A plant cryptochrome has multiple roles within the circadian clock and the sexual cycle of a unicellular photosynthetic organism.
Abstract
Cryptochromes are flavin-binding proteins that act as blue light receptors in bacteria, fungi, plants, and insects and are components of the circadian oscillator in mammals. Animal and plant cryptochromes are evolutionarily divergent, although the unicellular alga Chlamydomonas reinhardtii (Chlamydomonas throughout) has both an animal-like cryptochrome and a plant cryptochrome (pCRY; formerly designated CPH1). Here, we show that the pCRY protein accumulates at night as part of a complex. Functional characterization of pCRY was performed based on an insertional mutant that expresses only 11% of the wild-type pCRY level. The pcry mutant is defective for central properties of the circadian clock. In the mutant, the period is lengthened significantly, ultimately resulting in arrhythmicity, while blue light-based phase shifts show large deviations from what is observed in wild-type cells. We also show that pCRY is involved in gametogenesis in Chlamydomonas. pCRY is down-regulated in pregametes and gametes, and in the pcry mutant, there is altered transcript accumulation under blue light of the strictly light-dependent, gamete-specific gene GAS28. pCRY acts as a negative regulator for the induction of mating ability in the light and for the loss of mating ability in the dark. Moreover, pCRY is necessary for light-dependent germination, during which the zygote undergoes meiosis that gives rise to four vegetative cells. In sum, our data demonstrate that pCRY is a key blue light receptor in Chlamydomonas that is involved in both circadian timing and life cycle progression.
Light has major impacts on algal physiology and development. It serves as an energy source that drives photosynthesis, triggers photoorientation, entrains the circadian clock over light/dark cycles, controls metabolic pathways, and regulates developmental processes (for review, see Hegemann, 2008; Kianianmomeni and Hallmann, 2014). Diverse sensory photoreceptors are central to these processes, including CRYPTOCHROMES (CRYs), PHOTOTROPINS (PHOTs), AUREOCHROMES, RHODOPSINS, PHYTOCHROMES (PHYs) and UV-RESISTANT LOCUS8 (Hegemann, 2008; Petroutsos et al., 2016; Tilbrook et al., 2016).
In the green biflagellate alga Chlamydomonas reinhardtii (Chlamydomonas throughout), light controls the sexual cycle at three specific steps: (1) gamete formation; (2) maintenance of gamete mating competence; and (3) zygote germination (Pan et al., 1997; Huang and Beck, 2003; Goodenough et al., 2007). Vegetative Chlamydomonas cells become pregametes when the nitrogen source is removed from the medium, while their conversion to gametes is triggered mainly by blue light (Weissig and Beck, 1991; Saito et al., 1998). Gametes (mating type plus and mating type minus) then fuse to form zygotes, which germinate in the light on nitrogen-containing medium (Huang and Beck, 2003; Goodenough et al., 2007). The blue light photoreceptor PHOT was shown to be involved in the control of gametogenesis and germination in Chlamydomonas (Huang and Beck, 2003).
Light/dark cycles entrain the Chlamydomonas circadian clock. A clock-dependent rhythmic accumulation of cells in the light (also described as rhythm of phototaxis) was observed in Chlamydomonas. Automated methods have been developed to measure this rhythm (Bruce, 1970; Mergenhagen, 1984; Forbes-Stovall et al., 2014) and were even used to examine this phenomenon in space (low-gravity environment), where the rhythm persists for several days (Mergenhagen and Mergenhagen, 1987). Based on analyses of the rhythm of phototaxis, it was demonstrated that red, green, and blue light can reset the phase of the circadian clock (Johnson et al., 1991; Kondo et al., 1991; Forbes-Stovall et al., 2014). Over the last decade, several components of the Chlamydomonas circadian clock that influence its period, phase, and/or amplitude have been functionally characterized. These components include factors involved in RNA metabolism (Iliev et al., 2006; Dathe et al., 2012), a CASEIN KINASE1 (Schmidt et al., 2006), CONSTANS, a protein also involved in photoperiodic control (Serrano et al., 2009), and the RHYTHM OF CHLOROPLAST transcription factors ROC15, ROC40, ROC66, and ROC75 (Matsuo et al., 2008). Recently, ROC15 was demonstrated to be involved in the phase-resetting mechanism, acting together with the F-box protein ROC114 (Niwa et al., 2013).
CRYs and PHYs are plant photoreceptors that are known to entrain the circadian clock (Galvão and Fankhauser, 2015). Analyses of the full genome sequence of Chlamydomonas suggest that this alga does not encode the red light photoreceptor PHY (Mittag et al., 2005; Merchant et al., 2007). But Chlamydomonas has four candidate CRY proteins (Beel et al., 2012): an animal-like CRY (aCRY; Beel et al., 2012), a plant CRY named CPH1 (for Chlamydomonas Photolyase Homolog1; Reisdorph and Small, 2004), as well as two CRY-DASH proteins of so far unknown function (Beel et al., 2012). CRY proteins have a conserved photolyase homology region (PHR) and a C-terminal extension of various lengths. Their bound chromophore is FAD, with some that may additionally bind 5,10-methenyltetrahydrofolate as an antenna pigment (Sancar, 2003; Chaves et al., 2011).
Chlamydomonas aCRY is closely related to mammalian CRYs and DNA repair enzymes, the (6-4) photolyases. It is evolutionarily close to members of the Cryptochrome Photolyase Family1 of another green alga (Ostreococcus tauri) and the diatom Phaeodactylum tricornutum (Beel et al., 2012, 2013), which are sensory blue light receptors and have maintained photolyase activity (Coesel et al., 2009; Heijde et al., 2010). Recently, the first functional analyses of aCRY from Chlamydomonas were performed in vitro (Beel et al., 2012; Spexard et al., 2014; Nohr et al., 2016; Oldemeyer et al., 2016). aCRY absorbs light over the visible spectrum up to 680 nm as a consequence of the formation of the neutral radical form of flavin. In Chlamydomonas, aCRY influences transcript levels following pulses of blue, yellow, and red light (Beel et al., 2012). The transcripts impacted by aCRY encode proteins of chlorophyll and carotenoid biosynthesis, light-harvesting complexes, nitrogen metabolism, cell cycle control, and the circadian clock (Beel et al., 2012). Thus, it is thought that the aCRY photoreceptor is responsible for controlling blue and red light-regulated physiological processes in Chlamydomonas.
Plant CRYs such as CPH1 are closely related to photolyases that repair cyclobutane pyrimidine dimer lesions (Sancar, 2003; Beel et al., 2012; Fortunato et al., 2015). The response to light of the PHR domain of CPH1 (CPH1-PHR) has been resolved at the molecular level (Immeln et al., 2007, 2012; Langenbacher et al., 2009; Thöing et al., 2015). It also has been found that CPH1 is degraded rapidly in the light; this process appears to be mediated by the proteasome pathway (Reisdorph and Small, 2004). Recently, first experiments on the biological function of CPH1 were performed. Two RNA interference strains were created. One of them had a reduced level of CPH1 down to 46%, which did not affect properties of the circadian clock, while the other had a reduced level down to 24% and showed little effects on period but a 1- to 2-h shift of the phase of the phototaxis rhythm at one investigated time point (Forbes-Stovall et al., 2014). Within a short time frame, the strain reverted back to wild-type CPH1 levels and regained wild-type clock properties.
In this study, we use a stable insertional mutant containing 11% of the wild-type level of CPH1 to explore the biological functions of this photoreceptor in more detail. We observe significant changes in the period and in the blue light phase response curve over the circadian cycle in the mutant as well as severe perturbations of its life cycle with regard to germination, mating ability, and the maintenance of mating ability. We refer to CPH1 as Chlamydomonas plant CRY (pCRY) in the following.
RESULTS
A Mutant with Insertion in the Gene Encoding pCRY Exhibits Very Low pCRY Protein Levels
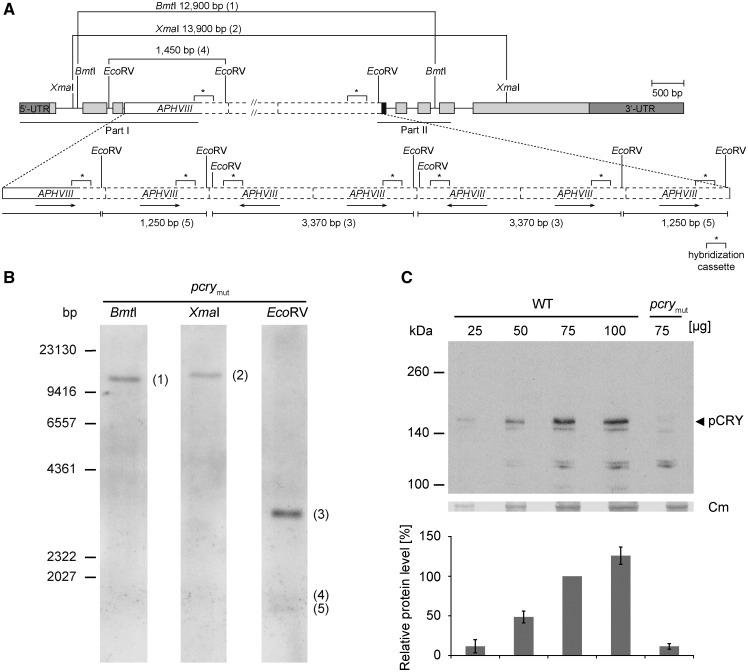
To examine the function of pCRY in Chlamydomonas, we generated a mutant by insertional mutagenesis in a manner used previously to generate an acry mutant (Beel et al., 2012). The APHVIII gene cassette, conferring paromomycin resistance, was inserted randomly into the Chlamydomonas genome (Gonzalez-Ballester et al., 2011), generating a library of 25,000 insertional mutants. PCR with specific primers that anneal to the APHVIII and pCRY genes was used to screen for strains disrupted for pCRY (see “Materials and Methods”). Initially, genomic DNA superpools from paromomycin-resistant transgenic lines (harboring the APHVIII cassette) were generated and screened for the disruption of pCRY. Superpools that yielded a PCR product indicative of pCRY disruption were analyzed further as described previously (Gonzalez-Ballester et al., 2011) until a single pcry mutant, designated CRMS102, was isolated. To determine the precise insertion site, genomic regions of CRMS102 flanking the APHVIII marker at the 5′ (part I) and 3′ (part II) ends were PCR amplified and sequenced (Supplemental Fig. S1, A–C). The analysis showed that the APHVIII cassette was inserted into intron 3 of pCRY and that the insertion was complicated by additional insertions and deletions at the same site (Supplemental Fig. S1C).
To determine the number of copies of the inserted cassette in the mutant genome, DNA gel-blot hybridizations were performed. In two cases, the restriction enzymes that were used (BmtI or XmaI) each cut within the pCRY gene, once upstream and once downstream of the insertion site, but not in the APHVIII cassette (Fig. 1A). Only a single band hybridized to the labeled APHVIII probe after restriction of the mutant genomic DNA with these enzymes, indicating the occurrence of a single disruption in intron 3 of pCRY (Fig. 1B). However, it became evident that more than one APHVIII cassette and/or other DNA fragments may have been inserted at the same site. The two restriction enzymes used (BmtI and XmaI) yielded hybridizing bands of ∼12,900 and 13,900 bp, respectively, which is much larger than would be expected if there was a single cassette inserted into the gene (3,139 and 4,317 bp for BmtI and XmaI, respectively), suggesting that an insertion of ∼11,300 bp was integrated into intron 3 of pCRY. Sequencing showed that both sides of the inserted DNA contained the APHVIII cassette (Supplemental Fig. S1). To investigate the possibility of multiple insertions of APHVIII in intron 3 of pCRY, we used a restriction enzyme (EcoRV) that cuts the genomic DNA upstream of the cassette in intron 2 of pCRY as well as within the cassette, immediately after the region to which the labeled probe hybridizes (Fig. 1, A and B). In this case, multiple hybridizing bands would be expected if integration of the cassette occurred multiple times within the same region. Indeed, we observed three hybridization bands. The estimated sizes may be explained by an event that resulted in seven integrations of the cassette in intron 3 of pCRY, with five oriented in the direction of the gene (5′→3′) and two in the reverse direction (3′→5′), as shown in Figure 1A.
Figure 1.
Characterization of a Chlamydomonas pcry mutant. A, The gene model of pcry in the insertional mutant CRMS102 is shown. This genetic scheme depicts one of the possibilities that are in agreement with the results from DNA sequencing (Supplemental Fig. S1) and Southern-blot analysis (in B). Boxes in light gray show exons, and black lines indicate introns. Boxes in dark gray depict the 5′ and 3′ untranslated regions. The black box indicates an insertion of 60 bp of unknown origin. The APHVIII cassette (white box) appears to be inserted seven times in intron 3 with different orientations (black arrows). Solid lines of the white box indicate sequenced parts along with small insertions or deletions, and dashed lines are nonsequenced regions. The labeled sequence-specific probe (299 bp) in the cassette used for Southern blotting is depicted as asterisks with square brackets. Sequenced regions in pCRY and the APHVIII cassette are indicated by black lines (middle, according to part I and part II in Supplemental Fig. S1C). Relevant restriction sites are shown. B, Southern-blot analysis of CRMS102 to determine the number of insertions of the APHVIII cassette. Genomic DNA (30 µg per lane) from the pcry mutant strain was digested with XmaI, BmtI, or EcoRV, as indicated. DNA fragments were separated on a 0.8% agarose gel and blotted onto a nylon membrane. For hybridization, a digoxigenin-labeled DNA fragment of the APHVIII cassette was used (see “Materials and Methods”). It was shown before that this probe does not give a signal with wild-type genomic DNA (Beel et al., 2012). C, Quantification of pCRY levels in the pcry insertional mutant. Cells were grown under an LD12:12 cycle and harvested in the night (LD22). Different amounts of proteins from crude extracts (25, 50, 75, and 100 µg per lane) of SAG73.72 wild-type (WT) and pcrymut cells were separated by 7% SDS-PAGE and used for immunoblots with anti-pCRY antibodies. The position of pCRY is indicated by the arrowhead. As a loading control, the polyvinylidene difluoride (PVDF) membrane was stained with Coomassie Brilliant Blue R 250 after immunochemical detection. From this stain, selected unspecified protein bands are shown (Cm). The quantified pCRY protein levels of three biological replicates are shown in the diagram at bottom.
The mutant line CRMS102 was generated in the genetic background of strain D66. For further functional characterization of the mutant, we backcrossed it into a wild-type strain that is routinely used for circadian biology (SAG73.72; see “Materials and Methods”). Finally, the protein level relative to the wild-type strain was analyzed in the pcry mutant strains (abbreviated from now on as pcrymut). For this purpose, we created anti-pCRY polyclonal antibodies that were generated to the first 504 of the 1,008 amino acids of pCRY (gene model Cre06.g295200; Chlamydomonas reinhardtii version 5.5, Phytozome 11). This region of the protein, containing the photolyase homologous region (pCRY-PHR), was fused to a 6× His tag at its C terminus, codon adapted, and expressed in Escherichia coli during isopropyl-β-d-1-thiogalactoside induction (Supplemental Fig. S2A). The soluble, yellow protein was purified by affinity chromatography. Proteins in the fractions of two elution peaks (Supplemental Fig. S2B, peaks P1 and P2) were analyzed by SDS-PAGE and immunoblotting using anti-His tag antibodies (Supplemental Fig. S2C) and Coomassie Blue staining of the resolved polypeptides on the polyacrylamide gel (Supplemental Fig. S2D). Both peaks were highly enriched for pCRY-PHR, but in peak P2, it was of higher purity. The identity of pCRY in peak P2 was further confirmed by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS; Supplemental Table S1). pCRY-PHR from peak fraction P2 was used in its native form for the production of antibodies.
As described earlier (Reisdorph and Small, 2004), full-length pCRY migrates more slowly than expected based on its predicted molecular mass of 105 kD. Based on immunological analyses of polypeptides resolved by SDS-PAGE, pCRY has an apparent mass of ∼155 kD, which was confirmed based on the strong reduction of the band in the pcry mutant (Fig. 1C). In the mutant, the pCRY level is reduced to ∼11% ± 4% (sd) compared with a level of 100% in wild-type cells (Fig. 1C). The presence of pCRY at a reduced level indicates that splicing of intron 3 bearing the large insertion is still possible at a low rate, although we cannot be sure that the protein synthesized in the mutant is exactly the same as the wild-type protein.
Chlamydomonas pCRY Accumulates during the Night as Part of a Complex
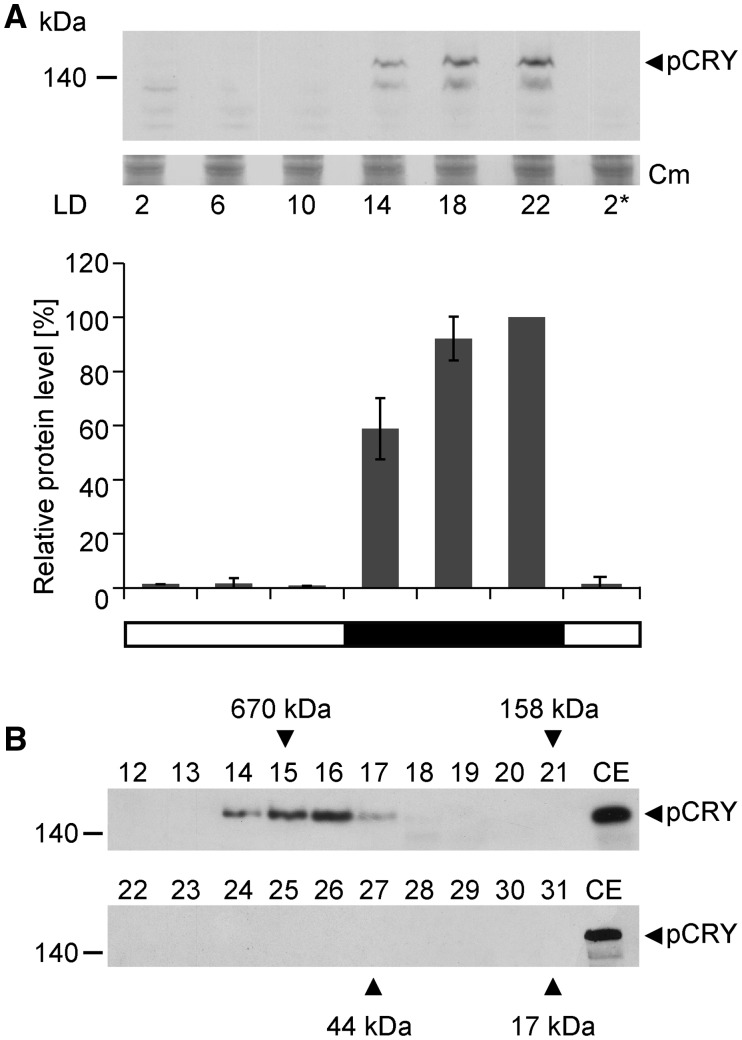
We examined the level of pCRY over an entire 12-h light/12-h dark cycle, measuring protein levels every 4 h (Fig. 2A). A far-red safety light was used during the preparation of protein extracts (see “Crude Extracts and Immunoblots” in “Materials and Methods”). During the day, pCRY was present at a very low level (∼1% compared with its highest accumulation), while at night, the protein level increased rapidly, with the highest accumulation at the end of the night at LD22. We also separated the soluble proteins of the crude extracts from cells harvested at night (LD22) by size-exclusion chromatography to determine if pCRY is part of a complex. The presence of pCRY in the fractions was assayed by immunoblots using the anti-pCRY antibody (Fig. 2B). All of the pCRY was detected only in one peak as a complex with an apparent mass of ∼500 kD. In another independent experiment (biological replicate), the peak position ranged from 400 to 500 kD. These data suggest that pCRY is present either as a homooligomer or in a heteromeric complex with yet unknown partner(s).
Figure 2.
Analysis of pCRY accumulation in Chlamydomonas over a diurnal cycle and complex formation of pCRY. A, Accumulation of pCRY in Chlamydomonas wild-type cells grown under an LD12:12 cycle. Cells were harvested at the indicated time points. The asterisk indicates the beginning of the next light period at LD2. Equal amounts of proteins from crude extracts (150 μg of protein per time point) were separated by 7% SDS-PAGE and immunoblotted using anti-pCRY antibodies. As a loading control, the PVDF membrane was stained with Coomassie Brilliant Blue R 250 (Cm) after immunochemical detection. From this stain, selected, unspecified protein bands are shown (middle). Quantified pCRY protein levels of three biological replicates are shown at bottom. B, Oligomeric state of pCRY in Chlamydomonas. Wild-type cells were harvested at night (LD22). Crude extracts of soluble proteins (1 mg) were loaded onto a Superdex 200 Increase 10/300 GL size-exclusion column, and 0.5-mL fractions of the elution were collected. Proteins from 100 µL of each fraction (numbers 12–31) were separated by 7% SDS-PAGE along with 100 µg of crude extract (CE) and used for immunoblotting with anti-pCRY antibodies. The molecular masses of the standard protein markers are indicated by arrowheads.
pCRY Affects Key Clock Properties
Influences on the Period
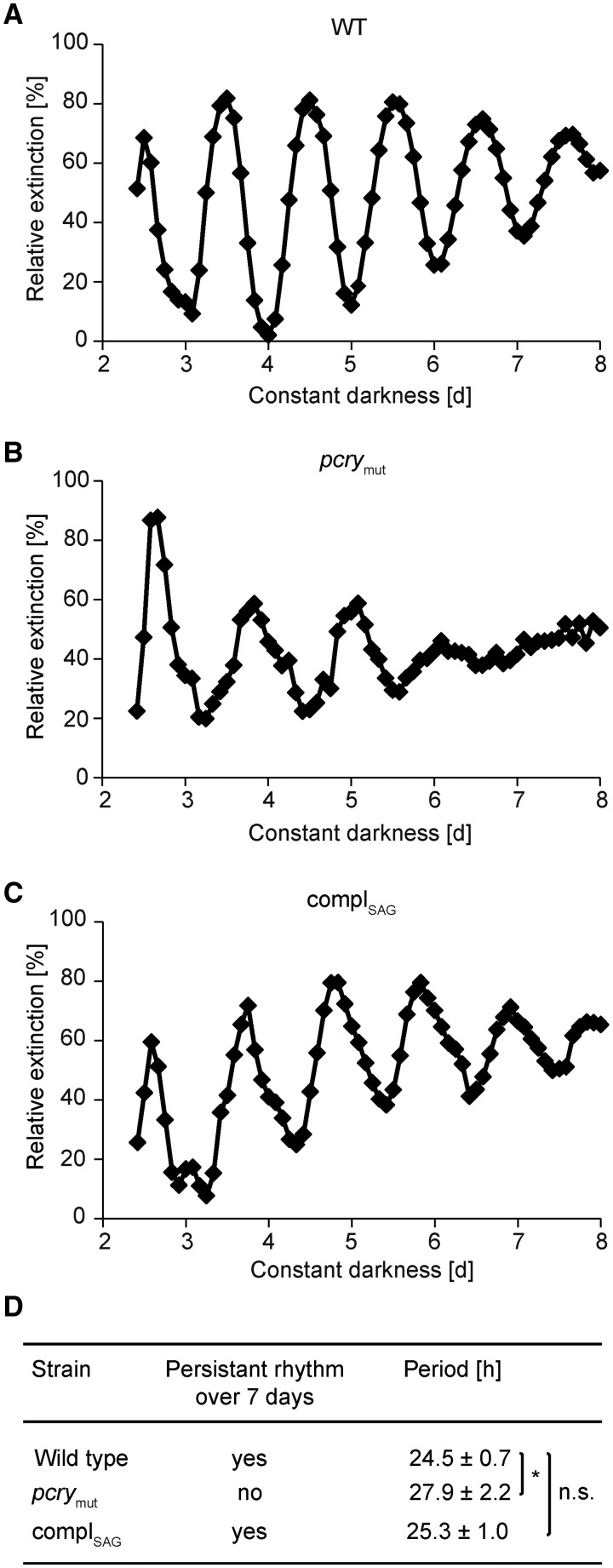
A study of the circadian rhythm of photoaccumulation of cells (phototaxis) using an automated counting system (Mergenhagen, 1984) was performed over several days under constant darkness with the wild-type strain SAG73.72 and pcrymut (Fig. 3, A and B). The wild-type strain showed a free-running period of 24.5 ± 0.7 h (Fig. 3D), which is in a similar range as reported previously (24.7 h in Iliev et al., 2006 and 24.5 h in Schmidt et al., 2006). The free-running period for pcrymut measured over the first 5 d is increased by ∼3 h (27.9 ± 2.2 h). Moreover, arrhythmicity was observed in the pcrymut strain on days 6 and 7, in contrast to the more sustained rhythmicity in wild-type cells.
Figure 3.
The period of the circadian rhythm of photoaccumulation is lengthened in pcrymut. A to C, Chlamydomonas was grown in Tris-acetate-phosphate (TAP) medium under an LD12:12 cycle. Cells were then transferred to minimal medium and maintained in constant darkness for the indicated amount of time before photoaccumulation of cells in a light beam was quantified. The ordinate shows cell density documented as relative extinction in percentage, and the abscissa shows time in constant darkness in days. Photoaccumulation curves of the wild type (WT; A), pcrymut (B), and the complemented mutant (C) are shown. D, Mean values of the free-running periods (n ≥ 5) of the wild type, pcrymut, and the complemented mutant are shown in hours ± sd. Significant differences were estimated by Student’s t test: *, P < 0.05; n.s., not significant.
To establish if the pcrymut phenotype is due to the reduced pCRY level and not a potential second-site lesion in the backcrossed line, we tried to rescue the mutant phenotype with an ectopically expressed wild-type copy of pCRY. For this purpose, we constructed vector pNM003 (Supplemental Fig. S3, A and B), which harbors 981 bp of the native pCRY promoter, followed by its genomic sequence with its predicted 5′ and 3′ untranslated regions. For the selection of transgenic lines, the APH7″ cassette, which provides resistance to hygromycin B, was used. Albeit more than 100 transgenic lines were screened by immunoblots and we obtained several lines expressing pCRY at a significantly higher level than in pcrymut, we were not able to generate a line with the same level of pCRY protein as found in wild-type cells. This may be due to the large gene and final vector size (more than 11 kb; Supplemental Fig. S3B). But we did succeed in generating a line with a level of ∼63% pCRY compared with 11% in the original mutant and 100% in wild-type SAG73.72 (Supplemental Fig. S4). We used the complemented strain, designated complSAG, to measure the circadian rhythmicity of phototaxis (Fig. 3C) and compared it with the rhythms of the wild type and pcrymut with regard to phase and period (Fig. 3, A and B; Supplemental Fig. S5). In contrast to pcrymut, the complSAG strain showed rhythmicity under constant darkness similar to that of wild-type cells with regard to period; it had a period of 25.3 ± 1 h, which is statistically indistinguishable from the period in wild-type cells (Fig. 3D). Our data demonstrate that the premature arrhythmicity and the significantly lengthened period of pcrymut are due to the reduced level of pCRY. Thus, pCRY may be a part of, or at least may be closely connected to, the central oscillator.
Influences on the Phase Response Curve Behavior
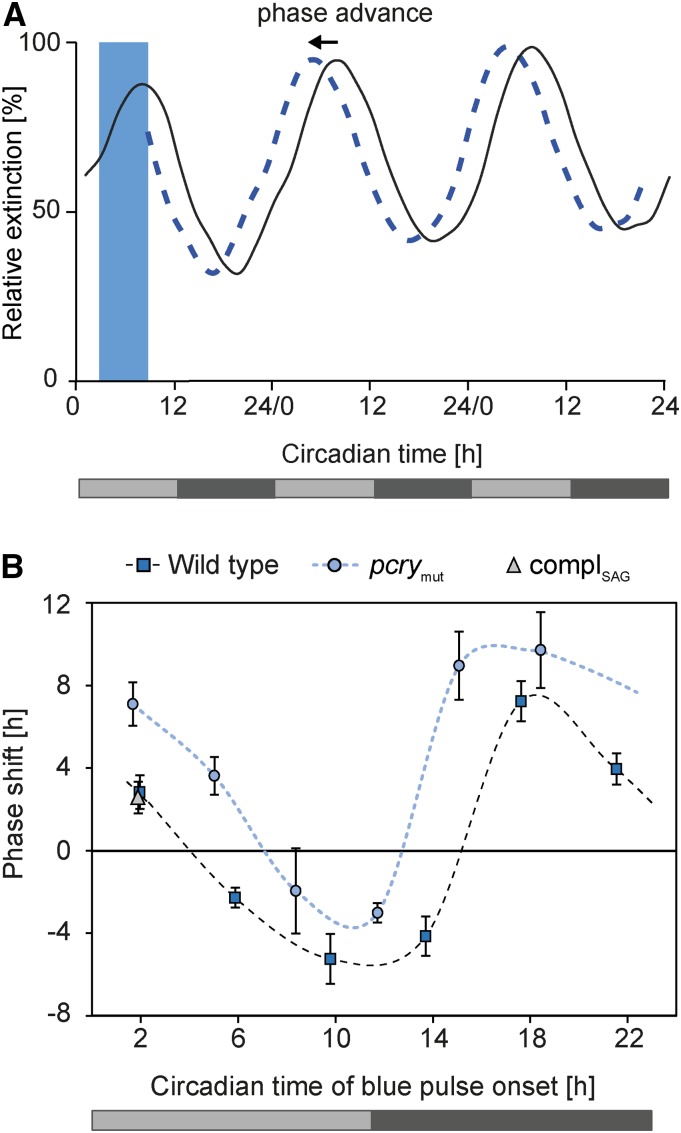
Prokaryotic and eukaryotic organisms can shift the phase of circadian rhythms when kept under constant conditions and exposed to either light or dark pulses (Johnson and Hastings, 1989; Kondo et al., 1993). Depending on the time of day, either phase advances or delays occur. To calculate phase advances and delays, the circadian time (CT) measurement is used, which adjusts the free-running period to a 24-h scale resulting in CT0 to CT24 values. To study the influence of pCRY on circadian-controlled phase response behavior, we imposed a blue light pulse-based phase response curve (see “Materials and Methods”) on wild-type cells and pcrymut with six pulses given over the circadian cycle at a wavelength of 465 nm (see “Materials and Methods”), close to the absorption maximum of pCRY (Immeln et al., 2007). Figure 4A shows an example of the protocol used and a schematic depicting a representative phase shift of wild-type cells. When wild-type cells are given a blue light pulse in the early subjective morning (CT2.0), the phase of the circadian clock is advanced. The impact of the six applied blue light pulses under constant darkness on the phase response curves of the wild-type and pcrymut strains is plotted with the calculated CT values (see “Materials and Methods”) in Figure 4B. Blue light pulses from the middle of subjective day (CT5.9) to the beginning of subjective night (CT13.7) resulted in phase delays of up to 5.3 h (CT9.8) in the wild type. A blue light pulse around subjective midnight (CT17.6) resulted in the strongest phase advance (7.2 h). At the end of subjective night (CT21.5), the phase advance was 4 h, and at early subjective morning (CT2), it was 2.8 h.
Figure 4.
Phase-response curves of photoaccumulation rhythms of Chlamydomonas wild type, pcrymut, and complSAG. A, Schematic photoaccumulation curve of wild-type cells after blue light illumination for 6 h (blue bar) starting at CT2 (dashed blue line) in comparison with cells kept in darkness (black line). The ordinate shows cell density documented as relative extinction in percentage, and the abscissa shows circadian time (see “Materials and Methods”). Light gray bars indicate subjective day, and dark gray bars indicate subjective night in constant darkness. B, Phase-response curves of photoaccumulation rhythms of the wild type (blue squares), pcrymut (blue circles), and the complemented mutant strain in the SAG73.72 background, named complSAG (gray triangles), after blue light pulses (see “Materials and Methods”; n ≥ 3). Blue light illumination was performed at CT2, CT5.9, CT9.8, CT13.7, CT17.6, and CT21.5 for the wild type and at CT1.7, CT5, CT8.4 CT11.7, CT15.1, and CT18.4 for the mutant. For complSAG, blue light illumination was done only at CT1.9. The ordinate shows phase shift in hours ± sd, and the abscissa shows circadian time in hours. The light gray bar shows the subjective day, and the dark gray bar shows the subjective night.
In the pcrymut strain (Fig. 4B, blue circles) significant changes in the phase response curve were observed compared with wild-type cells. While the wild type showed delays from around subjective midday until early subjective night (CT 13.7), the pcrymut strain only showed delays from subjective afternoon (CT8.4) until late subjective day (CT11.7). The largest differences (around 10 h) were observed in early subjective night (around CT14–CT15) between the wild type and the mutant. We also compared the complemented pcrymut strain (Fig. 4B, gray triangles) at CT1.9 and found their behavior to be similar to that of wild-type cells, confirming that the lesion in pCRY is responsible for these changes. These data reveal that pCRY also is part of the circadian input pathway that entrains the circadian clock.
pCRY Is Differentially Expressed over the Chlamydomonas Life Cycle
Light is not only an essential Zeitgeber for synchronizing the Chlamydomonas circadian clock, it also is important for establishing the sexual cycle (Fig. 5A; Huang and Beck, 2003; Goodenough et al., 2007). The conversion of pregametes to gametes as well as zygote germination require light (Fig. 5A). Blue light has been shown to play a major role in the pregamete-to-gamete conversion, while the blue light receptor PHOT was shown to be involved in both processes (Huang and Beck, 2003). We investigated whether the level of pCRY changes during the Chlamydomonas life cycle. For this purpose, cells were either grown in the presence of a nitrogen source in the medium (vegetative state) or shifted to nitrogen-free medium for initiation of the sexual cycle under different light/dark regimes, as depicted in Figure 5B. In vegetative cells kept in the dark, pCRY is detected readily (Fig. 5C). When a proteasome inhibitor was added to the cells 1 h prior to harvesting, the level of pCRY was elevated significantly. Surprisingly, while pCRY was nearly absent in pregametes, which were kept without a nitrogen source in the dark for 14 h, it did accumulate in pregametes that were preincubated with a proteasome inhibitor (Fig. 5C), suggesting that it is degraded via the proteasome pathway in pregametes in the dark. In the next step, pregametes were exposed for 6 h to light to produce mature gametes. pCRY was present at very low levels (1%; Fig. 5C) in the gametes. When gametes were incubated with proteasome inhibitor for 1 h prior to harvesting, the pCRY level was still quite low (14%). One possibility is that pCRY may be only slightly, if at all, subject to degradation by the proteasome pathway in gametes in contrast to pregametes, or most of the protein is degraded during the first 5 h of gamete development in a proteasome-dependent manner; however, the former appears to be the case (see below).
Figure 5.
Overview of the life cycle of Chlamydomonas and differential accumulation of pCRY in vegetative cells (V), prepregametes (pPG), pregametes (PG), G1 gametes (G1), and gametes (G). A, Life cycle of Chlamydomonas cells (modified from Huang and Beck, 2003; Goodenough et al., 2007), including the asexual and the sexual cycle with the different mating types (mt+, mating type plus; mt−, mating type minus). Light-dependent steps are indicated by the sun symbol. B, Treatment and harvesting schemes for the experiments shown in C, D, F, and G. Black bars indicate darkness and white bars indicate light. Application of a proteasome inhibitor is indicated by solid lines (1-h treatment) or dotted lines (2-h treatment), with the vertical arrows depicting the time of harvest and the horizontal tails depicting the length of the inhibitor treatment. Part 1, Scheme representing the harvesting conditions of V grown in nitrogen-containing medium (+N) under different light/dark regimes. Part 2, Scheme representing the harvesting conditions of pPG, PG, G1, and G induced by transfer into nitrogen-deprived medium (−N) under different light/dark regimes. C, Accumulation of pCRY in PG and G. Equal amounts of proteins from crude extracts (70 µg per lane) of CC-124 wild-type cells were resolved by SDS-PAGE on a 7% polyacrylamide gel and used for immunoblots with anti-pCRY antibodies. V were grown under an LD12:12 cycle in TAP medium (+N) and harvested in the night (LD22). For the induction of gametogenesis, cells were transferred to TAP medium without nitrogen (−N) at the end of the light phase (LD12) and kept in darkness for 14 h. By this time, PG were harvested. Afterward, the cells were illuminated with white light for 6 h, and G were harvested. Cells were treated with 10 µm proteasome inhibitor MG-132 (PI; carbobenzoxy-leucyl-leucyl-leucinal) for 1 h before harvesting (+PI) and compared with untreated cells (−PI). As a loading control, the PVDF membrane was stained with Coomassie Brilliant Blue R 250 (Cm) after immunochemical detection. From this stain, selected, unspecified protein bands are shown (middle). The quantified pCRY protein levels of three biological replicates are shown in the diagram at bottom along with the sd. D, Accumulation of pCRY in V during early subjective day. Immunoblotting and quantifications were carried out as described in C along with proteins of V harvested at CT2 (VCT2; see B, part 1), followed in one case by a 1-h light treatment (VCT2+L). Proteasome inhibitor was added as indicated. The black arrowhead indicates pCRY, and the white arrowhead indicates the Coomassie Brilliant Blue R 250-stained loading control. E, pCRY transcript accumulation analyzed by reverse transcription-quantitative PCR (RT-qPCR) for SAG73.72 wild-type cells. Cells were grown under an LD12:12 cycle in TAP medium, then transferred to TAP medium without nitrogen at the end of the light phase and kept in darkness for 14 h. The cells were then illuminated with blue light for 5 h to induce gametogenesis. Light-emitting diode (LEDs) with an energy fluence rate of 2.6 W m−2 were used. Total RNA was isolated, and equal amounts of RNA were used for the RT-qPCR with RACK1 as a reference gene. Changes in transcript levels after blue light illumination of gametes in comparison with dark-adapted pregametes are shown (n = 3 biological replicates; error bars represent sd). The asterisk indicates a significant difference estimated by Student’s t test (*, P < 0.05). F, Accumulation of pCRY in G1. Equal amounts of proteins from crude extracts (70 µg per lane) of CC-124 wild-type cells were resolved by SDS-PAGE on a 7% polyacrylamide gel and used for immunoblots with anti-pCRY antibodies. V were treated as described in C. Moreover, G1 were produced by exposing PG for 1 h to light. In some cases, a proteasome inhibitor was added in the dark (+PI) 2 h before harvesting the cells in the light. Quantification based on three biological replicates was done as in C. The black arrowhead indicates pCRY, and the white arrowhead indicates the Coomassie Brilliant Blue R 250-stained loading control. G, Immunoblots and quantifications were carried out as described in F along with proteins of pPG, PG, and G1 (see B, part 2). Proteasome inhibitor was added as indicated.
It seemed possible that the proteasome-triggered disappearance of pCRY in pregametes is influenced by the circadian clock. The 14-h duration of darkness used to create pregametes results in cells at time point CT2, representing subjective day (Fig. 5B, part 1). During a diurnal cycle, pCRY is degraded in the light (Reisdorph and Small, 2004). To check for this, vegetative cells were grown under the same light/dark regime as applied for pregametes (Fig. 5C) but in the presence of a nitrogen source in the medium. We observed that pCRY accumulates in vegetative cells harvested at CT2 (Fig. 5D). A 1-h light exposure of these cells caused the degradation of pCRY (Fig. 5D), as found with cells exposed to light within the diurnal cycle (Reisdorph and Small, 2004; Fig. 2A). Application of a proteasome inhibitor prevented the degradation of pCRY to a large extent (Fig. 5D). These data suggest that pCRY degradation is light driven in vegetative cells and not under circadian control.
We also analyzed the reason for the strongly reduced level of pCRY in gametes in more detail. The low level could be at least partially due to the absence of pCRY mRNA in gametes. Therefore, we compared pCRY transcript levels between gametes and pregametes (Fig. 5E) in the absence of a proteasome inhibitor. We found a reduction of transcript levels in gametes compared with pregametes, but only to a small extent (about one-third) that does not explain the very low level of pCRY in gametes.
We also considered whether the incubation time of only 1 h with the proteasome inhibitor is too short for the 6-h light exposure used to generate mature gametes (Fig. 5C) to prevent pCRY degradation. To distinguish between these events and to avoid an extended exposure of the cells over several hours to the inhibitor, which might cause side effects, we generated early gametes (so-called G1 gametes) by exposing the pregametes for 1 h to light (Fig. 5B, part 2). After this time, pCRY is already strongly degraded (down to ∼20%; Fig. 5F) compared with its level in vegetative cells. Proteasome inhibitor was then added 2 h prior to harvesting of the G1 cells, meaning that it was added to prepregametes that had been in the dark for 13 h. In the proteasome inhibitor-treated cells, the level of pCRY was still low (again ∼20%; Fig. 5F), suggesting that the low level of pCRY in gametes is indeed due to some other control mechanism of degradation. To further investigate the influence of proteasome degradation of pCRY in the pregamete-to-gamete switch, we conducted the following experiment. The pCRY level in prepregametes kept for 13 h in the dark was determined and compared directly with its level in prepregametes exposed for 1 h (state of pregametes) or for 2 h (state of G1 gametes) to the proteasome inhibitor. In prepregametes, pCRY accumulates at a low level. While a strong recovery of pCRY was observed in the proteasome inhibitor-treated pregametes, the proteasome inhibitor-treated G1 gametes contained only minimal levels of pCRY (Fig. 5G). These data further corroborate that there is a specific degradation mechanism in gametes independent from the proteasome pathway contributing to the low pCRY level there.
In summary, our results indicate that the proteasome is responsible for the degradation of pCRY in vegetative cells (which is induced by light) and pregametes (which is induced by nitrogen starvation). In contrast, the degradation of pCRY in gametes, which were induced by the illumination of nitrogen-starved pregametes, involves an additional, proteasome-independent mechanism.
pCRY Alters the Gamete-Specific Transcript Level of GAS28
Since pCRY is decreased in pregametes and gametes, and it is known that gamete-specific gene expression depends on light (von Gromoff and Beck, 1993; Rodriguez et al., 1999), we determined whether the knockdown of pCRY influences the expression of gamete-specific genes (GAS). We characterized transcript levels from previously studied GAS3, GAS18, GAS28, and GAS96 (von Gromoff and Beck, 1993; Rodriguez et al., 1999). In particular, GAS28 mRNA accumulation was reported to occur specifically during the pregamete-to-gamete conversion and was strictly light dependent (von Gromoff and Beck, 1993). The sequences and functions of the corresponding cDNAs/proteins were not known in the cases of GAS3, GAS18, and GAS96. Christoph Beck and Erika D. von Gromoff (University of Freiburg) kindly provided us with the corresponding cDNAs, which were sequenced (Supplemental Fig. S6, A–C), revealing that GAS3 encodes the small subunit of carbamoyl phosphate synthase, GAS18 encodes a flagellar associated P-type ATPase/cation transporter, and GAS96 encodes a protein with unknown function. GAS28 is known to encode a Hyp-rich glycoprotein that presumably is a cell wall constituent (Rodriguez et al., 1999; Hoffmann and Beck, 2005). Locus identifiers of these genes are listed in Supplemental Table S2.
We analyzed the impact of blue light on transcript accumulation of the selected GAS genes during the transition from pregametes to gametes in the wild type, the pcry mutant, and the partially complemented strain by RT-qPCR (see “Materials and Methods”; Supplemental Table S2). While GAS3 and GAS96 showed no significant induction after blue light treatment in the wild type and the pcrymut strain (Supplemental Fig. S7), GAS18 showed a minor increase (1.6-fold; Supplemental Fig. S7) and GAS28 showed a 3.7-fold increase in mRNA after illumination with blue light (Fig. 6A). The difference between the wild type and the pcrymut strain was low for GAS18 (1.5-fold reduction; Supplemental Fig. S7). In the case of GAS28, blue light induction was reduced significantly in the mutant (12.3-fold reduction compared with the wild type). There was partial restoration of the impact of blue light on the accumulation of this transcript in the partially complemented strain (Fig. 6A). These data suggest that pCRY is involved in altering in particular the transcript level of light-regulated GAS28.
Figure 6.
pCRY influences the transcript accumulation of the gamete-specific gene GAS28 under blue light illumination during gametogenesis, mating ability, and its maintenance as well as the germination efficiency. Asterisks indicate significant differences estimated by Student’s t test (*, P < 0.05; **, P < 0.01; and ***, P < 0.001; n.s., nonsignificant). A, pCRY transcript levels in pregametes (PG) and gametes (G). Transcript accumulation was quantified by RT-qPCR for the wild type (WT), pcry mutant (pcrymut), and complemented mutant (complSAG). Cells were grown under an LD12:12 cycle, then transferred to TAP medium without nitrogen at the end of the light phase and kept in darkness for 14 h. Afterward, the cells were illuminated with blue light for 5 h to convert PG to G. LEDs with an energy fluence rate of 2.6 W m−2 were used. Total RNA was isolated, and equal amounts of RNA were used for RT-qPCR with RACK1 as a reference gene. Changes in the transcript levels after blue light illumination (G) in comparison with dark-adapted cells (PG) are shown (n = 3 biological replicates; error bars represent sd). B, Germination of zygospores in percentage after 10 d. Crossings of mating type plus (mt+) and minus (mt−) of different combinations are shown: WT, wild-type strain; pcrymut, pcry mutant strain; pcrycompl, complemented strain of the pcry mutant (mt+). The germination of zygospores (see “Materials and Methods”) of wild-type strains CC-124/mt− (WT−) and CC-125/mt+ (WT+) is shown as a homozygote (WT+ × WT−) as well as the heterozygotes (WT+ × pcrymut− and pcrymut+ × WT−) and the homozygote of pcry mutant strains with mt+ and mt− (pcrymut+ × pcrymut−). In addition, the heterozygote of a complemented strain/mt+ (see “Materials and Methods”; Supplemental Fig. S8) and a wild-type CC-124/mt− (pcrycompl+ × WT−) is shown. C, Mating ability of the wild type, pcrymut, and the complemented strain pcrycompl. For the test, PG kept in the dark as well as PG illuminated for 1 h with light (G1) were used (see “Materials and Methods”). The mating ability of G1 of the wild type was set to 100% and used for comparison. D, Mating ability after dark treatment of the wild type, pcrymut, and pcrycompl. G1 gametes were put in the dark for 1 h to deactivate the mating ability. The gamete formation rate of G1 gametes of each strain was set to 100% separately.
pCRY Is a Positive Regulator of Zygote Germination and a Negative Regulator of Mating and Mating Maintenance
PHOT was shown to have a positive influence on zygote germination, mating ability, and mating maintenance based on an RNA interference line with a reduced PHOT level (Huang and Beck, 2003). While the rate of mating ability was reduced proportionally compared with the degree of PHOT silencing, germination was only partially lower, suggesting a role for yet another blue light receptor(s) in this process. We thus investigated at first the influence of pCRY on zygote germination (see “Materials and Methods”). The experiments were performed with strains CC-125 (mt+) and CC-124 (mt−) as mating partners (see “Materials and Methods”), which are often used for germination experiments (Suzuki and Johnson, 2002; Huang and Beck, 2003). For this purpose, we backcrossed the pcry mutant phenotype from strain CRMS102 into CC-124 and thereafter into CC-125 (see “Materials and Methods”). The progeny of this backcross has a reduced level of ∼11% (Supplemental Fig. S8). Approximately 90% of the zygotes germinated in the wild type mt+/mt− (CC-125 × CC-124) cross, only 60% of the zygotes germinated in the CC-125 × pcrymut and pcrymut × CC-124 crosses, while for the pcrymut homozygous zygotes, the rate of germination was only 25% (Fig. 6B). These results demonstrate that pCRY has a strong influence on germination. They were confirmed by analyses of the complemented strain in the background of CC-124/125 (abbreviated as pcrycompl; see “Materials and Methods”), in which the pCRY levels were in the range of 93% (Supplemental Fig. S8). The heterozygous zygotes in this complemented strain exhibited a germination rate of ∼80% (Fig. 6B). These data suggest a positive role of pCRY in germination, similar to PHOT.
We wondered whether pCRY also may have a role in mating ability and maintenance, as the results in Figure 6A indicate that the pregamete-to-gamete conversion also may be compromised in pcrymut. For this purpose, pregametes were exposed for 1 h to light to produce G1 gametes. The mating ability of wild-type G1 gametes was set to 100% and used for comparison. Wild-type pregametes exhibited a mating ability rate of 7% (Fig. 6C). It is known that some Chlamydomonas strains exhibit a low mating ability as pregametes (Saito et al., 1998). After light exposure for 1 h, the mating ability of the wild type increased strongly, as found before (Beck and Acker, 1992). In pcrymut, the mating ability of pregametes and gametes was significantly higher than that in the wild type (Fig. 6C). In the complemented strain pcrymut, the mating ability of pregametes and gametes was close to that of the wild type (Fig. 6C). These data suggest that pCRY is indeed involved in mating ability, but it acts as a negative regulator, opposite to PHOT. Thus, the reduction of the pCRY protein level results in an increase in mating ability.
It is known that incubation of gametes in the dark leads to a loss of their mating ability (Beck and Acker, 1992; Huang and Beck, 2003). We also examined whether pCRY has an effect on mating maintenance. Dark treatment of wild-type gametes resulted in a strong loss of mating ability down to 28% (Fig. 6D), consistent with previous studies (Beck and Acker, 1992; Pan et al., 1997; Huang and Beck, 2003). Dark treatment of pcrymut showed no significant loss of mating ability, indicating that pCRY is an essential component for the loss of the mating ability pathway. The complemented strain displayed a similar reduction in mating ability after dark treatment as the wild type (Fig. 6D), which corroborates the negative role of pCRY in the regulation of mating maintenance.
DISCUSSION
Algae can reside at various water depths, where they are exposed to ambient changes of light intensity and quality. It is known that the ratio of blue to red light increases with the depth, as red light and far-red light are strongly attenuated within the upper layer of the water column, while blue radiation penetrates deeper (Kirk, 1994). This may be one reason why algae use a broad variety of blue light receptors.
In plants, responses to blue light are mediated by different classes of photoreceptors including plant CRYs (Liu et al., 2010; Chaves et al., 2011), PHOTs (Christie, 2007), and members of the Zeitlupe family (Suetsugu and Wada, 2013). In stramenopile algae, an additional blue light flavoprotein photoreceptor, aureochrome, has been identified and shown to be needed to trigger photomorphogenesis (Takahashi et al., 2007), control the cell cycle (Huysman et al., 2013), and repress high light acclimation (Schellenberger Costa et al., 2013). Also, a novel CRY (named CryP) was found in diatoms, which regulates the expression of light-harvesting proteins (Juhas et al., 2014).
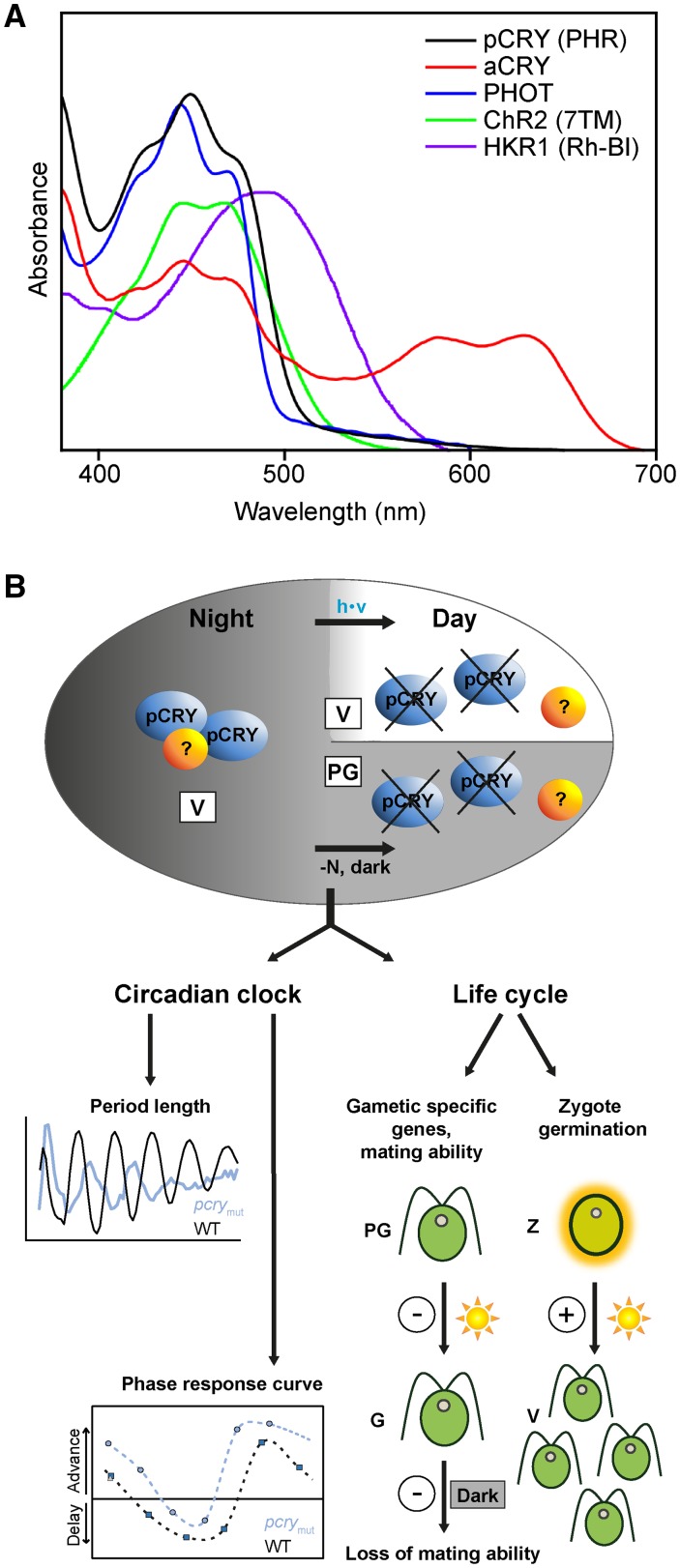
The green alga Chlamydomonas has a unique set of blue light receptors (Fig. 7A), including PHOT, aCRY, pCRY, CHANNELRHODOPSIN2 (ChR2; Nagel et al., 2003, 2005; Govorunova et al., 2004), and possibly HISTIDINE KINASE RHODOPSIN1 (HKR1; Luck et al., 2012). The similar spectral region of their absorption complicates any approach to distinguish their contributions to cellular responses by a selection of the light quality employed. To obtain detailed knowledge of the specific and overlapping functions of these photoreceptors, double, triple, or multiple mutants would be necessary. For example, a double phot pcry mutant would help disentangle the overlapping functionalities of the encoded blue light photoreceptors on the Chlamydomonas life cycle. While a null mutant for PHOT exists (Zorin et al., 2009), it was generated in a flagella-less strain that does not cross. Location and expression data on the photoreceptors also provide some information about their functions. ChR2 and HKR1 are solely localized in the eyespot (Schmidt et al., 2006, and refs. therein; Luck et al., 2012). PHOT is, in addition, present in the cell body and cilia (Huang et al., 2004; Schmidt et al., 2006). The locations of aCRY and pCRY are still under investigation, with pCRY being especially challenging because of its strong light-dependent degradation. Expression studies over an entire diurnal cycle now exist for aCRY (Beel et al., 2012) and pCRY (this work). While aCRY is present at high levels in vegetative cells during the day, its level declines at the end of the day. Reisdorph and Small (2004) and we (this work) showed that pCRY is degraded rapidly in the light (Figs. 2A and 5D) but is present during subjective day (CT2) in constant darkness (Fig. 5D), revealing light- but not clock-mediated degradation of pCRY. There is a steady increase of pCRY protein during the night in a diurnal cycle, with its maximum at the end of the night (LD22; Fig. 2A), and this accumulation in protein is in agreement with the increase of pCRY mRNA during the night (Zones et al., 2015). But even in the dark, a certain proportion of pCRY protein appears to be degraded by the proteasome pathway, as revealed by its increase in vegetative cells after proteasome inhibitor treatment (Fig. 5C). When working exclusively under a far-red safety light, pCRY can be found in a complex of ∼400 to 500 kD at night (Fig. 2B). This finding raises the possibility that pCRY forms homomers, as observed for aCRY in vitro (Oldemeyer et al., 2016), or it may be in a complex with other partners. Coimmunoprecipitation with proteins fractionated by size-exclusion chromatography should help resolve these possibilities.
Figure 7.
Overview of the variety of blue light receptors in Chlamydomonas and the functions of pCRY. A, Overview of absorption spectra of the unusually large number of blue light receptors found in Chlamydomonas to date. The overlap in the absorption in the region of blue light (around 450 nm) presents a challenge to study overlaps in function. UV/visible absorption spectra are shown for pCRY (first 504 amino acids, overexpressed in E. coli) compared with aCRY, PHOT, ChR2 (seven-transmembrane helix domain; taken from Hegemann, 2008), and HKR1 (UV-illuminated rhodopsin fragment; taken from Luck et al., 2012). Spectra are scaled for better visibility. Contributions at greater than 510 nm for pCRY and PHOT do not originate from absorption but are caused by scattering. B, Complex formation and degradation of pCRY and its influence on clock properties and on the life cycle of Chlamydomonas. The accumulation of pCRY during the night along with its complex formation is shown. Potential unknown interaction partners are indicated by question marks (?). The proteasome-induced degradation of pCRY at early day and in the dark upon the formation of pregametes (by removal of the nitrogen source from the medium) probably trigger a signal cascade resulting in the regulation of important clock features and crucial steps in the life cycle in Chlamydomonas. Positive and negative roles of pCRY in the life cycle are indicated by + and −, respectively.
The disappearance of pCRY in vegetative cells at the beginning of the day (Fig. 2A), as well as during subjective day after a light pulse (Fig. 5D) and in the dark in pregametes (Fig. 5C), appears to be mediated mainly by the proteasome pathway and may be critical for the signaling of pCRY with regard to circadian and life cycle control (Fig. 7B). Also in G1 and mature gametes, the pCRY level is reduced strongly, but the level is barely impacted by the administration of a proteasome inhibitor (Fig. 5, C, F, and G). However, the degradation of pCRY seems to be involved and mediated by a yet unknown gamete-specific degradation pathway. The difference in the pCRY transcript levels between pregametes and gametes is limited. About two-thirds of the level of pCRY mRNA in pregametes is still transcribed in gametes. Altogether, these data suggest a complex regulation of pCRY gene expression during the Chlamydomonas life cycle, involving, to a small extent, transcriptional, possibly posttranscriptional, such as a translational arrest, and mainly posttranslational control in the form of protein degradation in gametes.
The differential expression pattern of pCRY during gametogenesis indicated that it may be involved in the transition of pregametes to gametes, in which PHOT also was shown to play a role based on phenotypes of RNA interference lines (Huang and Beck, 2003). We found that transcript levels of two of the investigated GAS genes are blue light induced in the wild type and altered in the pcry mutant, with a profound change in the case of GAS28. GAS28 is known as one of the late-phase gametogenesis genes (switch from pregametes to gametes) that is strictly under light control (von Gromoff and Beck, 1993); it is only induced significantly after a light pulse applied to nitrogen-depleted cells kept in the dark (pregametes). In nitrogen-deprived cells kept in the light, induction of its expression is observed only after 7 h (von Gromoff and Beck, 1993). Consequently, it was not detectable when a protocol of 3 to 5 h of illumination of nitrogen-deprived cells was used (Lopez et al., 2015). In pcrymut, GAS28 is not only strongly reduced in gametes compared with the wild type but even more reduced in gametes than in pregametes. These data suggest that the absence of pCRY even activates GAS28 reduction. A dark function of pCRY must be assumed, and the degradation of pCRY is likely to start a signaling pathway, for example, by the release of a yet unknown interaction partner that influences GAS28 transcription.
Complex roles of pCRY also were found throughout the sexual cycle of Chlamydomonas. pCRY strongly affects gametogenesis. It acts as a negative regulator of mating ability and of the loss of mating ability in the dark, in contrast to PHOT, which has an opposite influence on these processes (Huang and Beck, 2003). This finding suggests again a dark function of pCRY. Moreover, pCRY influences light-dependent zygote germination in a positive manner, resulting in a reduced level of germination in the mutant (Figs. 6B and 7B). In this process, pCRY acts in concert with PHOT, which also has a positive effect on germination (Huang and Beck 2003). Hence, both pCRY and PHOT share important roles in the blue light-regulated life cycle of Chlamydomonas, being either complementary or opposite in their actions. Germination of Chlamydomonas also is controlled by the photoperiod, with increased rates under long-day compared with short-day conditions (Suzuki and Johnson, 2002). pCRY and PHOT are two promising candidates that might be involved in this regulation. They might act together, as found for the control of some light-regulated genes (Im et al., 2006; Beel et al., 2012) and in the germination process (Fig. 6B; Huang and Beck, 2003), or opposite, as in gametogenesis (Fig. 6, C and D; Huang and Beck, 2003).
Plant CRYs are critical photoreceptors that transmit light information to the circadian oscillator and help entrain the circadian clock (Harmer, 2009). The first data showing a role of pCRY on circadian function were obtained with RNA interference knockdown lines, as mentioned before (Forbes-Stovall et al., 2014). A reduction of pCRY down to 46% did not affect the period and phase of the circadian rhythm of phototaxis, while a reduction down to 24% caused little change in the period but a difference in the phase shift of the rhythm by 1 to 2 h (one time point analyzed; Forbes-Stovall et al., 2014). Here, we have studied the effects of pCRY on the phase of the circadian rhythm with six blue light pulses given over a 24-h time period in the wild type and the pcry mutant (Figs. 4 and 7B). Based on these phase response curve analyses, we found that wild-type Chlamydomonas exhibited phase delays from the middle of subjective day until the beginning of subjective night, with strong advances later in the night at CT17.6 and CT21.5 (Fig. 4B). This finding is in agreement with a blue light phase response curve generated from a dinoflagellate (Johnson and Hastings, 1989) and also with a white light phase response curve generated for Chlamydomonas (Niwa et al., 2013). In pcrymut, strong differences in phase shifts compared with wild-type cells were observed, with the largest deviations (∼10 h) at the beginning of subjective night (Fig. 4B). At subjective midday, the delay of the wild type was converted to an advance in the mutant, and a total differential between the mutant and the wild type of ∼4 h in shifting ability was observed. These data underline the importance of pCRY in the circadian input pathway, which entrains the Chlamydomonas clock. ROC15 also has a strong influence on the phase (Niwa et al., 2013), with the strongest differences in a white light phase response curve between roc15 and the wild type occurring in the late night phase, where the ∼4-h advance was essentially eliminated in the mutant. Also, in the SAG73.72 wild-type strain, an advance of about 4 h was observed in the late night phase after the blue light pulse (Fig. 4B). In pcrymut, this advance seems not to be eliminated. Thus, a functional coupling of pCRY and ROC15 within a pCRY-mediated blue light-triggered input pathway does not seem likely. Additionally, other photoreceptors or light qualities may be relevant for the ROC15-based signaling pathway. Notably, light-induced degradation of a ROC15 fusion protein was strongest in response to red light (Niwa et al., 2013). This raises the possibility that aCRY (Beel et al., 2012) also may act in the circadian input pathway. As mentioned in the introduction, red, green, and blue light are able to reset the phase of the Chlamydomonas circadian clock requiring photoreceptors that respond to various wavelengths. pCRY appears to be one of the key players in the blue light-mediated entrainment pathway of the clock.
We also found that the strongly reduced pCRY level down to 11% in pcrymut not only caused changes in phase but also period lengthening of about 3 h, ultimately resulting in arrhythmicity (Figs. 3 and 7B). The partially complemented strain rescued the arrhythmicity phenotype. The succinct effect on period and final arrhythmic behavior suggests that pCRY not only acts as a sensory blue light receptor to entrain the Chlamydomonas circadian clock but also may be connected with the oscillatory loop, as is known, for example, for the white collar complex in Neurospora crassa (Chen et al., 2010).
CONCLUSION
In summary, Chlamydomonas pCRY is an essential component of the circadian clock, maintaining its period and the blue light phase response behavior. At the same time, it is an important element that controls the sexual cycle of Chlamydomonas at its different stages. The pCRY protein accumulation pattern varies not only in a light/dark cycle in vegetative cells of Chlamydomonas but also during its developmental cycle. Moreover, pCRY is found in a protein complex during the night. The identification of other polypeptides that may be part of this complex and the elucidation of their functions will deepen our understanding of pCRY signaling pathway(s) in the future.
MATERIALS AND METHODS
Strains and Culture Conditions
The following wild types/parental strains of Chlamydomonas reinhardtii were used: SAG73.72 (mt+), D66 (cw15, nit2, mt+; Pootakham et al., 2010), CC-124 (nit1, nit2, agg1, mt−), and CC-125 (nit1, nit2, agg1+, mt+). Strain D66 was used for the insertional mutagenesis, with the putative mutants backcrossed into SAG73.72, CC-124, and CC-125 as outlined below. SAG73.72 was routinely used as our wild-type control and was grown under a 12-h-light/12-h-dark cycle (LD12:12) at an intensity of 75 μmol photons m−2 s−1 in TAP medium (Harris, 1989), unless indicated otherwise. CC-124 and CC-125 and their pcrymut derivatives were used for germination experiments. Cells were harvested at the indicated light/dark time points, where LD0 represents the beginning of the day and LD12 represents the beginning of the night.
Backcrossing of the pcry Mutant Strain to CC-124, SAG73.72, and CC-125
The mutant line CRMS102 was generated in a D66 (cw15, nit2, mt+) genetic background. It was backcrossed to SAG73.72 (mt+), which has a cell wall (CW) and an active nitrate reductase gene (NIT1) along with its transcriptional regulator (NIT2), as described (Beel et al., 2012). Therefore, CRMS102 was first crossed with the mt− strain CC-124 (nit1, nit2, agg1). One of the mt− progeny harboring the APHVIII resistance marker gene and lacking the phenotypically detectable (Harris, 1989) agg1 mutation of CC-124 was crossed with SAG73.72 (mt+). The tetrads from the germinating zygotes were selected on nitrate-containing medium supplemented with paromomycin (50 µg mL−1). The mutant strain used for further analyses was SAG73.72:pcry6C, which is abbreviated pcrymut. Growth on nitrate as the sole nitrogen source requires functional NIT1 and NIT2 genes. Gametogenesis, mating, zygote germination, and tetrad separation were performed as described by Jiang and Stern (2009). For the germination assays, pcrymut that resulted from the backcross with CC-124 was further backcrossed with CC-125, and the paromomycin-resistant progeny were analyzed for their mating type. These strains (presented only in Fig. 6, B–D) also are abbreviated as pcrymut.
Insertional Mutagenesis
Approximately 25,000 insertional mutants were generated using an APHVIII gene cassette, and the mutants were screened by PCR using cassette- and gene-specific primers, essentially as described previously (Pootakham et al., 2010; Gonzalez-Ballester et al., 2011; Beel et al., 2012). Target gene-specific primers were selected with primer design software (Clone Manager 9; Scientific and Educational Software) using the default setting for hairpin formation and dimer duplexing. Strain CRMS102 was identified with the PCR primer pair 1a/1b situated in pCRY (1a) and the cassette (1b; Supplemental Fig. S2). After identification of a transgenic line with the desired lesion, a single colony was isolated and cultured, the genomic DNA from the culture was isolated, regions covering the borders of the insertion cassette for CRMS102 were amplified with primer pairs 1a/1b and 2a/2b, and the products were sequenced.
DNA Gel-Blot Analysis
Genomic DNA was isolated from the pcry mutant CRMS102 as described (Lee et al., 1993), restricted, and the fragments were separated on an agarose gel and then transferred to a nylon membrane. A digoxigenin-labeled DNA probe was generated from a 304-bp fragment of the APHVIII cassette digested with BsgI and RsrII, as described previously (Beel et al., 2012). The labeled fragment was used for hybridization and detection with anti-digoxigenin antibodies. All steps were performed using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) according to the manufacturer’s protocol.
Complementation of pcrymut with pNM003
Vector pNM003 was used to complement pcrymut. This vector carries the complete pCRY genomic DNA sequence, including its putative promoter region of 981 bp and the 5′ and 3′ untranslated regions (chromosome 6, positions 6,837,075–6,844,247 [reverse] in Joint Genome Institute version 5.5). The Ampicillin resistance gene was used for selection in Escherichia coli and the Hygromycin B resistance gene from pHyg3 (Berthold et al., 2002) was used for selection in Chlamydomonas. The complete pNM003 sequence is presented in Supplemental Figure S3, along with a vector scheme. The mutant strain SAG73.72:pcry6C was transformed with XmnI-linearized pNM003 using the autolysin method as described earlier (Iliev et al., 2006). Cells were grown on TAP agar medium (Harris, 1989) containing 20 μg mL−1 hygromycin B. The selected rescued strain SAG73.72-6C:pNM003#178 (abbreviated as complSAG) was used for further experiments. For complementation of the mutant strain in the CC-124 and CC-125 backgrounds, SAG-6C:pNM003#178 was first backcrossed with CC-125 and then CC-124 (see procedure above). Complementation of the selected mt+ strain of this progeny (abbreviated as pcrycompl) was verified by immunoquantification of pCRY protein levels (Supplemental Fig. S8B).
Heterologous Expression, Purification, UV/Visible Spectroscopy and Antibody Production
The first 504 amino acids encoded by the codon-adapted pCRY (Immeln et al., 2007) were expressed in E. coli BL21-CodonPlus (DE3)-RP (Agilent Technologies) grown in LB medium (Luria/Miller; 10 g L−1 tryptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl) supplemented with 38 mg L−1 riboflavin, 25 mg L−1 kanamycin, and 30 mg L−1 chloramphenicol. At an OD600 of 0.5, the temperature was lowered from 37°C to 22°C. Expression was induced with 10 μm isopropyl-β-d-1-thiogalactoside when the cell cultures reached an OD600 of 0.7. After 20 h, the cells (from 750 mL) were harvested by centrifugation (4,000g, 5 min, 4°C) and resuspended on ice in 10 mL of 1× phosphate-buffered saline buffer, pH 7.4, containing 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4 and 1.8 mm KH2PO4. The suspension was centrifuged (10,000g, 5 min, 4°C) to collect the cells, and the supernatant was discarded. One gram of cells (wet weight) was suspended in 1 mL of lysis buffer (50 mm sodium phosphate buffer, pH 7.5, 300 mm NaCl, 10 mm imidazole, 10% [v/v] glycerol, and 0.5% [v/v] Triton X-100) containing 20 mm β-mercaptoethanol with one tablet of protease inhibitor per 10 mL of lysis buffer (complete EDTA-free; Roche). After complete resuspension, lysozyme was added to a final concentration of 1 mg mL−1 and incubated for 60 min on ice with shaking (200 rpm). Cells were then disrupted on ice by ultrasound with multiple 12-s pulses interrupted by 18-s breaks for a total of 15 min. The debris was removed by centrifugation at 50,000g (45 min, 4°C), the supernatant was filtered through a 0.22-µm membrane, and the proteins were resolved using a His affinity column (Novagen His.Bind Resin; Merck) and FPLC. The column was equilibrated in 50 mm sodium phosphate, pH 7.5, 100 mm NaCl buffer containing 10 mm imidazole, followed by gradual elution using a gradient from 10 to 500 mm imidazole (Supplemental Fig. S2B). Finally, the buffer was exchanged (50 mm sodium phosphate, pH 7.5, 100 mm NaCl, and 10% [v/v] glycerol) using ultrafiltration spin columns (10-kD exclusion size; Merck Millipore), and the protein solution was flash frozen in liquid nitrogen and stored at –80°C. Antibodies were generated in rabbits with 2 mg of native protein by Pineda Antikörper Service.
Characterization of Purified pCRY by Mass Spectrometry
For the identification of pCRY by LC-ESI-MS/MS, 10 μg of the heterologously expressed, purified protein was digested with 2.5 μg of sequencing-grade modified trypsin (Promega) at 37°C overnight. The digested protein was desalted with a self-made Tip column using Poros R2 10 μm column medium (Applied Biosystems), dried, dissolved in an aqueous solution containing 5% (v/v) dimethyl sulfoxide and 5% (v/v) formic acid, and then analyzed by LC-ESI-MS/MS according to Veith et al. (2009). A false discovery rate of 1% or less was set for data analysis.
Crude Extracts and Immunoblots
Extraction of Chlamydomonas crude protein and immunoblots for the detection of pCRY was performed according to Iliev et al. (2006) with the following modifications. Far-red safety lights (TO-66 high-power array; Roithner Lasertechnik) with a peak at 700 nm (with a full width at half-maximum [FWHM] of 22 nm) and a photon fluence rate of 15.2 μmol m−2 s−1 were used to prepare the protein extracts. Proteins were separated by SDS-PAGE, transferred to PVDF membranes, which were then blocked in a 5% (w/v) solution of milk powder in 20 mm Tris, pH 8, 150 mm NaCl with 0.1% (w/v) Tween 20, and probed with primary antibodies against pCRY for 18 h at 4°C and a dilution of 1:3,000. Horseradish peroxidase-conjugated anti-rabbit IgG was used as the secondary antibody, and peroxidase activity was detected by a chemiluminescence assay. PVDF membranes stained with Coomassie Brilliant Blue R 250 were used as both a loading control and for the verification of protein transfer. pCRY protein levels were quantified using ImageJ 1.48v (Wayne Rasband, National Institutes of Health). Expression levels were normalized to the wild-type signal, which was set to 100% (n = 3).
Size-Exclusion Chromatography
Size-exclusion chromatography was performed using an Äkta FPLC device (GE Healthcare) with a Superdex 200 Increase 10/300 GL column (GE Healthcare) at 4°C. For equilibration and elution, we used a buffer containing 50 mm phosphate (pH 7.4) and 150 mm NaCl. All of the following steps were performed under safety light (far red) conditions. Extracts of soluble proteins were made with the elution buffer containing a protease inhibitor cocktail (cOmplete Protease Inhibitor Cocktail; Roche) according to Zhao et al. (2004). Aliquots of 300 μL with a total protein amount of 1 mg were centrifuged at 10,000g for 10 min at 4°C and filtered through a 0.22-µm filter before loading onto the column. Elution profiles were recorded at 280 nm. Fractions of 500 µL were collected and denatured, and 100-µL aliquots were separated on a 7% polyacrylamide gel by SDS-PAGE and then analyzed by immunoblots. A set of protein standards (Bio-Rad; gel filtration standard no. 1511901) was used as a reference to determine the apparent molecular mass of pCRY in the samples.
Circadian Photoaccumulation (Phototaxis) and Phase Response Curves
Measurements for photoaccumulation were performed with a custom-made apparatus that was developed and described by Mergenhagen (1984). A light beam that passed through the liquid cell culture (in a 30-mL flat Falcon tube) was detected by a photocell, and the transmitted light was used to determine cell density (recorded as extinction in mV). The calculation was performed with the minimum value (few or no cells in the light path) set to zero, and measurements were taken every 2 h for a period of 20 min. The system for recording phototaxis was maintained in a temperature-controlled dark room at 20°C. The free-running periods were established from cells that were transferred to minimal medium (Harris, 1989) and kept in constant darkness for several days. The length of the period was determined based on the mean value ± sd of consecutive minima. Measurements were started at day 2 under constant darkness. For the wild type and the complemented mutant, up to five minima were routinely used for the calculation, and for the pcry mutant, up to four minima were used because of its arrhythmic behavior after 5 d.
For phase-response experiments, phase shifts were created in cultured cells by blue light of 6-h duration (Johnson et al., 1991) during the first day. For the blue light pulses, LEDs with the following settings were used: SuperFlux LED, energy fluence rate of 2.6 W m−2, peak at 465 nm (with an FWHM of 18.5 nm), and a photon fluence rate of 60 μmol m−2 s−1. Blue light pulses were applied at DD2, DD6, DD10, DD14, DD18, or DD22, and the subsequent photoaccumulation of cells was measured using our custom-made apparatus. Cells kept in the dark for 6 h at the indicated time points served as a reference to calculate the phase shift. For data evaluation, circadian time in hours was calculated with the real hour multiplied by a factor of 24 divided by the period (CT [h] = real hour [h] × (24/period [h]); Kondo et al., 1991). Thereby, the period was determined by comparing the mean values of three consecutive maxima and minima starting from day 3 in control and illuminated cells, respectively.
Generation of Pregametes and Gametes for RT-qPCR
Vegetative cells were transferred to TAP medium without NH4+ at the end of the light period (LD12) and maintained for 14 h in darkness to induce pregametes, which were taken as the control. The pregametes were then exposed for 5 h to blue light (Rodriguez et al., 1999) to elicit the transition to gametes. Blue light treatments were performed using LEDs with the following settings: SuperFlux LED, energy fluence rate of 2.6 W m−2, peak at 465 nm (with an FWHM of 18.5 nm), and a photon fluence rate of 10 μmol m−2 s−1. RT-qPCR was done as described previously (Beel et al., 2012). A quantity of 300 ng of total RNA was used in each reaction for one-step RT-qPCR; the reagents used to perform these reactions were commercially purchased (QuantiTect SYBR Green RT-PCR; Qiagen). RT-qPCR was performed using an Mx3005P instrument (Agilent Technologies), and the relative transcript abundances from target genes were calculated based on the 2-ΔΔCT method (Livak and Schmittgen, 2001). RACK1 (Mus et al., 2007) was taken as a constitutive reference transcript for normalization. Supplemental Table S2 lists all primers. These primers were designed to amplify a 100- to 250-bp region that bridges at least one intron in the genomic DNA (based on available gene models). Cycling conditions included an initial incubation at 95°C for 15 min followed by 40 cycles of 95°C for 20 s, 56°C for 25 s, and 72°C for 35 s. The specificity of the primers was determined from melt curves: a single, sharp peak in the melt curve was used as an indication that a single, specific DNA species was amplified.
Mating Ability and Mating Maintenance Test
Liquid cultures of vegetative cells (0.5 × 107 to 1 × 107 cells mL−1) were centrifuged (2,000g for 3 min) and resuspended in nitrogen-free TAP medium at LD12. Cells of the tester strain mt+ and the mating partner strain mt− were treated differentially. Vegetative cells of the tester strain were incubated in culture flasks (Nunc) with shaking at 70 rpm min−1 for 15 h in the dark to generate pregametes, while the vegetative cells of the mt− partner strain were incubated for 12 h in darkness followed by a 3-h light treatment (60 μmol m−2 s−1) to induce the formation of gametes. Pregametes of the tester strains were put under white light with a fluence of 60 μmol m−2 s−1 for 1 h (called G1 cells) to induce the mating ability. Mating ability was tested by mixing the tester strain with the mating partner, which was added in ∼2-fold excess to the tester strain. Then, the mixture was treated under dark for 1 h to allow mating (Beck and Acker, 1992). The mating efficiency was determined by recording the quadriflagellated and biflagellated cells using a microscope with phase contrast after fixation with 0.2% glutaraldehyde and calculating according to Beck and Acker (1992).
To test the mating maintenance of the strains, gametes with 1 h of illumination were put into a dark box for 1 h to deactivate their mating ability. The mating ability of the dark-inactivated gametes was examined as described.
Germination Assay
For the germination assay, a light intensity of 30 μmol m−2 s−1 was used. Vegetative cells of each mating type were cultivated separately on solid TAP medium for 3 d under LD12:12 and subsequently transferred to solid N10 medium with only one-tenth of the normal amount of nitrogen (Jiang and Stern, 2009). The cells were maintained on N10 for an additional 3 d under an LD12:12 regime to induce gametogenesis. The cells were suspended in mating buffer (0.6 mm MgCl2 and 1.2 mm HEPES, pH 6.8) at a cell density of approximately 1 × 107 cells mL−1 and shaken for 3 h in the light to obtain motile individual gametes (Suzuki and Johnson, 2002). Cells of two different mating types were mixed and incubated for 1 h in the light to allow mating. The mating mixture was applied to 3% Difco agar TAP plates and dried on a sterile bench. Dried plates were incubated under LD12:12 for 3 d. To analyze the germination rate, the unmated vegetative cells were scraped from the top of the plates with a razor blade, and then the agar blocks with the attached 3-d-old zygotes were cut and transferred to 1.5% Difco agar TAP plates. Single zygotes were distributed with a glass needle using a binocular microscope and treated for 30 s with chloroform to kill the unmated gametes surrounding the zygotes. The plates were returned to LD12:12 conditions, and germinated zygotes were counted as positive depending on whether one or more spores from a tetrad could form colonies. The germination rate reached a plateau level until day 10 after mating and was considered until then.
Accession Numbers
Sequences from this article can be found in the Phytozome 11.0 database (Chlamydomonas reinhardtii version 5.5) and are listed in Supplemental Table S2.
Supplemental Data
The following supplemental data are available.
Supplemental Figure S1. Sequences of the pcry mutant strain and primers for characterization.
Supplemental Figure S2. Sequence of the codon-adapted pCRY gene with a His tag for heterologous expression and purification from E. coli.
Supplemental Figure S3. Sequence and overview scheme of the complementation vector pNM003.
Supplemental Figure S4. Complemented strain of the pcry mutant in the background of SAG73.72.
Supplemental Figure S5. Comparative presentation of the phase and period of the circadian rhythm of photoaccumulation in the wild type, pcrymut, and the complemented mutant.
Supplemental Figure S6. Sequencing results of GAS3, GAS18, and GAS96.
Supplemental Figure S7. pCRY influences the transcript accumulation of GAS18, GAS3, and GAS96 only weakly (GAS18) or not significantly upon blue light illumination during gametogenesis.
Supplemental Figure S8. The pcry mutant in the background of CC-124/125 and its complementation.
Supplemental Table S1. LC-ESI-MS/MS analysis of heterologously expressed pCRY (first 504 amino acids) with a C-terminal 6× His tag.
Supplemental Table S2. Oligonucleotides used in RT-qPCR.
Supplementary Material
Acknowledgments
We thank Mark Heinnickel, David Dewez, and Danielle Ikoma for help with the insertional library, Dieter Mergenhagen for the donation of the automated photoaccumulation machine, Christoph Beck and Erika von Gromoff for supplying the GAS cDNAs, Wolfgang Mages for pHyg3, and Wolfram Weisheit for help with LC-ESI-MS/MS.
Glossary
- LC-ESI-MS/MS
liquid chromatography-electrospray ionization-tandem mass spectrometry
- RT-qPCR
reverse transcription-quantitative PCR
- TAP
Tris-acetate-phosphate
- FWHM
full width at half-maximum
- PVDF
polyvinylidene difluoride
- LED
light-emitting diode
Footnotes
This work was supported by the German Research Foundation within Research Group FOR1261 (grant nos. Mi373/12-2 and Mi373/15-1 to M.M. and grant no. TK 3580/1-2 to T.K.), the Jena School for Microbial Communication (fellowship to Y.Z.), the National Science Foundation (grant no. MCB 0951094 to A.G. for work on mutant isolation), and an Heisenberg Fellowship (grant no. TK 3580/4-1 to T.K.).
References
- Beck CF, Acker A (1992) Gametic differentiation of Chlamydomonas reinhardtii: control by nitrogen and light. Plant Physiol 98: 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel B, Müller N, Kottke T, Mittag M (2013) News about cryptochrome photoreceptors in algae. Plant Signal Behav 8: e22870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel B, Prager K, Spexard M, Sasso S, Weiss D, Müller N, Heinnickel M, Dewez D, Ikoma D, Grossman AR, et al. (2012) A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 24: 2992–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W (2002) An engineered Streptomyces hygroscopicus aph 7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412 [DOI] [PubMed] [Google Scholar]
- Bruce VG. (1970) The biological clock in Chlamydomonas reinhardtii. J Protozool 17: 328–334 [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364 [DOI] [PubMed] [Google Scholar]
- Chen CH, Dunlap JC, Loros JJ (2010) Neurospora illuminates fungal photoreception. Fungal Genet Biol 47: 922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM. (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58: 21–45 [DOI] [PubMed] [Google Scholar]
- Coesel S, Mangogna M, Ishikawa T, Heijde M, Rogato A, Finazzi G, Todo T, Bowler C, Falciatore A (2009) Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep 10: 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe H, Prager K, Mittag M (2012) Novel interaction of two clock-relevant RNA-binding proteins C3 and XRN1 in Chlamydomonas reinhardtii. FEBS Lett 586: 3969–3973 [DOI] [PubMed] [Google Scholar]
- Forbes-Stovall J, Howton J, Young M, Davis G, Chandler T, Kessler B, Rinehart CA, Jacobshagen S (2014) Chlamydomonas reinhardtii strain CC-124 is highly sensitive to blue light in addition to green and red light in resetting its circadian clock, with the blue-light photoreceptor plant cryptochrome likely acting as negative modulator. Plant Physiol Biochem 75: 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato AE, Annunziata R, Jaubert M, Bouly JP, Falciatore A (2015) Dealing with light: the widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J Plant Physiol 172: 42–54 [DOI] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C (2015) Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol 34: 46–53 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, Pootakham W, Mus F, Yang W, Catalanotti C, Magneschi L, de Montaigu A, Higuera JJ, Prior M, Galván A, et al. (2011) Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. Plant Methods 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Lin H, Lee JH (2007) Sex determination in Chlamydomonas. Semin Cell Dev Biol 18: 350–361 [DOI] [PubMed] [Google Scholar]
- Govorunova EG, Jung KH, Sineshchekov OA, Spudich JL (2004) Chlamydomonas sensory rhodopsins A and B: cellular content and role in photophobic responses. Biophys J 86: 2342–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Hegemann P. (2008) Algal sensory photoreceptors. Annu Rev Plant Biol 59: 167–189 [DOI] [PubMed] [Google Scholar]
- Heijde M, Zabulon G, Corellou F, Ishikawa T, Brazard J, Usman A, Sanchez F, Plaza P, Martin M, Falciatore A, et al. (2010) Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ 33: 1614–1626 [DOI] [PubMed] [Google Scholar]
- Hoffmann XK, Beck CF (2005) Mating-induced shedding of cell walls, removal of walls from vegetative cells, and osmotic stress induce presumed cell wall genes in Chlamydomonas. Plant Physiol 139: 999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Beck CF (2003) Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 100: 6269–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Kunkel T, Beck CF (2004) Localization of the blue-light receptor phototropin to the flagella of the green alga Chlamydomonas reinhardtii. Mol Biol Cell 15: 3605–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysman MJ, Fortunato AE, Matthijs M, Costa BS, Vanderhaeghen R, Van den Daele H, Sachse M, Inzé D, Bowler C, Kroth PG, et al. (2013) AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 25: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M (2006) A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol 142: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im CS, Eberhard S, Huang K, Beck CF, Grossman AR (2006) Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J 48: 1–16 [DOI] [PubMed] [Google Scholar]
- Immeln D, Schlesinger R, Heberle J, Kottke T (2007) Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J Biol Chem 282: 21720–21728 [DOI] [PubMed] [Google Scholar]
- Immeln D, Weigel A, Kottke T, Pérez Lustres JL (2012) Primary events in the blue light sensor plant cryptochrome: intraprotein electron and proton transfer revealed by femtosecond spectroscopy. J Am Chem Soc 134: 12536–12546 [DOI] [PubMed] [Google Scholar]
- Jiang X, Stern D (2009) Mating and tetrad separation of Chlamydomonas reinhardtii for genetic analysis. J Vis Exp 30: e1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Hastings JW (1989) Circadian phototransduction: phase resetting and frequency of the circadian clock of Gonyaulax cells in red light. J Biol Rhythms 4: 417–437 [DOI] [PubMed] [Google Scholar]
- Johnson CH, Kondo T, Hastings JW (1991) Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas. II. illuminated cells. Plant Physiol 97: 1122–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, von Zadow A, Spexard M, Schmidt M, Kottke T, Büchel C (2014) A novel cryptochrome in the diatom Phaeodactylum tricornutum influences the regulation of light-harvesting protein levels. FEBS J 281: 2299–2311 [DOI] [PubMed] [Google Scholar]
- Kianianmomeni A, Hallmann A (2014) Algal photoreceptors: in vivo functions and potential applications. Planta 239: 1–26 [DOI] [PubMed] [Google Scholar]
- Kirk JTO. (1994) Light and Photosynthesis in Aquatic Ecosystems, Ed 2 Cambridge University Press, Cambridge, UK [Google Scholar]
- Kondo T, Johnson CH, Hastings JW (1991) Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas. I. Cells in darkness. Plant Physiol 95: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH (1993) Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA 90: 5672–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbacher T, Immeln D, Dick B, Kottke T (2009) Microsecond light-induced proton transfer to flavin in the blue light sensor plant cryptochrome. J Am Chem Soc 131: 14274–14280 [DOI] [PubMed] [Google Scholar]
- Lee DH, Mittag M, Sczekan S, Morse D, Hastings JW (1993) Molecular cloning and genomic organization of a gene for luciferin-binding protein from the dinoflagellate Gonyaulax polyedra. J Biol Chem 268: 8842–8850 [PubMed] [Google Scholar]
- Liu B, Liu H, Zhong D, Lin C (2010) Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol 13: 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lopez D, Hamaji T, Kropat J, De Hoff P, Morselli M, Rubbi L, Fitz-Gibbon S, Gallaher SD, Merchant SS, Umen J, et al. (2015) Dynamic changes in the transcriptome and methylome of Chlamydomonas reinhardtii throughout its life cycle. Plant Physiol 169: 2730–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck M, Mathes T, Bruun S, Fudim R, Hagedorn R, Tran Nguyen TM, Kateriya S, Kennis JT, Hildebrandt P, Hegemann P (2012) A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J Biol Chem 287: 40083–40090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M (2008) A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev 22: 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergenhagen D. (1984) Circadian clock: genetic characterization of a short period mutant of Chlamydomonas reinhardii. Eur J Cell Biol 33: 13–18 [PubMed] [Google Scholar]
- Mergenhagen D, Mergenhagen E (1987) The biological clock of Chlamydomonas reinhardii in space. Eur J Cell Biol 43: 203–207 [PubMed] [Google Scholar]
- Mittag M, Kiaulehn S, Johnson CH (2005) The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol 137: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA 100: 13940–13945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Kateriya S, Adeishvili N, Hegemann P, Bamberg E (2005) Channelrhodopsins: directly light-gated cation channels. Biochem Soc Trans 33: 863–866 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Matsuo T, Onai K, Kato D, Tachikawa M, Ishiura M (2013) Phase-resetting mechanism of the circadian clock in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110: 13666–13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohr D, Franz S, Rodriguez R, Paulus B, Essen LO, Weber S, Schleicher E (2016) Extended electron-transfer in animal cryptochromes mediated by a tetrad of aromatic amino acids. Biophys J 111: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldemeyer S, Franz S, Wenzel S, Essen LO, Mittag M, Kottke T (2016) Essential role of an unusually long-lived tyrosyl radical in the response to red light of the animal-like cryptochrome aCRY. J Biol Chem 291: 14062–14071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JM, Haring MA, Beck CF (1997) Characterization of blue light signal transduction chains that control development and maintenance of sexual competence in Chlamydomonas reinhardtii. Plant Physiol 115: 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D, Tokutsu R, Maruyama S, Flori S, Greiner A, Magneschi L, Cusant L, Kottke T, Mittag M, Hegemann P, et al. (2016) A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537: 563–566 [DOI] [PubMed] [Google Scholar]
- Pootakham W, Gonzalez-Ballester D, Grossman AR (2010) Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol 153: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorph NA, Small GD (2004) The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol 134: 1546–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez H, Haring MA, Beck CF (1999) Molecular characterization of two light-induced, gamete-specific genes from Chlamydomonas reinhardtii that encode hydroxyproline-rich proteins. Mol Gen Genet 261: 267–274 [DOI] [PubMed] [Google Scholar]
- Saito T, Inoue M, Yamada M, Matsuda Y (1998) Control of gametic differentiation and activity by light in Chlamydomonas reinhardtii. Plant Cell Physiol 39: 8–15 [Google Scholar]
- Sancar A. (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103: 2203–2237 [DOI] [PubMed] [Google Scholar]
- Schellenberger Costa B, Sachse M, Jungandreas A, Bartulos CR, Gruber A, Jakob T, Kroth PG, Wilhelm C (2013) Aureochrome 1a is involved in the photoacclimation of the diatom Phaeodactylum tricornutum. PLoS ONE 8: e74451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M, Geimer S, Eitzinger N, Reissenweber T, Voytsekh O, et al. (2006) Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18: 1908–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F (2009) Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol 19: 359–368 [DOI] [PubMed] [Google Scholar]
- Spexard M, Thöing C, Beel B, Mittag M, Kottke T (2014) Response of the sensory animal-like cryptochrome aCRY to blue and red light as revealed by infrared difference spectroscopy. Biochemistry 53: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Wada M (2013) Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol 54: 8–23 [DOI] [PubMed] [Google Scholar]
- Suzuki L, Johnson CH (2002) Photoperiodic control of germination in the unicell Chlamydomonas. Naturwissenschaften 89: 214–220 [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yamagata D, Ishikawa M, Fukamatsu Y, Ogura Y, Kasahara M, Kiyosue T, Kikuyama M, Wada M, Kataoka H (2007) AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc Natl Acad Sci USA 104: 19625–19630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöing C, Oldemeyer S, Kottke T (2015) Microsecond deprotonation of aspartic acid and response of the α/β subdomain precede C-terminal signaling in the blue light sensor plant cryptochrome. J Am Chem Soc 137: 5990–5999 [DOI] [PubMed] [Google Scholar]
- Tilbrook K, Dubois M, Crocco CD, Yin R, Chappuis R, Allorent G, Schmid-Siegert E, Goldschmidt-Clermont M, Ulm R (2016) UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell 28: 966–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith T, Brauns J, Weisheit W, Mittag M, Büchel C (2009) Identification of a specific fucoxanthin-chlorophyll protein in the light harvesting complex of photosystem I in the diatom Cyclotella meneghiniana. Biochim Biophys Acta 1787: 905–912 [DOI] [PubMed] [Google Scholar]
- von Gromoff ED, Beck CF (1993) Genes expressed during sexual differentiation of Chlamydomonas reinhardtii. Mol Gen Genet 241: 415–421 [DOI] [PubMed] [Google Scholar]
- Weissig H, Beck CF (1991) Action spectrum for the light-dependent step in gametic differentiation of Chlamydomonas reinhardtii. Plant Physiol 97: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Schneid C, Iliev D, Schmidt EM, Wagner V, Wollnik F, Mittag M (2004) The circadian RNA-binding protein CHLAMY 1 represents a novel type heteromer of RNA recognition motif and lysine homology domain-containing subunits. Eukaryot Cell 3: 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones JM, Blaby IK, Merchant SS, Umen JG (2015) High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorin B, Lu Y, Sizova I, Hegemann P (2009) Nuclear gene targeting in Chlamydomonas as exemplified by disruption of the PHOT gene. Gene 432: 91–96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.