Light controls cytokinin signaling via transcriptional regulation of constitutively active sensor histidine kinase CKI1.
Abstract
In plants, the multistep phosphorelay (MSP) pathway mediates a range of regulatory processes, including those activated by cytokinins. The cross talk between cytokinin response and light has been known for a long time. However, the molecular mechanism underlying the interaction between light and cytokinin signaling remains elusive. In the screen for upstream regulators we identified a LONG PALE HYPOCOTYL (LPH) gene whose activity is indispensable for spatiotemporally correct expression of CYTOKININ INDEPENDENT1 (CKI1), encoding the constitutively active sensor His kinase that activates MSP signaling. lph is a new allele of HEME OXYGENASE1 (HY1) that encodes the key protein in the biosynthesis of phytochromobilin, a cofactor of photoconvertible phytochromes. Our analysis confirmed the light-dependent regulation of the CKI1 expression pattern. We show that CKI1 expression is under the control of phytochrome A (phyA), functioning as a dual (both positive and negative) regulator of CKI1 expression, presumably via the phyA-regulated transcription factors (TF) PHYTOCHROME INTERACTING FACTOR3 and CIRCADIAN CLOCK ASSOCIATED1. Changes in CKI1 expression observed in lph/hy1-7 and phy mutants correlate with misregulation of MSP signaling, changed cytokinin sensitivity, and developmental aberrations that were previously shown to be associated with cytokinin and/or CKI1 action. Besides that, we demonstrate a novel role of phyA-dependent CKI1 expression in the hypocotyl elongation and hook development during skotomorphogenesis. Based on these results, we propose that the light-dependent regulation of CKI1 provides a plausible mechanistic link underlying the well-known interaction between light- and cytokinin-controlled plant development.
Phytohormones from the cytokinin group regulate many fundamental physiological and developmental programs in plants, including embryo and seed development, germination, photomorphogenesis, plant growth and expansion, organ formation, vascular development, leaf senescence, plant immunity, and regulation of circadian rhythms (Werner and Schmülling, 2009; Kieber and Schaller, 2014). Cytokinin responses in Arabidopsis (Arabidopsis thaliana) are transduced via a multistep phosphorelay (MSP) pathway [for a recent review, see Hwang et al. (2012)]. In the MSP, the cytokinin signal is received by one of three sensor His kinases (HKs), AHK2, AHK3, and AHK4, which act as cytokinin receptors (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Activated HKs are supposed to phosphorylate His-containing phosphotransfer proteins (AHP1 through AHP5 in Arabidopsis; Mähönen et al., 2006), which act as positive regulators of cytokinin signaling (Hutchison et al., 2006) and are believed to transfer the P group to the nuclear-located final P acceptors, the B-type ARABIDOPSISRESPONSE REGULATORS (B-ARRs; Argyros et al., 2008). B-ARRs bind DNA and directly regulate the transcription of cytokinin-responsive genes (Sakai et al., 2001). There are also A-type ARRs (A-ARRs) that provide negative regulatory feedback in MSP signaling (Hwang and Sheen, 2001). A-ARRs are rapidly up-regulated upon cytokinin application even in the absence of translation, suggesting that they have roles as cytokinin primary response genes (Brandstatter and Kieber, 1998; D’Agostino et al., 2000). Thus, level of expression of A-ARRs can be regarded as a measure of MSP activity (Pernisová et al., 2009).
Sensor HK CKI1 was originally identified in an activation mutagenesis-based screen. Overexpression of CKI1 led to cytokinin-independent cell division and shoot formation in tissue culture, and thus it was proposed that CKI1 acts as a cytokinin receptor (Kakimoto, 1996). Subsequent studies have shown that although CKI1 is unable to either be activated by cytokinins or bind cytokinins (Yamada et al., 2001), it shares downstream signaling partners with the cytokinin signaling pathway (Hwang and Sheen, 2001; Hejátko et al., 2009; Deng et al., 2010). CKI1-mediated MSP signaling has been reported as regulating female gametophyte development (Pischke et al., 2002; Hejátko et al., 2003; Deng et al., 2010; Yuan et al., 2016), vascular tissue formation (Hejátko et al., 2009), and root elongation (Deng et al., 2010). Thus, although the CKI1 expression is hardly detectable (Hejátko et al., 2003, 2009), it seems to play an important role in MSP signaling and Arabidopsis development.
Light is one of the most important environmental signals for plants, modulating a broad spectrum of developmental processes. Regulation of plant development is strongly dependent on the spectral quality, quantity, direction, and periodicity of light (Chen et al., 2004; Kami et al., 2010). To date, several classes of photoreceptors have been identified in plants: phytochromes, cryptochromes, phototropins, and UVR8 and ZTL family proteins. Of these, phytochromes are the light receptors responsible for the majority of light perception (Quail et al., 1995; Rockwell et al., 2006; Li et al., 2011). Phytochromes are red/far-red absorbing photoconvertible (Pr inactive/Pfractive form) chromoproteins, and their photoconvertibility is mediated by phytochromobilin (PΦB), a covalently bound, light-absorbing cofactor molecule.
The existence of interactions between light- and cytokinin-mediated developmental regulations has been known for a long time. Light stimulates an increase in endogenous cytokinin levels (Mizuno et al., 1971; Qamaruddin and Tillberg, 1989; Kraepiel et al., 1994; Zubo et al., 2008). Cytokinins mediate photomorphogenic responses in etiolated Arabidopsis seedlings (Chory et al., 1994) and control expression and/or protein abundance of light-associated gene products, including the gene for the light receptor phytochrome A (phyA; Cotton et al., 1990; Bolle et al., 2000; Brenner et al., 2005, 2012). The interaction of one of the A-ARRs, ARR4, with another light sensor, phytochrome B (phyB), stabilizes phyB in its active (Pfr) form, resulting in plant hypersensitivity to red light (Sweere et al., 2001). On the other hand, it has been proposed that light-mediated up-regulation of cytokinin signaling interferes with auxin in the regulation of stem cell activity during shoot apical meristem (SAM) organogenesis (Yoshida et al., 2011) and that light through cytokinin signaling up-regulates WUSCHEL in SAM (Pfeiffer et al., 2016). However, the molecular mechanism of light-dependent control over cytokinin signaling and/or response remains elusive.
In a forward genetic screen, we identified LONG PALE HYPOCOTYL (LPH) as an upstream factor defining the spatiotemporal expression pattern of CKI1. The lph mutation defines hy1-7, a novel allele of HEMEOXYGENASE1 (HY1). HY1 is a key protein in the biosynthesis of a cofactor of the photoconvertible light receptors, and disruption of its activity in hy1-7 results in defective light perception. We show that CKI1 is controlled by light via phyA-mediated signaling. Deregulation of CKI1 expression in the hy1-7 and phyA mutant causes disturbance in MSP activity and developmental defects previously linked with the activities of CKI1 and the MSP pathway. These results indicate a conceptually novel mechanism involving signal-mediated control of a constitutively active sensor that further controls its cognate signaling pathway and provides a mechanistic link between light- and MSP-mediated regulation of plant development.
RESULTS
Forward Genetic Screening Identifies LPH as an Upstream Regulator of CKI1 Expression
Overexpression of CKI1 mimics cytokinin effects allowing de novo organogenesis (shooting) on hormone-free media (Kakimoto, 1996). Thus, control of CKI1 expression may be an intrinsic regulatory mechanism with important developmental impact. To identify regulatory components defining the spatiotemporal expression pattern of CKI1, we employed a forward genetic screen. Previously, the correlation between CKI1 promoter activity, as monitored by a GUS reporter in the ProCKI1:uidA line and the endogenous CKI1 expression pattern, has been confirmed by both in situ CKI1 mRNA localization and CKI1 immunolocalization (Hejátko et al., 2003, 2009). On the basis of those results, we mutagenized the ProCKI1:uidA line using ethyl-methane-sulfonate to search for genes involved in the determination of CKI1 expression. Screening for mutants with altered CKI1 expression was facilitated using an automated microscopy method (Dobisová and Hejátko, 2014).
Among the candidate mutant lines, we identified a chlorotic mutant, long pale hypocotyl (lph), revealing extensive alterations in the CKI1 expression pattern in both the shoot and root (Fig. 1A and Supplemental Fig. S1A). Typically, in short-d-grown seedlings carrying ProCKI1:uidA in the wild-type background (further in the text referred to just as “wild type”), we found CKI1 expression in the SAM, and only very weak activity was identifiable in the vasculature of cotyledons (Fig. 1A and Supplemental Fig. S1A). CKI1 expression was also detectable in the vascular tissue of the root/shoot junction, the more mature (upper) part of the root, and in the lateral root cap (LRC). In contrast, in the lph background, the expression of CKI1 was not detected in the SAM and root/shoot junction, while it was up-regulated in the vasculature of cotyledons and the root meristem transition zone, and higher CKI1 activity was also detected in the LRC (Fig. 1A). Further changes were detectable in the vasculature of true leaves and in the generative developmental phase. Increase in the CKI1 expression could be found in vascular tissue of all lph floral organs (Supplemental Fig. S1A ).
Figure 1.
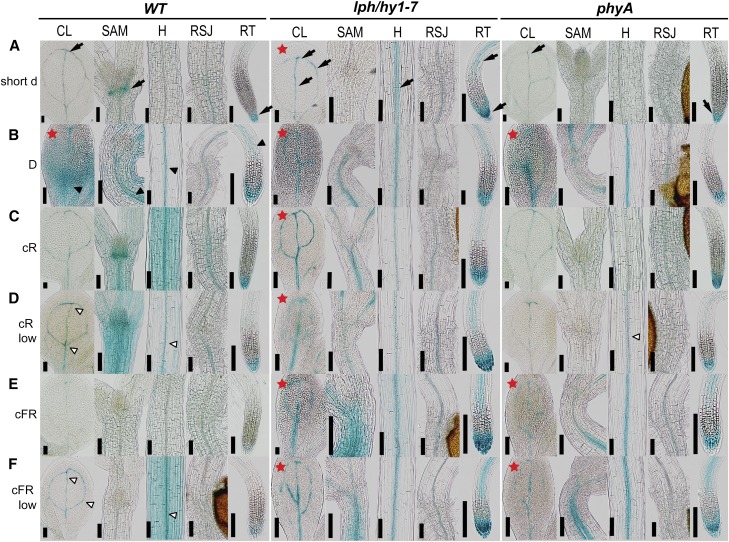
CKI1 expression under various light conditions (A to F) in wild-type, lph/hy1-7, and phyA seedlings. CKI1 expression was analyzed in 6-d-old ProCKI1:uidA seedlings grown under short day (short d; 8-h light/16-h darkness), darkness (D), continuous red (cR), and far-red (cFR) light (both 50 μmol m−2 s−1) and cR-low and cFR-low (both 2 μmol m−2 s−1). In the wild-type, developmental and light-controlled changes in CKI1 expression could be observed. Onset of etiolation leads to loss of CKI1 expression in the SAM, while it is up-regulated in the vasculature of hypocotyl and root transition zone. R light stimulates, but FR inhibits CKI1 activity. lph/hy1-7 disturbs light-dependent changes in CKI1 expression pattern and with some minor changes (see the main text), resembles the situation in the wild type under darkness. phyA largely phenocopies CKI1 expression in the lph/hy1-7. Note the absence of CKI1 activity in SAM of phyA under short-d and R conditions, suggesting phyA as a positive regulator of CKI1 in the absence of FR as the dominant light source. phyA mediates the negative effect of FR light on CKI1 activity indicated by persisting CKI1 expression in the phyA under FR conditions, suggesting the dual role of phyA in the control of CKI1. Arrows: Specific ProCKI1:uidA signal in individual tissues under the short d conditions. Red asterisk: Conditions leading to etiolation-specific CKI1 expression pattern. Black arrowhead: ProCKI1:uidA signal specific for etiolated seedling phenotype, shown only on wild type as an example. White arrowhead: CKI1 specific signal related to partially etiolation response under low light intensity. Scale bars: 100 μm. CL, cauline leaf; SAM, shoot apical meristem; H, hypocotyl; RSJ, root, shoot junction; RT, root tip.
The total amount of CKI1 mRNA in light-grown mutant seedlings was comparable to that in the wild type (Supplemental Fig. S1B), indicating that the mutation affects a factor determining the spatiotemporal pattern (specificity) rather than the overall strength of CKI1 expression. Besides the altered CKI1 expression pattern, the mutant was characterized by chlorosis and elongated hypocotyls. A segregation ratio of 3:1 (green and chlorotic plants) in the mapping population generated by crossing lph with Ler-0 suggests that the lph mutation is monogenic and recessive in nature.
lph Is a Novel Allele of HEME OXYGENASE1
To determine the molecular basis of the lph mutation, we employed map-based cloning. lph was found to be tightly linked to the SSLP marker nga1126, and, based on the phenotype of lph, HY1 was selected as the most likely candidate gene in this region. We found a point mutation in the genomic DNA of lph located close to the donor splicing site in the first intron of HY1 (Supplemental Fig. S2A). The corresponding region of the HY1 cDNA sequence was amplified (Supplemental Fig. S2B), and sequencing revealed the presence of an 81 bp deletion in the first exon of HY1 as a result of the formation of an alternative donor splicing site. This deletion corresponds to a loss of 27 amino acids from the N-terminal portion of HY1 and a G115D mutation (Supplemental Fig. S2, C and D). A genetic test for allelism (based on the phenotype of an F1 population after crossing lph with series of hy mutants; Supplemental Fig. S3) confirmed that lph is allelic with previously identified independent alleles of hy1 (hy1-1, hy1.6, and hy1-100). Based on these results, lph was redesignated hy1-7.
To determine whether there is a causal link between HY1 insufficiency and the control of CKI1 expression, ProCKI1:uidA was introgressed into a hy1-1 background. In the shoots of hy1-1/ProCKI1:uidA plants, we observed changes in CKI1 expression comparable to those in hy1-7/ProCKI1:uidA. However, in the root of hy1-1/ProCKI1:uidA, we detected a wild-type-like CKI1 expression pattern (Supplemental Fig. S3B). These findings are consistent with the fact that the hy1-7 phenotype exhibits the strongest defects in the shoot and the root development of all the tested hy1 mutants (Supplemental Fig. S3C and later in the text). In contrast, hy1-1 is a weaker allele, resembling the hy1-7 phenotype in the shoot, but exhibiting a wild-type-like root (Supplemental Fig. S3, C and D; and data not shown).
Overall, these data prove that there is a causal link between HY1 function and CKI1 expression, and show a positive correlation between changes in CKI1 expression and the strength of the hy1 phenotype.
Light Regulates Expression of CKI1
HY1 has been described as the key member of the heme oxygenase family (Davis et al., 1999; Muramoto et al., 1999). It is of crucial importance in the biosynthetic pathway of PΦB, a cofactor necessary for the formation of photoconvertible phytochromes and thus for proper light sensing (Parks and Quail, 1991; Rockwell et al., 2006). Light signaling deficiency, together with heme accumulation, also affects chlorophyll biosynthesis in hy1 (Chory et al., 1989; Terry and Kendrick, 1999). As indicated by the name that we originally used to designate the mutant line (lph), hy1-7 exhibits marked photomorphogenic defects, a phenotype that is consistent with the previously described role of HY1 in light perception (Davis et al., 1999; Muramoto et al., 1999). In hy1-7, total chlorophyll content is significantly decreased (Supplemental Fig. S4A) and hypocotyls of light-grown hy1-7 seedlings are longer in comparison to wild type (Supplemental Fig. S4, B and C). Taken together, the molecular nature of HY1, together with the previously reported interaction of light and cytokinin signaling pathways [reviewed by Zdarska et al. (2015)], led us to study the role of light in the control of CKI1 expression.
We inspected CKI1 expression under different light conditions. In addition to the previously tested short-d conditions (Fig. 1A), ProCKI1:uidA seedlings were grown in darkness (Fig. 1B) and in continuous red (cR) and far-red (cFR), both at photon fluxes of 50 μmol m−2 s−1 (Figs. 1, C and E; for the entire hypocotyl length see Supplemental Fig. S4E). In the etiolated (dark-grown) ProCKI1:uidA seedlings, the activity of the CKI1 promoter increased in the vasculature of the cotyledons, hypocotyl, and the transition zone of the root when compared with short-d conditions, while it was strongly down-regulated in the SAM. cR had mostly stimulatory effects on CKI1 activity: under cR illumination, CKI1 was up-regulated in the vasculature of the hypocotyl and cotyledons, in the SAM and in the nonvascular tissues of hypocotyl (both epidermis and ground tissues). In contrast, in cFR-grown seedlings, the CKI1 signal was almost absent from all tissues, suggesting a strong negative effect of cFR on CKI1 expression (Fig. 1E).
To investigate potential light dose-dependent effect on CKI1 expression, ProCKI1:uidA seedlings were grown in the presence of R and FR light of decreased intensity (cR-low and cFR-low, respectively, both at 2 μmol m−2 s−1). The overall expression pattern, when compared to that observed in the presence of cR and cFR, did not change, particularly in the SAM and root tip, suggesting wavelength-specific effects on CKI1 expression in those tissues. However, under the cR-low and cFR-low conditions we observed quantitative changes in the CKI1 activity in cotyledons and the hypocotyl (compare Fig. 1, D and F, and Supplemental Fig. S4E). In comparison to cR up-regulating CKI1 in both apical and basal parts of the hypocotyl, cR-low stimulated the CKI1 expression just under the SAM but not in the more distant (basal) hypocotyl portion. Under cFR-low (similarly to cFR), we observed inhibition of CKI1 in the SAM and the root tip. However, the CKI1 activity in cotyledons and the lower part of the hypocotyl was up-regulated when compared to short d (compare Fig. 1, A and F, and Supplemental Fig. S4E).
In contrast to wild type, the hy1-7 line exhibited dramatically decreased sensitivity of CKI1 to different light treatments. When compared to short-d conditions, partial light sensitivity of CKI1 expression in hy1-7 remained apparent only under cFR, leading to a slightly enhanced signal in the hypocotyl under the SAM (Fig. 1E and Fig. S4F). However, cFR light was unable to inhibit CKI1 expression in any tissue in the hy1-7 background. In general, irrespective of light conditions, the CKI1 expression pattern in hy1-7 resembled the CKI1 expression in the etiolated wild-type seedlings. However, some differences were found between the etiolated wild type and hy1-7. We have seen up-regulated CKI1 in the vasculature of cotyledons in hy1-7 while higher activity of CKI1 was apparent in the hypocotyl of wild type, and diffuse staining was apparent at the base of wild-type cotyledons (Fig. 1B and Supplemental Fig. S4F). These differences might have an important role in the changed behavior of wild type and hy1-7 during skotomorphogenesis (see below).
In conclusion, both light quality and quantity are important factors controlling the spatiotemporal specificity of CKI1 expression in Arabidopsis. As may be expected, considering the molecular nature of the mutation, hy1-7 inhibited light control over CKI1, leading to a changed, light-insensitive expression pattern, largely resembling the CKI1 expression observed in etiolated wild type irrespective of light growth conditions. The absence of CKI1 activity in the SAM of the dark-grown wild type and hy1-7 (under all light conditions) and expression of CKI1 in the SAM of R-grown seedlings implies the R light (not excluding the other components of the white light spectrum) as a positive regulator of CKI1 expression in the SAM. On the other side, FR light inhibits CKI1 expression in the SAM and columella/LRC.
CKI1 Expression Is Under Control of the phyA Signaling
The strong inhibitory effect of FR light on CKI1 expression suggested an important role for the light sensor phyA that is solely responsible for FR-mediated signaling in Arabidopsis (Nagatani et al., 1993; Reed et al., 1994). To further demonstrate the regulation of CKI1 by phyA in planta, we introduced the ProCKI1:uidA construct by crossing into the phyA background. Under short d, phyA partially phenocopied hy1-7, leading to the up-regulation of CKI1 in cotyledons with absence of CKI1 expression in the SAM. Further, again similarly to hy1-7, phyA impaired all the light-mediated changes of CKI1 activity observed in the wild type, i.e. cR-dependent up-regulation in the hypocotyl and cFR-mediated inhibition of CKI1 in the root tip (Fig. 1, C and E). The specific role of phyA in the cFR-mediated down-regulation of CKI1 in SAM cannot be determined, as there is no CKI1 activity in the SAM of phyA, irrespective of light conditions (as also seen in the case of hy1-7). However, in contrast to hy1-7, we did not see up-regulation of CKI1 in the vasculature of hypocotyl and root transition zone or in the LRC of short-d-grown phyA (compare wild type, hy1-7, and phyA in Fig. 1A). Based on the etiolated seedling phenotype of hy1-7 on short d and our further observations (discussed later in the text), these differences seem to be attributable to the etiolation-specific expression pattern of CKI1 (see below).
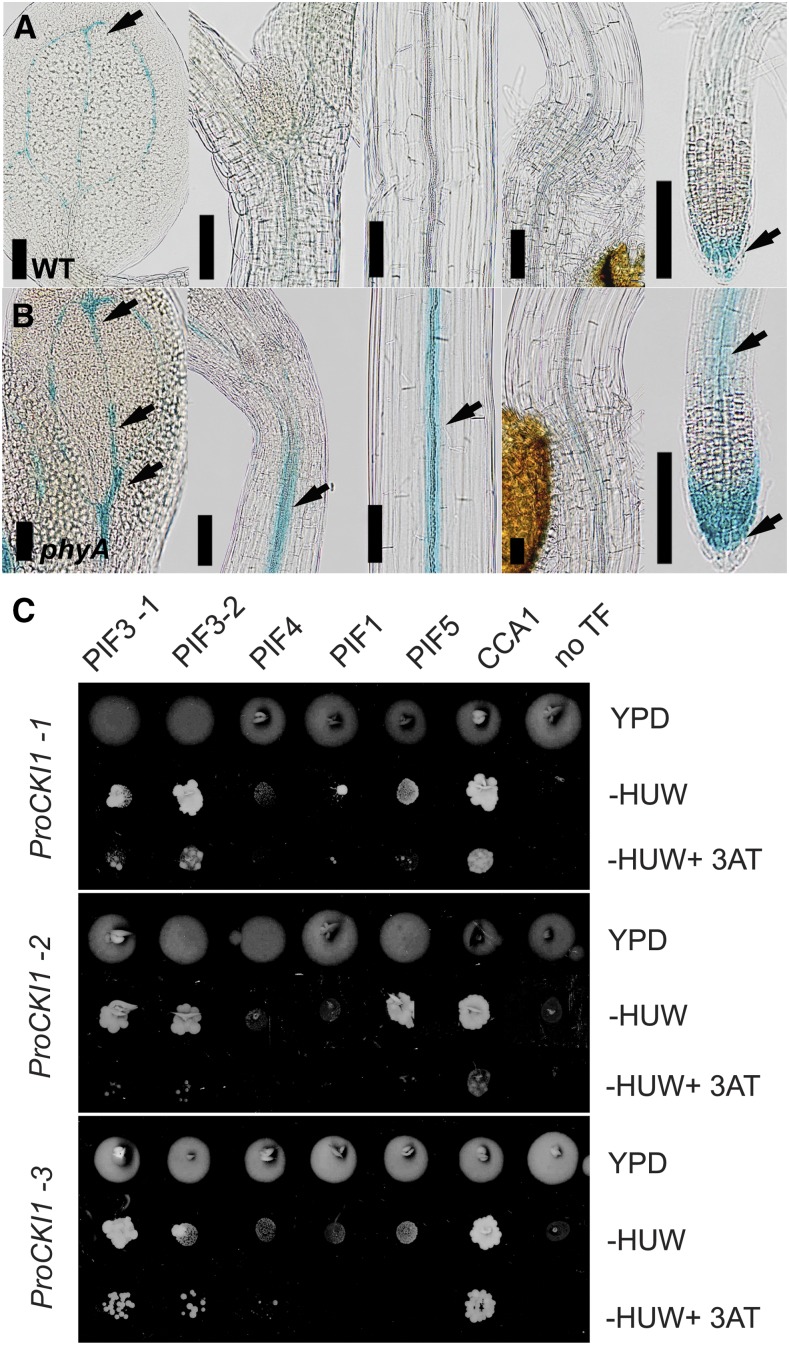
As the phyA is solely responsible for FR-mediated signaling, the etiolated phenotype is observed in FR-grown phyA. To confirm the importance of phyA in the FR-mediated inhibition of CKI1 expression, irrespective of possible etiolation-specific developmental regulations, we performed another type of experiment. Immediately after the dark adaptation phase, 4-d-old etiolated seedlings were grown for 2 d on cFR (Fig. 2, A and B). In the wild, under these conditions we detected only residual CKI1 activity (compare Figs. 1B and 2A), while in phyA, the CKI1 expression remained unchanged (compare Figs. 1B and 2B), thus confirming our previous results.
Figure 2.
CKI1 acts downstream of phyA-mediated signaling. A and B, In contrast to wild type, far-red light is unable to inhibit CKI1 expression in phyA (see also Fig. 1). To avoid possible bias due to developmental stage-specific changes, etiolated wild type and phyA carrying ProCKI1:uidA construct were treated with continuous far-red light (50 μmol m−2 s−1) immediately after the dark adaptation phase (4 d in darkness, 2 d on far-red). While in phyA, the CKI1 signal is still clearly detectable (arrows), only residual CKI1 activity can be detected in wild type, mostly in the vascular tissue of the cotyledons and in the LRC (arrows). Scale bars: 100 μm. C, Interactions between light-associated TFs and selected fragments of the CKI1 promoter (ProCKI1-1, ProCKI1-2, and ProCKI1-3) enriched in phyA-regulated motifs (see Supplemental Table S1 and Supplemental Fig. S5), identified by Y1H. Growth of yeast clones carrying the HIS3 reporter under the control of ProCKI1-1, ProCKI1-2, or ProCKI1-3 and selected TFs from the REGIA-REGULATORS collection was recorded after incubation for 6 d on either vector-selective media, interaction-selective media lacking His, uracil, and Trp or interaction-selective media supplemented with 10 mm 3AT, a competitive inhibitor of the HIS3 gene product. Only those yeast clones growing under the latter (more stringent) interaction-selective conditions were considered to be carrying interactors. PIF3 (both of the two independent clones available in the collection) specifically recognizes ProCKI1-1 and ProCKI1-2, while CCA1 interacts with all the fragments tested. HUW, His, uracil, and Trp; YPD, vector-selective media.
Under cR light, phyA shows a CKI1 expression pattern comparable to that of short-d conditions (compare phyA in Fig. 1, A and C, and Supplemental Fig. 4G). This is consistent with the fact that other phytochromes (probably dominantly phyB as the main R light receptor), are still active in phyA, and are able to inhibit the etiolation response. However, in contrast to wild type, even under cR light, phyB alone (or in cooperation with other phytochromes) is not sufficient to induce CKI1 expression in the SAM and/or in the hypocotyl in the absence of phyA (compare wild type and phyA in Fig. 1C and Supplemental Fig. S4, E and G). This, together with the down-regulation of CKI1 expression in the SAM of phyA under short-d conditions (Fig. 1A), implies the role of phyA as a positive regulator of CKI1 activity in the SAM in the absence of FR as the only light source.
To summarize, we show that white and R light up-regulates CKI1 promoter activity in a phyA-dependent way in the SAM and/or hypocotyl, respectively. Also, we show that CKI1 activity in LRC (and probably also in SAM) is under the phyA-mediated negative regulation by FR light.
phyA-controlled TFs Bind CKI1 Promoter
The aforementioned data clearly show that CKI1 is under control of phyA. Consistent with this, we found several phyA-responsive elements (Hudson and Quail, 2003) in the CKI1 promoter sequence. The presence of circadian rhythm-responsible elements, solid elements, and a large number of G-box and GATA motifs, is in a good accordance to our experimental data providing additional evidence that CKI1 is likely to be a light-regulated gene (Supplemental Table S1). To further substantiate the role of light-controlled TFs in the regulation of CKI1, we employed a Y1H assay to study the interaction of CKI1 promoter fragments enriched in these elements (Supplemental Fig. S5) with a selected subset of phy-regulated TFs (Obayashi et al., 2014). We found that a basic helix-loop-helix TF PHYTOCHROME INTERACTING FACTOR3 (PIF3) specifically recognizes two out of the three assayed CKI1 promoter regions, and the MYB-related TF CIRCADIAN CLOCK ASSOCIATED1 (CCA1) binds all three of the fragments (Fig. 2C). Thus, interaction of phytochrome-controlled TFs with CKI1 promoter indicates that CKI1 could be a direct target of phyA-mediated signaling.
CKI1 Expression Reveals Rhythmical Changes Integrating Both Light-Independent and Light-Regulated Control
It was previously shown that nuclear localization of phyA, and the activity of the phyA promoter, are under circadian control (Tóth et al., 2001; Kircher et al., 2002). Tóth et al. (2001) also showed that circadian oscillation within the phytochrome family is tissue specific, with the diurnal-controlled activity of phyA happening predominantly in the SAM. Our data showing binding of CCA1 to the CKI1 promoter implied possible circadian regulation of CKI1.
To investigate possible diurnal rhythms in CKI1 activity, we analyzed CKI1 expression in 6-d-old wild-type seedlings grown under long-d in various time intervals within a 48-h period. The time intervals for sample collection were set according to the published CCA1 oscillation pattern, where at the end of the light and night phase CCA1 reaches its expression minima and maxima, respectively (Gutiérrez et al., 2008; Flis et al., 2015; Supplemental Fig. S6). Certain variability in CKI1 expression pattern was observed even among seedlings collected in the frame of the individual time points. However, approximately 15 out of 20 seedlings were well synchronized, showing diurnal periodicity in the CKI1 expression (Supplemental Fig. S6). Under long-d conditions, CKI1 peaked in approximately 12-h intervals (with expression maxima at 12, 24, 36 and 48 h; Supplemental Fig. S6), thus covering both minima and maxima of CCA1. CKI1 predominantly fluctuated in the SAM and hypocotyl, implying possible tissue specificity of diurnal CKI1 regulation. During the light period, CKI1 activity was gradually increasing, reaching the maxima at the end of the light phase, followed by partial decrease of CKI1 expression early after dusk. Another maximum located just at the beginning of the following light phase and after partial decrease, the intensity of CKI1 expression was growing, again peaking just at the end of the light phase (Supplemental Fig. S6).
The diurnal oscillations of CKI1 could be caused either by alternation in light conditions or by activity of the internal circadian clock. To investigate the possibility of CKI1 regulation by circadian clock, several experiments were performed. As expected, the periodicity of CKI1 expression was partially impaired in seedlings grown for 6 d under continuous light. However, a certain level of periodicity still could be recognized, arguing in a favor of possible circadian regulation of CKI1 (Supplemental Fig. S6). Besides that, under continuous light conditions, we observed higher CKI1 activity in the SAM when compared to long-d (Supplemental Fig. S6), thus confirming our previous results indicating the positive role of light and the light dose, in the control of CKI1 in the SAM (Fig. 1; and Supplemental Table S2). In another experiment, the diurnal phase of 6-d-old long-d-grown seedlings was disturbed by placing the plants to the constant conditions (continuous light or dark) of the opposite phase (i.e. light phase was replaced by extended darkness; and vice versa, dark phase was replaced by continuous light). Under both the prolonged darkness and light phases, the trend in the CKI1 expression observed under short d was partially retained (Supplemental Fig. S6), further implying possibility of circadian regulation. However, both phase shifts also revealed light-controlled changes in CKI1 expression. Incubation in prolonged darkness apparently enhanced CKI1 expression in the hypocotyl close to the SAM and petioles, indicating a possibility of developmental-specific CKI1 expression pattern associated with the onset of etiolation. In the case of the prolonged light phase, after the initial decrease (circadian regulation), CKI1 expression in the SAM gradually increased, thus reflecting the light-mediated positive regulation observed in the continuous light grown seedlings.
In conclusion, CKI1 expression exhibits rhythmical changes, with an atypical 12-h (semidiurnal) cycle. However, besides the circadian regulation, changes in the light conditions seem to contribute to observed diurnal cycling of CKI1 expression.
CKI1 Shows Etiolation-Specific Expression Pattern
The observed up-regulation of CKI1 in the hypocotyl correlating with the onset of etiolation response under extended dark phase conditions (Supplemental Fig. S6) suggested possible developmental control over CKI1 expression, associated with dark adaptation. Accordingly, specific CKI1 expression pattern could be identified in all inspected genotypes, wild type, hy1-7, and phyA, when grown under conditions leading to the etiolation response (marked by an asterisk in Fig. 1). The etiolation-specific pattern could be characterized by up-regulated CKI1 in the vasculature of cotyledons, hypocotyl, and the root just above the transition zone and in columella/LRC (black arrowheads, Fig. 1). These developmental-specific effects seem to be responsible for most of the differences between hy1-7 and phyA (higher CKI1 expression in the vasculature of cotyledons, hypocotyl, and transition zone of the root in hy1-7) under short-d conditions, when hy1-7 (in contrast to phyA) shows an etiolation-like phenotype (Fig. 1 A and Supplemental Fig. S4, F and G). Up-regulated CKI1 activity in the hypocotyl vasculature could also be detected in the wild type and phyA under cR-low and wild type under cFR-low light, consistent with the increased hypocotyl length observed under these conditions, suggesting the onset of etiolation (Supplemental Fig. S4, D, E, and G). On the other side, the absence of CKI1 expression in the SAM of etiolated seedlings seems to be an effect of absent phyA signaling rather than an etiolation-specific developmental regulation, as the same effect (absence of CKI1 activity) could be observed in both short-d- and R-grown and thus deetiolated phyA (see above; and Fig. 1C and Supplemental Fig. S4G).
To sum up, CKI1 expression is under direct light (developmental-independent) control, particularly in the SAM and in columella/LRC. However, at the same time, CKI1 activity observed within the cotyledons, hypocotyl, and vasculature of the root transition zone seems to correspond to light-controlled developmental responses, particularly those associated with dark adaptation (onset of etiolation) and/or low light intensity, such as hypocotyl elongation and root shortening.
CKI1 Expression Controls Skotomorphogenesis
Changes in CKI1 expression associated with the initiation of etiolation (Supplemental Fig. S6), as well as the specific CKI1 expression pattern in the fully etiolated seedlings (Fig. 1), suggested a possible role of CKI1 during dark-adapted plant growth (skotomorphogenesis). To investigate this, we assayed the etiolated seedlings phenotype in hy1-7, phyA, and 35S:CKI1 plants. Hypocotyl elongation is one of the first responses to the lower-light intensity/darkness. Both hypocotyl growth dynamics, as well as final hypocotyl length in dark-grown seedlings, differed in opposite ways (i.e. being faster in hy1-7 and phyA, and slower in 35S:CKI1, when compared with respective wild-type controls; Supplemental Fig. S7, A, C, and D).
Besides elongated hypocotyls, dark-grown seedlings typically form an apical hook that protects the fragile shoot meristem during soil penetration (Raz and Ecker, 1999). Hook formation, and its opening, was previously demonstrated to be under hormonal and light control (Vriezen et al., 2004; Zádniková et al., 2010; Smet et al., 2014). Considering that, we investigated the potential impact of hy1-7 and phyA on the CKI1 expression during hook development and real-time kinetics of individual stages of the process, i.e. hook formation, maintenance, and opening (Raz and Ecker, 1999). Interestingly, the CKI1 expression pattern dynamically changed during the apical hook development (Supplemental Fig. S7B). In the wild-type seedlings at 30 h after germination (HAG), corresponding to the end of the hook formation stage, the CKI1 activity was rather weak and delimited to the hypocotyl vasculature. However, at 42 HAG, corresponding to the start of the hook opening, CKI1 was strongly up-regulated in the apical hypocotyl portion, including the bended (hook) part (Supplemental Fig. S7B). In hy1-7 and phyA, the CKI1 reporter activity at 30 HAG was comparable to the wild type, except of slightly higher CKI1 activity in the hypocotyl vasculature of hy1-7 (Supplemental Fig. S7B). However, at the later interval (42 HAG), both hy1-7 and phyA showed much weaker CKI1 expression in the apical portion of the hypocotyl proximally to SAM (Supplemental Fig. S7B). Accordingly, we observed dramatically prolonged maintenance phase in both hy1-7 and phyA (hook opens slower) when compared to that of wild type (Supplemental Fig. S7, E and F). Opposite differences in apical hook development were observed in the 35S:CKI1 line, where we found the hook opening phase shortened (the hook opens faster) in comparison to the wild type.
To sum up, the dynamic expression of CKI1 during etiolated seedling growth is deregulated in both hy1-7 and phyA, which corresponds with changes in hook development and hypocotyl elongation. Importantly, the developmental defects observed in dark-grown phyA mutant imply a light-independent role of phyA during skotomorphogenesis.
Light Signaling Controls Cytokinin Response in the Root
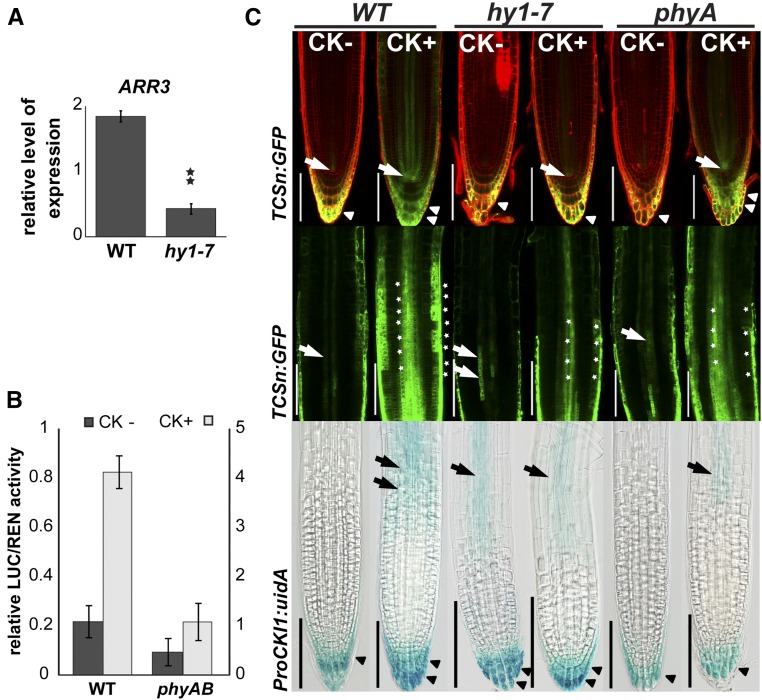
CKI1 acts through MSP and activates downstream responses, among them the transcription of type A-ARRs (Hwang and Sheen, 2001; Hejátko et al., 2009; Deng et al., 2010), the cytokinin primary response genes (D’Agostino et al., 2000). Altered CKI1 expression as a result of attenuated light perception in the hy1-7 mutant might, therefore, directly impact cytokinin responses, including A-ARRs transcription. Of eight A-ARRs tested, we found that ARR3 was strongly down-regulated in hy1-7 (Fig. 3A), suggesting a possible defect in cytokinin signaling due to disturbed light signaling. To further corroborate the role of phytochromes in the control of cytokinin signaling, we assayed cytokinin-mediated activation of MSP signaling using a TCS:LUC reporter (Müller and Sheen, 2008) in transiently transformed protoplasts prepared from a suspension culture of a double loss-of-function mutant defective in phyA and phyB, previously shown to be affected in the majority of phytochrome-mediated light responses (Reed et al., 1994). Our results show that in both mock- and cytokinin-treated samples, MSP activity was strongly reduced in phyA phyB protoplasts compared to that of wild type (Fig. 3B), suggesting decreased cytokinin responsiveness in the light signaling-defective mutant.
Figure 3.
hy1-7 exhibits disturbed MSP activity and attenuated cytokinin sensitivity. A, Relative quantification by qRT-PCR of ARR3 expression in 6-d-old wild-type and hy1-7 seedlings grown under short-d conditions. Data were normalized to UBIQUITIN 10. Statistically significant difference at α = 0.01 is shown (**; n = 3). B, Relative expression of the cytokinin-responsive MSP reporter (TCS:LUC) in protoplasts from suspension cultures of wild-type and the phyA phyB mutant line; error bars show sd. Impaired MSP signaling in phyA phyB is apparent in both control (values on y axis on the left) and cytokinin-induced (0.1 μm BA) protoplasts (values on y axis on the right). C, Disturbed MSP signaling and inducibility of CKI1 expression in root tips of 6-d-old seedlings by cytokinin (0.1 μm BA). Upper row: Expression of cytokinin-responsive MSP reporter TCSn:GFP in the root apical meristem. In the absence of exogenous cytokinins, MSP activity in wild type is detected in the stele close to the quiescent center (arrow) and in the LRC and columella (arrowhead). In hy1-7 and phyA, the MSP activity in the stele close to the QC center is missing. In hy1-7 and partially also in phyA, the TCSn:GFP signal is decreased in LRC and almost missing in the cells of columella. In wild type, exogenous cytokinin treatment strongly up-regulates the TCSn:GFP signal in the stele, LRC (double arrowhead) and columella. In contrast, only modest cytokinin-mediated up-regulation is apparent in LRC (double arrowhead) and columella of phyA, and only very weak increase of MSP activity is detectable in case of cytokinin-treated hy1-7 (single arrow/arrowhead). PI contrastaining was used (red signal). Middle row: MSP activity as measured by TCSn:GFP expression in the root including proximal meristem and transition zone of wild type, hy1-7, and phyA. In comparison to wild type (single arrow), both mutants show increased activity of MSP signaling in the root vasculature including the transition zone (double arrow) in the absence of exogenous cytokinins. However, both phyA and particularly hy1-7 show only weak increase in MSP activity upon exogenous cytokinin treatment in the vascular tissue and epidermis (*). Bottom row: Disturbed cytokinin inducibility of CKI1 as shown by ProCKI1:uidA activity. Cytokinin up-regulates expression of CKI1 in the root transition zone (arrow) and columella/LRC (double arrowhead) of wild type, but not in the cytokinin-insensitive hy1-7 line; cytokinin only partially activated CKI1 in the transition zone and columella/LRC of phyA. Note the higher level of expression in the vasculature of the transition zone and in the columella/LRC in hy1-7 in the absence of exogenous cytokinin. In phyA, the CKI1 expression is slightly decreased in the LRC (arrowhead) in the absence of cytokinins. Scale bars: 100 μm.
To study the possible spatial-specific changes of cytokinin signaling in the light signaling-defective lines, we introduced the cytokinin signaling reporter TCSn:GFP (Zürcher et al., 2013) by crossing into the hy1-7 and phyA mutants. In the mock-treated wild type, TCSn:GFP was active in columella/LRC and only weak signal was detectable in the (pro)vascular tissue close to the root tip. In comparison to that, we observed up-regulated cytokinin signaling (MSP output) in cell clusters of the root vasculature in both hy1-7 and phyA mutants in the absence of exogenous cytokinins (Fig. 3C). Exogenous cytokinin treatment led to the strong up-regulation of TCSn:GFP activity in the root tip of wild type, including the vasculature and epidermis of the transition zone and columella/LRC, as also described in Zürcher et al. (2013). In contrast, cytokinin induced almost no increase of TCSn:GFP in hy1-7 (weak up-regulation was apparent only in the root vasculature of hy1-7), and only partial activation was detectable in the vascular tissue and columella/LRC of phyA (Fig. 3C). Thus, in line with the protoplast assays, these data imply aberrant MSP signaling and disturbed ability of phyA and particularly hy1-7 to respond to cytokinins.
Interestingly, we found that not only light, but also cytokinins, control expression of CKI1. In the wild type, the presence of exogenous cytokinin led to CKI1 up-regulation in the vasculature of the root, particularly in the transition zone and in the columella/LRC (Fig. 3C). As mentioned previously, even in the absence of exogenous cytokinin, CKI1 was up-regulated in the vasculature of the transition zone in hy1-7, while wild-type-like expression (absent or very weak CKI1 activity) was observed in the transition zone of phyA (Fig. 3C). Consistent with the cytokinin insensitivity of TCSn:GFP reporter in hy1-7, CKI1 expression was not further up-regulated by cytokinin in the transition zone and columella/LRC of hy1-7 roots. Similar response, i.e. only partial up-regulation of CKI1, was observed in cytokinin-treated phyA (Fig. 3C).
Overall, we demonstrate that disrupting phytochrome-mediated light perception results in aberrant MSP signaling and disturbed ability of MSP pathway to respond to cytokinin. We also show that changes in CKI1 expression in light signaling-defective hy1-7 and phyA partially overlap with the changes in MSP output observed in those mutants.
Light-Regulated CKI1 Controls Root Growth
To further investigate the possible developmental consequences of disturbed cytokinin signaling in the root of light signaling mutants, wild-type, hy1-7, phyA, phyB, and phyA phyB seedlings were grown for two weeks on media supplemented with cytokinins. In the wild type, cytokinin treatment induced root hair formation and thickening of the differentiated (root hair-forming) portion of the root. In contrast, hy1-7 exhibited almost complete resistance to cytokinin in root hair formation and in primary root thickening (Fig. 4A and Supplemental Fig. S8A). Cytokinin-induced root hair formation was also disturbed in phyA and phyB single mutant lines. Interestingly, additive effects were apparent in phyA phyB double mutant, thus showing a role for both light receptors in cytokinin-regulated development (Fig. 4A). This is also in line with the previously documented role of phytochromes in root hair formation (De Simone et al., 2000).
Figure 4.
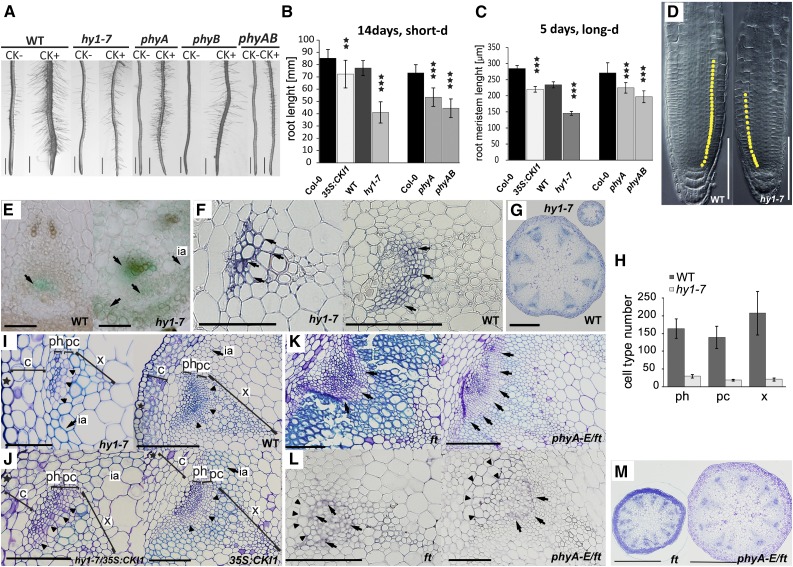
Changes in the spatiotemporal specificity of CKI1 expression are associated with changes in Arabidopsis meristematic activity. A, hy1-7 is insensitive to cytokinin with respect to root hair formation and root thickening (see also Supplemental Fig. S8). Additive loss of sensitivity to cytokinins is apparent in phyA, phyB, and phyA phyB mutants. Representative images show 14-d-old seedlings grown under short-d conditions in the presence (+) or absence (−) of cytokinin (0.1 μm BA). B to D, CKI1 is a negative regulator of root growth. Analysis of the root (B; n = 15) and the root apical meristem length (C; n = 20) in seedlings grown under white light. In comparison to wild type, hy1-7, like 35S:CKI1, has shorter roots (B) and reduced root apical meristem size (D); yellow dots in (D) indicate meristem cells. Note that the wild-type (ProCKI1:GUS) line is used as a control for hy1-7, while the Col-0 ecotype is used as a control for 35S:CKI1. Root length shortening, as well as root apical meristem reduction, is observed in the phytochrome mutants phyA and phyA phyB. Note additive effect of both phy mutations. E, ProCKI1:uidA expression profile in the vascular tissue of the inflorescence stem; arrows indicate CKI1 activity. In the wild type, CKI1 expression is predominantly localized in phloem adjacent to procambium, while in hy1-7, the CKI1 expression maximum is found mostly in the xylem. Weak activity in the phloem is also detectable. In hy1-7, up-regulation of CKI1 is apparent in the cortex, and ectopic CKI1 activity is detectable in the interfascicular arcs. F, Immunolocalization of CKI1 protein (arrows) in the vascular tissue of the inflorescence stem. In wild type, CKI1 is predominantly localized to the procambial cells, whereas the hy1-7 signal is shifted to the xylem cells. G, hy1-7 inflorescence stem show decrease in the number and size of VBs. Toluidine blue staining was used for VBs structure characterization. H, Clear reduction in numbers of all VB cell types is observed in hy1-7 (n = 3). I, Precocious lignification (light-blue stained cell walls, arrow) and reduction in the procambial cell layer (arrowheads) are observed in the basal portion of the inflorescence stem in hy1-7 when compared to wild type. Toluidine blue staining was used for VBs structure characterization. J, Partial rescue of hy1-7 phenotype by the 35S:CKI1, showing stronger, but dominantly wild-type-like (procambial) CKI1 localization (Hejátko et al., 2009). For the complementation of independent hy1-1 line, see Supplemental Fig. S9. In comparison to the parent hy1-7 line, hy1-7/35S:CKI1 reveals partial restoration of the procambial cell layer associated with increase of VB size and decreased lignification, particularly in the interfascicular arcs. Toluidine blue staining was used for VB structure characterization. K, Cross sections of the inflorescence stem of ft and phyA-E/ft mutant. In comparison to ft, phyA-E/ft reveals decrease in lignification (light-blue stained cell walls) and increased number of procambial cells (arrows). Toluidine blue staining was used for VB structure characterization. L, Immunolocalization of CKI1 protein (arrows) in the vascular tissue of ft and phyA-E/ft mutant line. Ectopic, mostly phloem localization of CKI1 in ft mutant is partially restored in the presence of phyA-E. However, ectopic CKI1 localization in the VB sheath cells (arrowheads) is still apparent in phyA-E/ft. M, Up-regulation of VBs number in ft is partially restored in the phyA-E/ft background. Toluidine-blue staining was used for VB structure characterization. Scale bars: 1 mm (A), 500 μm (G, M), 100 μm (D, E, F, I, J, K, L), and 50 μm (only for hy1-7 in E, F, and J). In all graphs, mean and sd are presented. Statistical significance determined by a t test at α-levels of 0.05, 0.01, and 0.001 (*, **, and ***, respectively) is shown. Star symbol (★), epidermal cell layer; c, cortex; ph, phloem; pc, procambium; x, xylem; ia, interfascicular arcs., long-d, long day; short d, short day.
Similar results, confirming the lowered cytokinin sensitivity of hy1-7, phyA, and phyA phyB, were observed in cytokinin-mediated root shortening, another well-established cytokinin sensitivity assay (Supplemental Fig. S8B). Root growth is largely dependent on the RAM activity. In particular, the equilibrium between cell division and cell differentiation in the root transition zone defines the size of the RAM and thus the extent of root growth (Dello Ioio et al., 2007). As mentioned in the previous section, in hy1-7 grown under control conditions (i.e. in the absence of exogenous cytokinins), the activity of the CKI1 promoter is enhanced in the vascular tissue of the root transition zone, which associates with the increase of MSP signaling in the root vasculature (Fig. 3C). We observed that this correlates with reduced root growth and smaller RAM, which is a phenotype reminiscent of that in plants overexpressing CKI1 (Fig. 4, B to D). Slightly up-regulated MSP signaling (weaker effect when compared with hy1-7) in the root vasculature of phyA in the absence of exogenous cytokinins (Fig. 3C), also correlates with the partial reduction of root length and RAM size (Fig. 4, B and C). In phyA phyB this effect becomes even stronger, indicating the role of phyB in the control of RAM size too (Fig. 4, B and C).
Altogether, disturbed cytokinin signaling and the reduced RAM size observed in hy1-7, phyA, and 35S:CKI1 lines, together with ectopic activation of MSP signaling in hy1-7 and partially also in phyA, suggests that the up-regulated CKI1 in both light-signaling mutants negatively affects RAM activity. This consequently inhibits root growth via up-regulation of MSP signaling in the root tip.
Altered CKI1 Expression Pattern Associates with Defects in Vascular Tissue Formation
The expression of CKI1 in vascular bundles (VBs) and the cortex of the inflorescence stem and its significance in VB development via control of procambial activity have been described previously (Hejátko et al., 2009). We therefore examined the impact of an altered CKI1 expression pattern in hy1-7 on VB development. In wild type, CKI1 transcription was mainly located in the phloem adjacent to the procambium. By contrast, the signal of CKI1 in hy1-7 was located mostly in the xylem, and weaker and less focused CKI1 activity was detectable in the phloem (Fig. 4E). These changes in CKI1 expression were confirmed by in situ CKI1 immunolocalization (Fig. 4F). Consistent with the observed changes in the CKI1 expression profile, we found VBs in hy1-7 to be severely affected in both number and architecture. The diameter of the hy1-7 inflorescence stem was drastically reduced, having a maximum of only six VBs in comparison to the more usual eight VBs in the wild type (Fig. 4G). In the individual VBs of hy1-7, the cell number of all three tissue types (phloem, procambium, and xylem) was substantially reduced (Fig. 4H). We also identified changes in the lignification of interfascicular arcs of both hy1-7 and 35S:CKI1 (Fig. 4, I and J).
Up-regulation of CKI1 in 35S:CKI1 was demonstrated to accelerate procambial activity (demonstrated by the thicker procambial layer; Fig. 4J) and thus the radial growth in the Arabidopsis inflorescence stem. Interestingly, stronger but mostly wild-type-like procambial localization of CKI1 has been demonstrated in 35S:CKI1, implying possible posttranscriptional regulations involved in the localization of CKI1 to the procambium (Hejátko et al., 2009). To confirm that changes in VB development observed in the hy1 background could be attributed to changes in the CKI1 expression pattern, we attempted to rescue the hy1-7 and independent hy1-1 allele phenotypes by crossing with the 35S:CKI1 line. In both hy1-7/35S:CKI1 and hy1-1/35S:CKI1, the VBs enlarged, the amount of the individual cell types per VB increased, and ectopic lignification decreased, thus presenting partial phenotypical rescue (Fig. 4J and Supplemental Fig. S9).
The importance of phyA in the number of developmental aspects was previously shown (Franklin and Quail, 2010). Its specificity within the developmental processes is highly dependent on the role and action of other light receptors (Cerdán and Chory, 2003; Franklin and Quail, 2010; Strasser et al., 2010). Thus, phyA-specific CKI1 regulation could differ in the early (seedling) and late (generative) plant development. Accordingly, when compared to wild type, we observed an increase in the VB number and diameter of the inflorescence stem in phyA, and the structure of VBs in phyA was also comparable to that of wild type (Supplemental Fig. S10). Thus, the role of phyA in the control over VB development seems to be minor and/or backed-up by other factors, most probably other phytochromes. To decipher the possible influence of other phytochromes on VB development, the quintuple mutant phyA-E in the flowering locus T (ft) mutant background was studied (Fig. 4, K to M). The phyA-E/ft was used because of the ability of ft to override the seedling developmental arrest observed in white light-grown phyA-E/FT (Strasser et al., 2010). Noteworthy, FT itself was found to be under phyB control (Cerdán and Chory, 2003). Firstly, we inspected the phenotype of ft mutant as a control to the phyA-E/ft line. Surprisingly, we observed increased number of VBs and highly up-regulated lignification in the ft inflorescence stems when compared to wild type. These developmental defects can be associated with ectopic, mostly phloem-specific and rather random localization of CKI1 (Fig. 4, K, L, and F). Interestingly, the phyA-E/ft mutants exhibit partial restoration of the VBs structure as compared to ft background. When compared with ft, the phyA-E/ft phenotype has a typical decreased number of VBs with thicker procambial layers, increased diameter of inflorescence stems, and fewer lignified xylem and interfascicular arc cells (Figs. 4, K to M). Accordingly, in the phyA-E/ft we found CKI1 to be arranged mostly to the phloem-adjacent procambial cells, thus partially overlapping with wild-type localization. However, persisting mislocalization of CKI1 into phloem and VB sheath was still apparent (Fig. 4L), confirming the role of phytochromes in the CKI1 control and VBs development.
In conclusion, altered CKI1 expression and CKI1 localization in hy1-7 largely correlates with phenotypic aberrations previously linked with CKI1 activity in the regulation of vascular development. Changes in the CKI1 localization and VB development could also be observed in multiple phyA-E/ft and ft mutant, providing further evidence that light signaling and light signaling-controlled processes impact CKI1-dependent plant development.
DISCUSSION
Light Controls CKI1 Expression
Here we identified HY1 as an upstream factor controlling the expression pattern of CKI1. HY1 in chloroplasts catalyzes cleavage of heme to form biliverdinIXa, which is further metabolized to the final product PΦB. PΦB is an essential cofactor covalently bound to phytochrome apoproteins, thus allowing formation of photoconvertible phytochromes (Weller et al., 1996; Emborg et al., 2006). In the Arabidopsis genome, four members of the heme oxygenase family (HY1, HO2, HO3, and HO4) have been identified. Further analysis showed that hy1 mutants contain no detectable holophytochrome and are almost insensitive to FR and R light. These findings suggest a dominant role for HY1 within the HO family in PΦB synthesis (Parks and Quail, 1991; Davis et al., 2001). In addition to attenuated light signaling, other developmental aberrations were reported in the heme oxygenase-deficient plants. Defects in chlorophyll biosynthesis and chloroplast development due to nonfunctional phytochrome signaling in hy1 mutants were described (Chory et al., 1989). Furthermore, chlorophyll biosynthesis in hy1 plants is also inhibited via a negative feedback loop mediated by heme accumulation (Terry and Kendrick, 1999). Based on our comparison of the lph/hy1-7 phenotype with the phenotypes of other hy1 mutants identified to date, it appears that hy1-7 represents one of the strongest alleles (Supplemental Fig. S3; Davis et al., 1999; Muramoto et al., 1999). Thus, besides aberrant light perception, light-independent defects could also be attributable to the hy1-7 phenotype, and only part of these defects being associated with changes in CKI1 expression. However, we have shown in two independent hy1 alleles (hy1-1 and hy1-7) that the phenotype strength correlates with the magnitude of changes in CKI1 activity. Furthermore, our data showing phenocopy of CKI1 expression in the independent phyA mutant provides evidence that compromised phyA signaling is the key factor contributing to the changes of CKI1 expression observed in hy1-7 seedlings.
Besides the role of phyA, the importance of phyB and/or other light receptors in the modulation of CKI1 expression cannot be completely excluded. In particular, the dual role of phyA in the control of CKI1 expression in the SAM (positive in the presence of white or R light while negative upon FR irradiation) might be explained by the existence of another light-controlled factor acting in synergy with phyA in a developmental-specific context. Interestingly, the light conditions under which CKI1 is active in the SAM can be explained by the presence of the two main Arabidopsis light receptors phyA and phyB in the nucleus (compare Fig. 1 and Supplemental Table S2). The necessity of coordinated action between phyA and another R/FR light receptor in the control of CKI1 activity is supported by the impaired ability of R light to up-regulate CKI1 in the SAM and hypocotyl of phyA (Fig. 1, C and D). Furthermore, it has been shown that not only phytochrome localization (cytoplasmic versus nuclear) but also the phytochrome content is essential for mediating light responses (Boylan and Quail, 1991; Whitelam et al., 1993). Under continuous light treatments most phyA is degraded, leading to the dramatic changes in the ratio among individual phytochromes (Sharrock and Clack, 2002). Under such conditions, we detected stronger CKI1 signal in SAM (compare Fig. 1, A and C, and Supplemental Fig. S6), implying that phyA abundance (possibly phyA to other phy ratio) could contribute to the control over CKI1 activity. That, however, remains to be identified.
According to this model, phyA translocates to the nucleus in a response to the light of several wave lengths (blue, R, FR; Hisada et al., 2000; Kim et al., 2000; Kircher et al., 2002) and (directly or indirectly) mediates phosphorylation of PIF3, a basic helix-loop-helix TF. In the response to FR light, phyA enters the nucleus and forms nuclear bodies, which, in their early form, colocalize with PIF3 (Bauer et al., 2004). PIF3 phosphorylation leads to its degradation and up-regulation of PIF3-inhibited gene expression (Al-Sady et al., 2006; Ni et al., 2013). PIF3 was found to be degraded under both R and FR conditions (Bauer et al., 2004), though with higher efficiency in the presence of R light (Al-Sady et al., 2006). Thus, PIF3 seems to integrate both R and FR signaling. Interestingly, PIF3 might act as both a positive and a negative transcriptional regulator (Sentandreu et al., 2011; Leivar and Monte, 2014). This is in line with our findings, suggesting a dual role of phyA in positive and negative control of CKI1 activity in the SAM, and with control of CKI1 expression in a response to both R (in the hypocotyl) and FR light (in the SAM and LRC). The spatial distribution of the phyA-mediated CKI1 expression also fits well with the phyA expression pattern, showing the strongest signal in the hypocotyl including SAM and in the LRC (Hall et al., 2001). Also, our data suggesting the role of PIF3 in the control over CKI1 are supported by an independent study reporting on PIF3 binding to the CKI1 promoter via ChIP-seq (Zhang et al., 2013). However, CKI1 expression at the whole-seedling level does not seem to be regulated by PIF3 (Tepperman et al., 2006; Zhang et al., 2013). That corresponds with our data showing an altered CKI1 expression pattern but only slightly and statistically insignificant changed total levels of the CKI1 transcript in hy1-7 (Supplemental Fig. S1B).
In parallel to PIF3, we detected binding of the CKI1 promoter to another light-associated TF, CCA1. CCA1 is a MYB-related TF involved in the regulation of circadian clock-regulated genes in Arabidopsis (Wang et al., 1997; Green and Tobin, 1999). Similarly to PIF3, CCA1 also acts downstream of phytochrome signaling (Tepperman et al., 2006) and may function as a transcription repressor or activator (Alabadí et al., 2001; Nagel and Kay, 2012). Interestingly, PIF3 has been supposed to up-regulate CCA1 via direct binding to its promoter (Martínez-García et al., 2000). Nevertheless, in pif3, CCA1 expression was largely unaffected, thus calling the importance of PIF3 in the regulation of CCA1 into question (Stephenson et al., 2009). In support of our Y1H results, the previously published CCA1 ChIP-Seq data (bioviz.org/igb/index.html; Nicol et al., 2009; Nagel et al., 2015) suggest that two of the three regions of the CKI1 promoter tested in our Y1H assay overlap with the CCA1-recognized genomic regions (Supplemental Fig. S11). The circadian cycling of CKI1 in SAM and adjacent apical hypocotyl portion suggest a possible role of CCA1 in this process (Supplemental Fig. S6). However, the observed semidiurnal (12 h) periodicity cannot be explained by the CCA1 expression itself, implying a more complex regulatory mechanism. These might include other factors such as both the morning- and evening-phased regulatory circuits (Pruneda-Paz and Kay, 2010) and/or light-controlled CKI1 expression, as apparent in both dark- and light-phase extension experiments. Alternatively, the semidiurnal periodicity might be explained as a lunar tides response, as described in the case of semidiurnal cycling of Arabidopsis root growth (Barlow et al., 2013).
Collectively, our data show that light quality and quantity are translated into changes in the spatiotemporal specificity of CKI1 expression via an as-yet unknown mechanism that includes the action of phyA and possibly other phytochromes, light receptors, or modulators of the light response. The diurnal oscillations in CKI1 activity seem to integrate both light-controlled and circadian (and/or other) regulations that might include further, yet elusive factors.
Light-Regulated Expression of CKI1 Provides a Molecular Link among Light, Cytokinin Signaling, and Meristematic Activity
Our data clearly show that the light-mediated regulation of CKI1 has important developmental consequences. We demonstrate that alterations in the spatiotemporal pattern of CKI1 expression in light perception-defective hy1-7 and phyA, as well as impaired light signaling in both single phyA phyB and double phyA phyB mutant, correspond with reduced cytokinin responsiveness of the MSP pathway and disturbed root cytokinin response. This implies a molecular link among phy-mediated control of the CKI1 expression, MSP pathway activity, and cytokinin-regulated development.
The phenotypic changes that we see due to disturbed expression of CKI1 in hy1-7 and phyA mutants are consistent with the previously identified role of cytokinin in the regulation of plant meristematic activity. A positive role for cytokinins and MSP-mediated cytokinin signaling in the regulation of procambial cell proliferation has been reported previously (Matsumoto-Kitano et al., 2008; Nieminen et al., 2008; Hejátko et al., 2009; Immanen et al., 2016). Manipulating CKI1 activity has been shown to affect procambial cell number, suggesting the role of CKI1 in the maintenance of procambial cell identity and/or regulation of procambial cell proliferation (Hejátko et al., 2009). The reduction in the size of the procambial layer in the VBs of hy1-7 is most probably a consequence of down-regulating CKI1 expression in the tissue adjacent to the procambium and a shift of the CKI1 expression maxima dominantly to the xylem. This seems to be confirmed by the ability of the 35S:CKI1 line, previously shown to reveal dominantly wild-type-like (i.e. procambial) CKI1 localization (Hejátko et al., 2009), to partially rescue the vascular phenotype of hy1-7 and hy1-1. Interestingly, the partial rescue of CKI1 localization in the phyA-E/ft background when compared to ft suggests that control over CKI1 expression integrates more complex regulatory interactions including individual phytochromes and/or phytochrome-controlled genes controlling inflorescence stem development, like FT.
In the RAM, cytokinin-activated MSP has been shown to inhibit root growth via induction of cell differentiation. Importantly, the vascular tissue of the root transition zone has been identified as the site of cytokinin action in this process (Dello Ioio et al., 2007). Thus, up-regulation of CKI1 in the vascular tissue of the transition zone in hy1-7 appears to mimic the cytokinin effects by activating MSP in this region, leading to a reduction in both the RAM size and root growth. This is further substantiated by the atypical activation of the MSP activity reporter TCSn:GFP in the root vasculature including the transition zone of hy1-7, which corresponds well with the position of ectopic CKI1 transcription observed in those tissues in the absence of exogenous cytokinins. The loss of cytokinin responsiveness of hy1-7 fits well to the previously published results demonstrating that CKI1 overexpression leads to the activation of MSP response that is, however, further insensitive to the exogenous cytokinin application (Hejátko et al., 2009). Finally, the negative role of CKI1 in RAM size and shortening of the main root is supported by the phenotype of the CKI1 overexpressing line, resembling the effects of exogenous cytokinin application (Figs. 3 and 4; Dello Ioio et al., 2007).
Consistent with our results showing the key effect of phyA on the expression of CKI1, we also observed RAM shortening in the phyA mutant line. In contrast to hy1-7, however, in phyA we were unable to see up-regulation of CKI1 in the transition zone of the root. Nonetheless, the additive effect of phyB to the RAM and root shortening in the phyA phyB background when compared to phyA suggests possible contribution of phyB to CKI1 expression, as discussed above. Thus, in this scenario, the up-regulation of CKI1 in the transition zone of the root below the detection limit (the CKI1 activity is very weak in general) would lead to the up-regulation of MSP activity in the root vasculature of phyA, as detected using the TCSn:GFP reporter and consequently RAM shortening.
LRC is a site of cytokinin biosynthesis (Aloni et al., 2004, 2005) and the functional importance of LRC in the control of RAM size has been demonstrated (Tsugeki and Fedoroff, 1999). Thus, the changes in CKI1 expression and defects in its cytokinin inducibility observed in the LRC of hy1-7 and phyA might also contribute to the changes in the RAM size of the light signaling mutants. Thus, although other mechanisms leading to RAM shortening in the cases of phyA and phyA phyB cannot be excluded, the ectopic expression of CKI1 overlapping with activation of MSP signaling in roots of both hy1-7 and phyA indicates that changes in the CKI1 expression in the light signaling mutants are at least partially responsible for the observed effects on RAM size.
Altogether, our findings show that transcriptional regulation of CKI1 by light impacts MSP activity, and affects MSP-mediated responses including cytokinin signaling. This fits well with the previously described role of CKI1 in the regulation of MSP activity (Hwang and Sheen, 2001; Hejátko et al., 2009; Deng et al., 2010), and implies that transcriptional regulation of CKI1 is an important mechanism mediating developmental regulations in Arabidopsis. As expected for this type of regulation, the spatiotemporal specificity of transcriptional control over CKI1, rather than changes in the total amount of the CKI1 transcript, is an important issue.
CKI1 and phyA Play a Role in Skotomorphogenesis
Our results show not only direct phytochrome-mediated control over CKI1 expression, but also developmental-specific expression of CKI1 associated with the light-controlled onset of etiolation. This implies a role for CKI1 during the dark-adapted developmental phase of plant growth (skotomorphogenesis) and/or the shade avoidance response. Surprisingly, we identified that in the absence of light, both hy1-7 and phyA show changes in hypocotyl growth and hook development when compared to wild type. This could show a potential role for (most probably) cytoplasmically located phyA and its importance within skotomorphogenesis. Cytoplasmic phytochrome signaling, and its developmental importance, has been unequivocally reported (Rösler et al., 2010; Hughes, 2013). The phyA response independent of light signaling has also been demonstrated (Correll and Kiss, 2005). Based on the differences in the rates of root growth between phyA phyB and wild type in the darkness, the authors suggested a role for the inactive (Pr) form of phytochromes in the root elongation. Nevertheless, these changes were not observed in other phyA mutants like phyA-201 (Ler ecotype), phyA-101, and phyA-105 (both RLD ecotype; Reed et al., 1994; Hoecker et al., 1998; Chen et al., 2015). Thus, we cannot exclude that the changes observed during skotomorphogenesis of phyA-211 could be allele/ecotype specific. In our study, we show similar changes in CKI1 expression and phenotype response (strong extension of the hook opening phase) in the dark-grown phyA (lacking the phyA protein) and hy1-7 (having a reduced amount of the photoconvertible phyA holoprotein). This suggests that the ability of phyA to be activated by light, rather than the presence of its inactive Pr form (as is the case in hy1-7), is necessary for proper CKI1 expression, hypocotyl elongation, and hook development in the dark. However, the light-independent role of photoconvertible phytochromes in the dark-adapted plants is still rather hypothetical. The effect of light used to activate seed germination (pregermination light) reported to affect hypocotyl elongation in dark-grown Arabidopsis seedlings (Alconada Magliano and Casal, 2004) could provide an alternative explanation.
CONCLUSIONS
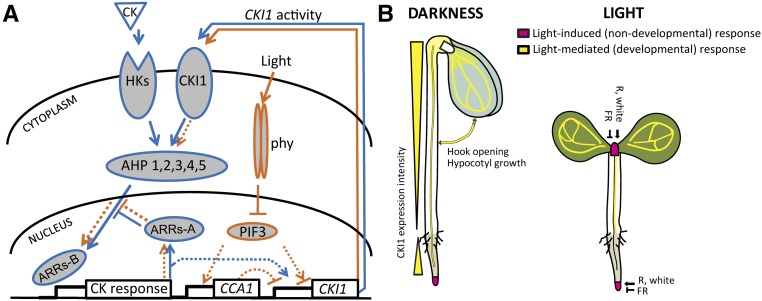
Identification of hy1-7 in a screen for upstream regulators of the CKI1 expression revealed an important molecular link between light and MSP signaling. Based on our findings, we propose a model in which the light-regulated expression of CKI1 controls MSP activity and thus sensitivity to cytokinins. In the model, light quality and quantity controls spatiotemporal specificity of CKI1 expression via an as-yet unknown mechanism that includes both positive and negative regulation of CKI1 via phyA-mediated signaling. The constitutive (cytokinin-independent) activity of CKI1 subsequently contributes to MSP signaling leading to changes in cytokinin sensitivity (Fig. 5A). This provides a direct molecular link between light signaling and several important aspects of cytokinin-mediated developmental regulation (including the cytokinin-regulated CKI1 expression), particularly the control of meristematic activity. Both light-mediated (developmental) and light-induced (nondevelopmental) effects participate in the control over CKI1 expression (Fig. 5B).
Figure 5.
Light-dependent CKI1 expression controls cytokinin signaling. A, Model of interaction between light and cytokinin signaling via light-controlled CKI1. Light initiates autophosphorylation and nuclear transport of phytochromes that interact with PIF3 and mediate its light-dependent degradation. PIF3, which is able to act as both positive and negative regulator, binds to the promoter of transcriptional repressor CCA1 and controls its expression. Both PIF3 and CCA1 bind directly to the CKI1 promoter. Via an as-yet unknown mechanism that includes the action of phyA, and possibly other light receptors, light quality is translated into changes in the spatiotemporal specificity of CKI1 expression that include both up- and down-regulation of CKI1 activity. Besides that, CKI1 is also under cytokinin control. The resulting changes in the expression of constitutively active CKI1 modulate MSP activity at the level of ARRs-A expression and regulate cytokinin sensitivity. The exact position at which cytokinin signaling is negatively regulated via ARRs-A is still not clear. For the sake of simplicity, it is shown here as inhibiting the final step in the MSP signaling. The nuclear transport of AHPs and phytochromes is not part of the model as well. (Color code: blue, CK signaling pathway and response; orange, phytochrome signaling pathway and light-mediated response.) B, CKI1 expression is both under direct (nondevelopmental) and indirect (developmental) light control. Light-induced (direct, nondevelopmental) control over CKI1 is apparent in the SAM and LRC, where R light, potentially together with other components of the white light, positively regulates CKI1 via phyA and possibly other light receptors. In parallel, phyA mediates negative regulation of CKI1 by FR light in those tissues. Light-mediated (indirect, developmental) regulation of CKI1 associates with etiolation, leading to up-regulation of CKI1 in the vasculature of cotyledons, hypocotyl, and root transition zone. The spatiotemporal specificity of CKI1 expression in etiolated seedlings controls apical hook opening, hypocotyl elongation, and root growth.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds of lines ProCKI1:uidA (Hejátko et al., 2003, 2009), the phyA-211 mutant (Reed et al., 1994; N6223), the phyA-211 phyB-9 double mutant (Strasser et al., 2010), 35S:CKI1(CKI1 2-2; Hejátko et al., 2009), Col-0 (the background ecotype of all the above lines; N1092), hy1-1 (NW67, background ecotype Ler; Koornneef et al., 1980), hy1-100 [also known as hy6 (cs236); background Col-0, LOS5; Chory et al., 1989] and hy1.62 (background Col-0, PoCA108; Vinti et al., 2000), and ft-1 (Lee et al., 2000) were used. ProCKI1:uidA was introduced to the phyA-211 and hy1-1 by crossing. The original phyA-211 cab3:uidA construct was outcrossed and does not interfere with the results. TCSn:GFP was introduced to phyA-211 and hy1-7 mutant lines by crossing, and two stable lines were used for phenotype analysis. Seeds were sterilized using 70% (v/v) ethanol and plated on full Murashige & Skoog salt media, 1% (w/v) plant agar, 1% (w/v) Suc (Duchefa Biochemie), supplemented when required with 0.1 μMBA (6-benzylaminopurine, dissolved in DMSO) or with 0.01% (v/v) DMSO (control). Plants were cultivated in growth chambers with controlled light, temperature (21°C/19°C, light/dark, respectively), and relative air humidity (50%). For histological analysis, and for GUS staining of leaves and inflorescences, we used flowering plants in phase II (when the first silique appears; Altamura et al., 2001). Light conditions were as follows: short d, photoperiod of 8/16 h (light/dark); long-d, photoperiod 16/8 h (light/dark); continuous light: photoperiod 24/0 (light/dark); (continuous) darkness (D): 24 h of induction by light (to initiate germination) followed by continuous cultivation under dark conditions. White light at an intensity of 150 μmol m−2 s−1 was used. For cultivation under light of defined quality, plants were cultivated in growth chambers (CLF floraLEDs; CLF Plant Climatics) equipped with diodes emitting 670 nm (red) and 740 nm (far-red), photoperiod 24/0 (light/dark), and light intensity 50 or 2 μmol m−2 s−1 for each wavelength.
Analysis of the lph Mutation
For chromosome linkage analysis (using at least two SSLP markers per chromosome), and for fine mapping with the nga1126 marker, we used 22 and 50 lph mutant plants, respectively, from the mapping population (lph plants from the F2 progeny of the lph cross with Ler-0 background). A genomic fragment of HY1 was isolated using primers HO1-u (5′-CAT TCA CCC TCT CAT CGT TAT CTT-3′) and HO1-l (5′-TGT ATT TGA GCT ATA AAA CGG CAG-3′). Sequencing of DNA was done with primers HO1-u, HO1-l, and HO1-ui (5′-CCT TCT TAT CTT CTT GTT ATG-3′). For analysis of cDNA length, and for cDNA sequencing, we used primers HO1-1exon (5′-GG TGA GAA AGA GAC TAA ATC-3′) and HO1-2exon (5′-TAG ATG TTG TAG AAG TGA CA-3′). In silico translation and protein comparison were performed with SDSC Biology WorkBench tools (http://workbench.sdsc.edu/).
Phenotypic Analysis
6- and 14-d-old seedlings were photographed and the root and hypocotyl parameters were evaluated using ImageJ (http://rsbweb.nih.gov/ij/). Measurement of the root meristem zone was performed on 5-d-old seedlings grown under long-d conditions. Cleared root tips were analyzed with a differential interference contrast microscope (model no. BX61; Olympus). Confocal microscopy was carried out on an inverted Observer.Z1 equipped with a LSM780 confocal unit and ×40 water immersion objective (Carl Zeiss). An appropriate set of filters was used for GFP imaging (excitation 488 nm, emission 507 nm). Roots of inspected lines were stained with propidium iodide (PI; 10 g/mL in water for 5 min) and observed using appropriate filter sets (PI excitation at 488 to 543 nm and emission at 610 nm). All measurements were done on at least two biological replicates and two technical replicates, each consisting of at least 15 samples. The mean and sd values or ses are shown on representative graphs. The hook opening assay and dynamics of hypocotyl growth were performed under an infrared light source (900 nm) as described by Zádniková et al. (2010). For the circadian experiment, plants were cultivated in growth chambers with controlled light (white light, 150 μmol m−2 s−), temperature (21°C/19°C, light/dark, respectively), and relative air humidity (50%) under long-d conditions: 16/8 h (light/dark) and moved to continuous light: 24/0 (light/dark) or darkness (D): 0/24 (light/dark) conditions. The end of the dark phase when the plants reached the age of 6 d was set as a time point 0 h. Possible circadian regulation was tested in long-d-grown seedlings by replacing the light phase with darkness (long-d > D) at the time point 0 or 24 h or vice versa replacing the dark phase by continuous light (long-d > continuous light) at the time point 16 or 40 h (see also Supplemental Fig. S6).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CKI1 (AT2G47430), ARR3 (AT1G59940), HY1 (AT2G26670), PHYA (AT1G09570), PHYB (AT2G18790), PIF3 (AT1G09530), CCA1 (AT2G46830), PIF1 (AT2G20180), PIF4 (AT2G43010), PIF5 (AT3G59060).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. CKI1 expression during wild-type and lph development-specificity in the vegetative and generative development.
Supplemental Figure S2. Molecular analysis of the lph/hy1-7 allele.
Supplemental Figure S3. Phenotypes of known mutations in the HY1 gene.
Supplemental Figure S4. Light-dependent phenotype of the hy1-7 mutant line.
Supplemental Figure S5. CKI1 promoter fragments, used in the Y1H binding assay.
Supplemental Figure S6. Circadian regulation of CKI1 expression.
Supplemental Figure S7. Early seedling development-correlation to CKI1 activity.
Supplemental Figure S8. Sensitivity of roots to the CK treatment.
Supplemental Figure S9. Partial phenotype rescue of hy1-1 by 35S:CKI.
Supplemental Figure S10. phyA phenotype in VB specific development.
Supplemental Figure S11. Normalized tag counts by location in the genome for CCA1 target CKI1.
Supplemental Table S1. Analysis of CKI1 promoter for the presence of phyA-responsive elements.
Supplemental Table S2. Schematic nuclear localization of phyA and phyB the main light receptors in relation to CKI1 expression in SAM.
Supplemental Materials and Methods. Supplemental methods.
Supplementary Material
Acknowledgments
We thank Dr. Enrique Lopéz Juéz (Royal Holloway University of London) for providing the hy1.62 and hy1-100 mutant lines, Pablo Cerdán for providing us with the double phyA phyB and hextuple phyA-E/ft mutant lines, and Bruno Mueller for the TCSn:GFP line and TCS:LUC construct. We also thank Hana Zimová for her valuable help and technical assistance, and Dr. Stephen Hunt (Qubit Systems, Canada) for careful reading of the manuscript, his revisions, and valuable comments.
Footnotes
1This research has been supported by the Ministry of Education, Youth and Sports of the Czech Republic under project no. CEITEC 2020 (LQ1601), and by LM2015062 from the Czech-BioImaging and the Czech Science Foundation (project nos. 15-22000S, 13-25280S and 13-39982S).
[OPEN]Articles can be viewed without a subscription.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alconada Magliano T, Casal JJ (2004) Pre-germination seed-phytochrome signals control stem extension in dark-grown Arabidopsis seedlings. Photochem Photobiol Sci 3: 612–616 [DOI] [PubMed] [Google Scholar]
- Aloni R, Langhans M, Aloni E, Dreieicher E, Ullrich CI (2005) Root-synthesized cytokinin in Arabidopsis is distributed in the shoot by the transpiration stream. J Exp Bot 56: 1535–1544 [DOI] [PubMed] [Google Scholar]
- Aloni R, Langhans M, Aloni E, Ullrich CI (2004) Role of cytokinin in the regulation of root gravitropism. Planta 220: 177–182 [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Altamura MM, Possenti M, Matteucci A, Baima S, Ruberti I, Morelli G (2001) Development of the vascular system in the inflorescence stem of Arabidopsis. New Phytol 151: 381–389 [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow PW, Fisahn J, Yazdanbakhsh N, Moraes TA, Khabarova OV, Gallep CM (2013) Arabidopsis thaliana root elongation growth is sensitive to lunisolar tidal acceleration and may also be weakly correlated with geomagnetic variations. Ann Bot (Lond) 111: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, Nagy F (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14: 1269–1278 [PMC free article] [PubMed] [Google Scholar]
- Boylan MT, Quail PH (1991) Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc Natl Acad Sci USA 88: 10806–10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T (2012) Gene regulation by cytokinin in Arabidopsis. Front Plant Sci 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen S, Lory N, Stauber J, Hoecker U (2015) Photoreceptor specificity in the light-induced and COP1-mediated rapid degradation of the repressor of photomorphogenesis SPA2 in Arabidopsis. PLoS Genet 11: e1005516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M (1994) A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Plant Physiol 104: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll MJ, Kiss JZ (2005) The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol 46: 317–323 [DOI] [PubMed] [Google Scholar]
- Cotton JL, Ross CW, Byrne DH, Colbert JT (1990) Down-regulation of phytochrome mRNA abundance by red light and benzyladenine in etiolated cucumber cotyledons. Plant Mol Biol 14: 707–714 [DOI] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD (2001) The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol 126: 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD (1999) The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA 96: 6541–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone S, Oka Y, Inoue Y (2000) Effect of light on root hair formation in Arabidopsis thaliana phytochrome-deficient mutants. J Plant Res 1131: 63–69 [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Deng Y, Dong H, Mu J, Ren B, Zheng B, Ji Z, Yang WC, Liang Y, Zuo J (2010) Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobisová T, Hejátko J (2014) Automated microscopy in forward genetic screening of Arabidopsis. Methods Mol Biol 1080: 53–66 [DOI] [PubMed] [Google Scholar]
- Emborg TJ, Walker JM, Noh B, Vierstra RD (2006) Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiol 140: 856–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flis A, Fernández AP, Zielinski T, Mengin V, Sulpice R, Stratford K, Hume A, Pokhilko A, Southern MM, Seaton DD, McWatters HG, Stitt M, et al. (2015) Defining the robust behaviour of the plant clock gene circuit with absolute RNA timeseries and open infrastructure. Open Biol 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96: 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, Coruzzi GM (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognár L, Tóth R, Nagy F, Millar AJ (2001) Conditional circadian regulation of PHYTOCHROME A gene expression. Plant Physiol 127: 1808–1818 [PMC free article] [PubMed] [Google Scholar]
- Hejátko J, Pernisová M, Eneva T, Palme K, Brzobohatý B (2003) The putative sensor histidine kinase CKI1 is involved in female gametophyte development in Arabidopsis. Mol Genet Genomics 269: 443–453 [DOI] [PubMed] [Google Scholar]
- Hejátko J, Ryu H, Kim GT, Dobesová R, Choi S, Choi SM, Soucek P, Horák J, Pekárová B, Palme K, Brzobohaty B, Hwang I (2009) The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell 21: 2008–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, Helariutta Y, Sussman MR, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisada A, Hanzawa H, Weller JL, Nagatani A, Reid JB, Furuya M (2000) Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell 12: 1063–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH (1998) SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Quail PH (2003) Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. (2013) Phytochrome cytoplasmic signaling. Annu Rev Plant Biol 64: 377–402 [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Immanen J, Nieminen K, Smolander OP, Kojima M, Alonso Serra J, Koskinen P, Zhang J, Elo A, Mähönen AP, Street N, Bhalerao RP, Paulin L, et al. (2016) Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr Biol 26: 1990–1997 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985 [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2014) Cytokinins. Arabidopsis Book 12: e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, Kircher S, Toth R, Adam E, Schäfer E, Nagy F (2000) Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J 22: 125–133 [DOI] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adám E, Schäfer E, Nagy F (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L) Heynh. Z Pflanzenphysiol 100: 147–160 [Google Scholar]
- Kraepiel Y, Jullien M, Cordonnier-Pratt MM, Pratt L (1994) Identification of two loci involved in phytochrome expression in Nicotiana plumbaginifolia and lethality of the corresponding double mutant. Mol Gen Genet 242: 559–565 [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X (2011) Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Higuchi M, Törmäkangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T (2006) Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol 16: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Matsumoto-Kitano M, Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Václavíková K, Miyawaki K, Kakimoto T (2008) Cytokinins are central regulators of cambial activity. Proc Natl Acad Sci USA 105: 20027–20031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Shimokor M, Komamine A (1971) Vessel element formation in cultured carrot-root phloem slices. Plant Cell Physiol 12: 823 [Google Scholar]
- Müller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM (1999) The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol 102: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112: E4802–E4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Kay SA (2012) Complexity in the wiring and regulation of plant circadian networks. Curr Biol 22: R648–R657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu SL, Chalkley RJ, Pham TN, Guan S, Maltby DA, Burlingame AL, Wang ZY, Quail PH (2013) Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr., Raja A, Loraine AE (2009) The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen K, Immanen J, Laxell M, Kauppinen L, Tarkowski P, Dolezal K, Tähtiharju S, Elo A, Decourteix M, Ljung K, Bhalerao R, Keinonen K, et al. (2008) Cytokinin signaling regulates cambial development in poplar. Proc Natl Acad Sci USA 105: 20032–20037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Okamura Y, Ito S, Tadaka S, Aoki Y, Shirota M, Kinoshita K (2014) ATTED-II in 2014: evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol 55: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH (1991) Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3: 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisová M, Klíma P, Horák J, Válková M, Malbeck J, Soucek P, Reichman P, Hoyerová K, Dubová J, Friml J, Zazímalová E, Hejátko J (2009) Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc Natl Acad Sci USA 106: 3609–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Janocha D, Dong Y, Medzihradszky A, Schöne S, Daum G, Suzaki T, Forner J, Langenecker T, Rempel E, Schmid M, Wirtz M, et al. (2016) Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. eLife 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischke MS, Jones LG, Otsuga D, Fernandez DE, Drews GN, Sussman MR (2002) An Arabidopsis histidine kinase is essential for megagametogenesis. Proc Natl Acad Sci USA 99: 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Kay SA (2010) An expanding universe of circadian networks in higher plants. Trends Plant Sci 15: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamaruddin M, Tillberg E (1989) Rapid effects of red light on the isopentenyladenosine content in scots pine seeds. Plant Physiol 91: 5–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D (1995) Phytochromes: photosensory perception and signal transduction. Science 268: 675–680 [DOI] [PubMed] [Google Scholar]
- Raz V, Ecker JR (1999) Regulation of differential growth in the apical hook of Arabidopsis. Development 126: 3661–3668 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and Phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösler J, Jaedicke K, Zeidler M (2010) Cytoplasmic phytochrome action. Plant Cell Physiol 51: 1248–1254 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sentandreu M, Martín G, González-Schain N, Leivar P, Soy J, Tepperman JM, Quail PH, Monte E (2011) Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23: 3974–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smet D, Žádníková P, Vandenbussche F, Benková E, van der Straeten D (2014) Dynamic infrared imaging analysis of apical hook development in Arabidopsis: the case of brassinosteroids. New Phytol 202: 1398–1411 [DOI] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD (2010) Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA 107: 4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Bäurle I, Kudla J, Nagy F, Schafer E, Harter K (2001) Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294: 1108–1111 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH (2006) phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J 48: 728–742 [DOI] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE (1999) Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol 119: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognár L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R, Fedoroff NV (1999) Genetic ablation of root cap cells in Arabidopsis. Proc Natl Acad Sci USA 96: 12941–12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinti G, Hills A, Campbell S, Bowyer JR, Mochizuki N, Chory J, López-Juez E (2000) Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J 24: 883–894 [DOI] [PubMed] [Google Scholar]
- Vriezen WH, Achard P, Harberd NP, van der Straeten D (2004) Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J 37: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Terry MJ, Rameau C, Reid JB, Kendrick RE (1996) The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXα. Plant Cell 8: 55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Schmülling T (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Mandel T, Kuhlemeier C (2011) Stem cell activation by light guides plant organogenesis. Genes Dev 25: 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu Z, Song X, Johnson C, Yu X, Sundaresan V (2016) The CKI1 histidine kinase specifies the female gametic precursor of the endosperm. Dev Cell 37: 34–46 [DOI] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, van der Straeten D, Friml J, et al. (2010) Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617 [DOI] [PubMed] [Google Scholar]
- Zdarska M, Dobisová T, Gelová Z, Pernisová M, Dabravolski S, Hejátko J (2015) Illuminating light, cytokinin, and ethylene signalling crosstalk in plant development. J Exp Bot 66: 4913–4931 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, Shakirova FM, Avalbaev AM, Kudryakova NV, Zubkova NK, Liere K, Kulaeva ON, Kusnetsov VV, Börner T (2008) Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B (2013) A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol 161: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.