The secretory ducts in the root of Thapsia garganica harbor the cytotoxin thapsigargin, and the cells lining these ducts express the first enzymes in the biosynthesis of thapsigargin.
Abstract
The Mediterranean plant Thapsia garganica (dicot, Apiaceae), also known as deadly carrot, produces the highly toxic compound thapsigargin. This compound is a potent inhibitor of the sarcoplasmic-endoplasmic reticulum Ca2+-ATPase calcium pump in mammals and is of industrial importance as the active moiety of the anticancer drug mipsagargin, currently in clinical trials. Knowledge of thapsigargin in planta storage and biosynthesis has been limited. Here, we present the putative second step in thapsigargin biosynthesis, by showing that the cytochrome P450 TgCYP76AE2, transiently expressed in Nicotiana benthamiana, converts epikunzeaol into epidihydrocostunolide. Furthermore, we show that thapsigargin is likely to be stored in secretory ducts in the roots. Transcripts from TgTPS2 (epikunzeaol synthase) and TgCYP76AE2 in roots were found only in the epithelial cells lining these secretory ducts. This emphasizes the involvement of these cells in the biosynthesis of thapsigargin. This study paves the way for further studies of thapsigargin biosynthesis.
Sesquiterpenoids are widely distributed across the plant kingdom and are recognized for their pharmacological properties and commercial value (Simonsen et al., 2013). Artemisinin, which today is the cornerstone of the treatment of malaria, is an outstanding example (Wiesner et al., 2003; Tu, 2011), along with fragrances such as patchoulol and santalol (Zhan et al., 2014). The genus Thapsia (Apiaceae) produces a variety of sesquiterpenoids including sesquiterpene lactones (Christensen et al., 1997; Drew et al., 2009). The most studied sesquiterpene from the genus is the sesquiterpene lactone thapsigargin, which is the predominant sesquiterpene lactone in Thapsia garganica and Thapsia gymnesica (Christensen et al., 1997). In the Mediterranean area, T. garganica has been used in traditional medicine for over 2,000 years for the treatment of pulmonary diseases, catarrh, fever, pneumonia, and as a counter irritant for the relief of rheumatic pains (Andersen et al., 2015b). The pharmacological effect of thapsigargin has been studied thoroughly, and it has been established that thapsigargin is an inhibitor of the sarcoplasmic-endoplasmic reticulum Ca2+-ATPase that leads to cell apoptosis (Thastrup et al., 1990). A prodrug (mipsagargin) based on thapsigargin toward solid cancer tumors is currently in clinical trials (Doan et al., 2015; Mahalingam et al., 2016).
An unusual feature of thapsigargin and related guaianolides is the presence of a β-disposed C-6-O bond and an α-disposed C-7-C-11 bond. In the majority of guaianolides from other plant families, the C-6-O bond is α-disposed and the C-7-C-11 bond is β-disposed (Christensen et al., 1997; Drew et al., 2009; Simonsen et al., 2013).
Thapsigargin is found in most parts of the plant T. garganica. Ripe fruits contain the highest amount of thapsigargin, with 0.7% to 1.5% of the dry weight, followed by roots (0.2%–1.2% of dry weight) and leaves (0.1% of dry weight; Smitt et al., 1995). It is well established that many Apiaceae species store lipophilic compounds such as phenyl propanoids and terpenoids in secretory ducts and vittae (Corsi et al., 1988; Poli et al., 1995; Maggi et al., 2015). We chose to examine the localization of sesquiterpenoids and their biosynthesis in T. garganica roots that have high amounts of thapsigargin. The roots also could be obtained commercially from plants grown in greenhouses, in contrast to the season-dependent harvest of fruits from the natural population. So far, fruits have not been obtained from greenhouse plants, even from plants more than 4 years old. By histochemical staining, we show that T. garganica roots contain secretory ducts in parenchymatic tissue and that these may harbor terpenoids. Matrix-assisted laser-desorption ionization-mass spectrometry imaging (MALDI-MSI) of the roots was used to show that thapsigargin is present in specific locations in the root and, furthermore, in a pattern likely to coincide with the location of secretory ducts.
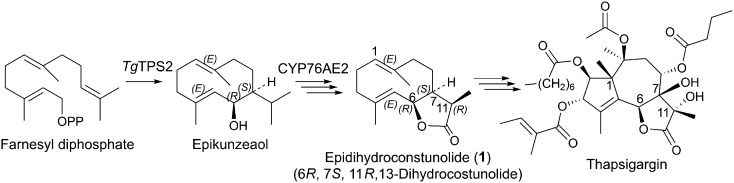
The site and route of thapsigargin biosynthesis have not yet been established. The first specific step in the biosynthesis of most sesquiterpenoids is catalyzed by sesquiterpene synthases (Bohlmann et al., 1998). Two sesquiterpene synthases have been described from T. garganica roots. These were expressed in Saccharomyces cerevisiae and biochemically characterized. It was shown that the sesquiterpenoid synthase TgTPS2 was of particular interest to thapsigargin biosynthesis. The major product of TgTPS2 is epikunzeaol, a germacrenol with a hydroxyl group at C-6 (Fig. 1; Pickel et al., 2012).
Figure 1.
Structures of epikunzeaol and epidihydrocostunolide (1). Both metabolites are suggested intermediates in the thapsigargin pathway in T. garganica.
Generally, the diversity of sesquiterpenoids is obtained by the catalytic activity of the sesquiterpene synthases and followed by modifications to the C15 backbone by cytochromes P450 (P450s), acyltransferases, and dehydrogenases, among others (Weitzel and Simonsen, 2015). The complexity of sesquiterpenoid structures, including chiral centers and regiospecific and stereospecific oxidation, makes chemical synthesis difficult, and synthesis approaches often result in low yields (Andrews et al., 2007; Ball et al., 2007; Chu et al., 2016). Attempts to find alternative sustainable production methods for sesquiterpenoids such as artemisinin by biological synthesis are actively pursued (Paddon et al., 2013). Sesquiterpenoid lactones are a subgroup of sesquiterpenoids, and compounds in this subgroup have been shown to have a potential for the treatment of various cancers (Curry et al., 2004; Simonsen et al., 2013). Despite recent advances, only a few steps in the complex biosynthetic routes of plant sesquiterpenoid lactones have been characterized. These include enzymes involved in the biosynthesis of artemisinin (Artemisia annua) and (+)-costunolide, a precursor for a range of other sesquiterpene lactones (in Cichorium intybus and Tanacetum parthenium). These plants are all from the family Asteraceae (Wen and Yu, 2011; Liu et al., 2014).
P450s are common participants in sesquiterpenoid biosynthesis, and especially P450s from the CYP71 clan have been shown to be involved in the biosynthesis of sesquiterpenoids (Luo et al., 2001; Diaz-Chavez et al., 2013; Liu et al., 2014; Weitzel and Simonsen, 2015; Yang et al., 2015; Takase et al., 2016). CYP71AV1, which is involved in artemisinin biosynthesis, is among the most well-studied P450s in sesquiterpenoid biosynthesis (Teoh et al., 2006). Here, we functionally characterize TgCYP76AE2, a P450 from the CYP71 clan, which was found in the root transcriptome of T. garganica. Through the transient coexpression in Nicotiana benthamiana of TgTPS2 and TgCYP76AE2, epikunzeaol is converted to epidihydrocostunolide, a likely precursor for more complex sesquiterpenoid lactones including thapsigargin. Within the Apiaceae, only P450s involved in the biosynthesis of the phenylpropanoid coumarin have been described (Larbat et al., 2007, 2009; Drew et al., 2013; Dueholm et al., 2015). This work presents the functional characterization of the first sesquiterpene-specific P450 found in the Apiaceae. In situ PCR was performed to investigate the specific site for the expression of TgTPS2 and TgCYP76AE2. Thereby, we were able to verify that TgTPS2 and TgCYP76AE2 are expressed in epithelial cells lining the secretory ducts containing thapsigargin.
RESULTS
Identification of P450s from Transcriptome Data
The root transcriptome data from T. garganica were mined to enable the discovery of possible sequential biosynthesis steps from epikunzeaol toward thapsigargin. The search was limited to P450s from the CYP71 clan, due to the previous findings in this clan of P450s involved in sesquiterpenoid biosynthesis. We investigated the occurrence of orthologous genes to CYP71s in the T. garganica transcriptome using BLAST searches. Eighteen full-length P450 genes were found to be distributed, with 12 belonging to the CYP71 family, one to the CYP706 family, and five to the CYP76 family. The P450s were named by David Nelson according to the current annotation system (Nelson, 2009); TgCYP71AH8 (KX826939), TgCYP71AJ5 (KP191555), TgCYP71AJ14 (KP191558), TgCYP71AS14 (KX845553), TgCYP71AT12 (KX826940), TgCYP71AU89 (KX845548), TgCYP71AU90 (KX845552), TgCYP71BK1 (KX826941), TgCYP71BK6 (KX845546), TgCYP71D183 (KX845554), TgCYP71D311 (KX845555), TgCYP71D319_ortholog (KX845550), TgCYP76AE1 (KX826942), TgCYP76AE2 (KX826943), TgCYP76AE8 (KX845545), TgCYP76AF7 (KX845549), TgCYP76B79 (KX845547), and TgCYP706C30_ortholog (KX845551).
Phylogeny
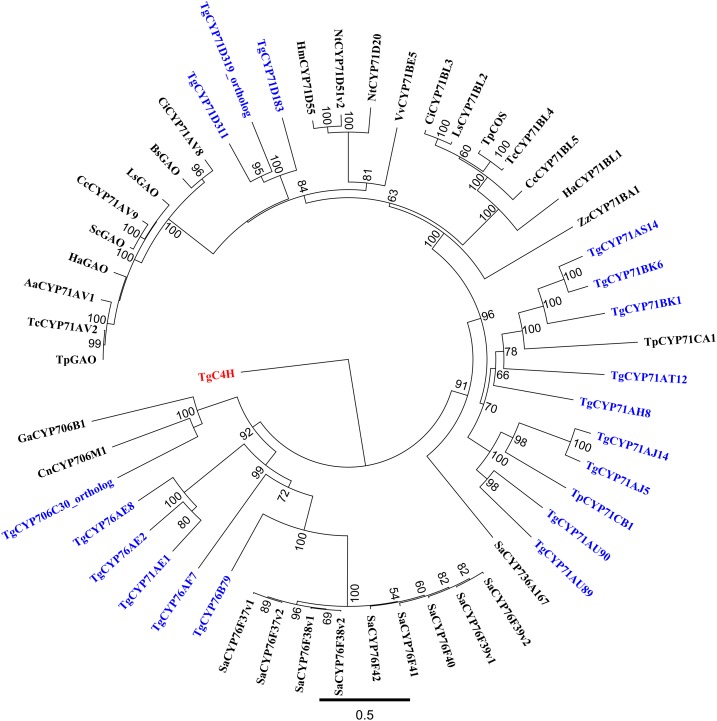
Phylogenetic analyses of 35 full-length genes from the CYP71 clade involved in sesquiterpenoid biosynthesis from several plant species and the 18 enzymes from T. garganica revealed that these enzymes are grouped in several subclades (Fig. 2). The analysis shows that there are blooms of genes within species and families (Hamberger and Bak, 2013), like the CYP76F bloom in Santalum album. Although the phylogeny only included P450s related to sesquiterpenoid metabolism, the sequences are from most families and subfamilies in the CYP71 clade. This shows that phylogenetic analyses are not useful to predict the specific functionality of P450s, as shown previously (Dueholm et al., 2015). The analyses merely serve to indicate the range of enzymes that have to be examined biochemically: in this case, 18 sequences.
Figure 2.
Tree showing the phylogeny of P450s from T. garganica (within the CYP71 clade) and cytochromes related to sesquiterpenoid biosynthesis. The nodes with Thapsia genes are marked in blue. The only two groups in the tree that can be assigned to a biochemical function are the germacrene A oxidase group CYP71AVx and the costunolide synthase group CYP71BLx. However, both groups are from related species within the Asteraceae family and, thus, cannot be used in general for functional identification. Only bootstrap values higher than 50 are shown in the tree.
Gas Chromatography-Mass Spectrometry and Liquid Chromatography-Mass Spectrometry Analyses of Extracts from N. benthamiana Expressing TgTPS2 and P450s, Including TgCYP76AE2
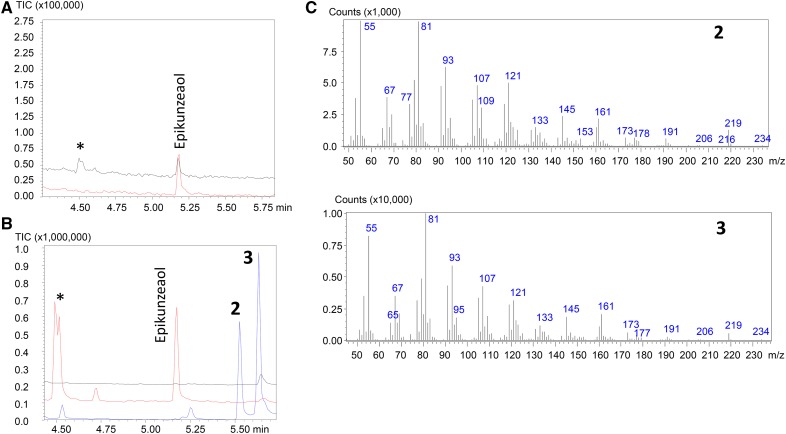
The cyclodeca-1(10),4-diene ring of epikunzeaol (Fig. 1) is susceptible to thermal Cope rearrangement at the high temperatures in the gas chromatography-mass spectrometry (GC-MS) injection port. Therefore, extracts of N. benthamiana expressing AtHMGR and TgTPS2 were analyzed with two injection port temperatures, 250°C and 160°C. At 250°C, the TgTPS2 product epikunzeaol is detected along with degradation compounds, whereas at 160°C, only epikunzeaol is detected (Fig. 3A). Similar rearrangements have been identified for other cyclodeca-1(10),4-diene products of sesquiterpene synthases (Andersen et al., 2015a). To enhance the level of precursors available for TgTPS2 in N. benthamiana, a truncated version of the Arabidopsis (Arabidopsis thaliana) HMGR (AttHMGR) was transiently coexpressed. AttHMGR was shown previously to enhance production levels of sesquiterpenoids (Cankar et al., 2015).
Figure 3.
GC-MS analysis of hexane extracts from N. benthamiana expressing AttHMGR and TgTPS2 with GC-MS injection port temperatures of 160°C (red) and 250°C (black; A) or expressing AttHMGR alone (black), AttHMGR and TgTPS2 (red), and AttHMGR, TgTPS2, and TgCYPAE2 (blue) with a GC-MS injection port temperature of 250°C (B). Asterisks denote epikunzeaol thermal rearrangement products. C, Mass spectra of the elemanolides 2 and 3.
In order to discover which P450s that could utilize epikunzeaol as a substrate, the 18 P450s from the CYP71 clan were transiently coexpressed with TgTPS2 and AttHMGR in N. benthamiana.
Of the tested P450s, only TgCYP76AE2 was evidently able to utilize epikunzeaol as a substrate. To test for further downstream pathway steps, the remaining P450s were coexpressed with AttHMGR, TgTPS2, and TgCYP76AE2. No new products or decline in substrate were detected. At this stage, it cannot be excluded that new products were not detected due to low expression or lack of expression of the 17 other P450s.
GC-MS analysis of the hexane extracts of N. benthamiana leaves expressing AttHMGR, TgTPS2, and TgCYP76AE2 is shown in Figure 3B, where two new products, 2 and 3, are observed. It was not possible to detect these products with the injection port at 160°C, which can be explained by a lower volatility of the new products in comparison with epikunzeaol. The coexpression of AttHMGR, TgTPS2, and TgCYP76AE2 in N. benthamiana resulted in a complete conversion of epikunzeaol (Fig. 3B).
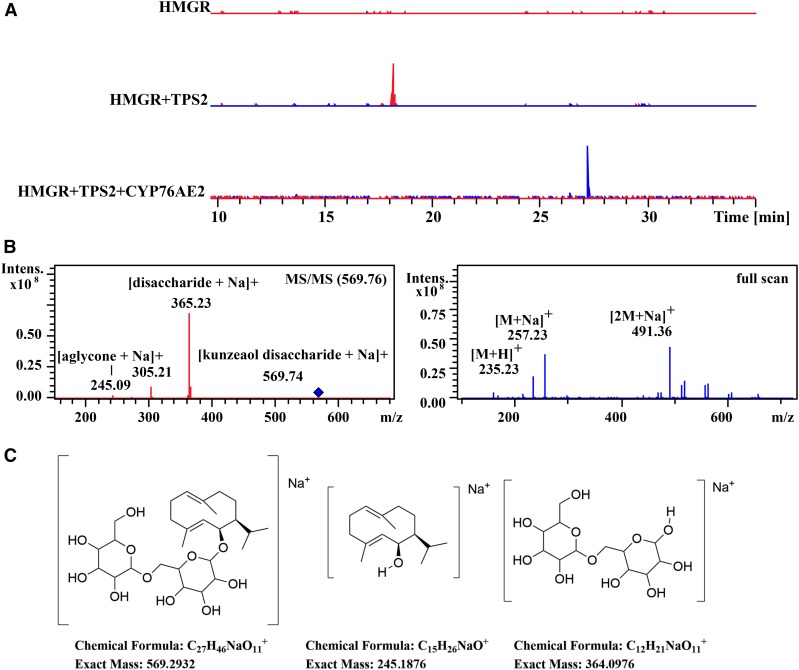
To expand the search for TgCYP76AE2 products or derivatives thereof not detectable by GC-MS, the N. benthamiana extracts was analyzed by analytical liquid chromatography-mass spectrometry (LC-MS). In contrast to the GC-MS analysis, epikunzeaol was not detected in the free form but as the aglycone in a glycoside of a disaccharide, and only one TgCYP76AE2 product was detected (Fig. 4). In the LC-MS analysis, product 1 was detected as the protonated molecular ion (m/z 235.23, [M+H]+), the sodium adduct (m/z 257.23, [M+Na]+), and the base peak equal to m/z 491.3126 ([2M+Na]+), corresponding to the sodiated dimer adduct.
Figure 4.
A, Extracted ion chromatograms of epikunzeaol (1) [epikunzeaol disaccharide+Na]+ m/z 569.74 (red) and epidihydrocostunolide [2M+Na]+ m/z 491.32 (blue) from the LC-MS analysis of the methanol extract of AttHMGR alone, AttHMGR plus TgTPS2, and AttHMGR plus TgTPS2 coexpressed with TgCYP76AE2. B, Tandem mass spectrometry (MS/MS) spectrum of the epikunzeaol peak at 18 min detected as the epikunzeaol disaccharide. The mass spectrometry spectrum of epidihydrocostunolide at 27 min shows an m/z of 491.3258 corresponding to a sodium adduct and a dimer of m/z 235.1741. C, Structure of epikunzeaol disaccharide including the fragmentation pattern.
Isolation of Epidihydrocostunolide (1) and the Two 1,3-Elemandien-12,6-Olides (2 and 3) by HPLC and Preparative GC-MS for NMR Analysis
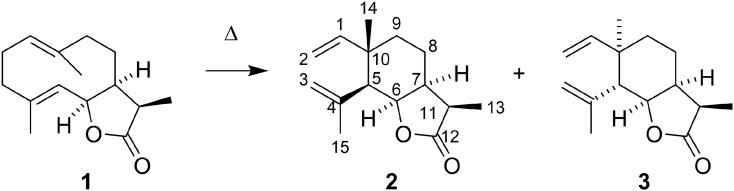
To determine the structure of the three new compounds (1, 2, and 3), these were isolated from the hexane extract of N. benthamiana leaves expressing AttHMGR, TgTPS2, and TgCYP76AE2. Compound 1 was isolated by semipreparative normal-phase HPLC, and the purity was confirmed by liquid chromatography-high resolution mass spectrometry. Compounds 2 and 3 were isolated using preparative GC-MS. To confirm that 2 and 3 are thermal Cope rearrangement products of 1, pure 1 was injected into the GC-MS apparatus (Fig. 5). No trace of compound 1 was seen in this GC-MS chromatogram, whereas the mass spectra of the peaks originating in compounds 2 and 3 were identical to those obtained from the compounds isolated by preparative GC-MS (Supplemental Fig. S2). This confirms the thermal Cope rearrangement of 1 into 2 and 3.
Figure 5.
Thermal rearrangement of 1 into the two compounds 2 and 3. The Cope rearrangement is well known for transforming germacradi-1(10),4-dienolides into elemanolides.
Structure Elucidation of Products 1, 2, and 3
The structures of 1, 2, and 3 were elucidated by the interpretation of 1H, 13C, and 2D COSY, HSQC, HMBC, and ROESY spectra. The molecular formula of 1 was established using high-resolution mass spectrometry to be C15H22O2 (observed m/z 235.1692, calculated for C15H22O2 [M+H]+ m/z 235.1693, 0.2 ppm error). Comparison of the 1H- and 13C-NMR spectra of 1 with that of dihydrocostunolide and in particular the 13C-NMR spectrum (Sanz et al., 1990; Barrero et al., 2002) revealed significant similarities. The structure elucidation was complicated, as all nuclei exhibited one major signal and a minor signal, as is clearly observed in the 13C-NMR spectrum (Supplemental Fig. S1). This is most likely an effect caused by the presence of two slowly interconverting conformers of the decadiene ring. However, closer inspection showed that the isolated germacranolide had to be a stereoisomer of the previously proposed structure of dihydrocostunolide. The ROE interactions between H-7, H-11, and H-6 indicated that these protons were all cis-disposed, thus supporting the proposed structure of 1.
From the preparative GC-MS isolation, two products (2 and 3) were obtained (Fig. 3B). Compound 2 had the molecular formula C15H22O2, as revealed by mass spectrometry. Inspection of the 13C-NMR spectrum indicated the presence of two signals for methylene groups at 110 and 116 and a signal for a quaternary carbon at 144 (Table I). Signals for a carbon atom at 111 ppm originating in a methylene group, the two protons of which resonate at 4.97 (doublet) and 4.98 (doublet) ppm, suggested the presence of a mono-substituted double bond. Signals at 116 ppm originating in a methylene group, the two protons of which resonate at 5.02 (singlet) and 5.07 ppm (singlet), suggested the presence of a di-substituted double bond. A signal for a carbonyl group at 179 ppm indicated the presence of a γ-lactone. These chemical shift values were very similar to those reported for the saussurea lactone (Barrero et al., 2002). Inspection of the 1H-NMR spectrum of 2 (Table II) and the reported spectrum of the saussurea lactone (Ando et al., 1983), however, revealed significant differences between the sizes of the three-bond coupling constants in 2 and in the saussurea lactone. In the saussurea lactone, the lactone ring is transfused with the cyclohexane ring, enabling an axial location of H-6 and H-7; consequently, coupling constants of approximately 10 Hz are expected. Assuming that the lactone ring and the cyclohexane ring of 2 are cis-fused, this would prevent an axial-axial coupling; consequently, smaller 3JHH couplings are expected. Inspection of the ROESY spectrum also revealed ROE correlation between H-6 and H-7, confirming the cis-fusion. An additional ROE correlation between H-7 and H-11 revealed that these protons also are cis-disposed. ROE correlations between CH3-15, CH3-14, and CH3-13 confirm that all of these methyl groups are β-disposed. Based on these data, the stereochemistry of 2, as shown in Figure 5, is suggested to be (3R,3aS,6S,7S,7aR)-3,6-dimethyl-7-(prop-1-en-2-yl)-6-vinylhexahydrobenzofuran-2(3H)-one.
Table I. 13C-NMR results for 1, 2, and 3.
Samples were analyzed at 300 K. The isolated product 1 was dissolved in MeCN-d3 (99.8 atom % D), while 2 and 3 were dissolved in CDCl3 (99.8 atom % D).
| C | 1 | 2 | 3 |
|---|---|---|---|
| δC |
δC |
δC |
|
| 1 | 128.7 | 148.7 | 147.2 |
| 2 | 26.4 | 110.9 | 111.2 |
| 3 | 40.6 | 116.4 | 114.4 |
| 4 | 137.7 | 143.5 | 144.6 |
| 5 | 127.1 | 54.8 | 51.9 |
| 6 | 79.3 | 81.2 | 81.2 |
| 7 | 46.2 | 40.6 | 37.2 |
| 8 | 23.7 | 18.6 | 18.9 |
| 9 | 37.7 | 37.6 | 32.1 |
| 10 | 139.4 | 39.0 | 37.7 |
| 11 | 41.5 | 41.3 | 40.6 |
| 12 | 180.6 | 179.3 | 178.9 |
| 13 | 10.9 | 9.2 | 10.2 |
| 14 | 22.2 | 18.6 | 25.5 |
| 15 | 17.4 | 26.0 | 26.1 |
Table II. 1H-NMR results for 1, 2, and 3.
Samples were analyzed at 300 K. The isolated product 1 was dissolved in MeCN-d3 (99.8 atom % D), while 2 and 3 were dissolved in CDCl3 (99.8 atom % D).
| H | 1 | 2 | 3 |
|---|---|---|---|
| δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | |
| 1 | 5.00 m, overlaid | 5.80 dd, J = 17.5, 10.8 Hz, 1H | 5.93 dd, J = 17.6, 11.0 Hz, 1H |
| 2a | 2.10 m, overlaid H | 4.97 d, J = 10.5 Hz, 1H | 4.98 d, J = 17.2 Hz, 1H |
| 2b | 2.27 m, overlaid H | 4.98 d, J = 18.0 Hz, 1H | 4.99 d, J = 11.7 Hz, 1H |
| 3a | 2.04 m, overlaid H | 5.02 s, 1H | 4.85 s, 1H |
| 3b | 2.23 m, overlaid H | 5.07 s, 1H | 4.99 s, 1H |
| 5 | 4.98 d, J = 10.1, overlaid H | 2.13 d, J = 2.6 Hz, 1H | 2.56 d, J = 4.0 Hz, 1H |
| 6 | 5.14 dd, J = 9.8, 4.6, 1H | 4.45 t, J = 3.3 Hz, 1H | 4.50 t, J = 4.8 Hz, 1H |
| 7 | 2.49 m, overlaid H | 2.29–2.36 m, 1H | 2.57–2.65 m, 1H |
| 8 | 1.59 s | ||
| 8a | 1.37 qd, J = 13.9, 2.90 Hz, 1H | 1.42–1.52 m, 1H | |
| 8b | 1.64 ddd, J = 13.5, 6.3, 3.3 Hz, 1H | 1.61–1.67 m, overlaid, 1H | |
| 9 | 1.56–1.61 m, overlaid, 2H | ||
| 9a | 1.70 m, overlaid H | 1.55 m, overlaid H | |
| 9b | 2.45 d, J = 13.0, 1H | 1.53 m, overlaid H | |
| 11 | 2.94 m, overlaid H | 2.76 quin, J = 7.0 Hz, 1H | 2.79 quin, J = 7.3 Hz, 1H |
| 13 | 1.13 s, 3H | 1.21 d, J = 7.3 Hz, 3 H | 1.22 d, J = 7.3 Hz |
| 14 | 1.53 s, 3H | 1.20 s, 3 H | 1.03 s, 3 H |
| 15 | 1.67 s, 3H | 1.87 s, 3 H | 1.85 s, 3 H |
The spectra of 3 were very similar to those of 2. In 3, however, a similar ROE correlation to that described above for compound 1 was present, except for ROE correlation between CH3-15 and CH3-13. Combined with similar 3JHH coupling constants, this suggests the stereochemistry of 3 shown in Figure 5 to be (3R,3aS,6R,7R,7aR)-3,6-dimethyl-7-(prop-1-en-2-yl)-6-vinylhexahydrobenzofuran-2(3H)-one. 1 is a new compound for which we suggest the name epidihydrocostunolide. The stable structures of 2 and 3 led to the full structure elucidation of 1.
Tissue Localization of Thapsigargin Biosynthesis in Roots
Histochemical staining and MALDI-MSI were performed to determine the localization of thapsigargin and its biosynthesis. Histochemical staining was used to indicate the presence of terpenoids in secretory ducts in root tissue. The location of thapsigargin was analyzed by MALDI-MSI. Based on these results, further investigations by in situ PCR were performed to observe if the expression of TgTPS2 and TgCYP76AE2 involved in the biosynthesis of thapsigargin showed colocalization with thapsigargin.
Histochemical Analysis of T. garganica Roots
The presence of special storage structures in the root possibly containing terpenoids was investigated by a histochemical analysis using NADI (dimethyl-p-phenyl- enediamine and a-naphthol) staining (David and Carde, 1964; Caissard et al., 2004; Jezler et al., 2013; Kromer et al., 2016; Muravnik et al., 2016; Stešević et al., 2016; Stojičić et al., 2016). The NADI reagent has been reported to give rise to a blue/purple color in the presence of oxygenated or lipophilic compounds (e.g. terpenoids) in oil secretory cells such as ducts, trichomes, and other specialized tissues. The mechanism is suggested to be a nonenzymatic reduction of the oxidized target and an oxidation of dimethyl-p-phenylenediamine followed by the formation of a radical, which reacts with α-naphthol to produce Indophenol Blue (Harwig, 1967; Takamatsu and Hirai, 1968).
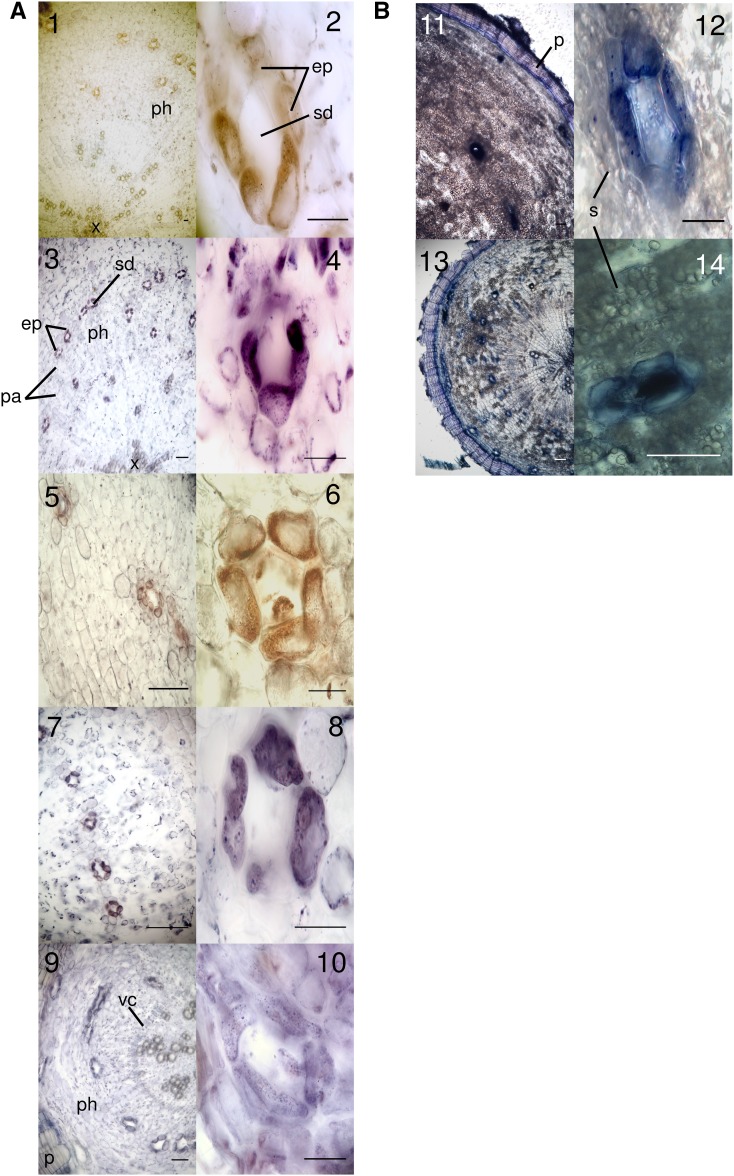
Prior to staining, T. garganica roots were cut into 60-µm cross sections. As in other Daucus spp., the roots showed a clear secondary growth (Havis, 1939; Korolev et al., 2000). White resin was observed oozing out of the root upon cutting. Figure 6 indicates the presence of oxygenated or lipophilic compounds (e.g. terpenoids) in the sections after staining with NADI, and secretory ducts are clearly visible as blue spots. These were located in concentric rings within the parenchymatic tissue, radiating out from the pith until just below the periderm, as also shown in Daucus spp. (Deutschmann, 1969). At higher magnification, it is evident that both the ducts themselves and the epithelial cells surrounding them were stained blue, indicating the presence of oxygenated or lipophilic compounds (e.g. terpenoids). Apart from the ducts in the parenchymatic tissue, the periderm also showed blue staining, which could be due to tannins in this tissue. In general, ducts were not found in cross sections from the crown or the very bottom of the roots but otherwise were distributed generally throughout the root.
Figure 6.
A, In situ PCR of TgCYP76AE2 and TgSTS2 in T. garganica root cross sections. All samples were stained with BM Purple. Blue/purple color indicates the presence of digoxigenin (DIG)-labeled cDNA, and brown indicates the absence of the amplified cDNA target. Images 1 and 2 are negative controls (TgCYP76AE2) where reverse transcription was omitted. Images 3 and 4 show the detection of TgCYP76AE2 mainly in epithelial cells surrounding secretory ducts within the vascular cambium. Images 5 and 6 are negative controls (TgTPS2) where reverse transcription was omitted. Images 7 and 8 show the detection of TgTPS2 mainly in epithelial cells surrounding secretory ducts within the vascular cambium, as with TgCYP76AE2. Images 9 and 10 are positive controls with 18S rRNA to show staining in all cell types. B, NADI staining of T. garganica root cross sections. In images 11 to 14, blue indicates the presence of oxygenated or lipophilic compounds (e.g. terpenoids) concentrated in epithelial cells and secretory ducts. ep, Epithelial cells; p, periderm; ph, phloem; pa, parenchyma cells; s, starch grains; sd, secretory ducts; vc, vascular cambium; x, xylem. Bars = 100 μm.
MALDI-MSI of T. garganica Roots
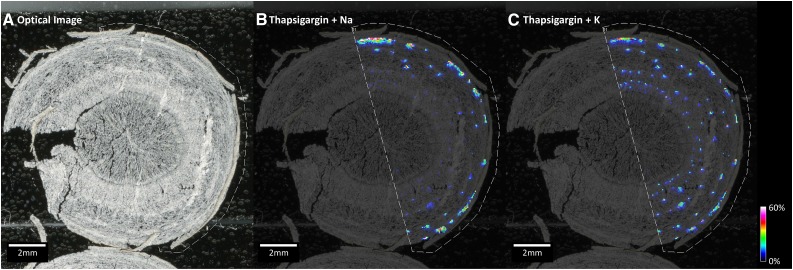
The spatial distribution of thapsigargin, the intermediates epikunzeaol and epihydrocostunolide, and related guaianolides was examined using high-resolution Fourier transform-ion cyclotron resonance-MALDI-MSI. MALDI-MSI is able to measure ionized chemicals as their m/z in a highly localized manner (Boughton et al., 2016). Root sections of T. garganica that had been confirmed previously to contain thapsigargin by HPLC were prepared by initial cryosectioning, mounting to slides on double-sided tape, freeze drying, and the application of 2,5-dihydroxybenzoic acid (DHB) matrix by sublimation (Jarvis et al., 2017). Prepared sections were analyzed by MALDI-MSI in the positive ionization mode across the mass range m/z 200 to 3,000. The results demonstrated complex mass spectra containing numerous ions that could be tentatively assigned as small molecule metabolites, sugars, and lipids (Supplemental Fig. S3). Specific ions were found to localize to the epidermis, parenchyma, stele, and a series of concentric spots correlating with the observed distribution of secretory ducts (Fig. 7). The acquired spectra were screened for thapsigargin, epikunzeaol, and epidihydrocostunolide by searching for the calculated m/z of proton, sodium, and potassium adducts. Ions corresponding to thapsigargin, [M+Na]+ ion m/z 673.3186 (calculated 673.31945, 1.26 ppm error) and [M+K]+ ion m/z 689.2924 (calculated 689.29339, 1.43 ppm error) were found. Proton adducts of thapsigargin were barely observed, and in general, the K adduct showed a much higher signal intensity relative to the Na adduct (signal intensity ratio of 2.5–3:1). The spatial distribution of both Na and K adducts showed a distinct distribution within the parenchymatic tissue (Fig. 7), where the distribution pattern correlated with concentric circles of secretory ducts visualized from histochemical staining (Fig. 6). Both epikunzeaol and epidihydrocostunolide were not detected in the images, indicating that concentrations were below the limit of detection using this methodology.
Figure 7.
MALDI-MSI analysis of a T. garganica taproot section. A, Optical image of a taproot section with sublimed DHB matrix. B, Distribution of thapsigargin Na adduct, [M+Na]+ m/z 673.3186 (calculated 673.31945, 1.26 ppm error). C, Distribution of thapsigargin K adduct, [M+K]+ m/z 689.2924 (calculated 689.29339, 1.43 ppm error). Images were normalized to root median square and scaled to 0% to 60% of maximum signal intensity using Compass FlexImaging 4.1 to enhance visualization. The results demonstrate thapsigargin to be localized to concentric circles similar to the pattern of secretory ducts.
The root sections also were screened for seven related guaianolides from T. garganica (Table III). The structures of the related guanolides and the MALDI imaging results from these are shown in Supplemental Figure S4. The seven structurally similar sesquiterpenoids, thapsigargicin, nortrilobolide, trilobolide, thapsivillosin I, and thapsivillosin L all showed distributions that coincided with thapsigargin, including a similar pattern of a more intense [M+K]+ adduct.
Table III. Calculated and observed masses (including error expressed in ppm) of Na and K adducts of thapsigargin, epikunzeaol, epidihydrocostunolide, and a selection of known guaianolides from T. garganica in the matrix-assisted laser-desorption ionization (MALDI) experiment.
| Compound | Molecular Formula | [M+Na]+ |
[M+K]+ |
||||
|---|---|---|---|---|---|---|---|
| Calculated | Observed | Error | Calculated | Observed | Error | ||
| Epikunzeaol | C15H26O | 245.18759 | – | – | 261.16152 | – | – |
| Epidihydrocostunolide | C15H22O2 | 257.15120 | – | – | 273.12514 | – | – |
| Nortrilobolide | C26H36O10 | 531.22007 | 531.22110 | −1.94 | 547.19401 | 547.19380 | 0.38 |
| Trilobolide | C27H38O10 | 545.23572 | 545.23370 | 3.70 | 561.20966 | 561.20730 | −0.01 |
| Thapsivillosin L | C30H42O12 | 617.25685 | – | – | 633.23079 | 633.23240 | −2.55 |
| Thapsivillosin I | C31H42O12 | 629.25685 | 629.25830 | −2.31 | 645.23079 | 645.2327 | −2.97 |
| Thapsivillosin J | C31H44O12 | 631.27250 | – | – | 647.24644 | – | – |
| Thapsigargicin | C32H46O12 | 645.28815 | 645.2864 | 2.71 | 661.26209 | 661.26040 | −0.01 |
| Thapsigargin | C34H50O12 | 673.31945 | 673.3186 | 1.26 | 689.29339 | 689.2924 | 1.43 |
| Thapsivillosin C | C35H52O12 | 687.34588 | – | – | 703.30904 | – | – |
Localization of mRNA for TgTPS2 and TgCYP76AE2
Following the specific localization of thapsigargin in the roots and likely in secretory ducts, it was investigated if the production of the compound takes place there as well. The cellular localization of transcripts encoding TgTPS2 and TgCYP76AE2, involved in the biosynthesis, was investigated by in tube in situ PCR. TgTPS2 and TgCYP76AE2 transcripts were found to show the same spatial expression pattern in the investigated tissue and were detected solely in the epithelial cells lining the secretory ducts (Fig. 6). The presence of the transcripts was visualized by the color reaction of the alkaline phosphates bound to the antibody specific to the DIG group incorporated during the amplification of the PCR product from the specific cDNA produced using the reverse specific primers recognizing TgTPS2 and TgCYP76AE2. In T. garganica roots, this reaction took place only in the epithelial cells of the secretory ducts when specific reverse primers were included. When no specific reverse primers were used, no color reactions were detected. This reaction clearly shows that the ducts follow the circular growth pattern of the parenchymatic tissue. The transcripts show a spatial distribution similar to the m/z corresponding to thapsigargin, as is seen in Figure 7.
DISCUSSION
In this study, TgCYP76AE2 was found to catalyze the oxidations that lead to the formation of a lactone ring, thereby converting epikunzeaol into epidihydrocostunolide (1; Fig. 1). The structures of 2 and 3 confirm the proposed structure of 1 and also finally confirm the structure of epikunzeaol produced by TgTPS2, as seen in Figure 1. The structure of kunzeaol published previously as the product of TgTPS2 did not benefit from the Cope rearrangement study included here; therefore, the stereochemistry was not final (Pickel et al., 2012). In general, Cope rearrangements are stereoselective, and only one product would be expected (Setzer, 2008; Adio, 2009). However, here, upon injection of 1 into a GC-MS device, two elemanolides, 2 and 3, were formed in almost equal amounts (Supplemental Fig. S2). This type of rearrangement has been studied for sesquiterpenoids belonging to the germacrenes and germacranolides. Germacrenes are characterized by their backbone structure, which is a 10-member open ring like that seen in epikunzeaol (Fig. 1). The conversion of germacranolides to elemanolides by Cope rearrangement has been used to structurally elucidate the heat-labile germacranolides (Fischer and Mabry, 1967; Takeda, 1974; Raucher et al., 1986; de Kraker et al., 2001; Barrero et al., 2002; Adio, 2009). At high temperatures or low pH, germacrenes via Cope rearrangement establish equilibrium with elemenes (Takeda, 1974; de Kraker et al., 2002; Pickel et al., 2012). Elemenes are characterized by the lack of a bond between C-2 and C-3 (Fig. 4). The unexpected formation of two products after the Cope rearrangement of 1 might be explained by the cis-fused lactone and instability of the 10-member decadiene ring. Most reported studies on Cope rearrangements have been performed on transfused germacranolides, where the preferred chair-chair conformation of the intermediate is easily accessible (Setzer, 2008; Adio, 2009). One example, however, with Cope rearrangement of a cis-fused germacranolide is described. In this example also, only one product is formed in high yield (Appendino and Gariboldi, 1983).
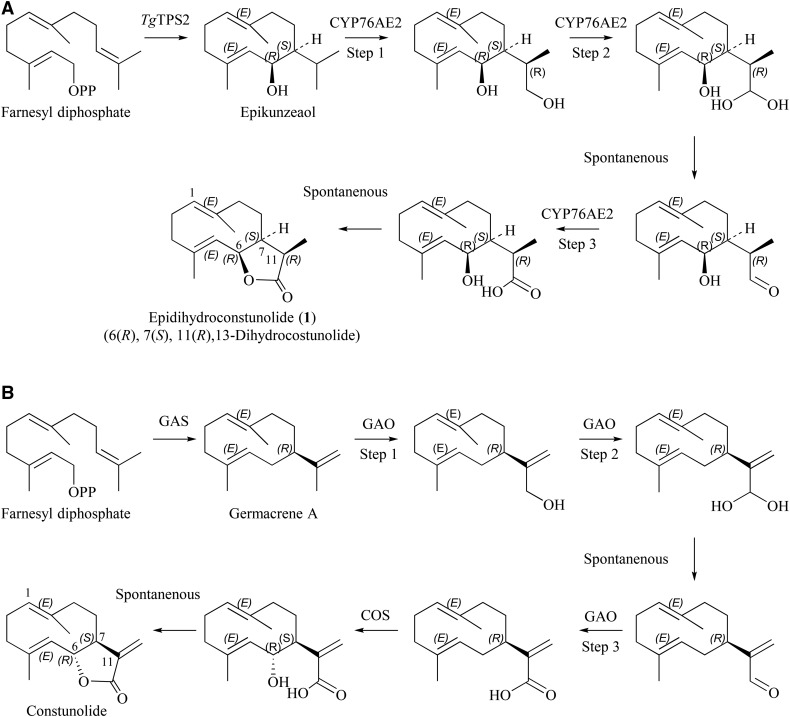
The biochemical lactone ring reaction observed here also is known from Asteraceae, where the biosynthesis of (+)-costunolide follows the same type of reaction. In order to establish the lactone ring in (+)-costunolide, two P450s are needed. First, three consecutive hydroxylations are performed on C-12 by one P450 (GAO), generating germacra-1(10),4,11(13)-trien-12-ol followed by germacra-1(10),4,11(13)-trien-12-al and resulting in germacra-1(10),4,11(13)-trien-12-oic acid (Nguyen et al., 2010; Cankar et al., 2011; Liu et al., 2011, 2014; Ramirez et al., 2013; Eljounaidi et al., 2014; Fig. 8). A second P450 (COS) hydroxylates germacra-1(10),4,11(13)-trien-12-oic acid at C-6, resulting in spontaneous lactone formation (Ikezawa et al., 2011; Liu et al., 2011; Ramirez et al., 2013; Eljounaidi et al., 2014). The lactone formation in T. garganica is simpler than the formation reported in the Asteraceae species, since epikunzeaol already has a hydroxyl group at C-6 and, therefore, requires only one P450. Thus, the triple oxidation on C-12 to the carboxylic acid leads to a spontaneous lactone ring formation. Consequently, it is not possible to detect the intermediates that are expected to be an alcohol, aldehyde, and acid. The suggested mechanism of epidihydrocostunolide biosynthesis in Figure 8 is based on the previously described mechanism for costunolide in Asteraceae (Nguyen et al., 2010; Cankar et al., 2011; Ikezawa et al., 2011; Liu et al., 2011; Ramirez et al., 2013; Eljounaidi et al., 2014).
Figure 8.
A, Proposed biosynthesis of 1. TgCYP76AE2 is suggested to catalyze three hydroxylations to obtain epidihydrocostunolide. The intermediates are based on knowledge from the costunolide pathway shown in B. B, The costunolide pathway. Step 1 is GAS (germacrene A synthase [Bouwmeester et al., 2002]), followed by three consecutive hydroxylations by GAO (germacrene A oxidase, CYP71AV2-9 [Nguyen et al., 2010; Cankar et al., 2011; Ramirez et al., 2013; Eljounaidi et al., 2014]). Finally, there is hydroxylation of C6 by COS (costunolide synthase, CYP71BL2-5 [Ikezawa et al., 2011; Liu et al., 2011; Ramirez et al., 2013; Eljounaidi et al., 2014]) to yield costunolide.
Neither of the two enzymes CYP71BL1 (GAO) and CYP71BL2 (COS), which catalyze the 8β- and 6α-hydroxylation of germacrene A acid, respectively, shows high identity to TgCYP76AE2, with 32.2% for GAO and 36.6% for COS (Ikezawa et al., 2011). This is despite Apiaceae and Asteraceae being closely related families, exemplifying limitations with the prediction of CYP functionality based on phylogenetic relationship. Sesquiterpenoid lactones are a broad class of compounds, and other mechanisms for the formation of a lactone ring have been reported. The biosynthesis of the lactone ring in the sesquiterpenoid artemisinin, for instance, is different from what is found for (+)-costunolide and epidihydrocostunolide. Here, an aldehyde dehydrogenase and an aldehyde reductase are involved in addition to a P450. These jointly participate in the formation of an acid that is then nonenzymatically converted into the lactone ring, as opposed to the (+)-costunolide and dihydrocostunolide biosynthesis, where ring formation is achieved solely by P450 catalysis (Teoh et al., 2006; Liu et al., 2011).
The P450s GAO and COS from Asteraceae that catalyze the lactone ring formation in (+)-costunolide are from the CYP71 family. While the CYP71 family of the CYP71 clan has had much focus, the CYP76 family is now emerging as another major participant in the biosynthesis of specialized metabolites, especially terpenoids. CYP76s have been found to participate in monoterpenoid, sesquiterpenoid, and diterpenoid biosynthesis (Collu et al., 2001; Guo et al., 2013; Weitzel and Simonsen, 2015). In sesquiterpenoid biosynthesis, CYP76F has been described in S. album as part of the santalol and bergamotol biosynthesis (Diaz-Chavez et al., 2013; Celedon et al., 2016). The CYP71 clan is becoming more of a continuum, and it is expected that future sequencing will add to this and make it even harder to distinguish between the families and subfamilies of this clan. Investigation of the P450s involved in sesquiterpenoid metabolism is expanding from the CYP71 family to the CYP71 clan, as seen in Figure 2, and in the future probably also beyond that.
Stereochemistry and Nonconjugation
A characteristic difference between the guaianolides from Asteraceae and Apiaceae, like thapsigargin, is the α-disposal of the C-6-O bond in Asteraceae whereas it is β-disposed in Apiaceae (Simonsen et al., 2013). This study supports that the β-C-6-O bond characteristic for thapsigargin is introduced already at the very first step of sesquiterpenoid biosynthesis with the formation of epikunzeaol. TgCYP76AE2 converts epikunzeaol to epidihydrocostunolide, which differs from dihydrocostunolide by the β-disposed C-6-O bond. Since no epimerization of C-6 is likely under the oxidative transformation of C-12 into a carboxylic acid, the previously suggested structure of epikunzeaol as the 6α-hydroxygermacrene must be considered unlikely (Pickel et al., 2012). The suggested structure was based only on the assignment of major signals in the spectrum, which might explain the erroneous assignment of stereochemistry.
The expression of genes involved in sesquiterpene biosynthesis in N. benthamiana has been reported to result in glycosylation or other conjugations of the produced sesquiterpenoids. For artemisinic acid in the artemisinin pathway, it was shown that this was conjugated to a diglucoside (van Herpen et al., 2010). When the genes in the costunolide biosynthesis were expressed in N. benthamiana, costunolide was conjugated to glutathione or Cys (Liu et al., 2011). Analysis of the products of TgCYP76AE2 did not reveal any conjugation and, therefore, were detectable by GC-MS. In contrast to costunolide, epidihydrocostunolide does not possess a C-11-C-13 double bond preventing a conjugation to a thiol group.
Downstream Pathway toward Thapsigargin
Costunolide has been suggested as a precursor of many of the studied sesquiterpene lactones in the Asteraceae (de Kraker et al., 2002). The finding of a P450 from T. garganica that synthesizes epidihydrocostunolide indicates that epidihydrocostunolide could have a similar role in Thapsia and that the unusual stereochemistry at C-6 is introduced at a very early stage. Further modification of epidihydrocostunolide to produce thapsigargin would require a mechanism to produce a five- and seven-ring closure of the 10-member ring of epidihydrocostunolide. The exact reaction and enzyme responsible are yet unknown. A putative route could be the oxidation-mediated ring closure similar to what has been described for the biosynthesis of lathyranes in Euphorbiacae species (Luo et al., 2016). Furthermore, several hydroxylations and various acetylations of the backbone are required. A number of these reactions are likely to be performed by P450s.
Specialized Tissue for the Storage of Sesquiterpenoids
Terpenoids often are located in specialized storage structures in plants, which include oil glands, secretory ducts, laticifers, trichomes, and vacuoles (Fahn, 1988; Chadwick et al., 2013). Cells adjacent to or harboring these compartments have been shown to be involved in the biosynthesis of terpenoids that are then transported into the storage compartment (Olsson et al., 2009; Lange, 2015). T. garganica is closely related to the genus Daucus and has a taproot like carrot (Daucus carota; Weitzel et al., 2014). In carrot, it was shown that lipophilic compounds including specialized metabolites were localized primarily in extracellular, long schizogenous hydrophobic oil ducts that were located in the periderm/pericyclic parenchyma tissue (Esau, 1940; Schuphan and Boek, 1960; Deutschmann, 1969; Garrod and Lewis, 1980). The secretory ducts in Daucus are highly organized and connected throughout the phloem of roots and exhibit a concentric ring pattern in horizontal sections (Deutschmann, 1969; Bowes and Mauseth, 2008). Likewise, a positive correlation between the number of ducts and the amount of terpenoids has been demonstrated (Garrod and Lewis, 1980; Senalik and Simon, 1986).
NADI was chosen as a possible stain for terpenoids. The use of NADI has been known since the 1880s and has since been used to detect oxidized compounds. David and Carde (1964) were the first to report the use of NADI for the staining of terpenoids; unfortunately, neither the exact mechanism nor the specificity was described. Throughout the years, the exact mechanism has been much debated, and the specificity for terpenoids is still uncertain. However, NADI does function as a useful method in the current setting to visualize secretory ducts whether the stain is specific toward oxidized and/or lipophilic compounds, including terpenoids.
The finding that transcripts from TgTPS2 and TgCYP76AE2 are found exclusively in epithelial cells surrounding secretory ducts in the middle part of the root supports their involvement in the biosynthesis of thapsigargin. Intermediates from the biosynthesis of thapsigargin were not identified in extracts from T. garganica in this study or in previous studies. This could signify a specific and efficient or possibly even a channeled biosynthetic pathway indicating metabolon formation of the enzymes (Møller, 2010). The presence of thapsigargin in highly specific locations also supports the function of ducts as storage compartments in the roots and possibly also ducts in the fruits of T. garganica, which might explain the high content in these two organs. Similar observations were made in stem cross sections from the conifer Picea sitchensis upon methyl jasmonate treatment (Zulak and Bohlmann, 2010). An antibody against the diterpene synthase levopimaradiene/abietadiene synthase allowed the detection of a fluorescence signal in the epithelial cells of cortical and traumatic resin ducts 2 d after methyl jasmonate treatment. In addition, this indicated the importance of these epithelial cells in the biosynthesis of specialized metabolites. In a subsequent study in P. sitchensis, the importance of the epithelial cells was described further (Abbott et al., 2010; Hamberger et al., 2011). Here, epithelial cells were isolated with laser microdissection, studied by RT-qPCR, and found to be enriched in a variety of CYP720s, including PsCYP720B4, involved in the biosynthesis of isopimaric acid and abietic acid. This approach is of high relevance to the further elucidation of the thapsigargin biosynthetic pathway.
The focus in this study has been directed toward localizing specific tissues in T. garganica that could biosynthesize and store sesquiterpenoids such as thapsigargin. As it was shown previously that the number of secretory ducts correlates with the amount of terpenoids and especially lipophilic terpenoids, cells lining these are an obvious target for biosynthesis studies (Senalik and Simon, 1986). The findings that thapsigargin is stored in secretory ducts in the roots and that the enzymes involved in the biosynthesis are present in the surrounding cells open up new possibilities. The data presented here suggest that future studies, including the identification of enzymes involved in specialized metabolism and specialized transporters in the cells lining the secretory ducts, would benefit from the transcriptomics of these.
CONCLUSION
The roots of the Mediterranean plant T. garganica, also known as deadly carrot, have been shown to have secretory ducts that likely contain the highly toxic compound thapsigargin and similar sesquiterpenoid lactones. Through the transient expression of TgTPS2 (epikunzeaol synthase) and TgCYP76AE2 in N. benthamiana, it was shown that TgTPS2 and TgCYP76AE2 convert epikunzeaol into epidihydrocostunolide, compounds that are possible intermediates in thapsigargin biosynthesis. Transcripts from TgTPS2 and TgCYP76AE2 were found in the epithelial cells lining the secretory ducts and nowhere else in the root. This emphasizes the involvement of these specific cells in the biosynthesis of thapsigargin and other sesquiterpene lactones in T. garganica.
The findings presented here enable the full elucidation of the biosynthesis of thapsigargin in the cells lining the secretory ducts. The findings also show the power of in situ PCR combined with MALDI-TOF.
MATERIALS AND METHODS
Plant Material
Thapsia garganica roots used for cDNA synthesis to obtain genes of interest were collected in June 2010, 25 km south-southwest of Bari, Italy (GPS 40.898625, 16.706139).
For NADI staining, in tube in situ PCR, and MALDI-MSI, 2-year-old T. garganica plants were used. These had been grown in Ibiza, Spain, and were purchased from ThapsIbiza.
Identification and Cloning of Genes
The open reading frame of P450s belonging to the CYP71 clade was found in the transcriptome of the T. garganica root (SRX096991; Pickel et al., 2012) based on BLAST searches using a set of CYP71 clan members as described previously (Dueholm et al., 2015). The P450s are available from the National Center for Biotechnology Information: TgCYP71AH8 (KX826939), TgCYP71AS14 (KX845553), TgCYP71AT12 (KX826940), TgCYP71AU89 (KX845548), TgCYP71AU90 (KX845552), TgCYP71BK1 (KX826941), TgCYP71BK6 (KX845546), TgCYP71D183 (KX845554), TgCYP71D311 (KX845555), TgCYP71D319_ortholog (KX845550), TgCYP76AE1 (KX826942), TgCYP76AE2 (KX826943), TgCYP76AE8 (KX845545), TgCYP76AF7 (KX845549), TgCYP76B79 (KX845547), and TgCYP706C30_ortholog (KX845551). TgCYP71AJ5 (KP191555) and TgCYP71AJ14 (KP191558) also were tested previously (Dueholm et al., 2015). The discovery of TgTPS2 (JQ290345) has been described (Pickel et al., 2012). Full-length gene sequences were obtained from a cDNA library of T. garganica root material.
Forward and reverse primers for all genes were designed with USER overhangs to enable cloning into a pEAQ USER-compatible version of the pEAQ-HT vector (Supplemental Table S1; Luo et al., 2016). pEAQ-HT harbors the viral suppressor p19 and was provided by George Lomonosonoff (Peyret and Lomonossoff, 2013). USER cloning was performed as depicted previously (Nour-Eldin et al., 2006). A truncated version of Arabidopsis (Arabidopsis thaliana) HMGR (GenBank no. J04537), described previously (Cankar et al., 2015), was provided by Katarina Cankar.
Furthermore, TcCYP71AV2 (KC441527.1), CiCYP71AV8 (HQ166835.1), CcCYP71AV9 (KF752448.1), LsCYP71BL2 (HQ439599.1), CiCYP71BL3 (JF816041.1), TcCYP71BL4 (KC441528.1), and CcCYP71BL5 (KF752451.1) were BLASTed into the T. garganica root transcriptome. Setting the expectation value to 1E-100, no hits were available.
Phylogeny
In total, 53 full-length sequences of functionally characterized P450s related to sesquiterpenoid biosynthesis and P450s from T. garganica were used to build the phylogenetic tree. The full list is in Supplemental Figure S4 as part of the alignment, and National Center for Biotechnology Information numbers for the Thapsia genes are given above.
All obtained full-length sequences were aligned using default options in MUSCLE and ClustalW (Edgar, 2004), as implemented in the software Geneious 10.0.5 (www.geneious.com), followed by manual modification. Phylogenetic analyses were conducted using maximum likelihood. Default options for PhyML based on the substation model LG, in Geneious 10.0.5, was chosen (Guindon et al., 2010). All maximum likelihood trees were obtained using 1,000 replicates of a random taxon addition sequence. All characters were included in the analyses. Clade support was assessed using nonparametric bootstrap resampling. Bootstrap analysis (Felsenstein, 1985) was carried out using 1,000 replicates. We defined bootstrap percentages (BS) less than 50% to be unsupported, BS between 50% and 74% as weak support, BS between 75% and 89% as supported, and BS greater than 90% as strong support. Alignments supporting the tree are given in Supplemental File S1 (for Fig. 2). The Thapsia gene cinnamate-4-hydroxylase was used to root the tree.
Expression of TgTPS2 and P450s in Nicotiana benthamiana
N. benthamiana plants were grown from seeds at 24°C/19°C (day/night) for 5 weeks before transformation. The transformation of Agrobacterium tumefaciens and infiltration of N. benthamiana with A. tumefaciens followed the protocol described by Bach et al. (2014). In short, 10 mL of LB medium containing kanamycin, rifampicin, and carbenicillin was inoculated with several A. tumefaciens colonies containing the plasmid of interest. Cultures were grown overnight at 28°C and 200 rpm. Cell pellets were washed twice with water before final resuspension in water followed by a dilution to OD600 = 0.5. Resuspended A. tumefaciens carrying plasmids containing AttHMGR, TgTPS2, or TgCYPs were mixed 1:1:1 and infiltrated into leaves of at least three N. benthamiana plants by use of a syringe. Plants were placed at 24°C/19°C (day/night) and harvested 5 d after infiltration. As controls, plants were infiltrated with A. tumefaciens carrying plasmids with no additional genes, AttHMGR, and AttHMGR plus TgTPS2.
The ∼100 plants needed for the purification of products 1, 2, and 3 were infiltrated by use of a vacuum. Three A. tumefaciens cultures containing AttHMGR, TgTPS2, or TgCYP76AE2 were grown overnight at 28°C and 200 rpm in 500 mL of LB medium (containing kanamycin, rifampicin, and carbenicillin) from 20-mL starter cultures. Cell pellets were washed twice with water before final resuspension in water followed by a dilution to OD600 = 0.5. Resuspended A. tumefaciens carrying plasmids containing AttHMGR, TgTPS2, or TgCYP76AE2 were mixed 1:1:1. Plants were submerged in a 1-L suspension of A. tumefaciens and infiltrated by use of a vacuum at 50 to 100 mbar for 1 min (Andersen-Ranberg et al., 2016).
GC-MS Detection of Sesquiterpenoids
Two leaf discs (Ø = 3 cm) from N. benthamiana expressing genes from T. garganica were extracted with 1.2 mL of hexane for GC-MS analysis to provide one sample; a minimum of three biological replicates were examined.
Hexane extracts were analyzed on a Shimadzu GCMS-QP2010 using an Agilent HP-5MS UI, 20 m, 0.18-mm-diameter × 0.18-μm-film thickness column kept at a pressure of 66.7 kPa giving a column flow of 1 mL min−1 (Drew et al., 2012). The injection port temperature was set to 160°C or 250°C to find the optimal temperature (Andersen et al., 2015a). The oven temperature was set to 60°C for 3 min and then increased to 160°C at a rate of 7°C min−1. The temperature was further increased to 300°C at a rate of 50°C min−1, held for 5 min, finally increased to 320°C at 50°C min−1, and maintained for 3 min. The carrier gas was H2, and the ionization electron energy was 70 eV. The ion source temperature was 230°C, with an interface temperature of 280°C. The total run time was 11.67 min. All data were analyzed using the Shimadzu software Lab Solutions, GCMS Solutions version 2.50 SU3, with the 2008 libraries provided by NIST and Wiley.
Analytical LC-MS for the Detection of Novel Sesquiterpenoids
Two leaf discs with a diameter of 3 cm were ground in liquid nitrogen to provide one sample, and a minimum of three biological replicates was examined. Samples were extracted with 1,200 µL of 80% (v/v) methanol and sonicated for 30 min. Before LC-MS analysis, samples were filtered through a 0.45-µm filter. Analytical LC-MS was carried out using an Agilent 1100 Series device (Agilent Technologies) coupled to a Bruker HCT-Ultra ion-trap mass spectrometer (Bruker Daltonik). A Gemini-NX column (Phenomenex; 3 μm, C18, 110A, 2 × 150 mm) maintained at 35°C was used for separation. The mobile phases were as follows: A, water with 0.1% (v/v) formic acid; and B, acetonitrile with 0.1% (v/v) formic acid. The gradient program was as follows: 0 to 1 min, isocratic 12% B; 1 to 33 min, linear gradient 12% to 80% B; 33 to 35 min, linear gradient 80% to 99% B; 35 to 38 min, isocratic 99% B; 38 to 47 min, isocratic 12% B. The flow rate was 0.2 mL min−1. The mass spectrometer was run in positive mode, and the mass range m/z 100 to 1,000 was acquired.
Preparative GC-MS Purification of Cope Rearranged Sesquiterpenoids (2 and 3)
For the isolation of compounds 2 and 3, a large-scale hexane extraction was made from N. benthamiana leaves expressing AttHMGR, TgTPS2, and TgCYP76AE2. The leaves from approximately 130 5-week-old plants were used. The sample was subjected to an initial separation on a silica column and eluted with hexane-ethyl acetate at 13% ethyl acetate. The final purification was done on an Agilent 7890B gas chromatograph installed with an Agilent 5977A inert MSD device, a GERSTEL AT 6890/7890 preparative fraction collector (PFC), and a GERSTEL CIS 4C bundle injection port. For separation by gas chromatography (GC), a RESTEK Rtx-5 column (30 m × 0.53 mm i.d. × 1 µm df) with H2 as the carrier gas was used. At the end of this column was a split piece with a split of 1:100 to the mass spectrometer and the PFC, respectively. A sufficient amount of sesquiterpene product for NMR analysis (0.5–1 mg) was obtained by 100 repeated injections of 5 µL of extract. The injection port was put in solvent vent mode with a carrier gas flow of 100 mL min−1 until 0.17 min. This was combined with an injection speed of 1.5 mL min−1. The purge flow was set to 3 mL min−1 from 0.17 to 2.17 min. The injection temperature was held at 40°C for 0.1 min, followed by ramping at 12°C s−1 to 320°C, which was held for 2 min. The column flow was set to 7.5 mL, which was held constant throughout the GC program. The GC program was set to hold at 60°C for 1 min, ramp 20°C min−1 to 320°C, which was held for 3 min. The temperature of the transfer line from the gas chromatograph to the PFC and of the PFC itself was set to 250°C. The PFC was set to collect the peak of products 2 and 3 by their retention times identified by mass spectrometry (MS). The mass spectrometer was set in scan mode from m/z 35 to 500 with a threshold of 150. The solvent cutoff was set to 4 min, and the temperatures of the MS source and the MS quadrupole were set to 300°C and 150°C, respectively. Traps were kept at −20°C and rinsed with chloroform-d (Euriso-top; 99.8 atom % D).
Purification of Epidihydrocostunolide (1)
For the isolation of compound 1, a large-scale hexane extraction was made from N. benthamiana leaves expressing truncated AtHMGR, TgTPS2, and TgCYP76AE2. The leaves from ∼100 5-week-old plants were used. The crude hexane extract was subjected to an initial preparative separation on a Biotage Isolera autoflasher using a 10-g, 50-μm diol column and eluted stepwise with hexane to hexane:ethyl acetate (80:20) with 2% increments of ethyl acetate. Final isolation of 1 was achieved by semipreparative HPLC on a 250- × 10-mm, 7-μm Nucleosil PEI column (Macherey-Nagel) eluted isocratically with hexane on a Waters 600 HPLC apparatus equipped with a Waters 996 PDA detector.
1H and 13C NMR Spectroscopic Analysis
NMR spectra were acquired using a 600-MHz Bruker Avance III HD NMR spectrometer (1H operating frequency of 600.13 MHz) equipped with a Bruker SampleJet sample changer and a cryogenically cooled gradient inverse triple-resonance 1.7-mm TCI probe head (Bruker Biospin) optimized for 13C and 1H. Samples were analyzed at 300 K. Proton spectra at 600.03 MHz were obtained using 30°C pulses and a spectral width of 12 kHz, collecting 16 scans with a length of 65,536 data points and with a relaxation delay of 1 s. Carbon spectra were acquired at 150.88 MHz with 30°C pulses and a spectral width of 36 kHz, collecting 256 scans with a length of 65,536 data points and with a relaxation delay of 2 s. The 13C nuclei were 1H decoupled using the Waltz-16 composite pulse-decoupling scheme. FIDs were exponentially multiplied with a line-broadening factor of 1 Hz before Fourier transformation. The two-dimensional experiments were recorded using Bruker standard parameter settings. The isolated product 1 was dissolved in MeCN-d3 (99.8 atom % D), while 2 and 3 were dissolved in CDCl3 (99.8 atom % D) prior to NMR analysis.
Histochemical Analysis Staining
Taproots (∼2 cm in diameter) were dug up just prior to sectioning. Cross sections of roots were cut in 60-µm-thick sections with a vibratome (Microm; HM 650 V). Histochemical analysis was performed with NADI reagent (1% naphthol + 1% dimethyl-p-phenylenediamine + 0.05 m phosphate buffer, pH 7.2) reported to stain terpenoids blue (David and Carde, 1964). Sections were immersed in 0.05 m phosphate buffer (pH 7.2) or NADI solution immediately after cutting and left for a minimum of 30 min before analysis. Samples were mounted on glass slides, and images of the sections were obtained with a Leica DMR HC microscope through ×20 and ×40 dry objectives and a ×100 oil-immersion objective.
MALDI-MSI
Prior to MALDI-MSI analysis, thapsigargin had been verified to be present in the roots by HPLC as described previously (Christensen et al., 1984; Pickel et al., 2012).
Tissue Preparation
Root tissue was snap frozen in a cooling bath (2-propanol and dry-ice), and tissue blocks from the middle part of the root were mounted to the chuck using Tissue-Tek O.C.T. compound. Tissues were sectioned on a cryosectioner (Leica 3050S cryostat). Sections were cut to 60 µm thick, ensuring that no O.C.T. came into contact with the sectioned tissue. Sections were transferred onto prechilled Menzel-Gläser Superfrost Plus 25-mm × 75-mm × 1-mm glass slides and gently adhered to premounted double-sided adhesive carbon tape (Agar Scientific). The frozen slide with sections was transferred into a chilled 50-mL Falcon tube and then freeze dried for 24 h using a freeze dryer (ScanVac CoolSafe) set to −95°C and an operating pressure of 1 mbar. Samples were stored in a vacuum desiccator prior to matrix deposition.
Matrix Deposition
Matrix DHB was sublimed onto tissue sections using a custom-built sublimation apparatus at temperatures of 130°C to 140°C, at vacuum pressures of less than 0.1 mbar, for a period of 15 min, generating a matrix coverage of 0.3 ± 0.1 mg cm−2. An ice slurry was used to cool the sample cold finger. Alternatively, DHB was deposited onto tissues by spray deposition using an HTX Imaging Sprayer (HTX Technologies). DHB in 50 mg mL−1 acetone:water (95:5) was sprayed at 30°C using four passes, a solvent flow rate of 150 µL min−1, and a nozzle velocity of 1,200 mm min−1, with alternate passes at 90° offset, 2-mm track spacing, and 1-mm offset for repeat passes; the nitrogen sheath gas pressure was set to 10 p.s.i. After deposition, samples were stored in a desiccator prior to analysis.
Optical Imaging
Optical images of tissue sections were acquired using an EPSON Photosmart 4400 flatbed scanner using EPSON Scan version 3.04A with a setting of 4,800 d.p.i.
Mass Spectrometer
For spatial MS analysis, a SolariX XR 7 Tesla Hybrid ESI/Fourier transform-ion cyclotron resonance MALDI-MSI system (Bruker Daltonik) was used. The instrument was operated in positive ion mode using optimized instrument settings across the mass range 100 to 2,000 m/z, with the instrument set to broadband mode with a time domain for acquisition of 2 M, providing an estimated resolving power of approximately 260,000 at 400 m/z using a total of one ICR cell fills. The instrument was calibrated to less than 1 ppm tolerance against elemental Red Phosphorous clusters using a quadratic calibration curve across the mass range 216 to 1,951 m/z. The laser was set between 38% and 50% power using the minimum spot size with Smart Walk enabled and a width of 40 µm, grid increment of 10, and offset of 1 using a random walk pattern, resulting in ablation spots of approximately 40 to 50 µm in diameter. A total of 500 to 750 shots were fired per spectrum at a frequency of 2 KHz within a 50- × 50-µm array.
Data Analysis
Acquired MS data were analyzed using Compass FlexImaging 4.1 (build 116; Bruker Daltonik). Images were either normalized to root median square or total ion chromatogram, and brightness optimization was employed to enhance the visualization of the distribution of selected compounds. Individual spectra were analyzed and recalibrated using Bruker Compass DataAnalysis 4.3 (build 110.102.1532) to internal lock masses of known DHB clusters: C14H9O6 = 273.039364 and C21H13O9 = 409.055408 m/z. Peak lists were generated using S/N threshold = 4 and a 0.15% base peak height threshold.
In Tube in Situ Reverse Transcription-PCR on Root Tissue Sections
The in situ PCR was performed as described previously (Athman et al., 2014) with some modifications. Root tissue was left for 4 h in ice-cold FAA (Formalin-Acetic Acid-Alcohol). Root cross sections (75 μm thick) were cut with a vibratome and transferred into tubes containing RNAsin (Promega; N2515) as the RNase inhibitor. A DNase treatment with DNase 1 and 10x Turbo DNase (Qiagen; 79254) was extended to overnight at 37°C. Sections were incubated for 15 min in 0.5 m EDTA at 70°C, rinsed with ice-cold nuclease-free water, and then incubated on ice for 15 min in 2 mg of pepsin in 0.1 m HCl prior to the reverse transcription step to remove cross-linking. Reverse transcription was carried out using the Sensicript Reverse Transcription Kit (Qiagen; 205213). The first cycling conditions were 5 min at 65°C followed by 1 min at 4°C. RNAsin and the reverse transcriptase enzyme were then added, and the following cycling parameters used: 60 min at 42°C, 95°C for 5 min, and 1 min at 4°C. The tubes were then placed directly on ice. The in situ PCR followed using TaKaRa Ex Taq (Clontech; RR001A), 5 mm dNTPs, and PCR DIG labeling mix (Sigma-Aldrich; 11585550910) in the PCR solution.
The negative controls followed the preparation of the test samples, but with the reverse transcriptase enzyme omitted. A positive control was carried out with the ribosomal 18S transcript. The primers used for the cDNA synthesis step (reverse only) and PCR (forward and reverse) are listed in Supplemental Table S2; the products amplified by these primers were sequenced to confirm their specificity. The cycling parameters were as follows: initial denaturation at 95°C for 2 min, 40 cycles of 95°C for 10 s, X°C for 30 s, and 72°C for 1 min (where X = 58°C for TgCYP76AE2 and 56°C for TgTPS2 and 18S). The final extension was at 72°C for 5 min and then held at 4°C. Sections were incubated on ice for 30 min in a blocking solution (1% bovine serum albumin in 1× phosphate buffer) to prevent background staining. Anti-DIG-AP Fab fragments were then added followed by a 60-min incubation at room temperature. BM Purple AP substrate (Sigma-Aldrich; 000000011442074001) was used for colorimetric staining. Sections were stained for 1.5 h and mounted in antifading solution (Citifluor; AF3-25). Slides were viewed with a Leica DMR HC microscope.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. 13C- and 1H-NMR spectra of products 1, 2, and 3.
Supplemental Figure S2. Cope rearrangement of 1 upon GC-MS analysis.
Supplemental Figure S3. MALDI-MSI analysis of a T. garganica taproot section and structures of metabolites.
Supplemental Figure S4. Alignment for the tree in Figure 2.
Supplemental Table S1. Primers for cloning into the USER version of the vector pEAQ.
Supplemental Table S2. In tube in situ primers.
Supplementary Material
Acknowledgments
We thank George Lomonosonoff (John Innes Research Centre) for the pEAQ-HT plasmid, Katarina Cankar (Wageningen University) for the truncated HMGR, and Metabolomics Australia (School of BioSciences, University of Melbourne), an NCRIS initiative under Bioplatforms Australia, for conducting MALDI-MSI.
Glossary
- MALDI-MSI
matrix-assisted laser-desorption ionization-mass spectrometry imaging
- GC-MS
gas chromatography-mass spectrometry
- LC-MS
liquid chromatography-mass spectrometry
- DHB
2,5-dihydroxybenzoic acid
- MALDI
matrix-assisted laser-desorption ionization
- DIG
digoxigenin
- BS
bootstrap percentages
- PFC
preparative fraction collector
- GC
gas chromatography
- MS
mass spectrometry
Footnotes
This work was supported by SpotLight, a grant from the Danish Council for Strategic Research (to T.B.A., H.T.S., and S.B.C.), by the Center for Synthetic Biology bioSYNergy, UCPH Excellence Program for Interdisciplinary Research (to J.A-R.), and by MEDPLANT, a Marie Curie Actions Initial Training Network (to K.A.M.-S.); NMR equipment used was purchased via from the Danish Research Council for Independent Research|Nature and Universe (grant no. 10-085264).
References
- Abbott E, Hall D, Hamberger B, Bohlmann J (2010) Laser microdissection of conifer stem tissues: isolation and analysis of high quality RNA, terpene synthase enzyme activity and terpenoid metabolites from resin ducts and cambial zone tissue of white spruce (Picea glauca). BMC Plant Biol 10: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adio AM. (2009) Germacrenes A-E and related compounds: thermal, photochemical and acid induced transannular cyclizations. Tetrahedron 65: 1533–1552 [Google Scholar]
- Andersen TB, Cozzi F, Simonsen HT (2015a) Optimization of biochemical screening methods for volatile and unstable sesquiterpenoids using HS-SPME-GC-MS. Chromatography 2: 277–292 [Google Scholar]
- Andersen TB, López CQ, Manczak T, Martinez K, Simonsen HT (2015b) Thapsigargin: from Thapsia L. to mipsagargin. Molecules 20: 6113–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen-Ranberg J, Kongstad KT, Nielsen MT, Jensen NB, Pateraki I, Bach SS, Hamberger B, Zerbe P, Staerk D, Bohlmann J, et al. (2016) Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angew Chem Int Ed Engl 55: 2142–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando M, Tajima K, Takase K (1983) Studies on the syntheses of sesquiterpene lactones. 8. Syntheses of saussurea lactone, 8-deoxymelitensin, and 11,12-dehydro-8-deoxymelitensin via a novel fragmentation reaction. J Org Chem 48: 1210–1216 [Google Scholar]
- Andrews SP, Ball M, Wierschem F, Cleator E, Oliver S, Högenauer K, Simic O, Antonello A, Hünger U, Smith MD, et al. (2007) Total synthesis of five thapsigargins: guaianolide natural products exhibiting sub-nanomolar SERCA inhibition. Chemistry 13: 5688–5712 [DOI] [PubMed] [Google Scholar]
- Appendino G, Gariboldi P (1983) The structure and chemistry of hallerin, a mixture of anomeric sesquiterpenoids from Laserpitium halleri Crantz subsp. halleri. J Chem Soc Perkin Trans 1: 2017–2026 [Google Scholar]
- Athman A, Tanz SK, Conn VM, Jordans C, Mayo GM, Ng WW, Burton RA, Conn SJ, Gilliham M (2014) Protocol: a fast and simple in situ PCR method for localising gene expression in plant tissue. Plant Methods 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach SS, Bassard JÉ, Andersen-Ranberg J, Møldrup ME, Simonsen HT, Hamberger B (2014) High throughput testing of terpenoid biosynthesis candidate genes using transient expression in Nicotiana benthamiana. In Rodríguez-Concepción M, ed, Plant Isoprenoids. Springer, New York, pp 245–255 [DOI] [PubMed] [Google Scholar]
- Ball M, Andrews SP, Wierschem F, Cleator E, Smith MD, Ley SV (2007) Total synthesis of thapsigargin, a potent SERCA pump inhibitor. Org Lett 9: 663–666 [DOI] [PubMed] [Google Scholar]
- Barrero AF, Oltra JE, Alvarez M, Rosales A (2002) Synthesis of (+)-8-deoxyvernolepin and its 11,13-dihydroderivative: a novel reaction initiated by sulfene elimination leads to the 2-oxa-cis-decalin skeleton. J Org Chem 67: 5461–5469 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughton BA, Thinagaran D, Sarabia D, Bacic A, Roessner U (2016) Mass spectrometry imaging for plant biology: a review. Phytochem Rev 15: 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester HJ, Kodde J, Verstappen FWA, Altug IG, de Kraker JW, Wallaart TE (2002) Isolation and characterization of two germacrene A synthase cDNA clones from chicory. Plant Physiol 129: 134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes BG, Mauseth JD (2008) Plant Structure: A Colour Guide, Ed 2 Manson Publishing, London [Google Scholar]
- Caissard JC, Meekijjironenroj A, Baudino S, Anstett MC (2004) Localization of production and emission of pollinator attractant on whole leaves of Chamaerops humilis (Arecaceae). Am J Bot 91: 1190–1199 [DOI] [PubMed] [Google Scholar]
- Cankar K, Jongedijk E, Klompmaker M, Majdic T, Mumm R, Bouwmeester H, Bosch D, Beekwilder J (2015) (+)-Valencene production in Nicotiana benthamiana is increased by down-regulation of competing pathways. Biotechnol J 10: 180–189 [DOI] [PubMed] [Google Scholar]
- Cankar K, van Houwelingen A, Bosch D, Sonke T, Bouwmeester H, Beekwilder J (2011) A chicory cytochrome P450 mono-oxygenase CYP71AV8 for the oxidation of (+)-valencene. FEBS Lett 585: 178–182 [DOI] [PubMed] [Google Scholar]
- Celedon JM, Chiang A, Yuen MMS, Diaz-Chavez ML, Madilao LL, Finnegan PM, Barbour EL, Bohlmann J (2016) Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J 86: 289–299 [DOI] [PubMed] [Google Scholar]
- Chadwick M, Trewin H, Gawthrop F, Wagstaff C (2013) Sesquiterpenoids lactones: benefits to plants and people. Int J Mol Sci 14: 12780–12805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SB, Andersen A, Smitt UW (1997) Sesquiterpenoids from Thapsia species and medicinal chemistry of the thapsigargins. Prog Chem Org Nat Prod 71: 129–167 [DOI] [PubMed] [Google Scholar]
- Christensen SB, Norup E, Rasmussen U, Madsen JØ (1984) Structure of histamine releasing guaianolides from Thapsia species. Phytochemistry 23: 1659–1663 [Google Scholar]
- Chu H, Smith JM, Felding J, Baran PS (2017) Scalable Synthesis of (−)-Thapsigargin. ACS Cent Sci; 25: 47–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collu G, Unver N, Peltenburg-Looman AM, van der Heijden R, Verpoorte R, Memelink J (2001) Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett 508: 215–220 [DOI] [PubMed] [Google Scholar]
- Corsi G, Pagni AM, Innocenti G (1988) Carum appuanum (Viv.) Grande (Umbelliferae). I. Histochemical and anatomical study. Int J Crude Drug Res 26: 129–136 [Google Scholar]
- Curry EA III, Murry DJ, Yoder C, Fife K, Armstrong V, Nakshatri H, O’Connell M, Sweeney CJ (2004) Phase I dose escalation trial of feverfew with standardized doses of parthenolide in patients with cancer. Invest New Drugs 22: 299–305 [DOI] [PubMed] [Google Scholar]
- David R, Carde J (1964) Histochimie-coloration differentielle des inclusions lipidiques et terpeniques des pseudophylles du pin maritime au moyen du reactif NADI. C R Hebd Seances Acad Sci 258: 1338 [Google Scholar]
- de Kraker JW, Franssen MC, Dalm MC, de Groot A, Bouwmeester HJ (2001) Biosynthesis of germacrene A carboxylic acid in chicory roots: demonstration of a cytochrome P450 (+)-germacrene a hydroxylase and NADP+-dependent sesquiterpenoid dehydrogenase(s) involved in sesquiterpene lactone biosynthesis. Plant Physiol 125: 1930–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kraker JW, Franssen MCR, Joerink M, de Groot A, Bouwmeester HJ (2002) Biosynthesis of costunolide, dihydrocostunolide, and leucodin: demonstration of cytochrome P450-catalyzed formation of the lactone ring present in sesquiterpene lactones of chicory. Plant Physiol 129: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschmann F. (1969) Anatomische Studien uber die Exkretgange in Umbelliferenwurzeln. Beitrage zur Biologie der Pflanzen 45: 409–440 [Google Scholar]
- Diaz-Chavez ML, Moniodis J, Madilao LL, Jancsik S, Keeling CI, Barbour EL, Ghisalberti EL, Plummer JA, Jones CG, Bohlmann J (2013) Biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol. PLoS ONE 8: e75053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan NT, Paulsen ES, Sehgal P, Møller JV, Nissen P, Denmeade SR, Isaacs JT, Dionne CA, Christensen SB (2015) Targeting thapsigargin towards tumors. Steroids 97: 2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew DP, Dueholm B, Weitzel C, Zhang Y, Sensen CW, Simonsen HT (2013) Transcriptome analysis of Thapsia laciniata Rouy provides insights into terpenoid biosynthesis and diversity in Apiaceae. Int J Mol Sci 14: 9080–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew DP, Krichau N, Reichwald K, Simonsen HT (2009) Guaianolides in Apiaceae: perspectives on pharmacology and biosynthesis. Phytochem Rev 8: 581–599 [Google Scholar]
- Drew DP, Rasmussen SK, Avato P, Simonsen HT (2012) A comparison of headspace solid-phase microextraction and classic hydrodistillation for the identification of volatile constituents from Thapsia spp. provides insights into guaianolide biosynthesis in Apiaceae. Phytochem Anal 23: 44–51 [DOI] [PubMed] [Google Scholar]
- Dueholm B, Krieger C, Drew D, Olry A, Kamo T, Taboureau O, Weitzel C, Bourgaud F, Hehn A, Simonsen HT (2015) Evolution of substrate recognition sites (SRSs) in cytochromes P450 from Apiaceae exemplified by the CYP71AJ subfamily. BMC Evol Biol 15: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eljounaidi K, Cankar K, Comino C, Moglia A, Hehn A, Bourgaud F, Bouwmeester H, Menin B, Lanteri S, Beekwilder J (2014) Cytochrome P450s from Cynara cardunculus L. CYP71AV9 and CYP71BL5, catalyze distinct hydroxylations in the sesquiterpene lactone biosynthetic pathway. Plant Sci 223: 59–68 [DOI] [PubMed] [Google Scholar]
- Esau K. (1940) Developmental anatomy of the fleshy storage organ of Daucus carota. Hilgardia 13: 175–209 [Google Scholar]
- Fahn A. (1988) Tansley Review No. 14. Secretory tissues in vascular plants. New Phytol 108: 229–257 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fischer NH, Mabry TJ (1967) Structure of tamaulipin-B, a new germacranolide and the thermal conversion of a trans-1,2-divinylcyclohexane derivative into a cyclodeca-1,5-diene system. Chem Commun 1235–1236 [Google Scholar]
- Garrod B, Lewis BG (1980) Probable role of oil ducts in carrot root tissue. Trans Br Mycol Soc 75: 166–169 [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L, Wang Y, Zhang X, Liu W, Peters RJ, Chen X, et al. (2013) CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA 110: 12108–12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Bak S (2013) Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci 368: 20120426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Ohnishi T, Hamberger B, Séguin A, Bohlmann J (2011) Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol 157: 1677–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwig J. (1967) The use of histochemical reagents for cytochrome oxidase in plant tissues. Bot Rev 33: 116–129 [Google Scholar]
- Havis L. (1939) Anatomy of the hypocotyl and roots of Daucus carota. J Agric Res 58: 557–564 [Google Scholar]
- Ikezawa N, Göpfert JC, Nguyen DT, Kim SU, O’Maille PE, Spring O, Ro DK (2011) Lettuce costunolide synthase (CYP71BL2) and its homolog (CYP71BL1) from sunflower catalyze distinct regio- and stereoselective hydroxylations in sesquiterpene lactone metabolism. J Biol Chem 286: 21601–21611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis DE, Ho YS, Lightfoot DJ, Schmöckel SM, Li B, Borm TJA, Ohyanagi H, Mineta K, Michell CT, Saber N, et al. (2017) The genome of Chenopodium quinoa. Nature 542: 307–312 [DOI] [PubMed] [Google Scholar]
- Jezler CN, Batista RS, Alves PB, Silva DdC, Costa LCdB (2013) Histochemistry, content and chemical composition of essential oil in different organs of Alpinia zerumbet. Cienc Rural 43: 1811–1816 [Google Scholar]
- Korolev AV, Tomos AD, Bowtell R, Farrar JF (2000) Spatial and temporal distribution of solutes in the developing carrot taproot measured at single-cell resolution. J Exp Bot 51: 567–577 [DOI] [PubMed] [Google Scholar]
- Kromer K, Kreitschitz A, Kleinteich T, Gorb SN, Szumny A (2016) Oil secretory system in vegetative organs of three Arnica taxa: essential oil synthesis, distribution and accumulation. Plant Cell Physiol 57: 1020–1037 [DOI] [PubMed] [Google Scholar]
- Lange BM. (2015) The evolution of plant secretory structures and emergence of terpenoid chemical diversity. Annu Rev Plant Biol 66: 139–159 [DOI] [PubMed] [Google Scholar]
- Larbat R, Hehn A, Hans J, Schneider S, Jugdé H, Schneider B, Matern U, Bourgaud F (2009) Isolation and functional characterization of CYP71AJ4 encoding for the first P450 monooxygenase of angular furanocoumarin biosynthesis. J Biol Chem 284: 4776–4785 [DOI] [PubMed] [Google Scholar]
- Larbat R, Kellner S, Specker S, Hehn A, Gontier E, Hans J, Bourgaud F, Matern U (2007) Molecular cloning and functional characterization of psoralen synthase, the first committed monooxygenase of furanocoumarin biosynthesis. J Biol Chem 282: 542–554 [DOI] [PubMed] [Google Scholar]
- Liu Q, Majdi M, Cankar K, Goedbloed M, Charnikhova T, Verstappen FW, de Vos RC, Beekwilder J, van der Krol S, Bouwmeester HJ (2011) Reconstitution of the costunolide biosynthetic pathway in yeast and Nicotiana benthamiana. PLoS ONE 6: e23255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Manzano D, Tanić N, Pesic M, Bankovic J, Pateraki I, Ricard L, Ferrer A, de Vos R, van de Krol S, et al. (2014) Elucidation and in planta reconstitution of the parthenolide biosynthetic pathway. Metab Eng 23: 145–153 [DOI] [PubMed] [Google Scholar]
- Luo D, Callari R, Hamberger B, Wubshet SG, Nielsen MT, Andersen-Ranberg J, Hallström BM, Cozzi F, Heider H, Lindberg Møller B, et al. (2016) Oxidation and cyclization of casbene in the biosynthesis of Euphorbia factors from mature seeds of Euphorbia lathyris L. Proc Natl Acad Sci USA 113: E5082–E5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P, Wang YH, Wang GD, Essenberg M, Chen XY (2001) Molecular cloning and functional identification of (+)-delta-cadinene-8-hydroxylase, a cytochrome P450 mono-oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J 28: 95–104 [DOI] [PubMed] [Google Scholar]
- Maggi F, Papa F, Giuliani C, Maleci Bini L, Venditti A, Bianco A, Nicoletti M, Iannarelli R, Caprioli G, Sagratini G, et al. (2015) Essential oil chemotypification and secretory structures of the neglected vegetable Smyrnium olusatrum L. (Apiaceae) growing in central Italy. Flavour Fragrance J 30: 139–159 [Google Scholar]
- Mahalingam D, Wilding G, Denmeade S, Sarantopoulas J, Cosgrove D, Cetnar J, Azad N, Bruce J, Kurman M, Allgood VE, et al. (2016) Mipsagargin, a novel thapsigargin-based PSMA-activated prodrug: results of a first-in-man phase I clinical trial in patients with refractory, advanced or metastatic solid tumours. Br J Cancer 114: 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller BL. (2010) Dynamic metabolons. Science 330: 1328–1329 [DOI] [PubMed] [Google Scholar]
- Muravnik LE, Kostina OV, Shavarda AL (2016) Glandular trichomes of Tussilago farfara (Senecioneae, Asteraceae). Planta 244: 737–752 [DOI] [PubMed] [Google Scholar]
- Nelson DR. (2009) The cytochrome p450 homepage. Hum Genomics 4: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DT, Göpfert JC, Ikezawa N, Macnevin G, Kathiresan M, Conrad J, Spring O, Ro DK (2010) Biochemical conservation and evolution of germacrene A oxidase in Asteraceae. J Biol Chem 285: 16588–16598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MH, Jensen JK, Halkier BA (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson ME, Olofsson LM, Lindahl AL, Lundgren A, Brodelius M, Brodelius PE (2009) Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70: 1123–1128 [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, et al. (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496: 528–532 [DOI] [PubMed] [Google Scholar]
- Peyret H, Lomonossoff GP (2013) The pEAQ vector series: the easy and quick way to produce recombinant proteins in plants. Plant Mol Biol 83: 51–58 [DOI] [PubMed] [Google Scholar]
- Pickel B, Drew DP, Manczak T, Weitzel C, Simonsen HT, Ro DK (2012) Identification and characterization of a kunzeaol synthase from Thapsia garganica: implications for the biosynthesis of the pharmaceutical thapsigargin. Biochem J 448: 261–271 [DOI] [PubMed] [Google Scholar]
- Poli F, Tirillini B, Tosi B, Sacchetti G, Bruni A (1995) Histological localization of coumarins in different organs of Smyrnium perfoliatum (Apiaceae). Phyton Annales Rei Botanicae A 35: 209–217 [Google Scholar]
- Ramirez AM, Saillard N, Yang T, Franssen MCR, Bouwmeester HJ, Jongsma MA (2013) Biosynthesis of sesquiterpene lactones in pyrethrum (Tanacetum cinerariifolium). PLoS ONE 8: e65030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucher S, Chi KW, Hwang KJ, Burks JE Jr (1986) Synthesis via sigmatropic rearrangement. 12. Total synthesis of (+)-dihydrocostunolide via tandem Cope-Claisen rearrangement. J Org Chem 51: 5503–5505 [Google Scholar]
- Sanz JF, Castellano G, Marco JA (1990) Sesquiterpene lactones from Artemisia herba-alba. Phytochemistry 29: 541–545 [Google Scholar]
- Schuphan W, Boek K (1960) Histologisch-chemische Untersuchungen in Speicherwurzeln der Möhre (Daucus carota L.) in Beziehung zu Rückständen nach Aldrin-und Dieldrin-Behandlung. Qualitas Plantarum et Materiae Vegetabiles 7: 213–228 [Google Scholar]
- Senalik D, Simon PW (1986) Relationship between oil ducts and volatile terpenoid content in carrot roots. Am J Bot 73: 60–63 [DOI] [PubMed] [Google Scholar]
- Setzer WN. (2008) Ab initio analysis of the Cope rearrangement of germacrane sesquiterpenoids. J Mol Model 14: 335–342 [DOI] [PubMed] [Google Scholar]
- Simonsen HT, Weitzel C, Christensen SB (2013) Guaianolide sesquiterpenoids: their pharmacology and biosynthesis. In Ramawat KG, Merillon JM, eds, Handbook of Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes. Springer-Verlag, Berlin, pp 3069–3098 [Google Scholar]
- Smitt UW, Jäger AK, Adsersen A, Gudiksen L (1995) Comparative studies in phytochemistry and fruit anatomy of Thapsia garganica and T. transtagana, Apiaceae (Umbelliferae). Bot J Linn Soc 117: 281–292 [Google Scholar]
- Stešević D, Božović M, Tadić V, Rančić D, Stevanović ZD (2016) Plant-part anatomy related composition of essential oils and phenolic compounds in Chaerophyllum coloratum, a Balkan endemic species. Flora 220: 37–51 [Google Scholar]
- Stojičić D, Tošić S, Slavkovska V, Zlatković B, Budimir S, Janošević D, Uzelac B (2016) Glandular trichomes and essential oil characteristics of in vitro propagated Micromeria pulegium (Rochel) Benth. (Lamiaceae). Planta 244: 393–404 [DOI] [PubMed] [Google Scholar]
- Takamatsu H, Hirai KI (1968) NADI reaction of myeloid leucocytes and vitamin K. Acta Histochem Cytochem 1: 1–10 [Google Scholar]
- Takase H, Sasaki K, Shinmori H, Shinohara A, Mochizuki C, Kobayashi H, Ikoma G, Saito H, Matsuo H, Suzuki S, et al. (2016) Cytochrome P450 CYP71BE5 in grapevine (Vitis vinifera) catalyzes the formation of the spicy aroma compound (−)-rotundone. J Exp Bot 67: 787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K. (1974) Stereospecific Cope rearrangement of the germacrene-type sesquiterpenes. Tetrahedron 30: 1525–1534 [Google Scholar]
- Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS (2006) Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett 580: 1411–1416 [DOI] [PubMed] [Google Scholar]
- Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, Dawson AP (1990) Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2+-ATPase. Proc Natl Acad Sci USA 87: 2466–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y. (2011) The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat Med 17: 1217–1220 [DOI] [PubMed] [Google Scholar]
- van Herpen TW, Cankar K, Nogueira M, Bosch D, Bouwmeester HJ, Beekwilder J (2010) Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS ONE 5: e14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel C, Rønsted N, Spalik K, Simonsen HT (2014) Resurrecting deadly carrots: towards a revision of Thapsia (Apiaceae) based on phylogenetic analysis of nrITS sequences and chemical profiles. Bot J Linn Soc 174: 620–636 [Google Scholar]
- Weitzel C, Simonsen HT (2015) Cytochrome P450-enzymes involved in the biosynthesis of mono- and sesquiterpenes. Phytochem Rev 14: 7–24 [Google Scholar]
- Wen W, Yu R (2011) Artemisinin biosynthesis and its regulatory enzymes: progress and perspective. Pharmacogn Rev 5: 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner J, Ortmann R, Jomaa H, Schlitzer M (2003) New antimalarial drugs. Angew Chem Int Ed Engl 42: 5274–5293 [DOI] [PubMed] [Google Scholar]
- Yang K, Monafared R, Wang H, Lundgren A, Brodelius P (2015) The activity of the artemisinic aldehyde Δ11(13) reductase promoter is important for artemisinin yield in different chemotypes of Artemisia annua L. Plant Mol Biol 88: 325–340 [DOI] [PubMed] [Google Scholar]
- Zhan X, Zhang YH, Chen DF, Simonsen HT (2014) Metabolic engineering of the moss Physcomitrella patens to produce the sesquiterpenoids patchoulol and α/β-santalene. Front Plant Sci 5: 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulak KG, Bohlmann J (2010) Terpenoid biosynthesis and specialized vascular cells of conifer defense. J Integr Plant Biol 52: 86–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.