.molcel.2008.10.017 [DOI] [PubMed] [Google Scholar]
- 45.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A 2013; 110:1404-9; PMID:23297232; http://dx.doi.org/ 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wedeles CJ, Wu MZ, Claycomb JM. Silent no more: Endogenous small RNA pathways promote gene expression. Worm 2014; 3:e28641; PMID:25254148; http://dx.doi.org/ 10.4161/worm.28641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siles-Lucas M, Morchon R, Simon F, Manzano-Roman R. Exosome-transported microRNAs of helminth origin: new tools for allergic and autoimmune diseases therapy? Parasite Immunol 2015; 37:208-14; PMID:25712154; http://dx.doi.org/ 10.1111/pim.12182 [DOI] [PubMed] [Google Scholar]
- 48.Koczera P, Martin L, Marx G, Schuerholz T. The ribonuclease a superfamily in humans: canonical rnases as the buttress of innate immunity. Int J Mol Sci 2016; 17.(8). pii:E1278 [DOI] [PMC free article] [PubMed] [Google Scholar]


