The immune regulator pipecolic acid is synthesized by ALD1-mediated conversion of l-Lys to 2,3-dehydropipecolic acid and consecutive reduction, to which Arabidopsis SARD4/ORNCD1 largely contributes.
Abstract
The nonprotein amino acid pipecolic acid (Pip) regulates plant systemic acquired resistance and basal immunity to bacterial pathogen infection. In Arabidopsis (Arabidopsis thaliana), the lysine (Lys) aminotransferase AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1) mediates the pathogen-induced accumulation of Pip in inoculated and distal leaf tissue. Here, we show that ALD1 transfers the α-amino group of l-Lys to acceptor oxoacids. Combined mass spectrometric and infrared spectroscopic analyses of in vitro assays and plant extracts indicate that the final product of the ALD1-catalyzed reaction is enaminic 2,3-dehydropipecolic acid (DP), whose formation involves consecutive transamination, cyclization, and isomerization steps. Besides l-Lys, recombinant ALD1 transaminates l-methionine, l-leucine, diaminopimelate, and several other amino acids to generate oxoacids or derived products in vitro. However, detailed in planta analyses suggest that the biosynthesis of 2,3-DP from l-Lys is the major in vivo function of ALD1. Since ald1 mutant plants are able to convert exogenous 2,3-DP into Pip, their Pip deficiency relies on the inability to form the 2,3-DP intermediate. The Arabidopsis reductase ornithine cyclodeaminase/μ-crystallin, alias SYSTEMIC ACQUIRED RESISTANCE-DEFICIENT4 (SARD4), converts ALD1-generated 2,3-DP into Pip in vitro. SARD4 significantly contributes to the production of Pip in pathogen-inoculated leaves but is not the exclusive reducing enzyme involved in Pip biosynthesis. Functional SARD4 is required for proper basal immunity to the bacterial pathogen Pseudomonas syringae. Although SARD4 knockout plants show greatly reduced accumulation of Pip in leaves distal to P. syringae inoculation, they display a considerable systemic acquired resistance response. This suggests a triggering function of locally accumulating Pip for systemic resistance induction.
Plants are able to recognize conserved molecular patterns of potentially pathogenic microorganisms and, thereupon, induce a basal immune program that is generally designated as pattern-triggered immunity or basal resistance (Macho and Zipfel, 2014). Basal resistance is able to attenuate the invasive spread of virulent pathogens in plant tissue but often is not strong enough to fully prevent disease development (Spoel and Dong, 2012). However, plants can reinforce their defensive capacities to virulent pathogen invasion after a previous microbial attack. A precedent localized leaf infection can thus activate a state of enhanced immunity to biotrophic and hemibiotrophic pathogens systemically in the entire foliage, a resistance phenomenon termed systemic acquired resistance (SAR; Fu and Dong, 2013).
Plants with activated SAR have undergone massive transcriptional and metabolic reprogramming at the systemic level and are primed for more rapid and effective defense activation during pathogen challenge (Jung et al., 2009; Návarová et al., 2012; Gruner et al., 2013; Bernsdorff et al., 2016). In Arabidopsis (Arabidopsis thaliana), several genes required for proper SAR activation have been identified previously (for review, see Shah and Zeier, 2013). Two of the Arabidopsis genes implicitly required for SAR are AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1) and FLAVIN-DEPENDENT-MONOOXYGENASE1 (FMO1). Both genes are systemically up-regulated in the plant upon localized pathogen inoculation (Song et al., 2004b; Mishina and Zeier, 2006; Návarová et al., 2012).
Plant metabolites are important regulatory components of plant basal resistance and SAR (Zeier, 2013). While the oxylipin jasmonic acid primarily promotes plant defenses directed against necrotrophic pathogens and herbivorous insects (Memelink, 2009), the phenolic salicylic acid (SA) is a key regulator of plant basal immunity to biotrophic and hemibiotrophic pathogens (Vlot et al., 2009). Upon pathogen attack, chorismate-derived SA accumulates in free and glycosidic forms systemically in the foliage and induces the expression of a set of SAR-related genes (Wildermuth et al., 2001; Attaran et al., 2009). For more than two decades, SA has been considered an important mediator of SAR in plants (Vernooij et al., 1994; Nawrath and Métraux, 1999; Wildermuth et al., 2001).
More recently, our own studies have revealed that the Lys-derived nonprotein amino acid pipecolic acid (Pip) acts as a crucial regulator of plant SAR (Návarová et al., 2012; Vogel-Adghough et al., 2013; Zeier, 2013; Bernsdorff et al., 2016). Upon inoculation of Arabidopsis with the hemibiotrophic bacterial pathogen Pseudomonas syringae, Pip accumulates to high levels in the inoculated leaves and to considerable amounts also in leaves distal from the site of attack (Návarová et al., 2012). The biosynthesis of Pip is thereby strictly dependent on a functional ALD1 gene (Návarová et al., 2012), which encodes an aminotransferase with in vitro substrate preference for Lys (Song et al., 2004a). Pip-deficient ald1 mutant plants are fully defective in SAR, in the induced systemic priming phenomenon associated with SAR, and also exhibit reduced basal resistance toward bacterial infection (Song et al., 2004b; Návarová et al., 2012). The immune defects of ald1 can be rescued by exogenous Pip, demonstrating that Pip accumulation in the plant is necessary for SAR, defense priming, and a complete basal resistance program (Návarová et al., 2012). In addition, elevation of the in planta levels of Pip to physiologically relevant amounts by exogenous application is sufficient to induce SAR-like resistance, to amplify pathogen-induced defense responses such as SA biosynthesis and defense gene expression, and to establish a primed state in wild-type plants (Návarová et al., 2012). Pip requires a functional FMO1 gene to exert its defense-amplifying and resistance-enhancing activities, indicating that the flavin monooxygenase FMO1 acts downstream of Pip in SAR activation (Návarová et al., 2012; Bernsdorff et al., 2016). Moreover, SAR induction and the establishment of a primed defense state in plants proceed by SA-dependent and SA-independent signaling pathways that both require Pip accumulation and functional FMO1. In addition, Pip and SA act both synergistically and independently from each other in Arabidopsis basal resistance to P. syringae attack (Bernsdorff et al., 2016).
It has been known since the 1950s that l-Pip is widely distributed in angiosperms (Morrison, 1953; Zacharius et al., 1954; Broquist, 1991). Besides its P. syringae-inducible accumulation in Arabidopsis, Pip was found to be biosynthesized to high levels in response to bacterial, fungal, or viral infection in several monocotyledonous and dicotyledonous plant species, including rice (Oryza sativa), potato (Solanum tuberosum), tobacco (Nicotiana tabacum), and soybean (Glycine max; Pálfi and Dézsi, 1968; Vogel-Adghough et al., 2013; Aliferis et al., 2014). Moreover, Arabidopsis plants with constitutively activated defenses and autophagy mutants that exhibit stress-related phenotypes exhibit constitutively elevated Pip levels (Návarová et al., 2012; Masclaux-Daubresse et al., 2014). Exogenously applied Pip also increases the resistance of tobacco plants to P. syringae pv tabaci infection and primes tobacco for early SA accumulation (Vogel-Adghough et al., 2013). Furthermore, transgenic rice plants overexpressing the Pip biosynthesis gene ALD1 exhibited increased resistance toward infection by the fungus Magnaporthe oryzae (Jung et al., 2016). Together, these findings suggest a conserved regulatory role for Pip in plant immunity. In addition, Pip might contribute to the control of root nodulation in legumes, since Lotus japonicus ALD1 is strongly expressed in nodules induced by the symbiotic rhizobium Mesorhizobium loti. A proper function of the ALD1 gene proved necessary for normal nodule development and the containment of the number of bacterial infection threads (Chen et al., 2014).
l-Pip is an endogenous metabolite in both plants and animals (Broquist, 1991). Soon after Pip was first identified as a plant natural product (Zacharius et al., 1954), its biosynthetic origin from l-Lys was uncovered by the finding that rats metabolize 14C-radiolabeled l-Lys to Pip (Rothstein and Miller, 1954). Notably, when administering Lys labeled with the heavy nitrogen isotope 15N at either the α- or ε-amino group, Rothstein and Miller (1954) found that the α-NH2 group was abstracted and the ε-NH2 group was retained in the conversion of Lys to Pip. These experiments supported the hypothesis that the metabolic pathway from Lys to Pip in mammals includes an aminotransferase reaction to the α-ketoacid ε-amino-α-ketocaproic acid (KAC), which is in chemical equilibrium with the dehydrated, cyclized ketimine compound Δ1-piperideine-2-carboxylic acid, alias 1,2-dehydropipecolic acid (DP). A subsequent reduction of the imine bond in the 1,2-DP molecule would then be required to generate l-Pip. Later, partially purified enzyme preparations from different mammalian tissues with imine reductase activities toward several cyclic ketimines and the ability to convert 1,2-DP to Pip were obtained (Meister and Buckley, 1957; Nardini et al., 1988; Hallen et al., 2013). More recently, the mammalian protein µ-crystallin (CRYM) was characterized as an NAD(P)H-dependent ketimine reductase that is able to reduce 1,2-DP to the supposed product l-Pip (Hallen et al., 2011, 2015).
Feeding studies with radioisotope-labeled Lys also showed that, like animals, plants synthesize l-Pip from Lys (Schütte and Seelig, 1967; Gupta and Spenser, 1969; Fujioka and Sakurai, 1997). However, contradictory results were reported with respect to the biochemical mechanism of the Lys-to-Pip conversion in plants. For example, Schütte and Seelig (1967) administered α-15NH2- and ε-15NH2-labeled Lys variants to bean (Phaseolus vulgaris) plants and observed a preferred incorporation of the α-amino group into Pip. This would favor a pathway that includes the formation of α-aminoadipate semialdehyde (AAS) and the cyclized imine Δ1-piperideine-6-carboxylic acid, alias 1,6-DP. AAS and 1,6-DP have been described as biosynthetic intermediates of another Lys-derived amino acid, α-amino adipic acid (Aad; Galili et al., 2001; Zeier, 2013). By contrast, after feeding δ-3H-,6-14C-radiolabeled Lys to bean plants, Sedum acre plants, or intact rats, Gupta and Spenser (1969) found that the 3H-14C ratios of the administered Lys precursor and the Pip metabolic product were similar, consistent with the assumption that, in plants and animals, the ε-amino group of Lys is incorporated into Pip via the formation of KAC and 1,2-DP.
The results of the above-described isotope-tracer studies strongly suggest that the first step in the Pip biosynthetic pathway represents a transamination of l-Lys. Since the above-mentioned, SAR-essential ALD1 protein was characterized as a plant aminotransferase with a substrate preference for l-Lys (Song et al., 2004a), and because our own findings demonstrated that functional ALD1 is necessary for Pip biosynthesis (Návarová et al., 2012), we previously suggested that ALD1 catalyzes this first transamination step in the biosynthesis of Pip from l-Lys (Návarová et al., 2012; Zeier, 2013). The presumed ALD1-derived DP product could then be reduced to Pip in a second step (Zeier, 2013), since ketimine reductase activities similar to those in animals have been described (Meister and Buckley, 1957). In addition, an Arabidopsis gene initially annotated as ORNITHINE CYCLODEAMINASE/μ-CRYSTALLIN (ORNCD1) and later designated as SAR-DEFICIENT4 (SARD4) shows high sequence similarity to the mammalian ketimine reductase CRYM (Zeier, 2013; Ding et al., 2016).
In this work, we show that ALD1 directly transfers the α-amino group of l-Lys to acceptor oxoacids such as pyruvate or α-ketoglutarate. Both in vitro assays with recombinant ALD1 protein and in planta studies with wild-type and ald1 mutant plants suggest that the end product of the ALD1-catalyzed transamination of l-Lys is the enamine Δ2-piperideine-2-carboxylic acid, alias 2,3-DP. A plausible mechanism of 2,3-DP formation includes ALD1-mediated transamination of l-Lys to KAC, dehydrative cyclization to 1,2-DP, and isomerization to 2,3-DP. Although ALD1 is able to accept several other amino acid substrates to generate corresponding transamination products in vitro, in planta analyses suggest that the dominant function for ALD1 in vivo is the conversion of l-Lys to 2,3-DP. Our findings also indicate that Arabidopsis SARD4 reduces DP intermediates to Pip in vitro and in planta. The accumulation of Pip in sard4 knockout plants is significantly attenuated but still considerable in P. syringae-inoculated leaves and is markedly reduced in the distal leaf tissue. Bacterial growth assays indicate that SARD4-generated Pip contributes to basal resistance to P. syringae infection and to the resistance level of SAR-induced plants. Nevertheless, a wild-type-like resistance increase is observed after SAR induction in sard4 knockout plants. These findings refine our previous model of SAR establishment, because they suggest that locally accumulating Pip substantially contributes to resistance induction in the systemic tissue.
RESULTS
The bacterial strain Pseudomonas syringae pv maculicola (Psm) robustly activates SAR in wild-type Arabidopsis plants (Mishina and Zeier, 2007). Two days after an inducing inoculation with Psm in lower (1°) leaves has occurred, SAR manifests itself by the accumulation of the immune signals Pip and SA both in inoculated (1°) and in upper, distal (2°) leaves and protects plants against subsequent pathogen challenge (Attaran et al., 2009; Návarová et al., 2012; Bernsdorff et al., 2016).
ALD1-Mediated l-Lys Conversion: Abstraction of the α-NH2 Group Leads to the Formation of Enaminic 2,3-DP in Vitro and in Planta
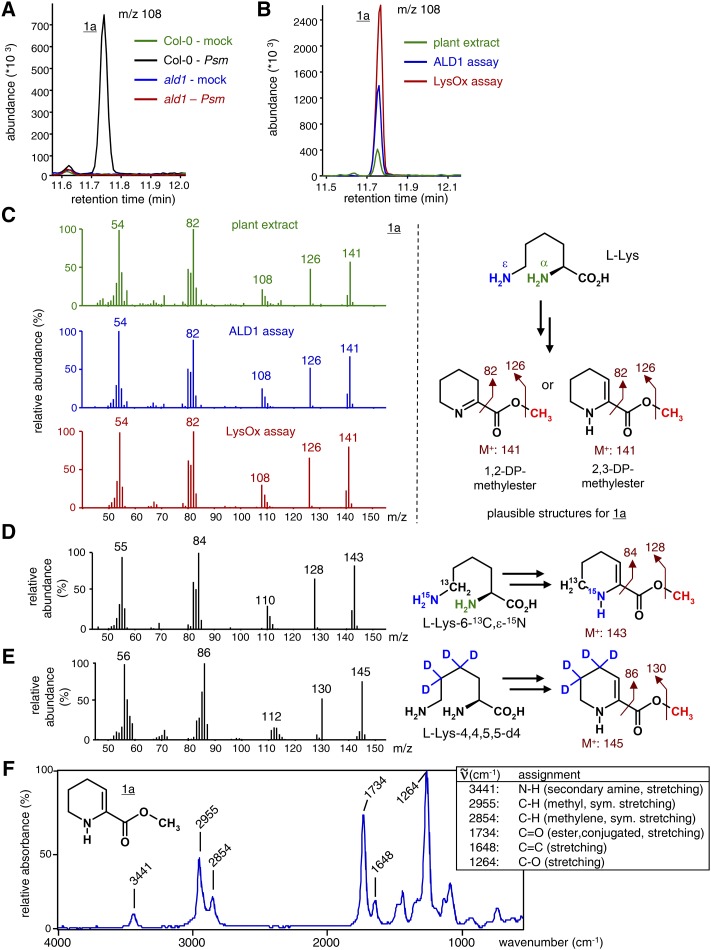
To experimentally verify the proposed scheme of Pip biosynthesis and identify possible ALD1-derived pathway intermediates (Zeier, 2013), we first performed comparative gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling of leaf extracts from Psm-inoculated and mock-treated wild-type Columbia-0 (Col-0) and ald1 mutant leaves. The applied analytical procedure (procedure A) includes a final derivatization step that converts free carboxylic acids into their methyl esters to optimize gas chromatographic separation (Schmelz et al., 2004; Mishina and Zeier, 2006). By comparatively analyzing individual ion chromatograms (mass-to-charge ratio [m/z] between 50 and 300) of extract samples from leaves of Psm-inoculated Col-0, Psm-inoculated ald1, mock-treated Col-0, and mock-treated ald1 plants, we identified a molecular species (1a) that was present in the samples from Psm-treated Col-0 leaves but barely detectable in the other samples (Fig. 1A). Hence, the plant compound associated with this substance peak accumulated in the Col-0 wild type but not in the ald1 mutant upon pathogen treatment, which parallels the accumulation characteristics of Pip (Návarová et al., 2012).
Figure 1.
ALD1-mediated l-Lys conversion: formation of 2,3-DP by abstraction of the α-NH2 group of l-Lys. The GC-MS (and GC-FTIR) analyses of plant extracts or assay samples included the methylation of carboxyl groups as a derivatization strategy (procedure A). A, Segment of overlaid ion chromatograms (m/z = 108) of GC-MS-analyzed extracts from mock- or Psm-inoculated leaves of Col-0 wild-type and ald1 mutant plants. Leaf samples were harvested 48 h post inoculation (hpi). A molecular species (1a) with m/z 108 (or m/z 126 or 141) is present exclusively in the Col-0-Psm samples. B, Segment of overlaid ion chromatograms (m/z = 108) of GC-MS-analyzed l-Lys conversion assays with ALD1 protein, l-Lys conversion assays with LysOx in the presence of catalase (Supplemental Fig. S2), and leaf extracts from Psm-inoculated Col-0 plants. Retention times of the molecular species with m/z 108 (or m/z 126 or 141) in the different samples are identical. C, At left, the mass spectra of the compound 1a derived from extracts of Psm-inoculated Col-0 plants (green), ALD1 in vitro assays (blue), and LysOx/catalase in vitro assays (red) are identical. At right, chemical structures of l-Lys and plausible structures for 1a, as deduced from the mass spectrometric information collected so far. The molecular ion (M+) and plausible ion fragments are indicated. The methyl group (red) is introduced by derivatization. D, The use of l-Lys-6-13C,ε-15N as a substrate in the ALD1 in vitro assay reveals retention of the ε-nitrogen and abstraction of the α-nitrogen in the transamination reaction leading to DP products. At left, a mass spectrum of the isotope-labeled product with shifts in fragmentation pattern by 2 mass units compared with unlabeled 1a. At right, isotope-labeled 2,3-DP-methylester is depicted as a plausible structure. The same result was observed for the LysOx/catalase assay. E, The use of l-Lys-4,4,5,5-d4 as a substrate in the ALD1 in vitro assay excludes the formation of DP isomers with double bonds in the 3,4-, 4,5-, or 5,6-position. At left, a mass spectrum of the isotope-labeled product with shifts in fragmentation pattern by 4 mass units compared with unlabeled 1a. At right, isotope-labeled 2,3-DP-methylester is depicted as a plausible structure. The same result was observed for the LysOx/catalase assay. F, GC-FTIR analysis of 1a indicates its enaminic structure, supports its identity as methylated 2,3-DP, and excludes the ketiminic 1,2-DP derivative as a possible structure. The IR spectrum of 1a is depicted (wave numbers from 4,000 to 600 cm−1). Assignments of IR absorption bands to functional groups are given in the box at right.
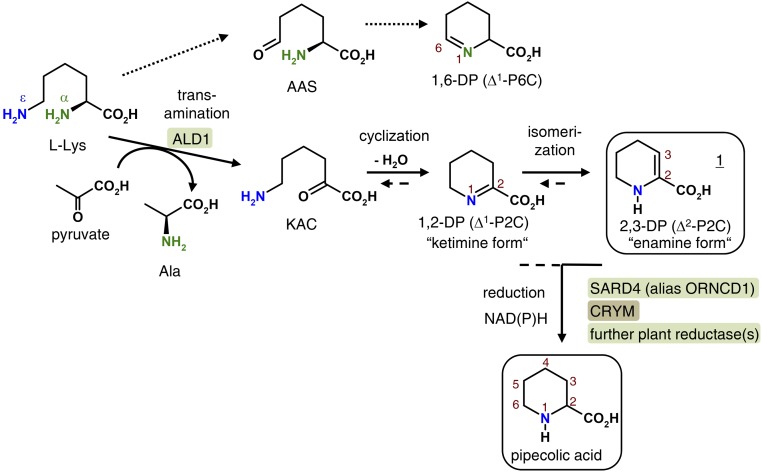
The mass spectrum of compound 1a showed a molecular ion of m/z 141 and a main fragment at m/z 82, which is characteristic for molecules with a 1-fold unsaturated piperideine ring (Fig. 1C). Overall, the mass spectral fragmentation pattern suggested that the chemical nature of 1a is the methyl ester of a piperideine carboxylic acid isoform (Fig. 1C), such as the ketimine 1,2-DP, the enamine 2,3-DP, or 1,6-DP (Fig. 2). 1,2-DP represents the previously proposed product of the ALD1-catalyzed l-Lys transamination (Zeier, 2013). The methyl group of 1a is thereby introduced after tissue extraction by derivatization, since 1a was not detected in nonderivatized samples.
Figure 2.
Summarized scheme of Pip biosynthesis from l-Lys in Arabidopsis. The biochemical pathway supported by the experimental data of this study is represented by arrows with solid lines. Detected Lys-derived compounds in extracts and in vitro assays are framed. Arrows with dashed lines represent likely biochemical scenarios, supported from literature findings. Arabidopsis ALD1 first catalyzes a transamination step that transfers the α-amino group of l-Lys to an acceptor oxoacid (preferentially pyruvate). Thereby, KAC and Ala are formed. A subsequent dehydrative cyclization of KAC produces the ketimine 1,2-DP, which isomerizes to the enamine 2,3-DP. 2,3-DP is detected in plant extracts and in vitro assays as the ALD1-derived product. Arabidopsis SARD4 (alias ORNCD1) then reduces a DP isomer to Pip. Since the human SARD4 homolog CRYM has been described as a ketimine reductase, it is possible that the reduction takes place via 1,2-DP, which is supposed to be in chemical equilibrium with 2,3-DP. Alternatively, a direct reduction of the detected intermediate 2,3-DP might take place. Since the sard4-5 knockout line is still able to biosynthesize (reduced amounts of) Pip, an alternative reductive mechanism capable of generating Pip from DP intermediates supposedly exists in plants. The hypothetical pathway illustrated with dotted arrows (top), proceeding via abstraction of the ε-amino group, and the formation of AAS and 1,6-DP can be ruled out on the basis of our data.
As a next step, recombinant ALD1 protein lacking the putative plastidial targeting sequence (Song et al., 2004a; Cecchini et al., 2015) and carrying a C-terminal His6 tag was overexpressed in Escherichia coli and then purified via immobilized metal ion affinity chromatography, followed by a desalting step to adjust the buffer conditions. l-Lys was tested in vitro as a substrate of recombinant ALD1 enzyme in the presence of various amino acceptors and pyridoxal-5′-phosphate (PLP) as a cofactor, using assay conditions similar to those employed previously by Song et al. (2004a; for details, see “Materials and Methods”). After stopping the reaction either by the addition of 0.1 m HCl or equal amounts of methanol or by heating of the reaction mixture (85°C for 10 min), substrates and ALD1 reaction products were derivatized to produce their respective methyl esters and analyzed by GC-MS as described before. In the presence of an oxoacid, such as pyruvate or α-ketoglutarate, ALD1 converted l-Lys to an in vitro reaction product whose GC retention time and mass spectrum were identical to the plant-derived substance 1a after derivatization (Fig. 1, B and C; Supplemental Fig. S1). Together, these data indicated that ALD1 catalyzes the conversion of l-Lys to the same DP isoform in vitro and in planta.
Next, the highly substrate-specific l-Lys α-oxidase (LysOx) from Trichoderma viride (Kusakabe et al., 1980) was used to generate 1,2-DP and compare it directly with the reaction product formed by ALD1 and the metabolite peak missing in ald1 plant extracts (Fig. 1, A–C). Like other members of the l-amino acid oxidases (EC 1.4.3.2.), LysOx from T. viridae is a flavoprotein with noncovalently bound FAD and catalyzes the stereospecific oxidative deamination of an l-amino acid, thereby producing ammonia and hydrogen peroxide as by-products (Lukasheva and Berezov, 2002; Pollegioni et al., 2013; Campillo-Brocal et al., 2015). The formal product of the oxygen-consuming and hydrogen peroxide-generating oxidation of the α-carbon atom of l-Lys is KAC, which is described to further cyclize to 1,2-DP if the reaction is conducted in the presence of catalase (Kusakabe et al., 1980; Supplemental Fig. S2). Biochemical assays with LysOx and l-Lys as substrates yielded, in the presence of catalase, a reaction product that was identical with 1a after derivatization (Fig. 1, B and C). Therefore, the ALD1 and LysOx/catalase biochemical assays both yielded the same Lys-derived piperideine carboxylic acid isoform as the one detected in plant extracts.
To further elucidate the biochemical mechanism and identify the end product of the ALD1-catalyzed Lys conversion reaction, we used isotope-labeled Lys precursors in the in vitro bioassays. Use of l-Lys-6-13C,ε-15N as an ALD1 substrate promised to clarify whether the transamination involves abstraction of the α- or ε-N and, thus, to discriminate between KAC and AAS as oxoacid intermediates and the DP cyclization products derived thereof (Fig. 2). With l-Lys-6-13C,ε-15N as the substrate, the product of the ALD1-catalyzed reaction showed a mass spectral fragmentation pattern that was incremented by 2 mass units compared with assays with unlabeled l-Lys, indicating that the ε-nitrogen is retained in the piperideine carboxylic acid product (Fig. 1D). This suggests that the ALD1-catalyzed aminotransferase reaction proceeds via abstraction of the α-amino group of Lys to form KAC, which, in turn, can cyclize to 1,2-DP; consequently, 1,6-DP is not the reaction product (Fig. 2). Furthermore, ketimine compounds can rearrange to enamine isomers (Nardini et al., 1988), and the ketimine 1,2-DP might in this way form enaminic 2,3-DP as the ultimate ALD1-derived product (Fig. 2). In any case, the so-far observed mass spectral fragmentation patterns are consistent with the structure of 1,2-DP, 2,3-DP, or, potentially, another piperideine-2-carboxylic acid isoform finally generated from KAC.
We then used tetradeuterated l-Lys-4,4,5,5-d4 as an ALD1 substrate in the in vitro assays. This yielded a product with a mass spectral fragmentation pattern that was incremented by 4 mass units compared with unlabeled l-Lys (Fig. 1E). This shift indicated that carbons 4 and 5 of the piperedeine ring are still saturated in the reaction product, essentially excluding that a piperideine-2-carboxylic acid isoform other than 1,2-DP or 2,3-DP was formed (Figs. 1E and 2). Notably, when l-Lys-6-13C,ε-15N or l-Lys-4,4,5,5-d4 was coapplied with Psm to Col-0 wild-type plants, we observed, in addition to substance 1a derived from endogenous Lys, the in planta formation of isotope-labeled versions of 1a that are identical to those detected in the ALD1 in vitro assay (Supplemental Fig. S3). This indicates that the mechanism of the ALD1-catalyzed reaction in vitro and in planta is the same.
To discriminate between the ketimine 1,2-DP and the enamine 2,3-DP as possible end products, we analyzed the ALD1 assay mixture with l-Lys as the substrate by gas chromatography-Fourier transform infrared (GC-FTIR) spectroscopy to obtain the infrared (IR) spectrum of 1a (Fig. 1F). If the substance was an enamine, the IR spectrum would exhibit an N-H stretching band at wave numbers of about 3,400 cm−1, whereas a spectrum of a ketiminic substance would not. A distinct band at 3,441 cm−1 in the IR spectrum of 1a demonstrated the presence of the N-H vibration (Fig. 1F), indicating, together with the above mass spectral information, that the substance is the methyl ester of the enamine 2,3-DP and not of the ketimine 1,2-DP. Another distinctive feature of the IR spectrum of 1a is a strong band at 1,734 cm−1, which is indicative of a carbonyl stretching vibration (Fig. 1F). The absorption of the C=O vibration in aliphatic carboxylic acid methyl esters such as methyl butyrate or methyl valerate occurs at approximately 1,760 cm−1 (Supplemental Fig. S4). The C=O band of α,β-unsaturated carbonyl compounds is usually shifted to lower wave numbers compared with the respective nonconjugated forms. For example, the carbonyl vibrations of methyl crotonate and methyl benzoate occur at 1,749 and 1,746 cm−1, respectively (Supplemental Fig. S4). The observed carbonyl absorption at 1,734 cm−1 of 1a, therefore, is consistent with the occurrence of an unsaturated ester group with a conjugated C=C double bond in the α,β-position, as is the case for 2,3-DP. Moreover, the medium absorption at 1,648 cm−1 can be assigned to the stretching vibration of the C=C double bond in the 2,3-DP methylester (Fig. 1F). In sum, the IR spectral analysis of substance 1a is consistent with the enaminic structure of 2,3-DP but not with the structure of the ketimine 1,2-DP. Together, our data indicate that the enamine 2,3-DP is the final end product of the ALD1-catalyzed conversion of l-Lys, both in vitro and in planta. We propose a plausible mechanism for 2,3-DP formation that includes consecutive transamination of l-Lys to KAC, dehydrative cyclization to 1,2-DP, and isomerization to 2,3-DP (Fig. 2).
Previous studies show that LysOx also catalyzes the formation of a DP dimer from l-Lys in the presence of catalase (Hope et al., 1967; Supplemental Fig. S2A). Indeed, when l-Lys was used as a substrate, we detected a substance with a mass spectrum identical to that of the spectroscopically well-characterized dimer in the LysOx/catalase assay (Supplemental Fig. S5; Hope et al., 1967). When using l-Lys-6-13C,ε-15N and l-Lys-4,4,5,5-d4 as substrates, the mass spectral fragmentation patterns of the isotope-labeled DP dimer variants were shifted by 4 and 8 mass units, respectively, consistent with a previous assumption that the DP dimer can be generated from l-Lys via KAC-derived DP monomers (Supplemental Fig. S5; Hope et al., 1967). In addition to 2,3-DP, formation of the DP dimer also was observed in l-Lys conversion assays with recombinant ALD1, but only if the reaction mixtures were incubated overnight (more than 12 h). Moreover, the DP dimer was not detected in plant extracts.
In the absence of catalase, LysOx catalyzed the formation of valerolactam from l-Lys (Supplemental Figs. S2A and S6). This is consistent with the previously reported LysOx-catalyzed generation of 5-aminovaleric acid from l-Lys (Kusakabe et al., 1980; Pukin et al., 2010), since 5-aminovaleric acid is able to spontaneously cyclize to valerolactam (Bird et al., 2012). The fragmentation patterns of the isotope-labeled valerolactam derivatives also suggest that, like 2,3-DP, valerolactam is produced from LysOx by abstraction of the α-NH2 group of Lys (Supplemental Fig. S6). 2,3-DP, the DP dimer, and valerolactam all exhibit a secondary amino group in their ring structures. Moreover, the dimer and 2,3-DP both contain enamine moieties. Consistent with these structural similarities, all three substances exhibit a very similar N-H stretching vibration around 3,430 to 3,440 cm−1 (Supplemental Fig. S7).
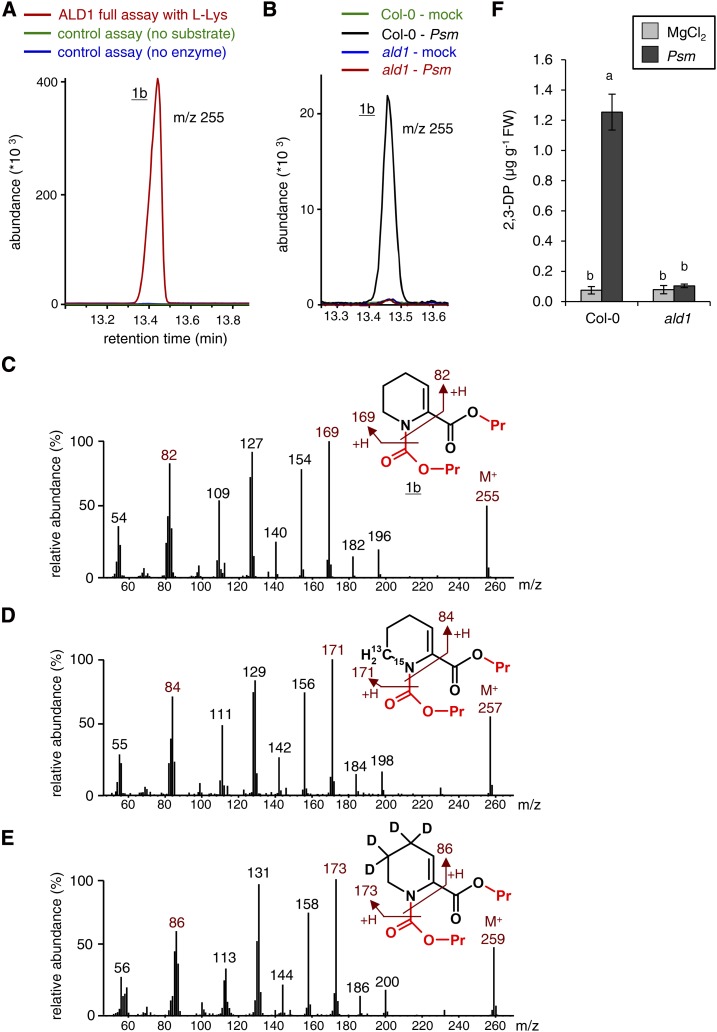
In addition to analytical procedure A, which includes a derivatization step to convert carboxylic acids into corresponding methyl esters (Schmelz et al., 2004; Mishina and Zeier, 2006), we applied an alternative procedure to prepare plant extracts and in vitro assays for GC-MS analysis. This method (procedure B) is based on propyl chloroformate derivatization, which converts amino groups into propyl carbamate and carboxyl groups into propyl ester derivatives (Kugler et al., 2006; Návarová et al., 2012). When applying analytical procedure B to l-Lys conversion assays with ALD1 or LysOx/catalase, we also identified a single product peak (1b) in the resulting GC-MS chromatograms (Fig. 3A). The substance corresponding to peak 1b accumulated in extract samples from Psm-inoculated wild-type leaves but not in samples derived from Psm-inoculated ald1 leaves or in samples from the leaves of mock-treated plants (Fig. 3, B and F). The mass spectrum of compound 1b exhibited a molecular ion of m/z 255 and a fragmentation pattern that was consistent with the structure of the enamine 2,3-DP, which is propyl carbamylated at its amino group and propyl esterified at its carboxyl group (Fig. 3C). When l-Lys-6-13C,ε-15N or l-Lys-4,4,5,5-d4 was used instead of l-Lys as the substrate in the ALD1 in vitro assays, or when the isotope-labeled l-Lys versions were coapplied with Psm to wild-type leaves, we detected the isotope-labeled variants of 1b (Fig. 3, D and E). For l-Lys-6-13C,ε-15N as the precursor, we found an increment in the mass spectral pattern of 2 mass units, whereas a shift of 4 mass units was observed for l-Lys-4,4,5,5-d4. As outlined before for methylated 1a, the now-observed labeling patterns are consistent with an ALD1-catalyzed deamination of l-Lys to KAC and the subsequent formation of 2,3-DP as an end product (Figs. 2 and 3, D and E). A derivatized 1,2-DP ketimine structure for the detected compound 1b can be excluded because the imine nitrogen is not prone to carbamylation, and a hypothetical propyl-esterified 1,2-DP product would generate a molecular ion of m/z 169 and not the observed M+ of m/z 255 in the mass spectrometric analysis.
Figure 3.
ALD1-mediated l-Lys conversion: formation of 2,3-DP as determined by GC-MS analysis using propyl chloroformate derivatization (procedure B). A, Segment of overlaid ion chromatograms (m/z = 255) of GC-MS-analyzed l-Lys conversion assays with ALD1 protein showing the propyl chloroformate-derivatized transamination product 1b. Red, ALD1 enzyme assay with l-Lys as an amino acid substrate and pyruvate as an acceptor oxoacid; green, control assay lacking ALD1 protein; blue, control assay lacking l-Lys as a substrate. B, Segment of overlaid ion chromatograms (m/z = 255) from extract samples of mock- or Psm-inoculated Col-0 wild-type and ald1 mutant leaves. Leaf samples were harvested 48 hpi. Substance 1b accumulates in the Col-0-Psm samples only. C, Mass spectrum and molecular structure of substance 1b derived from ALD1-mediated l-Lys conversion assays. The mass spectral information is consistent with a 2,3-DP derivative that is propyl carbamylated at its amino group and propyl esterified at its carboxyl group. The groups introduced by propyl chloroformate derivatization are labeled in red. The molecular ion (M+) and plausible ion fragments are indicated. D, The use of l-Lys-6-13C,ε-15N as a substrate in ALD1 in vitro assays results in the formation of an isotope-labeled variant of 1b with shifts in the mass spectral fragmentation pattern by 2 mass units compared with unlabeled 1b. E, The use of l-Lys-4,4,5,5-d4 as a substrate in ALD1 assays results in the formation of an isotope-labeled variant of 1b with shifts in the fragmentation pattern by 4 mass units compared with unlabeled 1b. F, Quantification of 2,3-DP in leaf extracts from Col-0 and ald1 plants. Leaves were mock (MgCl2) infiltrated or Psm inoculated and harvested at 48 hpi. Bars represent means ± sd of three biological replicate samples, each consisting of six leaves. FW, Fresh weight. A correction factor for the absolute quantification of 2,3-DP was estimated as described in “Materials and Methods.” Different letters above the bars denote statistically significant differences (P < 0.001, ANOVA and posthoc Tukey’s honestly significant difference [HSD] test).
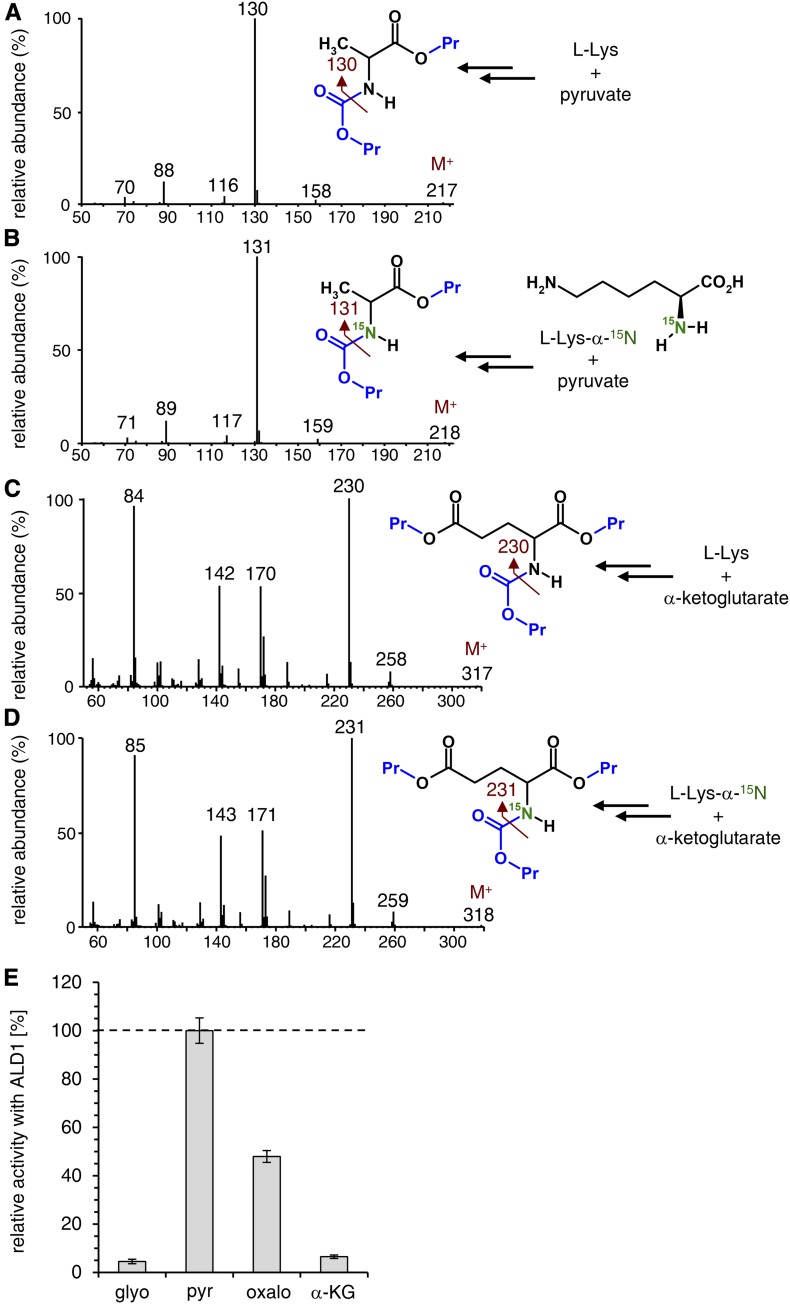
As a next step, we monitored the ALD1-mediated l-Lys transamination reaction from the perspective of the ketoacid that is supposed to accept the α-NH2 group of Lys to form the corresponding amino acid and KAC (Fig. 2). When pyruvate and α-ketoglutarate were used as amino group acceptors in the ALD1 bioassays, we observed, as expected, Ala and Glu as the product amino acids (Fig. 4, A and C). When l-Lys-α-15N was used as the amino acid substrate instead of l-Lys, α-15N-labeled Ala and Glu were formed, as indicated by shifts in the mass spectral patterns of the propyl chloroformate-derivatized amino acid products by 1 mass unit (Fig. 4, B and D). This directly shows that ALD1 transfers the α-NH2 group from l-Lys to acceptor ketoacids and confirms that the ε-NH2 group of l-Lys is retained in the ALD1-catalyzed aminotransferase reaction (Fig. 2). Moreover, we compared the relative activities of ALD1 to transaminate l-Lys in the presence of difference acceptor oxoacids, namely glyoxylate, pyruvate, oxaloacetate, and α-ketoglutarate (Fig. 4E). This comparison revealed that pyruvate is the preferred ketoacid (100% relative activity), followed by oxaloacetate (48%), α-ketoglutarate (6.5%), and glyoxylate (4.5%).
Figure 4.
ALD1 transfers the α-NH2 group from l-Lys to acceptor oxoacids. A, ALD1 in vitro assay with l-Lys as the substrate amino acid and pyruvate as the acceptor oxoacid results in the formation of Ala as the product amino acid. The mass spectrum of Ala after propyl chloroformate derivatization and the molecular structure of the product are shown. The M+ ion and the main fragment ion are indicated (dark red). The groups introduced by propyl chloroformate derivatization are labeled in blue. B, ALD1 assay with isotope-labeled l-Lys-α-15N as the amino acid substrate and pyruvate as the acceptor oxoacid results in the formation of α-15N-labeled Ala. For details, see A. C, ALD1 assay with l-Lys as the substrate amino acid and α-ketoglutarate as the acceptor oxoacid results in the formation of Glu as the product amino acid. For details, see A. D, ALD1 assay with isotope-labeled l-Lys-α-15N as the amino acid substrate and α-ketoglutarate as the acceptor oxoacid results in the formation of α-15N-labeled Glu. For details, see A. E, Relative activities of ALD1 toward l-Lys and glyoxylate (glyo), pyruvate (pyr), oxaloacetate (oxalo), or α-ketoglutarate (α-KG) as the acceptor oxoacid. The formation of the corresponding product amino acid (Gly for glyoxylate, Ala for pyruvate, Asp for oxaloacetate, and Glu for α-ketoglutarate) was determined after 30 min of incubation time for activity assessments.
ALD1 Catalyzes the Transamination of Various Amino Acids Other Than l-Lys in Vitro, But the Resulting Products Are Either Not Detectable in Plants or Not Relatable to an in Planta Function of ALD1
It has been shown previously that the ALD1 protein exerts in vitro aminotransferase activity toward several amino acids other than Lys in the presence of pyruvate or β-ketoglutarate as amino group acceptors (Song et al., 2004a). However, the resulting oxoacid or oxoacid-derived products of such alternative ALD1-catalyzed transamination reactions have not been characterized so far. Although it is now evident that the conversion of l-Lys to the Pip precursor 2,3-DP is a major in vivo function of ALD1 (Figs. 1–4; Návarová et al., 2012), it is not clear whether a possible activity of ALD1 toward other amino acids has biochemical and functional relevance in planta. To shed light on these unknown aspects of ALD1 biochemistry, we performed in vitro conversion assays with ALD1 protein and l-Lys or other possible amino acid substrates using pyruvate as the acceptor oxoacid (Table I). We assessed the in vitro activities of ALD1 toward the different amino acids by monitoring the formation of Ala from pyruvate. Moreover, we attempted to identify the corresponding oxoacid or oxoacid-derived in vitro reaction products by GC-MS analysis using either procedure A or procedure B (Table I).
Table I. ALD1 catalyzes the transamination of various amino acids other than l-Lys in vitro but these ALD1-mediated reactions are not detected in planta.
The in vitro activities of the ALD1 aminotransferase toward the indicated amino acids were assessed by incubation of ALD1 protein with the amino acid substrate and pyruvate as an acceptor oxoacid for 30 min. The molar concentrations of the product amino acid Ala and the given substrate amino acid were determined by GC-MS, and the percentage of transamination was calculated by forming the quotient [Ala]/([substrate amino acid] + [Ala]). The percentage values represent means ± sd of the values from three replicate assays. Relative in vitro activities were calculated from the transamination percentages by relating the mean transamination values to the one determined for l-Lys. The in vitro formation of product oxoacids or oxoacid-derived compounds was determined by the indicated GC-MS procedure. Retention times, molecular ions, and specific ion fragments in the mass spectra of the derivatized products are indicated. Underlined m/z values represent dominant masses in mass spectra, and ion chromatograms for the m/z values indicated in boldface have been used for substance quantification in planta. The molecular structures of several of the nonderivatized and derivatized products and the mass spectra of the latter are given in Figures 1C, 3C, and 5, C and D, as well as in Supplemental Figure S8. The detectability of oxoacid products in leaf extracts from Psm-inoculated or mock-treated Col-0 and ald1 plants (48 hpi) is indicated. The level of a detected product in leaf tissue is given in ng g−1 fresh weight (means ± sd; n = 3); nd, not detected. Asterisks denote statistically significant differences between Psm- and mock-treated samples of a genotype (P < 0.05, two-tailed Student’s t test). The results for additional tested amino acid substrates are summarized in Supplemental Table S1.
| Amino Acid | ALD1 in Vitro Assay |
Chemical Analyses |
In Planta Analyses |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Relative in Vitro Activity | Percentage Transamination (30 min) | Possible Product Oxoacid (or Derived Product) | Compound | In Vitro Detection of Product | GC-MS Procedure | Compound | M+ | Retention Time | Ion Fragments | In Planta Detection | Col-0 Mock | Col-0 Psm | ald1 Mock | ald1 Psm | |
| % | m/z | min | m/z | ||||||||||||
| l-Lys | 100 | 77.75 ± 6.58 | 2,3-DP | 1 | Yes | A | 1a | 141 | 11.6 | 141, 126, 108 | Yes | 41 ± 7 | 1,557 ± 240* | 39 ± 4 | 47 ± 9 |
| B | 1b | 255 | 13.8 | 255, 169, 127 | Yes | 75 ± 25 | 1,253 ± 119* | 79 ± 28 | 104 ± 12 | ||||||
| d-Lys | 0.1 | 0.05 ± 0.01 | 2,3-DP | 1 | No | A, B | – | – | – | – | – | – | – | – | |
| l,l-DAP | 16.3 | 12.64 ± 1.19 | 6-Carboxy-2,3-DP | 2 | Yes | A | 2a | 199 | 17.3 | 199, 140, 108 | No | nd | nd | nd | nd |
| meso-DAP | 12.9 | 10.01 ± 1.76 | Yes | B | 2b | 341 | 17.3 | 239, 168, 108 | No | nd | nd | nd | nd | ||
| l-Orn | 52.9 | 36.31 ± 8.89 | Δ2-Pyr2C | 3 | Yes | B | 3b | 241 | 13.0 | 241, 155, 113 | No | nd | nd | nd | nd |
| 2,4-DABA | 2.0 | 1.58 ± 0.21 | 4-Amino-2-oxo-butyric acid | No | B | – | – | – | – | – | – | – | – | ||
| l-Met | 78.5 | 61.04 ± 7.41 | α-Keto-Met | 4 | Yes | A | 4a | 162 | 12.0 | 162, 103, 61 | No | nd | nd | nd | nd |
| l-Leu | 57.0 | 44.34 ± 1.53 | α-Keto-Leu | 5 | Yes | A | 5a | 144 | 6.9 | 144,102, 85 | No | nd | nd | nd | nd |
| l-Ile | 0.5 | 0.36 ± 0.04 | α-Keto-Ile | No | A | 144 | 6.8 | 144, 85, 57 | No | nd | nd | nd | nd | ||
| l-Val | 0.3 | 0.23 ± 0.02 | α-Keto-Val | Yes | A | 130 | 4.9 | 130, 71, 59 | No | nd | nd | nd | nd | ||
| l-Phe | 10.6 | 8.25 ± 0.42 | Phenylpyruvic acid | 6 | Yes | A | 6a | 178 | 14.2 | 178, 119, 91 | Yes | 30 ± 8 | 79 ± 22* | 37 ± 6 | 87 ± 23* |
| l-Trp | 7.6 | 6.09 ± 0.63 | Indole-3-pyruvic acid | 7 | Yes | A | 7a | 217 | 23.3 | 217, 130, 77 | No | nd | nd | nd | nd |
| l-Glu | 12.4 | 9.61 ± 0.96 | α-Ketoglutaric acid | Yes | A | 174 | 12.2 | 143, 115, 87 | Yes | 5,981 ± 817 | 8,826 ± 1,064* | 6,507 ± 725 | 7,806 ± 784 | ||
| l-Aad | 11.9 | 9.23 ± 1.68 | α-Ketoadipic acid | 8 | Yes | A | 8a | 188 | 13.9 | 157, 129, 101 | Yes | 86 ± 15 | 147 ± 15* | 91 ± 5 | 181 ± 39* |
| l-Asn | 6.1 | 4.72 ± 0.21 | 2-Oxosuccinamic acid | Yes | A | 145 | 15.0 | 144, 115, 69 | No | nd | nd | nd | nd | ||
The in vitro assays with 500 nmol of l-Lys and 500 nmol of pyruvate as ALD1 substrates indicated that more than three-fourths of the employed l-Lys was consumed within 30 min of incubation time, resulting in the equimolar formation of the product amino acid Ala (Table I). As described above, 2,3-DP was detected as the Lys-derived product (Figs. 1 and 3; Table I). The transamination activity of ALD1 toward l-Lys was higher than toward any of the other employed substrate amino acids and set to 100% relative activity. By contrast, ALD1 did not accept the α-enantiomer d-Lys as a substrate, showing a relative in vitro activity of only 0.1% of the value for l-Lys (Table I). The observed residual activity might be attributed to impurities within the employed d-Lys (98% or greater; Sigma-Aldrich; L8021).
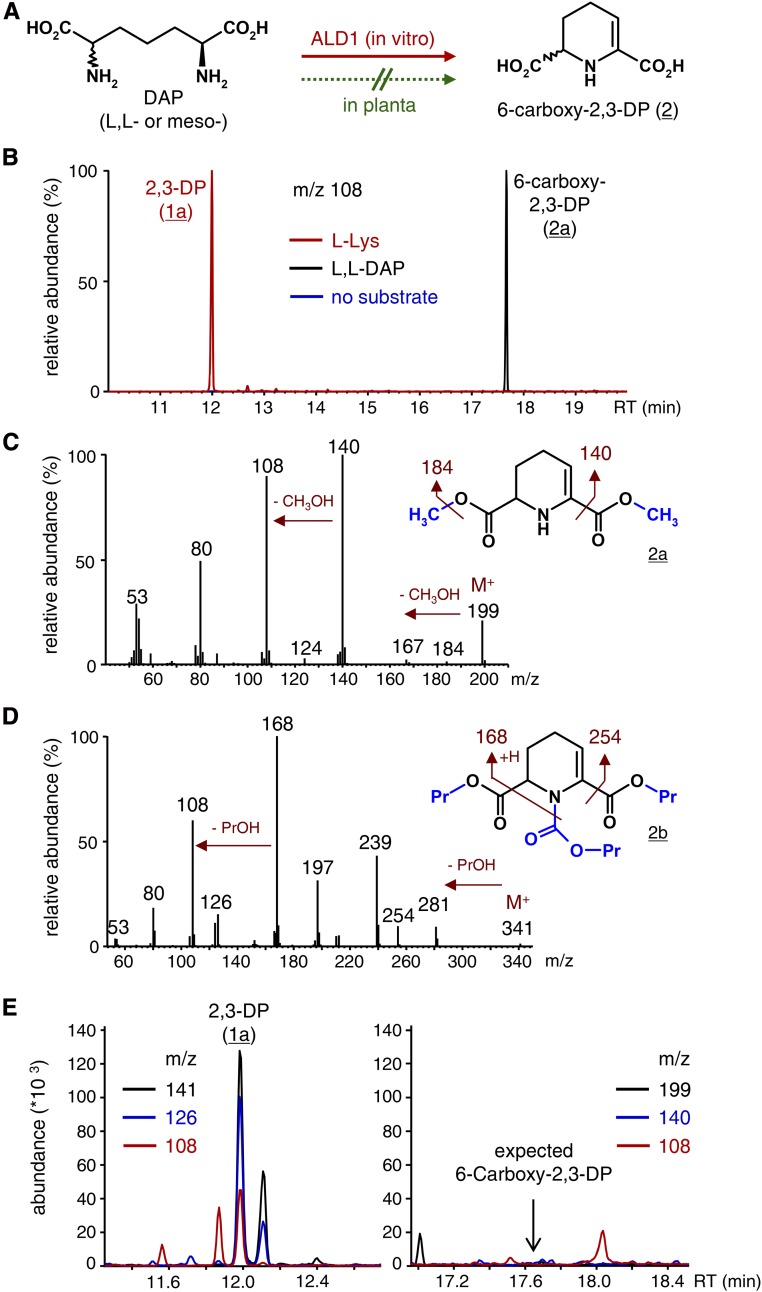
l,l-Diaminopimelate (DAP) and meso-DAP are diastereomeric Lys biosynthetic pathway intermediates. They only differ from l-Lys by the presence of an additional carboxyl group at the ε-carbon (Fig. 5A; Jander and Joshi, 2009). ALD1 accepted both l,l-DAP and meso-DAP as substrates, with relative activities of 16.3% and 12.9% compared with l-Lys (Table I). With both DAP diastereomers as substrates, we detected a transamination product that was identified as the 6-carboxylated variant of 2,3-DP (Fig. 5, A and B; Table I). The identity of the product as 6-carboxy-2,3-DP was supported by the mass spectral fragmentation pattern of the dimethylated derivative 2a (procedure A), which differed from those of methylated 2,3-DP (1a) by shifts of the M+ ion and main fragments by 58 mass units, indicating the presence of the additional methylated carboxyl moiety (Fig. 5C). In analogy, when applying the analytical procedure B, we detected chloroformate-derivatized 2b, whose mass spectrum differed from the spectrum of the corresponding 2,3-DP derivative 1b by 86 mass units, reflecting the additional propylated carboxyl group (Fig. 5D). The IR spectrum of dimethylated 6-carboxy-2,3-DP (2a) was strikingly similar to the IR spectrum of methylated 2,3-DP (1a), exhibiting an N-H stretching vibration at 3,429 cm−1, a C=C stretching bond at 1,650 cm−1, and a C=O bond at 1,739 cm−1 that was assigned to the methyl ester carbonyl in conjugation with the enaminic C=C double bond (Supplemental Fig. S7D). Together, these emphasize the enaminic structure of 2a. The additional nonconjugated carboxyl ester group of 2a compared with 1a discriminates both IR spectra, yielding a second carbonyl maximum at 1,755 cm−1 and an additional C-O stretching vibration at 1,210 cm−1 (Supplemental Figs. 4 and S7D). These analyses show that ALD1 catalyzes the in vitro conversion of l,l-DAP or meso-DAP to enaminic 6-carboxy-2,3-DP (Fig. 5A), which essentially parallels the l-Lys-to-2,3-DP conversion (Fig. 2). The use of a nonchiral GC column in our analyses did not allow discriminating between enantiomers. However, it is reasonable to assume that the S-enantiomer of 6-carboxy-2,3-DP is formed from l,l-DAP, whereas the R-enantiomer is supposed to be generated from meso-DAP (Fig. 5A).
Figure 5.
ALD1 catalyzes the conversion of DAP to 6-carboxy-2,3-DP in vitro, but the conversion does not occur to detectable levels in planta. A, Reaction scheme of the in vitro conversion of l,l-DAP or meso-DAP to 6-carboxy-2,3-DP (2) by ALD1. B, In vitro ALD1 activity assays with l-Lys as the amino acid substrate (red), l,l-DAP as the amino acid substrate (black), or no amino acid substrate (blue). Pyruvate served as the acceptor oxoacid in all cases. Overlaid ion chromatograms (m/z 108) are depicted after applying workup procedure A. The m/z 108 ion occurs both in the l-Lys-derived 2,3-DP product (1a) and in the l,l-DAP-derived 6-carboxy-2,3-DP product (2a). C, Mass spectrum and chemical structure of 2a obtained from 6-carboxy-2,3-DP (2) by procedure A. The two methyl groups (blue) are introduced by derivatization. The molecular ion (M+), plausible ion fragments, and fragment losses are indicated. D, Mass spectrum and chemical structure of 2b obtained from 6-carboxy-2,3-DP (2) by procedure B. The propyl and propyl carbamate groups introduced by derivatization are depicted in blue. The molecular ion (M+), plausible ion fragments, and fragment losses are indicated. E, Segments of overlaid ion chromatograms from extract samples of Psm-inoculated Col-0 wild-type leaves, as analyzed by GC-MS procedure A. Leaf samples were harvested 48 hpi. Whereas the presence of l-Lys-derived 2,3-DP (1a) in the extracts evokes characteristic ion chromatograms (m/z 141, 126, and 108) with the expected ratios of abundance (Fig. 1C) at a retention time (RT) of 12 min (left), 6-carboxy-2,3-DP (2a; m/z 199, 140, and 108; Fig. 5C) is not detected at the supposed retention time of 17.65 min (right).
We then analyzed plant extracts from Psm-inoculated Col-0 leaves for the occurrence of the in vitro observed ALD1 products in planta. To this end, we closely inspected the ion chromatograms of three characteristic fragment ions of a particular in vitro product at its specific retention time (Table I). Whereas in the extract samples, as documented above (Figs. 1 and 3), we observed distinct peaks with expected ratios of abundance in the characteristic ion chromatograms for the derivatized 2,3-DP forms (e.g. 1a; Fig. 1C) at the specific retention time (Fig. 5E), no 6-carboxy-2,3-DP (2a; Fig. 5C) could be detected (Fig. 5E). Therefore, albeit ALD1 is able to substantially convert DAP into 6-carboxy-2,3-DP in vitro, the conversion does not take place in planta to detectable levels.
In addition to l-Lys, we employed two other ω-NH2-substituted α-amino acids, l-Orn and 2,4-diaminobutyric acid (2,4-DABA), as possible ALD1 in vitro substrates. With 52.9% relative conversion, l-Orn was readily accepted by recombinant ALD1 as a substrate (Table I). Moreover, an l-Orn-derived in vitro product was identified, and the mass spectrum of the propyl chloroformate-derivatized compound (3b) strongly suggested its nature as Δ2-pyrroline-2-carboxylic acid (Pyr2C; 3), the pyrrolidine homolog of 2,3-DP (Table I; Supplemental Fig. S8, A and B). However, Δ2-Pyr2C was not detected in plant extracts of Psm- and mock-treated wild-type or ald1 plants (Table I). With 2% relative activity, ALD1 showed only a modest tendency to transaminate 2,4-DABA, and a possible in vitro transamination product could not be identified (Table I).
ALD1 also exhibited considerable in vitro transamination capacities for l-Met, l-Leu, and l-Arg, showing relative activities between 78.5% and 57% toward these amino acids (Table I; Supplemental Table S1). For l-Met and l-Leu, we identified the corresponding oxoacids α-keto-Met and α-keto-Leu as the in vitro transamination products (Table I; Supplemental Fig. S8, C–F). Again, these ketoacids were not detectable in extracts of Psm- or mock-treated plants (Table I). For l-Arg, an in vitro transamination product could not be identified with the employed analytical methods, most likely because the highly basic nature of the guanidine group of the putative product prevents GC analyses.
ALD1 also showed a medium in vitro transamination activity (between 6.1% and 12.4% relative activity) toward l-Phe, l-Trp, l-Glu, l-Aad, l-Asn, and l-His (Table I; Supplemental Table S1). For all of these amino acid substrates except l-His, we identified the corresponding oxoacids as in vitro ALD1 products by GC-MS analyses. As expected, the l-Glu transamination product and the common primary metabolite α-ketoglutarate were detected to high levels in plant extracts. However, no differences between Col-0 wild-type and ald1 mutant plants were observed (Table I). Interestingly, l-Phe-derived phenylpyruvic acid and l-Aad-derived α-ketoadipic acid were detectable in plant extracts to moderate amounts, and tendencies toward 2- to 3-fold higher levels in Psm-treated compared with mock-control leaves were registered (Table I; Supplemental Fig. S8, G, H, K, and L). However, both Col-0 and ald1 plants contained similar amounts of these substances, indicating that ALD1 does not contribute to their formation in planta (Table I). Moreover, l-Trp-derived indole-3-pyruvic acid (Supplemental Fig. S8, I and J) and l-Asn-derived 2-oxosuccinamic acid were not found in plant extracts (Table I). Finally, amino acids for which ALD1 showed either a very low or substantially no in vitro transamination activity included l-Ile, l-Val, l-Ala, l-Asp, l-Tyr, l-Ser, l-Thr, l-Cys, Gly, l-Pro, and l-Pip (Table I; Supplemental Table S1).
Together, these in vitro analyses show that l-Lys is the preferred amino acid substrate of the ALD1 aminotransferase. However, considerably high in vitro enzymatic activities also exist toward l-Met, l-Arg, l-Leu, and l-Orn, and clearly observable but more moderate conversion activities were measured for l,l-DAP, meso-DAP, l-Phe, l-Glu, l-Aad, l-Asn, and 2,4-DABA. Although most of the corresponding ketoacids or ketoacid-derived products could be identified in in vitro assays, they were, in contrast to the l-Lys-derived product 2,3-DP, generally not detectable in plant extracts. In the few cases in which we had detected transamination products in planta, they were, in contrast to 2,3-DP, equally produced in Col-0 wild-type and ald1 mutant plants. These results suggest that the transamination of l-Lys leading to 2,3-DP and Pip production is the major in vivo function of ALD1.
Arabidopsis SARD4 (Alias ORNCD1) Is Able to Catalyze the Reductive Step from DP to Pip
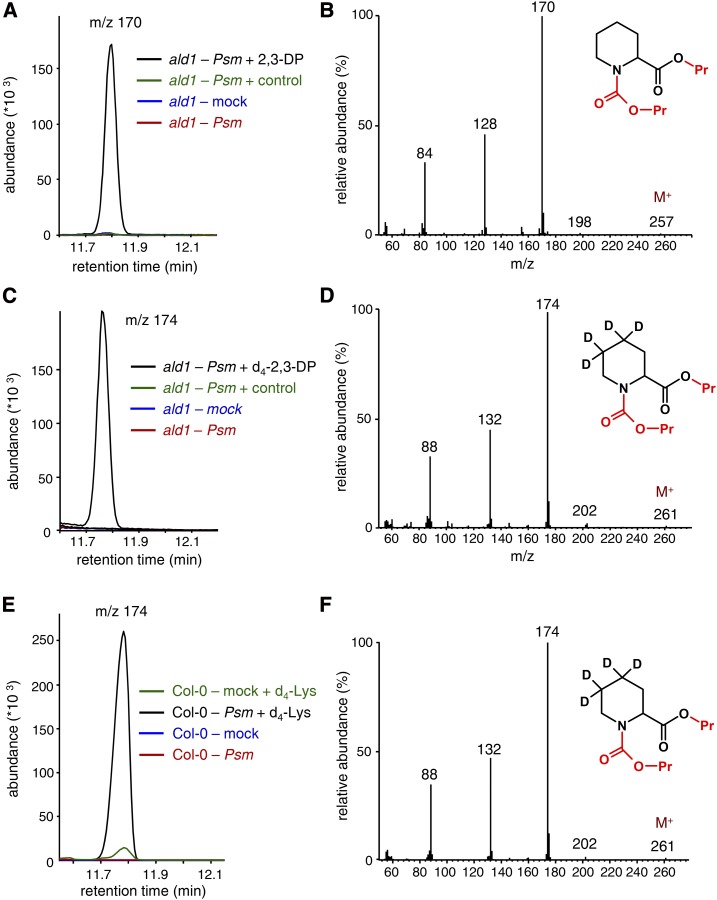
Our data so far indicate that ALD1 catalyzes the first transamination step of Pip biosynthesis from l-Lys, which results in the generation of 2,3-DP in planta. However, as postulated before (Návarová et al., 2012; Zeier, 2013), ALD1-initiated Pip generation requires a further reduction step that converts the ALD1-derived DP intermediate into Pip (Fig. 2). The ald1 mutant is unable to accumulate Pip upon P. syringae inoculation (Návarová et al., 2012; Bernsdorff et al., 2016; Fig. 6A), potentially because of its inability to generate the DP intermediate necessary for Pip production (Figs. 1 and 3). To examine whether exogenously applied 2,3-DP can restore the ability of ald1 to biosynthesize Pip, we coinfiltrated Psm and 2,3-DP, which was produced in vitro by the ALD1 bioassay, into ald1 leaves (Fig. 5A). As a consequence, the ald1 leaves were able to accumulate Pip (Fig. 6, A and B). By contrast, Pip accumulation did not occur when a blank assay mixture lacking 2,3-DP was coinfiltrated with Psm (Fig. 6A). Furthermore, we conducted the same coinfiltration experiment with tetradeuterated d4-2,3-DP that was produced from l-Lys-4,4,5,5-d4 in the ALD1 bioassays (Figs. 1E and 3F). In the presence of d4-2,3-DP, Psm-challenged ald1 plants synthesized a compound whose mass spectrum contained a fragmentation pattern shifted by 4 mass units compared with the pattern of the Pip spectrum, indicating that tetradeuterated d4-Pip was produced in leaves that were supplemented with d4-2,3-DP (Fig. 6, C and D). These data show that P. syringae-inoculated leaves of ald1 mutant plants are able to synthesize Pip, providing external addition of 2,3-DP to the plants.
Figure 6.
The ald1 mutant is able to convert exogenously supplied 2,3-DP to Pip in response to P. syringae inoculation. A, Segment of overlaid ion chromatograms (m/z = 170) from leaf extract samples of differently treated ald1 mutant plants. Leaves were coinfiltrated with Psm and 2,3-DP obtained from ALD1 in vitro assays (black; Fig. 3C). The assays containing 20 mm l-Lys as a substrate were run to completion (greater than 99% of Lys used), enzyme inactivated, and diluted 6-fold with 10 mm MgCl2. The mixture was then supplied with the bacterial suspension to a final OD600 of 0.005 and infiltrated into three leaves of 5-week-old ald1 plants. At 48 h later, the infiltrated leaves were harvested and processed by analytical procedure B. Coinfiltration of the assay mixture not containing ALD1 enzyme (green), MgCl2 (mock) infiltration alone (blue), and Psm treatment alone (red) served as the control treatments. The peak at 11.8 min is observed only in the Psm/2,3-DP sample (black) and corresponds to derivatized Pip. Similar results were obtained when 2,3-DP generated by the LysOx/catalase assay was used. B, Mass spectrum of propyl chloroformate-derivatized Pip (compare Návarová et al., 2012) derived from 2,3-DP-supplemented and Psm-inoculated ald1 leaves (peak at 11.8 min in A). The groups introduced by derivatization are labeled in red. C, Segment of overlaid ion chromatograms (m/z = 174) from leaf extract samples of differently treated ald1 mutant plants. Leaves were coinfiltrated with Psm and d4-labeled 2,3-DP obtained from ALD1 in vitro assays (black; Fig. 3E), coinfiltrated with Psm and the assay mixture not containing 2,3-DP (green), infiltrated with mock (10 mm MgCl2) solution alone (blue), or infiltrated with Psm alone. Further experimental details are identical to those described in A. The peak at 11.76 min observed only in the Psm/d4-2,3-DP sample (black) corresponds to derivatized d4-Pip. D, Mass spectrum of propyl chloroformate-derivatized d4-Pip derived from d4-2,3-DP-supplemented and Psm-inoculated ald1 leaves (peak at 11.76 min in C). E, Segment of overlaid ion chromatograms (m/z = 174) from leaf extract samples of differently treated Col-0 wild-type plants. Plants were coinfiltrated in leaves with 5 mm l-Lys-4,4,5,5-d4 (d4-Lys) and Psm (OD600 = 0.005; black), coinfiltrated with 5 mm d4-Lys and mock (10 mm MgCl2) solution (green), infiltrated with mock solution alone (blue), or infiltrated with Psm alone. The peak at 11.76 min accumulating in the Psm/d4-Lys sample (black) corresponds to derivatized d4-Pip. It is noteworthy that, in addition to labeled Pip, Psm/d4-Lys-treated Col-0 plants also accumulate endogenous, unlabeled Pip. F, Mass spectrum of propyl chloroformate-derivatized d4-Pip derived from d4-Lys-supplemented and Psm-inoculated Col-0 leaves (peak at 11.76 min in E).
In addition, we performed a similar experiment with the Col-0 wild type that is able to accumulate both 2,3-DP and Pip after Psm challenge (Figs. 1 and 3; Návarová et al., 2012). After coapplication of l-Lys-4,4,5,5-d4 and Psm to Col-0 leaves, we observed not only the production of d4-2,3-DP (Fig. 1E) but also a strong accumulation of d4-labeled Pip (Fig. 5D). The isotope-labeled compound thereby exhibited a mass spectrum identical to the spectrum of the d4-Pip detected in the d4-2,3-DP-supplemented ald1 plants (Fig. 5E). In sum, this set of experiments indicates that the inability of ald1 plants to generate 2,3-DP upon pathogen inoculation is causative for the failure of Pip biosynthesis in this mutant. Nevertheless, ald1 mutant plants possess the capacity to reduce 2,3-DP to Pip.
Arabidopsis At5g52810 (SARD4, alias ORNCD1) represents the Arabidopsis gene with the highest sequence similarity to CRYM (30.5% amino acid identity and 51.8% amino acid similarity; EMBOSS Needleman-Wunsch pairwise alignment, version 6.3.1, EMBL-EBI; http://www.ebi.ac.uk/Tools/psa/; Zeier, 2013), the ketimine reductase that is able to reduce 1,2-DP to Pip (Hallen et al., 2011, 2015). Similar to CRYM, SARD4 possesses a putative NAD(P)-binding domain with a predicted Rossmann fold, a structural motif frequently found in nucleotide-binding dehydrogenases (Rao and Rossmann, 1973; Zeier, 2013). To test our previous hypothesis that SARD4 is involved in catalyzing the reductive step in Pip biosynthesis (Zeier, 2013), recombinant C-terminally His-tagged SARD4 enzyme was purified via immobilized metal ion affinity chromatography and tested for reductase activity in a coupled assay setting with purified ALD1 enzyme. As a positive control, CRYM protein also was expressed in E. coli, purified, and tested analogously to SARD4, as described in detail in “Materials and Methods.”
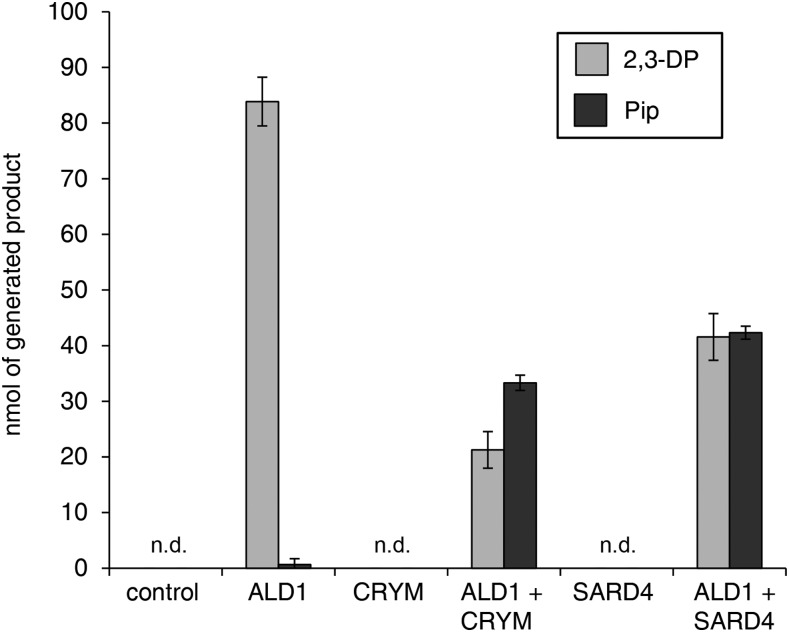
Thereby, we examined whether enzyme combinations of ALD1 and CRYM or ALD1 and SARD4 would be able to biosynthesize Pip from l-Lys in vitro. As outlined before, ALD1 protein alone was able to transaminate l-Lys in the presence of pyruvate to produce 2,3-DP in the bioassay (Fig. 7). When, in addition to l-Lys, ALD1, the acceptor oxoacid, and the cofactor PLP necessary for the transamination step, CRYM protein and NADPH (or NADH) were present in the reaction mixture, the levels of 2,3-DP were reduced markedly compared with the ALD1-only assay, and substantial amounts of Pip were generated (Fig. 7). Similarly, when ALD1 and SARD4 were coapplied in the bioassay, Pip was detected to high amounts in the enzyme mixture and the levels of 2,3-DP decreased compared with an individual ALD1 incubation (Fig. 7). Therefore, the combinatory assays support the existence of a Pip biosynthetic pathway that involves both an ALD1-mediated transamination step and a SARD4-catalyzed reductive step (Fig. 2).
Figure 7.
Coupled assays with ALD1 and the human reductase CRYM or its Arabidopsis ortholog SARD4 (ORNCD1) yield Pip in vitro with l-Lys as a substrate. Reactions were carried out in an assay mix (200 µL) containing purified recombinant enzymes (ALD1, CRYM, and SARD4) at a final concentration of 100 µg mL−1 with 20 mm l-Lys, 20 mm pyruvate, 5 mm MgCl2, 100 µm PLP, and 200 µm NADH in 20 mm Tris buffer, pH 8. All reactions contained 5% (v/v) glycerol for additional enzyme stability and were incubated at 37°C for 16 h. At the end of the incubation period, the reaction was stopped by inactivating the enzymes at 85°C for 10 min. The formation of 2,3-DP and Pip was monitored using GC-MS after derivatization of the assays with trimethylsilyl diazomethane (procedure A) or propyl chloroformate (procedure B), respectively. From left to right are no enzyme (control), ALD1 single-enzyme assay, CRYM single-enzyme assay, ALD1 and CRYM coincubation, SARD4 single-enzyme assay, and ALD1 and SARD4 coincubation. The amounts of generated 2,3-DP and Pip products are given in nmol. Bars represent means ± sd of three replicate incubations (n.d., not detected).
sard4 Mutant Plants Show Attenuated Pip Biosynthesis and Overaccumulate 2,3-DP
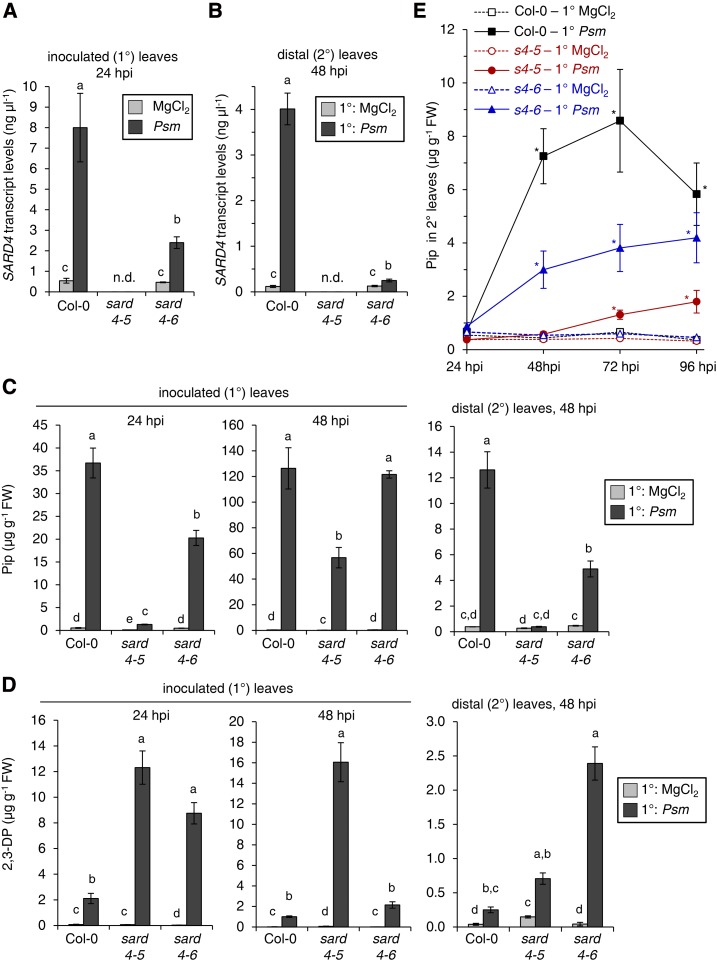
Consistent with a role for SARD4 in the pathogen-inducible biosynthesis of Pip, SARD4 transcripts accumulated in both inoculated (1°) and distal (2°) leaves of Col-0 plants upon Psm treatment (Fig. 8, A and B). To functionally investigate the relevance of SARD4 for in planta Pip production and plant immunity, we searched for Arabidopsis T-DNA insertion lines with defects in the At5g52810 gene. Using PCR-based genotyping (Alonso et al., 2003), we identified two lines, GK_428E01 (initially named orncd1-1, later renamed sard4-5 according to the nomenclature proposed by Ding et al. [2016]) and GK_696E11 (sard4-6, initially orncd1-2), with T-DNA insertions in the single exon of the SARD4 gene (Supplemental Fig. S9). Whereas the T-DNA insertion in the sard4-5 line leads to a full loss of gene function (knockout), as shown by a complete lack of transcripts, the sard4-6 line still showed clearly detectable SARD4 transcript levels that increased in the 1° inoculated leaves and in the distal leaves of P. syringae-treated sard4-6 mutant plants, albeit to markedly lower amounts than in the Col-0 wild type (Fig. 8, A and B).
Figure 8.
Transcript levels of SARD4 and metabolite levels of 2,3-DP and Pip in inoculated (1°) and distal (2°) leaves of wild-type Col-0, sard4-5, and sard4-6 mutant plants in response to P. syringae and mock treatments. A, SARD4 transcript levels in 1° leaves of Col-0, sard4-5, and sard4-6 plants infiltrated with Psm (OD600 = 0.005) or with 10 mm MgCl2 (mock treatment) at 24 h after treatment. Transcript values are given in µg µL−1 and represent means ± sd of at least three biological replicate samples from different plants, each replicate consisting of six leaves. Values for biological replicates were calculated as means of two technical replicates. Different letters above the bars denote statistically significant differences (P < 0.002, ANOVA and posthoc Tukey’s HSD test); n.d., no transcripts detected. The results were confirmed in two independent experiments. B, SARD4 transcript levels in untreated distal (2°) leaves of Col-0, sard4-5, and sard4-6 plants infiltrated in 1° leaves with Psm (OD600 = 0.005) or 10 mm MgCl2 (mock treatment) at 48 h after treatment. Other experimental details were as described for A. Different letters above the bars denote statistically significant differences (P < 0.002, ANOVA and posthoc Tukey’s HSD test); n.d., no transcripts detected. The results were confirmed in two independent experiments. C, Levels of Pip in treated 1° leaves of Col-0, sard4-5, and sard4-6 plants infiltrated with Psm (OD600 = 0.005) or 10 mm MgCl2 (mock treatment) at 24 h (left graph) and 48 h (middle graph) after treatment and in untreated distal (2°) leaves at 48 h after treatment of 1° leaves (right graph). Data represent means ± sd of at least three biological replicates from different plants, each replicate consisting of six leaves from two plants. Different letters above the bars denote statistically significant differences (P < 0.05, ANOVA and posthoc Tukey’s HSD test). The results were confirmed in two independent experiments. FW, Fresh weight. D, Levels of 2,3-DP in inoculated (1°) and distal (2°) leaves of Col-0, sard4-5, and sard4-6 plants. Experimental details were as described for C. The results were confirmed in two independent experiments. E, Levels of Pip in untreated 2° leaves of Col-0, sard4-5 (s4-5), and sard4-6 (s4-6) plants infiltrated with Psm (OD600 = 0.005) or 10 mm MgCl2 at 24, 48, 72, and 96 h after treatment. Data represent means ± sd of three biological replicates. Asterisks denote statistically significant differences between Psm- and mock-treated samples of a given plant genotype and time point (P < 0.01, two-tailed Student’s t test).
A possible explanation for this at first sight surprising difference of the mutant lines lies in the predicted sites of T-DNA insertions. The predicted insertion is relatively central within the exon for sard4-5 and within the first 100 bp downstream of the start codon (ATG, +1) for sard4-6 (Supplemental Fig. S9A; http://signal.salk.edu/cgi-bin/tdnaexpress). This is consistent with the sizes of bands resulting from PCR analyses with genomic DNA from sard4-5 and sard4-6 plants and a combination of T-DNA left border primers and gene-specific primers annealing in the 3′ untranslated regions (Supplemental Fig. S9B). The second ATG and possible start codon in frame with the coding sequence is located at position +166 of the nucleotide sequence, suggesting the possibility that a shorter but potentially functional version of SARD4 might be expressed in sard4-6.
We next determined the levels of Pip in the leaves of Col-0, sard4-5, and sard4-6 plants upon Psm inoculation or mock control treatments. The contents of Pip in the control plants were generally in the range between 0.1 to 0.6 µg g−1 fresh weight (Fig. 8C). In the Psm-inoculated leaves of the Col-0 wild type, Pip strongly accumulated to levels of about 35 µg g−1 fresh weight at 24 hpi and to levels of 125 µg g−1 fresh weight at 48 hpi (Fig. 8C). By contrast, inoculated leaves of sard4-5 showed a strongly diminished early increase of Pip at 24 hpi to levels of 1.3 µg g−1 fresh weight only. At 48 hpi, a considerable accumulation of Pip in the Psm-inoculated sard4-5 leaves to levels of about 55 µg g−1 fresh weight was detected (Fig. 8C), which accounted for about half of the values of the wild type at this later infection stage. At inoculation sites, the sard4-6 mutant showed a still marked but, compared with the wild type, attenuated Pip accumulation to 20 µg g−1 fresh weight at 24 hpi, which increased to wild-type-like levels of about 120 µg g−1 fresh weight at 48 hpi (Fig. 8C). The Pip levels in the distal 2° leaves increased in the wild type to 12 µg g−1 fresh weight at 48 hpi of 1° leaves. This systemic increase of Pip at 48 hpi was absent in the sard4-5 mutant, and sard4-6 showed a diminished accumulation to 4 µg g−1 fresh weight in the distal leaves at this time point (Fig. 8C). To test whether the systemic accumulation of Pip was fully compromised in sard4-5 or, as the local accumulation patterns suggested, only delayed, we performed a time-course analyses of Psm-induced Pip accumulation between 24 and 96 hpi in the systemic leaves of the plant lines under investigation (Fig. 8E). In the Col-0 wild type and the sard4-6 mutant, the systemic levels of Pip were increased significantly at 48 hpi onward in response to Psm inoculation, whereby Pip again accumulated to higher levels in the wild type than in the sard4-6 mutant. Notably, Psm inoculation triggered systemic increases in the sard4-5 mutant at 72 and 96 hpi. However, the degree of systemic Pip accumulation was greatly reduced in sard4-5, since Pip levels stayed below 2 µg g−1 fresh weight until 96 hpi (Fig. 8E).
As a next step, we analyzed the basal and P. syringae-induced levels of the Pip precursor 2,3-DP in the leaves of Col-0 and sard4 mutant plants, which can be converted into Pip in vitro by recombinant SARD4 protein (Fig. 7). The Col-0 wild type showed a significant increase of 2,3-DP both in the inoculated 1° leaves and in the distal 2° leaves upon Psm treatment (Fig. 8D). Remarkably, the levels of 2,3-DP in inoculated Col-0 plants were 1 to 2 orders of magnitude lower than the levels of Pip (Fig. 8, C and D). Compared with the wild type, both sard4-5 and sard4-6 mutants overaccumulated 2,3-DP in the locally inoculated and/or in the systemic leaf tissue, suggesting that they harbor defects in the SARD4-mediated 2,3-DP-to-Pip conversion (Fig. 8D).
Together, these metabolite analyses indicate that the gene knockout line sard4-5 has a reduced but not fully abrogated capacity to synthesize Pip after Psm attack in the locally inoculated and systemic leaf tissue. SARD4, therefore, significantly contributes to pathogen-induced Pip production but appears not to be the only reducing enzyme involved in Pip biosynthesis (Fig. 2). The sard4-6 mutant showed a pronouncedly weaker metabolic phenotype than the knockout line sard4-5, because it exhibited an attenuated but still marked ability to produce Pip in Psm-inoculated and systemic leaves. This confirms the notion that sard4-6 represents a knockdown line for the SARD4 gene.
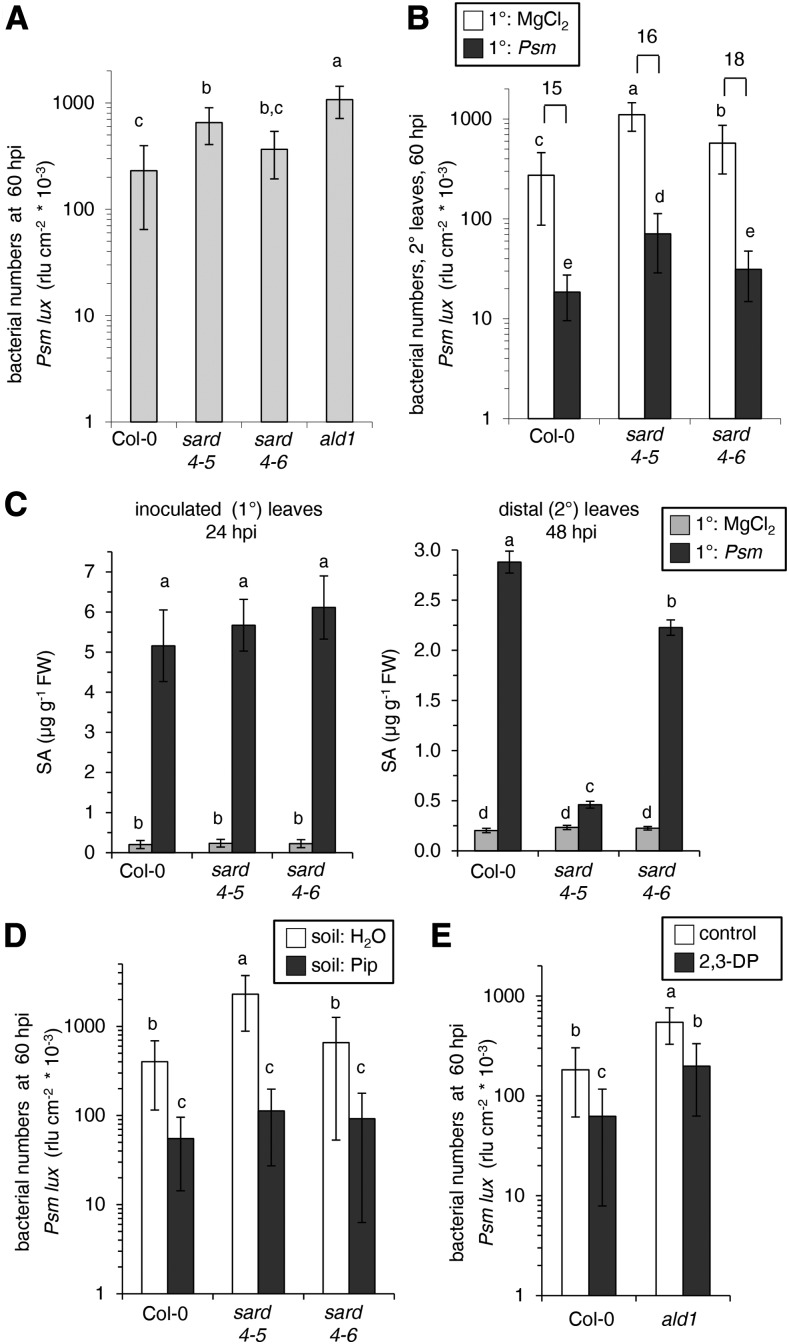
The sard4-5 Mutant Displays Attenuated Basal Resistance to P. syringae But Is Competent to Systemically Enhance Resistance in Response to Precedent Attack
We next compared basal resistance toward an attack of virulent P. syringae bacteria between Col-0, sard4-5, sard4-6, and ald1 plants. The ald1 mutant is fully blocked in the accumulation of 2,3-DP or Pip upon P. syringae attack (Fig. 3C; Návarová et al., 2012; Bernsdorff et al., 2016). The growth of a bioluminescent, virulent Psm strain that expresses the Photorhabdus luminescens luxCDABE operon (Psm lux) in inoculated leaves was thereby taken as a parameter to assess basal resistance. The use of the Psm lux strain allows a rapid quantification of bacterial numbers in leaves via luminescence measurement (Fan et al., 2008), which was performed at 60 hpi. After inoculating leaves with Psm lux suspensions of OD600 = 0.001, we determined significantly higher bacterial numbers in the leaves of sard4-5 compared with the leaves of the wild type at 60 hpi, indicating a higher susceptibility to bacterial attack of the mutant (Fig. 9A). Moreover, the ald1 mutant was more susceptible to Psm lux than sard4-5, and sard4-6 exhibited a resistance phenotype intermediate to those of Col-0 and sard4-5 (Fig. 9A). Therefore, the efficiency of basal resistance to P. syringae appeared to correlate with the amount of Pip produced during the plant-bacterium interaction (Fig. 8C; Návarová et al., 2012). In addition, we measured the levels of the phenolic defense hormone SA, whose accumulation in inoculated leaves is required for effective basal resistance to P. syringae (Wildermuth et al., 2001; Bernsdorff et al., 2016), at 24 h post Psm inoculation. At this time point, SA accumulated to similar levels in the leaves of Psm-inoculated Col-0, sard4-5, and sard4-6 plants (Fig. 9C).
Figure 9.
Basal resistance responses, SAR, Pip-induced resistance, and 2,3-DP-induced resistance in wild-type Col-0, sard4-5, sard4-6, and/or ald1 mutant plants. A, Basal resistance to P. syringae of Col-0, sard4-5, sard4-6, and ald1 plants. Naive plants (three leaves each) were inoculated with bioluminescent Psm lux (OD600 = 0.001), and bacterial growth was assessed 60 h later by luminescence and expressed as relative light units (rlu) per cm2 of leaf area. Data represent means ± sd of the growth values of at least 15 leaf replicates. Different letters above the bars denote statistically significant differences (P < 0.05, ANOVA and posthoc Tukey’s HSD test). The results were confirmed in three other independent experiments. B, SAR in Col-0, sard4-5, and sard4-6 plants. Three lower, 1° leaves per plant were infiltrated with either 10 mm MgCl2 or Psm (OD600 = 0.005), and three upper, 2° leaves were challenge infected with Psm lux (OD600 = 0.001) 2 d later. Growth of Psm lux in 2° leaves was assessed 60 h after 2° challenge inoculation by luminescence measurements. Experimental details and statistical analyses were as described for A. The results were confirmed in two other independent experiments. C, Levels of SA in treated 1° leaves of Col-0, sard4-5, and sard4-6 plants infiltrated with Psm (OD600 = 0.005) or 10 mm MgCl2 (mock treatment) at 24 h (left graph) and in untreated distal (2°) leaves at 48 h after treatment of 1° leaves (right graph). Data represent means ± sd of at least three biological replicates from different plants, each replicate consisting of six leaves from two plants. Different letters above the bars denote statistically significant differences (P < 0.05, ANOVA and posthoc Tukey’s HSD test). The results were confirmed in two other independent experiments. FW, Fresh weight. D, Pip-induced resistance in Col-0, sard4-5, and sard4-6 plants. Plants were each supplied with 10 mL of 1 mm Pip (dose of 10 µmol) or with 10 mL of water (control treatment) via the root system (Návarová et al., 2012), and three leaves per plant were challenge infected 1 d later with Psm lux (OD600 = 0.001). Bacterial growth was assessed 60 hpi and analyzed as described for A. The results were confirmed in two other independent experiments. E, 2,3-DP-induced resistance in Col-0 and ald1 plants. Three leaves per plant were coinfiltrated with 2,3-DP (∼2 mm) obtained by the ALD1 in vitro assay and Psm lux (OD600 = 0.001), and bacterial growth was assessed 60 h later. The determination of bacterial growth and statistical analysis were performed as described for A. The results were confirmed in two other independent experiments.
The ability of Arabidopsis plants to synthesize Pip upon bacterial attack is a prerequisite for the establishment of SAR (Zeier, 2013). Since sard4-5 mutant plants accumulated markedly reduced levels of Pip in the 2° tissue (Fig. 8, C and E), we hypothesized that they might be, like ald1, fully compromised in SAR. Hence, we conducted comparative SAR bioassays with Col-0 and both of the sard4 mutant lines (Fig. 9B). Therefore, plants were Psm inoculated or mock treated in three 1° leaves and, 2 d later, challenge infected in three distal 2° leaves with Psm lux. SAR establishment is characterized by a significantly lower bacterial multiplication in the course of the 2° challenge infection in Psm-pretreated compared with mock-pretreated plants, and this is generally accompanied by a distinctly visible reduction of chlorotic disease symptoms (Mishina and Zeier, 2007). In Col-0 plants, inducing inoculations with Psm limited the growth of Psm lux in challenged distant leaves by a factor of about 15 at 60 hpi, indicating strong SAR establishment in the wild type (Fig. 9B). This was associated with a marked reduction of disease symptom severity (Supplemental Fig. S10). Mock-pretreated sard4-5 mutant plants suffered from a higher bacterial multiplication compared with wild-type plants, again indicating that basal resistance is attenuated in sard4-5 (Fig. 9C). Remarkably, in response to Psm pretreatment, the sard4-5 mutant was able to establish a significant SAR response, which manifested itself by a wild-type-like factor of growth attenuation in challenge-infected 2° leaves (mock/Psm = 16; Fig. 9C) and by a pronounced reduction of the disease symptoms of 2° leaves (Supplemental Fig. S10). Consequently, however, the absolute resistance levels of SAR-induced wild-type plants were higher than those of SAR-induced sard4-5 plants (Fig. 9B). The sard4-6 mutant again displayed an intermediate phenotype: the resistance levels of mock-treated control plants were less strongly compromised compared with sard4-5, and the induction of SAR by 1° Psm treatment enhanced the resistance levels of sard4-6 to almost wild-type-like levels (Fig. 9B; Supplemental Fig. S10).
Finally, since SA accumulation in the 2° leaf tissue is one of the hallmarks of SAR (Vlot et al., 2009), we analyzed the levels of SA in 1° mock-treated and SAR-induced Col-0 and sard4 mutant plants. As expected, SA strongly accumulated in the nontreated 2° leaves of wild-type plants following a 1° Psm inoculation (Fig. 9C). Notably, the systemic accumulation of SA was reduced markedly, albeit not totally compromised, in sard4-5 (Fig. 9C). Furthermore, sard4-6 mutants accumulated SA in the systemic tissue to relatively high levels that reached about 80% of the respective wild-type values (Fig. 9C).
Taken together, the SAR assays show that sard4-5 mutant plants display a considerable SAR response following Psm treatment, although Pip accumulation in the systemic tissue is delayed and strongly reduced. Nevertheless, like naive plants, SAR-induced sard4-5 plants possess a significantly lower resistance level to bacterial infection than the corresponding wild-type plants. This goes hand in hand with a distinctly reduced systemic accumulation of SA after SAR activation.
Exogenous Feeding with 2,3-DP Increases Plant Basal Resistance to Bacterial Infection
Exogenous application of Pip to plants increases resistance to leaf infection by P. syringae in the Col-0 wild type and overrides the defects in basal resistance in Pip-deficient ald1 mutants (Návarová et al., 2012). Since sard4 mutants show attenuated Pip generation upon pathogen inoculation (Fig. 8C), we tested whether exogenously applied Pip would restore their basal resistance capacity to wild-type-like levels. Indeed, plants watered with Pip 1 d before Psm lux inoculation significantly increased resistance to bacterial infection in Col-0, sard4-5, and sard4-6 (Fig. 9D). Thereby, the bacterial growth in the leaves of the normally more susceptible sard4-5 line was restricted to the same absolute values than that of the wild type and the sard4-6 line (Fig. 9D). This indicates that the resistance defects of sard4 mutants are attributable to their attenuated biosynthesis of Pip during the plant-bacterium interaction (Fig. 8C).
We next examined the effect of exogenous treatment with the Pip precursor 2,3-DP on plant basal resistance toward P. syringae infection. When 2,3-DP generated by the ALD1 in vitro assay was coinfiltrated into leaves with Psm lux, bacterial growth was reduced by a factor of about 3 in both Col-0 and ald1 plants at 60 hpi compared with the coapplication of Psm lux and a control solution that did not contain 2,3-DP (Fig. 9E). This suggests that elevated levels of the Pip precursor 2,3-DP in leaves are sufficient to trigger an increased resistance response in Arabidopsis.
DISCUSSION
The accumulation of Pip in leaves in response to an inducing pathogen inoculation is a prerequisite for the establishment of SAR in plants. Pip amplifies pathogen-induced signals and promotes plants to a primed state to ensure effective activation of defense responses (Návarová et al., 2012; Vogel-Adghough et al., 2013). Pip-mediated SAR induction and defense priming thereby occur by signaling mechanisms that have both SA-dependent and SA-independent characteristics (Bernsdorff et al., 2016).
ALD1-Mediated Lys Conversion: Formation of Enaminic 2,3-DP
The biosynthetic precursor of Pip in plants and animals is l-Lys (Rothstein and Miller, 1954; Fujioka and Sakurai, 1997), and a first necessary step for the formation of Pip from l-Lys is the transamination of the precursor amino acid (Gupta and Spenser, 1969). Previous studies by Song et al. (2004a) identified the Arabidopsis ALD1 protein as an l-Lys aminotransferase. Moreover, our own findings demonstrated that functional ALD1 is required for the pathogen-inducible biosynthesis of Pip in plants (Návarová et al., 2012). Together, these findings strongly suggested that ALD1 is the plant aminotransferase responsible for Pip biosynthesis (Zeier, 2013). l-Lys carries amino groups at both the α- and ε-positions, leaving the possibility that the transamination reaction required for Pip biosynthesis might involve abstraction of either the α- or ε-amino group, which would result in the formation of KAC or AAS as oxoacid intermediates, respectively. KAC can cyclize by water loss to form the ketiminic DP derivative 1,2-DP, whereas AAS is able to form the ketimine 1,6-DP in an analogous manner (Gupta and Spenser, 1969; Galili et al., 2001; Fig. 2). Previous studies based on supplementation of plant tissue with isotope-labeled l-Lys precursors and analysis of the distribution of the isotope labels in the Pip product report controversial results about the mechanism of the Lys transamination reaction that lead to Pip formation. For instance, whereas Schütte and Seelig (1967) proposed a pathway via abstraction of the ε-amino group and the formation of AAS and 1,6-DP as intermediates, the findings of Gupta and Spenser (1969) were consistent with a reaction scheme that involves abstraction of the α-amino group and subsequent generation of KAC and 1,2-DP.
In this study, we used two different approaches to examine the ALD1-catalyzed conversion reaction of l-Lys involved in Pip biosynthesis. On the one hand, we performed comparative metabolite profiling of leaf extracts from wild-type Col-0 and ald1 knockout plants to identify ALD1-derived products in planta. On the other hand, we studied the biochemistry of purified, recombinant ALD1 protein in vitro. Using a combination of GC-MS and GC-FTIR analysis techniques, we identified enaminic 2,3-DP (1) as the final l-Lys-derived conversion product of ALD1 in vitro and in planta (Fig. 2). The use of the isotope-labeled l-Lys variants l-Lys-6-13C,ε-15N and l-Lys-α-15N in plant feeding experiments and in in vitro assays showed that ALD1 abstracts the α-amino group of l-Lys and directly transfers it to acceptor oxoacids. The ε-amino group, by contrast, remains in the DP product (Figs. 1, 3, and 4; Supplemental Fig. S3). Our analyses are consistent with the reaction scheme proposed previously by Gupta and Spenser (1969), in which transamination of the α-position of l-Lys forms KAC as an oxoacid intermediate in the biosynthesis of Pip. Following this ALD1-mediated aminotransferase reaction, we propose that a dehydrative cyclization of KAC to the ketimine 1,2-DP and a subsequent isomerization to the detected 2,3-DP end product take place (Fig. 2). The cyclization and isomerization steps in this reaction sequence might occur independently of the ALD1 protein in a spontaneous manner or be part of the enzymatic mechanism.
In addition to the 15N-isotope-labeling studies mentioned above, several experimental data indicate that the in vitro- and in planta-detected ALD1-derived product is enaminic 2,3-DP (1). First, the mass spectral fragmentation patterns of methylated 1a and propyl chloroformate-derivatized 1b are consistent with the piperideine-2-carboxylate structure (Figs. 1C and 3C). Second, the experiments in which l-Lys-4,4,5,5-d4 was fed to plants or used as an in vitro substrate exclude the formation of DP isomers with double bonds in the 3,4-, the 4,5-, or the 5,6-position of the piperideine ring (Figs. 1E and 3E). Third, the IR spectrum of 1a substantiates the enamine structure and rules out the ketimine form (Fig. 1F). Notably, the spectrum shows a distinct band at 3,441 cm−1, which indicates the presence of an N-H bond in the molecule. Similar N-H bonds and corresponding stretching vibrations are present in the IR spectra of valerolactam, the in vitro-detected DP dimer, and dimethylated 6-carboxy-2,3-DP (2a), the compound detected in ALD1 in vitro assays with DAP as the substrate amino acid (Supplemental Fig. S7). The IR spectrum of 1a further exhibits an absorption at 1,648 cm−1, which supports the presence of an enaminic C=C double bond. A band at nearly the same wave number (1,646 cm−1) was observed in resonance Raman spectra of 2,3-DP obtained as the enzymatic product of d-Lys and d-amino acid oxidase from hog kidney and has been assigned to the enaminic C(2)=C(3) stretching mode (Nishina et al., 1991). The enaminic C=C vibrations in IR spectra of differently substituted piperideine enamines have been recorded between 1,640 and 1,673 cm−1 (Leonard and Hauck, 1957). In addition, the absorption of the C=O stretching bond in the IR spectrum of 1a occurs at lower wave numbers (1,734 cm−1) than those of aliphatic, saturated carboxylic acid methyl esters and, therefore, is consistent with the occurrence of an enaminic C=C double bond in conjugation with the methyl carboxylate group in 1a (Fig. 1F; Supplemental Fig. S4). Fourth, propyl chloroformate derivatization of plant extracts and in vitro assay samples generated the N-propyl carbamate product 1b (Fig. 3C), which is supposed to be generated from nucleophilic attack of amine groups to the derivatization reagent propyl chloroformate. This further supports the notion that the amine 2,3-DP, but not the imine 1,2-DP, constitutes the ALD1-derived product. Hence, two different analytical methods and complementary mass spectral and IR spectroscopic information suggest that 2,3-DP is the final product generated from l-Lys by the action of the ALD1 protein.
The biochemical investigation of the ALD1-mediated l-Lys conversion by liquid chromatography-tandem mass spectrometry (MS/MS) analysis published recently by Ding et al. (2016) coincides with our work here with respect to the stereospecific abstraction of the α-amino group of l-Lys and the formation of an ALD1-generated dihydropipecolic acid end product via KAC as the oxoacid. Remarkably, KAC was detected neither by the liquid chromatography-mass spectrometry protocol of Ding et al. (2016) nor in the two GC-MS-based analytical procedures that we have applied here. On the basis of MS/MS fragmentation patterns after positive ionization, Ding et al. (2016) proposed 1,2-DP as the end product of the reaction. Our findings, however, indicate that the in vitro- and in planta-generated ALD1-derived product is 2,3-DP. As we have outlined for our own MS analyses of compound 1a (Fig. 1), the mass spectrometric information alone is not sufficient to distinguish between 1,2-DP and 2,3-DP forms (Fig. 1), and only the additional IR spectroscopic data gave direct evidence for the presence of the N-H group and the enaminic C=C double bound, discriminating the actual 2,3-DP end product from the 1,2-DP isomer. In our opinion, the MS/MS-derived information presented by Ding et al. (2016) alone might not be sufficient to allow the discrimination between both DP isomers, because the [M+H]+ ions generated from 1,2-DP and 2,3-DP after positive ionization would possess identical masses and could both yield the observed mass fragments, including the C4H7 fragment that was quoted as an indicator of 1,2-DP formation (Ding et al., 2016).
Chemical interconversions between 1,2-DP and 2,3-DP and structurally related ketimine/enamine tautomeric pairs can occur, and the relative stability of corresponding ketimine and enamine tautomers might depend on several factors, such as the pH in free solution or complexation with cofactors in an enzymatic environment (Nardini et al., 1988; Nishina et al., 1991). Alkaline conditions have been proposed to favor enamine formation (Nardini et al., 1988). Subcellular localization studies in Arabidopsis and rice show that ALD1 fusion proteins localize to chloroplasts (Cecchini et al., 2015; Jung et al., 2016), indicating that the site of ALD1 action is the organelle in which the substrate l-Lys is biosynthesized (Mazelis et al., 1976). With pH values around 7 and 8 in the stroma of dark-exposed and illuminated chloroplasts, respectively (Werdan et al., 1975), the site of ALD1-mediated l-Lys conversion is neutral to moderately alkaline, which might favor the formation of the 2,3-DP enamine. Our standard assay buffer for in vitro conversion studies also was adjusted to slightly basic conditions (pH 7.5–8). However, enamine formation by ALD1 in in vitro studies proved rather pH independent in our hands, because variations of the pH of the assay buffer between pH 5 and 9 always yielded the same products 1a and 1b in the two employed analytical procedures (data not shown). While analytical procedure A, which produced the methylated derivative 1a, involves a short acidic extraction step, analytical procedure B, yielding chloroformate-derivatized 1b, involves an acidic extraction step, followed by an alkaline washing step and derivatization under neutral conditions. Despite these methodological differences, both procedures detected derivatives of enaminic 2,3-DP as the end product of the ALD1-mediated reaction.
LysOx from T. viride in combination with catalase is reported to convert l-Lys to ketiminic 1,2-DP, which was characterized by complexation with o-aminobenzaldehyde and UV/visible spectroscopy of the yellow adduct in previous studies (Vogel and Davis, 1952; Misono et al., 1971; Kusakabe et al., 1980). We aimed at generating 1,2-DP by the LysOx/catalase reaction to directly compare it with the l-Lys-derived ALD1 product (Supplemental Fig. S2). Both LysOx/catalase and ALD1 assays yielded a yellow substance with the reported absorption maximum at 450 nm (Soda et al., 1968; Kusakabe et al., 1980), which sets it clearly apart from the spectra of the described o-aminobenzaldehyde complexes formed by the isomer 1,6-DP (λmax = 465 nm) or the five-ring analog Δ1-Pyr2C (λmax = 436 nm). Hence, these analyses suggest the ketimine tautomer 1,2-DP as the product of both the LysOx/catalase- and ALD1-catalyzed reactions. By contrast, as detailed above, our GC-MS and GC-FTIR analyses indicate that 2,3-DP is the final product of l-Lys conversion by LysOx/catalase and ALD1 (Figs. 1 and 3). Yet, the conclusiveness of the UV/visible spectroscopic detection method is not unambiguous, because heterocyclic enamines such as 2,3-DP also can undergo condensation with o-aminobenzaldehyde (Cervinka, 1988). Moreover, evidence has been provided that heterocyclic imines such as Δ1-piperideine, which builds the core structural element of 1,2-DP, can indeed react as their heterocyclic enamine tautomers in the o-aminobenzaldehyde reaction (Harada et al., 1970; Hickmott, 1982). Together, these findings strongly argue against the possibility to discriminate between 1,2-DP and 2,3-DP by the o-aminobenzaldehyde reaction and subsequent UV/visible spectroscopic analysis.
Do ALD1-Mediated Reactions beyond the Pip Biosynthetic Pathway Possess Physiological Relevance in Planta?
Recombinant ALD1 was characterized previously as an aminotransferase with substrate preference for l-Lys, but the protein also accepted several other amino acids as substrates in vitro. The corresponding transamination products, however, had not been identified (Song et al., 2004a). Since leaf petiole exudates of ALD1-overexpressing plants induced stronger defense responses than those from wild-type plants and Pip levels were similar in both kinds of exudates, it was proposed that ALD1 could activate defense responses by non-Pip metabolites (Cecchini et al., 2015). To shed light on the possible alternative roles of ALD1 beyond Lys catabolism, we examined the relative in vitro activity of ALD1 protein toward a series of other amino acids and attempted to identify the respective conversion products in vitro. Furthermore, we analyzed whether in vitro-identified conversion products would be detectable in plant extracts, accumulate upon P. syringae inoculation in the wild type, and fail to do so in the ald1 mutant (Table I; Supplemental Table S1). These features are common to the Lys-derived metabolites 2,3-DP and Pip (Figs. 1 and 3; Návarová et al., 2012).
Besides l-Lys being the preferred substrate, recombinant ALD1 exhibited marked relative aminotransferase activities toward l-Met, l-Leu, l-Arg, and l-Orn and more moderate but clearly detectable relative activities toward l,l-DAP, meso-DAP, l-Phe, l-Glu, l-Aad, l-Asn, and l-His (Table I; Supplemental Table S1). The activity range observed here overlaps largely with the set of ALD1-converted amino acid substrates described previously by Song et al. (2004a). A common feature of the accepted α-amino acid substrates is a side chain extending to at least the δ-position, whereby the chemical characteristics of individual side chain positions vary considerably. A branching of the side chain at the β-position, as is the case for l-Ile, appears to reduce the substrate affinity of ALD1 (Table I).
For all of the above-mentioned amino acid substrates except l-Arg and l-His, we were able to detect in vitro transamination products by GC-MS analyses. Whereas the corresponding α-ketoacids were identified as in vitro products for l-Met, l-Leu, l-Phe, l-Glu, l-Aad, and l-Asn as ALD1 substrates, cyclized products derivable from 2-ketoacid precursors were detected as DAP- and l-Orn-derived ALD1 conversion products (Table I; Supplemental Fig. S8). Out of the products detected in vitro, only l-Glu-derived α-ketoglutaric acid, a common primary metabolite involved in numerous metabolic pathways (Lancien et al., 2000), occurred to higher levels in plant extracts. Moreover, moderate levels of the l-Phe-derived phenylpyruvic acid and l-Aad-derived α-ketoadipic acid were identified (Table I). However, these transamination products occurred to similar amounts in wild-type and ald1 mutant plants, indicating that the in vivo biosynthesis of these compounds is not related to functional ALD1. The other identified in vitro products were not detected in Col-0 leaf extracts, irrespective of whether plants were P. syringae inoculated or not (Table I). This suggests that the corresponding catabolic pathways are not activated in plants to detectable levels. From dilution experiments with standard substances, we conservatively estimate that the detection limits of the applied GC-MS methods are well below 0.5 nmol g−1 fresh weight for the relevant compounds. Our results thus indicate that the transamination of l-Lys to 2,3-DP leading to Pip production is a major in vivo function of ALD1. Since possible transamination products of l-Arg and l-His were not amenable to our analyses, we cannot completely exclude a role for ALD1 in l-Arg and l-His catabolism in the plant. However, since exogenous application of Pip is virtually sufficient to complement the defects of the ald1 mutant in basal resistance to P. syringae and SAR (Návarová et al., 2012; Bernsdorff et al., 2016), we consider it unlikely that the l-Arg- or l-His metabolism significantly contributes to ALD1-controlled immune responses.
Why is l-Lys catabolism to 2,3-DP the supposedly dominant if not the only in planta function of ALD1, although the enzyme exerts considerable in vitro transaminase activity toward other substrates? First, the relative activity toward l-Lys is higher than that toward any of the other amino acids under investigation (Table I). Second, ALD1 resides in the chloroplasts (Cecchini et al., 2015), the subcellular compartment in which l-Lys biosynthesis takes place (Mazelis et al., 1976). Third, the levels of Lys strongly increase upon pathogen inoculation in parallel to the ALD1 transcript abundance (Návarová et al., 2012). Hence, ALD1-mediated l-Lys catabolism to 2,3-DP in planta is obviously favored by the spatial and temporal concurrence of enzyme and substrate. However, the biosyntheses of the readily accepted ALD1 substrates l-Met, l-Leu, DAP, and l-Orn are supposed to take place in chloroplasts as well (Jander and Joshi, 2009; Binder, 2010; Winter et al., 2015), and their transamination products α-keto-Met, α-keto-Leu, 6-carboxy-2,3-DP, and Δ2-Pyr2C, respectively, could be detected in in vitro ALD1 assays but not in planta (Table I; Fig. 5; Supplemental Table S1; Supplemental Fig. S8). The levels of Met, DAP, and Orn in P. syringae-inoculated leaves are markedly lower than the levels of Lys or not detectable at all (Návarová et al., 2012; Supplemental Table S3), indicating that their availabilities as in planta ALD1 substrates lag significantly behind the availability of Lys. However, like Lys, Leu strongly accumulates in the P. syringae-infected leaves (Návarová et al., 2012), making it another good candidate for an ALD1 in vivo substrate, in theory. The main organelle of Leu catabolism in plants is the mitochondrion (Binder, 2010), and possibly, rapid and efficient transport of Leu out of the chloroplast might account for our finding that ALD1-mediated transamination in the chloroplast does not occur to detectable levels.
l,l-DAP and meso-DAP, the 6-carboxylated variants of l-Lys, were converted by recombinant ALD1 into 6-carboxy-2,3-DP (Fig. 5), and it is reasonable to assume that the underlying reaction series conforms to the proposed l-Lys-to-2,3-DP conversion sequence (Fig. 2). The enaminic 6-carboxy-2,3-DP (2a and 2b) and 2,3-DP (1a and 1b) derivatives show strikingly overlapping mass and IR spectral properties, reflecting the fact that the two compounds only differ from each other by the presence or absence of the 6-carboxyl substitution (Figs. 1, 3, and 5; Supplemental Fig. S7). Both l,l-DAP and meso-DAP are intermediates in the biosynthetic pathway of l-Lys (Jander and Joshi, 2009). The aminotransferase with the closest sequence similarity to ALD1 in Arabidopsis is ABERRANT GROWTH AND DEATH2 (AGD2; Song et al., 2004a), and recombinant AGD2 has been reported to catalyze the interconversion of l-tetrahydrodipicolinate to l,l-DAP in vitro (Hudson et al., 2006). Since l-tetrahydrodipicolinate and 6-carboxy-2,3-DP are ketimine/enamine tautomers, the previous data and our results indicate that AGD2 and ALD1 both have in vitro transamination activities toward l,l-DAP. Nevertheless, the crystal structures of AGD2 and ALD1 show differences in their presumed substrate-binding sites, suggesting different biochemical functions in vivo (Watanabe et al., 2007; Sobolev et al., 2013).
SARD4-Catalyzed Generation of Pip from 2,3-DP
Although ald1 mutant plants fail to accumulate 2,3-DP and Pip after P. syringae inoculation, they are able to convert exogenously applied 2,3-DP into Pip in the presence of a pathogen stimulus (Fig. 6, A and B). Therefore, the capacity to reduce a DP intermediate to Pip is intact in the ald1 mutant. On the basis of protein sequence comparison, we previously proposed that Arabidopsis SARD4 (alias ORNCD1) might be involved in the reduction step required for Pip formation (Zeier, 2013). SARD4 is the closest homolog of the mammalian ketimine reductase CRYM that utilizes either NADH or NADPH as a cofactor (Hallen et al., 2011). CRYM preferentially catalyzes the reduction of cyclic ketimine monocarboxylate substrates such as 1,2-DP or its five-membered ring analog Δ1-Pyr2C (Hallen et al., 2015). Subcellular localization experiments indicate that, like ALD1, Arabidopsis SARD4 resides in plastids (Sharma et al., 2013). Moreover, SARD4 transcripts accumulate in both inoculated (1°) and distal (2°) leaves of Col-0 plants upon P. syringae attack (Fig. 8, A and B). These features made SARD4 a likely candidate reductase for a possible function in pathogen-inducible Pip biosynthesis that could act in combination with ALD1 in the chloroplast.
As outlined above, enzyme assays in which recombinant ALD1, the amino acid substrate l-Lys, the acceptor oxoacid pyruvate, and the cofactor PLP necessary for the transamination step were present resulted in the formation of 2,3-DP from l-Lys (Fig. 1; Table I). The same was true when the reducing cofactor NADPH (or NADH) was additionally present in these ALD1-only assays (Fig. 7). However, in coupled assays in which both ALD1 and SARD4 protein were combined with PLP, pyruvate, and reducing equivalents, substantial amounts of Pip were generated from l-Lys, and the detected levels of 2,3-DP were markedly lower than in the ALD1-only assay (Fig. 7). This indicates that the ALD1 product 2,3-DP is converted to Pip by SARD4 and NAD(P)H as the reducing cofactor in the enzyme mixture. Likewise, after coapplication of ALD1 and the mammalian ketimine reductase CRYM in the bioassay, Pip was formed to significant amounts in the enzyme mixture, and the levels of 2,3-DP decreased compared with an individual ALD1 incubation (Fig. 7). This confirms our hypothesis that the combined action of ALD1 and SARD4 is sufficient to realize the l-Lys-to-Pip conversion in vitro.
Interestingly, we also detected trace amounts of Pip in the ALD1 control assays lacking any of the reductase enzymes in the reaction mixture but containing all of the cofactors relevant for the coupled assay (Fig. 7). An explanation for this at first sight surprising observation is given by earlier studies showing that DP can be reduced nonenzymatically to Pip by pyridine nucleotides such NADH (Meister et al., 1957).
Our results further suggest that SARD4 and its mammalian homolog CRYM have comparable biochemical characteristics, because the ALD1/CRYM and ALD1/SARD4 combinations provided similar results. Since CRYM has been characterized as a reductase that accepts ketiminic substrates such as 1,2-DP (Hallen et al., 2015), we assume that the 2,3-DP detected in our assays and plant extracts is not the direct substrate for SARD4 (and CRYM). Instead, an equilibrium between 2,3-DP and 1,2-DP that is largely shifted to the detectable enaminic form could exist. Reductive conversion of the minor equilibrium component 1,2-DP to Pip by SARD4 (or CRYM) might constantly reestablish low amounts of 1,2-DP ketimine through an equilibrium shift (Fig. 2). The mechanism suggested here involving enaminic 2,3-DP as the dominant cyclic DP form and a rapid enamine-ketimine tautomerism in the course of reduction could indirectly favor Pip generation, because ketimines are prone to spontaneous hydrolysis to the corresponding open oxoacid (Nardini et al., 1988), which would destabilize the piperideine ring structure and, hence, disfavor a subsequent reduction to the saturated piperideine ring of Pip.
It is noteworthy in this context that, in coupled assays with ALD1 and SARD4 and with l-Orn as a substrate, the formation of significant amounts of Pro could be detected, which suggests that SARD4 also possesses Pyr2C reductase activity in vitro (Supplemental Fig. S11). Mammalian CRYM possesses a similar Pyr2C reductase activity (Hallen et al., 2015; Supplemental Fig. S11). Meister et al. (1957) already observed a Pyr2C-to-Pro reducing activity in plant extracts. Meister (1965) later hypothesized that this activity may reflect a lack of specificity of the enzyme catalyzing the reduction step in Pip formation. The commonly accepted core Pro biosynthetic pathway in planta, however, operates via pyrroline-5-carboxylate (Verslues and Sharma, 2010). Since the Δ2-Pyr2C metabolite was not detected in planta in our study and the leaf levels of Pro do not rise in response to P. syringae infection (Table I; Supplemental Fig. S8; Supplemental Table S3), it is unlikely that the SARD4-mediated Pyr2C-to-Pro reduction or the coupled ALD1/SARD4-mediated Orn-to-Pro conversion has biochemical or functional relevance in planta.
To examine the biochemical and physiological functions of SARD4 in planta, we examined the metabolite profiles and immune responses of the knockout line sard4-5 that completely lacks SARD4 transcription (Fig. 8, A and B). The local and systemic accumulation of Pip in P. syringae-inoculated plants is markedly delayed in this mutant (Fig. 8, C and E). Whereas at inoculation sites, still considerable levels of Pip are generated at 2 d post inoculation (dpi) in sard4-5, the systemic accumulation characteristically observed in the wild type at 2 dpi is absent. At later stages (3 and 4 dpi), however, a modest systemic increase is detectable (Fig. 8, C and E). In addition, 2,3-DP overaccumulated in the P. syringae-inoculated sard4-5 plants (Fig. 8D). These data and the outcome of the coupled ALD1/SARD4 assays suggest that SARD4 contributes significantly to the pathogen-inducible biosynthesis of Pip in Arabidopsis leaves by reducing DP intermediates to Pip. However, since the ald1 knockout mutant completely lacks Pip accumulation but sard4-5 does not (Návarová et al., 2012; Fig. 8, C and E), another reducing activity downstream of ALD1 supposedly exists that contributes to Pip formation from DP precursors (Fig. 2). This suggests the necessity to investigate the relevance of alternative plant dehydrogenases involved in Pip biosynthesis in future studies.
Parallel to sard4-5, the knockdown line sard4-6, which exhibited residual and pathogen-inducible expression of SARD4, was analyzed (Fig. 8, A and B). Together with the quantitative PCR (qPCR)-based transcriptional analysis, PCR-based genotyping suggested that a truncated SARD4 transcript version is expressed in sard4-6 (Fig. 8, A and B; Supplemental Fig. S9). The metabolite analyses showed that the ability to accumulate Pip in the local and systemic leaf tissue was still strong in sard4-6, although moderately reduced compared with the wild type (Fig. 8, C and E). This suggests that sard4-6 expresses a still active, albeit a presumably truncated, version of SARD4. In line with this interpretation, a study by Wang (2008) collecting data on the effect of T-DNA insertions in the exon region of Arabidopsis genes on transcript levels showed that, in 8% of all documented cases, reduced transcript levels still could be detected, and in 2% of cases, a truncated version of the protein was expressed. However, the sard4-6 mutant also overaccumulated 2,3-DP in the leaves of P. syringae-inoculated plants, which might be the result of reduced SARD4 gene expression or attenuated enzymatic activity (Fig. 8, A, B, and D). Although the sard4-6 mutant is a weaker allele than sard4-5, sard4-6 overaccumulated 2,3-DP to an even greater extent than sard4-5 in the systemic, 2° leaves (Fig. 8C). As exemplified by the higher systemic accumulation of SA in sard4-6 compared with sard4-5, the overall systemic response is presumably stronger in the sard4-6 mutant than in sard4-5 (Fig. 9C). Since an increased expression of ALD1 is an integral part of this systemic response (Bernsdorff et al., 2016), this would result in a significantly stronger systemic ALD1 expression and concomitant systemic biosynthesis of 2,3-DP in sard4-6 compared with sard4-5. Together with the markedly attenuated SARD4 levels in sard4-6 (Fig. 8B), this would then contribute to the observed strong overaccumulation of 2,3-DP in the systemic tissue of sard4-6.
In parallel to this study, the above-mentioned work of Ding et al. (2016) has characterized the role of SARD4 in Pip biosynthesis and SAR. A forward genetic screen identified Arabidopsis sard4 mutants with defects in SAR to the oomycete pathogen Hyaloperonospora arabidopsidis, and the mutated SARD4 gene was shown to be allelic with SARD4. Using the sard4-5 mutant (GK_428E01, alias orncd1-1) for in planta analyses and in vitro biochemical characterization of the SARD4 protein, the results of Ding et al. (2016) essentially coincide with the function of SARD4 described here as a reductase that significantly contributes to Pip biosynthesis in Arabidopsis. Notably, based on leaf samples collected at 2 d post P. syringae inoculation, Ding et al. (2016) reported the complete absence of systemic Pip increases in sard4-5 plants and, consequently, emphasized the strict necessity of functional SARD4 for the systemic accumulation of Pip. By contrast, our more extended time-course analysis identified a significant pathogen-induced systemic increase of Pip at later infection stages in sard4-5, even though we also did not observe systemic increases at 2 dpi and detected greatly reduced levels at 3 and 4 dpi (Fig. 8, C and E). Thus, although SARD4 provides a predominant contribution to the systemic accumulation of Pip, a delayed SARD4-independent pathway exists for this process.
Relevance of SARD4-Mediated Pip Biosynthesis for Basal Plant Immunity and SAR
Naive ald1 mutant plants are not able to accumulate Pip in response to pathogens and are more susceptible to P. syringae infection than wild-type plants. This resistance defect of ald1 is abolished after exogenous treatment of plants with Pip, indicating a significant function for Pip in plant basal immunity (Fig. 9B; Song et al., 2004b; Návarová et al., 2012; Bernsdorff et al., 2016). The P. syringae-inoculated sard4-5 plants exhibited a marked delay in local Pip accumulation, and naive or mock-pretreated sard4-5 mutants also showed significantly attenuated basal resistance to P. syringae in different experiments (Figs. 8B and 9, A, B, and D). Non pretreated or mock-pretreated sard4-6 plants, which are only moderately compromised in local Pip biosynthesis, allowed marginally higher bacterial multiplication in their leaves than the wild type after a single infection; this mild tendency, however, was not statistically significant in most of the experiments (Fig. 9, A, B, and D). Together, these data indicate that the effectiveness of basal resistance to P. syringae correlates with the magnitude of Pip accumulation in Col-0, ald1, sard4-5, and sard4-6 plants (Fig. 8C; Návarová et al., 2012). In addition, sard4-5 and sard4-6 mutant plants exogenously supplemented with Pip showed enhanced basal resistance to P. syringae similar to the resistance level observed in the Col-0 wild type (Fig. 9D). In sum, these data suggest that the accumulation of SARD4-derived Pip in infected leaves adds a significant contribution to the basal resistance capacity of these leaves to bacterial infection. In this context, it should be noted that Ding et al. (2016) observed no difference in basal resistance to P. syringae between the sard4-5 line and the wild type. As outlined above, we cannot confirm this finding, because sard4-5 showed significant defects in basal resistance to P. syringae in all of our experiments (Fig. 9, A, B, and D).
The defect of sard4-5 in basal resistance, however, was not associated with a weakening of pathogen-induced SA biosynthesis, as documented by a similar accumulation of the phenolic defense hormone in P. syringae-inoculated Col-0 and sard4-5 leaves at 24 hpi (Fig. 9C). The relation of SA and Pip in resistance induction was investigated recently, and it was established that the Pip-mediated amplification of SA-inducible defense gene expression such as PATHOGENESIS-RELATED1 is an important component of Pip/SA cross talk (Bernsdorff et al., 2016). Therefore, the diminished production of Pip in inoculated sard4-5 leaves, which is especially pronounced at earlier infection stages (24 hpi; Fig. 8C), might cause attenuated defense gene expression and, as a consequence, reduced disease resistance.
The ability of Arabidopsis plants to synthesize Pip upon pathogen attack is a prerequisite for the establishment of SAR (Shah and Zeier, 2013). Evidence for the necessity of elevated Pip levels for SAR comprises the findings that Pip strongly accumulates in the pathogen-inoculated 1° leaves and in distant 2° leaves. Furthermore, Pip-deficient ald1 mutants are fully compromised in SAR as well as in the systemic transcriptional response associated with SAR. Exogenous Pip, in turn, restores the ability of ald1 to establish SAR upon localized P. syringae inoculation (Návarová et al., 2012; Bernsdorff et al., 2016). The chemical complementation assays used in these previous studies were based on exogenous pretreatments of individual plants with a dose of 10 µmol of Pip via the root system, which resulted in an uptake of Pip into the shoot and an increase of the Pip levels in leaves to values of about 8 µg g−1 fresh weight (Návarová et al., 2012). Thus, the increases of Pip levels in leaves by this treatment were similar to the rises of Pip observed in the systemic (2°) leaves after biological SAR induction with P. syringae. Moreover, the resistance effects caused by biological SAR induction and by exogenous Pip treatments were quantitatively comparable (Fig. 8, C and E; Návarová et al., 2012; Bernsdorff et al., 2016). On this basis, we previously proposed a working model that highlighted a central role for the Pip that accumulates in the systemic (2°) leaf tissue for the induction of SAR (Návarová et al., 2012; Shah and Zeier, 2013; Zeier, 2013; Bernsdorff et al., 2016). However, both the metabolic phenotype of the ald1 mutant lacking Pip biosynthesis in the 1° inoculated and noninoculated 2° leaf tissue and the mode of the chemical complementation assay via the root that resulted in the rise of Pip in the whole shoot (and, therefore, in both 1° and 2° leaves) cannot rule out a decisive function for accumulating Pip in the 1° inoculated leaves from which SAR signaling to the systemic tissue is initiated. The question of whether the SAR-inducing action of Pip primarily takes place in the inoculated leaves, in the distant, systemic leaves, or to a similar extent in both types of leaf tissue is not trivial to address in our experience. For example, feeding experiments that include localized infiltration of Pip specifically to the 1° or the 2° leaves are difficult to interpret, because the infiltration of Pip solutions into leaves did not result in resistance induction within the treated leaves (Návarová et al., 2012). Possibly, in contrast to root tissue, leaf cells lack a proper uptake system for the polar amino acid across the plasma membrane (Tegeder, 2014).
The metabolic phenotype of the sard4-5 mutant, however, is informative in this context. Albeit in a markedly delayed manner, Pip accumulated to high levels in the P. syringae-inoculated leaves of sard4-5. By contrast, the systemic increase in the 2° leaves are absent at 2 dpi and rather weak at later infection stages (Fig. 8, C and E). Nevertheless, sard4-5 plants were able to induce a significant SAR response to P. syringae (Fig. 9B) and actually established a partial but diminished SAR also toward the oomycete pathogen H. arabidopsidis (Ding et al., 2016). In terms of relative bacterial growth attenuation between mock-pretreated and P. syringae-pretreated plants, the SAR effect in sard4-5 was similar to the effect in the wild type in our bacterial growth assays (Fig. 9B). However, the absolute resistance levels of both mock-treated plants (basal resistance) and of SAR-induced plants were lower for the sard4-5 mutant than for the wild-type plants (Fig. 9B). A possible interpretation of these findings is that Pip locally accumulating in the inoculated 1° leaves already exerts a significant triggering function for systemic resistance induction. The systemic Pip that accumulates in the 2° leaves after induction would then contribute to the absolute strength of acquired resistance in the systemic tissue. The sard4-5 mutants also accumulated markedly reduced amounts of SA in their systemic leaves compared with the wild type (Fig. 9C). This is consistent with our previous hypothesis that the systemic increases of Pip observed upon SAR induction in the wild type is necessary for a strong systemic accumulation of SA (Návarová et al., 2012; Bernsdorff et al., 2016). The phenotype of sard4-5, however, now allows us to refine our previous model and to introduce a spatial aspect of the action of Pip during SAR. This includes a SAR-inducing function of locally accumulating Pip in the inoculated (1°) leaves and, as worked out previously (Návarová et al., 2012; Bernsdorff et al., 2016), a role in defense amplification in the distant (2°) leaves.
Exogenous application of the Pip precursor 2,3-DP proved sufficient to induce a moderate resistance response in Arabidopsis leaves (Fig. 9E). It is not entirely clear whether this effect is related to a direct resistance-enhancing function of 2,3-DP or to the in planta production of immune-active Pip from the DP precursor (Fig. 6, A and B). In the wild type, a marked contribution of 2,3-DP to the overall pipecolate resistance pathway is unlikely, because the local and systemic accumulation of signaling-active Pip quantitatively exceeds the accumulation of the 2,3-DP intermediate by about 2 orders of magnitude (Fig. 8, C and D). However, 2,3-DP strongly overaccumulates in the sard4-5 and sard4-6 mutants and, therefore, might add a significant contribution to the local and systemic resistance responses in these plants (Fig. 8D). As mentioned before, enhanced defense-stimulating activity of leaf exudates from ALD1-overexpressing plants was reported recently, albeit the collected exudates were not enriched in Pip (Cecchini et al., 2015). Considering the consecutive ALD1/SARD4 mechanism for Pip production described in this study and by Ding et al. (2016; Fig. 2), it is conceivable that the 2,3-DP accumulating in the ALD1-overexpressing lines studied by Cecchini et al. (2015) might have mediated the observed resistance effects.
CONCLUSION
Our study has identified a pathogen-inducible Pip biosynthesis pathway starting from l-Lys in Arabidopsis and involving the consecutive action of the l-Lys aminotransferase ALD1 and the NAD(P)H-dependent reductase SARD4. These findings are consistent with results from a parallel study by Ding et al. (2016). Both studies show that ALD1 abstracts the α-amino group of l-Lys in vitro. On top of that, our data also demonstrate this l-Lys-α-aminotransferase function for ALD1 directly in planta. In addition, we show by in vitro analyses that the α-N of l-Lys is transferred directly to acceptor oxoacids (e.g. pyruvate or α-ketoglutarate) to produce the corresponding amino acids (Ala or Glu) within the ALD1-mediated transamination reaction. These findings are consistent with earlier studies that propose an l-Pip biosynthesis pathway via the ketoacid KAC as an intermediate in plants and animals (Rothstein and Miller, 1954; Gupta and Spenser, 1969) but inconsistent with a suggested plant metabolic pathway that involves abstraction of the ε-N of l-Lys and the formation of AAS as a biosynthetic intermediate (Schütte and Seelig, 1967; Fig. 2).
However, the ketoacid KAC is not the final product of the ALD1-catalyzed transamination of l-Lys. Whereas Ding et al. (2016), based solely on mass spectrometric information, suggested the cyclic ketimine 1,2-DP as the detectable product, our study indicates, by combined mass spectrometric and IR spectroscopic evidence and the application of two different analytical procedures, that the final in vitro and in planta product of the ALD1-catalyzed reaction is enaminic 2,3-DP. As a plausible mechanism of 2,3-DP formation, we propose ALD1-mediated transamination of l-Lys to KAC, dehydrative cyclization to 1,2-DP, and isomerization to the 2,3-DP tautomer (Fig. 2). Quantitative analyses of 2,3-DP in plant extracts showed that the Pip precursor 2,3-DP accumulates in both P. syringae-inoculated and systemic leaves of wild-type plants, but the absolute levels of 2,3-DP are about 2 orders of magnitude lower than those of Pip.
Our study also addresses the question of whether the ALD1-mediated transamination of amino acids other than l-Lys would have biochemical and physiological relevance in plants. Although ALD1 is able to transaminate several amino acids such as l-Met, l-Leu, l-Orn, and DAP to corresponding oxoacids or derived products in vitro, detailed metabolite analyses of plant extracts suggest that such transamination reactions do not occur in planta, indicating that l-Lys catabolism to 2,3-DP is the predominant if not the only in vivo function of ALD1. In planta analyses also show that ald1 knockout plants, albeit fully defective in Pip accumulation, have the capacity to reduce exogenous 2,3-DP into Pip, indicating that their Pip deficiency relies on the inability to form the 2,3-DP enamine.
Our combined in vitro assays with recombinant ALD1 and SARD4 and comparable assays conducted by Ding et al. (2016) reveal that SARD4 functions as a dehydropipecolate reductase and converts DP intermediates into Pip. Analyses of sard4 knockout plants further show that SARD4 contributes significantly to the pathogen-inducible formation of Pip in Arabidopsis but that another reducing activity must exist in the plant that accounts for a SARD4-independent Pip accumulation. In line with the data of Ding et al. (2016), we find substantial, but delayed and reduced, accumulation of Pip in P. syringae-inoculated leaves of sard4 knockout plants. However, whereas Ding et al. (2016) proposed a strict necessity of SARD4 for the systemic accumulation of Pip on the basis of samples from knockout plants collected at 2 dpi, we performed a more extended time-course analysis and identified a markedly reduced but significant pathogen-induced systemic increase of Pip in these plants later at 3 and 4 dpi, indicating that a residual SARD4-independent Pip biosynthesis in Arabidopsis is not restricted to inoculation sites. Furthermore, our comparative in vitro assays with Arabidopsis SARD4 and the mammalian reductase homolog CRYM directly illustrate the biochemical similarity of both enzymes. Interestingly, both reductases have not only the in vitro capacity to reduce the six-member heterocycle 2,3-DP into Pip but also can convert the five-member Pyr2C into Pro, albeit in planta analyses suggest that the latter reaction is not related to an in vivo function of SARD4.
The results of both this work and the study of Ding et al. (2016) suggest that functional SARD4 is necessary for a proper, wild-type-like SAR response. However, while Ding et al. (2016) report a full SAR defect of sard4 knockout plants, we observe a significant SAR response in these mutants. We show that sard4 plants exhibit a reproducibly lower basal resistance to P. syringae than the wild type but increase systemic resistance upon a localized inoculation to a wild-type-like degree (as expressed by fold changes of bacterial growth in the challenge-infected 2° leaves upon 1° mock versus 1° Psm treatment). This ultimately results in SAR-induced sard4 plants with a reduced absolute resistance level compared with SAR-induced wild-type plants, accompanied by markedly reduced systemic responses (such as SA accumulation) in the mutants. Finally, on the bases of the metabolic and immune phenotypes of sard4 knockout plants, we propose that a local action of Pip that accumulates in the 1° inoculated leaves exerts a significant triggering function contributing to systemic resistance induction.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in individual pots containing a mixture of soil (Klasmann-Deilmann; Substrat BP3), vermiculite, and sand (8:1:1) under strictly controlled conditions inside a growth chamber with a 10-h-day (9 am–7 pm; photon flux density of 100 µmol m−2 s−1)/14-h-night cycle. The relative humidity in the chambers was adjusted to 60%, and the temperatures during the light and dark periods were set to 21°C and 18°C, respectively. For experiments, 5- to 6-week-old plants with an unstressed, uniform appearance were used. The SARD4 (alias ORNCD1; At5g52810) T-DNA insertion lines sard4-5 (orncd1-1; GK_428E01) and sard4-6 (orncd1-2; GK_696E11) were obtained from the European Arabidopsis Stock Centre. Identification of homozygous individual mutants was performed according to the protocol of Alonso et al. (2003) using gene-specific and T-DNA left border primers (Supplemental Fig. S9; Supplemental Table S2). In addition, the SAR-deficient Arabidopsis ald1 mutant (ald1_T2; Salk_007673) was used in this study (Song et al., 2004b; Návarová et al., 2012). All mutant lines are in the Arabidopsis Col-0 background.
Cultivation of Bacteria and Plant Inoculation
Pseudomonas syringae pv maculicola strain ES4326 and Psm carrying the Photorhabdus luminescens luxCDABE operon were grown in King’s B medium supplemented with the appropriate antibiotics at 28°C under permanent shaking (Zeier et al., 2004; Fan et al., 2008). Overnight log-phase cultures were washed four times with 10 mm MgCl2 solution and diluted to different final OD600 levels for leaf inoculation. The diluted bacterial solutions as well as mixtures thereof with more complex solutions, containing metabolites of interest, were carefully pressure infiltrated from the abaxial side of the leaves using a needleless syringe, covering the whole surface of the respective leaf.
Assessment of Basal Resistance and SAR
To induce SAR, plants were infiltrated into three fully grown lower (1°) leaves with a suspension of Psm at OD600 = 0.005 between 10 and 11 am. Plants infiltrated with a 10 mm MgCl2 solution served as mock-treated controls. For the determination of systemic responses, upper (2°) leaves were harvested 48 h after the primary treatment.
For SAR assessment, three upper (2°) leaves of all pretreated plants were infiltrated with Psm lux at OD600 = 0.001 48 h after the primary treatment. Bacterial growth was quantified by measuring the bacterial bioluminescence in leaf discs (10 mm in diameter) of 2° leaves (one disc per leaf, three discs per plant) with a Serius FB12 luminometer (Berthold Detection Systems). Bacterial growth rates were expressed as relative light units per cm2 of leaf area (Fan et al., 2008). At least six to seven replicate plants per treatment and plant genotype were measured before a statistical analysis of the resulting values was performed.
To assess bacterial growth in naive 5-week-old plants (basal resistance), three fully grown leaves per plant were infiltrated with a suspension of Psm lux at OD600 = 0.001 between 10 and 11 am. At 60 h later, the bacterial growth in the infiltrated leaves was determined by measuring the bacterial bioluminescence in leaf discs as described above. The analysis of the resulting data was performed as explained for the SAR assessment.
Determination of 2,3-DP, SA, and Other Relevant Metabolites in Leaf Material and in Vitro Enzyme Assays (Analytical Procedure A)
Levels of free SA, 2,3-DP, and other metabolites listed in Table I in Arabidopsis leaf samples or enzyme assays were determined by solvent extraction of leaf material or enzyme assays followed by a vapor-phase extraction-based workup of the extracts and subsequent analysis of the resulting samples via GC-MS. In detail, shock-frozen leaf material (approximately 100–200 mg originating from up to six leaves) was pulverized in liquid N2 using a ball mill and homogenized with 600 µL of extraction buffer, consisting of water:1-propanol:HCl (1:2:0.005). In the case of enzyme assays, 50 to 100 µL per assay was used and extracted as described above. Following the addition of 100 ng of d4-SA as an internal standard and 1 mL of methylene chloride, the mixture was thoroughly rehomogenized and centrifuged at 14,000g for 1 min for optimal phase separation. For the analyses of free SA and 2,3-DP, the lower organic phase was removed, dried with Na2SO4, and incubated with 2 µL of 2 m trimethylsilyl diazomethane in hexane (Sigma-Aldrich) for 5 min at room temperature, driving the conversion of carboxylic acids into the corresponding methyl esters. To stop the methylation reaction, an excess of acetic acid (2 µL of a 2 m solution in hexane) was added to the vials, and the sample was then subjected to a vapor-phase extraction procedure at 70°C and 200°C using a steady stream of nitrogen and a volatile collector trap packed with Porapak-Q absorbent (VCT-1/4X3-POR-Q; Analytical Research Systems) according to Schmelz et al. (2004). The volatilized and trapped metabolites were then eluted from the absorbent with 1 mL of methylene chloride, and the sample volume was reduced to 30 µL in a stream of nitrogen prior to GC-MS analysis.
Four microliters of the processed sample mixture was then separated on a gas chromatograph (GC 7890A; Agilent Technologies) equipped with a fused silica capillary column (ZB5 MS; Zebron), and mass spectra were recorded with a 5975C mass spectrometric detector (Agilent Technologies) in the electron ionization mode as described before (Návarová et al., 2012). The quantitative determination of metabolites was performed by integrating peaks originating from selected ion chromatograms (for m/z values, see Table I) and relating the areas of a substance peak to the peak area of d4-SA (ion, m/z 124). Correction factors experimentally determined for each substance by the use of authentic substances were considered. The correction factor for the quantification of 2,3-DP (ion, m/z 108) was approximated by measuring the consumption of the amino acceptor α-ketoglutarate in the ALD1 assay reaction mix at distinct concentrations. Metabolite levels were related to either leaf fresh weight or total volume of an in vitro assay.
Determination of Amino Acids (Analytical Procedure B)
Levels of Pip and other relevant amino acids were determined using the EZ:faast free amino acid analysis kit for GC-MS (Phenomenex), which is based on the separation and identification of propyl chloroformate-derivatized amino acids by GC-MS (Kugler et al., 2006). To assess amino acid levels from Arabidopsis leaves, 50 mg of homogenized leaf material was extracted, derivatized, and analyzed as described previously in detail (Návarová et al., 2012). For the identification and quantification of amino acids from in vitro enzyme assays, the workup procedure was identical, with the exception that instead of 50 mg of leaf material, 50 µL of an individual assay was processed and analyzed. A correction factor for the quantification of 2,3-DP (ion, m/z 255) was established experimentally with reference to the internal standard l-Norvaline by measuring the consumption of l-Lys and the formation of Ala from the amino acceptor pyruvate in the ALD1 assay reaction mix at distinct substrate concentrations.
GC-FTIR Analysis of Metabolites
GC-FTIR spectra were acquired using a Hewlett-Packard 6890 Series gas chromatograph coupled with an IRD3 IR detector manufactured by ASAP Analytical. IR spectra were recorded in the mid-IR range from 4,000 to 600 cm−1 (wave number = reciprocal value of wavelength) with a resolution of 4 cm−1 and a scan rate of eight scans per second. The IRD3 method parameters were as follows: resolution = 16; apodization = triangle; phase correction = mertz; zero-fill = 1; co-add = 2. The temperature of the light pipe and the transfer lines were both set to 250°C. Nitrogen was used as the sweep gas. The gas chromatograph was operated in splitless mode using helium as the carrier gas, with a flow rate of 2 mL min−1 and a column head pressure of 9.54 p.s.i. The column used in the GC-FTIR studies was a fused silica capillary column (Zebron ZB-5; 30 m × 0.32 mm × 0.25 µm) purchased from Phenomenex. The gas chromatograph oven temperature program consisted of an initial temperature of 50°C for 3 min, ramped up to 240°C for 8 min, followed by a ramping step to 320°C over the course of 20 min (total time, 33.75 min). Depending on the nature of the sample, 1 to 4 µL was injected using an Agilent 7863 Series autoinjector. Please note that the derivatization of the samples (procedure A), the column, as well as the temperature program used in our GC-FTIR studies were all identical to the setup used in the complementing GC-MS studies, with the goal of simplifying the identification of relevant target peaks.
Plant Treatment with l-Lys and Isotope-Labeled Lys Varieties
For in planta labeling experiments, three to four mature leaves of 5-week-old soil-grown Arabidopsis wild-type Col-0 and ald1 mutant plants were infiltrated in the morning with Psm (OD600 = 0.005) or MgCl2 (mock controls) as described before. After 4 h, the same leaves were infiltrated with 5 mm solutions of l-Lys or one of the isotope-labeled varieties l-Lys-6-13C-ε-15N (204451-46-7; Sigma-Aldrich) and l-Lys-4,4,5,5-d4 (Cambridge Isotope Laboratories) prepared in HPLC-grade water. Water infiltrations served as control treatments. Infiltrated leaves were harvested at 48 hpi (counting from the first infiltration event with Psm or mock treatment) and subsequently extracted, derivatized, and analyzed by GC-MS according to analytical procedures A and B.
Plant Treatments with Pip
Treatments with Pip were performed essentially as detailed by Návarová et al. (2012). Ten milliliters of an aqueous 1 mm d,l-Pip solution (equates to 10 µmol) or 10 mL of water as a control treatment was exogenously applied to individual 5-week-old plants. For resistance assays, inoculation with Psm lux (OD600 = 0.001) was performed 24 h after Pip or water application, and bacterial growth was assessed as outlined above. At 48 h later, leaf samples were harvested and defense responses were quantified as described (see “Assessment of Basal Resistance and SAR” above).
Plant Treatment with 2,3-DP
Application of exogenous 2,3-DP to Arabidopsis Col-0 and ald1 plants was achieved by coinfiltrating mature leaves of 5-week-old plants with Psm and 2,3-DP obtained from ALD1 or LysOx/catalase in vitro assays. With regard to ALD1 assays, mixtures containing 20 mm l-Lys as a substrate were run to completion (greater than 99% of Lys used) as described in detail in the respective sections, heat inactivated, filtered to eliminate remaining protein (Vivaspin 6 centrifugal concentrator, molecular weight cut off 10,000 D; Sigma-Aldrich), and diluted 6-fold in 10 mm MgCl2. As a control, the same assay containing boiled, inactive ALD1 enzyme (no-enzyme control) was used in the coinfiltration assays. With respect to LysOx/catalase assays, LysOx- and catalase-containing reaction mixtures were run to completion, stopped, filtered as described above, and diluted. Controls consisted of full assays that lacked LysOx (catalase only). The bacterial suspension was added into ALD1 (or LysOx/catalase) assay mixture to a final OD600 of 0.005 and infiltrated into three leaves of 5-week-old plants. At 48 h later, infiltrated leaf samples were harvested for subsequent metabolite analysis and processed by analytical procedure B. Additional controls consisted of Psm and mock infiltrations without any of the ALD1 or LysOx/catalase assay components.
For basal resistance assays, Psm lux (OD600 = 0.001) and enzymatically synthesized 2,3-DP were coinfiltrated into three mature leaves of 5- to 6-week-old Col-0 or ald1 plants. The final concentration of 2,3-DP in these infiltration experiments was approximately 2 mm. Control experiments were performed as described in the last paragraph of the “Results” section. After 60 h, bacterial growth was assessed as outlined above.
Data Analyses
HPLC data were analyzed using ChemStation for LC 3D systems software version Rev.B.04.02 SP1 [208] (Agilent Technologies). GC-MS data were analyzed using MSD ChemStation software version E.02.01.1177 (Agilent Technologies). For analysis of Fourier transform IR data, Essential FTIR software version 3.10.037 was used (Operant).
Analysis of Gene Expression
Gene expression was assessed by quantitative real-time PCR analysis essentially as described by Návarová et al. (2012) and Bernsdorff et al. (2016), with the exception that an absolute standard curve was used (Pfaffl, 2004). In brief, 1 µg of total RNA was extracted with peqGOLD TriFast reagent (PeqLab) and used for cDNA synthesis using the GoScript reverse transcriptase (Promega) in combination with oligo(dT)18 primers according to the manufacturer’s instructions. Quantification of the targeted amplified cDNA templates was carried out with GoTaq qPCR master mix (Promega) according to the manufacturer’s instructions using the gene-specific primer pairs listed in Supplemental Table S2. qPCR was performed in triplicate using a Rotor-Gene Q system (Qiagen) with the following cycling program: 1 min at 95°C (enzyme activation); followed by 40 cycles of 20 s at 95°C (denaturation) and 20 s at 60°C (annealing/extension); and a final extension step at 72°C for 20 s. Data analysis was performed with Rotor-Gene Q software version 2.0.2 via absolute quantification of transcripts using an external standard curve. The standard curve was obtained using recombinant plasmid DNA, more precisely the SARD4 coding sequence cloned into the commercial pET32b vector (Invitrogen), quantified by multiple measurements in various dilutions and concentrations (0.02–20 ng). Unknown sample amounts were determined from the respective calibration data by interpolating the PCR signals into the external standard curve. The values for the correlation coefficient r2 for the standard curves had to be greater than 0.99 to be considered reliable. The resulting quantification data were depicted as concentrations in ng µL−1.
LysOx Assays and Biosynthesis of 2,3-DP
LysOx from T. viride (EC 1.4.3.14) was purchased from Sigma-Aldrich as lyophilized powder (70132-14-8) and used according to the manufacturer’s protocol (Kusakabe et al., 1980) with a few minor modifications. Lyophilized powder was resuspended in either water or 50 mm sodium phosphate buffer (pH 8) to obtain a stock solution with approximately 5 units mL−1 enzyme. The solution was homogenized, divided into aliquots, and immediately stored at −20°C until further use. The enzyme stock solution could be stored for several months without any significant loss in activity. As described in detail before (Supplemental Fig. S2), the substrate-specific LysOx from T. viride catalyzes the conversion of l-Lys to KAC, and if conducted in the presence of catalase to remove hydrogen peroxide formed during the oxidation step, 2,3-DP is formed as the main product of the reaction due to the spontaneous intramolecular cyclization of the KAC intermediate. Catalase from bovine liver was purchased as lyophilized powder from Sigma-Aldrich (9001-05-2) and had to be prepared immediately before use in cold deionized water (stock solution with 3,500 units mL−1 enzyme). At first, LysOx assays were carried out following the exact protocol provided by the manufacturer. However, to minimize the generation of two undesired by-products, 5-aminovaleric acid and the piperideine-2-carboxylic acid dimer (Supplemental Fig. S2), various adjustments to this protocol were made. At the end of the optimization process, the following conditions were chosen. Reactions were carried out with a final concentration of l-Lys of 10 mm in 25 mm sodium phosphate buffer (pH 8) in the presence of 0.5 units of LysOx and 875 units of catalase per milliliter of reaction mix. Assays were allowed to run to completion at 37°C (1 h to overnight). CaCl2 (1 mm) was added to the reaction mix to improve relative activity (Kusakabe et al.,1980). Each set of assays included at least a blank control (no enzyme) as well as controls lacking one of the two enzymes of the reaction mixture, catalase and LysOx, incubated under the same conditions.
Depending on the next steps, the assays were either stopped by the addition of methanol (equal amount), 0.1 m HCl (1/10th of the volume), or by heating the reaction mixture at 85°C for 15 min. Full assays and controls were usually run as replicates of three. Thin-layer chromatography analysis was regularly used to rapidly assess the success of an assay before proceeding to a consecutive step of an experiment such as coupled assays or feeding experiments. In this case, 2 µL of an overnight assay series was spotted onto a thin-layer chromatography silica gel 60 F254 plate (20 × 20 cm; Merck). The mobile phase consisted of either acetone:methanol:water (10:2:2) or butanol:acetic acid:water (4:1:1). To visualize the consumption of l-Lys, the plates were sprayed with ninhydrin reagent (0.5% (w/v) ninhydrin in ethanol) and heated to 100°C in an oven until individual spots were seen. The postchromatographic derivatization with ninhydrin stained l-Lys dark red/orange, whereas Pip was stained purple. Quantitative measurements of the enzyme activity were performed in all cases by following both the formation of the 2,3-DP product and the consumption of the substrate Lys via GC-MS procedures A or B.
Cloning of ALD1, SARD4 (ORNCD1), and CRYM
cDNA/DNA fragments corresponding to ALD1 (At2g13810), SARD4 (ORNCD1; At5g52810), and human CRYM (EC 1.5.1.25) were amplified using high-fidelity Phusion polymerase (New England Biolabs) according to the manufacturer’s recommendations. National Center for Biotechnology Information reference sequence accession numbers are as follows: ALD1 (NM_126957.1) and SARD4 (NM_124659.1). The full-length human CRYM open reading frame bacterial expression clone (Entrez gene identifier, 1428 transcript variant 1; longer isoform clone identifier, HsCD00333103) was purchased from Harvard Medical School plasmid stocks. Presumable targeting sequences within the gene sequences were identified via TargetP, SignalP, and related tools (Emanuelsson et al., 2007) and taken into account during the primer design. For ALD1, the nucleotides coding for the plastidial N-terminal targeting (−20 amino acids [Δ20-ALD1]) sequence have been omitted as described in prior studies (Song et al., 2004a; Hudson et al., 2006; Sobolev et al., 2013). The gene sequences of Δ20-ALD1, SARD4, and CRYM were introduced into the target vector pET-32b(+) (Novagen) via sticky-end cloning (Zeng, 1998) using NdeI and XhoI restriction sites. The recombinant proteins thus contained eight nonnative residues (LEHHHHHH) at the C terminus, including the poly-His tag. Primer sequences can be found in Supplemental Table S2. Plasmids containing the respective genes were transformed into chemically competent Escherichia coli BL21 Rosetta 2(DE3) pLysS cells and plated on lysogeny broth (LB) agar plates containing the appropriate selection markers. Transformants carrying the gene of interest were identified by colony PCR using the same gene-specific primers used for the initial amplification and were verified by plasmid sequencing.
Expression and Purification of Recombinant Enzymes
All genes expressed in this study were cloned into the pET-32b vector system, putting the genes under the control of the phage-derived isopropyl-β-d-1-thiogalactopyranoside-inducible T7 promoter (LaVallie et al., 1993). For expression of the recombinant proteins described in this study, a single colony was picked and cultured overnight in 3 mL of LB medium supplemented with the appropriate selection markers at 37°C and with constant shaking on an orbital shaker before inoculating and growing 500 mL of culture under the same conditions until the OD600 reached 0.5 to 0.8. At this point, the culture was briefly cooled down and the transgene expression was induced with 1 mm isopropyl-β-d-1-thiogalactopyranoside, followed by incubation overnight at 16°C to 25°C with constant shaking (240 rpm). It should be noted that best results were obtained with freshly transformed cells and incubation at reduced temperatures (16°C) after induction of transgene expression to minimize the precipitation of recombinant enzymes as insoluble inclusion bodies. The bacterial pellets were collected by centrifugation at 6,000g for 15 min at 4°C (Eppendorf R5810). Cells were resuspended in a minimum of extraction/binding buffer (50 mm sodium phosphate, pH 8, 500 mm NaCl, 10% glycerol, 20 mm imidazole, 5 mm β-mercaptoethanol, and 1 mm phenylmethylsulfonyl fluoride). The homogenate was transferred to a precooled mortar and ground in liquid nitrogen with a pestle until a homogenous powder was obtained. The powder was transferred to 2-mL Eppendorf tubes and allowed to thaw on ice. The resulting homogenate was then precipitated using a centrifuge at 16,000g for 30 min at 4°C. The cell-free supernatant containing the majority of soluble recombinant proteins was collected and filtered through a low-protein-binding nylon filter (0.22 µm) before being applied to a preequilibrated immobilized metal ion affinity chromatography column, such as a nickel-charged His GraviTrapaffinity column (GE Healthcare) or a cobalt-charged HisTALON Gravity column (Takara Bio; 1 mL).
After the binding step, the column was washed successively according to the respective manufacturer’s recommendations. The proteins were then eluted with 50 mm sodium phosphate buffer, pH 8, containing up to 500 mm NaCl and 200 mm imidazole, and collected in fractions of 0.75 mL. Under these conditions, most of the enzyme activity was found to be in four to five consecutive fractions, which were combined and desalted using a 5-mL desalting PD10 column (GE Healthcare) equilibrated with the respective buffers, 50 mm Tris (pH 8) and 100 mm potassium phosphate buffer (pH 8), both containing 10% (v/v) glycerol and 1 mm DTT for ALD1 and the reductases, respectively. Aliquots of the purified proteins were used for the quantification of total protein content by the Bradford method (Kruger, 1994) using the Bradford Assay reagent (Bio-Rad) according to the manufacturer’s protocol. Bovine serum albumin was used for the standard curve. Target enzyme purity was determined by SDS-PAGE on a 12% gel in accordance with the method of Laemmli (1970; data not shown). The purified recombinant protein stocks were used directly for enzyme assays, and the remaining protein was flash frozen in liquid nitrogen and stored at −80°C in 20% (v/v) glycerol supplemented with 1% (w/v) phenylmethylsulfonyl fluoride until further use.
ALD1 Enzyme Assays
ALD1 enzyme assays were performed based on the protocol from Song et al. (2004a) with a few minor modifications. In general, reactions were carried out in an assay mix (50 or 200 µL) containing purified recombinant Δ20-ALD1 enzyme at a final concentration of 20 to 100 µg mL−1 with 10 to 40 mm of a respective amino donor (e.g. l-Lys), 10 to 40 mm of an amino acceptor (e.g. pyruvate), 5 mm MgCl2, and 100 µm PLP in 50 mm Tris buffer, pH 8, supplemented with 5% (v/v) glycerol. Depending on the purpose of the experiment, such as maximizing the formation of possible ALD1 products with alternative amino acid substrates, assays were incubated at 37°C for up to 16 h. In general, the reaction was initiated by the addition of purified Δ20-ALD1 enzyme, which had usually been diluted in the corresponding assay buffer. At the end of the incubation period, reactions were stopped by one of the following methods: addition of an equal volume of methanol, addition of 10% (v/v) 0.1 m HCl to the reaction mixture, or boiling of the assays for 10 min at 85°C. In general, the reaction was stopped by inactivating the enzyme at 85°C for 10 min. These different methods for stopping the reaction were all tested to determine the effects of pH and solvents on the composition of the reaction products. Reactions without ALD1 enzyme or with heat-inactivated enzyme were systematically performed as controls. Quantitative measurements of ALD1 activity were made either by following the formation of ALD1 reaction products such as 2,3-DP or the transamination side products like l-Ala (from pyruvate) or by measuring the consumption of the respective substrates via GC-MS-based procedures A and B. All assays were repeated at least in triplicate.
Coupled Assays with Recombinant ALD1 Enzyme and CRYM/SARD4
Coupled enzymes were assayed by incubating freshly purified recombinant ALD1 enzyme alone or in combination with one of the two reductases, CRYM and SARD4, at a final concentration of 100 µg mL−1 per enzyme in a reaction mix containing 20 mm l-Lys (or 20 mm l-Orn), 20 mm pyruvate, 5 mm MgCl2, 100 µm PLP, and 200 µm NADH (or NADPH) in 20 mm Tris buffer, pH 8. All reaction mixtures contained 5% (v/v) glycerol for additional enzyme stability and were incubated at 37°C for 16 h. Reactions were stopped by inactivating the enzymes at 85°C for 10 min. The formation of 2,3-DP or Pip was monitored using GC-MS after derivatization of the assays with trimethylsilyl diazomethane (procedure A) or propyl chloroformate (procedure B), respectively. Control assays included reaction mixtures not containing any substrates as well as reaction mixtures incubating all three enzymes separately in the presence of all assay components. Again, all assays were repeated at least in triplicate.
Statistical Analyses
Statistical significance was assessed by ANOVA with type II sum of squares using the R statistical package (https://www.r-project.org/), the command “aov (Phenotype ∼ Treatment + Genotype + Treatment * Genotype, data=object1)” (with object1 being the data table loaded into the R workspace), and subsequent posthoc Tukey’s HSD test (Brady et al., 2015; Bernsdorff et al., 2016).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_126957.1 (ALD1), NM_124659.1 (SARD4), and NM_001888.4 (CRYM).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The DP isomer identified in plant extracts and the in vitro ALD1 DP product have identical GC retention times.
Supplemental Figure S2. Synthesis of 2,3-DP by l-Lys oxidase from T. viride.
Supplemental Figure S3. In planta formation of isotope-labeled 2,3-DP versions.
Supplemental Figure S4. Wave numbers of the IR absorption bands of the C=O stretching vibrations occurring in different carboxylic acid methyl esters, as determined by GC-FTIR analysis.
Supplemental Figure S5. In addition to 2,3-DP, the l-Lys-derived DP dimer can be detected in l-Lys oxidase assays carried out in the presence of catalase.
Supplemental Figure S6. Valerolactam is the main l-Lys-derived reaction product in l-Lys oxidase assays performed in the absence of catalase.
Supplemental Figure S7. IR spectra of valerolactam, DP dimer, 2,3-DP methyl ester, and 6-carboxy-2,3-DP dimethyl ester, as determined by GC-FTIR analyses.
Supplemental Figure S8. Structures of nonderivatized and derivatized ALD1 transamination products and their mass spectra, as determined by GC-MS analysis.
Supplemental Figure S9. Genomic organization of the SARD4 mutant alleles sard4-5 (orncd1-1; GK_428E01) and sard4-6 (orncd1-2; GK-696E11), and characterization of sard4 mutant lines.
Supplemental Figure S10. SAR bioassay with the Col-0 wild type, the sard4-5 mutant, and the sard4-6 mutant. Disease symptomology of representative plants.
Supplemental Figure S11. Coupled in vitro assays with Arabidopsis ALD1 and SARD4, or with ALD1 and the human reductase CRYM, yield Pro with l-Orn as a substrate.
Supplemental Table S1. ALD1 in vitro activity toward additional amino acids.
Supplemental Table S2. Primers used in this study.
Supplemental Table S3. Levels of free amino acids in the leaves of Psm-inoculated and mock-treated Arabidopsis Col-0 plants.
Supplementary Material
Acknowledgments
We thank Karin Kiefer and Holger Schnell for excellent technical assistance.
Glossary
- SAR
systemic acquired resistance
- SA
salicylic acid
- Pip
pipecolic acid
- KAC
ε-amino-α-ketocaproic acid
- DP
dehydropipecolic acid
- AAS
α-aminoadipate semialdehyde
- Aad
α-amino adipic acid
- GC-MS
gas chromatography-mass spectrometry
- Col-0
Columbia-0
- GC
gas chromatography
- PLP
pyridoxal-5′-phosphate
- GC-FTIR
gas chromatography-Fourier transform infrared
- IR
infrared
- DAP
diaminopimelate
- 2,4-DABA
diaminobutyric acid
- Pyr2C
pyrroline-2-carboxylic acid
- hpi
hours post inoculation
- MS/MS
tandem mass spectrometry
- qPCR
quantitative PCR
- dpi
days post inoculation
- HSD
honestly significant difference
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. ZE467/6-1).
Articles can be viewed without a subscription.
References
- Aliferis KA, Faubert D, Jabaji S (2014) A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS ONE 9: e111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Attaran E, Zeier TE, Griebel T, Zeier J (2009) Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 21: 954–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsdorff F, Döring AC, Gruner K, Schuck S, Bräutigam A, Zeier J (2016) Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28: 102–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. The Arabidopsis Book 8: e0137, doi/10.1199/tab.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird RG, Vaquero-Vara V, Zaleski DP, Pate BH, Pratt DW (2012) Chirped-pulsed FTMW spectra of valeric acid, 5-aminovaleric acid, and δ-valerolactam: a study of amino acid mimics in the gas phase. J Mol Spectrosc 280: 42–46 [Google Scholar]
- Brady SM, Burow M, Busch W, Carlborg Ö, Denby KJ, Glazebrook J, Hamilton ES, Harmer SL, Haswell ES, Maloof JN, et al. (2015) Reassess the t test: interact with all your data via ANOVA. Plant Cell 27: 2088–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broquist HP. (1991) Lysine-pipecolic acid metabolic relationships in microbes and mammals. Annu Rev Nutr 11: 435–448 [DOI] [PubMed] [Google Scholar]
- Campillo-Brocal JC, Lucas-Elío P, Sanchez-Amat A (2015) Distribution in different organisms of amino acid oxidases with FAD or a quinone as cofactor and their role as antimicrobial proteins in marine bacteria. Mar Drugs 13: 7403–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini NM, Jung HW, Engle NL, Tschaplinski TJ, Greenberg JT (2015) ALD1 regulates basal immune components and early inducible defense responses in Arabidopsis. Mol Plant Microbe Interact 28: 455–466 [DOI] [PubMed] [Google Scholar]
- Cervinka O. (1988) Heterocyclic enamines. In Cook AG, ed, Enamines: Synthesis: Structure, and Reactions, Ed 2 Marcel Dekker, New York, pp 442–529 [Google Scholar]
- Chen W, Li X, Tian L, Wu P, Li M, Jiang H, Chen Y, Wu G (2014) Knockdown of LjALD1, AGD2-like defense response protein 1, influences plant growth and nodulation in Lotus japonicus. J Integr Plant Biol 56: 1034–1041 [DOI] [PubMed] [Google Scholar]
- Ding P, Rekhter D, Ding Y, Feussner K, Busta L, Haroth S, Xu S, Li X, Jetter R, Feussner I, et al. (2016) Characterization of a pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell 28: 2603–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fan J, Crooks C, Lamb C (2008) High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J 53: 393–399 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Sakurai A (1997) Conversion of lysine to L-pipecolic acid induces flowering in Lemna paucicostata 151. Plant Cell Physiol 38: 1278–1280 [Google Scholar]
- Galili G, Tang G, Zhu X, Gakiere B (2001) Lysine catabolism: a stress and development super-regulated metabolic pathway. Curr Opin Plant Biol 4: 261–266 [DOI] [PubMed] [Google Scholar]
- Gruner K, Griebel T, Návarová H, Attaran E, Zeier J (2013) Reprogramming of plants during systemic acquired resistance. Front Plant Sci 4: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RN, Spenser ID (1969) Biosynthesis of the piperidine nucleus: the mode of incorporation of lysine into pipecolic acid and into piperidine alkaloids. J Biol Chem 244: 88–94 [PubMed] [Google Scholar]
- Hallen A, Cooper AJ, Jamie JF, Haynes PA, Willows RD (2011) Mammalian forebrain ketimine reductase identified as μ-crystallin: potential regulation by thyroid hormones. J Neurochem 118: 379–387 [DOI] [PubMed] [Google Scholar]
- Hallen A, Cooper AJ, Jamie JF, Karuso P (2015) Insights into enzyme catalysis and thyroid hormone regulation of cerebral ketimine reductase/μ-crystallin under physiological conditions. Neurochem Res 40: 1252–1266 [DOI] [PubMed] [Google Scholar]
- Hallen A, Jamie JF, Cooper AJ (2013) Lysine metabolism in mammalian brain: an update on the importance of recent discoveries. Amino Acids 45: 1249–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K, Mizoe Y, Furukawa J, Yamashita S (1970) Reactions of Δ1-piperideine derivatives with heterocumulenes. Tetrahedron 26: 1579–1588 [Google Scholar]
- Hickmott PW. (1982) Enamines: recent advances in synthetic, spectroscopic, mechanistic, and stereochemical aspects. II. Tetrahedron 38: 3363–3446 [Google Scholar]
- Hope DB, Horncastle KC, Aplin RT (1967) The dimerization of delta-1-piperidine-2-carboxylic acid. Biochem J 105: 663–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AO, Singh BK, Leustek T, Gilvarg C (2006) An ll-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol 140: 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Joshi V (2009) Aspartate-derived amino acid biosynthesis in Arabidopsis thaliana. The Arabidopsis Book 7: e0121, doi/10.1199/tab.0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung GY, Park JY, Choi HJ, Yoo SJ, Park JK, Jung HW (2016) A rice gene homologous to Arabidopsis AGD2-LIKE DEFENSE1 participates in disease resistance response against infection with Magnaporthe oryzae. Plant Pathol J 32: 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kruger NJ. (1994) The Bradford method for protein quantitation. Methods Mol Biol 32: 9–15 [DOI] [PubMed] [Google Scholar]
- Kugler F, Graneis S, Schreiter PPY, Stintzing FC, Carle R (2006) Determination of free amino compounds in betalainic fruits and vegetables by gas chromatography with flame ionization and mass spectrometric detection. J Agric Food Chem 54: 4311–4318 [DOI] [PubMed] [Google Scholar]
- Kusakabe H, Kodama K, Kuninaka A, Yoshino H, Misono H, Soda K (1980) A new antitumor enzyme, L-lysine alpha-oxidase from Trichoderma viride: purification and enzymological properties. J Biol Chem 255: 976–981 [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lancien M, Gadal P, Hodges M (2000) Enzyme redundancy and the importance of 2-oxoglutarate in higher plant ammonium assimilation. Plant Physiol 123: 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM (1993) A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology (NY) 11: 187–193 [DOI] [PubMed] [Google Scholar]
- Leonard NJ, Hauck FP (1957) Unsaturated amines. X. The mercuric acetate route to substituted piperidines, Δ2-tetrahydropyridines and Δ2-tetrahydroanabasines. J Am Chem Soc 79: 5279–5292 [Google Scholar]
- Lukasheva EV, Berezov TT (2002) L-Lysine alpha-oxidase: physicochemical and biological properties. Biochemistry (Mosc) 67: 1152–1158 [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2014) Plant PRRs and the activation of innate immune signaling. Mol Cell 54: 263–272 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Clément G, Anne P, Routaboul JM, Guiboileau A, Soulay F, Shirasu K, Yoshimoto K (2014) Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell 26: 1857–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelis M, Miflin BJ, Pratt HM (1976) A chloroplast-localized diaminopimelate decarboxylase in higher plants. FEBS Lett 64: 197–200 [DOI] [PubMed] [Google Scholar]
- Meister A. (1965) Intermediary metabolism of amino acids. In Meister A, ed, Biochemistry of the Amino Acids, Ed 2 Academic Press, New York, pp 593–1020 [Google Scholar]
- Meister A, Buckley SD (1957) Pyridine nucleotide-dependent reduction of the α-keto acid analogue of lysine to L-pipecolic acid. Biochim Biophys Acta 23: 202–203 [DOI] [PubMed] [Google Scholar]
- Meister A, Radhakrishnan AN, Buckley SD (1957) Enzymatic synthesis of L-pipecolic acid and L-proline. J Biol Chem 229: 789–800 [PubMed] [Google Scholar]
- Memelink J. (2009) Regulation of gene expression by jasmonate hormones. Phytochemistry 70: 1560–1570 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Misono H, Yamamoto T, Soda K (1971) Bacterial L-lysine-α-ketoglutarate aminotransferase. Bull Inst Chem Res Kyoto Univ 49: 128–165 [Google Scholar]
- Morrison RI. (1953) The isolation of L-pipecolinic acid from Trifolium repens. Biochem J 53: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M, Ricci G, Caccuri AM, Solinas SP, Vesci L, Cavallini D (1988) Purification and characterization of a ketimine-reducing enzyme. Eur J Biochem 173: 689–694 [DOI] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring A-C, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina Y, Sato K, Shiga K (1991) Isomerization of delta 1-piperideine-2-carboxylate to delta 2-piperideine-2-carboxylate on complexation with flavoprotein D-amino acid oxidase. J Biochem 109: 705–710 [DOI] [PubMed] [Google Scholar]
- Pálfi G, Dézsi L (1968) Pipecolic acid as an indicator of abnormal protein metabolism in diseased plants. Plant Soil 29: 285–291 [Google Scholar]
- Pfaffl MW. (2004) Quantification strategies in real-time PCR. In Bustin SA, ed, The Real-Time PCR Encyclopedia: A–Z of Quantitative PCR. International University Line, La Jolla, CA, pp 87–120 [Google Scholar]
- Pollegioni L, Motta P, Molla G (2013) L-Amino acid oxidase as biocatalyst: a dream too far? Appl Microbiol Biotechnol 97: 9323–9341 [DOI] [PubMed] [Google Scholar]
- Pukin AV, Boeriu CG, Scott EL, Sanders JPM, Franssen MCR (2010) An efficient enzymatic synthesis of 5-aminovaleric acid. J Mol Catal B Enzym 65: 58–62 [Google Scholar]
- Rao ST, Rossmann MG (1973) Comparison of super-secondary structures in proteins. J Mol Biol 76: 241–256 [DOI] [PubMed] [Google Scholar]
- Rothstein M, Miller LL (1954) The conversion of lysine to pipecolic acid in the rat. J Biol Chem 211: 851–858 [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT (2004) The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J 39: 790–808 [DOI] [PubMed] [Google Scholar]
- Schütte HR, Seelig G (1967) Zur Biosynthese der Pipecolinsäure in Phaseolus vulgaris. Z Naturforsch 22b: 824–826 [PubMed] [Google Scholar]
- Shah J, Zeier J (2013) Long-distance communication and signal amplification in systemic acquired resistance. Front Plant Sci 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Shinde S, Verslues PE (2013) Functional characterization of an ornithine cyclodeaminase-like protein of Arabidopsis thaliana. BMC Plant Biol 13: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V, Edelman M, Dym O, Unger T, Albeck S, Kirma M, Galili G (2013) Structure of ALD1, a plant-specific homologue of the universal diaminopimelate aminotransferase enzyme of lysine biosynthesis. Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda K, Misono H, Yamamoto T (1968) L-Lysine:alpha-ketoglutarate aminotransferase. I. Identification of a product, delta-1-piperideine-6-carboxylic acid. Biochemistry 7: 4102–4109 [DOI] [PubMed] [Google Scholar]
- Song JT, Lu H, Greenberg JT (2004a) Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, ABERRANT GROWTH AND DEATH2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JT, Lu H, McDowell JM, Greenberg JT (2004b) A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J 40: 200–212 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Tegeder M. (2014) Transporters involved in source to sink partitioning of amino acids and ureides: opportunities for crop improvement. J Exp Bot 65: 1865–1878 [DOI] [PubMed] [Google Scholar]
- Vernooij B, Friedrich L, Morse A, Reist R, Kolditz-Jawhar R, Ward E, Uknes S, Kessmann H, Ryals J (1994) Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6: 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Sharma S (2010) Proline metabolism and its implications for plant-environment interaction. The Arabidopsis Book 8: e0140, doi/10.1199/tab.0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vogel HJ, Davis BD (1952) Glutamic γ-semialdehyde and Δ1-pyrroline-5-carboxylic acid, intermediates in the biosynthesis of proline. J Am Chem Soc 74: 109–112 [Google Scholar]
- Vogel-Adghough D, Stahl E, Návarová H, Zeier J (2013) Pipecolic acid enhances resistance to bacterial infection and primes salicylic acid and nicotine accumulation in tobacco. Plant Signal Behav 8: e26366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH. (2008) How effective is T-DNA insertional mutagenesis in Arabidopsis? J Biochem Tech 1: 11–20 [Google Scholar]
- Watanabe N, Cherney MM, van Belkum MJ, Marcus SL, Flegel MD, Clay MD, Deyholos MK, Vederas JC, James MN (2007) Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana: a recently discovered enzyme in the biosynthesis of L-lysine by plants and Chlamydia. J Mol Biol 371: 685–702 [DOI] [PubMed] [Google Scholar]
- Werdan K, Heldt HW, Milovancev M (1975) The role of pH in the regulation of carbon fixation in the chloroplast stroma: studies on CO2 fixation in the light and dark. Biochim Biophys Acta 396: 276–292 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Winter G, Todd CD, Trovato M, Forlani G, Funck D (2015) Physiological implications of arginine metabolism in plants. Front Plant Sci 6: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharius RM, Thompson JF, Steward FC (1954) The detection, isolation and identification of L(-)-pipecolic acid in the non-protein fraction of beans (Phaseolus vulgaris). J Am Chem Soc 76: 2908–2912 [Google Scholar]
- Zeier J. (2013) New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ 36: 2085–2103 [DOI] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S (2004) Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]
- Zeng G. (1998) Sticky-end PCR: new method for subcloning. Biotechniques 25: 206–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.