Abstract
The ptm mutant of Arabidopsis does not show a genomes uncoupled mutant phenotype and PTM is therefore unlikely to function in chloroplast-to-nucleus signaling as previously reported.
Chloroplast development requires communication between the nucleus and the developing chloroplast to ensure that this process is optimized (Jarvis and López-Juez, 2013; Chan et al., 2016). This is especially true during de-etiolation as mis-regulation of chloroplast development can lead to seedling death from photo-oxidative damage. Retrograde signaling from the developing chloroplast (plastid) to the nucleus, which is termed biogenic signaling (Pogson et al., 2008), can be revealed using either the bleaching herbicide Norflurazon (NF), an inhibitor of carotenoid synthesis, or the plastid translation inhibitor, lincomycin (Lin) to damage the plastid. Under these conditions there is a strong downregulation of hundreds of nuclear genes (Koussevitzky et al., 2007; Aluru et al., 2009; Page et al., 2017). Despite decades of research, the biogenic retrograde signaling pathway is still very poorly understood. What we do know has mostly come from an innovative screen by the group of Joanne Chory in which genomes uncoupled (gun) mutants were identified that retained nuclear gene expression of chloroplast-related genes after NF treatment (Susek et al., 1993). This screen now defines the gun phenotype: increased expression, compared to wild type, of nuclear genes following chloroplast damage. In total, six original gun mutants have been described. GUN1 is a pentatricopeptide repeat protein with a still unknown function (Koussevitzky et al., 2007). The other GUNs are all related to the tetrapyrrole pathway (Mochizuki et al., 2001; Larkin et al., 2003; Woodson et al., 2011). Further analysis of these mutants has supported the idea that tetrapyrroles are important for plastid signaling (Vinti et al., 2000; Strand et al., 2003; Moulin et al., 2008; Mochizuki et al., 2008; Voigt et al., 2010), and our current understanding is that the synthesis of heme by ferrochelatase 1 results in a positive signal that promotes expression of nuclear-encoded chloroplast genes (Woodson et al., 2011; Terry and Smith, 2013).
Additional mutants identified through screens for a gun phenotype are the blue-light photoreceptor mutant cry1 (Ruckle et al., 2007) and the coe1 mutant lacking a functional mitochondrial transcription termination factor 4 (Sun et al., 2016). A number of happy on norflurazon (hon) mutants were also identified by screening seedlings grown on NF under lower light intensities (Saini et al., 2011). This identified one hon mutation in the ClpR4 subunit of the chloroplast-localized Clp protease complex (Saini et al., 2011). Other mutants with a gun phenotype have been identified via informed approaches to test potential signaling components. These include the transcription factor mutants abi4 (Koussevitzky et al., 2007), hy5 (Ruckle et al., 2007), and glk1glk2 (Waters et al., 2009). Interestingly, GOLDEN2-LIKE (GLK) overexpressing plants (Leister and Kleine, 2016) have also been reported to show gun phenotypes, perhaps reflecting the complex relationship between the anterograde signals by which the nucleus controls chloroplast development and retrograde signaling (Martin et al., 2016).
In 2011, Sun et al. identified a PHD transcription factor associated with the chloroplast envelope, called PTM, which they proposed mediates chloroplast signals to the nucleus through cleavage in response to changes in plastid status. Accumulation of the N terminus of the protein in the nucleus would then inhibit nuclear gene expression. Consistent with this, they reported that the ptm mutant has a gun phenotype with elevated expression compared to wild type of Lhcb on both NF and Lin. This was a significant result for the field as it defined a mechanism for plastid signaling, and is unsurprisingly included in numerous models for this pathway (e.g. Chan et al., 2016; Bobik and Burch-Smith, 2015; Terry and Smith, 2013; Barajas-López et al., 2013). Subsequent studies from the same group have suggested that PTM functions in retrograde signaling from the chloroplast to regulate flowering under high light (Feng et al., 2016) and in the integration of light and chloroplast retrograde signaling during de-etiolation (Xu et al., 2016). However, the demonstration that PTM shows a gun phenotype and is involved in retrograde signaling has yet to be supported by additional experimental data from other groups.
Given the potential importance of PTM for our understanding of plastid signaling, we have further examined the role of PTM in responses to NF and Lin in two different laboratories (See Supplemental Materials and Methods and Table S1). For the experiments at Southampton, it was necessary for us to isolate the same insertional ptm mutant allele described in Sun et al. (2011) from the SALK collection because this was no longer available from the authors. Isolation of the ptm mutant for this study, which we name here as ptm-1, is described in Supplemental Figure S1. Analysis of gene expression after NF treatment was then performed. As shown in Figure 1A, 5 µM NF treatment using the experimental conditions (1% Suc, 25 µmol m−2 s−1 white light (WL) for 7 d) of Woodson et al. (2011) resulted in no change in gene expression for a suite of five photosynthesis-related genes (including LHCB2.1 used by Sun et al. (2011) for their real-time PCR experiments) in ptm-1 compared to wild-type seedlings, whereas there was clear rescue of gene expression in the control gun5 and gun6 mutants. Next we repeated the experiment under identical conditions (2% Suc, 4 d dark followed by 3 d 120 µmol m−2 s−1 WL) to those reported in Sun et al. (2011). Under these conditions we also saw rescue of gene expression in gun5 and gun6, but not in ptm-1 (Fig. 1B). These studies were performed using ADF2 as a reference gene. To confirm that the lack of a gun phenotype in ptm1 was not related to the choice of reference gene, we also normalized the data using YLS8, which gave essentially identical results (Supplemental Fig. S2). Finally, we examined expression under conditions we have previously described (McCormac and Terry, 2004). With 3 d dark followed by 3 d 120 µmol m−2 s−1 WL, we also saw no gun phenotype for ptm-1 either in the presence or absence of Suc (Supplemental Fig. S3). Only under one particular set of conditions did we see any indication of a rescue of gene expression in ptm-1 after NF treatment. Under these conditions (1% Suc, 2 d dark followed by 3 d 100 µmol m−2 s−1 WL with a lower NF concentration of 1 µM), we saw a very small, but statistically significant increase for LHCB2.1 and HEMA1, but not for the other three genes tested (Supplemental Fig. S4). Given that under these conditions gun1-1 rescue was complete for both genes (> 300% for HEMA1), we do not believe this one exception supports a role for PTM in the plastid signaling pathway exposed by NF treatment.
Figure 1.
The ptm-1 mutant does not show a gun phenotype on Norflurazon (NF). Seedlings were grown on half-strength Linsmaier and Skoog medium (A) supplemented with 1% Suc and 0.8% agar (pH 5.7) with (dark gray bars) or without (light gray bars) 5 µM NF under continuous low white light (25 µmol m−2 s−1) for 7 d, or (B) supplemented with 2% Suc and 0.8% agar (pH 5.8) with (dark gray bars) or without (light gray bars) 5 µM NF under the following conditions: an initial 2 h WL treatment (120 µmol m−2 s−1) to stimulate germination, 4 d dark, 3 d WLc (120 µmol m−2 s−1). For A and B, genomes uncoupled 5 (gun5) and gun6 mutants were included as positive controls (known to rescue nuclear gene expression on NF). Expression was determined with qRT-PCR and is relative to wild-type −NF and normalized to ACTIN DEPOLYMERIZING FACTOR 2 (ADF2, At3g46000). Data shown are the means ±SEM of three independent biological replicates. Asterisks denote a significant difference versus wild type for the same treatment (−NF or +NF), Student’s t test (P < 0.05).
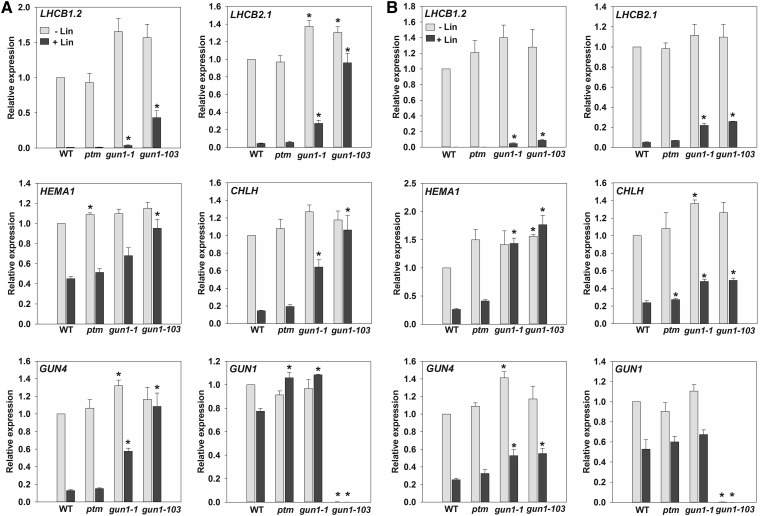
The ptm-1 mutant was also reported to result in elevated gene expression compared to wild-type seedlings when grown on Lin (Sun et al., 2011). We therefore also tested ptm-1 under these conditions. As shown in Figure 2, ptm-1 failed to result in elevated gene expression on Lin while gun1-1 (Koussevitzky et al., 2007) and gun1-103 (see methods) control seedlings both showed strong rescue of gene expression (Fig. 2). This was true whether seedlings were grown in the dark (Fig. 2A) or in the light (Fig. 2B) and was independent of the reference gene used (Supplemental Fig. S5).
Figure 2.
The ptm mutant does not show a gun phenotype on lincomycin (Lin). Seedlings were grown on half-strength Linsmaier and Skoog medium supplemented with 2% Suc and 0.8% agar (pH 5.8) with (dark gray bars) or without (light gray bars) 0.5 mm Lin in dark for 5 d (A), or (B) on half-strength Murashige and Skoog medium supplemented with 1% Suc and 1% agar (pH 5.8) with (dark gray bars) or without (light gray bars) 0.5 mm Lin under the following conditions: 2 d dark, 3 d WL (100 µmol m−2 s−1). For A and B, the genomes uncoupled, gun1-1, and gun1-103 mutants were included as positive controls (known to rescue gene expression on Lin). Expression is relative to wild-type −Lin and normalized to ACTIN2 (ACT2, At3g18780). Data shown are means +SEM of three independent biological replicates. Asterisks denote a significant difference versus wild type for the same treatment (−Lin or + Lin), Student’s t test (P < 0.05).
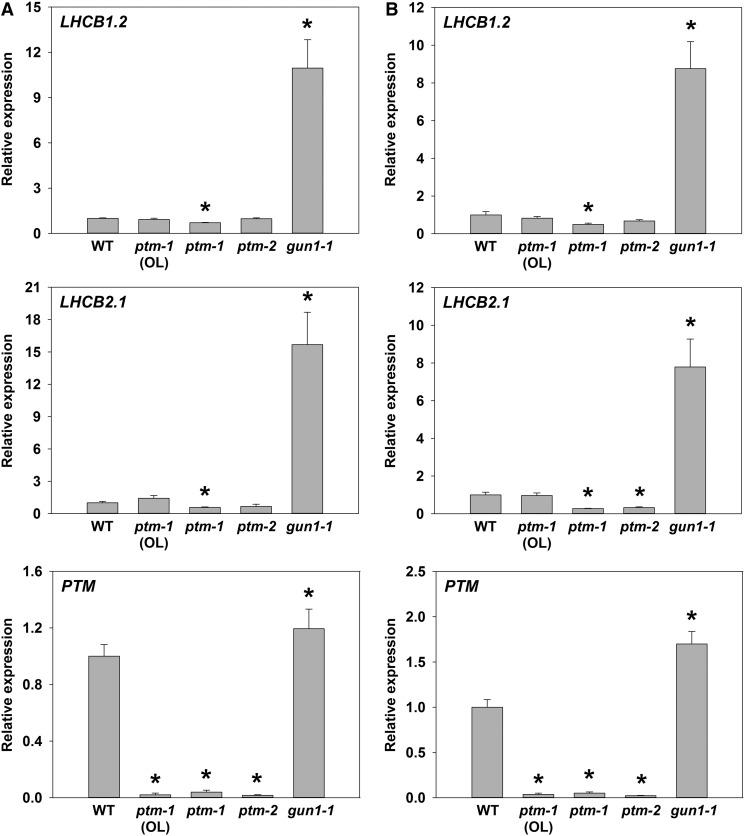
To verify further whether we could detect a gun mutant phenotype for ptm mutants, we also performed experiments in parallel in Kyoto. For this set of experiments, two ptm alleles were used. The original ptm mutant (ptm-1 OL) was obtained from Lixin Zhang (CAS, Beijing; Sun et al., 2011) and independently from the SALK collection (ptm-1) and, in addition, a second ptm allele, ptm-2, was also identified from the SALK collection (Supplemental Fig. S1). As shown in Figure 3, none of the ptm mutants showed an elevation of LHCB1.2 (although the primer set used is also likely to detect LHCB1.1 and LHCB1.3) or LHCB2.1 expression after NF or Lin treatment compared to wild type, while a strong increase was observed in the gun1-1 control.
Figure 3.
A second ptm mutant allele does not show a gun phenotype on Norflurazon (NF) or lincomycin (Lin). Seedlings were grown on Murashige and Skoog medium supplemented with 2% Suc and 0.8% agar (pH 5.8), and either (A) 2.5 µM NF or (B) 560 µM Lin. All seedlings were grown under continuous white light (WLc, 100 µmol m−2 s−1) for 4 d at 23°C. Three ptm mutant lines were tested: ptm-1 (OL) is the original line as used in Sun et al., 2011; ptm-1 is the same insertion line as ptm-1 (OL), Salk_013123, but obtained independently from the stock center; ptm-2 is a second insertion line, Salk_073799. The genomes uncoupled 1-1 (gun1-1) mutant was included as a positive control (known to rescue nuclear gene expression on NF and Lin). Expression was determined with qRT-PCR and is relative to wild-type +NF (A) or +LIN (B), respectively, and normalized to TUBULIN BETA CHAIN 2 (TUB2, At5g62690). Data shown are the means ±SEM of five independent biological replicates. Asterisks denote a significant difference versus wild-type +NF (A) or +LIN (B), respectively, Student’s t test (P < 0.05).
In conclusion, rigorous testing of the phenotype of ptm mutants on NF and Lin shows that the ptm mutant does not show elevated expression of photosynthetic genes compared to wild type. This was true whether using the conditions described in the original publication or other conditions used routinely to test plastid signaling responses. One possible difference between our study and that of Sun et al. (2011) is that they used RNA gel blot analysis for most of their experiments. The probe used should preferentially detect LHCB1.1, but might also be expected to detect LHCB1.2 and LHCB1.3, and possibly other LHCB genes. In our experiments, we have tested both LHCB1.1 and LHCB1.2, so it remains possible that changes in another LHCB gene could account for the observed phenotype in the original paper (Sun et al., 2011). However, Sun et al. (2011) also reported the same gene expression phenotype for ptm using real-time PCR and a primer pair that most closely matches LHCB2.1, and we did not detect an increase in expression for this gene in our experiments (with one exception). We therefore believe it is unlikely that differences in detection methods or genes tested can account for the observed differences in phenotype. Moreover, if PTM is to be considered an important player in plastid signaling, the gun phenotype of ptm should be robust enough to withstand this level of scrutiny. We have not tested other results reported by Sun et al. (2011). However, we note that the 3-fold elevation of expression of PTM on NF measured using PTM:GUS was not apparent in our experiments (Fig. 1 and Supplemental Fig. S3) and the reduction in PTM expression in gun1 after NF and Lin treatment was also not observed (Fig. 3). In fact, PTM expression was moderately (but significantly) elevated in gun1-1 in our study (Fig. 3). Whether our result has implications for other PTM signaling roles (Feng et al., 2016; Xu et al., 2016) is currently unknown, but should be the subject of further scrutiny.
The signaling pathway by which the status of the developing chloroplast is relayed to the nucleus is one of the few remaining plant signaling pathways that we know of, but for which we have little idea of the signaling components involved. We believe this study resolves one of the major discrepancies in plastid signaling research by eliminating a major role for PTM, and paves the way for more focused studies that build on recent progress on the role of tetrapyrroles and chloroplast protein homeostasis in plastid retrograde signaling (Woodson et al., 2011; Maruta et al., 2015; Ibata et al., 2016; Tadini et al., 2016).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primers used in this study.
Supplemental Figure S1. Characterization of the ptm T-DNA insertion mutants
Supplemental Figure S2. The phenotype of ptm-1 after NF treatment using the Sun et al. (2011) method normalized to YLS8
Supplemental Figure S3. The phenotype of ptm-1 after NF treatment using the McCormac & Terry (2004) method in the presence and absence of Suc
Supplemental Figure S4. The phenotype of ptm-1 after NF treatment using a modification of the McCormac & Terry (2004) method in the presence of Suc
Supplemental Figure S5. The phenotype of ptm-1 after Lin treatment normalized to YLS8
Supplemental Methods. Supplemental materials and methods.
Supplementary Material
Acknowledgments
We would like to thank Tania Garcia-Becerra for technical support. Thanks also to Joanne Chory and Jesse Woodson (SALK Institute) for the gun1-1, gun5, and gun6 mutants used in this study. N.M. thanks Lixin Zhang (Chinese Academy of Sciences) for the ptm (ptm-1 OL) mutant.
Footnotes
The work was supported by UK Biotechnology and Biological Sciences Research Council grants BB/J018139/1 to M.J.T. and BB/J018694/1 to A.G.S. and by JSPS KAKENHI grants JP 24570046 and JP 21570039 to N.M. S.M.K. was supported by the Gatsby Charitable Foundation.
Articles can be viewed without a subscription.
References
- Aluru MR, Zola J, Foudree A, Rodermel SR (2009) Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol 150: 904–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas-López J de D, Blanco NE, Strand Å (2013) Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim Biophys Acta 1833: 425–437 [DOI] [PubMed] [Google Scholar]
- Bobik K, Burch-Smith TM (2015) Chloroplast signaling within, between and beyond cells. Front Plant Sci 6: 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Feng P, Guo H, Chi W, Chai X, Sun X, Xu X, Ma J, Rochaix JD, Leister D, Wang H, Lu C, Zhang L (2016) Chloroplast retrograde signal regulates flowering. Proc Natl Acad Sci USA 113: 10708–10713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibata H, Nagatani A, Mochizuki N (2016) CHLH/GUN5 function in tetrapyrrole metabolism is correlated with plastid signaling but not ABA responses in guard cells. Front Plant Sci 7: 1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Leister D, Kleine T (2016) Definition of a core module for the nuclear retrograde response to altered organellar gene expression identifies GLK overexpressors as gun mutants. Physiol Plant 157: 297–309 [DOI] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ (2004) The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J 40: 672–685 [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA 105: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, McCormac AC, Terry MJ, Smith AG (2008) Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci USA 105: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Miyazaki N, Nosaka R, Tanaka H, Padilla-Chacon D, Otori K, Kimura A, Tanabe N, Yoshimura K, Tamoi M, Shigeoka S (2015) A gain-of-function mutation of plastidic invertase alters nuclear gene expression with sucrose treatment partially via GENOMES UNCOUPLED1-mediated signaling. New Phytol 206: 1013–1023 [DOI] [PubMed] [Google Scholar]
- Page MT, McCormac AC, Smith AG, Terry MJ (2017) Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol 213: 1168–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini G, Meskauskiene R, Pijacka W, Roszak P, Sjögren LLE, Clarke AK, Straus M, Apel K (2011) ‘happy on norflurazon’ (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J 65: 690–702 [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2: 477. [DOI] [PubMed] [Google Scholar]
- Sun X, Xu D, Liu Z, Kleine T, Leister D (2016) Functional relationship between mTERF4 and GUN1 in retrograde signaling. J Exp Bot 67: 3909–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M, Rothbart M, Hedtke B, et al. (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170: 1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Smith AG (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinti G, Hills A, Campbell S, Bowyer JR, Mochizuki N, Chory J, López-Juez E (2000) Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J 24: 883–894 [DOI] [PubMed] [Google Scholar]
- Voigt C, Oster U, Börnke F, Jahns P, Dietz KJ, Leister D, Kleine T (2010) In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol Plant 138: 503–519 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chi W, Sun X, Feng P, Guo H, Li J, Lin R, Lu C, Wang H, Leister D, Zhang L (2016) Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat Plants 2: 16066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.