ABSTRACT
The mammalian gastrointestinal (GI) tract is a complex organ system with a twist—a significant portion of its composition is a community of microbial symbionts. The microbiota plays an increasingly appreciated role in many clinically-relevant conditions. It is important to understand the details of biofilm development in the GI tract since bacteria in this state not only use biofilms to improve colonization, biofilm bacteria often exhibit high levels of resistance to common, clinically relevant antibacterial drugs. Here we examine the initial colonization of the germ-free murine GI tract by Enterococcus faecalis—one of the first bacterial colonizers of the naïve mammalian gut. We demonstrate strong morphological similarities to our previous in vitro E. faecalis biofilm microcolony architecture using 3 complementary imaging techniques: conventional tissue Gram stain, immunofluorescent imaging (IFM) of constitutive fluorescent protein reporter expression, and low-voltage scanning electron microscopy (LV-SEM). E. faecalis biofilm microcolonies were readily identifiable throughout the entire lower GI tract, from the duodenum to the colon. Notably, biofilm development appeared to occur as discrete microcolonies directly attached to the epithelial surface rather than confluent sheets of cells throughout the GI tract even in the presence of high (>109) fecal bacterial loads. An in vivo competition experiment using a pool of 11 select E. faecalis mutant strains containing sequence-defined transposon insertions showed the potential of this model to identify genetic factors involved in E. faecalis colonization of the murine GI tract.
KEYWORDS: antibiotic resistance, competitive fitness, intestinal microbiota, opportunistic pathogen
Introduction
The mammalian gastrointestinal (GI) tract hosts a large and diverse consortium of microbial symbionts increasingly recognized as important factors in a wide-range of host disease states—including numerous clinically-relevant human conditions. The gut microbe population is generally considered to be largely bacterial and contains 1013–1014 cells of roughly 1,000 different species in adult humans.1 This bacterial colonization was originally thought to largely be a phenomenon of the large bowel; significant bacterial presence in the small intestine was historically considered to be a pathophysiological state.2 More recent research has demonstrated significant microbial colonization throughout the normal mammalian small intestine, though the specific composition — particularly in human hosts — remains poorly characterized.2,3 The ecology of the intestinal tract varies dramatically along its length as the microenvironment changes.1,4 This environmental variability affects both the species heterogeneity and the absolute numbers of bacteria: in the human GI tract the duodenum is home to only a few species (mostly Gram-positive, aerotolerant organisms such as lactic acid bacteria and staphylococci) with bacterial counts of ∼103–104 CFU/ml while the distal colon is home to hundreds of species (largely Gram-negative, obligate anaerobes like Bacteroides) with densities in the 1012–1013 CFU/ml range (Fig. 1).5 How an individual's microbiome is initially seeded and how it develops over time is a fundamental question and an area of active research.6-9
Figure 1.
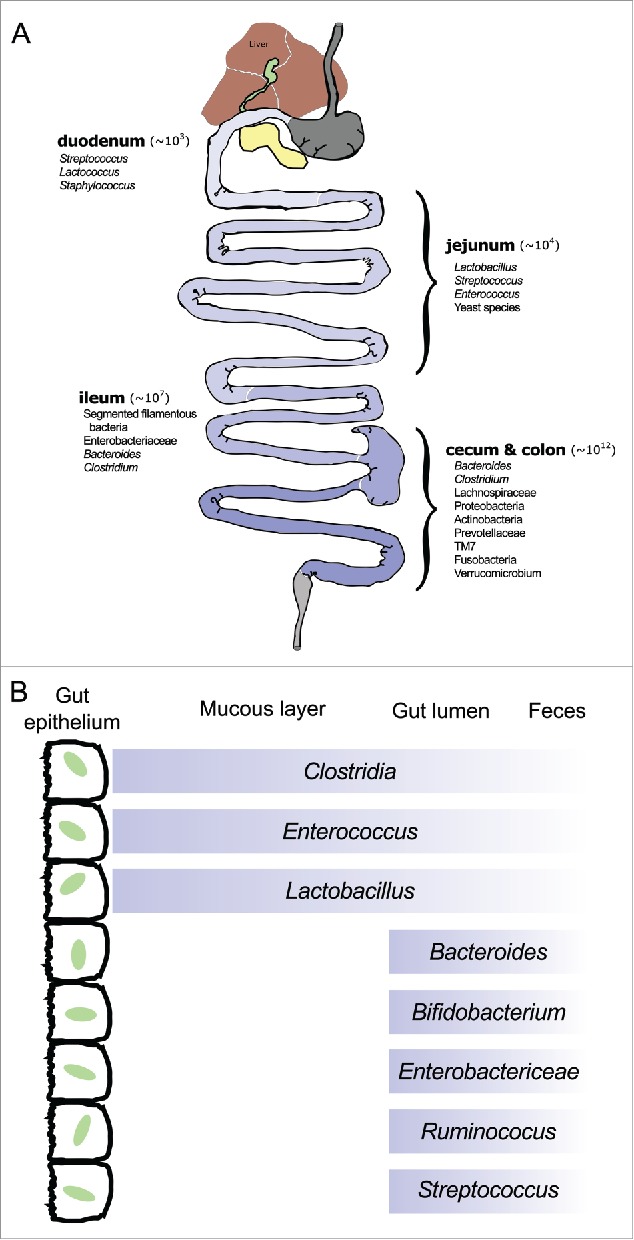
(A) Anatomy of the murine gastrointestinal tract with general mammalian microbial spatial distribution pattern.1,5 Values in parentheses indicate average total bacterial cells per gram of host tissue.1 (B) Cross-section of murine GI tract demonstrating localization of bacterial species relative to the gut epithelium (after Sekirov et al. 2010).
Bacterial biofilms are complex microbial communities attached to or associated with a surface that are encased in a bacterially-derived extracellular matrix. This matrix is structurally and compositionally complex, typically containing proteins, lipids, nucleic acids, and polysaccharides. The extracellular matrix is also a dynamic developmental structure that varies due to a multitude of factors. Clinically, biofilms are of pressing interest due to the dramatic increase in the difficulty of clearing chronic bacterial infections in the biofilm state. It has been estimated that up to 65% of clinical infections involve biofilms,10 and these bacterial populations can be up to 1,000-fold more resistant to common antibiotic therapies than those in the planktonic (free-living) state.11 An important factor driving the intensive study of how and why bacterial communities form biofilms and interact with the host GI tract lies in the necessity of laying the foundations for new methods of antibacterial control. We need supplemental and adjunctive therapies tailored against specific bacteria in the biofilm state to reduce our dependence on classic antibacterial agents and to slow the rise of drug-resistant infections.12
Enterococcus faecalis is a Gram-positive commensal bacterium common in the gastrointestinal tract of animals—from insects to humans—and a clinically-relevant opportunistic pathogen of increasing concern.13 E. faecalis is also one of the first colonizers of the infant GI tract.14,15 Little is known about the mechanisms enterococci employ to facilitate GI tract colonization; this is true in the normal state as well as in dysbiotic hosts.16 Due mostly to the transient, flow-through nature of the GI tract, whether E. faecalis forms biofilms in the mammalian gut has been a matter of some controversy.17
Here we present a germ-free murine gastrointestinal model for investigating initial bacterial colonization of the mammalian gut epithelium and demonstrate its utility to study spatiotemporal relationships in E. faecalis attachment and colonization (Fig. 2a). Lack of a pre-existing normal microbiota in these animals allowed for rapid development of this fitness model. These initial studies provided several sets of results: biofilm morphology and colonization distribution throughout the intestinal tract, absence of a gross inflammatory response to E. faecalis colonization, and the relative fitness of a defined mutant pool in mammalian gut colonization. To our knowledge, these data are the first direct microscopic imaging demonstrating that colonization of the germ-free murine GI tract by E. faecalis involves biofilm microcolony formation. We also report data from experiments comparing the relative fitness of selected mutants in the mouse GI tract, providing a proof of principle for future screens for fitness determinants using a TnSeq approach.
Figure 2.
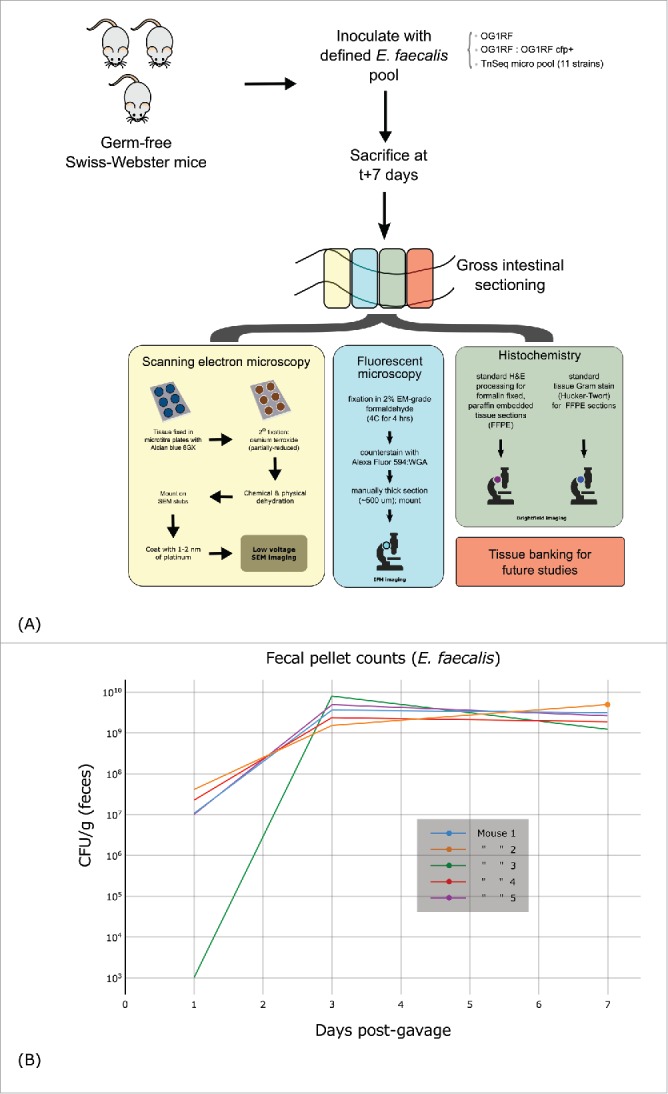
(A) Experimental overview of sample preparation and processing. (B) Time series of E. faecalis recovered from fecal samples of initially germ free mice after inoculation by oral gavage (CFUs are per g of recovered feces).
Results
E. faecalis colonization of the murine GI tract leads to biofilm microcolonies remarkably similar to those observed in vitro
We have published extensively on in vitro E. faecalis biofilm formation on a variety of substrates—from cellulose18 to surgical sutures19 to a range of polymers20-22—and under a wide range of growth conditions (Fig. 3A–D). We have also done work with ex vivo models of tissue attachment by E. faecalis in porcine cardiac valves,23 and in an in vivo model of osteomyelitis using rat orthopedic implant infections.24 In the present study, we found that the overall morphology of in vivo E. faecalis OG1RF25 biofilm microcolonies in our germ-free murine model (Fig. 4) was strikingly similar to that observed in vitro: bacterial microcolonies had a low profile (rarely more than ∼25 cells deep), appear to be composed of approximately 102–104 cells, and were almost completely covered in a sweater-like extracellular matrix. Under optimal in vitro growth conditions like the CDC biofilm reactor system (constant nutrients, moderate flow),26 E. faecalis OG1RF will generally colonize an abiotic surface and grow to near confluence within 24 hours (Fig. 3A). As a monoculture in this germ-free mouse model, however, the enterococcal microcolonies appeared to distribute themselves discretely over the epithelial surface (Fig. 4). Data presented here are representative for mice sacrificed 7 d post-inoculation.
Figure 3.
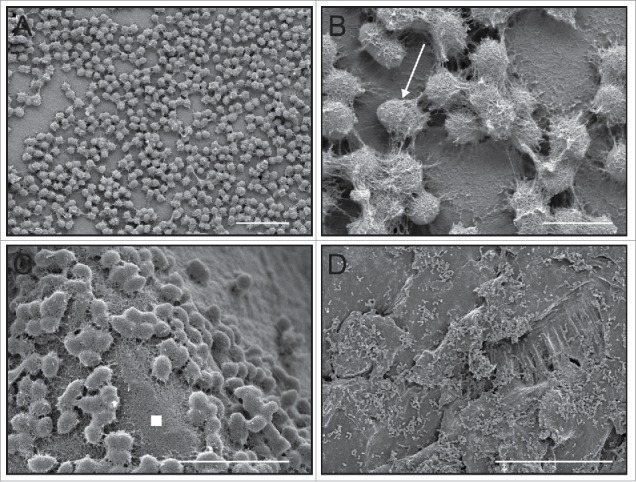
Four previously-unpublished low-voltage scanning electron microscopy (LV-SEM) of E. faecalis microcolony and biofilm formation under a range of on in vitro conditions similar to our previously published work demonstrating both the rapidity in which attachment and extracellular matrix production begins as well as the general structural morphology of the ECM when the matrix is properly preserved using a cationic dye stabilization system. (A) Aclar fluoropolymer coupons,20 24 hrs, CDC biofilm reactor system, bar = 6 µm;51 (B) higher magnification from (A) showing the E. faecalis diplococci surrounded by the sweater-like ECM (arrow), bar = 1 µm; (C) note that the biofilm ECM not only covers cells, but extends into interstitial space between cell clusters (white filled square), regenerated cellulose membrane, 24 hrs, bar = 5 µm;50 (D) polycarbonate coupon, 24 hrs, bar = 50 µm.
Figure 4.
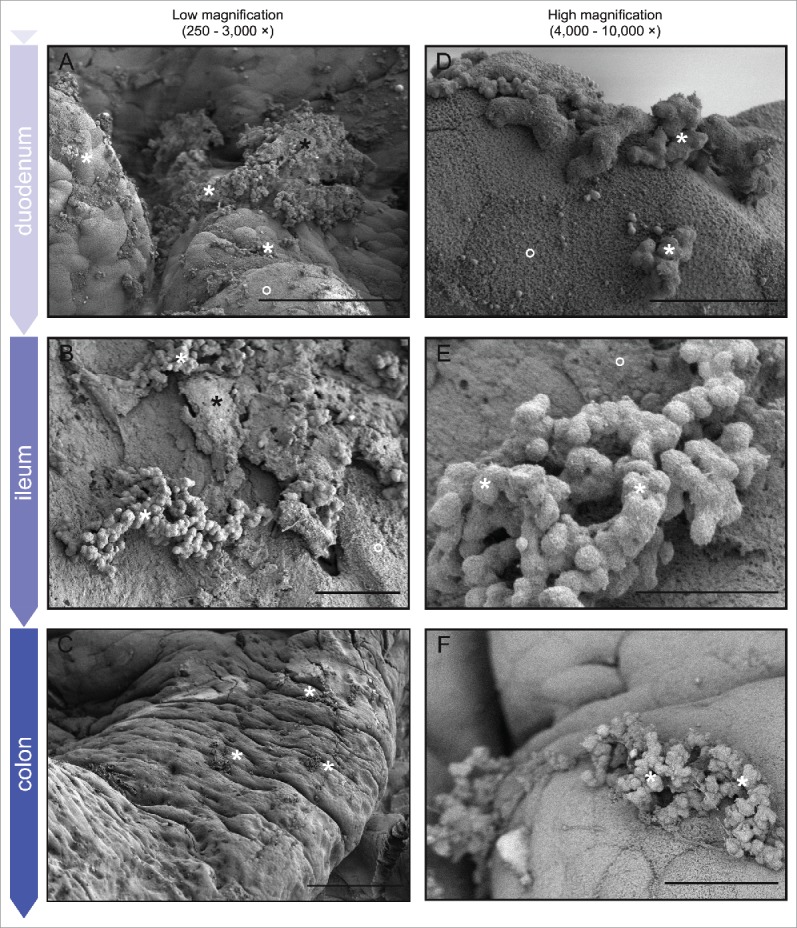
LV-SEM of representative murine GI tract tissue sacrificed 7 d post-gavage with E. faecalis. Rows show samples from each of the major divisions of the lower GI tract from the proximal duodenum through the distal colon; columns show increasing micrograph magnification (left to right). For space reasons low magnification micrographs demonstrating anatomic structure and micrographs of surface colonization in the jejunum and cecum are presented separately in Fig S1. White stars = E. faecalis microcolonies; black stars = retained fecal material; open circles = mucinous material (host).57 Left column: The traditional biofilm ECM structure of E. faecalis microcolonies can be seen throughout the lower GI tract. Note the relatively large distances between many microcolonies even 7 d post-inoculation (mostly clearly in C).Right column: Biofilm microcolonies exhibit wide size variation, from a few tens of cells (D) to several hundred or more (E). Note also the generally bland nature of the underlying host epithelium (D, F)Bars: A = 50 µm; B, C, F = 10 µm, D = 1 mm, E = 200 µm.
E. faecalis colonization is notable for consistent distribution through the GI tract: small and large bowel colonization are grossly similar by low-voltage scanning electron microscopy (LV-SEM)
Along with staphylococci, the upper portions of the mammalian gastrointestinal tract are largely colonized by lactic acid bacteria, including the enterococci (Fig. 1). As seen in Fig. 4, when inoculated into germ-free mice, E. faecalis colonization of the GI epithelium was consistent throughout the small and large bowel. There were no appreciable gross changes in the microcolony morphology or overall distribution in E. faecalis OG1RF by LV-SEM when comparing the upper small bowel (duodenum) with the lower large bowel (colon). Specifically, there were no significant visible differences in microcolony number (compare Fig. 4A to E, K, & N, for example), microcolony distribution (B, F K), or microcolony morphology (C, I L) apparent. While there is little mucous present – typical in newly-colonized germ-free animals without a preexisting microbiota— the luminal surface of the underlying host epithelium appears largely normal (E, L, O).
E. faecalis colonization and biofilm microcolony formation on the gut epithelium does not appear to induce an obvious gross inflammatory response by the host
The surface of the murine gut epithelium was largely undisturbed in the mono-colonized mice; there was a notable lack of gross acute inflammatory changes at both the SEM (Fig. 4) and histological (Fig. 5A) levels. At both microscopic scales there also appears to be a notable lack of cell-based immune responses to the nascent biofilm. Whether this is a specific attribute of E. faecalis colonization or a normal germ-free murine immune response early in bacterial colonization will require further follow-up.
Figure 5.
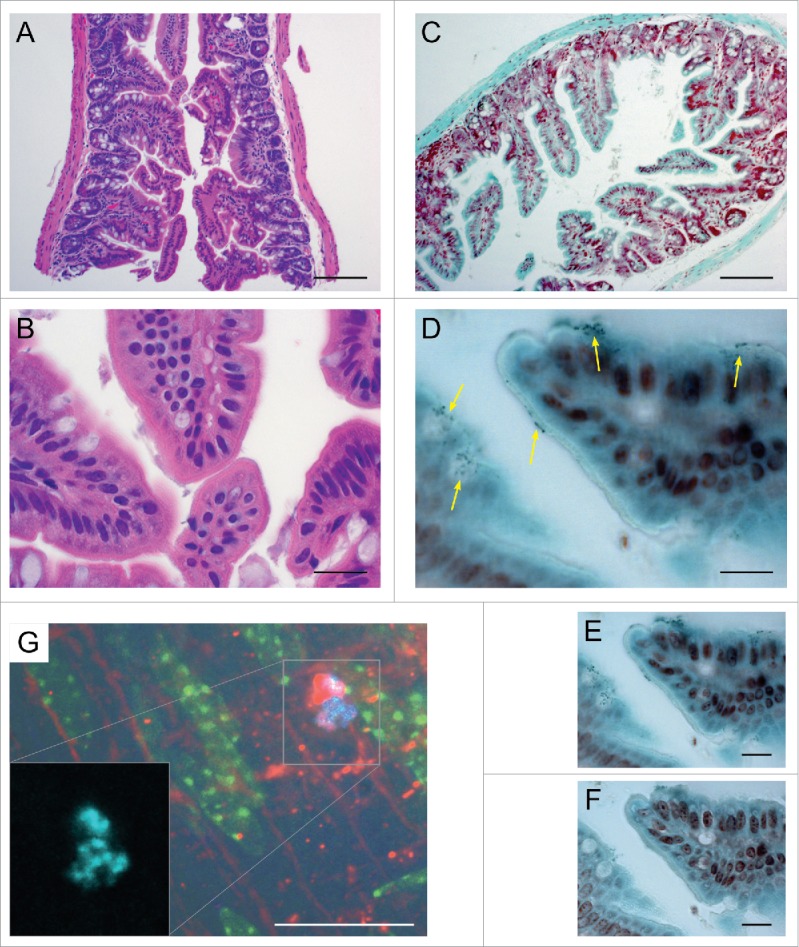
Histological and IFM examination of murine GI tissue from representative sections adjacent to those prepared for SEM.A-F: Histology of the colonized murine terminal ileum. (A). H&E demonstrating normal ileal morphology; note marked lack of obvious inflammatory response. (B). Higher magnification of A. (C). Tissue Gram stain (Hucker-Twort) at low magnification. (D). Higher magnification of C showing numerous small, Gram-positive organisms (yellow arrows showing clusters of small, dark blue cells at the luminal border of epithelial cells. (E). Additional image of same section ∼1 μm deeper in the tissue. (F). Additional image of same section ∼2 μm deeper where the murine epithelium is in focus. G: IFM of the colonized proximal murine colon. (G). Immunofluorescent microscopy of the murine proximal colon colonized by the constitutive CFP-expressing strain OG1RF cfp+. Blue (cyan) = CFP-positive E. faecalis cells; red = Alexa Fluor 594. WGA ; green = autofluorescence from the murine epithelium. The red WGA lectin antibody primarily labels the host epithelial layer as well, but some labeling can be seen around bacterial cells that have begun to produce polysaccharide-rich extracellular matrix. Bars: A, C = 100 µm; B, D - F = 20 µm.
Immunofluorescent and histological microscopy are consistent with LV-SEM data
In mice colonized with a 50:50 inoculum of our OG1RF parental strain and an isogenic strain harboring a constitutively expressing cyan fluorescent protein reporter, thick tissue sections from gross sections adjacent to the SEM and histological samples were imaged to provide additional validation. The gut tissue appeared largely normal for germ-free animals (Fig. 5A); qualitatively there appeared to be a decrease in immunological tissue throughout the gut as is typical for these animals.27 Tissue Gram stains (Hucker-Twort) of adjacent mouse intestine sections demonstrate numerous collections of small, dark blue cells mostly at or near the luminal surface of the epithelium (Fig. 5B–D; terminal ileum shown here as a representative example) consistent with Gram-positive bacteria; no other bacterial morphologies were observed. The microcolony distribution did not appear to be biased to the base of the villi. The cyan fluorescent protein (CFP)-labeled E. faecalis strain successfully colonized the murine colonic epithelium under the same experimental conditions (Fig. 5E).
Competitive fitness of selected mutants in a CDC Biofilm Reactor is different from that observed in the GI tract of matched, parental-strain mice
We compared the relative fitness of 11 previously identified transposon mutants inoculated as a pool into either germ-free mice or a CDC biofilm reactor. Several of these mutants had previously demonstrated altered biofilm forming ability in vitro, or altered virulence in an experimental endocarditis model, while others had no known phenotype (Table 2). The relative abundance of each mutant in the input versus output pools was determined by high-density Illumina-based sequencing of Tn-chromosome junctions in genomic DNA (Materials and Methods). Despite being seeded from the same inocula, the output populations obtained from the in vitro CDC biofilm reactor had markedly different compositions from those recovered from murine fecal pellets (Table 2); this illustrates that the phenotypes most important for biofilm formation in vitro are not directly applicable to fitness in the GI tract. There was no consistent correlation between the behavior of mutants previously reported to have altered virulence in non-GI tract infection models and those conferring fitness effects in the mouse intestine. For example, disruption of the transcription factor ahrC previously showed reduced virulence in experimental endocarditis and a fitness defect in the mouse UTI model.28 Using this mouse GI tract model, we observed decreased abundance of an ahrC mutant while there was no significant loss in biofilm colonization using the CDC biofilm reactor model. In addition, disruption of pyrC did not attenuate virulence in an endocarditis model, but exhibited a strong fitness defect in the current mouse GI tract model.28
Table 2.
E. faecalis OG1RF-derived mutants selected from an existing transposon mutageneis screen for pooling and infection.
| Mouse modelb |
CDC bioreactorc |
||||||
|---|---|---|---|---|---|---|---|
| Gene | Annotated gene function | Virulencea (animal model) | Source | rank | (% counts/input) | rank | (% counts/input) |
| pyrC | dihydroorotase | +++(rabbit endocarditis; murine UTI) | Frank 2013 | 1 | 27.4 | 4 | 146.8 |
| sepF | hypothetical protein | ++(rabbit endocarditis; murine UTI) | Frank 2013 | 2 | 54.3 | 1 | 43.9 |
| argR3/ahrC | arginine repressor | -(rabbit endocarditis) | Frank 2013 | 3 | 58.8 | 6 | 166.2 |
| Inter_442 | 3 nt upstream of arginine repressor (argR2) | no data | n/a | 4 | 79.3 | 9 | 187.5 |
| Inter_29 | intergenic region | no data | n/a | 5 | 89.7 | 10 | 193.7 |
| Inter_28 | intergenic region | no data | n/a | 6 | 99.4 | 8 | 182.7 |
| Inter_442 | 34 nt upstream of arginine repressor (argR2) | no data | n/a | 7 | 108.6 | 11 | 228.0 |
| lyzl6/atlA | cell wall lysis (peptidoglycan hydrolase) | +++(rabbit endocarditis) | Frank 2013 | 8 | 112.4 | 2 | 69.6 |
| eut6 | ethanolamine ammonia-lyase - small subunit | -(mouse GI) | Fox 2009; Zhao 2013 | 9 | 128.0 | 3 | 137.5 |
| argR | arginine repressor | ++(rabbit endocarditis; murine UTI) | Frank 2013 | 10 | 147.1 | 7 | 181.5 |
| ebrA/gntR | GntT family transcriptional regulator | +++(rabbit endocarditis; murine UTI) | Frank 2013 | 11 | 170.4 | 5 | 150.4 |
Note.
Virulence of mutant strain based on published in vivo pathogenesis studies. Virulence attributes similar to parent (+++), slightly attenuated (++), attenuated (−), or in vivo experiments were not performed (no data).
Ranking of mutants from least (1) to most (11) fit using fecal samples of 3 mice obtained from day 7 post-infection using the mouse GI model.
Ranking of mutants from poor (1) to strong (11) biofilm formers after 24h growth using the CDC Biofilm Reactor model system. All rankings were based on the percentage of sequencing counts per mutant compared to input counts of the entire population.
We also observed an interesting increase in the morphological variability of the adherent microcolony extracellular matrix (ECM) seen in the SEM micrographs of the mutant pools that was not visible in those only containing the parental OG1RF strain (Fig. 6). The relationship of this variation to the composition of this mutant pool will be an important topic for future studies. These results provide an important foundation for future TnSeq-based screens for GI tract fitness determinants on a genomic scale (Dale et al., in preparation).
Figure 6.
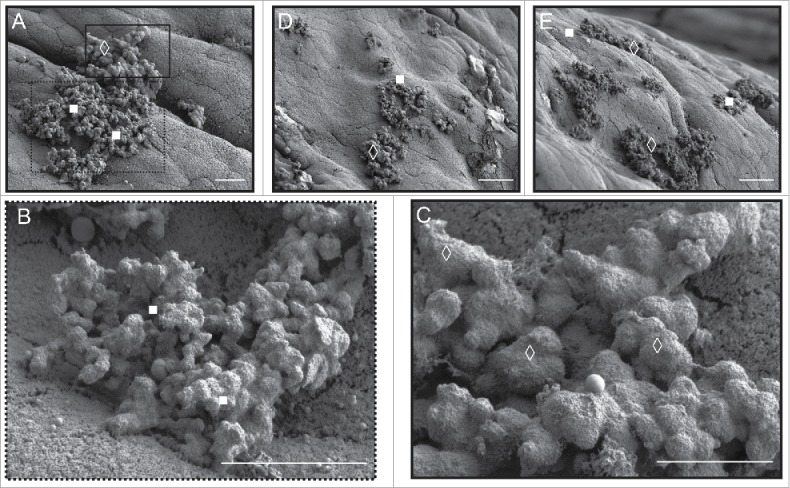
Morphological variation in bacterial extracellular matrix apparently specific to biofilm microcolonies in mice gavaged with a small, TnSeq-derived mutant pool of 11 E. faecalis. (A) E. faecalis microcolonies attached to murine colonic epithelium. Under higher magnification, the foreground cluster (white filled squares; higher magnification in B) appears to have a rougher, less voluminous extracellular matrix compared to the background microcolony ECM (white open diamonds; higher magnification in C). Additional examples of other areas of the colon exhibit the same duality (D, E). Murine colon colonized by the parental OG1RF E. faecalis strain only appears to form the classic ECM form predominantly seen in (C) – compare with Figure 4E. Bars: A - C = 5 µm; D, E = 30 µm.
Discussion
Enterococci are among the first bacterial organisms to colonize the mammalian GI tract and one of the few microbes that establish above, below, and throughout the GI mucous layer.1 To better understand this gut microbiota pioneer, and as a first step in moving our extensive in vitro biofilm characterization work into GI animal models, here we demonstrate that E. faecalis readily colonizes naïve gut epithelium in germ-free Swiss-Webster mice using 3 different microscopic techniques: standard histochemical labeling, immunofluorescent, and low-voltage scanning electron microscopy (LV-SEM). We show that these biofilm microcolonies are strikingly morphologically similar at the scanning electron microscopy level to our published in vitro experiments; a pilot project to demonstrate the ability to screen transposon mutant libraries in this murine infection using high-density sequencing shows promise as well.
Choosing an animal model is often complicated, especially in GI tract systems where the roles the microbiota play must be accounted for in addition to the usual anatomic and physiologic considerations. This is further complicated by colonization variations due to time (developmental changes as the host matures and diets change) and immunology (as the host immune system responds and acclimates to the microbiota). Finally, the GI tract forms a complex protective mucous layer (up to 50 µm thick) that develops as an immunological response to bacterial colonization over the first few weeks of life.
An additional experimental issue comes with the choice of whether to study the organism of interest in isolation as a monoculture or as part of a microbiological community. The former can be easier to decipher while the latter is obviously more representative of the normal state. Finally, in monoculture systems, a question of pre-existing host colonization arises: should one use germ-free uncolonized animals, specific-pathogen free mice, or antibiotic-cleared hosts? We were also confronted with an additional interesting dilemma in this work: as lactic acid bacteria, enterococci are naturally among the very first bacterial species to colonize the neonatal GI tract. Because major factors in post-natal gut development, including production of a functional mucous layer, occur days to weeks after microbial colonization begins, enterococci are exposed to an epithelial surface few other organisms ever experience. This may be a factor in both initial enterococcal colonization as well as the apparent long-term persistence of these organisms. Enterococci are also extraordinarily adaptable organisms with highly flexible metabolisms and the ability to grow under a wide range of environmental conditions—factors that help to explain their recovery throughout the GI tract. In this model system, we have chosen to use germ-free mice for 3 reasons: ease of establishing a new model, examination of initial colonization in a typically early acquired bacterial strain, and as a base for extending this work to include germ-free mice transplanted with a defined human microbiome.
When surveying the entire intestinal tract by LV-SEM, E. faecalis biofilm colonization appears to be similar throughout, with similar distributions and microcolony morphologies from the proximal duodenum to the distal colon. Previous work reporting enterococcal localization within the fully-developed adult human gut has varied widely over time: most recent work has suggested they are present, albeit in small numbers (≤ 1% of the total population), throughout the GI tract.29 Notably, after 7 d post-inoculation, the E. faecalis biofilms remained in the small microcolony state and did not cover large areas of the host epithelium, even in the absence of any other microbial competition. Finally, the bacterial ECM was noticeably altered in a fraction of the biofilm microcolonies present on the epithelial surface inoculated with the mutant pool. We would not suggest using electron microscopy as a primary screening tool since any number of biologically-relevant confounding factors, including the rescue of mutant phenotypes by parent or other mutant strains within the pool may complicate the findings. However, our work does suggest that combining multiple imaging modalities (immunofluorescent microscopy [IFM] and SEM) and examining small numbers of selected mutants expressing different markers may allow for a more definitive understanding of these complex bacterial communities.
Examination of conventional histological sections demonstrates both the overall normality of the epithelium by H&E (hematoxylin and eosin; Fig. 5A) as well as the presence of small, Gram-positive cocci at the luminal surfaces (tissue Gram stain; Fig. 5B–D). The histological results not only provide validation of our SEM observations, they also serve as a proof-of-concept for using traditional formalin-fixed, paraffin-embedded (FFPE) and tissue Gram staining as a screening method for our later planned work. In addition, the success of our CFP-labeled constructs will allow investigation into subsets of interesting mutants from future TnSeq screens via competitions involving wild-type and several labeled mutants.
The output populations from the transposon mutant pool isolates differ significantly between the CDC biofilm reactor and the mouse model: frankly, this is not surprising given the dramatic environmental and immunologic differences between the in vitro and in vivo systems. Importantly, the gut microenvironment complexity is not limited to the longitudinal dimension but has also been reported to have a large latitudinal component as well: many bacteria are restricted to the luminal side of the host mucous layer while a few (some clostridia, lactobacilli, and enterococci) exist both above and below the layer.1,30 The finding that enterococci can colonize the intestinal epithelium and establish semi-permanent colonization below the mucous layer becomes even more interesting in light of 2 more factors: early host colonization and antibiotic resistance.
Enterococci are also remarkably antibiotic resistant—or at least tolerant to normal clinical levels—to a large collection of drugs. Intrinsically, most enterococci are resistant to aminoglycosides, natural and semi-synthetic penicillins, most cephalosporins, clindamycin, trimethoprim-sulfamethoxazole and, for E. faecalis, quinupristin-dalfopristin, as well as tolerant of single-therapy β-lactams and vancomycin.31 Clinically-relevant enterococci are also extraordinarily proficient at acquiring resistance genes via conjugation. From a practical GI colonization standpoint, this can be seen clinically in increasing levels of enterococcal isolation from stool samples in neonates in the intensive care unit of hospitals (who are frequently given one or more antibiotics),32 as well as hospitalized adults on intravenous antibiotics.33 This model system can readily be adapted to experiments examining effects of antibiotics, either pre- or post-enterococcal colonization, e.g., examining how wild-type or mutant E. faecalis strains in a humanized microbiota respond when antibiotic pressure on the community increases the relative enterococcal fitness.
Why haven't these findings of epithelial attachment and biofilm microcolony formation in E. faecalis colonization been reported before? We think there are 2 likely factors: 1) lack of effective biofilm ECM preservation techniques leading to gross bacterial microcolony loss from the surface; and, 2) advances in SEM technology (softer, low-voltage imaging techniques) allowing for better visualization.34
Historically, conventional SEM sample prep – with multiple rounds of fixation, chemical and physical dehydration, and metal coating – has led to concerns of significant loss of host mucous layers and bacterial biofilm from the epithelial surface and thus an artificially depleted picture of the luminal gut milieu. As we have discussed in our previous E. faecalis biofilm work,18 we have developed a cationic dye-based extracellular matrix stabilization technique using Alcian Blue 8GX that dramatically improved biofilm matrix preservation to the point that it approaches cryo-SEM levels of retention.20 The high degree of morphological similarity between our previous in vitro work and these in vivo studies further bolsters our confidence in this technique. Finally, Alcian Blue has also long been used to preserve acid mucopolysaccharides in the mammalian gut at both the histological35 and EM levels36-38 – a serendipitous coincidence. Recent advances in SEM instrumentation are also particularly important here: gut epithelium – particularly in the small bowel with its highly complex surface structure – is notoriously difficult to image well under the scanning electron beam. The recognition that low-voltage imaging (generally below 5 kV) can lead to significant improvement in resolution and reduced surface charging in biological samples was made 25 y ago;39 we are now in the era of very-low (<1.5 kV) and ultra-low (<500 V) imaging. Combined with electron beam deceleration techniques and improvements in detector technology, these formerly obscured bacterial structures can finally be visualized in situ.
We think that microbiome studies have been overly reliant on examining microbial colonization of the GI tract by using fecal pellet sequencing to the exclusion of other techniques. Sequencing is an invaluable tool – we use it here to provide baseline longitudinal data for E. faecalis levels in the gut lumen – but the technology has inherent limitations, too. For example, fecal population sampling assumes that the bacterial population in the gut lumen is representative of that attached to the epithelial surface—an assumption that has been incompletely investigate, particularly in bacteria like enterococci that appear to persist both above and below the gut mucous layer.1,40 There are also important bacterial physiology and development details that may be lost in fecal sequencing: luminal and feces-associated E. faecalis cells may not be in the same planktonic or biofilm state as those attached to the epithelial surface. Given the grossly different antibiotic resistance profiles found in these 2 states it seems important to determine this before presuming fecal pellet cells are representative of the colonizing population. Finally, spatial information at both the macro (Where in the GI tract did colonization happen?) and micro (Is colonization uniform on the gut surface? What is the relationship between microcolonies?) levels is lost in fecal sequencing – factors that will be increasingly important as we unravel the details of interspecies relationships in the gut microbiome.41
To put this research in its historical context, it is important to note that, alone, this germ-free mouse GI tract model is not especially novel: pioneering work in characterization and the dynamics of the mammalian microbiota was done in the late 1960s and early 1970s, notably by the Costello, Dubos, and Schaedler labs at Rockefeller University and continuing with Savage's work at the Universities of Illinois and Tennessee.4,15,30,40,42,43 Nor did these labs limit themselves to fecal enumeration – within the significant technological limitations of the time they did some careful light and electron microscopy revealing the earliest bacterial spatial and distributional information in the developing murine gut. What is new is the fact that E. faecalis can and does form biofilm microcolonies throughout the GI tract and that the biofilm extracellular matrix produced is remarkably morphologically similar to matrix seen via in vitro assays at the ultrastructural level.
Finally, enterococci have a documented role as an early colonizer in naïve mammalian guts, as well as prominent roles in dysbiotic GI tracts such as those perturbed by antibiotic treatment.44 But in the normal adult gut they appear to be a small (≤1%) constituent of the total microbiota by traditional culture or sequencing surveillance – what role does E. faecalis have in these hosts? As colonizers of the submucosal layer, do they have an outsized role in modulating host immune surveillance? Is their prominence in the jejunal portion of the small bowel simply a function of enterococcal resilience to environmental insults (and thus they outcompete other bacteria) or a more specialized reason? Unusually, E. faecalis has also been shown to increase gut translocation under certain antibiotic stimuli: is this again a function of location near the epithelial surface or is there a more specific mechanism? We are investigating these and similar questions with the correlative multimodal microscopy techniques and new genetic tools presented here. This germ free murine GI model system allows for a wide variety of experimental questions to be asked at the genetic, developmental, and physiologic levels and may help to address some of the disparities in understanding the bacterial microbiome in mammalian hosts at both the basic and translational levels.
Materials and methods
Strains and plasmids
Strains and plasmids are shown in Table 1. The E. faecalis plasmid-free parent strain OG1RF25 and mutant derivatives were used in all in vitro and in vivo experiments.
Cyan fluorescent protein ( CFP) construct
Table 1.
E. faecalis strains used in this study.
| Strain | Description | Source | |
|---|---|---|---|
| OG1RF | Parent strain, Rifr, Fusr | Dunny 1978 | |
| OG1RF cfp+ | OG1RF::p23 cfp | This work | |
| Transposon mutants (all in OG1RF background) | |||
| Strain | Description | Coordinate (Tn insertion site) | Source |
| 12H1 | Intergenic region 28 | 32179 | This work |
| 90A1 | Intergenic region 29 | 35226 | This work |
| 22H9 | argR (OG1RF_10098) | 109994 | This work |
| 74A8 | Intergenic region 442 | 427629 | This work |
| 17M8 | Intergenic region 442 | 427660 | Kristich 2008 |
| 25G5 | lyzl6/atlA (OG1RF_10533) | 559358 | Kristich 2008 |
| 20K19 | argR3/ahrC (OG1RF_10717) | 741838 | Kristich 2008 |
| 30B8 | sepF (OG1RF_10732) | 758083 | Kristich 2008 |
| 29E11 | eutC (OG1RF_11343) | 1406287 | This work |
| 33F7 | pyrC (OG1RF_11429) | 1491339 | This work |
| 26A10 | ebrA/gntR (OG1RF_11518) | 1577332 | This work |
Using a previously described genetic markerless insertion system,45 a codon-optimized CFP sequence recently reported by Bergé et al.46 to be stable in Gram positive cocci was inserted into the chromosome of E. faecalis OG1RF under constitutive expression using the lactococcal P23 promoter system47 creating OG1RF::p23 cfp (OG1RF cfp+). From a series of CFP-expressing transconjugants, mutants were empirically screened by IFM to select those with the best combination of fluorescent intensity, stability, and resistance to photobleaching; candidates were checked for basic metabolic and phenotypic growth to ensure no significant differences from parental strain prior to general experimental use.
Transposon mutant pool generation for TnSeq
To examine the differences between in vitro and in vivo gene expression, a small, defined pool of 11 mutant strains was generated from an existing saturated mariner transposon library.48-50 Equal numbers of cells from each of the 11 strains were combined to make an input pool for TnSeq experiments. Separate aliquots of the same input pool were used to inoculated a CDC Biofilm Reactor (see below) and gavaged into germ-free mice.
In vitro biofilm growth
E. faecalis biofilms were cultivated on polycarbonate coupon surfaces using the CDC Biofilm Reactor system (BioSurface Technologies) largely as we have reported previously.51 Briefly, a sterile bioreactor was setup using 100% MM9YEG, a semi-defined medium based on M9 supplemented with 1% casamino acids, 0.3% yeast extract, 20 mM glucose, 1 mM MgSO4, and 0.1 mM CaCl2 and inoculated with 2.8 × 109 total organisms (∼1.1×108 CFU per 11 defined transposon mutants from Table 2) under gentle agitation without flow to allow for establishment. After 4 hours at 37°C, fresh 10% MM9YEG growth medium began flowing into the reactor at 8 ml min−1 (complete vessel volume replacement approximately once per hour). Growth under flow continued for 20 additional hours (24 hours total), after which biofilm cells were mechanically removed from the coupons using a previously validated vortexing protocol and processed as below.
In vivo mouse model
Germ-free (GF) Swiss-Webster mice were maintained and bred in sterile isolators (Class Biologically Clean, Ltd.) until euthanasia. All experiments were approved by Mayo Clinic IACUC (protocol # A1115) and complied with BSL-2 guidelines from the Centers for Disease Control and Prevention (CDC). GF mice were fed an autoclaved standard diet (Purina LabDiet 5K67) and isolator sterility was confirmed with negative cultures (absence of growth) from swabs of GF isolators and GF fecal pellets in Sabouraud Dextrose Media, Brain-Heart Infusion Media, and Nutrient Broth Media at 37°C for 7 d under aerobic and anaerobic conditions, as well as by conducting PCR assays using universal 16S rRNA primers to screen for bacterial contamination prior to starting experiments.
As summarized in Figure 2b, mice were orally gavaged with ∼5 × 106 bacteria (100 μL of a ∼5 × 107 suspension) and maintained for 7 d prior to sacrifice by CO2 asphyxiation followed by cervical dislocation. Necropsies were done rapidly to reduce any post-mortem bacterial growth. The entire intestinal tract from immediately below the pyloric valve to the terminal colon was removed, divided into rough anatomic assignments: the small bowel was measured without significant distention, and divided to approximate the duodenum, jejunum, and ileum. Each of the 5 gross sections (small bowel plus cecum and colon) were divided at ∼1 cm intervals in sets of 4 samples destined for SEM, histology, IFM, etc. Gross tissue sections were gently rinsed individually and placed in ice-cold potassium phosphate buffered saline (KPBS; 10 mM phosphate, 0.9% sodium chloride). Tissues were re-processed en masse with further gentle washing in cold KPBS, dissection, and preparation for fixation. Fecal counts (per gram of feces) are shown in Figure 2b. A total of 9 animals in 2 non-overlapping experiments were used.
DNA extractions
E. faecalis planktonic and biofilm cell pellets obtained from CDC biofilm reactors were extracted using the DNeasy Blood and Tissue Kit (Qiagen) following the pretreatment protocol for Gram-positive bacteria. Extractions from fecal pellets were performed as described by Yu and Morrison with modifications.52 Briefly, fecal pellets (approximately 50 mg per sample) were lysed for 15 min at 37°C using 250 µl fresh lysis buffer (500 mM NaCl, 50 mM Tris-HCl pH 8, 50 mM EDTA, 30 mg/ml lysozyme, 500 U mutanolysin, 4% SDS). An additional 700 µl lysis buffer was added to the sample and transferred to 0.5 mm ZR BashingBead Lysis Tubes (Zymo Research) and homogenized for 1 min using a FastPrep FP120 (Thermo Savant). Samples were incubated at 70°C for 15 min with gentle agitation every 5 min followed by centrifugation at 4°C for 5 min at 16,000 × g. Supernatant was transferred to a fresh tube, 300 µl fresh lysis buffer was added to the lysis tube and homogenization was repeated for 1 min. Nucleic acids were precipitated from the supernatant using ammonium acetate. RNA was removed by incubating resuspended nucleic acids for 15 min at 37°C using 4 µl of 100 mg/ml DNase-free RNase. Proteins were removed by incubating the sample at 70°C for 10 min using 30 µl Proteinase K (Qiagen DNeasy Blood and Tissue Kit) and 900 µl Fecal DNA Binding Buffer (Zymo Research). Nucleic acids were purified by adding equal volumes of AL Buffer (Qiagen DNeasy Blood and Tissue Kit) and 100% EtOH followed by centrifugation using DNeasy Mini spin columns according to the manufacturer protocol. Samples were further purified using the Zymo-Spin IV-HRC Spin filter (Zymo Research) according to the manufacturer. Extracted DNA underwent PCR amplification of the transposon insertion junctions and was sequenced by the University of Minnesota Genomics Center using the MiSeq 150 platform (Illumina, Inc.) with paired-end reads prior to mapping and quantification for overall fitness calculations.53
Imaging
All image processing (histological, fluorescent, and SEM) was done in accordance with this journal's policies and adhere to the larger Journal of Cell Biology standards originally outlined by Rossner and Yamada54 as well as the standards set by the Microscopy Society of America.55
LV-SEM imaging
Processing for SEM was done largely as we have previously reported.20,56 Briefly, samples were fixed for ∼22 hours in 2% formaldehyde + 2% glutaraldehyde in 135 mM sodium cacodylate buffer system (all Electron Microscopy Sciences [EMS]) with 4% sucrose and 0.15% Alcian blue 8GX (Sigma-Aldrich) to preserve the biofilm extracellular matrix. After extensive washing, secondary fixation in a partially-reduced 1% osmium tetroxide (EMS) solution was done for 75 minutes, followed by further washing and chemical dehydration in a progressive ethanol series [25%→50%→70%→80%→95% (×2) →100% (×3)]. Samples were physically dried in a critical-point CO2-based system (Samdri 780A, Tousimis), mounted on aluminum stubs with conductive carbon adhesive (EMS) and cyanoacrylate glue (3M), and coated with 1–2 nm of platinum in an ion beam sputter coater (VCR, South Bay Technology) prior to imaging. Low-voltage imaging (0.1 – 2.5 kV) was done using a Hitachi S-4700 (paired upper and lower secondary electron (SE) detectors) and a Hitachi SU-8230 (paired upper and lower SE or low-angle backscatter electron [LA-BSE] with SE suppression detectors; beam deceleration for images acquired with beam energies below 0.5 kV). All micrographs were acquired as lossless TIFF images.
SEM micrographs are representative of approximately 690 micrographs comprising samples taken from between 7 and 13 germ-free animals and 51 SEM samples (depending on anatomical location: there are more replicates for the ileum and colon segments, for example) spread over multiple experiments.
IFM labeling
Matched adjacent gut tissue samples were fixed in a 2% EM-grade formaldehyde solution (EMS) with 8% sucrose in a 10 mM KPBS buffer for 4 hours at 4°C. After rinsing, thick sections (∼500 μm) were manually prepared and counterstained with Alexa Fluor 594: Wheat Germ Agglutinin (WGA; Molecular Probes / Thermo Fisher) prior to mounting on charged glass slides (Superfrost Plus, Thermo Fisher) in Prolong Diamond (Molecular Probes, Inc.). Slides were allowed to cure for 48 hours at 4°C prior to final imaging. Images acquired as a widefield Z-stack with an step size below the sampling limit (100× 1.45 NA objective; Nikon, Inc.) on a Cascade 1K EM-CCD camera (Photometrics, Inc.) using individual CFP/GFP/Texas Red filter sets (Semrock, Inc.) followed by deconvolution with Huygens (Scientific Volume Imaging, Inc.). Image shown is a maximum intensity projection of the deconvolved stack.
Histochemical staining
Matched tissue samples were fixed in 10% normal buffered formalin (Thermo Fisher) containing 8% sucrose for 48 hours at 4°C, rinsed extensively with KPBS, and stored in 70% ethanol at 4°C. Routine paraffin block embedding and staining with H&E or tissue Gram stain (Hucker-Twort, Newcomer Supply) was done by the Comparative Pathology Shared Resource facility at the University of Minnesota. Imaging was done with a Plan-Apochromat 20× (0.8 N.A.) or Plan-Neofluar 100×, (1.3 N.A.) objective on an AxioImager M.1 using an Axiocam MRc 5 camera and AxioVision (ver. 4.7.2) or Zen (ver. 2.1); software (all Zeiss).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Sequencing and expression analysis was done by the University of Minnesota Genomics Center. Histopathological processing was done by the Comparative Pathology Shared Resource (CPSR) facility at the UMN Masonic Cancer Center. In addition, we acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing resources that contributed to the research results reported within this paper.
Funding
Support for this work was provided by several National Institutes of Health (NIH) grants: to G.M.D. (NIAID AI058134 and AI120601) and to R.P. (R01 AR056647 and R01 AI91594). During portions of this work, A.M.T.B. received support from an NIH Medical Scientist Training Grant (NIGMS GM008244), as well as funding through a T32 postdoctoral training grant at the University of Minnesota Department of Pulmonary, Allergy, and Critical Care (NHLBI HL007741). J.L.D was supported by grant T90 DE 0227232 from the National Institute of Dental & Craniofacial Research. Parts of this work were carried out in the Characterization Facility, University of Minnesota, which receives partial support from National Science Foundation (NSF) through the MRSEC program. The Hitachi SU-8320 SEM was provided by a major research instrumentation grant (NSF MRI DMR-1229263).
References
- [1].Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev [Internet] 2010. [cited 2014December17]; 90:859-904. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20664075; PMID:20664075; http://dx.doi.org/ 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- [2].Quigley EMM. Small intestinal bacterial overgrowth: what it is and what it is not. Curr Opin Gastroenterol [Internet] 2014. [cited 2016January4]; 30:141-6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24406476; PMID:24406476; http://dx.doi.org/ 10.1097/MOG.0000000000000040 [DOI] [PubMed] [Google Scholar]
- [3].Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol [Internet] 1996. [cited 2016January7]; 4:430-5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8950812; PMID:8950812; http://dx.doi.org/ 10.1016/0966-842X(96)10057-3 [DOI] [PubMed] [Google Scholar]
- [4].Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med [Internet] 1968. [cited 2015December31]; 127:67-76. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2138434&tool=pmcentrez&rendertype=abstract; PMID:4169441; http://dx.doi.org/ 10.1084/jem.127.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol [Internet] 2013. [cited 2014September15]; 14:660-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23778793; PMID:23778793; http://dx.doi.org/ 10.1038/ni.2611 [DOI] [PubMed] [Google Scholar]
- [6].Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. Biphasic assembly of the murine intestinal microbiota during early development. ISME J [Internet] 2013. [cited 2016April27]; 7:1112-5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3660675&tool=pmcentrez&rendertype=abstract; PMID:23535917; http://dx.doi.org/ 10.1038/ismej.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Blaser M, Bork P, Fraser C, Knight R, Wang J. The microbiome explored: recent insights and future challenges. Nat Rev Microbiol [Internet] 2013. [cited 2016May2]; 11:213-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23377500; PMID:23377500; http://dx.doi.org/ 10.1038/nrmicro2973 [DOI] [PubMed] [Google Scholar]
- [8].Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A [Internet] 2011. [cited 2014July12]; 108(Suppl):4578-85. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3063592&tool=pmcentrez&rendertype=abstract; PMID:20668239; http://dx.doi.org/ 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Human Microbiome Project Consortium A framework for human microbiome research. Nature [Internet] 2012. [cited 2014July10]; 486:215-21. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3377744&tool=pmcentrez&rendertype=abstract; PMID:22699610; http://dx.doi.org/ 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Potera C. Forging a link between biofilms and disease. Science [Internet] 1999. [cited 2015December2]; 283:1837-1839. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10206887; PMID:10206887; http://dx.doi.org/ 10.1126/science.283.5409.1837 [DOI] [PubMed] [Google Scholar]
- [11].Hoyle BD, Costerton JW. Bacterial resistance to antibiotics: the role of biofilms. Prog drug Res Fortschritte der Arzneimittelforschung Progreès des Rech Pharm [Internet] 1991. [cited 2015December16]; 37:91-105. Available from: http://www.ncbi.nlm.nih.gov/pubmed/1763187 [DOI] [PubMed] [Google Scholar]
- [12].Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol [Internet] 2015. [cited 2015December30]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/26714431; PMID:26714431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sava IG, Heikens E, Huebner J. Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect [Internet] 2010. [cited 2015December17]; 16:533-40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20569264; PMID:20569264; http://dx.doi.org/ 10.1111/j.1469-0691.2010.03213.x [DOI] [PubMed] [Google Scholar]
- [14].Pacheco AR, Barile D, Underwood MA, Mills DA. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci [Internet] 2015. [cited 2015November30]; 3:419-45. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4349412&tool=pmcentrez&rendertype=abstract; PMID:25387230; http://dx.doi.org/ 10.1146/annurev-animal-022114-111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dubos R, Schaedler RW, Costello R. Association of germfree mice with bacteria isolated from normal mice. J Exp Med [Internet] 1965. [cited 2015December31]; 122:77-82. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2138033&tool=pmcentrez&rendertype=abstract; PMID:14325475; http://dx.doi.org/ 10.1084/jem.122.1.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lebreton F, Willems RJL, Gilmore MS. Enterococcus diversity, origins in nature, and gut colonization [Internet]. In: Gilmore MS, Clewell DB, Ikeda Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. (Boston, MA: Massachusetts Eye and Ear Infirmary; ) - via NCBI Bookshelf; 2014. Available from: http://www.ncbi.nlm.nih.gov/books/NBK190427/ [PubMed] [Google Scholar]
- [17].Macfarlane S, Bahrami B, Macfarlane GT. Mucosal biofilm communities in the human intestinal tract. Adv Appl Microbiol [Internet] 2011. [cited 2015December22]; 75:111-43. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21807247; PMID:21807247; http://dx.doi.org/ 10.1016/B978-0-12-387046-9.00005-0 [DOI] [PubMed] [Google Scholar]
- [18].Erlandsen SL, Kristich CJ, Dunny GM. Ultrastructure of Enterococcus faecalis biofilms. Biofilms 2004; 1:131-7; http://dx.doi.org/ 10.1017/S1479050504001206 [DOI] [Google Scholar]
- [19].Henry-Stanley MJ, Hess DJ, Barnes AMT, Dunny GM, Wells CL. Bacterial contamination of surgical suture resembles a biofilm. Surg Infect (Larchmt) [Internet] 2010. [cited 2011February15]; 11:433-9. Available from: http://www.liebertonline.com/doi/abs/10.1089/sur.2010.006; PMID:20673144; http://dx.doi.org/ 10.1089/sur.2010.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Barnes AMT, Ballering KS, Leibman RS, Wells CL, Dunny GM. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. MBio [Internet] 2012. [cited 2014October26]; 3:e00193-12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22829679; PMID:22829679; http://dx.doi.org/ 10.1128/mBio.00193-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dale JL, Cagnazzo J, Phan CQ, Barnes AMT, Dunny GM. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob Agents Chemother 2015; 59:4094-105; PMID:25918141; http://dx.doi.org/ 10.1128/AAC.00344-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Frank KL, Barnes AMT, Grindle SM, Manias DA, Schlievert PM, Dunny GM. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect Immun [Internet] 2012. [cited 2012June21]; 80:539-49. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22144481; PMID:22144481; http://dx.doi.org/ 10.1128/IAI.05964-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chuang ON, Schlievert PM, Wells CL, Manias DA, Tripp TJ, Dunny GM. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immun 2009; 77:539-48; PMID:18955479; http://dx.doi.org/ 10.1128/IAI.01034-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Frank KL, Vergidis P, Brinkman CL, Greenwood Quaintance KE, Barnes AMT, Mandrekar JN, Schlievert PM, Dunny GM, Patel R. Evaluation of the Enterococcus faecalis biofilm-associated virulence factors AhrC and Eep in rat foreign body osteomyelitis and in vitro biofilm-associated antimicrobial resistance. PLoS One 2015; 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 1978; 75:3479-83; PMID:98769; http://dx.doi.org/ 10.1073/pnas.75.7.3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Donlan RM, Piede JA, Heyes CD, Sanii L, Murga R, Edmonds P, El-Sayed I, El-Sayed MA. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl Environ Microbiol [Internet] 2004. [cited 2015December29]; 70:4980-8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=492445&tool=pmcentrez&rendertype=abstract; PMID:15294838; http://dx.doi.org/ 10.1128/AEM.70.8.4980-4988.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Umesaki Y, Setoyama H. Structure of the intestinal flora responsible for development of the gut immune system in a rodent model. Microbes Infect [Internet] 2000. [cited 2016January9]; 2:1343-51. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11018451; PMID:11018451; http://dx.doi.org/ 10.1016/S1286-4579(00)01288-0 [DOI] [PubMed] [Google Scholar]
- [28].Frank KL, Guiton PS, Barnes AMT, Manias DA, Chuang-Smith ON, Kohler PL, Spaulding AR, Hultgren SJ, Schlievert PM, Dunny GM. AhrC and Eep are biofilm infection-associated virulence factors in Enterococcus faecalis. Infect Immun [Internet] 2013. [cited 2014June5]; 81:1696-708. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3648002&tool=pmcentrez&rendertype=abstract; PMID:23460519; http://dx.doi.org/ 10.1128/IAI.01210-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol [Internet] 2005. [cited 2015December14]; 54:1093-101. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16192442; PMID:16192442; http://dx.doi.org/ 10.1099/jmm.0.45935-0 [DOI] [PubMed] [Google Scholar]
- [30].Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol [Internet] 1977. [cited 2015December26]; 31:107-33. Available from: http://www.ncbi.nlm.nih.gov/pubmed/334036; PMID:334036; http://dx.doi.org/ 10.1146/annurev.mi.31.100177.000543 [DOI] [PubMed] [Google Scholar]
- [31].Kristich CJ, Rice LB, Arias CA. Enterococcal infection—treatment and antibiotic resistance [Internet] In: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. 2014. page 87-134.Available from: http://www.ncbi.nlm.nih.gov/pubmed/24649502 [Google Scholar]
- [32].Arrieta M-C, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol [Internet] 2014. [cited 2015February7]; 5:427. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4155789&tool=pmcentrez&rendertype=abstract; PMID:25250028; http://dx.doi.org/ 10.3389/fimmu.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, et al.. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest [Internet] 2010. [cited 2015November27]; 120:4332-41. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2993598&tool=pmcentrez&rendertype=abstract; PMID:21099116; http://dx.doi.org/ 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Alhede M, Qvortrup K, Liebrechts R, Høiby N, Givskov M, Bjarnsholt T. Combination of microscopic techniques reveals a comprehensive visual impression of biofilm structure and composition. FEMS Immunol Med Microbiol [Internet] 2012. [cited 2016January6]; 65:335-42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22429654; PMID:22429654; http://dx.doi.org/ 10.1111/j.1574-695X.2012.00956.x [DOI] [PubMed] [Google Scholar]
- [35].Ito S. The enteric surface coat on cat intestinal microvilli. J Cell Biol [Internet] 1965. [cited 2015December19]; 27:475-91. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2106768&tool=pmcentrez&rendertype=abstract; PMID:4223093; http://dx.doi.org/ 10.1083/jcb.27.3.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Behnke O, Zelander T. Preservation of intercellular substances by the cationic dye alcian blue in preparative procedures for electron microscopy. J Ultrastruct Res [Internet] 1970. [cited 2015December18]; 31:424-8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/4196083; PMID:4196083; http://dx.doi.org/ 10.1016/S0022-5320(70)90159-0 [DOI] [PubMed] [Google Scholar]
- [37].Fassel TA, Edmiston CE. Bacterial biofilms: strategies for preparing glycocalyx for electron microscopy. Methods Enzymol [Internet] 1999. [cited 2015December18]; 310:194-203. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10547793; PMID:10547793; http://dx.doi.org/ 10.1016/S0076-6879(99)10017-X [DOI] [PubMed] [Google Scholar]
- [38].Scott JE. Histochemistry of Alcian blue. 3. The molecular biological basis of staining by Alcian blue 8GX and analogous phthalocyanins. Histochemie [Internet] 1972. [cited 2015December18]; 32:191-212. Available from: http://www.ncbi.nlm.nih.gov/pubmed/4117063; PMID:4117063; http://dx.doi.org/ 10.1007/BF00306028 [DOI] [PubMed] [Google Scholar]
- [39].Pawley JB, Erlandsen SL. The case for low voltage high resolution scanning electron microscopy of biological samples. Scanning Microsc [Internet] 1989. [cited 2015December21]; 3:163-78. Available from: http://www.ncbi.nlm.nih.gov/pubmed/2694267 [PubMed] [Google Scholar]
- [40].Dubos R, Schaedler RW, Costello R. The development of the bacterial flora in the gastrointestinal tract of mice. J Exp Med [Internet] 1965. [cited 2015December31]; 122:59-66. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2138024&tool=pmcentrez&rendertype=abstract; PMID:14325473; http://dx.doi.org/ 10.1084/jem.122.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Trosvik P, de Muinck EJ. Ecology of bacteria in the human gastrointestinal tract-identification of keystone and foundation taxa. Microbiome [Internet] 2015. [cited 2015October15]; 3:44. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4601151&tool=pmcentrez&rendertype=abstract; PMID:26455879; http://dx.doi.org/ 10.1186/s40168-015-0107-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dubos R, Schaedler RW, Costello R, Hoet P. Indigenous, normal, and autochthonous flora of the gastrointestinal tract. J Exp Med [Internet] 1965. [cited 2015December31]; 122:67-76. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2138034&tool=pmcentrez&rendertype=abstract; PMID:14325474; http://dx.doi.org/ 10.1084/jem.122.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Savage DC. Associations of indigenous microorganisms with gastrointestinal mucosal epithelia. Am J Clin Nutr [Internet] 1970. [cited 2015December31]; 23:1495-501. Available from: http://www.ncbi.nlm.nih.gov/pubmed/5475367; PMID:5475367 [DOI] [PubMed] [Google Scholar]
- [44].Hendrickx APA, Top J, Bayjanov JR, Kemperman H, Rogers MRC, Paganelli FL, Bonten MJM, Willems RJL. Antibiotic-driven dysbiosis mediates intraluminal agglutination and alternative segregation of enterococcus faecium from the intestinal epithelium. MBio [Internet] 2015. [cited 2015November13]; 6. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4659461&tool=pmcentrez&rendertype=abstract; PMID:26556272; http://dx.doi.org/ 10.1128/mBio.01346-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vesić D, Kristich CJ. A Rex family transcriptional repressor influences H2O2 accumulation by Enterococcus faecalis. J Bacteriol [Internet] 2013. [cited 2016January14]; 195:1815-24. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3624565&tool=pmcentrez&rendertype=abstract; PMID:23417491; http://dx.doi.org/ 10.1128/JB.02135-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bergé MJ, Kamgoué A, Martin B, Polard P, Campo N, Claverys J-P. Midcell recruitment of the DNA uptake and virulence nuclease, EndA, for pneumococcal transformation. PLoS Pathog [Internet] 2013. [cited 2014July29]; 9:e1003596. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3764208&tool=pmcentrez&rendertype=abstract; PMID:24039578; http://dx.doi.org/ 10.1371/journal.ppat.1003596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].van der Vossen JM, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol [Internet] 1987. [cited 2016January14]; 53:2452-7. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=204128&tool=pmcentrez&rendertype=abstract; PMID:2447829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods [Internet] 2009. [cited 2014May23]; 6:767-72. Available from: http://dx.doi.org/10.1038/nmeth.1377; PMID:19767758; http://dx.doi.org/ 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].van Opijnen T, Lazinski DW, Camilli A. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol [Internet] 2015. [cited 2016June21]; 36:1E.3.1-24. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25641100; PMID:25641100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kristich CJ, Nguyen VT, Le T, Barnes AM, Grindle S, Dunny GM. Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl Environ Microbiol [Internet] 2008; 74:3377-86. Available from: http://aem.asm.org/cgi/reprint/74/11/3377; PMID:18408066; http://dx.doi.org/ 10.1128/AEM.02665-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cook L, Chatterjee A, Barnes A, Yarwood J, Hu W-SW-S, Dunny G. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol Microbiol [Internet] 2011. [cited 2012June21]; 81:1499-510. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3187857&tool=pmcentrez&rendertype=abstract; PMID:21843206; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07786.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques [Internet] 2004. [cited 2016January5]; 36:808-12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15152600; PMID:15152600 [DOI] [PubMed] [Google Scholar]
- [53].van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol [Internet] 2013. [cited 2016June21]; 11:435-42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23712350; PMID:23712350; http://dx.doi.org/ 10.1038/nrmicro3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Rossner M, Yamada KM. What's in a picture? The temptation of image manipulation. J Cell Biol [Internet] 2004. [cited 2012March12]; 166:11-5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2172141&tool=pmcentrez&rendertype=abstract; PMID:15240566; http://dx.doi.org/ 10.1083/jcb.200406019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mackenzie J., Burke MG, Carvalho T, Eades A. Ethics and digital imaging. Micros Today [Internet] 2006; 14:40-1. Available from: http://www.microscopy-today.com/jsp/print_archive/print_archive.jsf# [Google Scholar]
- [56].Erlandsen SL, Kristich CJ, Dunny GM, Wells CL. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J Histochem Cytochem [Internet] 2004. [cited 2012March15]; 52:1427-35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15505337; PMID:15505337; http://dx.doi.org/ 10.1369/jhc.4A6428.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Macfarlane S, Woodmansey EJ, Macfarlane GT. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl Environ Microbiol [Internet] 2005. [cited 2016May23]; 71:7483-92. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1287682&tool=pmcentrez&rendertype=abstract; PMID:16269790; http://dx.doi.org/ 10.1128/AEM.71.11.7483-7492.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


