Naturally co-occurring abiotic stresses that mimic field conditions revealed different transcriptional signatures and functional pathways among common stress- and stress combination unique-genes.
Abstract
Climate-change-driven stresses such as extreme temperatures, water deficit, and ion imbalance are projected to exacerbate and jeopardize global food security. Under field conditions, these stresses usually occur simultaneously and cause damages that exceed single stresses. Here, we investigated the transcriptional patterns and morpho-physiological acclimations of Brachypodium dystachion to single salinity, drought, and heat stresses, as well as their double and triple stress combinations. Hierarchical clustering analysis of morpho-physiological acclimations showed that several traits exhibited a gradually aggravating effect as plants were exposed to combined stresses. On the other hand, other morphological traits were dominated by salinity, while some physiological traits were shaped by heat stress. Response patterns of differentially expressed genes, under single and combined stresses (i.e. common stress genes), were maintained only among 37% of the genes, indicating a limited expression consistency among partially overlapping stresses. A comparison between common stress genes and genes that were uniquely expressed only under combined stresses (i.e. combination unique genes) revealed a significant shift from increased intensity to antagonistic responses, respectively. The different transcriptional signatures imply an alteration in the mode of action under combined stresses and limited ability to predict plant responses as different stresses are combined. Coexpression analysis coupled with enrichment analysis revealed that each gene subset was enriched with different biological processes. Common stress genes were enriched with known stress response pathways, while combination unique-genes were enriched with unique processes and genes with unknown functions that hold the potential to improve stress tolerance and enhance cereal productivity under suboptimal field conditions.
In the next half-century, a significant increase in agricultural production (e.g. food, feed, fibers, and bio-energy) will be needed in order to meet global food demands of the growing human population (Myers et al., 2017). This challenge is further exacerbated under the projected climate change scenarios, which predict an aggravation in the intensity and frequency of extreme events, such as temperature fluctuations, water deficit and salt toxicity (Sillmann et al., 2013; Munns and Gilliham, 2015). Under these conditions, yields of staple cereal crops (e.g. bread wheat [Triticum aestivum], rice [Oryza sativa], and maize [Zea mays]), which provide over 50% of human calories, are predicted to decrease (Reynolds et al., 2016). Thus, elucidating the mechanisms underlying plant-resilience to suboptimal field conditions and developing stress-tolerant crops is the most promising strategy to ensure global food security.
Under natural and agricultural systems, plants have to cope with multiple environmental stresses simultaneously and acclimate to ever changing environments. Nevertheless, the majority of studies on plant responses to abiotic stresses focus on short and extreme single stresses at early plant vegetative stages, with plant survival or recovery rates as a measurement for plant stress tolerance. These studies have identified and characterized numerous stress-responsive genes; however, their introgression into crop-plants has achieved limited success in the field (reviewed by Reguera et al., 2012). Plant acclimations to stress conditions (Giordano 2013) vary depending on the length and severity of the stress, the plant species and genotype, the plant developmental stage and the examined tissue or organ (Mittler and Blumwald, 2010). The complexity of the plant response is further augmented under combinations of stresses, since it involves the integration of different metabolic pathways and the cross-talk between different sensors and signal transduction pathways (Mittler, 2006). Although recently, there has been an increase in the number of studies addressing stress combinations (Suzuki et al., 2014, Zandalinas et al., 2017 and references therein), studies that focus on gradual and long-term stress combinations and the effects of combined stresses on plant productivity are scanty.
Plant acclimation to combinations of environmental stresses elicited specific physiological, molecular, and metabolic responses that could not be inferred from single stress treatments, especially if the combined stress resulted in antagonistic responses (reviewed by Suzuki et al., 2014; Prasch and Sonnewald, 2015). Tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana) plants displayed an antagonistic interaction to the combination of heat and drought. Under heat stress, plant stomata were opened to cool the leaves by transpiration, but when heat was combined with drought, stomata remained closed, resulting in a higher leaf temperature and a lower photosynthesis rate (Rizhsky et al., 2002, 2004). These distinct physiological responses were correlated with changes at the transcriptional and metabolic levels (Rizhsky et al., 2002). On the other hand, tomato (Solanum lycopersicon) plants showed a higher degree of tolerance to a combination of heat and salt stress compared with plants under salt stress alone (Rivero et al., 2014). This beneficial effect is unexpected since enhanced transpiration to cool the leaf would result in an enhanced ion uptake, with concomitant ion toxicity (Mittler, 2006). Transcriptional profiling of Arabidopsis plants under single stresses (cold, heat, high light intensity, salinity, and flagellin) and their double combinations indicated that a significant change in the transcriptome induced by double stress combinations could not be predicted from the corresponding single stresses (Rasmussen et al., 2013). These and other studies (reviewed by Suzuki et al., 2014) support the notion that plant responses to stress combinations are regulated through interactions, either positive or negative, of different signaling pathways. Yet, little is known about the genetic and physiological patterns among partially overlapping stress combinations and the genetic determinants facilitating plant acclimations.
Brachypodium distachyon (L.) Beauv. is a wild temperate model grass with close evolutionary relationships to wheat and barley (International Brachypodium Initiative, 2010). In recent years, B. distachyon has emerged as a powerful model plant to accelerate improvement of stress related traits in cereal crops (Lv et al., 2014; Priest et al., 2014; Shaar-Moshe et al., 2015; Des Marais et al., 2017). Here, we employed a system biology approach to study the morpho-physiological and transcriptional patterns associated with acclimations of B. distachyon to combinations of salinity, drought, and heat stresses. We hypothesized that combination-unique acclimations, which occur only among combined stresses, have distinct transcriptional and morpho-physiological patterns compared with common stress acclimations, which are shared among single and combined stresses. The aims of the current study were to (1) characterize the morpho-physiological acclimations under combinations of stresses and decipher their underlying genetic factors, (2) identify transcriptional patterns that differ between common stress- and combination unique-genes, and (3) detect biological processes associated with physiological traits which differ between the two gene subsets.
Comprehensive morpho-physiological analyses of B. distachyon plants demonstrated a gradual decrease in plant performances as more stresses were combined, as well as dominance of a specific stress treatment that shaped plant acclimations. Investigation of the genetic interplay between the transcriptional patterns of common stress and combination unique differentially expressed genes (DEGs) revealed specific transcriptional signatures within the two gene subsets. Enrichment analysis of coexpressed networks identified functional stress-modules and potential hub genes among the two gene subsets that could shed light on both central and novel acclimation mechanisms to combinations of salinity, drought, and heat stresses. The results provide a wide perspective of the physiological and transcriptional modifications under four partially overlapping stress combinations and highlight the unique patterns and functional pathways that are differentially regulated. In addition, the results manifest the significance of the experimental design for breeding efforts toward development of resilience cereal crops under the projected climatic changes.
RESULTS
Developing Stress Combination Assays
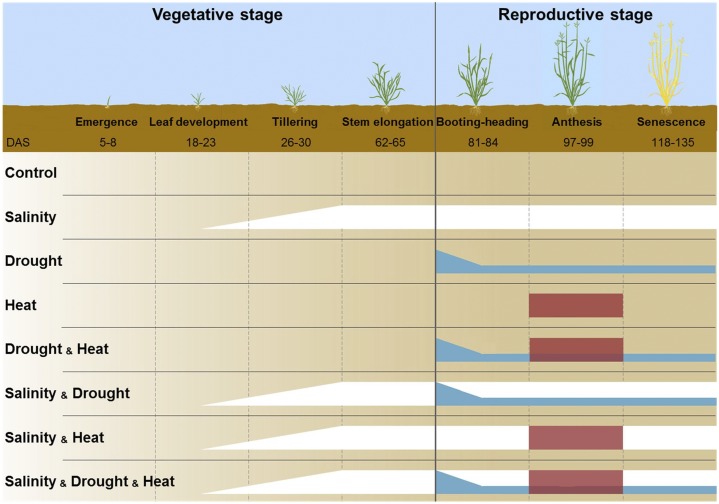
To study the acclimation of B. distachyon to co-occurring environmental stresses, combinatorial stress assays were established. Soil-grown plants were exposed to control conditions, single stresses (salinity [S], drought [D] or heat [H]), double-stress combinations (S&D, S&H and D&H), and a triple-stress combination (S&D&H). To mimic the natural Mediterranean-like field conditions, we applied prolonged and progressive salinity and drought stresses. Heat stress, which developed rapidly and fluctuated during day and night, was imposed at anthesis for four days, similarly to a typical Mediterranean spring heatwave. Stress combinations were established by integrating single stresses (Fig. 1). With the exception of single heat stress, both drought and salinity stresses and any combination of each single stress resulted in plant biomass reduction (Supplemental Fig. S1). Similarly, a reduction in shoot and root growth was observed 4 d after anthesis under all stress treatments, except for single heat stress (Fig. 2). Spike size and morphology, as well as leaf size, were also altered under the different stresses and their combinations (Fig. 2, A–D).
Figure 1.
An illustration of Brachypodium distachyon developmental stages and stress combination strategy. Plant developmental stages under the Mediterranean-like conditions and the corresponding number of days after sowing (DAS) ranging among treatments are indicated. Control plants were grown under optimal conditions throughout the experiment. Salinity (indicated by white color) was applied gradually starting at five leaf stage and maintained throughout the experiment. Drought (indicated by blue color) was applied at booting stage by gradually withholding irrigation and maintaining soil water content at 40%. Heat (indicated by red color) was applied at anthesis for four days. Double- and triple-stress combinations were generated by applying the single stresses at the same developmental stages and stress intensities.
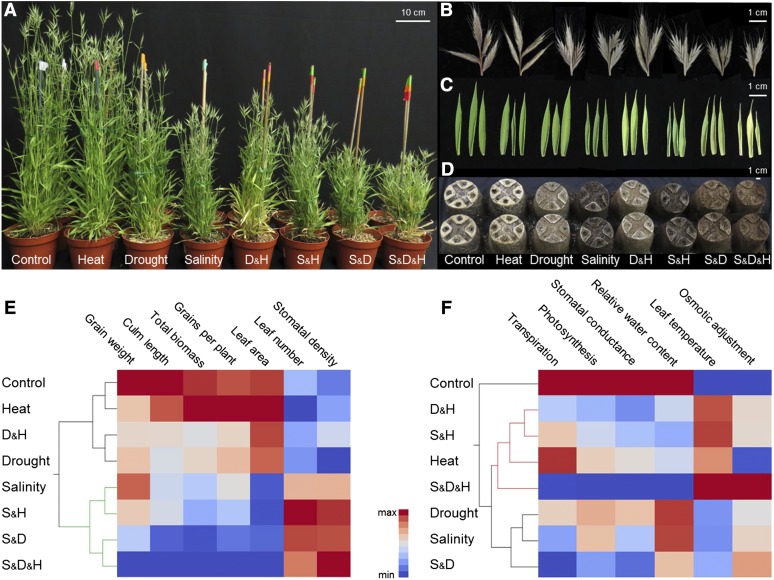
Figure 2.
Effects of single and combined stresses on plant development and productivity. Brachypodium distachyon plants were grown under control, salinity, drought, heat, and their combinations: salinity and drought (S&D), salinity and heat (S&H), drought and heat (D&H), salinity, drought and heat (S&D&H). A, Shoot and B, spike morphology, C, second leaf, and D, root development. Pictures A, C, and D are representative of plants and organs four days after anthesis. Picture B is representative of spikes at the end of the growing season. E, Hierarchical clustering of morphological and yield parameters. F, Hierarchical clustering of physiological parameters. Red and blue colors indicate high and low values compared with the mean of each trait, respectively.
Plant Acclimation to Stress Was Shaped by a Specific Stress Treatment or by a Gradual Aggravation under Combined Stresses
Plant acclimations to environmental stresses are consequences of stress-induced changes in several processes resulting in an array of phenotypical differences. To study these differences, we conducted a morpho-physiological characterization of plant acclimations to single and combined stress. Traits were assembled into morphological/yield-related traits and physiological categories and analyzed using hierarchical clustering. The analysis of morphological and yield parameters grouped all salinity treatments to one branch, separating them from all other stresses (Fig. 2E; Supplemental Fig. S2). Within the salinity and nonsalinity branches, heat treatments clustered either with control or with the corresponding stresses (for example, the combination of heat and salinity was clustered with single salinity stress; Fig. 2E). Among the different stresses, the combination of salinity and drought and the triple combination caused detrimental impacts on plant performances (e.g. reduced culm length [−59.1% and −61.8%], biomass [−61.9% and −63%], and grain number [−73.2% and −82%], respectively; Fig. 2E; Supplemental Fig. S2A).
An altered treatment partition was detected by the hierarchical clustering of physiological traits. While control conditions formed an isolated branch, stress treatments were separated into two sub-branches based on heat and nonheat treatments. Among heat treatments, the double stresses, S&H and D&H, were clustered together and separated from both heat and the triple combination. D&H and S&D&H showed a similar pattern of significant decreases in relative water content (RWC; −4.7% and −10%, respectively), transpiration rate (−61.8% and −89.5%, respectively), and photosynthesis (−75.1% and −99.9%, respectively), compared with control (Supplemental Fig. S2B). S&D&H showed stronger stress effects, which were opposite to control for all the physiological traits (Fig. 2F; Supplemental Fig. S2B). It is worth noting that at the time of sampling (i.e. four days after anthesis), heat treatments did not induce distinct visual effects in the plants and that these changes became apparent 2 weeks later (i.e. leaf senescence; Supplemental Fig. S3).
Heat Treatments Resulted in a Major Impact on Plant Transcriptome
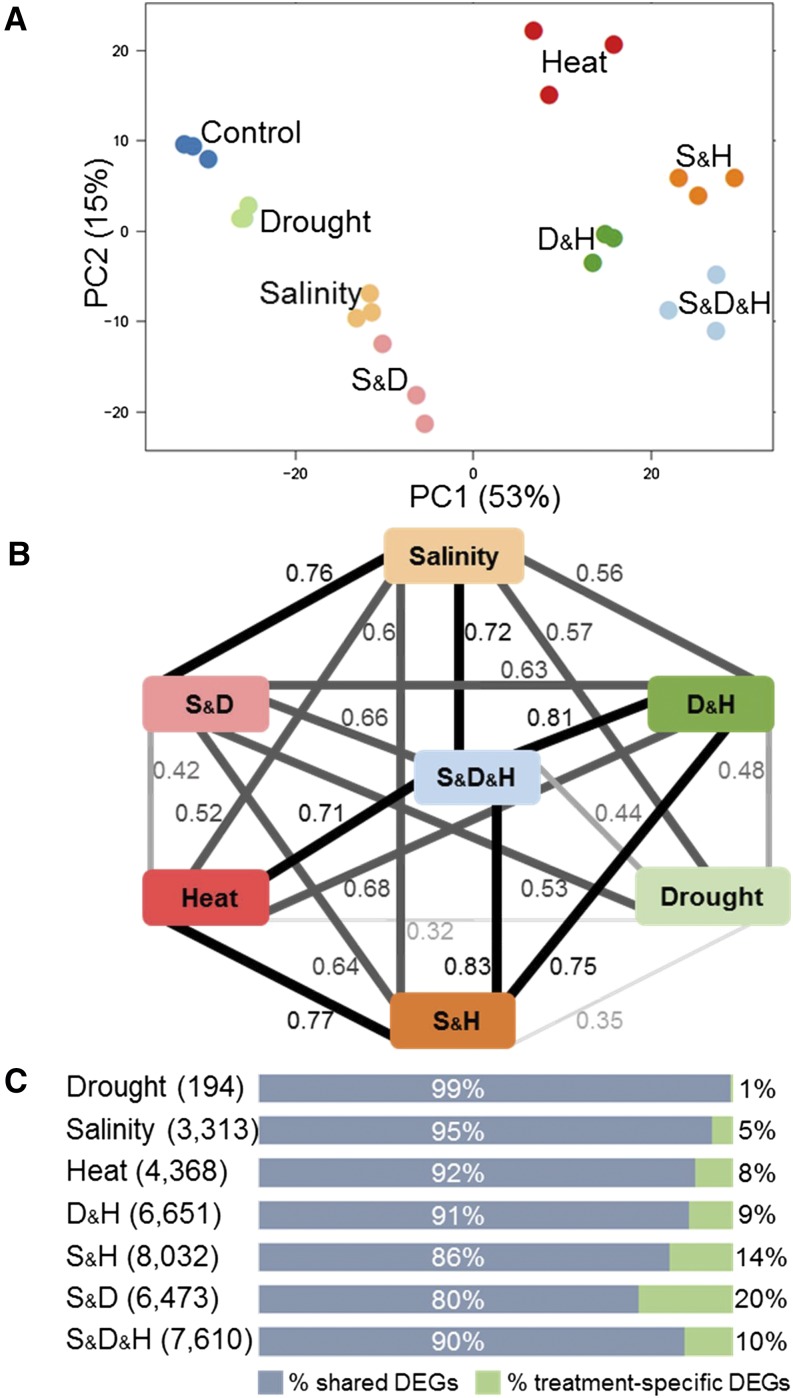
We used RNA-seq to examine global transcriptional changes in plants subjected to single and combined stresses. A principal component analysis (PCA), which detected expression patterns regardless to significance or fold change thresholds, revealed a high similarity among the three biological replicates within each treatment. Among treatments, a clear separation between heat and nonheat treatments was observed on the first principal component (PC1), explaining 53% of the total variance (Fig. 3A). Correlation coefficient (r) analysis of log2 fold change (FC) values further indicated the similarities among heat treatments (0.68 ≤ r ≤ 0.83). Yet, as opposed to the PCA, the triple-stress combination was found to have high correlation coefficients with all treatments (average r = 0.75), except with drought stress (r = 0.44). Between drought and its corresponding double stresses, a lower correlation (average r = 0.5) was found compared with salinity and heat and their corresponding double stresses (average r = 0.68 and 0.72, respectively; Fig. 3B).
Figure 3.
Stress transcriptome profile. A, Principal component (PC) analysis of rlog transformed gene expression data, generated by RNA-sequencing. Each treatment contains three biological repeats and is indicated by a different color. B, A correlation map of fold change values across stress treatments. Correlation coefficient (r) between two samples is designated on each edge. Black edges indicate r ≥ 0.7 and gray edges indicate r < 0.7. C, Percentage of shared (blue) and treatment-specific (green) significantly differentially expressed genes (DEGs) within each stress treatment (FDR ≤ 0.05, FC ≥ |0.5|). Number of DEGs in each treatment is indicated in brackets. Drought and heat (D&H), salinity and heat (S&H), salinity and drought (S&D), and salinity, drought, and heat (S&D&H).
To examine the uniqueness of plant transcriptional acclimation to combined stresses, the percentage of treatment-specific differentially expressed genes (DEGs) and the percentage DEGs that were altered in at least two stresses (i.e. shared DEGS) were assessed. Stress combination treatments showed a significant 2.8-fold increase in treatment-specific DEGs compared with single stress (13.3% versus 4.7%, respectively, P < 0.02; Figure 3C). Notably, S&D had the highest percentage of treatment-specific DEGs, which was in agreement with the PCA that separated S&D from all other stress combinations and with the low correlation coefficient that was found for S&D as compared with the other stress combinations (Fig. 3).
Integration between Transcriptional Changes and Physiological Acclimations
It has been suggested that the number of DEGs is associated with the complexity of the stress and its intensity (e.g. Prasch and Sonnewald, 2013). Our results showed that in B. distachyon, the percentage of DEGs varied extensively in response to single stresses, ranging between 0.7% and 16% of the B. distachyon transcriptome (3313, 194, and 4368 DEGs under salinity, drought, and heat stresses, respectively). Drought stress resulted in the lowest DEG number (Fig. 3C), which is in accordance with a previous report (Verelst et al., 2013). In general, a prominent increase in the percentage of DEGs was found under combinations of stresses, where 24–30% of the transcriptome was modulated (Fig. 3C; Supplemental Table S1). However, focusing on DEG numbers may not be sufficient to explain the severity and complexity of the stress. For example, the highest number of DEGs was detected under the combination of salinity and heat (8032 DEGs) rather than the triple stress combination (7610 DEGs; Fig. 3C).
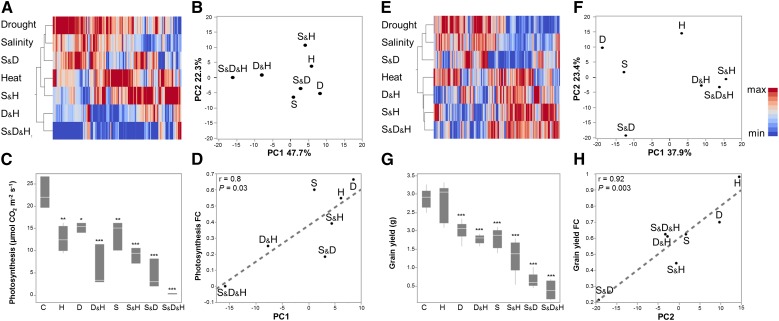
To find a more informative measurement for stress intensity and complexity, and gain a better understanding on plant acclimations to different stresses, we integrated transcriptional patterns and physiological traits. Functional gene categories of photosynthesis and stress responses (MapMan annotations; Thimm et al., 2004) were selected for this analysis. PCA was conducted to detect categories’ first and second principle components (PC), which showed maximal data variation and best summarize category's expression patterns. Each functional category displayed a unique order of stress severity and treatments clustering. Photosynthesis was adversely affected mainly by combinations of drought and heat (i.e. D&H and S&D&H), whereas stress-response genes and grain yield clustered based on single versus combined stresses, indicating gradual changes in the expression levels of stress-associated genes and yield parameters, concomitant with an increase in stress intensity (Fig. 4). Among 157 photosynthesis-related DEGs that were differentially regulated in at least one stress treatment, PC1, which explained 47.7% of the total variance, was highly correlated with leaf photosynthetic rate (r = 0.8, P = 0.03; Fig. 4A). Similarly, among 501 stress response DEGs differentially regulated in at least one stress treatment, PC2, which explained 23.4% of the variance, showed a high correlation with grain yield (r = 0.92, P = 0.004; Fig. 4B). Thus, expression modifications of stress-response genes at anthesis may have long-term mitigating effects on plant productivity at the end of the growing season.
Figure 4.
Association between transcriptome and physiology. A to D, Correlation between expression pattern of photosynthesis associated genes and photosynthesis measurements. E to F, Correlation between expression pattern of stress associated genes and grain yield. Transcripts were assigned to each category based on MapMan bin allocation. A and E, hierarchical clustering heat-map of gene expression of each category. First (B) and second (F) principle components (PC) that were detected by principal component analysis of gene expression. Box plots of (C) photosynthesis and (G) grain yield under control and stress treatments. Values are means ± sd (n = 3 and 6 for photosynthesis and grain yield, respectively) Significant differences, detected by one-way ANOVA followed by Dunnet’s test, are denoted with asterisks (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001). Correlation between first (D) or second (H) PC for photosynthesis or stress associated genes, respectively, and fold change (FC) values of the corresponding trait. Red and blue colors indicate high and low values compared with the mean of each gene, respectively. Conditions are as follows: control (C), heat (H), drought (D), salinity (S), drought and heat (D&H), salinity and heat (S&H), salinity and drought (S&D), and salinity, drought and heat (S&D&H).
Common Stress DEGs Are Characterized by Partial Response Mode Consistency
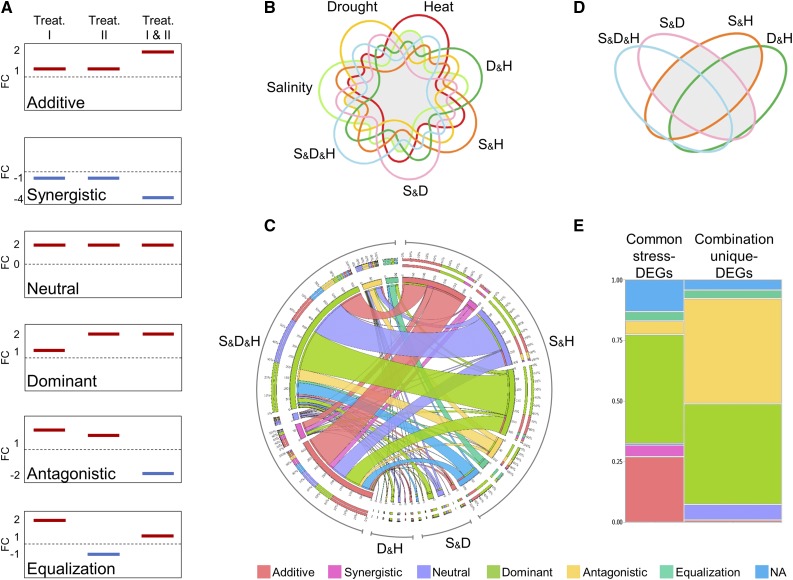
To assess the extent of similarity in gene expression under single and combined stresses, and the involvement of these DEGs in plant acclimations to stress combinations, we conducted a transcriptional pattern analysis. Six response modes, which broadly encapsulate the major patterns among single and combined stresses, were defined based on transcript FC values: additive, synergistic, neutral, dominant, antagonistic, and equalization (Fig. 5A). This analysis focused on two gene subsets: (1) DEGs shared among single and combined stresses (i.e. common stress DEGs) and (2) genes that were differentially expressed only among combinations of stresses (i.e. combination unique DEGs). A seven-way Venn diagram was used to detect common stress DEGs that were defined as DEGs under at least two single stresses and included 1,550 transcripts (Fig. 5B; Supplemental Fig. S4). These genes were assigned to the six transcriptional response modes and yielded four distinct expression patterns of double- and triple-stress combinations (Supplemental Fig. S5; Supplemental Table S2).
Figure 5.
Transcriptional patterns among common stress and combination unique differentially expressed genes (DEGs). A, Schematic illustration of gene expression among six response modes. Common stress DEGs were assigned to each mode by comparing fold change values ± sd under the triple or double stress combinations to the corresponding single stresses. Stress combination unique DEGs were assigned to each mode by comparing fold change values ± sd under the triple stress combination to the corresponding double stress combinations. Additive and synergistic modes, expression level under combined stresses is equal to or higher than the summation of the corresponding single stresses, respectively; neutral mode, similar expression level across the examined stresses; dominant mode, single (or double) stress(es) prioritized the expression level under combined stresses; antagonistic mode, opposite expression direction or control level under stress combinations compared with the corresponding single (or double) stresses; equalization mode, DEGs with opposite expression that their expression under combined stresses is the summation of expression under the corresponding single stresses. Genes with expression patterns that did not suit the six response modes were designated as not assigned (NA). B, A seven-way Venn diagram of DEGs was used to detect 1,550 common stress-DEGs. Gray areas indicate intersections that were included in the analysis, in which genes from at least two single stresses are differentially expressed. C, Response mode consistency of common stress-DEGs across the four response patterns. Inner circle shows the number of genes within each response mode across the triple or double stress combinations, which are indicated by the outer black circle. Middle circle shows the percentage of each response mode that was found upon comparing the consistency of gene expression patterns between triple and double stress combinations. Ribbon size corresponds to the percentage of genes from double stress combinations that were found in the triple stress combination. D, A four-way Venn diagram of DEGs was used to detect 2,561 combination unique DEGs. Gray areas indicate intersections that were included in the analysis, in which genes from at least two double stresses are differentially expressed. E, Response mode partition among common stress- and combination unique-DEGs. Drought and heat (D&H), salinity and heat (S&H), salinity and drought (S&D), and salinity, drought and heat (S&D&H).
In general, the six modes can be classified into expected (i.e. neutral, additive, and equalization) and unexpected (i.e. synergistic, dominant, and antagonistic) responses. Among the response pattern of the triple-stress combination, most DEGs corresponded to unexpected modes (63%) compared with an average of 40% of the DEGs among response patterns of double-stress combinations. The transcriptional pattern of S&D was depleted with antagonistic and synergistic modes, while D&H was enriched with additive mode. The transcriptional pattern of the triple-stress combination displayed an increment in dominant mode and a decrease in neutral mode (Supplemental Fig. S5). Examination of response mode consistency of each DEG among the four transcriptional patterns of combined stresses revealed that, on average, only 37% of the DEGs maintained the same response mode (Fig. 5C; Supplemental Table S2). Although these were common stress-DEGs, the majority of the genes did not maintain the same mode under the partially overlapping stress combinations.
Response Patterns of Stress Combination Unique DEGs Versus Common Stress DEGs
To test the hypothesis that combination unique acclimations have distinct transcriptional patterns compared with common stress acclimations, we defined combination unique DEGs. A four-way Venn diagram, which included unique DEGs among S&D, S&H, D&H, and S&D&H, was used to detect DEGs common to at least two double stresses (Fig. 5D; Supplemental Fig. S6). This analysis identified 2,561 combination unique DEGs, which were assigned to the six response modes (Supplemental Table S3). A comparison between the response mode partition of common stress DEGs (represented by the expression pattern of the triple-stress combination) and combination unique DEGs revealed significant differences. While common stress DEGs were enriched with additive (27%) and synergistic (5%) modes (Pχ2 < 0.001), combination unique DEGs were enriched with neutral (6%) and antagonistic (44%) modes (Pχ2 < 0.001; Figure 5E).
Pathway and Coexpression Analyses Detected Functional Differences between Common Stress and Combination Unique DEGs
To explore the biological functions among common stress and combination unique DEGs, we performed an enrichment analysis of biological pathways based on MapMan annotations. This analysis detected seven enriched categories among common stress DEGs, including transport (i.e. transport of peptides, amino acids, and major intrinsic proteins) and metabolism of amino acids, carbohydrates (i.e. raffinose family of oligosaccharides and trehalose), secondary metabolites (i.e. isoprenoids and phenylpropanoids), and hormones. Among combination unique DEGs, only one category of photosynthesis was enriched (Supplemental Fig. S7).
A weighted gene coexpression network analysis (WGCNA) of the two gene subsets, in which an a priori partition to response modes was not defined, was used to uncover relationships between genes based on their correlation patterns and identify core functional clusters (i.e. modules) of highly coexpressed genes (Langfelder and Horvath, 2008). We detected 12 and 22 modules among common stress and combination unique DEGs, respectively, including on average, 95% of the DEGs in each subset (Supplemental Fig. S8, A and B). Enrichment analysis of response modes within the modules revealed that 75% and 91% of the modules were enriched with at least one response mode among common stress- and combination unique-DEGs, respectively. Within each module, on average, 58% of the genes were assigned to one response mode. These results demonstrated the coherence between response pattern and WGCNA (Supplemental Fig. S9, A and B).
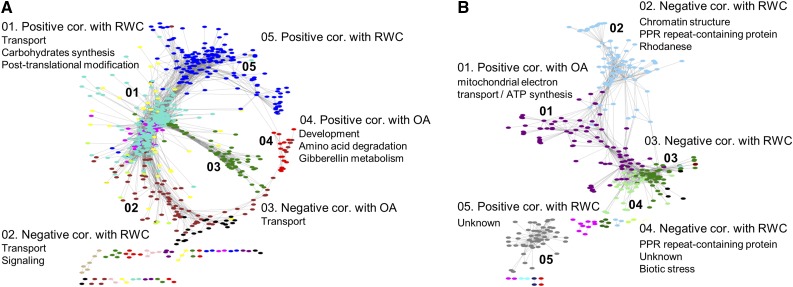
WGCNA was further used to identify associations between gene expression and stress acclimation physiological traits (i.e. relative water content [RWC] and osmotic adjustment [OA]). Among common stress DEGs, six and three modules correlated significantly between module eigengene (ME, the first principal component of a given module; Langfelder and Horvath, 2007) and RWC or OA, respectively (r ≥ 0.6, P ≤ 0.05; Supplemental Fig. S10A; Supplemental Table S4). Among combination unique DEGs, six and five modules were significantly correlated with RWC or OA, respectively (r ≥ 0.6, P ≤ 0.05; Supplemental Fig. S10B; Supplemental Table S5). Module membership (i.e. the correlation of ME and gene expression profile) quantify the similarity of all genes to each module and implies highly connected intramodular hub genes. On average, 80% of the aforementioned modules had a high correlation (r > 0.5) between module membership and gene significance (i.e. correlation of gene expression and the physiological traits; Supplemental Figure S11). This may indicate that central genes within these modules were also associated with RWC or OA.
Modules, significantly correlated with RWC or OA, were subjected to enrichment analysis of biological pathways (FDR ≤ 0.05). By allocating the common stress DEGs into functional modules, we were able to detect additional processes among four of the modules, including posttranslational modification (module 01), signaling (module 02), transport (module 03), and development (module 04) (Fig. 6B; Supplemental Table S6). Among combination unique DEGs, four modules that showed correlation with physiological traits were also enriched with biological pathways. Most of these pathways were annotated as not assigned or unknown (modules 04 and 05), and none of them was detected among common stress DEGs. These pathways included numerous genes that encoded PPR repeat-containing proteins (modules 02 and 04), DNA synthesis and chromatin structure (module 02), and mitochondrial electron transport/ATP synthesis genes (module 01; Fig. 6C; Supplemental Table S7). Notably, photosynthesis, which was the only process detected among combination unique DEGs, was not detected by WGCNA (Fig. 6, A and C).
Figure 6.
Functional analysis of common stress and combination unique DEGs. Major modules (indicated by numbers) detected by weighted gene coexpression network analysis among A, common stress and B, combination unique DEGs. These modules were significantly correlated (cor.) with leaf relative water content (RWC) or osmotic adjustment (OA) (FDR ≤ 0.05, r ≥ |0.6|), and eight of them were enriched with biological processes.
Representative hub genes that were assigned to enriched pathways and showed significant correlation with RWC or OA were selected for qPCR validation. Altogether, the six genes selected from modules of common stress and combination unique DEGs, showed similar expression patterns and comparable expression levels with RNA-seq analysis (Supplemental Fig. S12; Supplemental Table S8). Among common stress DEGs, BdWAK2 (BRADI3G49170; module 01, Fig. 6B), was significantly down-regulated across all stress treatments, except for drought. BdWAK2 expression showed a positive correlation with RWC, which was consistent with the loss of cell expansion upon reduction in WAK protein levels in Arabidopsis (Wagner and Kohorn, 2001). AtWAK2 was shown to function as a cell-wall-associated kinase required for invertase activity, a key factor in turgor maintenance in growing cells (Kohorn et al., 2006). Expression patterns of BdTIP1;2 (BRADI2G62520) and BdPIP2;2, (BRADI5G15970) encoding two aquaporins that were assigned to module 03 and presented negative correlation with OA (i.e. their expression was affected mainly by heat treatments rather than salinity and drought) were also validated. Among enriched modules of combination unique DEGs, two genes from module 02, BdPTAC3 (BRADI3G28060) and BdPRORP1 (BRADI5G27596), that exhibited negative correlation with RWC were validated. BdPTAC3 and BdPRORP1 are involved in DNA organization and quality control and RNA cleavage and decay at the plastid nucleoids, respectively (Majeran et al., 2012). The two genes were significantly up-regulated across combined stresses, except for S&D. In addition, we examined the expression of an unknown gene (BRADI4G33310) from module 04 that displayed a negative correlation with RWC. Its Arabidopsis ortholog (AT5G24350, AtMIP2) was recently shown to be part of a unique complex on the ER that is responsible for efficient transport of seed storage proteins (Li et al., 2013). Expression levels of BdMIP2 were significantly up-regulated across combined stresses, with the exception of S&D (Supplemental Figure S12).
DISCUSSION
The Near Eastern Mediterranean region, where Brachypodium has evolved (Vogel et al., 2006), is characterized by a long, hot, dry summer and a short, mild, wet winter (Loss and Siddique, 1994). In this region, soil salinity is a growing problem due to both natural and anthropogenic effects (Munns and Gilliham, 2015). Salinity, drought, and heat usually develop as gradual and chronic stresses that occur simultaneously (e.g. salinity and drought) or as a combination of chronic and transient stresses (e.g. terminal drought and heatwaves). These stresses reach a high level of severity at the reproductive stage, which is one of the most sensitive stages in determining yield under stress for temperate grasses (Barnabas et al., 2008). To gain knowledge on temperate grass acclimations to naturally co-occurring stresses, we developed an experimental strategy that mimicked the Mediterranean-like field conditions. Plant acclimations to abiotic stresses are affected by the plant species, the developmental stage of the plant and the nature of the stress. Therefore, the stress protocol (i.e. progressive or short stress, climatic conditions and growth substrate of the plants) as well as sampling dates and plant organ were carefully selected and managed. All single and combined stresses were maintained at sublethal levels to simulate naturally occurring long-term stresses and to ensure grain production (Fig. 2; Supplemental Fig. S2). We used yield-related traits, rather than plant survival, to assess plant acclimation to stress (Peleg et al., 2011).
Our working hypothesis was that combination unique acclimations show specific morph-physiological pattern compared with common stress acclimations. However, hierarchical clustering of morphological and physiological traits revealed two different responses: increased stress intensity under combinations of stresses compared with single stresses, and dominance of either salt or heat treatments (Fig. 2, E and F; Supplemental Fig. S2). Several physiological traits (e.g. photosynthesis rate and stomatal conductance) and yield-related traits (e.g. total biomass and grains per plant) declined more under combined stresses compared with single stresses; on the other hand, morphological traits were most influenced by salt treatments. The long exposure of the plants to elevated salt concentrations, starting at an early developmental stage, resulted in smaller plants (Munns, 2002). Salt toxicity and reduced soil osmotic potential resulted in a reduction in the amount of energy acquired by the plants (by reducing photosynthesis rate and leaf area) and in the redistribution of energy from growth and grain filling to stress defense mechanisms, which subsequently decreased yields (Munns and Gilliham, 2015; Fig. 2; Supplemental Fig. S2). Under heat treatments, RWC values were significantly lower than the control plants. This reduction can explain the decrease in stomatal conductance and photosynthesis rates, especially under combinations of heat stress (Fig. 2E; Supplemental Fig. S2B). This may be associated with the limited ability of the plants to fulfill the high atmospheric demand as indicated by leaf VPD (∼2.5-fold higher under heat stress as compared with control conditions).
In general, B. distachyon plants demonstrated a high phenotypic plasticity, i.e. the ability of a single genotype to produce a range of phenotypes under different environments (Bradshaw, 1965; Fig. 2; Supplemental Fig. S2). Plasticity, which is genetically controlled, can contribute to species’ evolution since it provides a defense against rapid climate change and assists in rapid adaptation (Nicotra et al., 2010). Physiological plasticity and its importance in plant acclimation to environmental stress conditions was previously reported for various wild grasses that originated from the eastern Mediterranean region, such as wild emmer wheat (T. turgidum spp. dicoccoides [Körn.] Thell.; Peleg et al., 2005) and wild barley (Hordeum spontaneum Koch; Volis et al., 2002).
To better understand the transcriptional changes that are involved in plant acclimations to single and combined stresses, we analyzed the transcriptional patterns under stress. Similar to our findings, combined application of heat, drought, and virus in Arabidopsis highlighted heat as a major stress factor that resulted in a higher number of DEGs compared with the other treatments and with a profound effect on the plant transcriptome (Prasch and Sonnewald, 2013). These changes can result from alleviation of loci silencing within heterochromatin under heat stress through heat-induced reduction of nucleosome occupancy (Lang-Mladek et al., 2010). It has been postulated that the decondensation of heterochromatin is important for transcriptional activation of heterochromatin-embedded targets that may be involved in heat tolerance (Pecinka et al., 2010).
Under combinations of stresses, plants undergo changes in gene expression that are in part common with the corresponding single stresses (i.e. common stress DEGs) and are in part unique to stress combinations (i.e. combination unique DEGs). The extent and composition of combination unique DEGs varied among different studies, but it has been shown that the multifactorial nature of stress combinations limits the ability to predict plants’ transcriptional responses based on the corresponding single stresses (Prasch and Sonnewald, 2013; Rasmussen et al., 2013; Sewelam et al., 2014; Suzuki et al., 2016). Investigation of the transcriptional changes of 1,550 common stress DEGs yielded four different response patterns of triple- and double-stress combinations that varied in response mode partitions (Supplemental Fig. S5). This analysis enabled us, for the first time, to quantify the extent of gene expression consistency and the maintenance of expression patterns among partially overlapping stress combinations. Interestingly, comparison between the transcriptional patterns of the triple- and the three double-stress combinations showed that only 37% of the common stress DEGs maintained the same response mode (Fig. 5C; Supplemental Table S2). These results suggest the fine-tuning in the mode of action of DEGs common to single and combined stresses under different stress combinations, limiting our ability to deduce the function and contribution of specific genes to stress acclimation based on the corresponding single stresses.
Comparison between common stress DEGs and combination unique DEGs (2,561 genes) revealed a significant shift from increased stress intensity (i.e. enrichment in additive and synergistic modes) to opposite responses (i.e. enrichment in antagonistic mode), respectively (Fig. 5E). The majority (99%) of the combination unique DEGs that were assigned to the antagonistic mode were differentially expressed under at least two double-stress combinations, while their expression under the triple-stress combination remained unchanged (Supplemental Table S3). Moreover, among stress combination unique DEGs, only 53% of the genes were differentially expressed under the triple-stress combination (Supplemental Fig. S6), as opposed to 90% among common stress-DEGs (Supplemental Fig. S4). The enrichment in antagonistic mode under combined stresses can result from different signaling pathways that interact and inhibit one another (Suzuki et al., 2014), a process that may be further exacerbated under the triple-stress combination. It is also possible that the increased stress intensity under the triple-stress combination resulted in a partial shutdown of stress combination unique pathways so that mainly conserved stress pathways (i.e. common stress DEGs) were activated. Alternatively, since 10% of the genes under the triple-stress combination were uniquely expressed only under this combination (Fig. 3C), specific pathways regulating plant acclimation to stress may be activated (Atkinson and Urwin, 2012). The partition of DEGs to expected and unexpected responses further supports the specificity of plant acclimations to each stress combination. Rasmussen et al. (2013) found that, on average, 61% of the genes expressed among various double stresses were assigned to unexpected responses. Here, we show that among common stress DEGs, unexpected modes constituted, on average, 40% and 63% of the plant response to double- and triple-stress combinations, respectively, whereas among combination unique DEGs, 88% of the genes were assigned to unpredictable modes (Fig. 5E; Supplemental Fig. S5). These results indicated that the predictability of the transcriptional response was diminished as the complexity of the stress and the specificity of gene expression increased.
Common stress and combination unique DEGs differ not only in their transcriptional signature but also between their functional pathways. Although this difference may result from distinct strategies that the plants adopted to cope with combined stresses, we cannot exclude the possibility that it is a result of gaps in our knowledge about the effects of combined stresses given the scarcity of studies that focused on the response(s) of plants to stress combinations. Enrichment analysis of common stress DEGs, complemented by WGCNA, detected processes that are known to be involved in environmental stress acclimation. For example, degradation and transport of amino acids, metabolism of the raffinose family of oligosaccharides, trehalose (Krasensky and Jonak, 2012), isoprenoids and phenylpropanoids (Wahid et al., 2007; Tattini et al., 2015), posttranslational modifications, and signaling (Cabello et al., 2014; Zhu, 2016; Fig. 5A; Figure 6B; Supplemental Table S6). These processes are suggested as core stress processes that are functionally conserved under both single and combined stresses. Among combination unique DEGs, we identified specific and unique enriched pathways that were not detected among common stress-DEGs, for example, DNA synthesis and chromatin structure (module 02). Investigation of the genes within this module revealed that 28% of them (55 genes) were predicted to be involved in chloroplast nucleoid metabolism and function (Supplemental Table S9). Nucleoids are comprised of multiple copies of the plastid chromosome that are organized in DNA-protein complexes (Sakai et al., 2004). The proteins in these complexes are involved in DNA- and RNA-associated functions, posttranscriptional and translational processes, as well as additional unknown functions. Mutant analyses and transgenic approaches of numerous nucleoid-associated genes revealed a plethora of phenotypes, which affected not only chloroplast development and plastid metabolism, but also hormone metabolism, plant morphology and stress responses (reviewed by Melonek et al., 2016). In maize leaves, nucleoid-enriched proteome shifted the predominating function from RNA metabolism in undeveloped plastids to translation and homeostasis in mature chloroplasts (Majeran et al., 2012). Interestingly, our results showed that the majority of the nucleoid-associated genes (47%), involved in known functions, were assigned to RNA metabolic functions. This result suggests that combinations of stresses may cause perturbations at the steady-state level and functionality of the chloroplast nucleoids and may partially explain the low photosynthesis rates and decreased plant performances under combined stresses compared with single stresses. This and other processes (e.g. mitochondrial electron transport), which are specifically involved in plant acclimations to stress combinations, can serve as novel targets for the development of resilient crops to naturally co-occurring long-term stresses.
Conclusions
The current study aims to bridge the gap between genomics and physiological acclimations to naturally co-occurring long-term abiotic stresses by employing a system biology approach. Dissection of plant transcriptome into common stress and combination unique DEGs revealed specific transcriptional signatures and functional processes within each gene subset. The increment in stress intensity among common stress DEGs correlated well with the pattern detected by morpho-physiological analyses and with the enriched pathways that are involved in canonic stress processes. Among combination unique DEGs, response patterns shifted toward antagonistic responses, indicating the limited ability to predict plant responses when several stresses are combined and the unique acclimations to each of the partially overlapping stress combinations. The processes that are specifically involved in plant acclimations to combined stresses are proposed as a basis for future research and breeding efforts toward developing crop plants better adapted to the predicted climatic changes.
MATERIALS AND METHODS
Plant Growth Conditions and Stress Treatments
Seeds of B. distachyon accession 21-3 were obtained from the National Small Grains Collection (NSGC). Seeds were sown in trays containing soil mixture (50% Baltic Sphagnum peat moss 0–20 mm, 8–10 g/g water capacity, 30% coir pith 0–8 mm and 2 g L−1 Multicote Agri controlled release fertilizer [N:P:K 14% nitrogen, 14% phosphorus and 14% potassium, eight months longevity], Tuff Merom Golan, Israel) and stored in darkness at 4°C for 48 h, followed by 5 d at 15°C to synchronize and establish germination, respectively. Subsequently, trays were transferred to a Phytotron. Uniform seedlings were transplanted into preweighted 1L pots (one plant per pot) and irrigated to runoff three times a week. Plants were grown under Near Eastern Mediterranean climate conditions (22°C day/ 16°C night, 10 h light /14 h dark) and transferred to a long day regime (15 h light/ 9 h dark) 10 weeks after germination. All plants were fertilized with 1 g L−1 N:P:K (20-20-20) + micronutrients 8 weeks after germination.
A split-plot factorial (treatment) complete random design, with eight treatments and six replicates was employed (Fig. 1). Control (C): plants were grown at 22°C day/16°C night throughout the experiment. Salinity (S): starting at five-leaf stage, plants were progressively exposed to salinity by two irrigations of 20 mm NaCl, followed by five irrigations with 50 and then 80 mm NaCl. Runoff electric conductivity was monitored weekly and target concentration of 100 mm NaCl, which was achieved within four weeks, was kept throughout the experiment. Drought (D): starting at booting stage (BBCH scale 45; Hong et al., 2011) approximately 12 weeks after germination. Plants were gradually exposed to drought by withholding irrigation. Each pot was weighted and relative soil water content was maintained at 40% for 17d. Heat (H): At anthesis (BBCH scale 65), plants were transferred at 20:00 p.m. to a preheated greenhouse at the Phytotron (34°C day/28°C night) for four days. Double (S&D, S&H, D&H)- and triple (S&D&H)-stress combinations were assembled by merging single stresses at the same developmental stages and stress intensities as described before. Plants that were subjected to combinations of salinity and drought (i.e. S&D and S&D&H) were irrigated with tap water once drought stress was imposed.
Morphological and Yield Components Characterization
Documentation and measurements of morphological traits were conducted four days after anthesis, which corresponded to four days of heat stress, 17 d of drought stress, and 11 weeks of salt stress. For culm length, leaf area, and leaf number assessment, four representative tillers from each plant were selected (n = 6). Second leaves were scanned and leaf area was analyzed with the automated digital image software Easy-Leaf-Area (https://github.com/heaslon/Easy-Leaf-Area). Stomatal density was evaluated based on third leaf epidermal tissues as described by Geisler et al. (2000). Briefly, leaves were embedded in a dental cartridge (eliteHD+, Zhermack Clinical, Badia Polesine, Italy), which was subsequently coated with transparent nail polish to produce a mirror image of adaxial epidermis tissue (n = 4). Samples were placed on cover slides and photographed under a bright-field inverted microscope (Axiovert 200M; Zeiss, Jena, Germany) with a Hitachi HV-D30 CCD camera (Hitachi Kokusai Electric Inc. Tokyo, Japan). Stomata number, within four random areas of each sample, was counted and an average stomatal density was calculated. At the end of the growing season (BBCH scale 99), spikes and shoots were separately harvested from each plant. Samples were oven-dried (75°C for 96 h) and weighted to determine plant biomass (n = 5). Subsequently, grain samples (n = 5) were threshed, counted, and weighted.
Physiological Characterization
Measurements of physiological traits were conducted in a complete random design 4 d after anthesis, except for leaf temperature, which was measured 3 d after anthesis. The LI-6400 portable photosynthesis system with a 6400-40 leaf-chamber fluorometer (Li-Cor Inc., Lincoln, NE) was used to measure photosynthetic rate, transpiration, and stomatal conductance. Leaf gas exchange measurements were conducted on the abaxial surface of the mid portion of the third leaf between 9:00-12:00 a.m. (n = 3). Light intensity was kept at 1,000 µmol m−2 s−1, with a constant air flow rate of 500 µmol s−1 and a reference CO2 concentration of 400 µmol CO2 mol−1 air. Block temperature was maintained at 27°C and 37°C, and the average registered leaf temperatures ranged between 26.3°C (± 0.5 SE) and 36.3°C (± 0.1 SE) at the controlled and heated greenhouses, respectively. Since B. distachyon leaves did not cover the measurement cell, the measured area of each leaf was marked and scanned, and photosynthesis and stomatal conductance measurements were divided by the corrected leaf area. Average leaf vapor pressure deficit (VPD) at control conditions and heat treatments were 1.8 kPa and 4.6 kPa, respectively. Relative water content (RWC) and osmotic adjustment (OA) measurements were performed on the third leaves at midday (n = 5), as described previously (Shaar-Moshe et al., 2015). Leaf temperature was evaluated with an infrared thermometer laser (52224-A, Mastercool Inc., Randolph, NJ) at 10:00 a.m. (n = 6).
RNA Extraction and Sequencing
Flag and second leaf samples were collected four days after anthesis between 9:00–10:00 a.m. from six independent plants and immediately frozen in liquid nitrogen. Total RNA was extracted using Plant/Fungi Total RNA Purification Kit (Norgen Biotek Corp., Canada) with on-column DNase treatment (Qiagen, Germany). Sample contamination and RNA integrity were assessed using ND-1000 spectrophotometer (Thermo Fisher Scientific) and Agilent RNA 6000 Nano Chip on an Agilent 2100 BioAnalyzer (Agilent Technologies Inc., Germany), according to the manufacturer’s protocol. For each condition, three biological repeats with the highest RNA integrity values and minimal intrarelative water content variations were chosen for RNA sequencing. Twenty-four single end (50 bp) bar-coded cDNA libraries were prepared for multiplex sequencing on the Illumina HiSeq2500 sequencer (Technion Genome Center, Haifa, Israel) using the TruSeq RNA sample preparation kit ver.2 (Illumina, San Diego, CA), according to manufacturer’s standard protocols.
Data Processing and Analysis
FastQ files, generated from raw data obtained from Illumina sequencing, were processed and filtered with the following publicly available tools. Quality evaluation of each sample was manually inspected using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Barcode removal and filtering of low quality reads were executed using the command line tools cutadapt (https://cutadapt.readthedocs.io/en/stable/; -e 0.1, -O 5, -m 15) and FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit; fastq_quality_trimmer: -l 35, -t 28, fastq_quality_filter: -q 20, -p 90), respectively. Sample sequences were aligned to the B. distachyon reference genome (Bd21-3 v.1; http://plants.ensembl.org) using the Bowtie2 aligner (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) within TopHat2 (http://ccb.jhu.edu/software/tophat/index.shtml; -i 20 -I 10000; Kim et al., 2013), producing on average a 24-fold (± 6.2 SE) sequence coverage. The Bioconductor package GenomicFeatures was used to retrieve transcript-related features from B. distachyon GTF file (ftp://ftp.ensemblgenomes.org/pub/release-31/plants/gtf/brachypodium_distachyon), and a read count table of all samples was produced with the Bioconductor package GenomicAlignments (Lawrence et al., 2013). Differential expression analysis of count data and data visualization were conducted with the DESeq2 package (Love et al., 2014). We used the function rlogTransform to transform the data to regularized logarithm (rlog) values. This transformation is similar to a log2 transformation for genes with high counts, while for genes with low counts, it shrinks together the values toward the genes’ averages across all samples (Love et al., 2014). A PCA was performed to explore the transcriptome and visualize sample-to-sample distances. To detect DEGs, a 5% false discovery rate (FDR) correction for multiple comparisons was determined (Benjamini and Hochberg, 1995), and a minimal 0.5 log2FC threshold was applied. These thresholds resulted in detection of 13,580 genes that were differentially regulated in at least one stress treatment. Venn diagrams were created using the publicly available software Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn). Functional annotation of DEGs to biological processes and pathways was conducted with MapMan tool (http://mapman.gabipd.org/web/guest/mapman), which allows genes to be tentatively assigned, even when their function is only roughly known (Thimm et al., 2004). Pathway enrichment analysis was performed with the functional enrichment analysis software tool FunRich (FDR ≤ 0.05; http://www.funrich.org; Pathan et al., 2015), using MapMan annotation as reference data.
Expression Patterns of Common Stress and Combination Unique DEGs
A seven-way Venn diagram that included significant DEGs within each stress treatment was used to identify DEGs common to at least two single stresses. These genes were defined as common stress DEGs. Stress combination unique DEGs were detected by first subtracting DEGs common between each combined stress and its corresponding single stresses. Next, a four-way Venn diagram, which included the unique DEG lists of S&D, S&H, D&H, and S&D&H, was used to identify DEGs common to at least two double-stress combinations. All together 1,550 and 2,561 genes were identified as common stress and combination unique DEGs, respectively. To determine gene expression patterns among the two gene subsets, log2FC values of nonsignificant transcripts (FDR > 0.05), were replaced with zeros, assuming minimal effect of random FC variation. Likewise, log2FC values < 0.5 were replaced with zeros, to minimize background noise. log2FC values of each gene, under the different treatments, were used to assign genes to six predefined transcriptional modes: additive, synergistic, neutral, dominant, antagonistic, and equalization sds (sd) of stress combination log2FC values were used to differentiate between modes. Common stress DEGs yielded four expression patterns that were generated by comparing between FC values of triple- or double-stress combinations and the corresponding single stresses. Combination unique DEGs resulted in one expression pattern, generated by comparing between FC values of the triple-stress combination and double-stress combinations. For simplicity, definition of each response mode is explained using the common stress DEGs. Transcripts assigned to additive, synergistic, or neutral modes were either up- or down-regulated among the single and combined stresses (i.e. similar expression pattern). Additive and synergistic modes included transcripts with stress combination log2FC values (± sd) equal to or higher than log2FC summation of the corresponding single stresses, respectively. Neutral mode included transcripts that maintained similar log2FC values (± sd of stress combination) among single and combined stresses. Transcripts with opposite expression patterns (e.g. up-regulated in one treatment and down-regulated in another treatment) were assigned to antagonistic or equalization modes. Antagonistic mode included transcripts with opposite log2FC values or control levels under stress combinations compared with the corresponding single stresses, whereas equalization mode included transcripts that their log2FC value under stress combination was equivalent (± sd) to the summation of log2FC of the corresponding single stresses. Dominant mode comprised of transcripts from both similar and opposite expression patterns that their log2FC value under single stress(es) was equal to log2FC value under the stress combination (± sd). Transcripts that did not match the aforementioned modes were assigned to not assigned (NA) mode. Circos online tool (http://circos.ca; Krzywinski et al., 2009) was used to visualize mode consistency across the four patterns of common stress DEGs.
Weighted Gene Coexpression Network Analysis
Weighted gene coexpression network analysis (WGCNA) was conducted based on Langfelder and Horvath (2008) and the WGCNA package R scripts available at (https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/index.html). Coexpression networks were constructed based on common stress and combination unique DEGs. RNA-seq count data of these genes was rlog-transformed using the function rlogTransform, which is implemented in DESeq2 package (Love et al., 2014). These data were used to define a coexpression similarity matrix using the absolute values of Pearson correlation coefficient between pairwise gene expression profiles. The coexpression similarity was transformed into connection strengths (i.e. adjacency matrix) by raising values to a power of 14 and 13, for common stress and combination unique DEGs, respectively, which best approximate scale-free topologies (model fit > 0.8; Supplemental Fig. S13, A and B). Gene sets with expression pattern that were highly correlated across samples (i.e. modules) were determined based on topological overlap dissimilarity measure in conjunction with linkage hierarchical clustering using the blockwise Modules function. Similar transcripts were merged into modules based on a height cut of 0.15 with a minimum module size of 25 genes. Module eigengene that best represents module expression pattern and gene significance, the correlation between gene expression, and RWC or AO, were used to define quantitative measures of module membership (P ≤ 0.05). Intramodular connectivity was subsequently used to identify module hub genes. Modules, significantly correlated with physiological traits (r ≥ 0.6, P ≤ 0.05), were exported for visualization in Cytoscape (http://www.cytoscape.org; Shannon et al., 2003) using the exportNetworkToCytoscape function, implemented in WGCNA and adjacency thresholds of 0.15 and 0.25 for common stress and combination unique DEGS, respectively. Within Cytoscape, networks were constructed using Spring Embedded layout method. Final figures generated based on Cytoscape edge thresholds did not include all genes and their connectivities as detected by WGCNA. However, gene annotations based on MapMan bin allocation and enrichment analysis performed with FunRich software comprised all genes within modules that were significantly correlated with physiological traits.
Quantitative PCR
First strand cDNA, of RNA extracted from leaf samples (n = 6), was synthesized using qScript cDNA Synthesis Kit (Quanta Biosciences Inc.) following the manufacturer’s instructions. qPCR was carried out using PerfeCTa SYBR Green FastMix (Quanta Biosciences Inc.) on PikoReal RT-PCR system (Thermo Fisher Scientific Inc.). Gene-specific primers were designed using Primer-BLAST software (Ye et al., 2012; Supplemental Table S8). An efficiency value of 100% ± 10% was confirmed for each set of primers, and 100% efficiency was assumed upon calculating transcript expression levels. Template cDNA was diluted by 16-fold, based on a standard curve of four serial dilution points. The 2-ΔΔCT method (Livak and Schmittgen, 2001) was used to normalize and calibrate transcript values relative to the housekeeping gene S-adenosyl-Met decarboxylase (SamDC, BRADI2G02580; Hong et al., 2008) that was highly expressed and showed minimal variation across control and stress conditions. Results are presented as log2FC.
Statistical Analyses
Physiological measurements and qPCR assay were analyzed statistically using JMP pro 12 statistical package (SAS Institute, Cary, NC). Bartlett’s test was used to examine homoscedasticity among treatments. PCA, based on FC values of genes assigned to photosynthesis or stress responses, was used to identify minimal number of principal components that accounted for most of the gene expression variation. Associations between physiological traits and first and second principle components (eigenvalues > 1), as well as among FC values within each treatment, were studied using Pearson correlation analysis. Differences between control and stress treatments within qPCR assay and morpho-physiological measurements were detected using one-way ANOVA followed by Dunnet’s test at P ≤ 0.05. Heat-maps based on hierarchical clustering of morpho-physiological traits were conducted using Ward’s method. Differences in mode probabilities across and between response patterns were determined with an χ2 test at P ≤ 0.05.
Accession Numbers
Raw sequencing files of mRNA sequencing are available at the short read archive of the National Center for Biotechnology Information (https://trace.ncbi.nlm.nih.gov/Traces/sra) under accession number PRJNA360513.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Plant biomass production among three independent stress assays.
Supplemental Figure S2. Characterization of morpho-physiological traits.
Supplemental Figure S3. Effects of single and combined stresses on plant development 17 d after anthesis.
Supplemental Figure S4. Intersections among 1,550 common stress DEGs across single and combined stresses.
Supplemental Figure S5. Response mode partition of common stress DEGs among the four expression patterns of triple- and double-stress combinations.
Supplemental Figure S6. Intersections among 2,561 stress combination unique DEGs across combined stresses.
Supplemental Figure S7. Enriched biological pathways found among common stress (outer) and combination unique (inner) DEGs.
Supplemental Figure S8. Relationships among eigengenes of (A) common stress modules (B) and stress combination unique modules.
Supplemental Figure S9. Enriched response modes among (A) common stress modules and (B) stress combination unique modules detected by coexpression network analysis.
Supplemental Figure S10. Module-trait relationships among (A) common stress modules and (B) stress combination unique modules.
Supplemental Figure S11. Correlation between module membership and gene significance among (A) common stress modules and (B) stress combination unique-modules that significantly correlated with relative water content osmotic adjustment.
Supplemental Figure S12. Relative expression of selected (A) common stress DEGs and (B) stress combination unique DEGs based on qPCR validation and RNA-seq analysis.
Supplemental Figure S13. Evaluation of scale free topology of coexpression networks that were constructed based on (A) common stress DEGs and (B) stress combination unique DEGs.
Supplemental Table S1. Significantly differentially expressed genes in at least one stress treatment and their annotations based on MapMan tool.
Supplemental Table S2. Classification of common stress DEGs to response modes.
Supplemental Table S3. Classification of stress combination unique DEGs to response modes.
Supplemental Table S4. Common stress modules showing significant correlation with relative water content or osmotic adjustment.
Supplemental Table S5. Stress combination unique modules showing significant correlation with relative water content or osmotic adjustment.
Supplemental Table S6. Significantly enriched biological pathways within common stress modules.
Supplemental Table S7. Significantly enriched biological pathways within stress combination unique modules.
Supplemental Table S8. List of primers used for the qPCR assay.
Supplemental Table S9. Candidate chloroplast nucleoids associated-genes in module 02.
Supplementary Material
Acknowledgments
We thank U. Landau for help with script writing and advice, and N. Teboul and Oz Shaar-Moshe for the illustrations in Figure 1. We thank Drs. R. Hayuka, A. Perry, Y. Gadri, and A. Oksenberg for technical assistance with the experiments. L.S.-M. is indebted to the Israeli President’s Scholarship for Scientific Excellence and Innovation.
Footnotes
This research was partially supported by the United States-Israel Binational Science Foundation (BSF; grant no. 2011310 to Z.P. and E.B.) and the Hebrew University of Jerusalem.
Articles can be viewed without a subscription.
References
- Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523–3543 [DOI] [PubMed] [Google Scholar]
- Barnabás B, Jäger K, Fehér A (2008) The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ 31: 11–38 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bradshaw AD. (1965) Evolutionary significance of phenotypic plasticity in plants. Adv Genet 13: 115–155 [Google Scholar]
- Cabello JV, Lodeyro AF, Zurbriggen MD (2014) Novel perspectives for the engineering of abiotic stress tolerance in plants. Curr Opin Biotechnol 26: 62–70 [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Lasky JR, Verslues PE, Chang TZ, Juenger TE (2017) Interactive effects of water limitation and elevated temperature on the physiology, development and fitness of diverse accessions of Brachypodium distachyon. New Phytol 214: 132–144 [DOI] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M. (2013) Homeostasis: an underestimated focal point of ecology and evolution. Plant Sci 211: 92–101 [DOI] [PubMed] [Google Scholar]
- Hong SY, Park JH, Cho SH, Yang MS, Park CM (2011) Phenological growth stages of Brachypodium distachyon: codification and description. Weed Res 51: 612–620 [Google Scholar]
- Hong SY, Seo PJ, Yang MS, Xiang F, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N (2006) An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J 46: 307–316 [DOI] [PubMed] [Google Scholar]
- Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63: 1593–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Mladek C, Popova O, Kiok K, Berlinger M, Rakic B, Aufsatz W, Jonak C, Hauser MT, Luschnig C (2010) Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol Plant 3: 594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ (2013) Software for computing and annotating genomic ranges. PLOS Comput Biol 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shimada T, Takahashi H, Koumoto Y, Shirakawa M, Takagi J, Zhao X, Tu B, Jin H, Shen Z, Han B, Jia M, et al. (2013) MAG2 and three MAG2-INTERACTING PROTEINs form an ER-localized complex to facilitate storage protein transport in Arabidopsis thaliana. Plant J 76: 781–791 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loss SP, Siddique KHM (1994) Morphological and physiological traits associated with wheat yield increases in Mediterranean environments. Adv Agron 52: 229–276 [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv DW, Subburaj S, Cao M, Yan X, Li X, Appels R, Sun DF, Ma W, Yan YM (2014) Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol Cell Proteomics 13: 632–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ (2012) Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SS, Smith MR, Guth S, Golden CD, Vaitla B, Mueller ND, Dangour AD, Huybers P (2017) Climate change and global food systems: Potential impacts on food security and undernutrition. Annu Rev Public Health (in press) 10.1146/annurev-publhealth-031816-044356 [DOI] [PubMed] [Google Scholar]
- Melonek J, Oetke S, Krupinska K (2016) Multifunctionality of plastid nucleoids as revealed by proteome analyses. Biochim Biophys Acta 1864: 1016–1038 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11: 15–19 [DOI] [PubMed] [Google Scholar]
- Mittler R, Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol 61: 443–462 [DOI] [PubMed] [Google Scholar]
- Munns R. (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239–250 [DOI] [PubMed] [Google Scholar]
- Munns R, Gilliham M (2015) Salinity tolerance of crops - what is the cost? New Phytol 208: 668–673 [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F, van Kleunen M (2010) Plant phenotypic plasticity in a changing climate. Trends Plant Sci 15: 684–692 [DOI] [PubMed] [Google Scholar]
- Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, Bacic A, Hill AF, et al. (2015) FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 15: 2597–2601 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z, Apse MP, Blumwald E (2011) Engineering salinity and water-stress tolerance in crop plants: getting closer to the field. Adv Bot Res 57: 405–443 [Google Scholar]
- Peleg Z, Fahima T, Abbo S, Krugman T, Nevo E, Yakir D, Saranga Y (2005) Genetic diversity for drought resistance in wild emmer wheat and its ecogeographical associations. Plant Cell Environ 28: 176–191 [Google Scholar]
- Prasch CM, Sonnewald U (2013) Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol 162: 1849–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch CM, Sonnewald U (2015) Signaling events in plants: Stress factors in combination change the picture. Environ Exp Bot 114: 4–14 [Google Scholar]
- Priest HD, Fox SE, Rowley ER, Murray JR, Michael TP, Mockler TC (2014) Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS One 9: e87499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera M, Peleg Z, Blumwald E (2012) Targeting metabolic pathways for genetic engineering abiotic stress-tolerance in crops. Biochim Biophys Acta (BBA)-. Gene Regulatory Mechanisms 1819: 186–194 [DOI] [PubMed] [Google Scholar]
- Reynolds MP, Quilligan E, Aggarwal PK, Bansal KC, Cavalieri AJ, Chapman SC, Chapotin SM, Datta SK, Duveiller E, Gill KS, Jagadish KSV, Joshi AK, et al. (2016) An integrated approach to maintaining cereal productivity under climate change. Glob Food Secur 8: 9–18 [Google Scholar]
- Rivero RM, Mestre TC, Mittler R, Rubio F, Garcia-Sanchez F, Martinez V (2014) The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ 37: 1059–1073 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Takano H, Kuroiwa T (2004) Organelle nuclei in higher plants: structure, composition, function, and evolution. Int Rev Cytol 238: 59–118 [DOI] [PubMed] [Google Scholar]
- Sewelam N, Oshima Y, Mitsuda N, Ohme-Takagi M (2014) A step towards understanding plant responses to multiple environmental stresses: a genome-wide study. Plant Cell Environ 37: 2024–2035 [DOI] [PubMed] [Google Scholar]
- Shaar-Moshe L, Hübner S, Peleg Z (2015) Identification of conserved drought-adaptive genes using a cross-species meta-analysis approach. BMC Plant Biol 15: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillmann J, Kharin VV, Zwiers FW, Zhang X, Bronaugh D (2013) Climate extremes indices in the CMIP5 multimodel ensemble: Part 2. Future climate projections. J Geophys Res Atmos 118: 2473–2493 [Google Scholar]
- Suzuki N, Bassil E, Hamilton JS, Inupakutika MA, Zandalinas SI, Tripathy D, Luo Y, Dion E, Fukui G, Kumazaki A, Nakano R, Rivero RM, et al. (2016) ABA is required for plant acclimation to a combination of salt and heat stress. PLoS One 11: e0147625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014) Abiotic and biotic stress combinations. New Phytol 203: 32–43 [DOI] [PubMed] [Google Scholar]
- Tattini M, Loreto F, Fini A, Guidi L, Brunetti C, Velikova V, Gori A, Ferrini F (2015) Isoprenoids and phenylpropanoids are part of the antioxidant defense orchestrated daily by drought-stressed Platanus × acerifolia plants during Mediterranean summers. New Phytol 207: 613–626 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Verelst W, Bertolini E, De Bodt S, Vandepoele K, Demeulenaere M, Pè ME, Inzé D (2013) Molecular and physiological analysis of growth-limiting drought stress in Brachypodium distachyon leaves. Mol Plant 6: 311–322 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Leong OM, Hayden DM (2006) Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ Cult 84: 199–211 [Google Scholar]
- Volis S, Mendlinger S, Ward D (2002) Differentiation in populations of Hordeum spontaneum Koch along a gradient of environmental productivity and predictability: plasticity in response to water and nutrient stress. Biol J Linn Soc Lond 75: 301–312 [Google Scholar]
- Wagner TA, Kohorn BD (2001) Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13: 303–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61: 199–223 [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandalinas SI, Mittler R, Balfagón D, Arbona V, Gómez-Cadenas A (2017) Plant adaptations to the combination of drought and high temperatures. Physiol Plant (in press) 10.1111/ppl.12540 [DOI] [PubMed] [Google Scholar]
- Zhu J-K. (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.