ABSTRACT
Skin constantly encounters external elements, including microbes. Culture-based studies have identified fungi present on human skin and have linked some species with certain skin diseases. Moreover, modern medical treatments, especially immunosuppressants, have increased the population at risk for cutaneous and invasive fungal infections, emphasizing the need to understand skin fungal communities in health and disease. A major hurdle for studying fungal flora at a community level has been the heterogeneous culture conditions required by skin fungi. Recent advances in DNA sequencing technologies have dramatically expanded our knowledge of the skin microbiome through culture-free methods. This review discusses historical and recent research on skin fungal communities – the mycobiome – in health and disease, and challenges associated with sequencing-based mycobiome research.
KEYWORDS: fungal community, Malassezia, mycobiome, skin
Introduction
Skin is the outermost compartment of the body and often the first point of contact for an extensive number of environmental microbes. The skin surface is a unique niche, serving as a habitat for a distinct community of microorganisms.1 While metagenomic analyses have shown that the majority of annotated reads from samples obtained from most skin sites are bacterial, some sites may have relatively high proportions of fungal or viral sequences.2,3 Understanding skin fungal communities as well as investigating interactions between bacterial and fungal communities are essential, especially given the association of fungi with various skin disorders, such as seborrheic dermatitis, atopic dermatitis, and dermatophytosis.4-9
Culturing has long been used to isolate and identify fungi on skin. However, this approach may not fully capture the diversity and/or relative composition of fungal communities, as optimal culture conditions and growth rates vary widely between species.10 Several molecular approaches such as restriction fragment length polymorphism (RFLP) analyses have been used to complement culturing; however, these low-throughput methods only take into account a portion of the community and likely underestimate the diversity of fungi present. Development of high-throughput sequencing techniques and microbiome analysis methods are revolutionizing our understanding of the “mycobiome”—here defined as the overall fungal habitat, including fungi, “their genomes (i.e. genes), and the surrounding environmental conditions.”11 This review focuses on efforts to examine the skin mycobiome in healthy individuals and alterations associated with various skin conditions and disorders.
“Normal” skin mycobiome
Cultivation methods recover primarily Malassezia (formerly known as Pityrosporum),12 as well as occasionally Candida and a variety of other species on healthy human skin.13 Initial studies used culture-based techniques to speciate Malassezia, relying on morphology and lipid dependence to differentiate isolates12 and identifying new species based on distinct physiological and biochemical features.14 Culturing surveys commonly have isolated M. globosa, M. restricta, and M. sympodialis from healthy human skin.14 However, cultivation-based studies can be biased by culture media and growth conditions, so results from different studies may not be comparable. Malassezia isolates can require different conditions for optimal growth, and some species (such as M. furfur and M. sympodialis) grow more quickly than others (M. restricta and M. obtusa), which may skew the relative abundances of species observed.15
Molecular methods have facilitated investigations of Malassezia spp. Genomic differences, which have been shown to correspond to morphological distinctions, allow for inter-species resolution.16 Analyses based on nested PCR or RFLP have suggested that M. globosa, M. restricta, M. sympodialis, and M. furfur inhabit the skin of healthy individuals.17,18 More recent studies performing culture-independent sequencing analyses of skin fungal communities rely on amplicon sequencing using fungi-specific phylogenetic markers such as the 18S rRNA gene, internal transcribed spacers, and the 28S rRNA gene.19 Researchers have used a single region or combination of regions, with primers targeting the D1/D2 region of the 28s rRNA gene,20 the 18S rRNA gene with the 5.8S/ITS2 region for Malassezia speciation,21 ITS1-5.8S-ITS2 and the 28S rRNA gene,22 or ITS1 and the 18S rRNA gene.23 These cultivation-independent studies from individuals residing in varying geographic regions also showed Malassezia predominates on most skin sites20-22,24 except the feet.23 Eleven of the fourteen Malassezia species have been identified by sequencing swabs from healthy human skin, indicating the range of colonization possible.23 Here, we discuss the distribution of skin fungi in association with body site, age, and gender.
Body site
Quantitative culturing has shown that Malassezia colonization is particularly high in sebaceous sites such as the scalp and the forehead.25 Several Malassezia species are present in statistically significantly different proportions at spatially distinct body sites. M. globosa was present in higher abundances on head sites (scalp and forehead) and in lower abundances on the back, while relative abundances of M. sympodialis were distributed in the inverse pattern.26 Results from culture-independent studies also indicated that fungal communities are strongly influenced by body location, with different sites harboring distinct populations.20,23 Using ITS1 sequencing, M. restricta and M. globosa patterns were similar among different individuals sampled at the same site. M. restricta predominated on head and facial sites whereas M. globosa was predominant on trunk sites. Other body sites demonstrated more mixed Malassezia communities, and feet were populated by diverse fungal communities including Aspergillus, Cryptococcus, Rhodotorula, and Epicoccum23 (Fig. 1).
Figure 1.
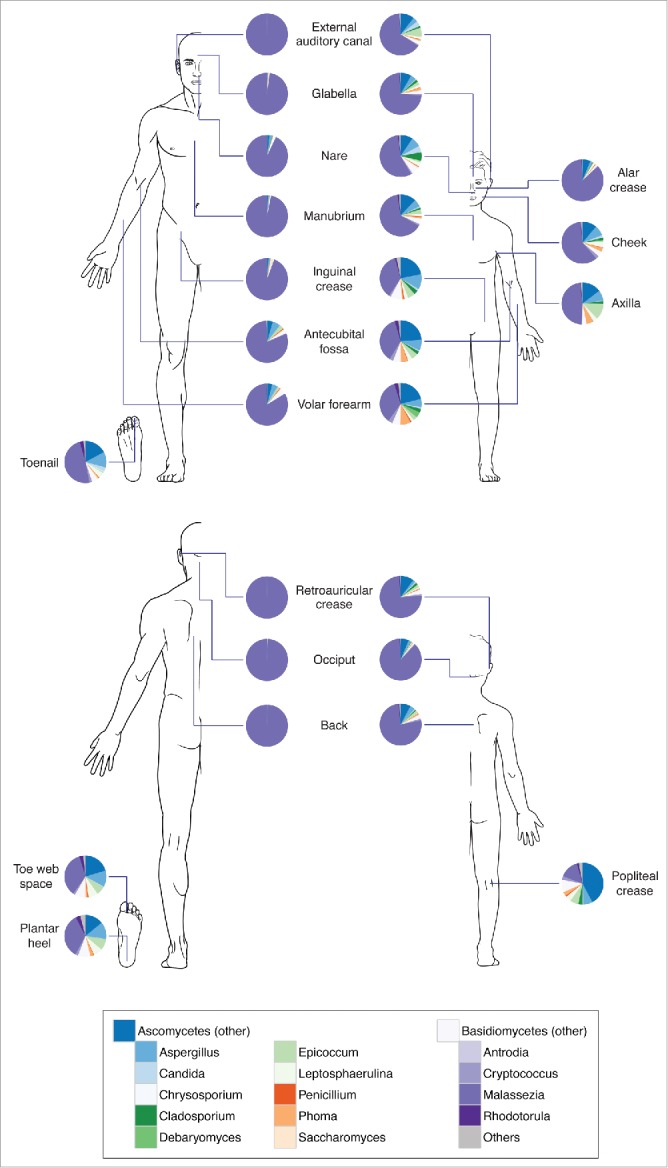
Composition of skin fungal communities distributed over various skin sites and in different age groups. Phylum- and genus-level classification of fungi colonizing skin of healthy adults (left, age 18–39 years) and children (right, age 8–13 years). Data adapted from Ref.27.
The site selectivity of Malassezia species may be explained by genomic differences within the Malassezia genus that can affect niche adaptation. The Malassezia genus lacks the fatty acid synthase gene, so species are lipid-dependent, explaining their predominance on sebaceous sites. However, different species vary in their lipase and phospholipase genes with resultant variation in lipid preferences.27 Given that lipid profiles vary at different skin sites,28,29 these preferences may underlie the observed adaptation to different sites, although more research is needed to clarify how genetics may drive niche preference.
Age
Shifts in fungal colonization occur with age, with young subjects colonized by lower levels of Malassezia and colonization generally increasing in adolescence to adulthood.15,26,30 Analysis based on ITS1 sequencing has also shown low relative abundance of Malassezia accompanied by high fungal diversity on the skin of preadolescent children aged 8–13 y as compared to adults31 (Fig. 1). These observed shifts may be explained by the increase in sebaceous activity during adolescence which provides a more favorable niche for lipophilic Malassezia to colonize.32 The variation in fungal communities across different age groups may contribute to the age predilection of several fungal infections. For example, tinea capitis and tinea corporis are typically caused by a variety of non-Malassezia fungi and are more prevalent in children, whereas tinea versicolor, associated with Malassezia spp., is more common in adults.6,7,33
At the species level, culture-based methods have shown that M. globosa predominated in children whereas M. sympodialis was typically absent; in contrast, M. globosa was less common than M. sympodialis in adults.26 Culture-independent analyses also showed that M. globosa was less abundant in adult subjects, particularly at facial sites, with M. restricta predominating.15,23,31 Changes in the composition of the secreted lipids may explain these shifts over time, given the different lipid preferences of individual species.27,34
Gender
Sugita et al. noted that female subjects had a reduced abundance of Malassezia at the age of 19–29 years as compared to male subjects of the same age or female subjects of different ages. The authors attributed this finding to the prevalent use of cosmetics, which contain compounds that may inhibit cutaneous fungal growth.15 In atopic dermatitis patients, culture-independent methods showed that females had a statistically significantly reduced abundance of Malassezia spp. on their faces, but not their upper trunk skin, as compared to males.35 However, another study failed to identify significant differences in Malassezia colonization based on gender, although results were based on culturing and cosmetic use was not discussed.36
Mycobiome and pathogenesis of skin diseases
The major commensal skin fungi: Malassezia spp
A goal of studying the human skin mycobiome is to understand the role of fungal communities in health and disease. In addition to being the most prevalent fungus identified on human skin, the genus Malassezia has been extensively studied to understand its connection to skin diseases. Various reports have proposed that Malassezia spp. are associated with several dermatologic conditions, including tinea versicolor (also known as pityriasis versicolor), atopic dermatitis, and seborrheic dermatitis.4,5
Tinea versicolor is characterized by the appearance of round lesions that are hyper-or hypopigmented than the surrounding skin.4 Lesions preferentially appear on sebaceous skin areas, such as the back and chest. Studies of the pathogenesis of tinea versicolor typically implicate Malassezia. Reports have shown that affected lesions have higher loads of M. globosa, M. sympodialis, and M. furfur as compared to non-lesional sites and the skin of healthy individuals.37-40 Mechanistically, it has been suggested that Malassezia proteins such as malassezin and pityriacitrin contribute to hypo- or hyperpigmentation of the lesion by inducing apoptosis of melanocytes and directly absorbing UV light.41-43
Atopic dermatitis is a common inflammatory skin disease characterized by chronic relapsing itchy red skin. Many factors contribute to the development and progression of atopic dermatitis, including genetics, skin barrier, environmental factors, and skin bacteria.44-47 Although several culture-based studies have described lack of association between atopic dermatitis and Malassezia spp.,25,40,48,49 others have shown an association between Malassezia and atopic dermatitis. A molecular-based study (nested PCR) showed that Malassezia spp. are more frequently detected and are more diverse in patients than healthy subjects.17 Some studies also demonstrated higher Malassezia-specific serum IgE in atopic dermatitis patients50-52 and systemic antifungal ketoconazole treatment in patients with relatively high Malassezia-specific IgE levels was associated with clinical improvement.50 Other studies have described IgE or reactive T cells that recognize Malassezia antigens, e.g. HSP70, MnSOD, and thioredoxin, in patients with atopic dermatitis.4,53-55
Seborrheic dermatitis is characterized by recurrent, relapsing inflammation with yellowish greasy scaling observed in sebaceous areas of the head and trunk, specifically the scalp (dandruff). Several studies have linked seborrheic dermatitis and dandruff with Malassezia. Culturing showed Malassezia spp. were more frequently isolated from the lesions of seborrheic dermatitis.40 Also, data suggest a causal link between Malassezia and dandruff, since various antifungal reagents—zinc salts, selenium salts and azoles—are highly effective in the treatment of dandruff.56,57 Improvement of the disease by anti-dandruff shampoo use is accompanied by reduced Malassezia levels.57 Although the role of Malassezia in this disease is not well understood, the higher prevalence of seborrheic dermatitis in immunodeficient HIV patients58-60 (10–30 times higher than the general population) suggests an immune response against Malassezia might be important for controlling the disease.
While culture-based studies have shown that Malassezia spp. are more frequently isolated from the lesions of tinea versicolor, atopic dermatitis, and seborrheic dermatitis, recent culture-independent analyses have shown that Malassezia are a part of the normal skin flora.23,27,61 Therefore, Malassezia spp. are likely to be opportunistic microorganisms that may elicit disease with changes in host immunity or growth phase—yeast versus mycelial form—of the fungus.25,62 Higher levels of anti-Malassezia antibodies have been observed in the serum of atopic dermatitis50 and seborrheic dermatitis63 patients (IgE and IgG, respectively). Microscopic studies illustrated that the mycelial forms of Malassezia were frequently observed in lesional sites of tinea versicolor, but the yeast form was present on non-lesional skin and on healthy controls.37,64,65 In addition, several Malassezia metabolites or virulence factors, e.g., indole compounds41 and lipases,66 have been proposed to play roles in the pathogenesis of certain skin diseases. Some indole derivatives from M. furfur have been shown to act as aryl hydrocarbon receptor agonists involved in various cellular processes such as immune signaling, cell cycle, and apotosis.67 More recently, whole genome sequencing of several Malassezia species was performed, which provides insights into niche selection and virulence.27
Mycobiome alteration and disease
While most studies examining the relationship between microbes and disease focus on culturing and implicate a single microbial species, microbial communities can be studied at a global level with sequencing-based approaches. In general, microbiome studies have focused primarily on bacteria with less of an emphasis on fungi. Studies of the bacterial skin communities have shown that diversity is inversely correlated with disease severity in atopic dermatitis and psoriasis.47,68,69 In contrast, higher fungal diversity has been associated with atopic dermatitis61 and psoriasis70 as compared to controls. Atopic dermatitis patients also had increased relative abundances of Candida spp., Cryptococcus spp. and Malassezia globosa in comparison to healthy individuals, and psoriatic skin lesions had increased relative abundances of Malassezia restricta. However, the relevance of these mycobiome diversity and community differences to disease pathophysiology remains unknown. Therefore, additional studies in these and other skin diseases with proper controls will be important for understanding mycobiome shifts.
Role of host factor in skin mycobiome homeostasis and disease
Host factors, such as genetics and immune status, can influence microbiome composition and, in turn, diseases. Patients with primary immunodeficiency syndromes often suffer from recurrent infections, including fungal infections.71-73 Chronic candidiasis is frequently observed in patients with mutations in immune modulatory genes, such as AIRE (autoimmune regulator), dectin-1 (C-type lectin receptor), and CARD9 (Caspase recruitment domain family 9).74-76 Signal transducer and activator of transcription 3 (STAT3) is a key transcription factor involved in immune signaling, and is activated by interleukin-6 (IL-6), IL-10, IL-23, IL-21 and IL-11.77 Heterozygous mutations in STAT3 causes hyper-IgE syndrome (HIES), a primary immunodeficiency disease characterized by high serum IgE, atopic dermatitis-like symptoms, and systemic infections.78-80 Oh et al. demonstrated that STAT3-HIES patients exhibited increased mycobiome richness, with reduced Malassezia and increased Aspergillus and Candida even on unaffected skin.81 This observation corresponds to clinical manifestations of STAT3-HIES, which often include fungal infections such as mucocutaneous candidiasis and pulmonary Aspergillus and Scedosporium infections. Since monogenic disorders that alter host immune function can result in susceptibility to fungal colonization and infection, studying mycobiome alterations in this context may enhance our understanding of host-microbial interactions.
Abnormal fungal infections can also occur in patients with acquired immunodeficiency, such as individuals with AIDS or those on immunosuppressive medications. The incidence of fungal infections has surged in the last few decades with the annual number of deaths from invasive mycoses in the United States increasing about 320% from 1980 to 1997,82,83 mainly due to the rise in immunocompromised individuals in the population.84-87 Cutaneous fungal infections in immunocompromised patients are caused by common opportunistic pathogens such as Candida, Aspergillus, and Cryptococcus. However, severely immunocompromised patients can sometimes be infected by fungi that usually do not colonize humans, such as Trichosporon spp, Fusarium spp., and Emmonsia spp..88 Although various fungal infections were reported in acquired immunodeficiency patients, no skin mycobiome studies have been done in these individuals. Individuals with acquired immunodeficiency may be colonized by a diverse group of fungi, similar to the primary immunodeficiency patients.81 Overall, host immune responses appear to be critical for maintaining a “healthy” fungal flora and preventing opportunistic infection; understanding the mycobiome alterations that occur in immunodeficient individuals may provide insights into the source and/or pathogenesis of these fungal infections.
Further directions of sequence-based mycobiome studies
In recent decades, our understanding of the microbiome has expanded with the development of and rapid technical advances in sequencing technology. Large-scale studies, such as the National Institutes of Health Common Fund's Human Microbiome Project, have not only provided profiles of the microbial communities at various body sites in healthy volunteers, but have also standardized pipelines for analysis.1 However, more investigations of the fungal communities on various body sites and standardized methods for studying the mycobiome are needed.
Many fungi require specific cultivation conditions for optimal growth and isolation, hampering precise quantitative analyses; sequencing-based approaches enable culture-free and high-throughput investigations of the skin fungal communities. However, there are several challenges and limitations to consider for sequencing-based mycobiome analysis. Given the low microbial biomass present on skin, samples are particularly prone to contaminants showing up in sequencing results. Sample handling and reagent contamination can affect results, so negative controls must be processed in parallel to confirm sample integrity.89 Additionally, standardized and high-yield protocols for extracting microbial DNA from the skin mycobiome are important. Use of physical cell disruption method (bead-beating) combined with chemical methods (detergents and enzymes) during DNA extraction facilitates breaking cell walls of fungi and accessing more fungal genomes, and can possibly lessen the bias toward a particular taxonomic group.90
Currently, targeted amplicon sequencing is widely used for mycobiome analysis; therefore, the selection of marker genes for taxonomic classification and the robustness of reference databases is critical for developing standardized mycobiome pipelines. Commonly used marker genes are internal transcribed spacer regions (ITS1/2) and rRNA (18S, 5.8S and 28S rRNA) genes.21,22,61,91 Advantages and disadvantages for each marker are evident: ITS exhibits high sequence variation among different taxa, providing greater taxonomic resolution but poor alignment. The 18S rRNA gene is well conserved and easier to align for better phylogenetic analysis, but allows for less taxonomic resolution. Although current reference databases for ITS1 (UNITE,92 ITSoneDB93) and 18S rRNA (SILVA94) are useful for classification, these databases include a limited number of fungi. It is estimated that approximately 1.5–5.1 million fungal species exist, whereas existing databases include less than 100,000 species.95,96 Bioinformatic methods that address these difficulties are under active development. Fouquier et al. developed a hybrid pipeline that uses ITS1 and 18S rRNA sequences together to ensure both taxonomic resolution and phylogenetic analysis,97 and a newer ITS1 database (ISHAM98; International Society of Human and Animal Mycology) and sequence analysis pipeline specific for mycobiome studies (CloVR-ITS99) are available.
An additional limitation of sequencing-based approaches includes the inability to differentiate between live and dead organisms. Several studies have addressed this challenge by developing methods that prevent DNA amplification and sequencing of dead cells.100-101 Furthermore, longitudinal surveys in the same study population can help to distinguish between long-term colonizers vs. transient microorganisms.
Several studies of the human microbiome have suggested that a homeostatic balance exists between a host and its commensal microbes. Disturbance of this homeostasis may result in disease; therefore, investigating skin mycobiome-host interactions is important for understanding the pathophysiology of various skin conditions and infections. Utilizing various “omic” approaches will be important for understanding and deciphering host-microbe and microbe-microbe interactions. Likewise, inter-kingdom interactions on skin is another intriguing area of research. Targeted species analysis has shown that there are diverse interactions between fungi and bacteria. For example, Candida albicans and Staphylococcus aureus can co-colonize and form mixed biofilms, potentially protecting them from antifungal and antibiotic reagents.102 Additionally, Pseudomonas aeruginosa was shown to inhibit C. albicans colonization on burn wounds.103 Therefore, inter-kingdom interactions within the microbiome are another crucial area to be explored. Metagenomic shotgun sequencing currently enables examination of the bacterial communities, mycobiome, and virome in a single sequencing reaction. Development of additional methods and pipelines for incorporating proteomics, transcriptomics, metabolomics, and lipidomics will widen our perspective regarding the biological and clinical implications of the mycobiome. Ultimately, studying skin mycobiome-host interactions will expand our understanding of disease pathogenesis and ability to find novel therapies for various skin conditions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Julia A. Segre, Mark C. Udey, and laboratory members for helpful discussions.
Funding
This work is supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institutes, National Institutes of Health and a grant of the Korean Health Technology R&D Project, Ministry of Health andWelfare, Republic of Korea (HI15C1095, J-HJ). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- [1].Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207-14; PMID:22699609; http://dx.doi.org/ 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, Segre JA. Biogeography and individuality shape function in the human skin metagenome. Nature 2014; 514:59-64; PMID:25279917; http://dx.doi.org/ 10.1038/nature13786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell 2016; 165:854-66; PMID:27153496; http://dx.doi.org/ 10.1016/j.cell.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 2012; 25:106-41; PMID:22232373; http://dx.doi.org/ 10.1128/CMR.00021-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL Jr. Skin diseases associated with Malassezia species. J Am Acad Dermatol 2004; 51:785-98; PMID:15523360; http://dx.doi.org/ 10.1016/j.jaad.2003.12.034 [DOI] [PubMed] [Google Scholar]
- [6].Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses 2008; 51 (Suppl 4):2-15; PMID:18783559; http://dx.doi.org/ 10.1111/j.1439-0507.2008.01606.x [DOI] [PubMed] [Google Scholar]
- [7].Seebacher C, Bouchara JP, Mignon B. Updates on the epidemiology of dermatophyte infections. Mycopathologia 2008; 166:335-52; PMID:18478365; http://dx.doi.org/ 10.1007/s11046-008-9100-9 [DOI] [PubMed] [Google Scholar]
- [8].Clavaud C, Jourdain R, Bar-Hen A, Tichit M, Bouchier C, Pouradier F, El Rawadi C, Guillot J, Ménard-Szczebara F, Breton L, et al.. Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS One 2013; 8:e58203; PMID:23483996; http://dx.doi.org/ 10.1371/journal.pone.0058203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 2002; 296:2229-32; PMID:12077418; http://dx.doi.org/ 10.1126/science.1070784 [DOI] [PubMed] [Google Scholar]
- [10].St-Germain G, Summerbell R. Identifying Fungi: A clinical laboratory handbook 1st ed., Star Publishing Company, 2011 [Google Scholar]
- [11].Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome 2015; 3:31; PMID:26229597; http://dx.doi.org/ 10.1186/s40168-015-0094-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Roberts SO. Pityrosporum orbiculare: incidence and distribution on clinically normal skin. Br J Dermatol 1969; 81:264-9; PMID:5813527; http://dx.doi.org/ 10.1111/j.1365-2133.1969.tb13978.x [DOI] [PubMed] [Google Scholar]
- [13].Mok WY, Barreto da Silva MS. Mycoflora of the human dermal surfaces. Can J Microbiol 1984; 30:1205-9; PMID:6509388; http://dx.doi.org/ 10.1139/m84-191 [DOI] [PubMed] [Google Scholar]
- [14].Gueho E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek 1996; 69:337-55; PMID:8836432; http://dx.doi.org/ 10.1007/BF00399623 [DOI] [PubMed] [Google Scholar]
- [15].Sugita T, Suzuki M, Goto S, Nishikawa A, Hiruma M, Yamazaki T, Makimura K. Quantitative analysis of the cutaneous Malassezia microbiota in 770 healthy Japanese by age and gender using a real-time PCR assay. Medical Mycol 2010; 48:229-33; PMID:19462267; http://dx.doi.org/ 10.3109/13693780902977976 [DOI] [PubMed] [Google Scholar]
- [16].Guillot J, Gueho E. The diversity of Malassezia yeasts confirmed by rRNA sequence and nuclear DNA comparisons. Antonie Van Leeuwenhoek 1995; 67:297-314; PMID:7778898; http://dx.doi.org/ 10.1007/BF00873693 [DOI] [PubMed] [Google Scholar]
- [17].Sugita T, Suto H, Unno T, Tsuboi R, Ogawa H, Shinoda T, Nishikawa A. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J Clin Microbiol 2001; 39:3486-90; PMID:11574560; http://dx.doi.org/ 10.1128/JCM.39.10.3486-3490.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oh BH, Song YC, Lee YW, Choe YB, Ahn KJ. Comparison of nested PCR and RFLP for identification and classification of malassezia yeasts from healthy human skin. Ann Dermatol 2009; 21:352-7; PMID:20523823; http://dx.doi.org/ 10.5021/ad.2009.21.4.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Khot PD, Ko DL, Fredricks DN. Sequencing and analysis of fungal rRNA operons for development of broad-range fungal PCR assays. Applied Environmental Microbiol 2009; 75:1559-65; PMID:19139223; http://dx.doi.org/ 10.1128/AEM.02383-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sugita T, Yamazaki T, Makimura K, Cho O, Yamada S, Ohshima H, Mukai C. Comprehensive analysis of the skin fungal microbiota of astronauts during a half-year stay at the international space station. Medical Mycol 2016; 54:232-9; PMID:26773135; http://dx.doi.org/ 10.1093/mmy/myv121 [DOI] [PubMed] [Google Scholar]
- [21].Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol 2006; 44:2933-41; PMID:16891514; http://dx.doi.org/ 10.1128/JCM.00785-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang L, Clavaud C, Bar-Hen A, Cui M, Gao J, Liu Y, Liu C, Shibagaki N, Guéniche A, Jourdain R, et al.. Characterization of the major bacterial-fungal populations colonizing dandruff scalps in Shanghai, China, shows microbial disequilibrium. Exp Dermatol 2015; 24:398-400; PMID:25739873; http://dx.doi.org/ 10.1111/exd.12684 [DOI] [PubMed] [Google Scholar]
- [23].Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Becker J, et al.. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013; 498:367-70; PMID:23698366; http://dx.doi.org/ 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Leung MH, Chan KC, Lee PK. Skin fungal community and its correlation with bacterial community of urban Chinese individuals. Microbiome 2016; 4:46; PMID:27558504; http://dx.doi.org/ 10.1186/s40168-016-0192-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gupta AK, Kohli Y, Summerbell RC, Faergemann J. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Medical Mycol 2001; 39:243-51; PMID:11446527; http://dx.doi.org/ 10.1080/mmy.39.3.243.251 [DOI] [PubMed] [Google Scholar]
- [26].Gupta AK, Kohli Y. Prevalence of Malassezia species on various body sites in clinically healthy subjects representing different age groups. Medical Mycol 2004; 42:35-42; PMID:14982112; http://dx.doi.org/ 10.1080/13693780310001610056 [DOI] [PubMed] [Google Scholar]
- [27].Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, Saunders CW, Reeder NL, Reilman RA, Scheynius A, et al.. Genus-Wide comparative genomics of Malassezia Delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genetics 2015; 11:e1005614; PMID:26539826; http://dx.doi.org/ 10.1371/journal.pgen.1005614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Michael-Jubeli R, Bleton J, Baillet-Guffroy A. High-temperature gas chromatography-mass spectrometry for skin surface lipids profiling. J Lipid Res 2011; 52:143-51; PMID:20952798; http://dx.doi.org/ 10.1194/jlr.D008094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Warren R, Wertz PW, Kirkbride T, Brunner M, Gross MC. Comparative analysis of skin surface lipids of the labia majora, inner thigh, and forearm. Skin Pharmacol Physiol 2011; 24:294-9; PMID:21734438; http://dx.doi.org/ 10.1159/000328731 [DOI] [PubMed] [Google Scholar]
- [30].Faergemann J, Fredriksson T. Age incidence of Pityrosporum orbiculare on human skin. Acta Dermato-Venereologica 1980; 60:531-3; PMID:6162342 [PubMed] [Google Scholar]
- [31].Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WL, Segre JA, Kong HH. Diverse human skin fungal communities in children converge in adulthood. J Invest Dermatol 2016(Epub ahead of print); PMID:27476723; https://dx.doi.org/448169 10.1016/j.jid.2016.05.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pochi PE, Strauss JS, Downing DT. Age-related changes in sebaceous gland activity. J Invest Dermatol 1979; 73:108-11; PMID:448169; http://dx.doi.org/ 10.1111/1523-1747.ep12532792 [DOI] [PubMed] [Google Scholar]
- [33].Kyriakis KP, Terzoudi S, Palamaras I, Pagana G, Michailides C, Emmanuelides S. Pityriasis versicolor prevalence by age and gender. Mycoses 2006; 49:517-8; PMID:17022772; http://dx.doi.org/ 10.1111/j.1439-0507.2006.01285.x [DOI] [PubMed] [Google Scholar]
- [34].Yamamoto A, Serizawa S, Ito M, Sato Y. Effect of aging on sebaceous gland activity and on the fatty acid composition of wax esters. J Invest Dermatol 1987; 89:507-12; PMID:3668294; http://dx.doi.org/ 10.1111/1523-1747.ep12461009 [DOI] [PubMed] [Google Scholar]
- [35].Akaza N, Akamatsu H, Sasaki Y, Takeoka S, Kishi M, Mizutani H, Sano A, Hirokawa K, Nakata S, Matsunaga K. Cutaneous Malassezia microbiota in atopic dermatitis patients differ by gender and body part. Dermatology 2010; 221:253-60; PMID:20924162; http://dx.doi.org/ 10.1159/000320234 [DOI] [PubMed] [Google Scholar]
- [36].Prohic A, Simic D, Sadikovic TJ, Krupalija-Fazlic M. Distribution of Malassezia species on healthy human skin in Bosnia and Herzegovina: correlation with body part, age and gender. Iran J Microbiol 2014; 6:253-62; PMID:25802709 [PMC free article] [PubMed] [Google Scholar]
- [37].Crespo Erchiga V, Ojeda Martos A, Vera Casano A, Crespo Erchiga A, Sanchez Fajardo F. Malassezia globosa as the causative agent of pityriasis versicolor. Br J Dermatol 2000; 143:799-803; PMID:11069459; http://dx.doi.org/ 10.1046/j.1365-2133.2000.03779.x [DOI] [PubMed] [Google Scholar]
- [38].Gupta AK, Kohli Y, Faergemann J, Summerbell RC. Epidemiology of Malassezia yeasts associated with pityriasis versicolor in Ontario, Canada. Medical Mycol 2001; 39:199-206; PMID:11346269; http://dx.doi.org/ 10.1080/mmy.39.2.199.206 [DOI] [PubMed] [Google Scholar]
- [39].Dutta S, Bajaj AK, Basu S, Dikshit A. Pityriasis versicolor: socioeconomic and clinico-mycologic study in India. Int J Dermatol 2002; 41:823-4; PMID:12453015; http://dx.doi.org/ 10.1046/j.1365-4362.2002.01645.x [DOI] [PubMed] [Google Scholar]
- [40].Nakabayashi A, Sei Y, Guillot J. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Medical Mycol 2000; 38:337-41; PMID:11092380; http://dx.doi.org/ 10.1080/mmy.38.5.337.341 [DOI] [PubMed] [Google Scholar]
- [41].Mayser P, Schafer U, Kramer HJ, Irlinger B, Steglich W. Pityriacitrin – an ultraviolet-absorbing indole alkaloid from the yeast Malassezia furfur. Arch Dermatol Res 2002; 294:131-4; PMID:12029500; http://dx.doi.org/ 10.1007/s00403-002-0294-2 [DOI] [PubMed] [Google Scholar]
- [42].Machowinski A, Kramer HJ, Hort W, Mayser P. Pityriacitrin–a potent UV filter produced by Malassezia furfur and its effect on human skin microflora. Mycoses 2006; 49:388-92; PMID:16922790; http://dx.doi.org/ 10.1111/j.1439-0507.2006.01265.x [DOI] [PubMed] [Google Scholar]
- [43].Kramer HJ, Podobinska M, Bartsch A, Battmann A, Thoma W, Bernd A, Kummer W, Irlinger B, Steglich W, Mayser P. Malassezin, a novel agonist of the aryl hydrocarbon receptor from the yeast Malassezia furfur, induces apoptosis in primary human melanocytes. Chem Bio Chem 2005; 6:860-5; PMID:15812864; http://dx.doi.org/ 10.1002/cbic.200400247 [DOI] [PubMed] [Google Scholar]
- [44].Cramer C, Link E, Horster M, Koletzko S, Bauer CP, Berdel D, von Berg A, Lehmann I, Herbarth O, Borte M, et al.. Elder siblings enhance the effect of filaggrin mutations on childhood eczema: results from the 2 birth cohort studies LISAplus and GINIplus. J Allergy Clin Immunol 2010; 125:1254-60 e5; PMID:20513523; http://dx.doi.org/ 10.1016/j.jaci.2010.03.036 [DOI] [PubMed] [Google Scholar]
- [45].von Mutius E. The environmental predictors of allergic disease. J Allergy Clin Immunol 2000; 105:9-19; PMID:10629447; http://dx.doi.org/ 10.1016/S0091-6749(00)90171-4 [DOI] [PubMed] [Google Scholar]
- [46].Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, et al.. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet 2006; 38:441-6; PMID:16550169; http://dx.doi.org/ 10.1038/ng1767 [DOI] [PubMed] [Google Scholar]
- [47].Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Murray PR, et al.. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012; 22:850-9; PMID:22310478; http://dx.doi.org/ 10.1101/gr.131029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Broberg A, Faergemann J, Johansson S, Johansson SG, Strannegard IL, Svejgaard E. Pityrosporum ovale and atopic dermatitis in children and young adults. Acta Dermato-Venereologica 1992; 72:187-92; PMID:1357856 [PubMed] [Google Scholar]
- [49].Sandstrom Falk MH, Tengvall Linder M, Johansson C, Bartosik J, Back O, Sarnhult T, Wahlgren CF, Scheynius A, Faergemann J. The prevalence of Malassezia yeasts in patients with atopic dermatitis, seborrhoeic dermatitis and healthy controls. Acta Derm Venereol 2005; 85:17-23; PMID:15848985; http://dx.doi.org/ 10.1080/00015550410022276 [DOI] [PubMed] [Google Scholar]
- [50].Back O, Scheynius A, Johansson SG. Ketoconazole in atopic dermatitis: therapeutic response is correlated with decrease in serum IgE. Arch Dermatol Res 1995; 287:448-51; PMID:7625855; http://dx.doi.org/ 10.1007/BF00373427 [DOI] [PubMed] [Google Scholar]
- [51].Savolainen J, Lintu P, Kosonen J, Kortekangas-Savolainen O, Viander M, Pene J, Kalimo K, Terho EO, Bousquet J. Pityrosporum and Candida specific and non-specific humoral, cellular and cytokine responses in atopic dermatitis patients. Clin Exp Allergy 2001; 31:125-34; PMID:11167960; http://dx.doi.org/ 10.1111/j.1365-2222.2001.00958.x [DOI] [PubMed] [Google Scholar]
- [52].Kim TY, Jang IG, Park YM, Kim HO, Kim CW. Head and neck dermatitis: the role of Malassezia furfur, topical steroid use and environmental factors in its causation. Clin Exp Dermatol 1999; 24:226-31; PMID:10354185; http://dx.doi.org/ 10.1046/j.1365-2230.1999.00460.x [DOI] [PubMed] [Google Scholar]
- [53].Andersson A, Rasool O, Schmidt M, Kodzius R, Fluckiger S, Zargari A, Crameri R, Scheynius A. Cloning, expression and characterization of two new IgE-binding proteins from the yeast Malassezia sympodialis with sequence similarities to heat shock proteins and manganese superoxide dismutase. Eur J Biochem 2004; 271:1885-94; PMID:15128298; http://dx.doi.org/ 10.1111/j.1432-1033.2004.04098.x [DOI] [PubMed] [Google Scholar]
- [54].Balaji H, Heratizadeh A, Wichmann K, Niebuhr M, Crameri R, Scheynius A, Werfel T. Malassezia sympodialis thioredoxin-specific T cells are highly cross-reactive to human thioredoxin in atopic dermatitis. J Allergy Clin Immunol 2011; 128:92-9 e4; PMID:21489611; http://dx.doi.org/ 10.1016/j.jaci.2011.02.043 [DOI] [PubMed] [Google Scholar]
- [55].Zargari A, Midgley G, Back O, Johansson SG, Scheynius A. IgE-reactivity to seven Malassezia species. Allergy 2003; 58:306-11; PMID:12708978; http://dx.doi.org/ 10.1034/j.1398-9995.2003.00082.x [DOI] [PubMed] [Google Scholar]
- [56].Shuster S. The aetiology of dandruff and the mode of action of therapeutic agents. Br J Dermatol 1984; 111:235-42; PMID:6235835; http://dx.doi.org/ 10.1111/j.1365-2133.1984.tb04050.x [DOI] [PubMed] [Google Scholar]
- [57].Pierard GE, Arrese JE, Pierard-Franchimont C, DED P. Prolonged effects of antidandruff shampoos - time to recurrence of Malassezia ovalis colonization of skin. Int J Cosmet Sci 1997; 19:111-7; PMID:18507638; http://dx.doi.org/ 10.1111/j.1467-2494.1997.tb00174.x [DOI] [PubMed] [Google Scholar]
- [58].Smith KJ, Skelton HG, Yeager J, Ledsky R, McCarthy W, Baxter D, Turiansky GW, Wagner KF, Turianski G. Cutaneous findings in HIV-1-positive patients: a 42-month prospective study. Military medical consortium for the advancement of retroviral research (MMCARR). J Am Acad Dermatol 1994; 31:746-54; PMID:7929920; http://dx.doi.org/ 10.1016/S0190-9622(94)70236-5 [DOI] [PubMed] [Google Scholar]
- [59].Berger RS, Stoner MF, Hobbs ER, Hayes TJ, Boswell RN. Cutaneous manifestations of early human immunodeficiency virus exposure. J Am Acad Dermatol 1988; 19:298-303; PMID:2971687; http://dx.doi.org/ 10.1016/S0190-9622(88)70175-9 [DOI] [PubMed] [Google Scholar]
- [60].Goodman DS, Teplitz ED, Wishner A, Klein RS, Burk PG, Hershenbaum E. Prevalence of cutaneous disease in patients with acquired immunodeficiency syndrome (AIDS) or AIDS-related complex. J Am Acad Dermatol 1987; 17:210-20; PMID:2957397; http://dx.doi.org/ 10.1016/S0190-9622(87)70193-5 [DOI] [PubMed] [Google Scholar]
- [61].Zhang E, Tanaka T, Tajima M, Tsuboi R, Nishikawa A, Sugita T. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol 2011; 55:625-32; PMID:21699559; http://dx.doi.org/; http://dx.doi.org/ 10.1111/j.1348-0421.2011.00364.x [DOI] [PubMed] [Google Scholar]
- [62].McGinley KJ, Leyden JJ, Marples RR, Kligman AM. Quantitative microbiology of the scalp in non-dandruff, dandruff, and seborrheic dermatitis. J Invest Dermatol 1975; 64:401-5; PMID:237965; http://dx.doi.org/ 10.1111/1523-1747.ep12512335 [DOI] [PubMed] [Google Scholar]
- [63].Ashbee HR, Ingham E, Holland KT, Cunliffe WJ. Cell-mediated immune responses to Malassezia furfur serovars A, B and C in patients with pityriasis versicolor, seborrheic dermatitis and controls. Exp Dermatol 1994; 3:106-12; PMID:7952921; http://dx.doi.org/ 10.1111/j.1600-0625.1994.tb00267.x [DOI] [PubMed] [Google Scholar]
- [64].Prohic A, Ozegovic L. Malassezia species isolated from lesional and non-lesional skin in patients with pityriasis versicolor. Mycoses 2007; 50:58-63; PMID:17302750; http://dx.doi.org/ 10.1111/j.1439-0507.2006.01310.x [DOI] [PubMed] [Google Scholar]
- [65].Tarazooie B, Kordbacheh P, Zaini F, Zomorodian K, Saadat F, Zeraati H, Hallaji Z, Rezaie S. Study of the distribution of Malassezia species in patients with pityriasis versicolor and healthy individuals in Tehran, Iran. BMC Dermatol 2004; 4:5; PMID:15119958; http://dx.doi.org/ 10.1186/1471-5945-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Parry ME, Sharpe GR. Seborrhoeic dermatitis is not caused by an altered immune response to Malassezia yeast. Br J Dermatol 1998; 139:254-63; PMID:9767239; http://dx.doi.org/ 10.1046/j.1365-2133.1998.02362.x [DOI] [PubMed] [Google Scholar]
- [67].Gaitanis G, Magiatis P, Stathopoulou K, Bassukas ID, Alexopoulos EC, Velegraki A, Skaltsounis AL. AhR ligands, malassezin, and indolo[3,2-b]carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J Invest Dermatol 2008; 128:1620-5; PMID:18219281; http://dx.doi.org/ 10.1038/sj.jid.5701252 [DOI] [PubMed] [Google Scholar]
- [68].Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One 2008; 3:e2719; PMID:18648509; http://dx.doi.org/; http://dx.doi.org/ 10.1371/journal.pone.0002719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Alekseyenko AV, Perez-Perez GI, De Souza A, Strober B, Gao Z, Bihan M, Li K, Methé BA, Blaser MJ. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome 2013; 1:31; PMID:24451201; http://dx.doi.org/ 10.1186/2049-2618-1-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Takemoto A, Cho O, Morohoshi Y, Sugita T, Muto M. Molecular characterization of the skin fungal microbiome in patients with psoriasis. J Dermatol 2015; 42:166-70; PMID:25510344; http://dx.doi.org/ 10.1111/1346-8138.12739 [DOI] [PubMed] [Google Scholar]
- [71].Alcais A, Abel L, Casanova JL. Human genetics of infectious diseases: between proof of principle and paradigm. J Clin Invest 2009; 119:2506-14; PMID:19729848; http://dx.doi.org/ 10.1172/JCI38111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Casanova JL, Abel L. Primary immunodeficiencies: a field in its infancy. Science 2007; 317:617-9; PMID:17673650; http://dx.doi.org/; http://dx.doi.org/ 10.1126/science.1142963 [DOI] [PubMed] [Google Scholar]
- [73].Parvaneh N, Casanova JL, Notarangelo LD, Conley ME. Primary immunodeficiencies: a rapidly evolving story. J Allergy Clin Immunol 2013; 131:314-23; PMID:23374262; http://dx.doi.org/ 10.1016/j.jaci.2012.11.051 [DOI] [PubMed] [Google Scholar]
- [74].Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, Lalioti MD, Mullis PE, Antonarakis SE, et al.. Positional cloning of the APECED gene. Nature Genetics 1997; 17:393-8; PMID:9398839; http://dx.doi.org/ 10.1038/ng1297-393 [DOI] [PubMed] [Google Scholar]
- [75].Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, et al.. Human dectin-1 deficiency and mucocutaneous fungal infections. N Eng J Med 2009; 361:1760-7; PMID:19864674; http://dx.doi.org/ 10.1056/NEJMoa0901053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Glocker EO, Hennigs A, Nabavi M, Schaffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, et al.. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Eng J Med 2009; 361:1727-35; PMID:19864672; http://dx.doi.org/; http://dx.doi.org/ 10.1056/NEJMoa0810719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].O'Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med 2015; 66:311-28; PMID:25587654; http://dx.doi.org/ 10.1146/annurev-med-051113-024537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, et al.. STAT3 mutations in the hyper-IgE syndrome. N Eng J Med 2007; 357:1608-19; PMID:17881745; http://dx.doi.org/ 10.1056/NEJMoa073687 [DOI] [PubMed] [Google Scholar]
- [79].Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, et al.. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 2007; 448:1058-62; PMID:17676033; http://dx.doi.org/ 10.1038/nature06096 [DOI] [PubMed] [Google Scholar]
- [80].Tangye SG, Cook MC, Fulcher DA. Insights into the role of STAT3 in human lymphocyte differentiation as revealed by the hyper-IgE syndrome. J Immunol 2009; 182:21-8; PMID:19109129; http://dx.doi.org/ 10.4049/jimmunol.182.1.21 [DOI] [PubMed] [Google Scholar]
- [81].Oh J, Freeman AF, Program NCS, Park M, Sokolic R, Candotti F, Holland SM, Segre JA, Kong HH. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res 2013; 23:2103-14; PMID:24170601; http://dx.doi.org/ 10.1101/gr.159467.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].McNeil MM, Nash SL, Hajjeh RA, Phelan MA, Conn LA, Plikaytis BD, Warnock DW. Trends in mortality due to invasive mycotic diseases in the United States, 1980–1997. Clin Infect Dis 2001; 33:641-7; PMID:11486286; http://dx.doi.org/ 10.1086/322606 [DOI] [PubMed] [Google Scholar]
- [83].Low CY, Rotstein C. Emerging fungal infections in immunocompromised patients. F1000 Med Rep 2011; 3:14; PMID:21876720; http://dx.doi.org/ 10.3410/M3-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Richardson MD. Changing patterns and trends in systemic fungal infections. J Antimicrob Chemother 2005; 56 (Suppl 1):i5-i11; PMID:16120635; http://dx.doi.org/ 10.1093/jac/dki218 [DOI] [PubMed] [Google Scholar]
- [85].Hobson RP. The global epidemiology of invasive Candida infections–is the tide turning? J Hosp Infect 2003; 55:159-68; quiz 233; PMID:14572481; http://dx.doi.org/ 10.1016/j.jhin.2003.08.012 [DOI] [PubMed] [Google Scholar]
- [86].Marr KA, Carter RA, Crippa F, Wald A, Corey L. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin Infect Dis 2002; 34:909-17; PMID:11880955; http://dx.doi.org/ 10.1086/339202 [DOI] [PubMed] [Google Scholar]
- [87].Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 2007; 20:133-63; PMID:17223626; http://dx.doi.org/ 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Guegan S, Lanternier F, Rouzaud C, Dupin N, Lortholary O. Fungal skin and soft tissue infections. Curr Opin Infect Dis 2016; 29:124-30; PMID:26915073; http://dx.doi.org/ 10.1097/QCO.0000000000000252 [DOI] [PubMed] [Google Scholar]
- [89].Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014; 12:87; PMID:25387460; http://dx.doi.org/ 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 2011; 13:47-58; PMID:22179717; http://dx.doi.org/23698366 10.1038/nrg3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, et al.. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013; 498:367-70; PMID:23698366; http://dx.doi.org/ 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Koljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, et al.. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. N Phytologist 2005; 166:1063-8; PMID:15869663; http://dx.doi.org/ 10.1111/j.1469-8137.2005.01376.x [DOI] [PubMed] [Google Scholar]
- [93].Santamaria M, Fosso B, Consiglio A, De Caro G, Grillo G, Licciulli F, Liuni S, Marzano M, Alonso-Alemany D, Valiente G, et al.. Reference databases for taxonomic assignment in metagenomics. Briefings Bioinformatics 2012; 13:682-95; PMID:22786784; http://dx.doi.org/ 10.1093/bib/bbs036 [DOI] [PubMed] [Google Scholar]
- [94].Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007; 35:7188-96; PMID:17947321; http://dx.doi.org/ 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].O'Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R. Fungal community analysis by large-scale sequencing of environmental samples. Applied Environmental Microbiol 2005; 71:5544-50; PMID:16151147; http://dx.doi.org/ 10.1128/AEM.71.9.5544-5550.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hawksworth DL. The Fungal Dimension of Biodiversity - Magnitude, Significance, and Conservation. Mycol Res 1991; 95:641-55; http://dx.doi.org/ 10.1016/S0953-7562(09)80810-1 [DOI] [Google Scholar]
- [97].Fouquier J, Rideout JR, Bolyen E, Chase J, Shiffer A, McDonald D, Knight R, Caporaso JG, Kelley ST. ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses. Microbiome 2016; 4:11; PMID:26905735; http://dx.doi.org/ 10.1186/s40168-016-0153-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinali G, Arthur I, Normand AC, Giraldo A, et al.. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database–the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Medical Mycol 2015; 53:313-37; PMID:25802363; http://dx.doi.org/ 10.1093/mmy/myv008 [DOI] [PubMed] [Google Scholar]
- [99].White JR, Maddox C, White O, Angiuoli SV, Fricke WF. CloVR-ITS: Automated internal transcribed spacer amplicon sequence analysis pipeline for the characterization of fungal microbiota. Microbiome 2013; 1:6; PMID:24451270; http://dx.doi.org/ 10.1186/2049-2618-1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Nocker A, Camper AK. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Applied Environmental Microbiol 2006; 72:1997-2004; PMID:16517648; http://dx.doi.org/ 10.1128/AEM.72.3.1997-2004.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Nocker A, Richter-Heitmann T, Montijn R, Schuren F, Kort R. Discrimination between live and dead cellsin bacterial communities from environmental water samples analyzed by 454 pyrosequencing. Int Microbiol 2010; 13:59-65; PMID:20890840 [DOI] [PubMed] [Google Scholar]
- [102].Rogers GB, Cuthbertson L, Hoffman LR, Wing PA, Pope C, Hooftman DA, Lilley AK, Oliver A, Carroll MP, Bruce KD, et al.. Reducing bias in bacterial community analysis of lower respiratory infections. ISME J 2013; 7:697-706; PMID:23190732; http://dx.doi.org/ 10.1038/ismej.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Baena-Monroy T, Moreno-Maldonado V, Franco-Martinez F, Aldape-Barrios B, Quindos G, Sanchez-Vargas LO. Candida albicans, Staphylococcus aureus and Streptococcus mutans colonization in patients wearing dental prosthesis. Med Oral Patol Oral Cir Bucal 2005; 10 (Suppl 1):E27-39; PMID:15800465 [PubMed] [Google Scholar]
- [104].Gupta N, Haque A, Mukhopadhyay G, Narayan RP, Prasad R. Interactions between bacteria and Candida in the burn wound. Burns 2005; 31:375-8; PMID:15774298; http://dx.doi.org/ 10.1016/j.burns.2004.11.012 [DOI] [PubMed] [Google Scholar]


