MAP18 physically interacts with ROP2 and competes with AtRhoGDI1/SCN1 for binding to ROP2 during root hair growth in Arabidopsis.
Abstract
Establishment and maintenance of the polar site are important for root hair tip growth. We previously reported that Arabidopsis (Arabidopsis thaliana) MICROTUBULE-ASSOCIATED PROTEIN18 (MAP18) functions in controlling the direction of pollen tube growth and root hair elongation. Additionally, the Rop GTPase ROP2 was reported as a positive regulator of both root hair initiation and tip growth in Arabidopsis. Both loss of function of ROP2 and knockdown of MAP18 lead to a decrease in root hair length, whereas overexpression of either MAP18 or ROP2 causes multiple tips or a branching hair phenotype. However, it is unclear whether MAP18 and ROP2 coordinately regulate root hair growth. In this study, we demonstrate that MAP18 and ROP2 interact genetically and functionally. MAP18 interacts physically with ROP2 in vitro and in vivo and preferentially binds to the inactive form of the ROP2 protein. MAP18 promotes ROP2 activity during root hair tip growth. Further investigation revealed that MAP18 competes with RhoGTPase GDP DISSOCIATION INHIBITOR1/SUPERCENTIPEDE1 for binding to ROP2, in turn affecting the localization of active ROP2 in the plasma membrane of the root hair tip. These results reveal a novel function of MAP18 in the regulation of ROP2 activation during root hair growth.
Root hairs function in water and nutrient uptake, interaction with microbes, and anchoring plants in the soil (Dolan et al., 1994; Ketelaar and Emons, 2001, 2009; Libault et al., 2010). Root hairs initiate at defined sites in trichoblasts and root hair-forming cells in the root epidermis, and their growth takes place exclusively at the tip. Tip growth characteristics of root hairs are similar to those of pollen tubes, fungal hyphae, moss protonema, and neurites. Under laboratory conditions, root hairs are dispensable for plant growth, enabling the isolation and characterization of root hair mutants. The availability of genetic analysis together with the relatively simple methodology required to image root hair development and intracellular components make root hairs an attractive model system for studying tip growth (Foreman and Dolan, 2001; Carol and Dolan, 2002; Cole and Fowler, 2006; Lee and Yang, 2008). Unidirectional root hair growth depends upon tip-focused calcium and reactive oxygen species gradients, intracellular pH gradient, cytoskeleton dynamics, and vesicle trafficking to deliver new membrane and cell wall materials to the growing tip (Ketelaar and Emons, 2001, 2009; Carol and Dolan, 2002; Carol et al., 2005; Ovečka et al., 2005; Diet et al., 2006; Jones et al., 2007; Takeda et al., 2008; Bloch et al., 2011; Pei et al., 2012; Yoo et al., 2012). Accumulating evidence has implicated Rho-related GTPases from plants (ROPs) in regulating signaling networks in the orchestration of ion gradients, cytoskeleton dynamics, and vesicle trafficking during root hair growth and development (Kost, 2008; Lee and Yang, 2008; Yalovsky et al., 2008).
Arabidopsis (Arabidopsis thaliana) ROPs include a protein family of 11 members that have been divided into either four subgroups named types I, II, III, and IV by phylogenetic analysis (Yang, 2002) or into two subgroups called type I and type II based on the amino acid sequences of their C-terminal hypervariable domains (Winge et al., 2000). Previous studies have shown that several ROPs can affect root hair growth (Molendijk et al., 2001; Jones et al., 2002; Bloch et al., 2005; Sorek et al., 2010). ROP2, ROP4, and ROP6 were shown to accumulate at the root hair initiation site (Molendijk et al., 2001; Jones et al., 2002; Payne and Grierson, 2009). Overexpression of constitutively active (CA) mutant forms of ROP2, ROP4, ROP6, and ROP11 induced isotropic growth, resulting in balloon-shaped root hairs (Molendijk et al., 2001; Jones et al., 2002; Bloch et al., 2005). Overexpression of ROP2 induced the formation of additional root hairs and root hair branching, implicating ROP2 in root hair differentiation (Molendijk et al., 2001; Jones et al., 2002). Overexpressing a dominant negative (DN) mutant of ROP2 resulted in tip growth inhibition (Jones et al., 2002). ROP2 expression led to the activation of NADPH oxidases AtRbohC/ROOT HAIR-DEFECTIVE2 that generate reactive oxygen species, which in turn stimulate calcium ion influx into the cytoplasm and change calcium ion gradients. Changes in intracellular Ca2+ levels have been suggested to regulate actin dynamics (Molendijk et al., 2001; Jones et al., 2002, 2007; Carol et al., 2005; Lee et al., 2008; Takeda et al., 2008). ROP11 and ROP6 also were shown to inhibit endocytosis in root hairs in an actin-independent manner (Bloch et al., 2005; Sorek et al., 2010).
As a binary molecular switch that transmits extracellular signals to intracellular biological events, the rapid conversion of ROPs between the GTP-bound form (active) and the GDP-bound form (inactive) enables a rapid response to cellular processes (Nibau et al., 2006; Yang and Fu, 2007). Three major classes of regulatory proteins, guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (GDIs), play important roles in regulating the activities of the RHO family of proteins in various cells (Gu et al., 2004; Klahre and Kost, 2006; Fu et al., 2008; Kost, 2008; Nagawa et al., 2010; Cherfils and Zeghouf, 2013). RhoGDIs have long been considered negative regulators that function by sequestering inactive Rho proteins within the cytoplasm. Recently, RhoGDIs also were implicated in the recycling of inactive Rho protein from the plasma membrane (PM) and providing protection from degradation in the cytosol (Klahre et al., 2006; Fu et al., 2008; Lee and Yang, 2008; Boulter et al., 2010; Boulter and Garcia-Mata, 2010). Translocation and activation of ROP2 could be inhibited by coexpression with RhoGDI1 in guard cells (Jeon et al., 2008). Hwang et al. (2010) demonstrated that the global inhibition of ROP1 activity by RopGAP1 and RhoGDI1 could balance the lateral propagation of apical ROP1 activation induced by overexpressing ROP1 in pollen tubes. NtRhoGDI2 has a role in recycling NtRac5 to the apex to maintain the polarity of ROP signaling and pollen tube growth (Klahre et al., 2006). Three Arabidopsis GDIs (RhoGDI1, RhoGDI2a, and RhoGDI2b) have been suggested to antagonistically interact with phosphatidylinositol 4,5-bisphosphate in cellular homeostasis and the maintenance of pollen tube growth through ROP signaling (Feng et al., 2016). AtRhoGDI1/SUPERCENTIPEDE1 (SCN1) mutants have trichoblasts with multiple root hair initiation tips, indicating that AtRhoGDI1/SCN1 spatially restricts the site of root hair growth to a single spot (Carol et al., 2005). The multiple root hair phenotype of scn1 confirms the role of RhoGDI-dependent Rho recycling in the maintenance of cell polarity (Marée et al., 2006; Jilkine et al., 2007).
MICROTUBULE-ASSOCIATED PROTEIN18 (MAP18), also known as PLASMA MEMBRANE-ASSOCIATED CA2+ BINDING PROTEIN2 (PCaP2), was first identified as a microtubule-associated protein that contributes to cortical microtubule organization and the regulation of polarized diffuse growth in vegetative tissues (Wang et al., 2007). MAP18 contains the developmentally regulated plasma membrane protein (DREPP) domain and belongs to the DREPP family (Logan et al., 1997). Recently, we found that MAP18 plays a key role in modulating actin filaments in growing cells at the tip, thereby regulating pollen tube growth direction (Zhu et al., 2013) as well as the proper positioning of the nucleus in growing root hairs (Zhang et al., 2015). Either down-regulation or overexpression of MAP18 could lead to the disorganization of actin filaments in the apex and subapical regions of root hairs, resulting in aberrant root hair growth patterns (Zhang et al., 2015). Similarly, MAP18/PCaP2 has been shown to be expressed in root hairs and affects development (Kato et al., 2010, 2013).
Genetic analyses have indicated that ROP2 and MAP18 may function in the same pathway or coregulate similar signaling processes. ROP2 loss-of-function mutants and map18 knockdown mutants have shorter root hairs (Jones et al., 2002; Zhang et al., 2015). Furthermore, overexpression of MAP18 or ROP2 induced the formation of multiple root hair tips or root hair branching (Jones et al., 2002; Zhang et al., 2015). However, whether and how MAP18 and ROP2 coordinately regulate root hair polarity and growth remain unclear. In this study, we demonstrate that MAP18 and ROP2 genetically and functionally interact to affect root hair development. Furthermore, we prove that MAP18 preferably binds to the inactive form of ROP2 via its N-terminal end, leading to ROP2 activation. We further demonstrate that MAP18 interferes in the interaction between inactive ROP2 and AtRhoGDI1/SCN1. Collectively, this study reveals a novel function of MAP18 in the regulation of ROP2 activity and root hair growth.
RESULTS
MAP18 Genetically and Functionally Interacts with ROP2 in Root Hairs
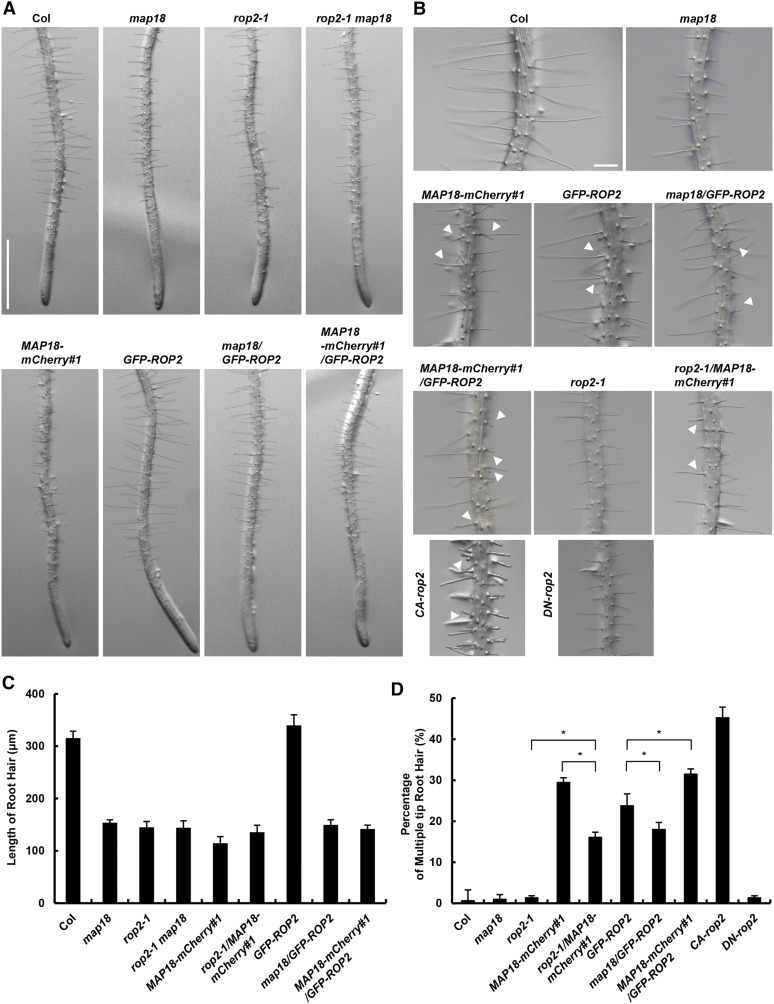
To elucidate whether MAP18 and ROP2 coordinately regulate root hair tip growth, we first compared the root hair phenotypes of the map18 knockdown mutant (SALK_021652; Zhu et al., 2013), the rop2-1 knockout mutant (SALK_055328; Jeon et al., 2008), and the rop2-1 map18 double mutant. Consistent with our previous report (Zhang et al., 2015), map18 seedlings exhibited short root hairs, with an average length of about half of the wild-type length (Fig. 1, A and C). Similarly, the average length of rop2-1 root hairs was reduced significantly (Fig. 1, A and C). Next, we generated a rop2-1 map18 double mutant to examine the genetic interaction between MAP18 and ROP2. The rop2-1 map18 double mutant seedlings also exhibited short root hairs that were significantly shorter than the wild-type hairs but not significantly different from those of map18 and rop2-1 single mutants (Fig. 1, A and C). The nonadditive phenotype of the rop2-1 map18 double mutant suggests that MAP18 and ROP2 might function in the same pathway.
Figure 1.
MAP18 genetically interacts with ROP2. Representative images of root hair morphologies of various 5-d-old seedlings are shown. A, The map18, rop2-1, and rop2-1 map18 mutants have obvious shorter root hairs compared with wild-type Columbia-0 (Col) seedlings. The root hairs of GFP-ROP2 are longer than wild-type hairs, while MAP18-mCherry#1, map18/GFP-ROP2, and MAP18-mCherry#1/GFP-ROP2 have shorter root hairs. Bar = 1 mm. B, The branching root hair phenotype in various lines. White arrowheads indicate the branching tips. MAP18-mCherry#1, GFP-ROP2, MAP18-mCherry#1/GFP-ROP2, map18/GFP-ROP2, and rop2-1/MAP18-mCherry#1 all exhibited obvious multiple tips or branching root hairs, and CA-rop2 had more multiple tips or branching root hairs, whereas root hair branching was hardly detected in the wild type, map18, and DN-rop2. Bar = 200 μm. C, Quantitative analysis of the average root hair length of various lines. The root hair length of the rop2-1 map18 double mutant was as short as that of the rop2-1 and map18 single mutants. Overexpression of GFP-ROP2 caused longer root hairs, but there were shorter root hairs in map18/GFP-ROP2. D, Quantitative analysis of multiple-tip root hairs in various lines. The percentage of branched root hairs was higher in GFP-ROP2 than in the wild type. However, the frequency of branched root hairs was decreased in map18/GFP-ROP2 and increased in MAP18-mCherry#1/GFP-ROP2 lines compared with GFP-ROP2 plants. The percentage of branched root hairs was higher in MAP18-mCherry#1 than in rop2-1/MAP18-mCherry#1. More than 150 root hairs from at least 20 growing roots for each line were measured. Data are presented as means ± sd. Asterisks indicate significant differences at P < 0.05, by ANOVA.
To further elucidate the functions of MAP18 and ROP2 during root hair growth, we analyzed the effects of GFP-ROP2 (Supplemental Fig. S1A) and MAP18 (MAP18-mCherry#1) overexpression (Supplemental Fig. S2) on root hair morphology. Consistent with previously reported data on overexpressing ROP2 (Jones et al., 2002) and MAP18 (Zhang et al., 2015) lines, GFP-ROP2 and MAP18-mCherry overexpression led to the formation of multiple root hair tips and root hair branching, respectively (Fig. 1B, white arrowheads). The phenotypes induced by either ROP2 or MAP18 suggest that they both regulate root hair polarity growth and elongation. Interestingly, the expression of constitutively active ROP2 (CA-rop2) induced a higher number of multiple root hair initiation tips or root hair branches, whereas the expression of a dominant negative ROP2 (DN-rop2) significantly inhibited root hair elongation but did not cause root hair branching (Fig. 1, B and D). This evidence indicates that ROP2 activity is critical for both root hair elongation and polarity maintenance, which are two different (although related) processes during root hair growth and development.
We then introduced GFP-ROP2 into a map18 mutant or MAP18-mCherry#1 plants by crossing (Fig. 1A). While GFP-ROP2 root hairs were longer than wild-type hairs (Fig. 1C), map18/GFP-ROP2 root hairs were significantly shorter and displayed an average length that was about half that of wild-type root hairs (Fig. 1, B and C). In addition, while map18/GFP-ROP2 seedlings still developed branched root hairs, the percentage was significantly lower compared with GFP-ROP2 (Fig. 1D). MAP18-mCherry#1/GFP-ROP2 had more branched/multiple root hairs at the tip than GFP-ROP2 plants (Fig. 1D). These results indicate that MAP18 is involved in ROP2 function in root hair growth, both in elongation and maintaining the growth point. We introduced MAP18-mCherry#1 into a rop2-1 mutant by crossing (Fig. 1B). The frequency of root hairs with branched/multiple tips in rop2-1/MAP18-mCherry#1 was lower than in MAP18-mCherry#1 seedlings but higher than in rop2-1 and the wild type (Fig. 1D). These data suggest that the effect of MAP18 on maintaining the growth point may be partially dependent on ROP2. Interestingly, overexpression of MAP18 in a wild-type background (MAP18-mCherry#1), in a ROP2 overexpression background (MAP18-mCherry#1/GFP-ROP2), or in a rop2-1 mutant (rop2-1/MAP18-mCherry#1) all led to significantly shorter root hairs compared with wild-type plants (Fig. 1, A–C), which suggests that a ROP2-independent mechanism also was involved, and the effect of MAP18 on root hair elongation was partially dependent on ROP2 as well.
We next examined whether the genetic interactions between MAP18 and ROP2 result from a physical interaction between MAP18 and ROP2 proteins.
Physical Interaction between MAP18 and ROP2 in Vitro and in Vivo
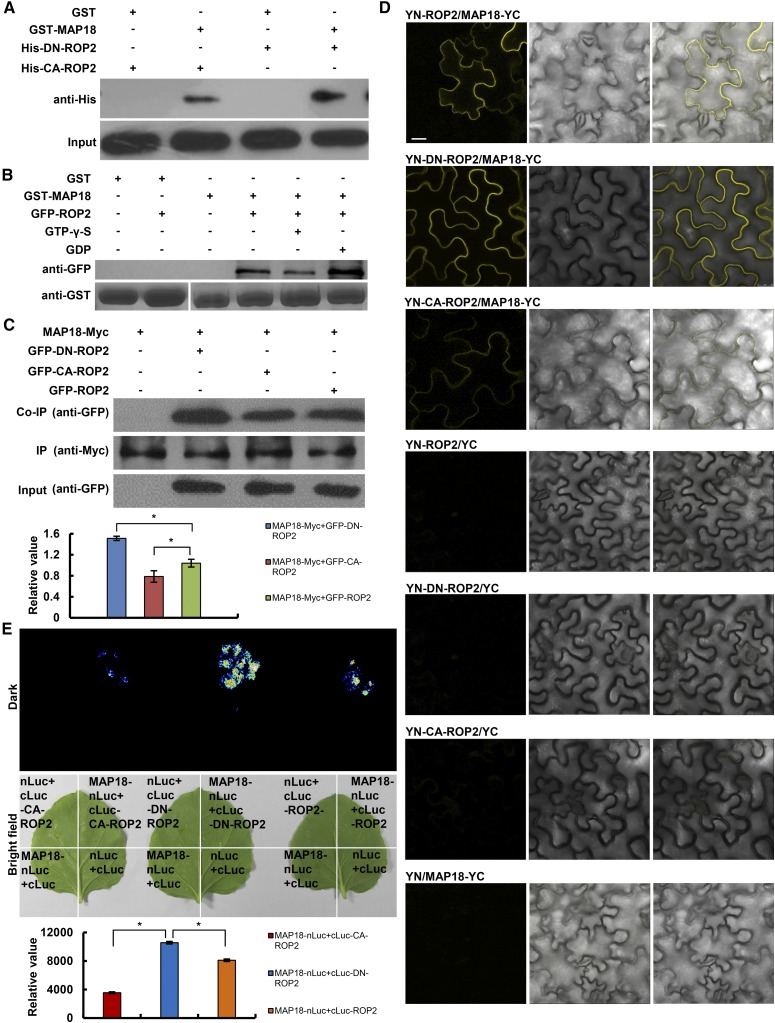
To study the mechanistic basis for the functional interactions between MAP18 and ROP2, we examined whether the proteins could interact physically with each other. In vitro pull-down assays with Escherichia coli expressing recombinant proteins demonstrated that GST-MAP18 preferentially interacted with a poly-His-tagged dominant negative ROP2 mutant harboring a D121A mutation (DN-ROP2) compared with constitutively active CA-ROP2 (G15V; Fig. 2A). Next, we performed pull-down assays with total protein extracts prepared from GFP-ROP2 plants (Fig. 2B). To further examine the relationship between ROP2 activation status and its interaction with MAP18, GDP or GTP-γ-S (a nonhydrolyzable analog of GTP) was included in the pull-down reaction mixture. The interaction between GST-MAP18 and GFP-ROP2 was stronger in the presence of GDP than in the presence of GTP-γ-S (Fig. 2B), which is in agreement with data showing that GST-MAP18 had a stronger interaction with DN-ROP2 than with CA-ROP2 (Fig. 2A). Next, we performed a coimmunoprecipitation (Co-IP) assay using an Arabidopsis protoplast expressing MAP18-Myc and GFP-ROP2, GFP-DN-ROP2, or GFP-CA-ROP2. MAP18-Myc coimmunoprecipitated with the wild type and with constitutively active and dominant negative ROP2 variants. In agreement with the data from the pull-down assay (Fig. 2, A and B), the interaction with DN-ROP2 was significantly stronger (Fig. 2C).
Figure 2.
Analysis of the interaction between MAP18 and ROP2. A, In vitro binding assays were performed using recombinant E. coli-expressed GST-MAP18 and ROP2 variants. Pull down of His-CA-ROP2 or His-DN-ROP2 by GST-MAP18 was detected by anti-His antibodies. The bottom gel shows that similar amounts of His-CA-ROP2 and His-DN-ROP2 were used in the binding assays. Nonfused GST was used as a negative control. B, GFP-ROP2 proteins extracted from transgenic seedlings were pulled down by GST-MAP18. Total protein extracted from GFP-ROP2 seedling roots were preloaded with GTP-γ-S or GDP for pull-down assays. Anti-GFP antibody was used for detection. The bottom gel shows input bait proteins detected by anti-GST antibody, and similar amounts of extracted proteins were used in the binding assays. Nonfused GST was used as a negative control. C, Co-IP analysis shows that MAP18-Myc protein could interact with GFP-ROP2, GFP-DN-ROP2, or GFP-CA-ROP2 in vivo. Three biological repeats were performed, and one representative result is shown. The histogram indicates the ratios of GFP-DN-ROP2/CA-ROP2/ROP2 proteins to MAP18-Myc protein levels. The interaction of MAP18-Myc with DN-ROP2 was significantly stronger than that with GFP-ROP2 or GFP-CA-ROP2. Data are presented as mean values from three independent experiments ± sd. Asterisks indicate significant differences at P < 0.05, by ANOVA. D, Interaction between MAP18 and different forms of ROP2 tested using the BiFC assay. Images are from N. benthamiana leaves transformed with different combinations of pSPYCE and pSPYNE constructs. Left to right columns show YFP (BiFC) fluorescence, corresponding bright-field images, and merged images. Bar = 10 μm. E, Interaction between MAP18 and different forms of ROP2 tested using the LCI assay. Fluorescence and bright-field images were collected from N. benthamiana leaves transformed with various combinations of vectors as indicated. nLuc+cLuc-ROP2/CA-ROP2/DN-ROP2, MAP18-nLuc+cLuc, and nLuc+cLuc were used as negative controls. Relative fluorescence values (bottom) show that the binding capacity of MAP18 with DN-ROP2 was more than that with ROP2 and CA-ROP2. Data are presented as means ± sd based on three independent experiments. Asterisks indicate significant differences at P < 0.05, by ANOVA.
The interaction between MAP18 and ROP2 also was confirmed in vivo by bimolecular fluorescence complementation (BiFC). We fused the C-terminal half of eYFP (YC) to the C terminus of MAP18 (MAP18-YC) and fused the N-terminal half of YFP (YN) to the N terminus of ROP2, DN-ROP2, or CA-ROP2 (YN-ROP2, YN-DN-ROP2, or YN-CA-ROP2). MAP18-YC was transiently coexpressed with YN-ROP2, YN-DN-ROP2, YN-CA-ROP2, or empty vector controls in Nicotiana benthamiana leaf epidermis cells. Reconstituted YFP fluorescence signals were detected in cells that coexpressed MAP18 with either ROP2 or DN-ROP2 but were detected weakly in cells that coexpressed MAP18 and CA-ROP2. No YFP signal was detected in the negative controls (Fig. 2D). Additionally, a luciferase complementation imaging (LCI) assay also was performed by transient expression in N. benthamiana leaves, revealing a stronger interaction between MAP18 and DN-ROP2 in vivo compared with ROP2 or CA-ROP2 (Fig. 2E). In vitro and in vivo protein interaction assays indicate that MAP18 can interact physically with ROP2, preferentially with the inactive form of the GTPase.
Preferential binding of MAP18 to the inactive form of ROP2 suggests that it possibly functions as a regulator rather than a downstream effector of ROP2.
MAP18 Activates ROP2 Function during Root Hair Tip Growth
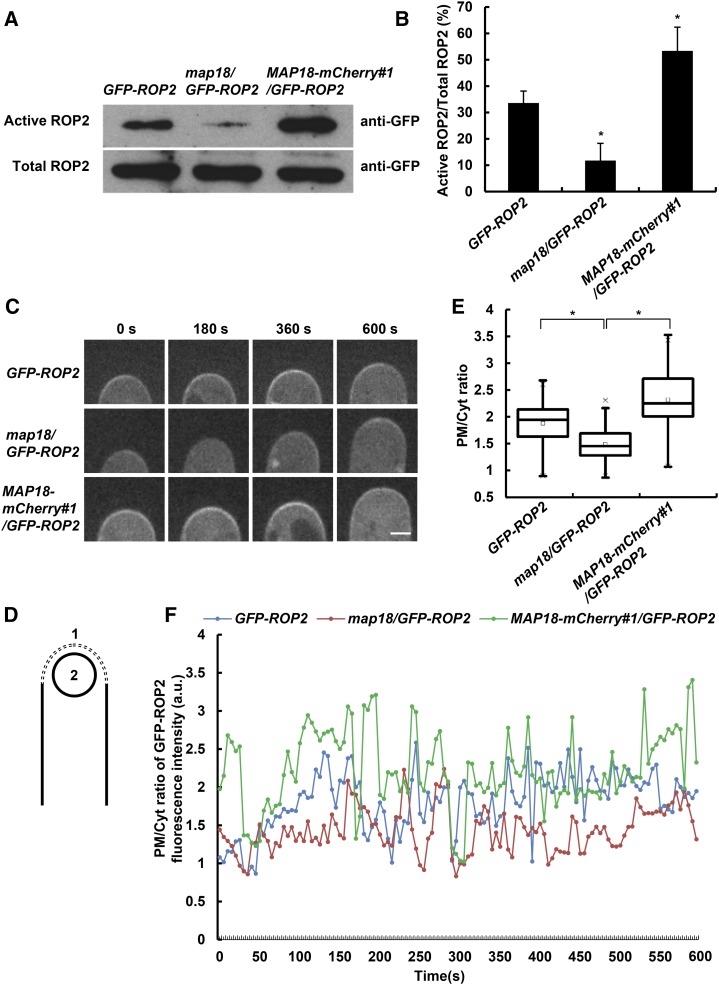
It has been shown that levels of activated Rho GTPases can be estimated by comparing levels of the GTPase that are pulled down with an effector-binding domain fused to GST, such as GST-PBD (PAK-binding domain), with their overall levels (Pellegrin and Mellor, 2008). We utilized a modification of this method using the ROP effector RIC1 fused to maltose-binding protein (MBP-RIC1) according to Lin et al. (2012) to examine whether MAP18 regulates ROP2 activation status. ROP2 activation assays were performed with plant protein extracts from roots containing GFP-ROP2. The amount of GFP-ROP2 that precipitated with MBP-RIC1 was lower when GFP-ROP2 was prepared from map18 mutants and was higher when protein extracts were prepared from MAP18-mCherry-overexpressing plants (MAP18-mCherry#1; Fig. 3, A and B). The MBP-RIC1 binding assays suggest that MAP18 is important for ROP2 activation.
Figure 3.
MAP18 positively regulates ROP2 activity. A, Total GFP-ROP2 extracted from transgenic seedling roots and active GFP-ROP2 pulled down by MBP-RIC1. Five independent experiments were performed and similar results were obtained; a representative data set is shown. B, Quantitative analysis of data from A. ROP2 activity level was determined as the amount of GTP-bound ROP2 versus the amount of total GFP-ROP2. ROP2 activity decreased significantly in map18/GFP-ROP2 and increased in MAP18-mCherry#1/GFP-ROP2 lines. Data are mean values from five independent experiments ± sd. Asterisks indicate significant differences at P < 0.05, by ANOVA. C, Time-lapse images of growing root hairs expressing GFP-ROP2 in the wild-type, map18, or MAP18-mCherry#1 background. PM-associated GFP-ROP2 at the growing root hair tip was weakened in map18 but enhanced in MAP18-mCherry#1. Bar = 5 μm. D, Cartoon depicting how the fluorescence intensity of the PM/Cyt ratio was measured. A dashed line was drawn along the apical PM of the root hair (region 1), and a circle was drawn in the cytosol of the root hair apex (region 2). E, Quantitative analysis of GFP-ROP2 fluorescence intensity (PM/Cyt ratio) in wild-type, map18 mutant, and MAP18-mCherry#1 root hairs. Data were collected from 50 root hairs from 15 seedlings per data set. Data are presented as box plots that reflect 25%, 50%, 75%, and the maximum/minimum of total values. Asterisks indicate significant differences at P < 0.05, by ANOVA. F, Representative result showing that the PM/Cyt ratio of GFP-ROP2 fluorescence intensity oscillated during root hair growth. The PM/Cyt ratio of GFP-ROP2 signal was higher in MAP18-mCherry#1/GFP-ROP2 lines but lower in map18/GFP-ROP2 lines compared with GFP-ROP2. a.u., Arbitrary units.
The polar accumulation of ROPs in trichoblasts is correlated with local auxin gradients and precedes root hair initiation (Fischer et al., 2006). Root hair tip PM ROP2 localization persists throughout root hair tip growth (Jones et al., 2002). To further substantiate the relationship between ROP2 activation and MAP18, we tracked the dynamics of PM-localized GFP-ROP2 in growing root hairs of a map18 mutant or MAP18-overexpressing plants using spinning-disk confocal microscopy. In agreement with the published data (Jones et al., 2002; Fischer et al., 2006), GFP-ROP2 was associated with the apical PM in growing root hairs (Fig. 3C) and absent from the tips of fully grown root hairs (Supplemental Fig. S1B). In addition, the short root hair phenotype of rop2-1 also was rescued by the expression of ROP2pro::GFP-ROP2 (ROP2pro::GFP-ROP2/rop2 #1 line, in which the relative expression level of ROP2 was similar to that of the wild type), indicating that ROP2 fused to GFP could functionally replace endogenous ROP2 (Supplemental Fig. S1, C–E). Next, a time series of GFP-ROP2 localization at the PM was acquired. The fluorescence intensity of GFP-ROP2 at the PM was lower in the map18 mutant and higher in MAP18-mCherry-overexpressing plants (Fig. 3C). We analyzed the plasma membrane/cytosol (PM/Cyt) ratio of GFP-ROP2 fluorescence intensity at the apical region of growing root hairs (schematic diagram shown in Fig. 3D). Compared with control GFP-ROP2 plants, the average PM/Cyt ratio of GFP-ROP2 signals was significantly higher in MAP18-mCherry#1/GFP-ROP2 root hairs, which suggests an increase of ROP2 activity by overexpressing MAP18. This is consistent with the phenotype shown in Figure 1: MAP18-mCherry#1/GFP-ROP2 has more branched/multiple-tip root hairs than GFP-ROP2 plants (Fig. 1, B and D). The average PM/Cyt ratio of GFP-ROP2 signals was lower in map18/GFP-ROP2 root hairs (Fig. 3E). This indicates a decrease of ROP2 activity when MAP18 was down-regulated, resulting in shorter root hairs and a lower frequency of branched root hairs compared with control GFP-ROP2 plants (Fig. 1). To further support this conclusion, we tracked 30 growing root hairs for a period of 10 min each and observed that the GFP-ROP2 signal oscillated over time in all root hairs. We analyzed the PM/Cyt ratio of GFP-ROP2 signals at each time point, with a representative result shown in Figure 3F. In general, the PM/Cyt ratio was mostly higher in MAP18-mCherry#1/GFP-ROP2 root hairs but lower in map18/GFP-ROP2 root hairs compared with control GFP-ROP2 root hairs.
A GFP-tagged deletion mutant of the ROP effector RIC4 (RIC4ΔC-GFP) has been used to monitor ROP activity in pollen tubes. RIC4ΔC-GFP expression did not interfere with pollen tube growth, and membrane localization was correlated with ROP activity (Hwang et al., 2005). We adopted RIC4ΔC-GFP as a marker for active ROP2 in growing root hairs by introducing it into wild-type and map18 plants. As shown in Supplemental Figure S3, similar to GFP-ROP2 distribution, RIC4ΔC-GFP signals were stronger at the apical PM in growing control RIC4ΔC-GFP root hairs compared with map18/RIC4ΔC-GFP root hairs and disappeared from the PM when root hairs stopped growing. The average RIC4ΔC-GFP PM/Cyt ratio at the apex was lower in map18/RIC4ΔC-GFP than in RIC4ΔC-GFP control root hairs, while RIC4ΔC-GFP expression levels were similar (Supplemental Fig. S3, A, C, and D). This observation was consistent with what we observed in root hairs expressing GFP-ROP2 (Fig. 3, C and F).
The MBP-RIC1 binding assay and distribution of GFP-ROP2 and RIC4ΔC-GFP in growing root hairs (Fig. 3; Supplemental Fig. S3) strongly suggested that MAP18 is important for ROP2 activities in the PM at the root hair tip.
ROP2 Activation by MAP18 Is Independent of Microtubules or Actin Filaments
Previous studies have demonstrated that MAP18 is able to associate with both cortical microtubules and F-actin. It has been demonstrated that MAP18 regulates the polarized diffuse growth of vegetative tissue cells by destabilizing microtubules (Wang et al., 2007) and modulates apical fine F-actin dynamics in the tips of growing pollen tubes and root hairs through calcium-dependent F-actin-severing activity (Zhu et al., 2013; Zhang et al., 2015). To investigate whether MAP18 regulates microtubule organization in growing root hairs, we observed microtubule arrangement in root hairs expressing GFP-MBD driven by the promoter UBQ (Marc et al., 1998) in the wild type, the map18 mutant, and MAP18 overexpression lines. In wild-type growing root hairs, microtubules were organized as bundles along the longitudinal axis in the shank region. Short microtubule fragments were observed at the subapical region, and few microtubules were detected in the apical domain (Supplemental Fig. S4A). These findings are similar to our observations of map18 and MAP18 overexpression in root hairs (Supplemental Fig. S4A). We then measured the skewness of the fluorescence intensity distribution in microtubules to evaluate the extent of microtubule bundling in the shank region of root hairs. As shown in Supplemental Figure S4B, microtubule bundling due to map18 and MAP18 overexpression in root hairs was similar to that in wild-type hairs. We measured the percentage of microtubule occupancy in root hairs, and no significant differences were detected among wild-type, map18, and MAP18 overexpression root hairs (Supplemental Fig. S4C). Therefore, loss of function or overexpression of MAP18 did not significantly impact microtubule organization in growing root hairs.
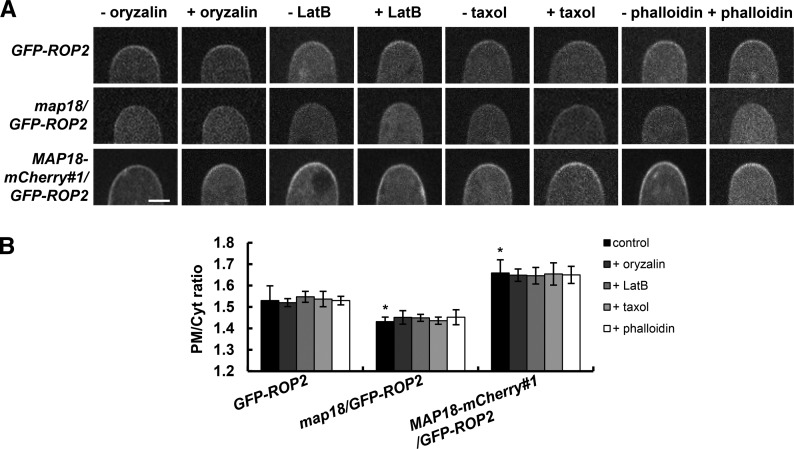
It was plausible that MAP18 might regulate ROP2 activity through its action on microfilaments. To test this assumption, we conducted a pharmacological analysis to test whether microtubules and F-actin are involved in MAP18-dependent ROP2 activation in growing root hairs. No differences in GFP-ROP2 distribution in growing root hairs were observed following treatments with the microtubules and the F-actin depolymerizing drugs oryzalin and latrunculin B (LatB) for 10 min (Fig. 4). Similarly, treatment with microtubules or F-actin stabilizing drugs (taxol and phalloidin) for 60 min also did not affect the distribution of GFP-ROP2 in growing root hairs (Fig. 4). The concentration and treatment duration of various drugs we adopted were shown to be effective in stabilizing or destabilizing microtubules or actin filaments, respectively (Supplemental Fig. S5; Zhang et al., 2015) but were not sufficient to cause the growth-defective phenotype of root hairs.
Figure 4.
MAP18 regulation of ROP2 activity is likely independent of the cytoskeleton. A, PM-associated GFP-ROP2 at the tips of different growing root hairs treated with or without microtubules and the F-actin depolymerizing drugs oryzalin and LatB for 10 min or microtubules and the F-actin stabilizing drugs taxol and phalloidin for 60 min. The PM distribution of GFP-ROP2 in the wild-type, map18, or MAP18 overexpression background was not influenced by various drug treatments. B, Quantification of the average PM/Cyt ratio of GFP-ROP2 fluorescence at the apex in growing root hairs shown in A. Data represent means ± sd, and at least 20 measurements from three independent experiments were included. Asterisks indicate significant differences at P < 0.05, by ANOVA.
One day of drug treatment with oryzalin, LatB, taxol, or phalloidin caused short and branched root hairs (Supplemental Fig. S6A), which is consistent with previous reports (Bibikova et al., 1999; Ketelaar et al., 2003), and none of the drugs affected the pull down of GFP-ROP2 by MBP-RIC1 in GFP-ROP2, map18/GFP-ROP2, and MAP18-mCherry#1/GFP-ROP2 plants (Supplemental Fig. S6, B and C), which is in agreement with the observations of GFP-ROP2 distribution. This evidence suggests that root hair growth and the maintenance of the growth point are precisely regulated by multiple regulatory mechanisms and that MAP18-mediated ROP2 activation is likely not dependent on microtubules or F-actin.
MAP18 Regulates ROP2 Activity by Interfering with the Interaction between AtRhoGDI1/SCN1 and ROP2
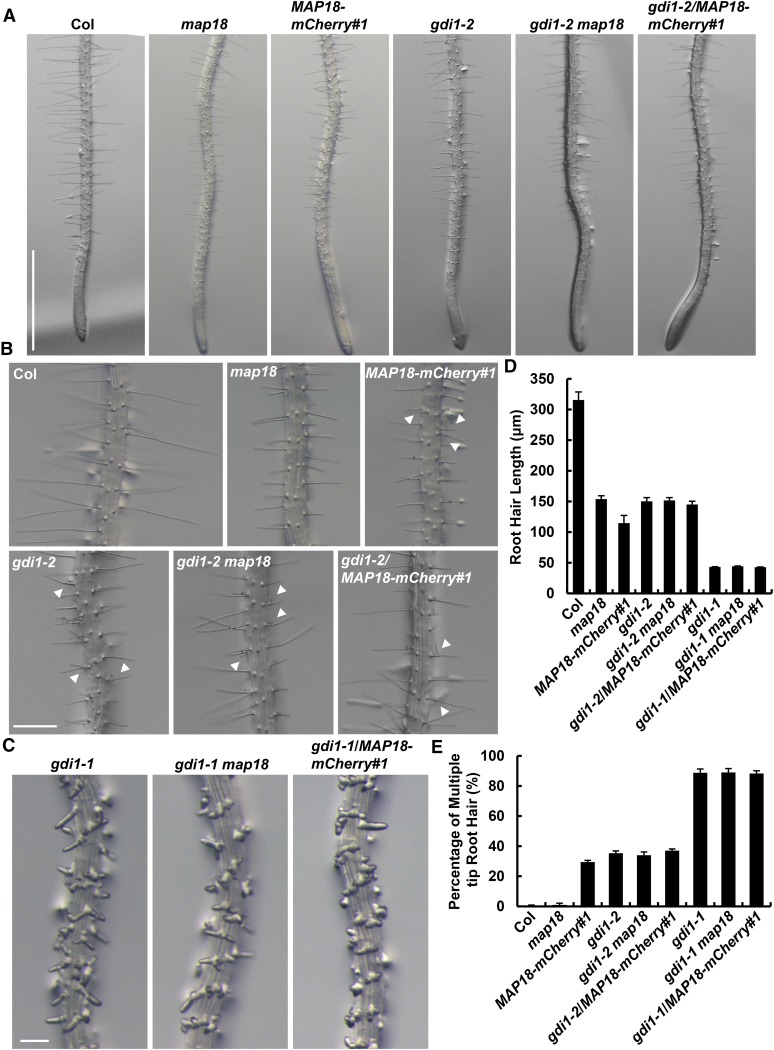
RhoGDI1 scn1 mutant plants develop multiple short root hairs (Carol et al., 2005) similar to MAP18-mCherry-overexpressing plants (Fig. 1). This raises the possibility that AtRhoGDI1/SCN1 might be involved in MAP18-mediated ROP2 activation. To examine this hypothesis, we analyzed the phenotypes of two T-DNA insertion lines in AtRhoGDI1/SCN1 (SALK_129991 and SALK_035400) that were designated as gdi1-1 and gdi1-2, respectively (Supplemental Fig. S7A). Quantitative reverse transcription (qRT)-PCR analysis showed reductions in AtRhoGDI1/SCN1 expression levels by about 90% and 60% in gdi1-1 and gdi1-2, respectively (Supplemental Fig. S7B).
Analysis revealed that gdi1-1 root hairs exhibited multiple tips and were shorter (Fig. 5C; Supplemental Fig. S7C), similar to the phenotypes of scn1-1 and scn1-3 AtRhoGDI1/SCN1 mutant alleles (Carol et al., 2005). As expected from higher AtRhoGDI1/SCN1 mRNA expression levels in gdi1-2 compared with gdi1-1, the root hair phenotype (average root hair length of 150 μm and only 35% developing multiple tips; Fig. 5, D and E) of this allele was much weaker compared with the severely distorted gdi1-1 root hairs (average root hair length of 43 μm and 89% developing multiple tips; Fig. 5, D and E). Interestingly, the phenotype of gdi1-2 root hairs resembles that of MAP18-mCherry-overexpressing plants (Fig. 5, A and B). The short root hair phenotypes of gdi1-1 and gdi1-2 were both rescued by expressing an AtGDI1promotor::GDI1 construct. Fifteen out of 18 transformed gdi1-1 lines and 22 out of 25 gdi1-2 transformed lines recovered the short root hair phenotype. Two of these lines (COM#3 for gdi1-1 and COM#7 for gdi1-2) were selected for phenotypic analysis, and representative images are shown in Supplemental Figure S7, C to E. We concluded that the defects observed in gdi1-1 and gdi1-2 root hairs resulted from the down-regulation of AtRhoGDI1/SCN1 expression and that AtRhoGDI1/SCN1 is required for normal root hair growth.
Figure 5.
MAP18-ROP2 functionally interacts with AtRhoGDI1/SCN1 to regulate root hair growth and morphology. A, Representative images of root hairs in Columbia-0 (Col), map18, MAP18-mCherry#1, gdi1-2, gdi1-2 map18 double mutant, and gdi1-2/MAP18-mCherry#1 plants. Bar = 1 mm. B, The branching root hair phenotype in Columbia-0, map18, MAP18-mCherry#1, gdi1-2, gdi1-2 map18, and gdi1-2/MAP18-mCherry#1 plants. White arrowheads mark the branching root hairs. Bar = 200 μm. C, Representative images of root hairs in gdi1-1, gdi1-1 map18 double mutant, and MAP18-mCherry#1 crossed with gdi1-1. Root hairs of gdi1-1 showed an obvious short and branching phenotype. The root hairs of various lines crossed with the gdi1-1 mutant were similar to those of gdi1-1 no matter whether there was a decrease or increase in the expression of MAP18. Bar = 200 μm. D, Quantitative analysis of the average root hair length shown in A. The gdi1-2 root hairs were shorter than wild-type hairs but similar to those of the gdi1-2 map18 double mutant and gdi1-2/MAP18-mCherry#1. The gdi1-1 root hairs were much shorter than gdi1-2 and wild-type hairs but similar to those of the gdi1-1 map18 double mutant and gdi1-1/MAP18-mCherry#1. E, Developing multiple tips of gdi1-2 were weaker compared with those of gdi1-1 root hairs. However, a similar percentage of branching root hairs was detected in gdi1-2 map18 and gdi1-2/MAP18-mCherry#1 compared with the gdi1-2 single mutant as well as in the gdi1-1 background. More than 150 root hairs from at least 20 growing roots for each line were measured. Data are presented as means ± sd.
To learn more about the relationship between MAP18 and AtRhoGDI1/SCN1 in root hair growth, we generated gdi1-1 map18 double mutants, gdi1-1/MAP18-mCherry#1 and gdi1-2 map18 double mutants, and gdi1-2/MAP18-mCherry#1 plants (Fig. 5). As expected, the root hair phenotypes of gdi1-1 map18 and gdi1-1/MAP18-mCherry#1 seedlings both resembled gdi1-1, but root hairs were obviously shorter than map18 or MAP18-mCherry#1 seedlings and significantly different from map18 root hairs, which hardly produced any multiple tips (Fig. 5, C–E). In addition, gdi1-2 map18 and gdi1-2/MAP18-mCherry#1 root hairs both displayed branched and short root hairs, similar to the gdi1-2 single mutant (Fig. 5, A, B, D, and E). Root hair phenotypes of gdi1-2 or gdi1-1 with map18 or MAP18-mCherry#1 double mutants suggest that AtRhoGDI1/SCN1 may function together with MAP18 to regulate ROP2 activity during root hair development.
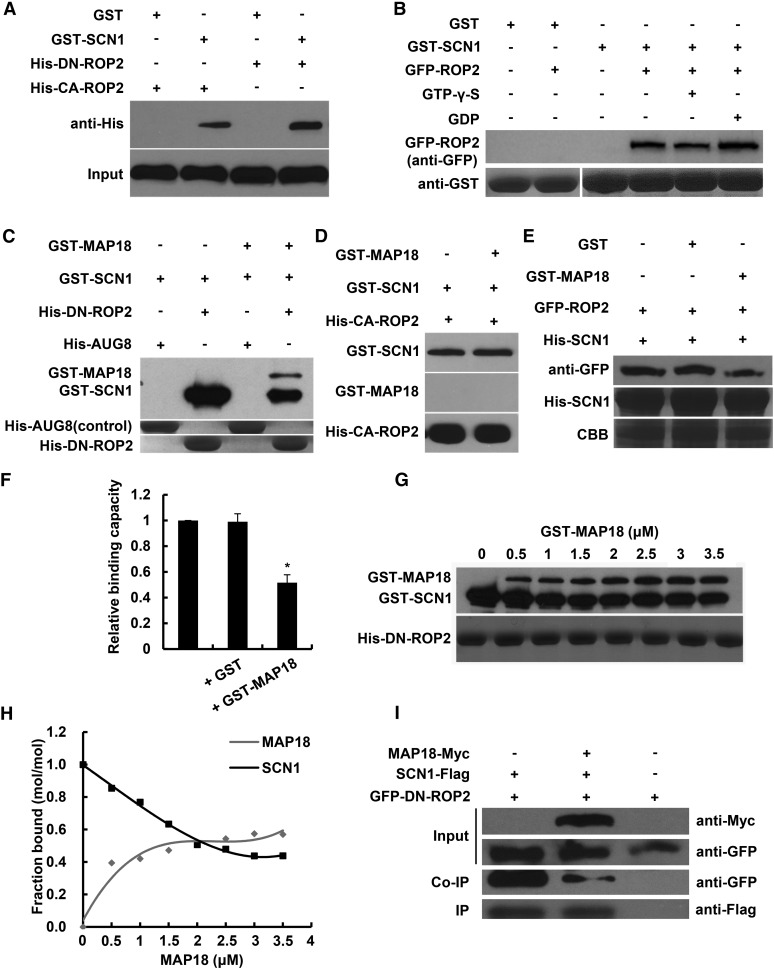
To study the molecular mechanisms that underlie the interaction between AtRhoGDI1/SCN1 and MAP18, we next examined whether they could mutually affect each other’s interaction with ROP2. It was reported previously that AtRhoGDI1/SCN1 interacts with ROP4 and ROP6 in yeast (Bischoff et al., 2000). In vitro pull-down assays demonstrated that recombinant E. coli expressing GST-AtRhoGDI1/SCN1 can interact with His-DN-ROP2 and His-CA-ROP (Fig. 6A). Under physiological conditions, the interaction between RhoGDI and the RHO family of proteins was shown to depend on geranylgeranylation of the GTPase (Hoffman et al., 2000; Grizot et al., 2001; Garcia-Mata et al., 2011). Since recombinant ROPs expressed in E. coli are not geranylgeranylated (Sorek et al., 2010), it was important to establish that RhoGDI1 interacts with geranylgeranylated ROP2 purified from plants. To this end, we prepared protein extracts from GFP-ROP2 seedlings and examined whether GFP-ROP2 could be pulled down by GST-AtRhoGDI1/SCN1. Results of the pull-down assays demonstrated that GFP-ROP2 was precipitated by GST-AtRhoGDI1/SCN1 in the presence of GDP or GTP-γ-S (Fig. 6B).
Figure 6.
MAP18 competes with AtRhoGDI1/SCN1 to bind ROP2. A, In vitro binding assays were performed using recombinant E. coli-expressed GST-SCN1 and ROP2 variants. His-CA-ROP2 or His-DN-ROP2 pulled down by GST- SCN1 was detected by anti-His antibody. The bottom gel shows that similar amounts of His-CA-ROP2 and His-DN-ROP2 were used in the binding assays. Nonfused GST was used as a negative control. B, Total proteins extracted from GFP-ROP2 seedling roots were preloaded with GTP-γ-S or GDP, and anti-GFP antibody was used for the detection of proteins pulled down by GST-SCN1. The bottom gel shows input bait proteins detected by anti-GST antibody. Similar amounts of extracted proteins were used in the binding assays. Nonfused GST was used as a negative control. C, Pull-down analysis shows that both GST-SCN1 and GST-MAP18 could bind with His-DN-ROP2. The interaction of GST-SCN1 with His-DN-ROP2 was interfered with by the addition of GST-MAP18. His-AUG8 (Cao et al., 2013) was used as a negative control. Experiments were repeated at least three times, and a representative result is shown. D, GST-MAP18 does not affect the interaction between AtRhoGDI1/SCN1 and CA-ROP2 in vitro. E, The capacity of His-SCN1 pull-down GFP-ROP2 proteins extracted from transgenic seedlings is inhibited by GST-MAP18. The middle gel shows His-SCN1, and the bottom gel shows input bait proteins stained by Coomassie Brilliant Blue (CBB). Anti-GFP was used for the detection of ROP2 protein pulled down by His-SCN1. Nonfused GST was used as a negative control. F, Quantification analysis of the relative ROP2-binding capacity of His-SCN1. GFP-ROP2 pulled down by His-SCN1 was decreased by the addition of GST-MAP18. Data represent means ± sd from at least three repeats. Relative amounts of binding GFP-ROP2 protein were normalized to input bait protein and were arbitrarily defined as 1. The asterisk indicates a significant difference at P < 0.05, by ANOVA. G, His-DN-ROP2 was incubated with various concentrations of recombinant GST-MAP18. Lanes 1 to 8 show 0, 0.5, 1, 1.5, 2, 2.5, 3, and 3.5 μm GST-MAP18, 2 μm GST-SCN1, and 2 μm His-DN-ROP2. Experiments were repeated at least three times, and a representative result was shown. H, Quantification analysis of G. The amounts of GST-MAP18 or GST-SCN1 bound to His-DN-ROP2 were determined by blots and scanning of stained gels. Binding to His-DN-ROP2 was saturated at a stoichiometry of 0.58 mol of GST-MAP18 per mol of His-DN-ROP2. The amount of GST-SCN1 binding to His-DN-ROP2 was decreased gradually with the increase of GST-MAP18. I, GFP-DN-ROP2 and AtRhoGDI1/SCN1-Flag coexpressed in the protoplasts with or without MAP18-Myc. Co-IP assay showed that MAP18 competes with AtRhoGDI1/SCN1 for binding to ROP2 in vivo. GFP-DN-ROP2 coprecipitated with AtRhoGDI1/SCN1-Flag by anti-GFP antibody was reduced in the presence of MAP18-Myc.
We next examined whether MAP18 affects the interaction between AtRhoGDI1/SCN1 and ROP2. Control pull-down assays indicate that GST-MAP18 did not interact with AtRhoGDI1/SCN1 (Supplemental Fig. S8). However, the levels of GST-AtRhoGDI1/SCN1 precipitated by recombinant His-DN-ROP2 were reduced in the presence of MAP18 (Fig. 6C; Supplemental Fig. S9). Interestingly, the addition of GST-MAP18 did not affect the interaction between AtRhoGDI1/SCN1 and CA-ROP2 (Fig. 6D). These results suggest that MAP18 competitively reduces the interaction between AtRhoGDI1/SCN1 and inactive ROP2 but not active ROP2.
Moreover, GFP-ROP2 (extracted from GFP-ROP2 plants) precipitated by His-AtRhoGDI1/SCN1 also was reduced when MAP18 was present (Fig. 6, E and F). Increased levels of GST-MAP18 in the pull-down reaction mixture (0–3.5 μm) were correlated with a corresponding reduction in the levels of GST-AtRhoGDI1/SCN1 that were precipitated by His-DN-ROP2 (Fig. 6, G and H; Supplemental Fig. S10). To further substantiate that MAP18 competes with AtRhoGDI1/SCN1 on ROP2, GFP-DN-ROP2 and flag-tagged AtRhoGDI1/SCN1 (SCN1-Flag) were coexpressed in Arabidopsis protoplasts in the absence or presence of Myc-tagged MAP18 (MAP18-Myc). In agreement with the results presented in Figure 6, C, E, and G, GFP-DN-ROP2 levels precipitated by the flag-tagged AtRhoGDI1/SCN1 were reduced when coexpressed with MAP18-Myc (Fig. 6I). The data presented in Figure 6 and Supplemental Figure S8 indicate that MAP18 competitively reduces the interaction between AtRhoGDI1/SCN1 and E. coli- or plant-expressed ROP2.
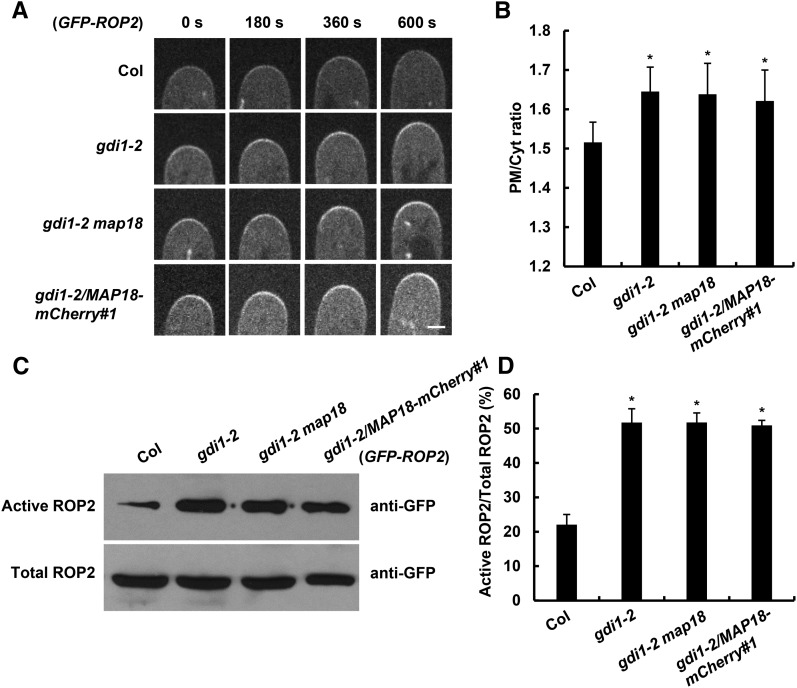
To examine whether MAP18-dependent ROP2 activation is associated with the competitive inhibition of AtRhoGDI1/SCN1-ROP2 interaction by MAP18, we determined the ROP2 activation status in gdi1 mutants (all in a GFP-ROP2 background). First, we observed and analyzed PM-associated GFP-ROP2 at the root hair tip. Because of a strong defect phenotype in gdi1-1, the fluorescence signal of GFP-ROP2 was difficult to observe at the severely distorted root hair tips. Hence, we chose the gdi1-2 allele with a relatively weaker phenotype for further analyses.
PM-associated GFP-ROP2 levels at the root hair growing tip were higher in gdi1-2, gdi1-2 map18, and gdi1-2/MAP18-mCherry#1 seedlings (Fig. 7, A and B). In agreement with GFP-ROP2 distribution, higher GFP-ROP2 levels were precipitated by MBP-RIC1 in gdi1-2 compared with GFP-ROP2 control plants in ROP activity detection assays (Fig. 7C). Remarkably, GFP-ROP2 levels that were precipitated by MBP-RIC1 in gdi1-2 map18 and gdi1-2/MAP18-mCherry#1 were similar to those of gdi1-2 single mutants (Fig. 7, C and D), while active ROP2 levels in map18 single mutants were lower compared with the GFP-ROP2 control (Fig. 3, A and B). The higher ROP2 activity in double mutants was consistent with their root hair phenotypes: gdi1-2 map18 and gdi1-2/MAP18-mCherry#1 root hairs displayed branched and short root hairs similar to gdi1-2 (Fig. 5, A and B). These results indicate that MAP18-dependent ROP2 activation is associated with MAP18’s competition with AtRhoGDI1/SCN1 for interaction with ROP2 and suggest that MAP18 may counteract with AtRhoGDI1/SCN1 to regulate ROP2 activity during root hair development.
Figure 7.
MAP18 positively regulates ROP2 activity by interfering with the interaction between AtRhoGDI1/SCN1 and ROP2. A, Time-lapse images of growing root hairs expressing GFP-ROP2 in the wild-type Columbia-0 (Col), gdi1-2, gdi1-2 map18, or gdi1-2/MAP18-mCherry#1 background. PM-associated GFP-ROP2 at the tips of growing root hairs was enhanced in all gdi1-2, gdi1-2 map18, and gdi1-2/MAP18-mCherry#1 plants. Bar = 5 μm. B, Quantitative analysis of the PM/Cyt ratio of GFP-ROP2 fluorescence intensity at the apex of growing root hairs. The PM/Cyt ratio is higher in gdi1-2, gdi1-2 map18, and gdi1-2/MAP18-mCherry#1. Data represent means ± sd, and at least 20 root hairs from three independent experiments were measured for each data set. Asterisks indicate significant differences at P < 0.05, by ANOVA. C, Total GFP-ROP2 and active GFP-ROP2 pulled down by MBP-RIC1 were analyzed in control, gdi1-2, gdi1-2 map18 double mutant, and gdi1-2/MAP18-mCherry#1 plants (all expressing GFP-ROP2). Five independent experiments were performed, and one representative result is shown. D, Quantitative analysis of data from C. Relative ROP2 activity was determined as the amount of active ROP2 pulled down by MBP-RIC1 versus the amount of total GFP-ROP2. The ROP2 activity increased significantly in the gdi1-2 mutant (asterisks indicate significant differences at P < 0.05, by ANOVA), and neither knocking down nor overexpression of MAP18 could further affect the ROP2 activity in gdi1-2. Data are mean values from five independent experiments ± sd.
This evidence indicates that MAP18 regulates ROP2 activity through competition with AtRhoGDI1/SCN1 to bind the inactive form of ROP2.
The N23 Domain Is Necessary for MAP18 to Interact with ROP2
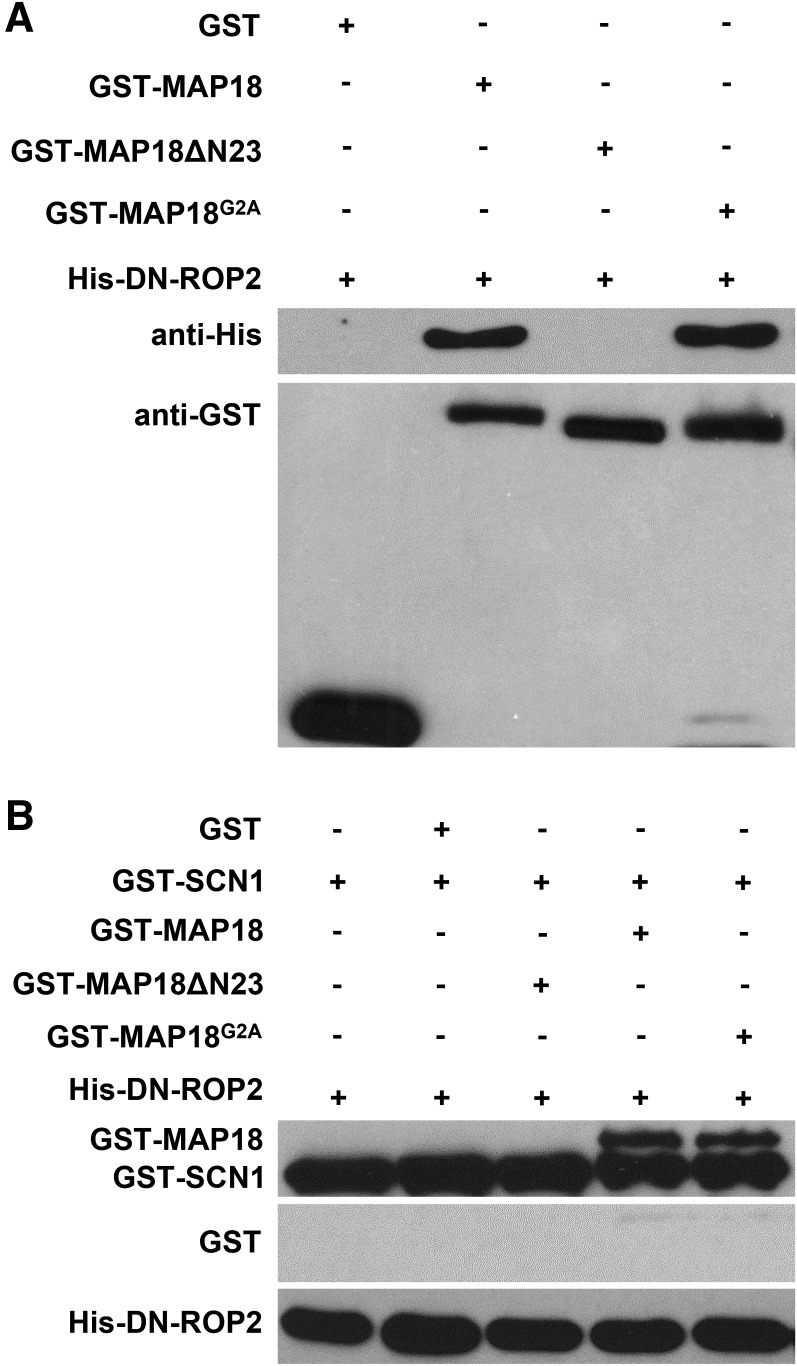
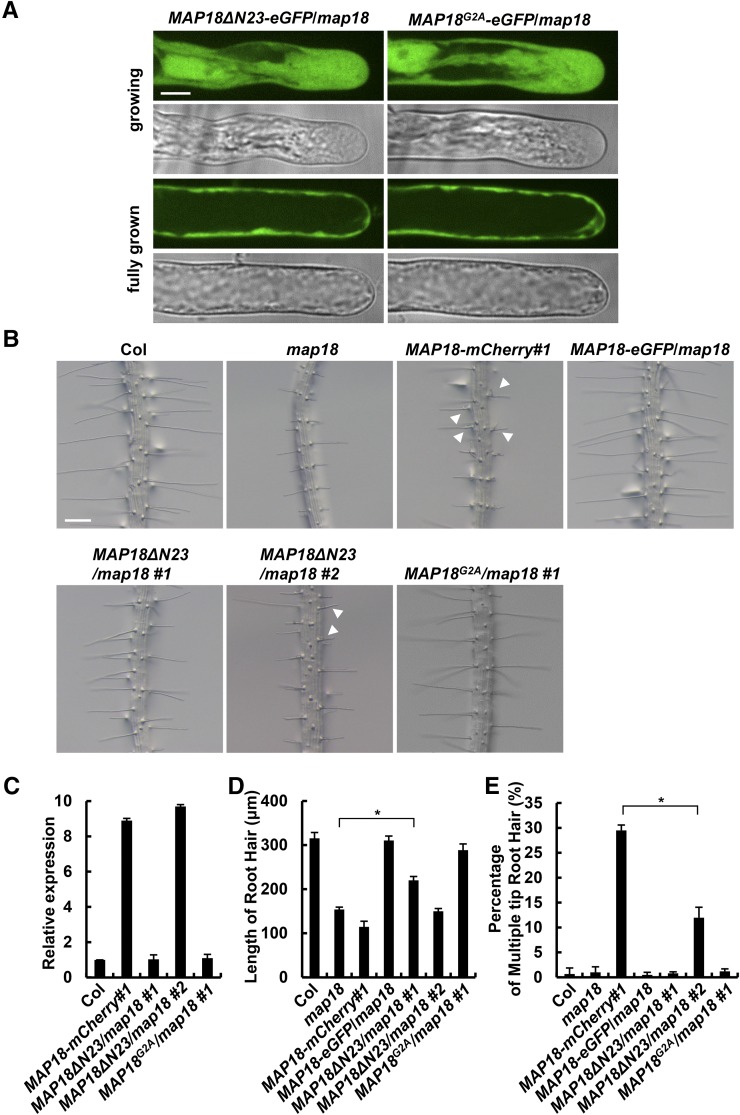
To determine the ROP2-binding sites in MAP18, we generated truncated or mutant proteins with site-directed mutations. MAP18 has an N-terminal basic domain composed of 23 amino acids (the N23 domain). The N23 domain functions in binding to phosphatidylphosphoinositide phosphates (PtdInsPs) and Ca2+/calmodulin and is responsible for the localization of MAP18 to the PM (Kato et al., 2013). The second Gly residue (Gly-2) has been demonstrated as the site of N-myristoylation, which also is required for membrane anchoring. G-to-A mutation at this site abolished the PM localization of MAP18 (Kato et al., 2010). To examine whether the N23 domain also is required for the interaction between MAP18 and ROP2, we generated a truncated MAP18 mutant protein lacking the N-terminal 23 amino acids (MAP18ΔN23) and a mutant harboring a G2A point mutation (MAP18G2A).
Purified recombinant mutant proteins were used for an in vitro pull-down assay. We found that non-PM-localized GST-MAP18G2A was still able to interact with His-DN-ROP2, similar to GST-MAP18 (Fig. 8A). Consistently, GST-MAP18G2A interfered with the interaction between AtRhoGDI1/SCN1 and DN-ROP2 (Fig. 8B). MAP18ΔN23-truncated mutants failed to interact with His-DN-ROP2 (Fig. 8A), suggesting that the N23 domain of MAP18 is essential for the interaction of MAP18 with ROP2 in vitro. Similarly, the interaction between AtRhoGDI1/SCN1 and DN-ROP2 was not affected by the addition of MAP18ΔN23 (Fig. 8B), indicating a failure of MAP18ΔN23 to compete with AtRhoGDI1/SCN1 for interaction with ROP2.
Figure 8.
The N23 domain is necessary for interaction between MAP18 and ROP2. A, An in vitro binding assay was performed to test the interaction between GST-MAP18 variants and DN-ROP2. His-DN-ROP2 pulled down by GST-MAP18 and GST-MAP18G2A was detected by anti-His antibody. GST-MAP18ΔN23 failed to interact with His-DN-ROP2. The bottom gels show input bait proteins, and similar amounts of GST-MAP18, GST-MAP18ΔN23, and GST-MAP18G2A were used in the binding assay. Nonfused GST was used as a negative control. B, In vitro pull-down assay showing that MAP18 and MAP18G2A, but not MAP18ΔN23, competed with AtRhoGDI1/SCN1 for ROP2 binding. Anti-GST antibody was used for detection. Nonfused GST was used as a negative control.
To examine whether MAP18 binds to ROP2 via the N23 domain of MAP18 in vivo, we introduced GFP-fused MAP18ΔN23 and MAP18G2A into the map18 mutant. We first observed the localization pattern of MAP18 in growing root hairs. In growing map18 root hairs stably expressing MAP18ΔN23-eGFP and MAP18G2A-eGFP, the fluorescence signal was localized exclusively to the cytoplasm (Fig. 9A), which is different from what was observed in MAP18-eGFP/map18 growing hairs (Supplemental Fig. S2D). Hence, in agreement with previously published data, the N-terminal N23 domain and Gly-2 are required for the PM association of MAP18 (Kato et al., 2010, 2013). In addition, PM-localized GFP-ROP2 is decreased significantly in MAP18ΔN23-overexpressing root hairs compared with MAP18-mCherry#1 (overexpression of full-length MAP18 with a similar expression level to MAP18ΔN23; Fig. 9C; Supplemental Fig. S11), further suggesting that the overexpression of MAP18ΔN23 could not promote ROP2 activity like full-length MAP18 because the N23 domain is required for the interaction with ROP2 (Fig. 8). Next, we examined the root hair morphology of MAP18ΔN23/map18 #1 and MAP18G2A/map18 #1 seedlings, in which the relative expression levels of mutant MAP18 proteins were similar to MAP18 expression levels in the wild type (Fig. 9C). As expected, the expression of MAP18ΔN23 did not fully recover the short root hair phenotype of map18, indicating the importance of the N23 domain as well as the MAP18-ROP2 interaction in the MAP18 regulation of root hair growth (Fig. 9, B and D). However, MAP18ΔN23/map18 #1 root hairs were still significantly longer than map18 root hairs (Fig. 9, B and D), suggesting that MAP18 regulation of root hair growth also can be independent of ROP2. In agreement, we demonstrated previously that MAP18 regulates the organization and dynamics of apical F-actin during root hair tip growth (Zhang et al., 2015).
Figure 9.
MAP18ΔN23 cannot fully rescue root hair defects in a map18 mutant. A, MAP18ΔN23-eGFP and MAP18G2A-eGFP were localized exclusively to the cytoplasm in MAP18ΔN23/map18 and MAP18G2A/map18 root hairs, respectively. Bar = 10 μm. B, Representative images of MAP18ΔN23/map18 #1, MAP18ΔN23/map18 #2, and MAP18G2A/map18 #1 root hairs. Expression of MAP18G2A mostly rescued the elongation defect of map18 root hairs. However, although MAP18ΔN23/map18 #1 root hairs were longer than map18 hairs, the expression of MAP18ΔN23 could not fully rescue the short root hair phenotype of map18. MAP18ΔN23/map18 #2 seedlings still developed short and branched root hairs. Bar = 200 μm. C, The relative expression levels of MAP18 variants in MAP18ΔN23/map18 #1 and MAP18G2A/map18 #1 were similar to the MAP18 level in wild-type Columbia-0 (Col), while that in MAP18ΔN23/map18 #2 was higher than the MAP18 level in the wild type but was similar to the MAP18 level in MAP18-mCherry#1. D, Quantitative analysis of the average root hair lengths of different lines. MAP18ΔN23/map18 #1 root hairs was shorter than wild-type hairs but longer than map18 hairs. MAP18ΔN23/map18 #2 root hairs were shorter than MAP18ΔN23/map18 #1 hairs but longer than MAP18-mCherry#1 hairs. MAP18G2A/map18 #1 mostly rescued the root hair defects in the map18 mutant. E, Quantitative analysis of multiple-tip root hairs in various lines. The percentage of branched/multiple-tip root hairs was higher in MAP18ΔN23/map18 #2 than in the wild type but lower than that of MAP18-mCherry#1. More than 150 root hairs from at least 20 growing roots for each line were measured. Data are presented as means ± sd. Asterisks indicate significant differences at P < 0.05, by ANOVA.
Furthermore, we investigated the root hair morphology of MAP18ΔN23/map18 #2 seedlings, in which the relative expression level of the mutant MAP18 was much higher than the expression level of MAP18 in the wild type but was similar to the expression in MAP18-mCherry#1 (Fig. 9C). Overexpression of MAP18ΔN23 also led to the short root hair and branched root hair phenotype, similar to the overexpression of full-length MAP18 (MAP18-mCherry#1; Fig. 9, B and D), although the percentage of branched root hairs was significantly lower compared with MAP18-mCherry#1 (Fig. 9E). These results indicate that the maintenance of growth point by MAP18 was at least partially dependent on the interaction with ROP2.
However, 89% of root hairs from MAP18G2A/map18 #1 seedlings had a similar length to that of the wild type (Fig. 9, B and D). The percentage of multiple-tip root hairs in MAP18G2A/map18 #1 was very close to that of the wild type (Fig. 9E), indicating that MAP18G2A rescues the root hair defect in the map18 mutant and that the PM localization of MAP18 at the shank region is not absolutely required for its function in root hair growth.
To summarize, these results suggest that interaction with ROP2, but not PM binding, is important for MAP18 function in ROP2 signaling during root hair growth.
DISCUSSION
In this study, we showed that MAP18 genetically and functionally interacts with ROP2 signaling in root hair growth, both in maintaining growth polarity and sustaining root hair elongation. MAP18 preferably binds to the inactive form of ROP2 and promotes the activation of ROP2 in root hairs.
Further investigation elucidated that MAP18 interferes with the interaction between the inactive form of ROP2 and AtRhoGDI1/SCN1, which contributes to the subcellular distribution of active ROP2. RhoGDIs have been subjected to intensive studies in yeast and mammalian systems, and the availability of viable RhoGDI null mutants in Arabidopsis has enabled ROP function to be studied in RhoGDI-deficient backgrounds. In this study, we examined the function of MAP18 in the regulation of ROP2 activity during root hair growth using the gdi1 mutant background.
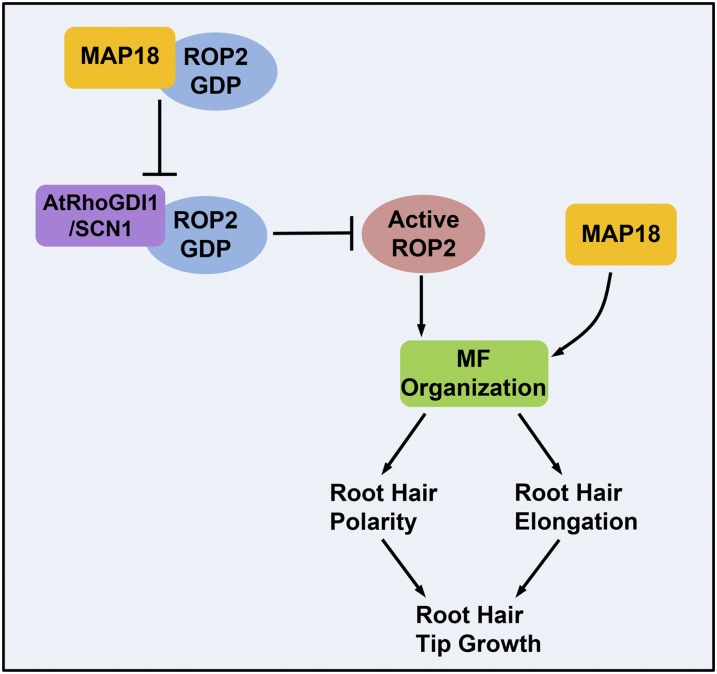
Based on the data presented, we propose the following model. MAP18 functions as a RhoGDI displacement factor that competes with AtRhoGDI1/SCN1 for interaction with GDP-ROP2. MAP18 interferes with the interaction between GDP-ROP2 and AtRhoGDI1/SCN1, resulting in more activated GTP-ROP2 on the apical PM. The apical polarized activated ROP2 triggers downstream signaling to maintain root hair polarity and elongation. Besides that, our genetic analysis suggested that MAP18 also may regulate root hair growth in a ROP2-independent manner, likely through directly modulating microfilament organization and dynamics at the root hair apex, as we demonstrated in our previous report (Zhang et al., 2015; Fig. 10). This work demonstrates a novel function of MAP18 in maintaining the growth point and modulating root hair elongation by regulating ROP2 activity during root hair tip growth.
Figure 10.
A working model depicting how MAP18 regulates ROP2 activity in root hair growth. MAP18 and ROP2 cooperate during root hair growth, both in maintaining the growth point and sustaining elongation. MAP18 functions as a RhoGDI displacement factor that competes with AtRhoGDI1/SCN1 for interaction with GDP-ROP2. MAP18 interferes with the interaction between GDP-ROP2 and AtRhoGDI1/SCN1, promoting the redistribution of activated GTP-ROP2 on the apical PM. Active ROP2 triggers downstream signaling to maintain the root hair growth point. In addition, MAP18 may regulate the maintenance of the root hair growth point by modulating F-actin dynamics and organization. During root hair elongation, MAP18 directly regulates fine F-actin dynamics and organization at the root hair apex. MAP18 also promotes ROP2 activity, which triggers downstream signaling to regulate root hair growth. MF: microfilaments.
Cooperation of MAP18 and ROP2 Signaling during Root Hair Growth
Overexpression ROP2 or MAP18 leads to the formation of multiple root hair tips and root hair branching, respectively. However, overexpression of MAP18 leads to the development of significantly shorter root hairs compared with wild-type plants. In contrast, root hairs due to the overexpression of ROP2 are longer than wild-type root hairs. These root hair phenotypes suggest that maintaining the root hair growth point and sustaining root hair elongation are two different functions in regulating root hair growth.
We propose a hypothesis that MAP18 has a dual function in the regulation of root hair growth: (1) maintaining the root hair growth point and sustaining elongation, through promoting ROP2 activity; and (2) influencing actin organization in the apical and subapical regions.
It has been proposed that the organization and dynamics of F-actin are able to affect the activity of ROP GTPases by participating in vesicle trafficking during pollen tube tip growth (Nagawa et al., 2010). In addition, well-ordered transverse microtubules seem to inhibit ROP2 activity at the neck of pavement cells (Fu et al., 2005). We reported previously that MAP18 modulated the dynamics of microtubules and F-actin in different cell types (Wang et al., 2007; Zhu et al., 2013; Zhang et al., 2015). However, using pharmacological analysis, we demonstrated that changes in ROP2 activity in a map18 mutant or MAP18-overexpressing root hairs were not further affected by the stabilization or disruption of either microtubules or F-actin. Further analysis of ROP2-binding activity in various MAP18 mutant proteins with a mutation in VEEKK motifs (MAP18 mutant proteins are described by Zhu et al. [2013]) indicated that MAP18 binding to ROP2 was unrelated to its Ca2+-dependent F-actin-severing activity (Supplemental Fig. S12C). These results demonstrate the existence of a MAP18-mediated activation of ROP2 in root hairs that is independent of the cytoskeleton, although we did not completely exclude the possibility that MAP18 also might indirectly affect ROP2 activity through the regulation of F-actin organization and dynamics in the root hair tip.
ROP2 has been reported to regulate the formation of fine F-actin at the root hair tips (Jones et al., 2002). MAP18 also was found to be able to interact with CA-ROP2 or GTP-ROP2 in vitro, with a much weaker binding capability compared with its interaction with inactive DN-ROP2 and GDP-ROP2. In addition, the GDP-bound form of a small GTPase (Bud1p) showed a positive influence on cellular functions through associations with Bem1 in determining budding yeast cell polarity (Park et al., 1997). This raises the question of whether MAP18 also can be a downstream effector of ROP2 signaling to regulate apical F-actin during root hair tip growth. Because loss of function of ROP2 did not affect the localization pattern of MAP18 in root hairs or alter the actin-severing activity of MAP18 in vitro (Supplemental Fig. S13), we suspect that active ROP2 is not required for the MAP18 regulation of F-actin in root hairs. During root hair growth, fine F-actin dynamics and organization at the root hair apex are regulated by MAP18 directly or modulated by active ROP2, in which activation is promoted by MAP18. Nevertheless, more direct evidence will be needed to make a conclusion.
Nonconventional Mechanism for Regulating ROP2 Activity during Root Hair Growth
As a binary molecular switch, ROPs rapidly cycle between GTP-bound (active) and GDP-bound (inactive) forms. There are conserved positive and negative regulatory proteins for small GTPases in eukaryotic genomes that respond to different signals. Three major classes of regulatory proteins, GEFs, GAPs, and GDIs, all play important roles in regulating ROP activities and functions (Gu et al., 2004; Fu et al., 2008; Kost, 2008; Nagawa et al., 2010; Cherfils and Zeghouf, 2013). In Arabidopsis, ROPGEF4 and ROPGEF10 were shown to function downstream of the receptor-like protein kinase FERONIA in regulating root hair elongation and initiation, respectively (Duan et al., 2010; Huang et al., 2013). AtRhoGDI1/SCN1 was shown to regulate root hair growth through spatially restricting the site of root hair growth to a single site (Carol et al., 2005). This study shows that MAP18 regulates root hair development by modulating the RhoGDI1-ROP2 interaction.
Biochemical studies have shown PRENYLATED RAB ACCEPTOR1 (PRA1) to be a RabGDI displacement factor (GDF; Hutt et al., 2000; Seabra and Wasmeier, 2004). Our studies provide a nonconventional mechanism for regulating ROP2 activity through competition with RhoGDI1, suggesting that MAP18 functions as a GDF of RhoGDI1 in Arabidopsis. This provides new insights in understanding the mechanism underlying the regulation of ROP GTPase activity. It also brings up interesting questions regarding whether there are other proteins involved in regulating ROP signaling, through either competition with or modification of well-known upstream regulators of ROPs, including GEFs, GDIs, and GAPs. Answering these questions will contribute to a better understanding of the mechanisms in ROP signaling and plant cell polar growth.
Different Domain of MAP18 Contributes to Its Distinct Function in Plant Cell Polar Growth
Promoter GUS expression analysis and data from the Genevestigator database (https://genevestigator.com/gv/) indicate that MAP18 is widely expressed in the root, hypocotyl, and cotyledon. Notably, it is highly expressed in rapidly elongating cells, such as root hairs and pollen tubes. In pollen tubes, it may serve as a GDF of RhoGDI1, RhoGDI2a, and RhoGDI2b. MAP18 was identified previously as a microtubule-associated protein involved in the regulation of anisotropic growth and cortical microtubule organization in vegetative tissues by destabilizing microtubules (Wang et al., 2007). Recently, we reported that MAP18 also possesses calcium-dependent actin filament-severing activity and regulates F-actin dynamics in tip-growing cells. It is involved in regulating pollen tube directional growth (Zhu et al., 2013) and in proper nuclear positioning during root hair elongation (Zhang et al., 2015). In this study, we revealed a novel function of MAP18 in the regulation of ROP2 activity in the root hair that is independent of the cytoskeleton. These findings suggest that MAP18 plays different roles in different cell types. The multifunctional MAP18 is likely able to have distinct functions by interacting with various partners.
According to analyses of amino acid sequences from MAP18/PCaP2 and its homolog PCaP1/MDP25, MAP18 could be divided into highly conserved N-terminal (N23 domain) and C-terminal regions containing seven repeats of a VEEKK motif (Wang et al., 2007; Kato et al., 2010; Li et al., 2011; Zhu et al., 2013). It was reported that the N23 domain of MAP18 is responsible for its binding to PtdInsPs and is necessary for localization to the PM. Additionally, the N23 domain also is sufficient for MAP18 binding to the Ca2+-calmodulin complex (Kato et al., 2013). However, the interaction of MAP18 with the Ca2+-calmodulin complex weakened the interaction between MAP18 and PtdInsPs (Kato et al., 2010). Previously published data have implicated the N23 domain of MAP18 in the regulation of root hair tip growth. It was reported that overexpression of the N23 domain created multiple bulges from single root epidermal cells and inhibited root hair elongation. The severity of the root hair phenotype was correlated with N23 expression levels (Kato et al., 2013). In this study, we revealed that MAP18 binds to ROP2 through its N23 domain. Therefore, the N23 domain of MAP18 is required not only to bind to PtdInsPs and the Ca2+-calmodulin complex but also to interact with ROP GTPases. Our findings show that expression of the MAP18ΔN23 mutant protein did not fully recover the short root hair phenotype of map18, further indicating the importance of the N23 domain in the MAP18 regulation of root hair growth. However, how exactly this domain integrates the Ca2+-calmodulin complex, PtdInsPs, and ROP signaling for the proper functioning of MAP18 is still unknown and will require further investigation.
Notably, the N23 domain does not participate in MAP18 binding to cytoskeletal elements (Supplemental Fig. S14). The truncated MAP18 protein without an N-terminal domain is still able to bind microtubules and sever actin filaments, suggesting that the residual C terminus of MAP18 is responsible for binding to cytoskeletal elements (Supplemental Fig. S12, A and B). In the C-terminal domain of MAP18, there are seven repeats of the VEEKK motif (Wang et al., 2007), which are conserved in SB401 from Solanum berthaultii and in MAP1B from mouse. Both of these proteins have been reported to be able to bind to microtubules and F-actin (Gordon-Weeks and Fischer, 2000; Mack et al., 2000; Huang et al., 2007; Riederer, 2007). Hence, the C-terminal region with VEEKK motifs may play a key role in functioning on microtubules and actin filaments. It has been reported that mutations in any one of the first two VEEKK motifs (M1 or M2) abolishes the F-actin-severing activity of MAP18 in the presence of Ca2+ (Zhu et al., 2013). However, mutations in a single VEEKK motif did not affect the interaction between MAP18 and microtubules or F-actin. It is possible that several VEEKK motifs contribute to the interaction between MAP18 and cytoskeletal elements, and mutation in a single individual VEEKK motif may not affect MAP18 binding to microtubules or F-actin (Zhu et al., 2013). These data do not exclude interactions between MAP18 and microtubules and F-actin via a VEEKK-independent mechanism.
In summary, these results indicate that different MAP18 domains contribute in different ways to MAP18 physiological functions in plant cell growth. Our previous studies along with this study reveal that the N23 domain of MAP18 is crucial for MAP18 in regulating ROP2 activity and that the C terminus of MAP18 participates in modulating actin filaments in the presence of Ca2+ in root hair development.
Further investigation will be needed to elucidate the roles of different MAP18 domain functions in the regulation of pollen tube tip growth and polarized diffuse growth in vegetative tissues.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used for all wild-type and mutant background tissue in this study.
Arabidopsis lines with T-DNA insertions were obtained from the Arabidopsis Biological Resource Center. The map18 knockdown mutant (SALK_021652) has been characterized previously (Zhu et al., 2013), and the rop2-1 knockout mutant (SALK_055328) was reported previously (Jeon et al., 2008). The gdi1-1 knockout mutant (SALK_129991) and the gdi1-2 knockdown mutant (SALK_035400) were identified. A PCR-based approach was used to identify homozygous lines. qRT-PCR analysis was performed to test whether gdi1 homozygous plants generated AtRhoGDI1/SCN1 transcripts. Total RNA was extracted from 10-d-old-seedlings using the RNAprep pure Plant Kit (Tiangen) according to the manufacturer’s instructions. SYBR Premix Ex Taq (Takara Bio) was used for amplification. The primer sets for PCR and qRT-PCR analysis are listed in Supplemental Table S1.
MAP18pro::MAP18-eGFP transgenic plants has been described previously (Zhu et al., 2013). MAP18pro::MAP18-mCherry, MAP18pro::MAP18ΔN23-eGFP, and MAP18pro::MAP18G2A-eGFP were generated (see below) and transformed into map18 mutant plants. MAP18pro::MAP18ΔN23 was generated (see below) and transformed into GFP-ROP2 plants. GDI1pro::GDI1 was generated (see below) and transformed into gdi1-1 or gdi1-2 mutant plants for complemented lines. ROP2pro::GFP-ROP2 was generated (see below) and transformed into rop2-1 mutant plants for complemented lines. 35S::GFP-ROP2, 35S::CA-rop2, and 35S::DN-rop2 seedlings, which have been described previously (Jones et al., 2002), were obtained from Zhenbiao Yang. 35S::GFP-fABD2-GFP, an F-actin reporter described by Wang et al. (2008), was obtained from Xuechen Wang, and GFP-MBD driven by the promoter UBQ (Marc et al., 1998) was obtained from Geoffrey O. Wasteneys. Super::RIC4ΔC-GFP was constructed (see below) and transformed into wild-type and map18 mutant plants to generate transgenic lines expressing RIC4ΔC-GFP.
Plants were grown in soil at 22°C with a photoperiod of 16 h of light/8 h of dark. Seeds were sterilized for 15 min in 5% (v/v) sodium hypochlorite and treated in growth medium at 4°C in the dark for 2 d before transferring to the growth room. Growth medium contained one-half-strength Murashige and Skoog salts with 0.9% (w/v) plant tissue culture (TC) agar (PhytoTechnology Laboratories). The 4-d-old seedlings were then transferred to different plates for various treatments.
Measurement of Root Hair Length
Root hairs were observed in 5-d-old seedlings grown on one-half-strength Murashige and Skoog agar medium. The region from tip to 4 mm of each primary root tip was examined as described by Zhang et al. (2015). Statistics were performed with SPSS statistics software (version 17.0) using one-way ANOVA followed by lsd and Student-Newman-Keuls posthoc analysis (*, P < 0.05).
Plasmid Construction and Protein Purification
The CA-ROP2 and DN-ROP2 site-directed mutagenesis coding sequence (Li et al., 2001) was amplified by PCR and cloned into the BamHI and SalI sites of vector pET30a+ (Novagen) to generate His-tagged recombinant His-CA-ROP2 and His-DN-ROP2, respectively. The cDNA sequence of SCN1 was obtained from The Arabidopsis Information Resource (SCN1, AT3G07880.1; http://www.arabidopsis.org) and then constructed into the pGEX-4T vector (Amersham Biosciences) for the preparation of recombinant GST-SCN1 protein or constructed into the pET30a+ vector for the preparation of recombinant His-SCN1. The recombinant proteins were transformed into Escherichia coli strain BL21 (DE3) and induced to express and then purified according to Cao et al. (2013) for His tag and Wang et al. (2007) for GST tag. The recombinant truncated protein MAP18ΔN23 and a G2A point mutation (MAP18G2A) were amplified by PCR and then constructed into the pGEX-4T vector for the preparation of GST-MAP18ΔN23 and GST-MAP18G2A proteins. PCR-based site-directed mutagenesis was performed according to the method described by Zhu et al. (2013). The coding sequence of mCherry was amplified by PCR and cloned into the BamHI and SmaI sites of vector pCAMBIA1300 (Cambia) to generate MAP18-mCherry reporter constructs driven by the MAP18 promoter. The AtRhoGDI1/SCN1 promoter gene and the coding sequence of AtRhoGDI1/SCN1 were amplified by PCR and cloned into pGoldenGate-MCY2 vector (Emami et al., 2013) to generate GDI1pro::GDI1. The coding sequence and promoter gene of ROP2 and the GFP coding sequence were amplified by PCR and cloned into the vector pCAMBIA1300 (Cambia) to generate GFP-tagged recombinant ROP2pro::GFP-ROP2. RIC4ΔC was amplified by PCR according to Hwang et al. (2005) and cloned into the vector Super::pCAMBIA1300 (Cambia) to generate GFP-tagged recombinant RIC4ΔC-GFP.
Protein concentrations were determined by the Bio-Rad Protein Assay Kit II (5000002). The list of primer sets used in this work is included in Supplemental Table S1.
Pull-Down Assays
For in vitro pull-down assays, GST (10 μg; used as a negative control for GST-MAP18), GST-MAP18 (10 μg), or GST-SCN1 (10 μg) was preincubated with glutathione-Sepharose 4B resin (Amersham Pharmacia) in 200 μL of phosphate-buffered saline (PBS; 10 mm Na2HPO4, 1.8 mm KH2PO4, 140 mm NaCl, and 2.7 mm KCl, pH 7.4) for 30 min at 4°C and mixed with 10 μg of His-CA-ROP2 or His-DN-ROP2. After 1 h of incubation at 4°C, the pellet was washed three times with PBS. Protein concentrations were determined by a Bio-Rad protein assay kit. Proteins in the pellet were separated by 12% (w/v) SDS-PAGE and transferred to a nitrocellulose membrane. Protein gel blotting was performed using anti-His antibody (1:5,000 dilution; monoclonal anti-polyhistidine [Sigma-Aldrich; H1029-0.2ML]), and anti-mouse antibodies conjugated with horseradish peroxidase were used to detect anti-His antibodies (1:10,000 dilution; Abmart).
For semi-in vivo pull-down assay, GST-MAP18 or GST-SCN1 was used to pull down GFP-ROP2 from seedlings. The protein extracts were prepared from 10-d-old seedling roots expressing GFP-ROP2 as described previously (Lin et al., 2012) with minor modifications. Briefly, total protein was extracted using extraction buffer (100 mm HEPES, 10 mm MgCl2, 10 mm EDTA, 5% (v/v) glycerol, protease inhibitor [Complete Tablets EDTA-free, EASYpacy, 4693132001; Roche], 1 mm phenylmethylsulfonyl fluoride, 10 mm DTT, and 0.5% (v/v) Trion X-100, pH 7.4). Then, protein extracts were pretreated with 100 μm GTP-γ-S (G8634; Sigma-Aldrich) or 1 mm GDP (G7127; Sigma-Aldrich) for 15 min at 22°C. MgCl2 was then added to the mixture at a final concentration of 60 mm. GST, GST-MAP18, or GST-SCN1 was preincubated with glutathione-Sepharose 4B resin for 30 min at 4°C and then incubated with extract proteins for another 2 h at 4°C, and the pellet was washed three times with PBS. Proteins in the pellet were separated by 12% SDS-PAGE and transferred to a nitrocellulose membrane. Protein gel blotting was performed using anti-GFP antibody (1:3,000 dilution; GFP tag [7G9] mouse monoclonal antibody [M20004M; Abmart]).
For the interference assay, His-DN-ROP2 was preincubated with Ni-NTA agarose (30210; Qiagen) in 200 μL of PBS for 30 min at 4°C and mixed with 10 μg of GST-MAP18 and/or GST-SCN1 for 1 h. Proteins in the pellet were washed by 1 mL of PBS three to five times, separated by 12% SDS-PAGE, and transferred to a nitrocellulose membrane. Protein gel blotting was performed using anti-GST antibody (1:50,000 dilution; monoclonal anti-GST [Sigma-Aldrich; G1160-0.2ML]) to detect the binding amount of GST-MAP18 or GST-SCN1.
BiFC Assay
For BiFC analysis, ROP2, CA-ROP2, or DN-ROP2 was tagged with nYFP in the vector pSPYNE(R)173 and MAP18 was fused with cYFP in the pSPYCE (M) vector as described previously (Waadt et al., 2008). These constructs were driven by the 35S promoter and transiently expressed in Nicotiana benthamiana leaves using Agrobacterium tumefaciens strain GV3101 according to Waadt et al. (2008).
Co-IP Assay
For the MAP18 and ROP2 interaction assay, total proteins were extracted from Arabidopsis protoplasts coexpressing Super::MAP18-Myc and 35S::GFP-ROP2, Super::MAP18-Myc and 35S::GFP-DN-ROP2, Super::MAP18-Myc and 35S::GFP-CA-ROP2, or Super::MAP18-Myc alone. The protein extracts were incubated with anti-Myc agarose beads (anti-c-Myc agarose affinity gel antibody produced in rabbit [Sigma-Aldrich; A7470-1ML]). Proteins bound to the beads were detected with anti-GFP antibody.
For the interference assay, total proteins were extracted from Arabidopsis protoplasts coexpressing Super::SCN1-Flag and 35S::GFP-DN-ROP2, Super::MAP18-Myc and 35S::GFP-DN-ROP2 with Super::SCN1-Flag, or 35S::GFP-DN-ROP2 alone. The protein extracts were incubated with anti-Flag agarose beads (anti-Flag M2-agarose from mouse [Sigma-Aldrich; A2220-5ML]) and detected with anti-Myc (monoclonal anti-c-Myc antibody produced in mouse [Sigma-Aldrich; M4439-100UL]), anti-Flag (monoclonal anti-Flag antibody produced in mouse [Sigma-Aldrich; F3165-1MG]), or anti-GFP antibody.
LCI Assay
ROP2, CA-ROP2, or DN-ROP2 was cloned into 35S::cLuc vector and MAP18 was cloned into 35S::nLuc vector for LCI assay. A. tumefaciens strain GV3101 containing various pairs of constructs was infiltrated into N. benthamiana leaves according to the protocol described previously (Walter et al., 2004). Firefly LCI assay was performed as described (Chen et al., 2008).
Biochemical Assay of ROP2 Activity
GFP-tagged active ROP2 was pulled down by the use of MBP-RIC1 according to Lin et al. (2012). The protein extracts were prepared from root tissue of 10-d-old 35S::GFP-ROP2 transgenic Arabidopsis seedlings and analyzed by immunoblot assay with anti-GFP antibody.
All blots and stained gels were scanned at 1,200-dpi (dots per inch) resolution, and band intensities were quantified using ImageJ to obtain the ratios of the amount of GTP-bound ROP2 relative to the amount of total GFP-ROP2 (GDP-bound and GTP-bound forms). Each assay was repeated five times.
Microtubule Binding and F-Actin-Severing Assays
To analyze the binding activity of various recombinant mutant proteins of MAP18 with microtubules, the preparation of purified porcine brain tubulin and cosedimentation experiments were performed as described previously (Mao et al., 2005; Wang et al., 2007).
To visualize the F-actin-severing activity of various recombinant mutant proteins of MAP18, Alexa 488-phalloidin-labeled F-actin was prepared, and experiments were performed according to previously published protocols (Zhu et al., 2013).
Spinning-Disk Confocal Microscopy and Quantitative Analyses
Seedlings expressing MAP18-mCherry/-eGFP or GFP-ROP2 were observed on a spinning-disk confocal microscope as described by Zhang et al. (2015).
Images were acquired using iQ software (Andor Technology). GFP was excited using 488-nm argon lasers, and emission was collected through 525- ± 5.5-nm filters. mCherry was excited using 561-nm argon lasers, and emission was collected through 593- ± 7-nm filters.
To quantify the localization of GFP-ROP2, mean fluorescence intensity of GFP signal at the apical PM (along the PM with length of 30 μm) and cytosol (area of 40 μm2) was measured using ImageJ software.
Drug Treatment
Oryzalin (3,5-dinitro-N4,N4-dipropylsulfanilamide; Sigma-Aldrich; 36182), a microtubule-specific depolymerized drug; taxol (paclitaxel [taxol equivalent]; Invitrogen; P3456), a microtubule polymerization stabilizer; LatB (Sigma-Aldrich; L5288), an actin polymerization inhibitor; and phalloidin (Invitrogen; P3457), an actin polymerization promoter, were used. Stocks were made in dimethyl sulfoxide and diluted at least 1,000-fold in our study.
For short-time treatment, 5-d-old seedlings were transferred to liquid one-half-strength Murashige and Skoog medium containing 500 nm oryzalin or 100 nm LatB for 10 min or containing 7 μm taxol or 3 μm phalloidin for 60 min.
For long-time treatment, 4-d-old seedlings were transferred to one-half-strength Murashige and Skoog medium containing oryzalin, LatB, taxol, or phalloidin at final concentrations of 150 nm, 50 nm, 5 μm, or 200 nm for 1 d, respectively.
Accession Numbers
Sequence data from this article can be found in Arabidopsis Genome Initiative database under the following accession numbers: MAP18 (Arabidopsis MAP18, At5g44610), ROP2 (Arabidopsis ROP2, Rac-like GTP-binding protein ARAC4, At1g20090), SCN1 (Arabidopsis SCN1, AtRhoGDI1/SCN1, At3g07880), and RIC4 (Arabidopsis RIC4, ROP-interactive CRIB motif-containing protein4, At5g16490).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression level, root hair phenotype, and localization pattern of GFP-ROP2.
Supplemental Figure S2. Expression level, root hair phenotype, and localization pattern of MAP18-mCherry.
Supplemental Figure S3. PM-localized RIC4ΔC-GFP is decreased significantly in map18 root hairs.
Supplemental Figure S4. Microtubule organization in map18 mutant and MAP18 overexpression growing root hairs.
Supplemental Figure S5. Short-time treatment with taxol or phalloidin, which cause microtubules or F-actin bundling but do not induce phenotypic changes of root hairs.
Supplemental Figure S6. MAP18 regulation of ROP2 activity is likely independent of microtubules or F-actin.
Supplemental Figure S7. Isolation of gdi1 knockdown and knockout mutants.
Supplemental Figure S8. MAP18 does not interact with AtRhoGDI1/SCN1 in vitro.
Supplemental Figure S9. The original full size image of Figure 6C.
Supplemental Figure S10. The original full size image of Figure 6G.
Supplemental Figure S11. PM-localized GFP-ROP2 is decreased significantly in MAP18ΔN23-overexpressing root hairs.
Supplemental Figure S12. N23 does not influence F-actin-severing activity and microtubule binding capacity of MAP18.
Supplemental Figure S13. The subcellular localization and F-actin-severing activity of MAP18 are not affected by loss of function of ROP2.
Supplemental Figure S14. The N23 fragment does not bind to microtubules in vitro.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Zhenbiao Yang (University of California, Riverside) for providing the Arabidopsis seeds expressing 35S::GFP-ROP2, 35S::CA-rop2, and 35S::DN-rop2; Geoffrey O. Wasteneys (University of British Columbia) for providing the seeds of Arabidopsis expressing UBQ::GFP-MBD; Xuechen Wang (China Agricultural University) for providing the seeds of Arabidopsis expressing 35S::GFP-fABD2-GFP; and Jianmin Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing plasmids for LCI.
Glossary
- Co-IP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- LCI
luciferase complementation imaging
- PM
plasma membrane
- PM/Cyt
plasma membrane/cytosol
- LatB
latrunculin B
- qRT
quantitative reverse transcription
- PtdInsPs
phosphatidylphosphoinositide phosphates
- PBS
phosphate-buffered saline
Footnotes
This work was supported by the Joint NSFC-ISF Research Program, funded by the National Natural Science Foundation of China and the Israel Science Foundation (grant no. 31361140354 to Y.F.), the Natural Science Foundation of China (grant no. 31325001 to Y.F. and grant nos. 31371352 and 31571384 to L.Z.), and the Beijing Higher Education Young Elite Teacher Project (grant no. YETP0303 to L.Z.).
Articles can be viewed without a subscription.
References
- Bibikova TN, Blancaflor EB, Gilroy S (1999) Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J 17: 657–665 [DOI] [PubMed] [Google Scholar]
- Bischoff F, Vahlkamp L, Molendijk A, Palme K (2000) Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol Biol 42: 515–530 [DOI] [PubMed] [Google Scholar]
- Bloch D, Lavy M, Efrat Y, Efroni I, Bracha-Drori K, Abu-Abied M, Sadot E, Yalovsky S (2005) Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol Biol Cell 16: 1913–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D, Monshausen G, Singer M, Gilroy S, Yalovsky S (2011) Nitrogen source interacts with ROP signalling in root hair tip-growth. Plant Cell Environ 34: 76–88 [DOI] [PubMed] [Google Scholar]
- Boulter E, Garcia-Mata R (2010) RhoGDI: a rheostat for the Rho switch. Small GTPases 1: 65–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K (2010) Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol 12: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Wang L, Zheng M, Cao H, Ding L, Zhang X, Fu Y (2013) Arabidopsis AUGMIN subunit8 is a microtubule plus-end binding protein that promotes microtubule reorientation in hypocotyls. Plant Cell 25: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Carol RJ, Dolan L (2002) Building a hair: tip growth in Arabidopsis thaliana root hairs. Philos Trans R Soc Lond B Biol Sci 357: 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93: 269–309 [DOI] [PubMed] [Google Scholar]
- Cole RA, Fowler JE (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9: 579–588 [DOI] [PubMed] [Google Scholar]
- Diet A, Link B, Seifert GJ, Schellenberg B, Wagner U, Pauly M, Reiter WD, Ringli C (2006) The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-l-rhamnose synthase. Plant Cell 18: 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Duckett DM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami S, Yee MC, Dinneny JR (2013) A robust family of Golden Gate Agrobacterium vectors for plant synthetic biology. Front Plant Sci 4: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng QN, Kang H, Song SJ, Ge FR, Zhang YL, Li E, Li S, Zhang Y (2016) Arabidopsis RhoGDIs are critical for cellular homeostasis of pollen tubes. Plant Physiol 170: 841–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Ikeda Y, Ljung K, Serralbo O, Singh M, Heidstra R, Palme K, Scheres B, Grebe M (2006) Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr Biol 16: 2143–2149 [DOI] [PubMed] [Google Scholar]
- Foreman J, Dolan L (2001) Root hairs as a model system for studying plant cell growth. Ann Bot (Lond) 88: 1–7 [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y, Kawasaki T, Shimamoto K, Yang Z (2008) ROP/RAC GTPases. In Yang Z, ed, Intracellular Signaling in Plants. Annual Plant Reviews, Vol 33 Wiley-Blackwell, Oxford, pp 64–99 [Google Scholar]
- Garcia-Mata R, Boulter E, Burridge K (2011) The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol 12: 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Weeks PR, Fischer I (2000) MAP1B expression and microtubule stability in growing and regenerating axons. Microsc Res Tech 48: 63–74 [DOI] [PubMed] [Google Scholar]
- Grizot S, Fauré J, Fieschi F, Vignais PV, Dagher MC, Pebay-Peyroula E (2001) Crystal structure of the Rac1-RhoGDI complex involved in NADPH oxidase activation. Biochemistry 40: 10007–10013 [DOI] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr Opin Plant Biol 7: 527–536 [DOI] [PubMed] [Google Scholar]
- Hoffman GR, Nassar N, Cerione RA (2000) Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100: 345–356 [DOI] [PubMed] [Google Scholar]
- Huang GQ, Li E, Ge FR, Li S, Wang Q, Zhang CQ, Zhang Y (2013) Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytol 200: 1089–1101 [DOI] [PubMed] [Google Scholar]
- Huang S, Jin L, Du J, Li H, Zhao Q, Ou G, Ao G, Yuan M (2007) SB401, a pollen-specific protein from Solanum berthaultii, binds to and bundles microtubules and F-actin. Plant J 51: 406–418 [DOI] [PubMed] [Google Scholar]
- Hutt DM, Da-Silva LF, Chang LH, Prosser DC, Ngsee JK (2000) PRA1 inhibits the extraction of membrane-bound rab GTPase by GDI1. J Biol Chem 275: 18511–18519 [DOI] [PubMed] [Google Scholar]
- Hwang JU, Gu Y, Lee YJ, Yang Z (2005) Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol Biol Cell 16: 5385–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JU, Wu G, Yan A, Lee YJ, Grierson CS, Yang Z (2010) Pollen-tube tip growth requires a balance of lateral propagation and global inhibition of Rho-family GTPase activity. J Cell Sci 123: 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon BW, Hwang JU, Hwang Y, Song WY, Fu Y, Gu Y, Bao F, Cho D, Kwak JM, Yang Z, et al. (2008) The Arabidopsis small G protein ROP2 is activated by light in guard cells and inhibits light-induced stomatal opening. Plant Cell 20: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilkine A, Marée AF, Edelstein-Keshet L (2007) Mathematical model for spatial segregation of the Rho-family GTPases based on inhibitory crosstalk. Bull Math Biol 69: 1943–1978 [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Yang Z, Smirnoff N (2007) NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J Exp Bot 58: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Aoyama T, Maeshima M (2013) The Ca2+-binding protein PCaP2 located on the plasma membrane is involved in root hair development as a possible signal transducer. Plant J 74: 690–700 [DOI] [PubMed] [Google Scholar]
- Kato M, Nagasaki-Takeuchi N, Ide Y, Tomioka R, Maeshima M (2010) PCaPs, possible regulators of PtdInsP signals on plasma membrane. Plant Signal Behav 5: 848–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, de Ruijter NCA, Emons AMC (2003) Unstable F-actin specifies the area and microtubule direction of cell expansion in Arabidopsis root hairs. Plant Cell 15: 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Emons AMC (2001) The cytoskeleton in plant cell growth: lessons from root hairs. New Phytol 152: 409–418 [DOI] [PubMed] [Google Scholar]
- Ketelaar T, Emons AMC (2009) The actin cytoskeleton in root hairs: a cell elongation device. In Emons AMC, Ketelaar T, eds, Root Hairs. Springer-Verlag, Berlin, pp 211–232 [Google Scholar]
- Klahre U, Becker C, Schmitt AC, Kost B (2006) Nt-RhoGDI2 regulates Rac/Rop signaling and polar cell growth in tobacco pollen tubes. Plant J 46: 1018–1031 [DOI] [PubMed] [Google Scholar]
- Klahre U, Kost B (2006) Tobacco RhoGTPase ACTIVATING PROTEIN1 spatially restricts signaling of RAC/ROP to the apex of pollen tubes. Plant Cell 18: 3033–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B. (2008) Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol 18: 119–127 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Szumlanski A, Nielsen E, Yang Z (2008) Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. J Cell Biol 181: 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Yang Z (2008) Tip growth: signaling in the apical dome. Curr Opin Plant Biol 11: 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol 126: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang X, Qin T, Zhang Y, Liu X, Sun J, Zhou Y, Zhu L, Zhang Z, Yuan M, et al. (2011) MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 23: 4411–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Brechenmacher L, Cheng J, Xu D, Stacey G (2010) Root hair systems biology. Trends Plant Sci 15: 641–650 [DOI] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, et al. (2012) A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol 22: 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Domergue O, Teyssendier de la Serve B, Rossignol M (1997) A new family of plasma membrane polypeptides differentially regulated during plant development. Biochem Mol Biol Int 43: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Mack TG, Koester MP, Pollerberg GE (2000) The microtubule-associated protein MAP1B is involved in local stabilization of turning growth cones. Mol Cell Neurosci 15: 51–65 [DOI] [PubMed] [Google Scholar]
- Mao T, Jin L, Li H, Liu B, Yuan M (2005) Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol 138: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao T, McCubbin AG, Cyr RJ (1998) A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marée AF, Jilkine A, Dawes A, Grieneisen VA, Edelstein-Keshet L (2006) Polarization and movement of keratocytes: a multiscale modelling approach. Bull Math Biol 68: 1169–1211 [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Yang Z (2010) RHO GTPase in plants: conservation and invention of regulators and effectors. Small GTPases 1: 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY (2006) RAC/ROP GTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci 11: 309–315 [DOI] [PubMed] [Google Scholar]
- Ovečka M, Lang I, Baluška F, Ismail A, Illeš P, Lichtscheidl IK (2005) Endocytosis and vesicle trafficking during tip growth of root hairs. Protoplasma 226: 39–54 [DOI] [PubMed] [Google Scholar]
- Park HO, Bi E, Pringle JR, Herskowitz I (1997) Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci USA 94: 4463–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RJ, Grierson CS (2009) A theoretical model for ROP localisation by auxin in Arabidopsis root hair cells. PLoS ONE 4: e8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei W, Du F, Zhang Y, He T, Ren H (2012) Control of the actin cytoskeleton in root hair development. Plant Sci 187: 10–18 [DOI] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H (2008) Rho GTPase activation assays. Curr Protoc Cell Biol 38: 14.8.1–14.8.19 [DOI] [PubMed] [Google Scholar]
- Riederer BM. (2007) Microtubule-associated protein 1B, a growth-associated and phosphorylated scaffold protein. Brain Res Bull 71: 541–558 [DOI] [PubMed] [Google Scholar]
- Seabra MC, Wasmeier C (2004) Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol 16: 451–457 [DOI] [PubMed] [Google Scholar]
- Sorek N, Segev O, Gutman O, Bar E, Richter S, Poraty L, Hirsch JA, Henis YI, Lewinsohn E, Jürgens G, et al. (2010) An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr Biol 20: 914–920 [DOI] [PubMed] [Google Scholar]
- Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L (2008) Local positive feedback regulation determines cell shape in root hair cells. Science 319: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M (2007) Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Yoo CM, Blancaflor EB (2008) Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol 177: 525–536 [DOI] [PubMed] [Google Scholar]
- Winge P, Brembu T, Kristensen R, Bones AM (2000) Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics 156: 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol 147: 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fu Y (2007) ROP/RAC GTPase signaling. Curr Opin Plant Biol 10: 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CM, Quan L, Cannon AE, Wen J, Blancaflor EB (2012) AGD1, a class 1 ARF-GAP, acts in common signaling pathways with phosphoinositide metabolism and the actin cytoskeleton in controlling Arabidopsis root hair polarity. Plant J 69: 1064–1076 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kang E, Yuan M, Fu Y, Zhu L (2015) PCaP2 regulates nuclear positioning in growing Arabidopsis thaliana root hairs by modulating filamentous actin organization. Plant Cell Rep 34: 1317–1330 [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Kang E, Xu Q, Wang M, Rui Y, Liu B, Yuan M, Fu Y (2013) MAP18 regulates the direction of pollen tube growth in Arabidopsis by modulating F-actin organization. Plant Cell 25: 851–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.