A Cuphea diacylglycerol acyltransferase CpuDGAT1 functions in concert with a specialized lysophosphatidic acyltransferase for production of medium-chain fatty acid-rich oils.
Abstract
Seed oils of many Cuphea sp. contain >90% of medium-chain fatty acids, such as decanoic acid (10:0). These seed oils, which are among the most compositionally variant in the plant kingdom, arise from specialized fatty acid biosynthetic enzymes and specialized acyltransferases. These include lysophosphatidic acid acyltransferases (LPAT) and diacylglycerol acyltransferases (DGAT) that are required for successive acylation of medium-chain fatty acids in the sn-2 and sn-3 positions of seed triacylglycerols (TAGs). Here we report the identification of a cDNA for a DGAT1-type enzyme, designated CpuDGAT1, from the transcriptome of C. avigera var pulcherrima developing seeds. Microsomes of camelina (Camelina sativa) seeds engineered for CpuDGAT1 expression displayed DGAT activity with 10:0-CoA and the diacylglycerol didecanoyl, that was approximately 4-fold higher than that in camelina seed microsomes lacking CpuDGAT1. In addition, coexpression in camelina seeds of CpuDGAT1 with a C. viscosissima FatB thioesterase (CvFatB1) that generates 10:0 resulted in TAGs with nearly 15 mol % of 10:0. More strikingly, expression of CpuDGAT1 and CvFatB1 with the previously described CvLPAT2, a 10:0-CoA-specific Cuphea LPAT, increased 10:0 amounts to 25 mol % in camelina seed TAG. These TAGs contained up to 40 mol % 10:0 in the sn-2 position, nearly double the amounts obtained from coexpression of CvFatB1 and CvLPAT2 alone. Although enriched in diacylglycerol, 10:0 was not detected in phosphatidylcholine in these seeds. These findings are consistent with channeling of 10:0 into TAG through the combined activities of specialized LPAT and DGAT activities and demonstrate the biotechnological use of these enzymes to generate 10:0-rich seed oils.
Fatty acid and triacylglycerol (TAG) biosynthetic pathways in species of the Cuphea genus are among the most highly divergent in the plant kingdom (Graham, 1989). Seed oils of Cuphea species such as C. avigera var pulcherrima and C. viscosissima accumulate the medium-chain length fatty acids octanoic (8:0) and decanoic (10:0) acids to >90 mol % of TAG fatty acids (Graham, 1989; Kim et al., 2015b). By contrast, seed oils of most plants, including those of the major commercial oilseed crops, are enriched in fatty acids with 16- and 18-carbon atoms. The Cuphea medium-chain fatty acids are produced by variations of the plastid-localized fatty acid synthase that results in the release of medium-chain fatty acids from acyl carrier protein by specialized FatB-type thioesterases before their extension to C16 and C18 (Dehesh et al., 1996a, 1996b; Leonard et al., 1997; Slabaugh et al., 1998). In addition to their synthesis, medium-chain fatty acids must accumulate to high levels at the three stereospecific positions of the glycerol backbone to generate TAGs with >90 mol % of medium-chain fatty acids found in C. avigera var pulcherrima and C. viscosissima. These reactions are catalyzed by the ER-localized glycerol 3-P acyltransferase (GPAT; EC 2.3.1.15), lysophosphatidic acid acyltransferase (LPAT; EC 2.3.1.51), and diacylglycerol acyltransferase (DGAT; EC 2.3.1.20) that use acyl-CoA substrates for successive acylation of the three stereospecific positions of glycerol intermediates in the Kennedy pathway of TAG biosynthesis (Kennedy, 1961). The production of TAGs with >90 mol % medium-chain fatty acids requires specialized acyltransferases with high specificities for medium-chain acyl-CoAs and medium-chain fatty acid-containing glycerol intermediates, as previously shown by enzymatic assays and radiolabeling using C. lanceolata seed microsomes (Bafor et al., 1990; Vogel and Browse, 1996). These studies are consistent with direct channeling of 10:0 in C. lanceolata seeds through the Kennedy pathway acyltransferases for TAG synthesis (Bafor et al., 1990), rather than through phosphatidylcholine (PC) as occurs in most seeds for polyunsaturated fatty acid synthesis (Slack et al., 1978; Stymne and Appelqvist, 1978; Bates et al., 2009).
We have previously identified a LPAT from C. viscosissima (CvLPAT2) that has high activity with 10:0-CoA (Kim et al., 2015a). The CvLPAT2 activity differs from that of typical ER-localized LPATs that instead primarily use C18 substrates with Δ9 unsaturation, and have only low activity with saturated fatty acyl-CoA substrates (Ichihara et al., 1987; Sun et al., 1988; Oo and Huang, 1989). Transgenic expression of CvLPAT2 along with a 10:0 producing C. viscosissima FatB (CvFatB1) in camelina (Camelina sativa) seeds resulted in the production of TAGs with 10:0 in the sn-2 position, which were absent from this position in seeds expressing only CvFatB1 (Kim et al., 2015a).
Previous analyses of DGAT activities in Cuphea sp. seed microsomes have also indicated substrate preference for medium-chain acyl-CoAs and medium-chain fatty acid-containing diacylglycerols (DAGs; Cao and Huang, 1986; Bafor et al., 1990). These findings point to a role for DGAT as a major contributor to the high levels of medium-chain fatty acids accumulated in TAGs of many Cuphea sp. Two major forms of DGAT are associated with TAG biosynthesis in seeds DGAT1 and DGAT2, and a third DGAT, DGAT3, has also been ascribed a role in TAG biosynthesis (Saha et al., 2006; Cao, 2011; Liu et al., 2012). DGAT1 and DGAT2, in contrast to the soluble DGAT3, are ER-localized but are structurally distinct (Shockey et al., 2006). Typical DAGAT1 polypeptides are >150 amino acids larger than DGAT2 and have eight to ten predicted transmembrane domains versus three to six in DGAT2 (Cao, 2011; Liu et al., 2012). Both DGAT1 and DGAT2 enzymes from a variety of species have been linked to the preferential accumulation of unusual fatty acids in seed TAGs (Burgal et al., 2008; Li et al., 2010; Aymé et al., 2015). Recently, a palm (Elaeis guineensis) kernel DGAT1 was shown to promote increased accumulation of the medium-chain fatty acid lauric acid (12:0) relative to the Arabidopsis (Arabidopsis thaliana) DGAT1 when expressed in the oleaginous yeast Yarrowia lipolytica. This preferential activity for lauric acid (12:0) was proposed to be related to variations in the N-terminal region observed in diverse DGAT1 polypeptides (Aymé et al., 2015). In addition, a DGAT2 from castor bean (Ricinus communis) was shown to increase accumulation of the hydroxy fatty acid ricinoleic acid in TAG when expressed in transgenic Arabidopsis seeds (Burgal et al., 2008). Similarly, an ironweed (Vernonia galamensis) DGAT2 was shown to increase accumulation of the epoxy fatty acid vernolic acid in TAG when expressed in transgenic soybean seeds (Li et al., 2010).
Here we report the identification of a cDNA for a seed-specific DGAT1 enzyme designated CpuDGAT1 from the transcriptome of developing C. avigera var pulcherrima seeds that is active with 10:0-CoA and didecanoin (10:0-DAG). In addition, we show that recombinant expression of this enzyme enhances 10:0 accumulation in camelina seeds engineered for CvFatB1 expression. Consistent with channeling of 10:0 through the Kennedy pathway, coexpression of the CpuDGAT1 and CvLPAT2 enzymes not only increased total 10:0 accumulation in camelina seed TAG, but also doubled the total content of 10:0 in the sn-2 position in TAG relative to expression of CvLPAT2 alone. This was achieved with no detectable accumulation of 10:0 in PC of engineered camelina seeds. Results of analyses of fatty acid content, seed weight, and germination assays of engineered camelina seeds are also reported. Overall, these findings expand the functional diversity of DGAT1 enzymes and demonstrate the concerted capacity of specialized DGAT and LPAT activities for conferring enhanced medium-chain fatty acid accumulation in an engineered oilseed crop.
RESULTS
Identification of a Seed-specific DGAT1 in C. avigera var pulcherrima
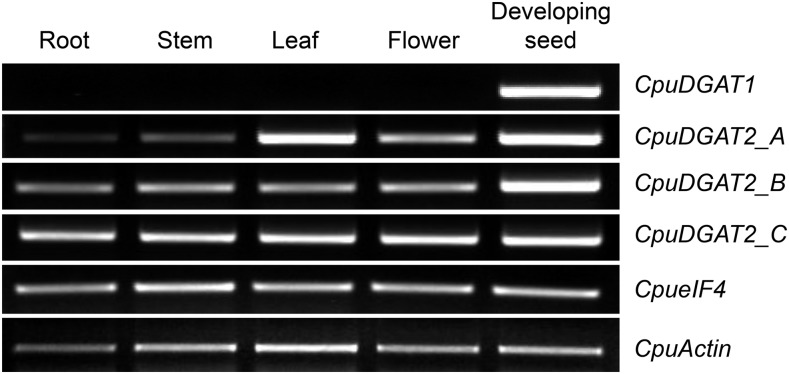
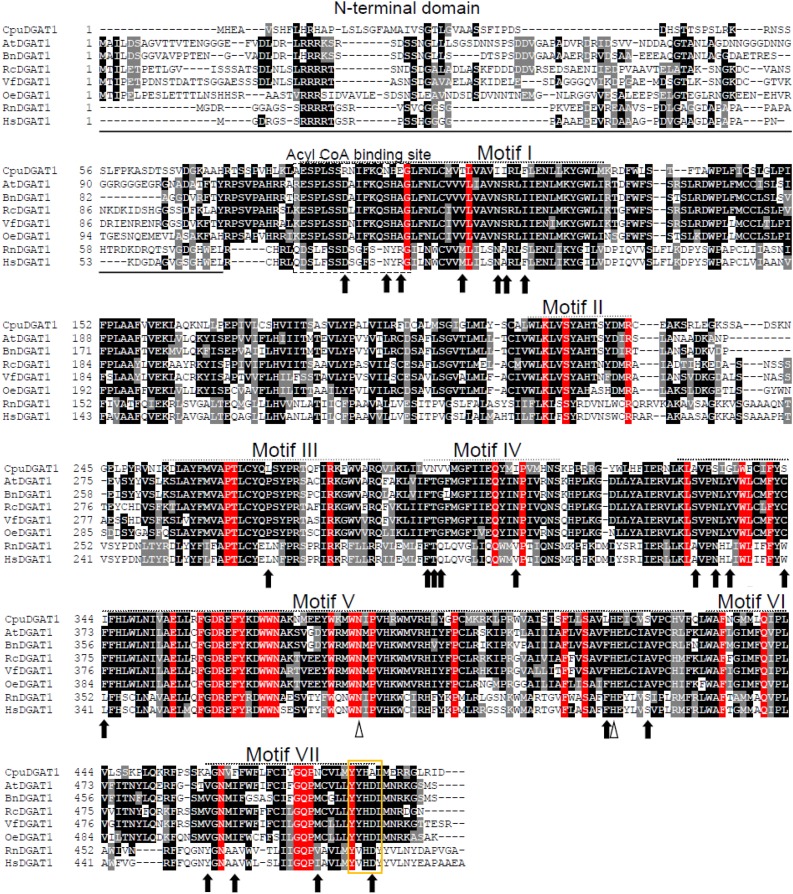
The previously described transcriptome of developing C. avigera var pulcherrima seeds (Kim et al., 2015b) was mined for candidate genes encoding DGATs that are specialized for medium-chain fatty acid metabolism. Sequences for one DGAT1-like gene designated CpuDGAT1 and three DGAT2-like genes designated CpuDGAT2_A, CpDGAT2_B, and CpuDGAT2_C, were identified from these studies. Based on RT-PCR analysis, CpuDGAT1 was determined to be seed specific, whereas CpuDGAT2_A, CpuDGAT2_B, and CpuDGAT2_C were found to be constitutively expressed in C. avigera var pulcherrima (Fig. 1). Based on its expression pattern that reflects the seed-specific occurrence of medium-chain fatty acids in C. avigera var pulcherrima, CpuDGAT1 was chosen for functional characterization. CpuDGAT1 encodes a 493 amino acid polypeptide with 59%, 54%, and 50% amino acid sequence identity with DGAT1 polypeptides from Arabidopsis, Brassica napus, and E. guineensis and ≤ 39% identity with mammalian DGAT1s (Supplemental Fig. S1). As shown in Figure 2, CpuDGAT1 has seven motifs that are conserved among dicot and monocot DGAT1 polypeptides, including catalytic active site residues (Asn-381) and (His-417) in Motif VII that are characteristic of members of the membrane-bound O-acyltransferase (MBOAT) enzyme family. The 78-amino acid terminus of CpuDGAT1 upstream of Motif I shares little or no identity with other plant DGAT1s and contains approximately 30 fewer amino acids than found in the N-terminal region of the Arabidopsis DGAT1 (AtDGAT1).
Figure 1.
Expression of CpuDGAT1, CpuDGAT2_A, CpuDGAT2_B, and CpuDGAT2_C transcripts in different organs of C. avigera var pulcherrima. RT-PCR analysis of DGAT1 and DGAT2 transcripts in different organs of C. avigera var pulcherrima. Eukaryotic initiation factor 4A (eIF4) and actin genes were used as internal controls. PCR products were obtained with gene-specific primers for CpuDGAT1, CpuDGAT2_A, CpuDGAT2_B, or CpuDGAT2_C (Supplemental Table S1).
Figure 2.
Alignment of deduced amino acid sequence of CpuDGAT1 with selected plant and mammalian homologs. Amino acid alignment of CpuDGAT1 with plant and mammalian DGAT1 homologs. Amino acid sequences were obtained from the NCBI. DGAT sequences are shown and NCBI accession numbers are as follows: CpuDGAT1, C. avigera var pulcherrima, CpuDGAT1 (KU055625); AtDGAT1, Arabidopsis, AtDGAT1 (CAB44774); BnDGAT1, B. napus, BnDGAT1 (AF164434); RcDGAT1, R. communis RcDGAT1 (XP002514132); VfDGAT1, Vernicia fordii, VfDGAT1 (ABC94471); OeDGAT1, Olea europaea, OeDGAT1(AAS01606); EgDGAT1, E. guineensis DGAT1 (XP010924968); RnDGAT1, Rattus norvegicus, RnDGAT1 (NP445889); HsDGAT1, Homo sapiens, HsDGAT1 (NP036211). The residues blocked on red background are 100% conserved bases in seven known motifs (in dotted boxes) of DGAT1s. Black arrows show the CpuDGAT1 residues differing from those of other plant DGAT1s within the seven motifs. White triangles show the catalytic active site residues (Asn-381) and (His-417) found in all MBOAT family enzymes (Chang et al., 2011). The ER retrieval motif is indicated with an orange box (within motif VII).
Functional Characterization of CpuDGAT1
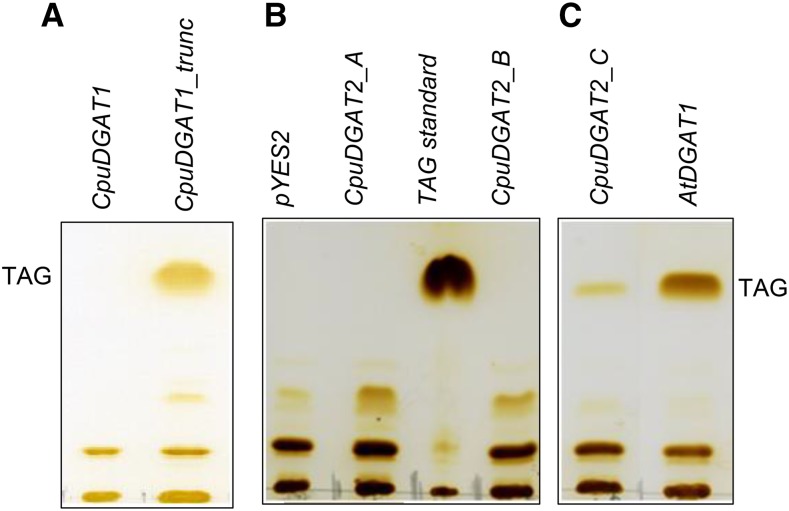
CpuDGAT1 was initially expressed in the TAG-deficient Saccharomyces cerevisiae H1246 mutant (Sandager et al., 2002) to determine if it encodes an active DGAT1. No TAG production was detected by TLC analysis of S. cerevisiae H1246 cells expressing CpuDGAT1 (Fig. 3A). Given the variant nature of the CpuDGAT1 N-terminus, a truncated form lacking the first 70 amino acids was tested and found to restore TAG biosynthesis to S. cerevisiae H1246 cells (Fig. 3A). Although no TAG production was detected by TLC analysis, expression of CpuDGAT1 was able to rescue lipotoxicity of S. cerevisiae H1246 cells grown in exogenous 250 μM 8:0 or 10:0, but not 18:1. By contrast, expression of AtDGAT1 rescued lipotoxicity associated with supplementation of 18:1 in media, but not with 8:0 or 10:0 supplementation (Supplemental Fig. S2). Among the three DGAT2-like genes, only CpuDGAT2_C functionally complemented TAG biosynthesis in S. cerevisiae H1246 cells (Fig. 3, B and C).
Figure 3.
Analysis of neutral lipids from S. cerevisiae H1246 strain expressing Cuphea and Arabidopsis DGATs. Thin layer chromatographic analysis of neutral lipids of S. cerevisiae H1246 strain expressing Cuphea DGAT2 cDNAs (CpuDGAT1_A, CpuDGAT2_B, and CpuDGAT2_C), full-length CpuDGAT1, CpuDGAT1 lacking the coding sequence for the first 70 amino acids (CpuDGAT1trunc), and the Arabidopsis DGAT1 (AtDGAT1) in the vector pYes2. CpuDGAT1trunc, CpuDGAT2_C, and AtDGAT1 expression functionally complement TAG biosynthesis in S. cerevisiae H1246 mutant. A, Neutral lipids from yeast cells expressing CpuDGAT1 and CpuDGAT1trunc. B, Neutral lipids from control yeast cells (pYes2) and yeast cells expressing CpuDGAT2_A and CpuDGAT2_B. C, Neutral lipids from yeast cells expressing CpuDGAT2_C and AtDGAT1.
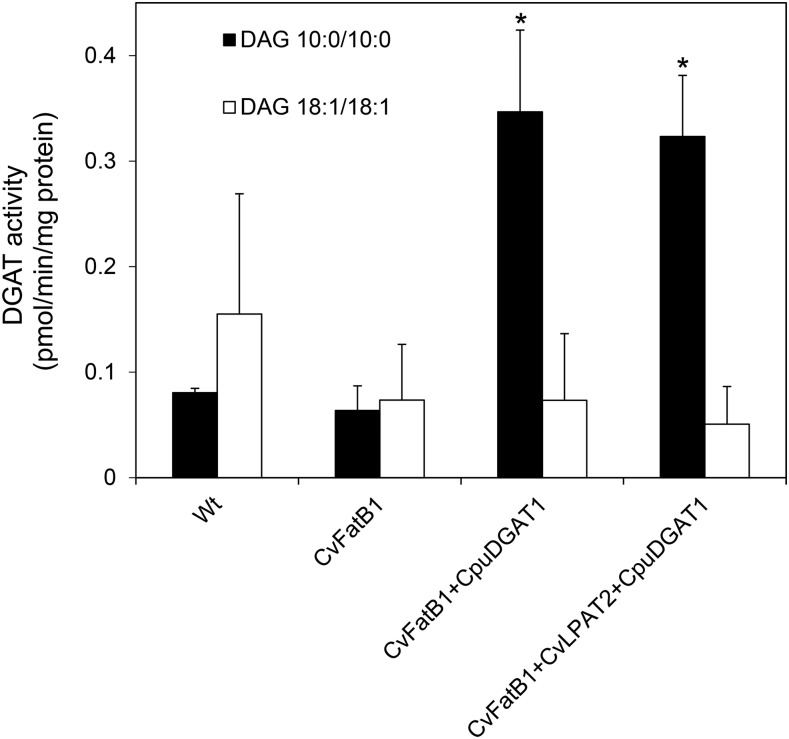
As an alternative approach for characterizing the function of CpuDGAT1, extracts of developing wild-type camelina seeds or transgenic camelina seeds expressing CvFatB1, CvFatB1+CpuDGAT1, or CvFatB1+CvLPAT2+CpuDGAT1 (described below) were assayed for DGAT activity using oleoyl (18:1)-CoA and diolein or decanoyl (10:0)-CoA and didecanoin as substrates. Extracts of seeds expressing CpuDGAT1 had approximately 4-fold higher activity with the 10:0 substrates than those lacking CpuDGAT1, but no significant difference in activity was detected with 18:1 substrates among seed extracts of the different lines (Fig. 4). As additional evidence that CpuDGAT1 is associated with medium-chain fatty acid metabolism, Agrobacterium infiltration of Nicotiana benthamiana leaves with cDNAs for CpuDGAT1, the 8:0- and 10:0-producing FatB from C. hookeriana, ChFatB2, and the Arabidopsis transcription factor Wrinkled1 (AtWRI1), all under control of CamV35S promoters, resulted in the accumulation of TAGs containing 8:0 and 10:0, which were not detected in leaves infiltrated with expression cassettes containing cDNAs for AtWRI1 only or AtWRI1 and ChFatB2 (Supplemental Fig. S3).
Figure 4.
DGAT activity in extracts of developing seeds from transgenic camelina plants expressing CpuDGAT1. Measurement of DGAT activity in crude extracts from developing wild-type camelina seeds (22 DAF) and developing camelina seeds expressing CvFatB1 alone, coexpressing CvFatB1 and CpuDGAT1 (CvFatB1+CpuDGAT1), or coexpressing CvFatB1, CvLPAT2, and CpuDGAT1 (CvFatB1+CvLPAT2+CpuDGAT1). Results are for assays using [1-14C] 10:0-CoA and DAG species: 10:0/10:0 and 18:1/18:1, respectively. Values are mean ± sd (n = 3). Asterisks denote statistically significant differences (*P < 0.0001) as compared to wild type, as determined by two-tailed Student’s t test. Wt, wild type.
Seed-specific CpuDGAT1 Overexpression Enhances 10:0 Content in Camelina Seeds
CpuDGAT1 was further examined for its ability to enhance 8:0 and 10:0 accumulation in camelina seeds resulting from expression of CvFatB1 encoding a specialized Cuphea thioesterase (Kim et al., 2015b). We also examined the ability of CpuDGAT1 to increase the accumulation of these fatty acids when coexpressed with the previously described CvLPAT2 that encodes an LPAT specialized for in planta accumulation of 8:0 and 10:0 (Kim et al., 2015a). These transgenes were expressed in camelina under control of strong seed-specific promoters. Seeds obtained from independent transgenic lines at the T2 generation were screened for expression of the transgenes (Supplemental Fig. S4) and for fatty acid composition (Supplemental Fig. S5). Increases in amounts of 10:0 were observed in seeds of these lines (Supplemental Fig. S5). Amounts of 10:0 ranged from 4 mol % to 13.5 mol % in T2 single seeds of lines from CvFatB1+CpuDGAT1 expression, which was nearly 2-fold higher than from expression of CvFatB1 alone (Supplemental Fig. S5 A; Kim et al., 2015b). Amounts of 10:0 ranged from 12 mol % to 21.5 mol % in T2 single seeds of lines from CvFatB1+CvLPAT2+CpuDGAT1 expression, which was up to 3-fold higher than from expression of CvFatB1 alone (Supplemental Fig, S5; Kim et al., 2015b).
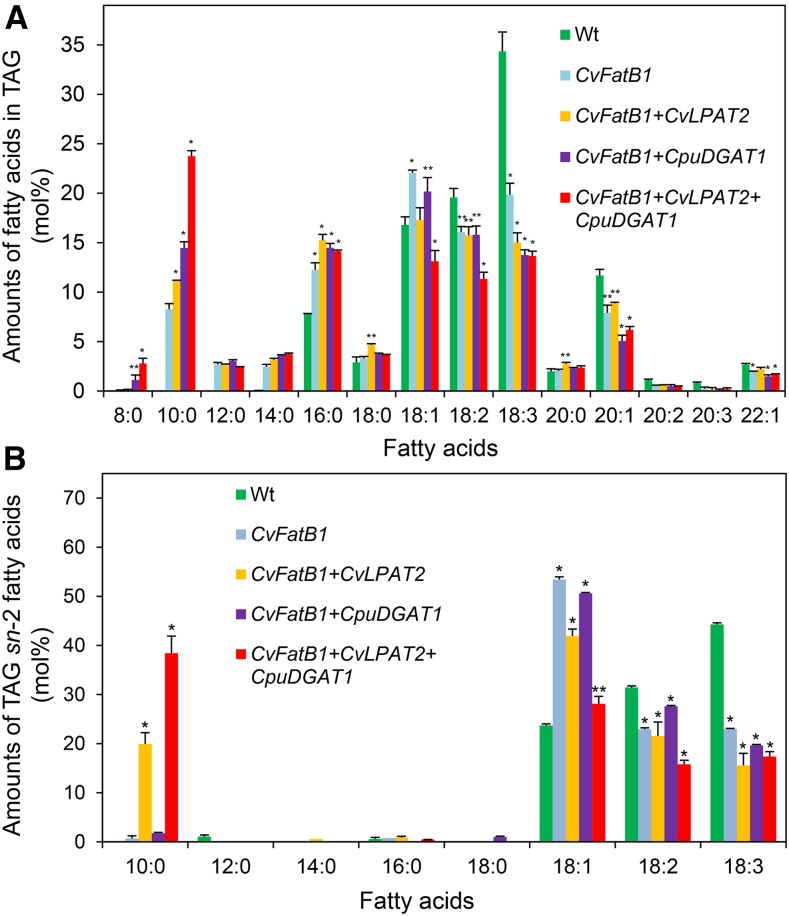
Subsequent analyses of fatty acid compositions were conducted on seeds from homozygous T4 lines. The lines chosen were those that were found to accumulate the highest amounts of 10:0 from screening of seeds at the T2 generation. The amount of 10:0 in seed TAGs from the CvFatB1+CpuDGAT1 line was 14.5 mol % versus 8.0 mol % of 10:0 in seeds from CvFatB1 expression alone. In addition, seed TAGs of the CvFatB1+CvLPAT2+CpuDGAT1 line contained 23.7 mol % of 10:0 mol % versus 11.1 mol % of this fatty acid in seed TAGs of the CvFatB1+CvLPAT2 line (Fig. 5A). Also notable was the detection of 8:0 in seed TAGs at amounts of 2.8 mol % in the CvFatB1+CvLPAT2+CpuDGAT1 line, which were not detected in seed TAGs from CvFatB1 expression alone. In addition, seed TAGs from all lines expressing CvFatB1 had increased amounts of 12:0, 14:0, and 16:0 with partially compensating decreases in amounts of linoleic (18:2), α-linolenic (18:3), and gondoic (20:1) acids (Fig. 5A).
Figure 5.
Fatty acid composition of TAG and sn-2 position of TAG in mature seeds of wild-type camelina and camelina lines engineered for expression of CvFatB1 alone or with combinations of CpuDGAT1 and CvLPAT2. A, Fatty acid composition of TAG in wild-type camelina seeds and camelina seeds engineered for expression of CvFatB1 alone, with CvLPAT2 (CvFatB1+CvLPAT2) or CpuDGAT1 (CvFatB1+CpuDGAT1), and with the combination of CvLPAT2 and CpuDGAT1 (CvFatB1+CvLPAT2+CpuDGAT1). Values shown are the means of mol % of each fatty acid and ± sd of the mean for three biological replicates. Asterisks denote statistically significant difference from wild type (or CvFatB1 in the case of 10:0) values at *P < 0.001, **P < 0.03 based on two-tailed Student’s t test. B, Fatty acid composition of TAG sn-2 position of wild-type camelina seeds and camelina seeds engineered for expression of CvFatB1 alone, with CvLPAT2 (CvFatB1+CvLPAT2) or CpuDGAT1 (CvFatB1+CpuDGAT1), and with the combination of CvLPAT2 and CpuDGAT1 (CvFatB1+CvLPAT2+CpuDGAT1). Values shown are the means of mol % of each fatty acid and ± sd of the mean for three biological replicates. Asterisks denote statistically significant difference from wild type (or CvFatB1 in the case of 10:0) values at *P < 0.002, **P < 0.02 based on two-tailed Student’s t test. Wt, wild type.
Coexpression of CpuDGAT1 and CvLPAT2 Promotes Accumulation of 10:0 at the TAG sn-2 Position
Stereospecific analyses of fatty acid species at the sn-2 position of seed TAGs of engineered lines was conducted using a standard TAG lipase digestion protocol to assess the effects of CpuDGAT1 expression on the TAG stereospecific accumulation of 10:0. Very low amounts (≤2 mol %) of 10:0 were detected in the TAG sn-2 position from seeds expressing CvFaB1 CvFatB1 alone or with CpuDGAT1. More strikingly, the amount of 10:0 detected in the TAG sn-2 position was 38.4 mol % in seeds of the CvFatB1+CvLPAT2+CpuDGAT1 line, which was nearly 2-fold the amount found in the TAG sn-2 position of the CvFatB1+CvLPAT2 (Fig. 5B). Seeds from all of the transgenic lines had large reductions in 18:3 amounts in the TAG sn-2 position relative to TAGs from wild-type seeds, but oleic acid (18:1) amounts were approximately 2-fold higher in the TAG sn-2 position in CvFatB1, CvFatB1+CvLPAT2, and CvFatB1+CpuDGAT1 seeds, relative to wild-type seeds (Fig. 5B).
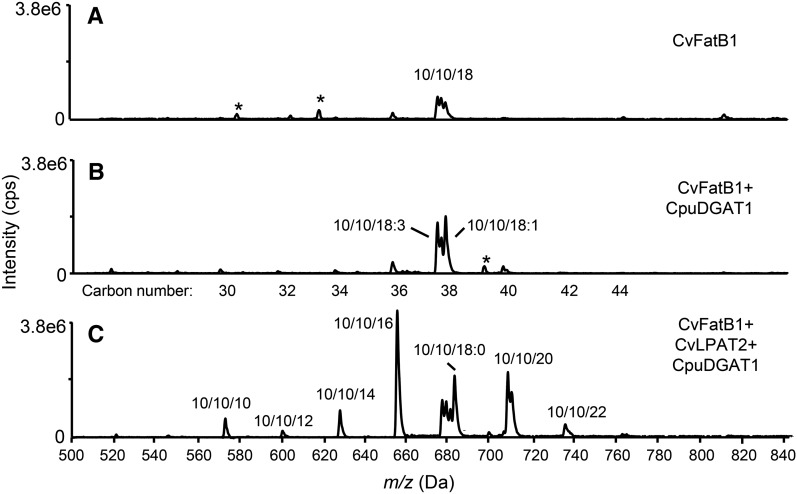
To confirm the positional distribution of 10:0 in TAG, liquid chromatography (LC) electrospray ionization-mass spectrometry/mass spectrometry (ESI-MS/MS) was used to identify the TAG molecular species in the engineered camelina seeds. TAG containing 10:0 in all three stereospecific positions (tridecanoin) was detectable in seeds of the CvFatB1+CvLPAT2+CpuDGAT1 line, but not detectable in seeds of the CvFatB1+CpuDGAT1 line (Fig. 6). TAGs from the CvFatB1+CvLPAT2+CpuDGAT1 seeds were also more enriched in molecular species containing 10:0/10:0 in combination with the saturated fatty acids 12:0, myristic acid (14:0), palmitic acid (16:0), and stearic acid (18:0) as well as with C20 and C22 very long-chain fatty acids (Fig. 6). These results support the channeling of DAG containing 10:0 in the sn-2 position for TAG biosynthesis due to the apparent selectivity of CpuDGAT1 for these molecular species.
Figure 6.
ESI-MS/MS profiling of 10:0/10:0 DAG-containing TAG molecular species in seeds of camelina lines engineered for expression of CvFatB1 alone or with combinations of CpuDGAT1 and CvLPAT2. TAG species from camelina seeds engineered for expression of CvFatB1 alone (A), with CpuDGAT1 (B), or with CvLPAT2 and CpuDGAT1 (C) were analyzed by ESI-MS/MS. Shown are precursor 383.3 m/z precursor scans of TAG species containing 10:0/10:0 DAGs. The labeled TAG species indicate fatty acid composition, but the stereospecific arrangement cannot be determined from the analyses. Asterisks indicate unknown compounds.
CpuDGAT1 and CvLPAT2 Coexpression Increases 10:0 Accumulation in DAG, but Little 10:0 Accumulation Is Detected in PC
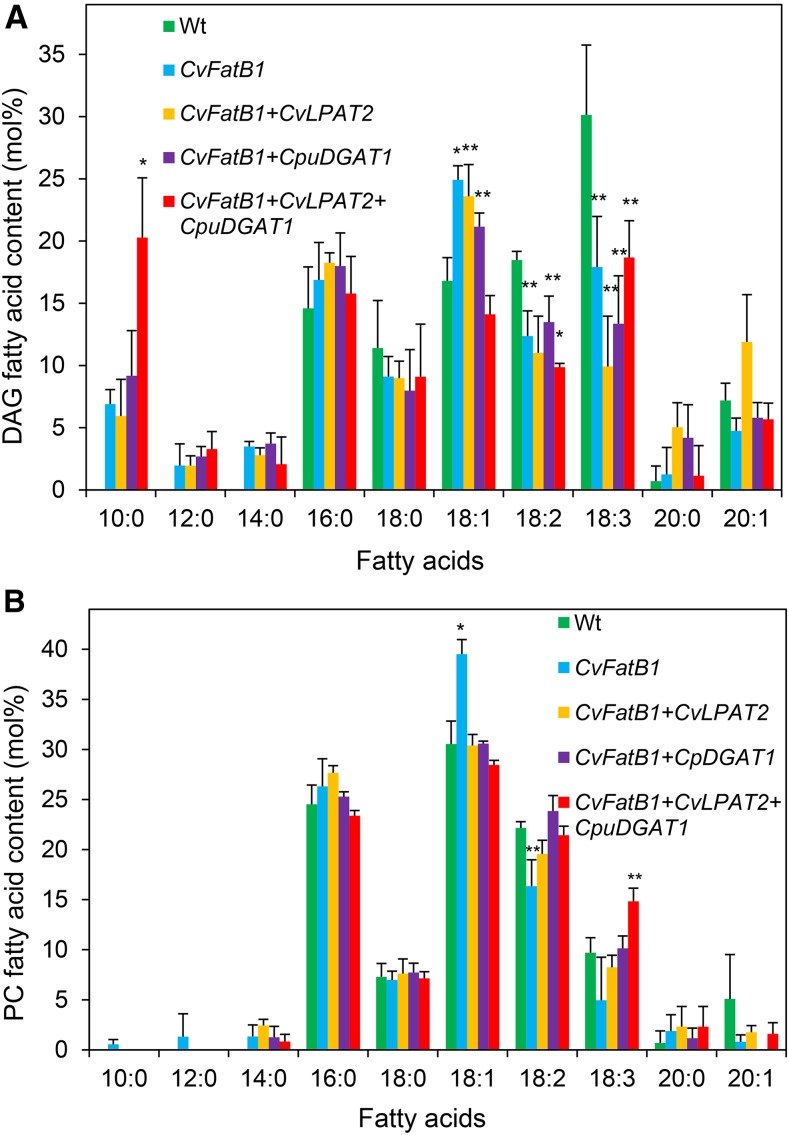
The fatty acid composition of DAG, a substrate for PC and TAG synthesis, was examined in camelina seeds expressing CvFatB1 in combination with CpuDGAT1 or CpuDGAT1 and CvLPAT2 to gain insights into how CpuDGAT1 mediates fatty acid flux in seeds. In seeds from all of the transgenic lines, 10:0, 12:0, and 14:0 were detected in DAG, but absent from DAG of wild-type seeds (Fig. 7A). The largest difference observed in DAG from seeds of the transgenic lines was the 2-fold higher amounts of 10:0 in DAG in CvFatB1+CvLPAT2+CpuDGAT1 seeds compared to DAG of seeds from other transgenic lines including CvFATB1+CpuDGAT1 (Fig. 7A). These results suggest that, although CpuDGAT1 does not directly participate in DAG synthesis, the coordinate activities of CvLPAT2 and CpuDGAT1 pull increased flux of 10:0 through DAG. Despite the presence of 20 mol % of 10:0 in DAG of CvFatB1+CvLPAT2+CpuDGAT1 seeds, no 10:0 was detected in PC of mature seeds (Fig. 7B), and in CvFatB1+CpuDGAT1 developing seeds at 22 d after flowering (DAF; Supplemental Fig. S6). This observation indicates that 10:0-DAG is either excluded from PC or is efficiently removed by acyl-editing.
Figure 7.
Fatty acid composition of DAG and PC in mature seeds of wild-type and engineered camelina lines expressing CvFatB1 alone or in combinations with CvLPAT2 and CpuDGAT1. A, Fatty acid composition of DAG in wild-type camelina seeds and camelina seeds engineered for expression of CvFatB1 alone, with CvLPAT2 (CvFatB1+CvLPAT2) or CpuDGAT1 (CvFatB1+CpuDGAT1), and with the combination of CvLPAT2 and CpuDGAT1 (CvFatB1+CvLPAT2+CpuDGAT1). B, Fatty acid composition of PC in wild-type camelina seeds and camelina seeds engineered for expression of CvFatB1 alone, with CvLPAT2 (CvFatB1+CvLPAT2) or CpuDGAT1 (CvFatB1+CpuDGAT1), and with the combination of CvLPAT2 and CpuDGAT1 (CvFatB1+CvLPAT2+CpuDGAT1). Values shown are the means of mol % of each fatty acid and ± sd of the mean for three biological replicates. Asterisks denote statistically significant difference from wild-type values or CvFatB1 for 10:0 values at *P < 0.01, **P < 0.05 based on two-tailed Student’s t test. Wt, wild type.
10:0-CoA Is a Minor Component of acyl-CoA Pools in Developing Seeds Coexpressing CpuDGAT1 and CvLPAT2
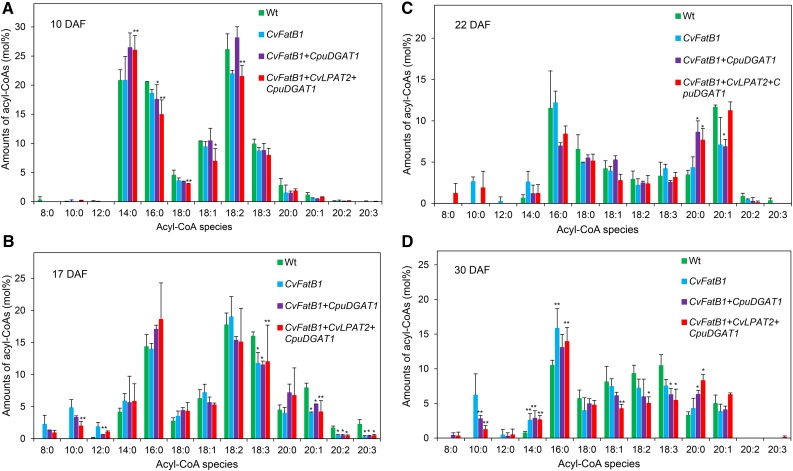
Acyl-CoA pools were monitored in seeds at four developmental stages (10, 17, 22, and 30 DAF) in wild-type camelina plants and plants engineered for expression of CvFatB1 alone or with CpuDGAT1 or CpuDGAT1 and CvLPAT2 (Fig. 8, A to D). In all of these lines, seed oil content was highest at 22 DAF, and 17 DAF corresponded to approximately the midstage of seed oil accumulation. In addition, seeds of the CvFatB1+CpuDGAT1+CvLPAT2 lines had 10:0 amounts of 24 mol % at 17 DAF and 30 mol % at 22 DAF (Supplemental Fig. S7). Despite these levels of 10:0 in the seed oil, amounts of 10:0-CoA were typically <5% of the total acyl-CoA pools in the seeds of the transgenic lines at all stages of seed development. In addition, relative amounts of 10:0-CoA were generally lower in developing seeds of CvFatB1+CpuDGAT1+CvLPAT2 lines compared to seeds expressing only CvFatB1. This was most pronounced at 17 DAF and 30 DAF. At these developmental stages, relative amounts of 10:0-CoA in CvFatB1+CpuDGAT1+CvLPAT2 seeds were 50% (17 DAF) and 80% (30 DAF) lower than those in CvFatB1 seeds (Fig. 8, B and D). These results suggest that CpuDGAT1 and CvLPAT2 efficiently metabolizes 10:0-CoA in developing seeds and that accumulation of 10:0 in the engineered seeds is limited by the supply of 10:0-CoA.
Figure 8.
Acyl-CoA species in camelina developing seeds engineered for expression of CvFatB1 alone and with CpuDGAT1 and CvLPAT2+CpuDGAT1. Acyl-CoA species of camelina developing seeds at 10, 17, 22, and 30 DAF from wild-type, and transgenic lines expressing CvFatB1 alone, with CpuDGAT1, and with CvLPAT2+CpuDGAT1 were analyzed by LC-MS/MS. The data are means of mol % of each acyl-CoA species ± sd of three biological replicates. Asterisks denote statistically significant difference from wild type (or CvFatB1 in the case of 10:0) values at *P < 0.02, **P < 0.05 based on two-tailed Student’s t test. A, Acyl-CoA species of camelina seeds at 10 DAF. B, Acyl-CoA species of camelina seeds at 17 DAF. C, Acyl-CoA species of camelina seeds at 22 DAF. D, Acyl-CoA species of camelina seeds at 30 DAF. Wt, wild type.
CpuDGAT1 and CvLPAT2 Coexpression Results in 10:0 Production with Modest Effects on Seed Properties
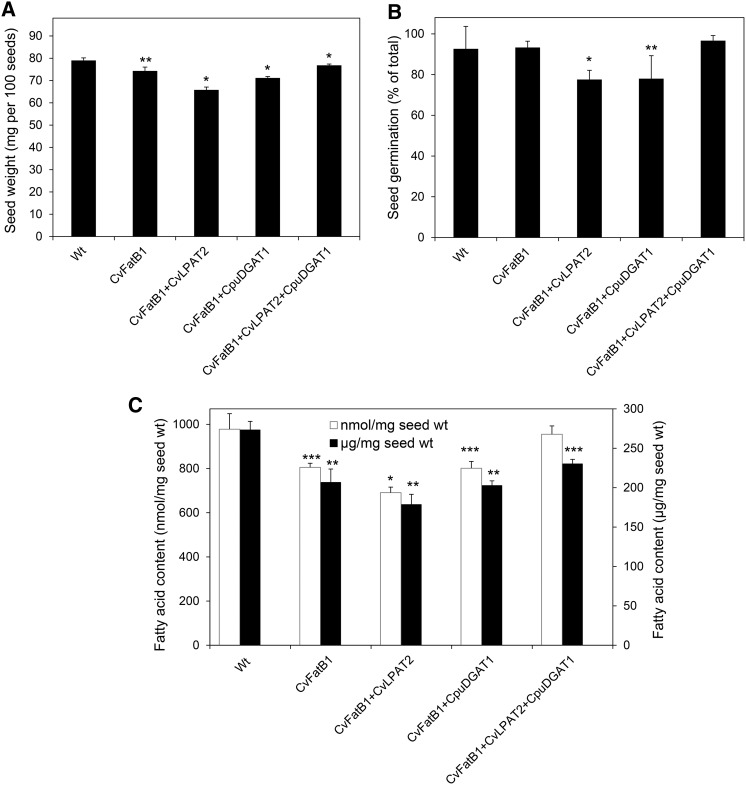
Seed weight, fatty acid content, and germination were examined for wild-type camelina plants and camelina lines engineered for expression of CvFatB1 alone or in combination with CpuDGAT1, CvLPAT2, or CpuDGAT1+CvLPAT2 (Fig. 9). For these studies, plants were grown under the same greenhouse conditions. The average weight of 100 seeds from wild-type plants and CvFatB1+CvLPAT2+CpuDGAT1 differed by <3% (79.0 mg wild-type versus 76.8 mg CvFatB1+CvLPAT2+CpuDGAT1; Fig. 9A). The largest difference in seed weight between the transgenic lines and wild-type plants was observed in the CvFatB1+CvLPAT2 line, which had approximately 17% lower 100-seed weight than wild-type plants. Weight for 100 seeds from CvFatB1 was 10% lower, and for that from CvFatB1+CpuDGAT1, it was 6% lower than those from wild-type plants (Fig. 9A). Germination of seeds from wild-type plants and CvFatB1 and CvFatB1+CvLPAT2+CpuDGAT1 lines were not significantly different, with germination efficiency of seeds ranging from 93% to 97% in these lines. Seed germination, however, was reduced by approximately 16% in CvFatB1+CvLPAT2 and CvFatB1+CpuDGAT1 lines relative to wild-type plants (Fig. 9B; Supplemental Fig. S8). Seed fatty acid content was calculated on a weight and molar basis (Fig. 9C), as well as moles and mg per seed (Supplemental Fig. S9). The latter calculation was done because of the lower Mr of 10:0 in seeds of transgenic plants relative to the C16-C20 fatty acids found in seeds of wild-type plants. Using both calculations, fatty acid content was significantly reduced in seeds of lines expressing CvFatB1 alone as well as lines expressing CvFatB1 in combination with CvLPAT2 or with CpuDGAT1. The largest differences in fatty acid content relative to that in wild-type seeds was detected in seeds coexpressing CvFatB1+CvLPAT2 (Fig. 9C). Seed fatty acid content was reduced in these lines by 30% on a molar basis and 34% on a weight basis compared to wild-type plants. This may reflect a limited ability of the native camelina DGAT to use DAG species containing 10:0 in the sn-2 position. By contrast, the fatty acid content of seeds coexpressing CvFatB1+CvLPAT2+CpuDGAT1 was not significantly different from that of wild-type seeds on a molar basis, but was 16% lower on a weight basis relative to the fatty acid content of wild-type seeds. Overall, these findings suggest that apparent CvLPAT2- and CpuDGAT1-mediated channeling of 10:0 through the Kennedy pathway for TAG synthesis results in no or modest effects on seed composition and performance.
Figure 9.
Seed weight, germination efficiency, and total fatty acid content of wild-type camelina seeds and seeds of engineered camelina lines expressing CvFatB1 alone or in combinations with CvLPAT2 and CpuDGAT1. Shown are 100-seed weight (A), germination efficiency (B), and total fatty acid content (C) of mature seeds from wild-type camelina and camelina seeds engineered for expression of CvFatB1 alone, with CvLPAT2 (CvFatB1+CvLPAT2) or CpuDGAT1 (CvFatB1+CpuDGAT1), and with the combination of CvLPAT2 and CpuDGAT1 (CvFatB1+CvLPAT2+ CpuDGAT1). Values shown for 100-seed weight are the means and ± sd of the mean for four biological replicates. Values for seed fatty acid content are the means and ± sd of the mean for three biological replicates. Asterisks indicate statistically significant difference (*P < 0.005, **P < 0.025) as compared to the wild type based on two-tailed Student’s t test. Values for germination efficiency are the means and ± sd of the mean for three biological replicates. Asterisks denote statistically significant difference (*P < 0.005, ***P < 0.05) in germination efficiency of transgenic seeds as compared to the CvFatB1+ CvLPAT2+CpuDGAT1 seeds based on two-tailed Student's t test. Wt, wild type.
DISCUSSION
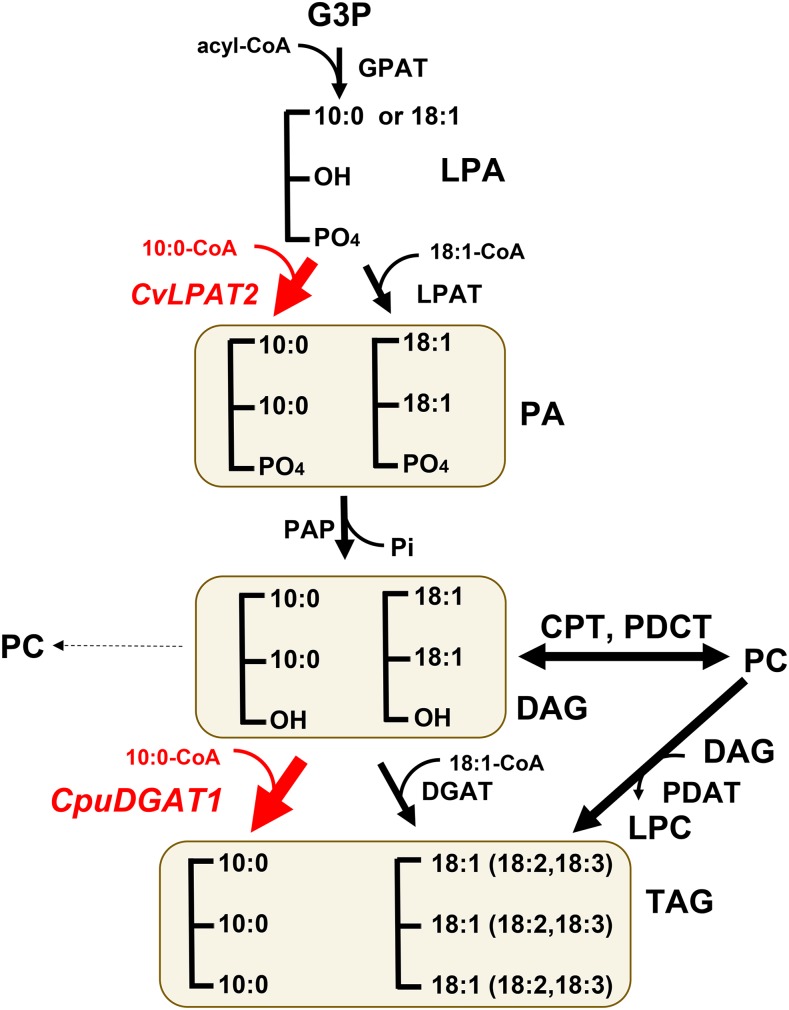
We identified a cDNA for C. avigera var pulcherrima DGAT1 (CpuDGAT1) from transcriptomic data that is active in vitro and in planta with 10:0-containing DAG and 10:0-CoA. We also showed that the coexpression of CpuDGAT1 with the CvLPAT2 in a 10:0-producing camelina background not only enhances the total accumulation of 10:0 in TAG but also doubles the content of 10:0 in the TAG sn-2 position relative to CvLPAT2 expression alone. These results provide direct genetic evidence for metabolic cooperation of LPAT and DGAT for channeling of 10:0 through the Kennedy pathway for TAG synthesis (Bafor et al., 1990; Fig. 10). This metabolic route is coupled with an apparent inability of the PC biosynthetic enzymes to use 10:0-containing DAGs. In addition to 10:0, CpuDGAT1 was also found to enhance accumulation of 8:0 in TAG in N. benthamiana agrobacterium-infiltration studies (Supplemental Fig. S3), indicating that this enzyme is readily able to use 8:0-containing substrates, if sufficient pools of these substrates are available. These findings contribute to an increased understanding of the extreme metabolic specialization that enables many Cuphea species to accumulate medium-chain fatty acids, including 8:0 and 10:0, to amounts >90 mol % in TAG.
Figure 10.
A proposed pathway for 10:0-rich TAG assembly in transgenic camelina seeds coexpressing CvLPAT2 and CpuDGAT1. Acyl-CoA species of 10:0 and 18:1 are sequentially esterified to glycerol-3-P (G3P) at sn-1 and sn-2 positions by GPAT and LPAT, forming LPA and PA. The release of P from PA by phosphatidic acid phosphatase forms DAG. The subsequent acylation of DAG at sn-3 catalyzed by DGAT forms TAG. DAG can be converted to PC via the action of DAG/CPT and/or PDCT. DAG molecules containing 10:0 at the sn-2 position arise from the specialized activity of the Cuphea LPAT CvLPAT2. DAGs containing 10:0 at the sn-2 position are selectively used for acylation with 10:0-CoA at the sn-3 position by activity of the specialized Cuphea diacylglycerol acyltransferase CpuDGAT1 to form TAG species enriched in 10:0. DAG molecules containing 10:0 appear to be poor substrates for PC synthesis, or 10:0 is rapidly removed from PC formed from 10:0-DAG species. As such, 10:0 accumulation in TAG appears to proceed primarily by the Kennedy pathway rather than through PC via activity of enzymes including PDCT and phospholipid/diacylglycerol acyltransferase, which catalyzes the transfer of fatty acids at the sn-2 position of PC to the sn-3 position of DAG producing TAG and lysophosphatidylcholine. The PC route of fatty acid flux for TAG synthesis involves primarily flux of 18:1 for desaturation to 18:2 and 18:3. LPC, lysophosphatidylcholine; PAP, phosphatidic acid phosphatase; PDAT, phospholipid/diacylglycerol acyltransferase.
The primary sequences of enzymes that are associated with the biosynthesis and metabolism of medium-chain fatty acids in seeds of Cuphea species provide tools for dissecting the structural basis for the substrate specificities of enzyme classes such as FatB thioesterases, LPATs, and DGATs. In the case of the DGAT1 enzyme, structural characterization of the substrate properties of plant and mammalian members of this enzyme class in plants and mammals has been conducted, despite the challenges associated with their occurrence as integral membrane proteins. Of most significance to the findings reported here, a cytosolic N-terminal region encompassing a predicted acyl-CoA binding domain has been linked to substrate specificities of mouse and B. napus DGAT1 (Weselake et al., 2006; Siloto et al., 2008; McFie et al., 2010). The predicted acyl-CoA binding domain of CpuDGAT1 has at least two residues that differ from conserved residues in other known plant DGAT1s, including Asn-97 (Ser in other plant DGAT1s) and Glu-99 (Ala in other plant DGAT1s) as well as one residue Arg-91 that differs from the Asp conserved in other plant DGAT1 and the human and mouse DGAT1s. The importance of these and other likely amino acid differences between CpuDGAT1 and other plant and mammalian DGAT1s for determination of substrate specificity remains to be determined. In addition, although the mechanism was not studied, the ability of CpuDGAT1 (Supplemental Fig. S2), in contrast to AtDGAT1, to rescue the lipotoxicity of exogenous 8:0 and 10:0 to S. cerevisiae strain H1246 may provide the basis for screening for engineered DGAT1s with activity for 8:0- and 10:0-CoA, using procedures similar to those previously described (Siloto et al., 2009).
The use of camelina in these studies allowed for examination of acyltransferase properties in a 10:0-null background. Just as importantly, expression of genes in camelina allowed for agronomic and metabolic assessment of 10:0 production in an oilseed crop. To this end, we showed that the coexpression of a 10:0-producing FatB thioesterase along with CpuDGAT1 and CvLPAT2 results in the accumulation of 10:0 to 25 mol % of TAG with no measurable effect on seed germination and weight and modest effect on seed oil content for plants grown under greenhouse conditions. It is possible that the accumulation of higher concentrations of 10:0 in TAG can be achieved by inclusion of a specialized GPAT with increased activity for 10:0-CoA to acylate the sn-1 position of the glycerol backbone in the Kennedy pathway. Although such activity has been previously detected in seeds of one Cuphea species (Bafor and Stymne, 1992), we did not detect a GPAT9 gene with seed-specific expression in seed transcriptomes of C. viscosissima and C. avigera var pulcherrima. Down-regulation of the native LPAT and DGAT activities in the ER of camelina seeds may also enhance amounts of 10:0 accumulation. These enzymes maintain an apparent metabolically separate pathway for TAG synthesis involving typical C16 and C18 fatty acid-containing substrates. In addition, suppression of native camelina acyl-ACP synthetase activity may reduce the possible recycling of 10:0 to fatty acid synthase, as indicated by recent studies in Arabidopsis (Tjellström et al., 2013). Furthermore, the relatively low content of 10:0 in acyl-CoA pools in the developing engineered camelina seeds (Fig. 8) suggests that plastid production of 10:0 is the major limiting factor for accumulation of this fatty acid in the transgenic seeds. Although divergent FatBs, such as CvFatB1, are primary determinants of 10:0 content in Cuphea seeds, it is likely that additional specialization is present in Cuphea fatty acid synthase to generate elevated amounts of the FatB 10:0-ACP substrate in C. viscosissima seeds or FatB 8:0-ACP substrate in C. avigera var pulcherrima seeds. For example, the Cuphea β-ketoacyl-ACP synthase IV has been previously implicated in contributing to production of high levels of medium-chain fatty acids in seeds of different Cuphea species (Dehesh et al., 1998; Leonard et al., 1998; Slabaugh et al., 1998). In addition, fatty acyl-CoA synthetases with specificities for medium-chain fatty acyl-CoAs cannot be excluded as a factor limiting 10:0 accumulation in camelina seeds, given the reported importance of fatty acyl-CoA synthetases with specificity for CoA esters of 16:0, 18:0 and longer chain fatty acids for seed oil accumulation (Schnurr et al., 2002; Jessen et al., 2015).
A notable observation from the studies with camelina is the absence of detectable amounts of 10:0 in PC in mature seeds of 10:0-producing lines expressing CvLPAT2 and CpuDGAT1 alone or together. These findings suggest that 10:0-containing DAGs are precluded from PC synthesis by CDP-choline phosphotransferase (CPT) or phospholipid:DAG cholinephosphotransferase (PDCT) activities or that 10:0 is efficiently edited or removed from PC by lipolytic activities. Even in developing seeds at 20 DAF, amounts of 10:0 detected in PC in seeds expressing CvFatB1 alone or in combination with CpuDGAT1 were approximately 1 mol % of the total PC fatty acids (Supplemental Fig. S6). This finding differs from results in engineered B. napus seeds that accumulated 10:0 in PC to approximately 12 mol % during seed development, but at seed maturity, 10:0 accounted for 1.3 mol % of PC fatty acids (Wiberg et al., 2000). Our findings can be interpreted that 10:0 flux largely occurs through the Kennedy pathway for TAG synthesis rather than being directed first through PC, as is the typical route for fatty acids such as 18:1 (Bates, 2016; Fig. 10). However, confirmation of this requires more detailed flux analyses. Other possibilities to explain the lack or low amounts of 10:0 in PC include rapid editing of PC for selective removal of 10:0 or direct transacylation of 10:0-DAG to TAG, precluding flux through PC.
Overall, our findings point to a viable path for metabolic engineering of 10:0-rich oils in existing oilseed crops for biofuel and industrial applications that appears to only be limited currently by the ability of fatty acid synthase to generate 10:0 substrate. Our studies also provide information to guide understanding of the structural basis for extreme functional divergence in the DGAT1 family. Finally, our findings highlight the synergy of LPAT and DGAT substrate specificities for generating preferential intermediates to drive production of novel oil compositions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Studies were conducted with camelina (Camelina sativa var Suneson) that was grown under greenhouse conditions with supplemental lighting as described in Kim et al. (2015b). Except where indicated, seeds analyzed were from homozygous transgenic lines in the T4 generation.
RNA Isolation
Total RNA was isolated from developing seeds, leaves, stems, flowers, and roots of Cuphea avigera var pulcherrima and cDNA for semiquantitative PCR amplification of CpuDGAT1, CpuDGAT2_A, CpuDGAT2_B, and CpuDGAT2_C was prepared from DNAase-digested total RNA as described in Kim et al. (2015a). Total RNA from developing seeds of transgenic camelina lines was isolated using the method described in Bekesiova et al. (1999).
Cloning of DGATs from C. avigera var pulcherrima
The CpuDGAT1 (1482 bp), CpuDGAT2_A (1005 bp), CpuDGAT2_B (948 bp), and CpuDGAT2_C (1005 bp) gene sequences were identified among the lipid biosynthesis genes in the C. avigera var pulcherrima 454 transcriptome sequence data (Kim et al., 2015b). The open reading frames corresponding to the identified DGATs were PCR amplified from C. avigera var pulcherrima cDNA using the gene-specific primers listed in Supplemental Table S1.
Preparation of CpuDGAT1 Yeast and Plant Expression Constructs
For yeast expression studies, the open-reading frames of the full-length CpuDGAT1, CpuDGAT1 truncated for the 70-amino acid N-terminal coding sequence (CpuDGAT1trunc), CpuDGAT2_A, CpuDGAT2_B, CpuDGAT2_C, and the Arabidopsis DGAT1 were amplified using oligonucleotides provided in Supplemental Table S1 and cloned into the corresponding restriction sites of the yeast expression vector pYES2 under control of the GAL1 promoter.
For preparation of plant expression vectors, the open-reading frame for CpuDGAT1 was amplified using oligonucleotides in Supplemental Table S1 and subcloned into the NotI site of pKMS3 (Nguyen et al., 2013) downstream of the seed-specific soybean GLYCININ-1 promoter and upstream of the GLYCININ-1 3′ UTR. This cassette was subsequently digested from pKMS3 with AscI and cloned into the MluI site of the previously described pBinGlyRed3_CvFatB1 (Kim et al., 2015b) yielding the plasmid pBinGlyRed3_CvFatB1+CpuDGAT1. The CvFATB1 coding sequence in this vector is under control of the GLYCININ-1 promoter. An AscI cassette containing the CvLPAT2 ORF under control of the seed-specific promoter for the soybean oleosin gene, generated as described in Kim et al. (2015a), was ligated into the AscI site of pBinGlyRed3_CvFatB1+CpuDGAT1 to form pBinGlyRed3_CvFatB1+CpuDGAT1+CvLPAT2. The backbone of the vector is derived from pCAMBIA0380 and was engineered with the DsRed marker gene under the control of the constitutively expressed cassava mosaic virus promoter (Nguyen et al., 2013) for selection of transgenic seeds by fluorescence (Lu and Kang, 2008).
Camelina Transformation and Selection
The binary vector containing a cassette for seed-specific expression of CvFatB1, CvLPAT2, and CpuDGAT1 was introduced into Agrobacterium tumefaciens by electroporation. Transformation of camelina plants was performed using the floral vacuum infiltration method and transgenic mature seeds were selected using DsRed protein fluorescence, the selection marker, as described in Lu and Kang (2008). Expression of transgenes in developing seeds was confirmed by RT-PCR (Supplemental Fig. S4) using gene-specific primers (Supplemental Table S1).
Phylogenetic Analysis
An unrooted phylogenetic tree of CpuDGAT1, CpuDGAT2_A, CpuDGAT2_B, and CpuDGAT2_C deduced amino acid sequences, along with other amino acid sequences homologous to DGAT1 or DGAT2, was generated using the neighbor-joining program in MEGA4 (http://www.megasoftware.net/mega4/; Tamura et al., 2007) with 1000 bootstrap replicates (Supplemental Fig. S1).
Yeast Transformation and Selection
Yeast expression constructs were introduced into Saccharomyces cerevisiae strain H1246 (W303; MATα are1-Δ::HIS3 are2-Δ::LEU2 dga1::KanMX4 lro1-Δ::TRP1 ADE2 met ura3; Sandager et al., 2002) using the PEG/lithium acetate method (Gietz et al., 1995). Transformed cells were selected on minimal medium (YSM) containing 2% (w/v) dextrose and lacking uracil (Invitrogen). YSM containing 2% (w/v) raffinose was inoculated with transformed yeast cells and grown at 28°C for 24 h with shaking (350 rpm). For induction, YSM containing 2% (w/v) Gal was seeded at OD600 ≈ 0.2 with the cells grown in raffinose. Cells were then grown at 28°C for 40 h. For fatty acid feeding experiments, cultures were grown for 2.5 h in YSM containing 2% Gal before adding 1% (w/v) Tergitol-40 and 250 μM of 8:0, 10:0, or 18:1. Cells were harvested by centrifugation, washed twice with 0.1% NaHCO3, freeze-dried, and used for fatty acid and TAG analysis. Lipids were extracted from cell pellets as described in Bligh and Dyer (1959) and Zhang et al. (2013). Neutral lipids including TAG were resolved on Silica Gel 60 (Merck) TLC plates using a solvent consisting of 70:30:1 v/v) heptane:ethyl ether:acetic acid and detected with iodine vapor.
Agrobacterium Infiltration of Nicotiana benthamiana
The ChFatB2 and CpuDGAT1 open-reading frames were cloned in the binary vector pXZP393 and Arabidopsis thaliana WRINKLED1 (AtWri1; At3G54320 synthetically produced by Eurofins MW Operon/Eurofins Genomics) was cloned in the binary vector pART27 downstream of a 35S promoter using the GATEWAY recombination system (Invitrogen). p19 (Qu and Morris, 2002), V2 (Naim et al., 2012), and GFP (Wood et al., 2009) are described elsewhere. A. tumefaciens GV3101:pMP90 (Koncz and Schell, 1986), harboring the individual vectors, were grown in LB broth supplemented with appropriate antibiotics at 28°C overnight. Coexpression was made with AtWRI1, GFP, and p19 or V2 with Cuphea candidate genes, at a concentration of OD600 ≈ 0.2 for each culture. Before infiltration, 100 μM acetosyringone was added to the mixture of cells.
A. tumefaciens infiltration was performed as described in Wood et al. (2009). Wild-type N. benthamiana plants were grown at 26°C, 60% relative humidity, and with a 12 h photoperiod (250 μmol/m2/s) for 6 weeks. Infiltration was made on young developing leaves on freshly watered plants until an area of at least 7 cm2 was infiltrated. The plants were returned to the growth cabinet for an additional 5 d, after which the infiltrated leaf areas were excised. When the infiltrated area was not visible, a fluorescent protein flashlight (DFP-1; NightSea) was used to track the GFP expression. The harvested leaf tissues were flash frozen and freeze dried.
Lipids were extracted (Bligh and Dyer, 1959), separated with TLC, and analyzed using gas chromatography (model no. GC-17A; Shimadzu) as described in Doan et al. (2012) and Grimberg et al. (2015). Fatty acid profiles and quantities of TAG were calculated using reference mixture Me 63 (LGC Standards) and internal standard triheptadecanoin (50 to 100 nmol).
Collection of Developing Seeds from Transgenic Camelina Lines for Fatty Acid Profiling
Seeds from transgenic camelina lines CvFatB1, CvFatB1+CpuDGAT1, and CvFatB1+CvLPAT2+CpuDGAT1 were grown in a greenhouse at the same time. Fully opened flowers were tagged for developing seeds to be collected at time points of 10, 17, 22, and 30 DAF.
Acyl-CoA Analyses
Acyl-CoAs were extracted and analyzed by mass spectrometry as described in Larson et al. (2002) and Kim et al. (2013) from camelina seeds at 10 and 17 DAF that were flash frozen in liquid N2 and stored at −80°C until used.
DGAT Assay
DGAT activity assays were conducted according to a method described in Zhang et al. (2013). [1-14C] Acyl-CoA substrates (50 to 60 mCi/mmol) were prepared as described in Zhang et al. (2013) or purchased (American Radiolabeled Chemicals). Assays were done using extracts from developing seeds (22 DAF). Seed extracts were prepared by grinding seeds in 50 mm HEPES pH 7.5, 50 mm NaCl, 20% glycerol containing 10 μL/mL protease inhibitor (for plants; Sigma-Aldrich). The homogenates were filtered through Miracloth (Calbiochem). Protein concentration in seed extracts was measured using a bicinchoninic acid assay (Smith et al., 1985) with purchased reagents (Sigma-Aldrich).
Total Fatty Acid Quantification and Lipid Analyses
Total oil was quantified by gas chromatographic analysis of fatty acid methyl esters generated from camelina seeds and a triheptadecanoin internal standard using the methodology described in Kim et al. (2015b). Total lipids were extracted for analysis of TAG, DAG, and PC fatty acid composition (Bligh and Dyer, 1959; Zhang et al., 2013). Camelina seeds (30 mg) in 13 × 100-mm glass tubes were ground using a TH Homogenizer (Omni) in 3 mL methanol:chloroform (2:1 v/v). After 1 h incubation at 25°C, 1 mL of chloroform and 1.9 mL of water were added. The solution was mixed thoroughly and spun at 4000 rpm in a clinical centrifuge for 10 min. The organic phase containing total lipids was transferred to a new glass tube, and used for separation of TAG, DAG, and PC using solid phase extraction columns (Supelco Supel Clean LC-Si SPE columns; Sigma-Aldrich). Lipid extracts dried under N2 were redissolved in 1 mL of heptane and loaded onto solid phase extraction columns preequilibrated with heptane. The TAG fraction was eluted with 5 mL of heptane:ethyl ether 80:20 (v/v). DAG was subsequently eluted with 3 mL chloroform:acetone 80:20 (v/v). Columns were washed with 6 mL of acetone followed by elution of phospholipids with 5 mL methanol:chloroform:water 100:50:40 (v/v/v). To the phospholipid fraction, 1.3 mL chloroform and 1.3 mL water were added. After mixing and centrifugation, the phospholipids recovered in the organic layer were dried under N2. The phospholipids were redissolved in 100 μL chloroform and separated by TLC on Silica 60 plates (Merck) in a solvent system consisting of chloroform:methanol:water:30% ammonium hydroxide (65:35:3:2.5 v/v/v/v). Bands from the TLC plates corresponding to PC were scraped and transferred to 13 × 100-mm glass tubes for fatty acid compositional analyses. Transesterification of total lipids, TAG, DAG, and PC fractions was done in 1 mL of 2.5% sulfuric acid in methanol by heating at 90°C for 30 min. Upon cooling, 1 mL H2O and 0.5 mL heptane were added followed by vortexing and centrifuging at 4000 rpm for 5 min. Heptane phase-containing FAMEs was transferred to autosampler vials. FAMEs were analyzed on a model no. 7890A gas chromatograph (Agilent Technologies) fitted with a 30-m length × 0.25 mm i.d. HP-INNOWax Column (Agilent Technologies) using instrument conditions as described in Kim et al. (2015b).
The purified TAG described above was also used for stereospecific analyses as described in Cahoon et al. (2006) and Kim et al. (2015b).
Neutral Loss ESI-MS/MS Analysis of TAG Species
MS analyses were conducted using a model no. 4000 QTRAP Linear Ion Trap Quadrupole Mass Spectrometer (Applied Biosystems) to characterize TAG molecular species according to the method described in Kim et al. (2015a).
Accession Numbers
GenBank Accession Numbers are as follows: CpuDGAT1, KU055625; CpuDGAT2_A, KU055626; CpDGAT2_B, KU055627; CpuDGAT2 C, KU055628.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Oligonucleotides used in these studies.
Supplemental Figure S1. Unrooted phylogram of C. avigera var pulcherrima DGAT1 and DGAT2s with other hypothetical and functionally characterized DGATs.
Supplemental Figure S2. Lipotoxicity resistance of yeast cells expressing CpuDGAT1 to medium-chain fatty acids.
Supplemental Figure S3. Fatty acid composition of TAG from N. benthamiana leaves infiltrated with AtWRI1 alone, with ChFatB2, or CpuDGAT1.
Supplemental Figure S4. Transcript analysis of Cuphea genes expressed in transgenic camelina lines.
Supplemental Figure S5. Medium-chain fatty acid profile in seeds of T2 camelina lines expressing CpuDGAT1.
Supplemental Figure S6. Fatty acid composition of phosphatidylcholine in developing seeds (22 d after flowering) of wild-type camelina seeds and camelina seeds engineered for expression of CvFatB1 alone and with CpuDGAT1 (CvFatB1+CpuDGAT1). Wt, wild type.
Supplemental Figure S7. Decanoic acid accumulation during seed development in wild-type and transgenic camelina seeds expressing CvFatB1 alone and with CpuDGAT1 or CvLPAT2+CpuDGAT1.
Supplemental Figure S8. Growth of camelina transgenic lines expressing CvFatB1 alone and with CpuDGAT1 or CvLPAT2+CpuDGAT1.
Supplemental Figure S9. Total fatty acid content of wild-type camelina seeds and camelina seeds engineered for expression of CvFatB1 alone or in combination with CvLPAT2 and CpuDGAT1.
Supplementary Material
Acknowledgments
We thank Tara Nazarenus for technical assistance and for maintenance of camelina.
Footnotes
The research was supported by the Center for Advanced Biofuel Systems, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Basic Energy Sciences under Award no. DE-SC0001295, and the National Science Foundation (Plant Genome no. IOS-13-39385 to E.B.C.).
Articles can be viewed without a subscription.
References
- Aymé L, Jolivet P, Nicaud J-M, Chardot T (2015) Molecular characterization of the Elaeis guineensis medium-chain fatty acid diacylglycerol acyltransferase DGAT1-1 by heterologous expression in Yarrowia lipolytica. PLoS One 10: e0143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafor M, Jonsson L, Stobart AK, Stymne S (1990) Regulation of triacylglycerol biosynthesis in embryos and microsomal preparations from the developing seeds of Cuphea lanceolata. Biochem J 272: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafor M, Stymne S (1992) Substrate specificities of glycerol acylating enzymes from developing embryos of two Cuphea species. Phytochemistry 31: 2973–2976 [Google Scholar]
- Bates PD. (2016) Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim Biophys Acta 1861(9 Pt B): 1214–1225 [DOI] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekesiova I, Nap J-P, Mlynarova L (1999) Isolation of high quality DNA and RNA from leaves of the carnivorous plant Drosera rotundifolia. Plant Mol Biol Report 17: 269–277 [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Dietrich CR, Meyer K, Damude HG, Dyer JM, Kinney AJ (2006) Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochemistry 67: 1166–1176 [DOI] [PubMed] [Google Scholar]
- Cao H. (2011) Structure-function analysis of diacylglycerol acyltransferase sequences from 70 organisms. BMC Res Notes 4: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YZ, Huang AH (1986) Diacylglycerol acyltransferase in maturing oil seeds of maize and other species. Plant Physiol 82: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Sun J, Chang T-Y (2011) Membrane-bound O-acyltransferases (MBOATs). Front Biol (Beijing) 6: 177–182 [Google Scholar]
- Dehesh K, Edwards P, Fillatti J, Slabaugh M, Byrne J (1998) KAS IV: a 3-ketoacyl-ACP synthase from Cuphea sp. is a medium chain specific condensing enzyme. Plant J 15: 383–390 [DOI] [PubMed] [Google Scholar]
- Dehesh K, Edwards P, Hayes T, Cranmer AM, Fillatti J (1996a) Two novel thioesterases are key determinants of the bimodal distribution of acyl chain length of Cuphea palustris seed oil. Plant Physiol 110: 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh K, Jones A, Knutzon DS, Voelker TA (1996b) Production of high levels of 8:0 and 10:0 fatty acids in transgenic canola by overexpression of Ch FatB2, a thioesterase cDNA from Cuphea hookeriana. Plant J 9: 167–172 [DOI] [PubMed] [Google Scholar]
- Doan TTP, Domergue F, Fournier AE, Vishwanath SJ, Rowland O, Moreau P, Wood CC, Carlsson AS, Hamberg M, Hofvander P (2012) Biochemical characterization of a chloroplast localized fatty acid reductase from Arabidopsis thaliana. Biochim Biophys Acta 1821: 1244–1255 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Graham SA. (1989) Cuphea: a new plant source of medium-chain fatty acids. Crit Rev Food Sci Nutr 28: 139–173 [DOI] [PubMed] [Google Scholar]
- Grimberg Å, Carlsson AS, Marttila S, Bhalerao R, Hofvander P (2015) Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol 15: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara K, Asahi T, Fujii S (1987) 1-Acyl-sn-glycerol-3-phosphate acyltransferase in maturing safflower seeds and its contribution to the non-random fatty acid distribution of triacylglycerol. Eur J Biochem 167: 339–347 [DOI] [PubMed] [Google Scholar]
- Jessen D, Roth C, Wiermer M, Fulda M (2015) Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol 167: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EP. (1961) Biosynthesis of complex lipids. Fed Proc 20: 934–940 [PubMed] [Google Scholar]
- Kim HJ, Silva JE, Iskandarov U, Andersson M, Cahoon RE, Mockaitis K, Cahoon EB (2015a) Structurally divergent lysophosphatidic acid acyltransferases with high selectivity for saturated medium chain fatty acids from Cuphea seeds. Plant J 84: 1021–1033 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Silva JE, Vu HS, Mockaitis K, Nam J-W, Cahoon EB (2015b) Toward production of jet fuel functionality in oilseeds: identification of FatB acyl-acyl carrier protein thioesterases and evaluation of combinatorial expression strategies in Camelina seeds. J Exp Bot 66: 4251–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jung JH, Lee SB, Go YS, Kim HJ, Cahoon R, Markham JE, Cahoon EB, Suh MC (2013) Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol 162: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Larson TR, Edgell T, Byrne J, Dehesh K, Graham IA (2002) Acyl CoA profiles of transgenic plants that accumulate medium-chain fatty acids indicate inefficient storage lipid synthesis in developing oilseeds. Plant J 32: 519–527 [DOI] [PubMed] [Google Scholar]
- Leonard JM, Knapp SJ, Slabaugh MB (1998) A Cuphea β-ketoacyl-ACP synthase shifts the synthesis of fatty acids towards shorter chains in Arabidopsis seeds expressing Cuphea FatB thioesterases. Plant J 13: 621–628 [DOI] [PubMed] [Google Scholar]
- Leonard JM, Slabaugh MB, Knapp SJ (1997) Cuphea wrightii thioesterases have unexpected broad specificities on saturated fatty acids. Plant Mol Biol 34: 669–679 [DOI] [PubMed] [Google Scholar]
- Li R, Yu K, Hatanaka T, Hildebrand DF (2010) Vernonia DGATs increase accumulation of epoxy fatty acids in oil. Plant Biotechnol J 8: 184–195 [DOI] [PubMed] [Google Scholar]
- Liu Q, Siloto RM, Lehner R, Stone SJ, Weselake RJ (2012) Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog Lipid Res 51: 350–377 [DOI] [PubMed] [Google Scholar]
- Lu C, Kang J (2008) Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep 27: 273–278 [DOI] [PubMed] [Google Scholar]
- McFie PJ, Stone SL, Banman SL, Stone SJ (2010) Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the N terminus in dimer/tetramer formation. J Biol Chem 285: 37377–37387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim F, Nakasugi K, Crowhurst RN, Hilario E, Zwart AB, Hellens RP, Taylor JM, Waterhouse PM, Wood CC (2012) Advanced engineering of lipid metabolism in Nicotiana benthamiana using a draft genome and the V2 viral silencing-suppressor protein. PLoS One 7: e52717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Silva JE, Podicheti R, Macrander J, Yang W, Nazarenus TJ, Nam JW, Jaworski JG, Lu C, Scheffler BE, Mockaitis K, Cahoon EB (2013) Camelina seed transcriptome: a tool for meal and oil improvement and translational research. Plant Biotechnol J 11: 759–769 [DOI] [PubMed] [Google Scholar]
- Oo K-C, Huang AH (1989) Lysophosphatidate acyltransferase activities in the microsomes from palm endosperm, maize scutellum, and rapeseed cotyledon of maturing seeds. Plant Physiol 91: 1288–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Morris TJ (2002) Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol Plant Microbe Interact 15: 193–202 [DOI] [PubMed] [Google Scholar]
- Saha S, Enugutti B, Rajakumari S, Rajasekharan R (2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol 141: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandager L, Gustavsson MH, Ståhl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S (2002) Storage lipid synthesis is non-essential in yeast. J Biol Chem 277: 6478–6482 [DOI] [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, de Boer G-J, Browse JA (2002) Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol 129: 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RM, Madhavji M, Wiehler WB, Burton TL, Boora PS, Laroche A, Weselake RJ (2008) An N-terminal fragment of mouse DGAT1 binds different acyl-CoAs with varying affinity. Biochem Biophys Res Commun 373: 350–354 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Truksa M, Brownfield D, Good AG, Weselake RJ (2009) Directed evolution of acyl-CoA:diacylglycerol acyltransferase: development and characterization of Brassica napus DGAT1 mutagenized libraries. Plant Physiol Biochem 47: 456–461 [DOI] [PubMed] [Google Scholar]
- Slabaugh MB, Leonard JM, Knapp SJ (1998) Condensing enzymes from Cuphea wrightii associated with medium chain fatty acid biosynthesis. Plant J 13: 611–620 [DOI] [PubMed] [Google Scholar]
- Slack CR, Roughan PG, Balasingham N (1978) Labelling of glycerolipids in the cotyledons of developing oilseeds by [1-14C] acetate and [2-3H] glycerol. Biochem J 170: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Stymne S, Appelqvist LA (1978) The biosynthesis of linoleate from oleoyl-CoA via oleoyl-phosphatidylcholine in microsomes of developing safflower seeds. Eur J Biochem 90: 223–229 [DOI] [PubMed] [Google Scholar]
- Sun C, Cao YZ, Huang AH (1988) Acyl coenzyme a preference of the glycerol phosphate pathway in the microsomes from the maturing seeds of palm, maize, and rapeseed. Plant Physiol 88: 56–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tjellström H, Strawsine M, Silva J, Cahoon EB, Ohlrogge JB (2013) Disruption of plastid acyl:acyl carrier protein synthetases increases medium chain fatty acid accumulation in seeds of transgenic Arabidopsis. FEBS Lett 587: 936–942 [DOI] [PubMed] [Google Scholar]
- Vogel G, Browse J (1996) Cholinephosphotransferase and diacylglycerol acyltransferase. Substrate specificities at a key branch point in seed lipid metabolism. Plant Physiol 110: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ, Madhavji M, Szarka SJ, Patterson NA, Wiehler WB, Nykiforuk CL, Burton TL, Boora PS, Mosimann SC, Foroud NA, Thibault BJ, Moloney MM, et al. (2006) Acyl-CoA-binding and self-associating properties of a recombinant 13.3 kDa N-terminal fragment of diacylglycerol acyltransferase-1 from oilseed rape. BMC Biochem 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberg E, Edwards P, Byrne J, Stymne S, Dehesh K (2000) The distribution of caprylate, caprate and laurate in lipids from developing and mature seeds of transgenic Brassica napus L. Planta 212: 33–40 [DOI] [PubMed] [Google Scholar]
- Wood CC, Petrie JR, Shrestha P, Mansour MP, Nichols PD, Green AG, Singh SP (2009) A leaf-based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnol J 7: 914–924 [DOI] [PubMed] [Google Scholar]
- Zhang C, Iskandarov U, Klotz ET, Stevens RL, Cahoon RE, Nazarenus TJ, Pereira SL, Cahoon EB (2013) A thraustochytrid diacylglycerol acyltransferase 2 with broad substrate specificity strongly increases oleic acid content in engineered Arabidopsis thaliana seeds. J Exp Bot 64: 3189–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.