ABSTRACT
Bacteria possess numerous peptide transporters for importing peptides as nutrients. However, these peptide transporters are now consistently reported to play a role in the virulence of various bacterial pathogens. Their ability to transport peptides has implications in antibacterial therapy as well. Therefore, it would be instrumental to have complete knowledge about the role of peptide transporters in mediating this cross connection between metabolism and pathogenesis. Studies on various peptide transporters in bacterial pathogens have improved our understanding of this field. In this review, we have given an overview of the functioning of bacterial peptide transporters and their contribution in virulence of major bacterial pathogens.
KEYWORDS: ABC transporters, antimicrobial therapy, bacterial pathogens, metabolism, peptide transporters, PTR transporters, virulence
Potential of peptide transporters in antimicrobial therapy
The general strategy of complete elimination of the pathogen for antimicrobial therapy is associated with the drawback of emergence of resistance. An alternate strategy involves targeting of metabolism associated genes. This may slow down the growth but does not affect survival and therefore leave little scope for the development of resistance. Metabolism associated genes of pathogens have been consistently linked with virulence of major pathogens,1-9 such as, role of carbon metabolism related pathways like glycolysis10 and TCA cycle11 in virulence of Salmonella Typhimurium and requirement of metabolism associated pathways for protection of Mycobacterium tuberculosis from host immune system.9 Salmonella is even known to modulate host to exploit the limited supply of nutrients.8,12
Coupling of nutrition with virulence reveals metabolism associated genes that can be targeted for antimicrobial therapy. Peptide transporters represent one such class of genes known to be required for virulence of Gram negative as well as Gram positive pathogens.13-15 Uptake of extracellular peptides is important for the growth of bacteria in peptide rich medium as they are a major source of amino acids, carbon and nitrogen. Peptides can also perform several alternate cellular functions ranging from modulation of stability of membrane proteins to regulation of function of 2 component systems.16 This has led researchers to search for novel transporters in the genomes of pathogens like M. tuberculosis.17
Hijacking of bacterial transporters for delivering antimicrobial agents is an upcoming strategy to circumvent infectious diseases.18 They can also act as potential therapeutic targets for treating infectious diseases or development of vaccines.19 Emergence of antimicrobial peptides as potent therapeutic agents against bacterial pathogens20-22 adds to the importance of these transporters in treating infectious diseases. Clearly, the knowledge of peptide transporters and their connection with virulence is essential in microbiological research today. This review gives an overview of the role of peptide transporters in virulence of major pathogens while describing the distinct features of the major categories of peptide transporters on one platform. It further describes the methodology required to identify and characterize unknown peptide transporters and their regulatory network in the context of pathogenesis.
The distinct mode of function of bacterial peptide transporters
Peptide transporters are located in the plasma membrane of Gram positive bacteria and inner membrane of Gram negative bacteria to acquire specific peptides from the periplasm. There are 2 major categories of peptide transporters known so far depending on the mode of transport. The proton motive force driven transporters (POT or PTR family transporters) capitalize on import of protons for the transport23 whereas the ATP binding cassette containing transporters (ABC transporters) hydrolyse ATP by coordination between multiple proteins24 (Table 1). The ability of these proteins to transport peptides is demonstrated frequently.13,25,26
Table 1.
Difference between PTR and ABC transporters in bacteria.
| PTR peptide transporters | ABC peptide transporters | |
|---|---|---|
| mRNA | Monocistronic | Polycistronic |
| Functional protein | Single protein as monomer | Multiple proteins as subunits |
| Source of energy for transport | Proton import | ATP hydrolysis |
| Common peptide substrates | Dipeptides, Tripeptides | Dipeptides, Tripeptides, Oligopeptides |
Proton coupled peptide transporters
The proton coupled transporters are also known for their pharmacological importance as they aid in uptake of drugs that have steric resemblance with substrate peptides.27 They are also known as PTR (peptide transporter)23 or POT (proton dependent oligopeptide transporter) transporters.28 These transporters provide amino acids and nitrogen in all living organisms except archaea.29 They belong to the major facilitator family (MFS) of transporters as they typically contain 12–18 transmembrane domains which derive their energy of transport from the import of proton. Despite their huge size (600 to 900 amino acids),30 they are commonly reported to transport small peptides like di- and tripeptides (Table 1).13,27 In Escherichia coli, the PTR transporters YdgR and YhiP were also reported to transport tetraalanine, whereas YjdL could transport the amino acid alanine as well as trialanine.26 Therefore, these transporters behave promiscuously toward their substrates, while being conserved in their structure, from bacteria to mammals.27
Most of the knowledge of the functioning of the PTR family of peptide transporters comes from the studies on the mammalian peptide transporters PepT1 and PepT2. While PepT1 is present in the small intestine to obtain broad range of peptides from dietary proteins,31 PepT2 works in kidney 32 to prevent loss of specific peptides in urine.33 The structural studies are extended to bacterial transporters namely PepTso in Shewanella oneidensis34 PepTst in Streptococcus thermophiles35 and GkPOT in Geobacillus kaustophilus.36 The crystal structures of these bacterial peptide transporters showed high resemblance with their mammalian counterparts.27 The transport of peptides by PTR transporters depends on the ionization state of the peptides where the inward electrochemical gradient of protons provides the energy for transport.37,38 The stoichiometry remains as one proton for cationic or neutral dipeptide, 2 protons for anionic dipeptide and 3 protons for anionic tripeptide, where the extra protons neutralize the charge on the peptide.29,39,40 Conserved sequences on the transmembrane domains allow specific binding with the peptides and thereby effective transport, like the ExxERFxYY motif on the transmembrane helix 1 (TM1) facilitates hydrogen bond formation with the carboxy terminus of the dipeptide. Similarly, the presence of tyrosine and tryptophan residues on specific transmembrane domains and FWALF motif in TM7 affect the Vmax and Km of the transport of peptides.27,35,36 As proven by the study on YdgR peptide transporter of E. coli,38 PTR transporters function in monomeric form. Several residues were found to be significant for binding of proton such as arginine, tyrosine and glutamine in TM1, lysine in TM4 and glutamine in TM7 and TM10.27
The procedure of peptide uptake involves major conformational changes in the transporter (Fig. 1). As studied in GkPOT, at first the transporter exists in an outward open conformation, stabilized by a salt bridge between Lys136 on TM4 and Glu413 on TM10. In this state, a proton binds to Glu310 (or Glu300 in PepTst) on TM7. This induces conformational change allowing the peptide in the periplasm to bind to the transporter that attains the peptide bound occluded conformation. A salt bridge is formed between Glu310 on TM7 and Arg43 on TM1 leading to deprotonation of Glu310 and breaking of salt bridge between Lys136 and Glu413. This causes conformational transition to the inward open state, releasing the peptide in the cytoplasm.35,36 The reverse phenomenon of conformational change from inward open state to outward open state remains to be elucidated. Though the substrate specificity is wide for these transporters, they can recognize the size and charge of the peptides and have specific affinity for a particular peptide, for example, PepTst shows higher affinity for hydrophobic peptides than charged peptides.35 There are many orphan transporters in this family, to be characterized yet, whose peptide substrates are not known.
Figure 1.
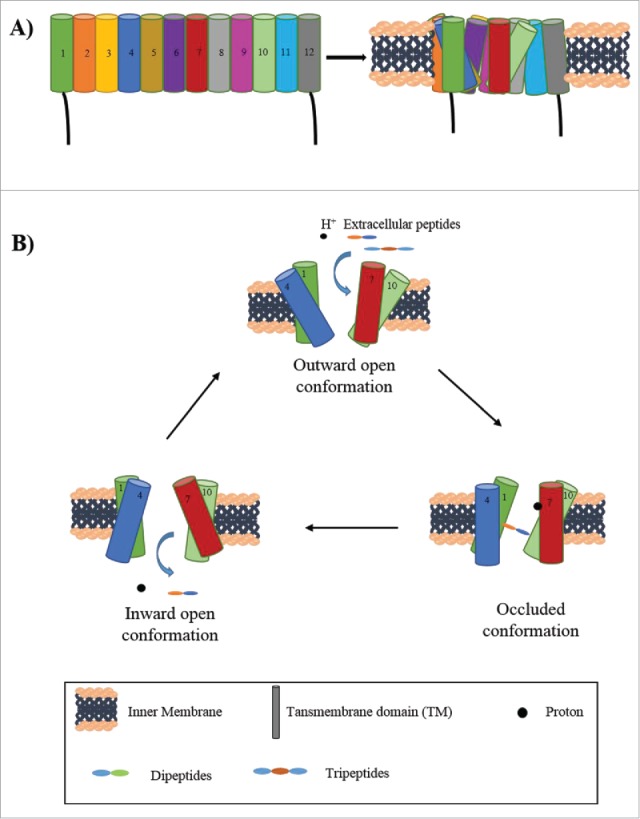
Proton coupled peptide transporters (PTR or POT transporters). A. General structure of PTR transporters. A representation of single PTR transporter showing number of transmembrane domains (TM 1–12) spanning the inner membrane and the final arrangement of these domains into monomeric functional unit. The arrangement of the transmembrane domains is not accurate. For exact structure of PTR transporters refer to Newstead, S. 2015 [27]. B. Mechanism of peptide transport by PTR transporters. The outward open conformation allows for the binding of proton and the peptide. Upon binding of the proton to TM7, the peptide binds to TM 1 and 10, forming the occluded conformation. Finally the inward open state leads to release of peptide and proton in the cytoplasm.
ABC peptide transporters
The ATP-binding cassette containing or ABC peptide transporters are members of the largest transport system superfamilies i.e. ABC-ATPase superfamily. These are widely present among living organisms starting from primitive life forms to higher eukaryotes, such as humans and other mammals. These transporters are multi-subunit protein pumps that couple ATP hydrolysis with the movement of peptides across the inner membrane.41,42 In Gram negative species, the substrate peptides are usually accessible through an extracytoplasmic receptor, called solute-binding protein (SBP), whereas the Gram positive species contain a lipoprotein subunit that extend beyond the extracellular face of the cell membrane.43 These transporters are also involved in peptide export as observed in the case of pheromone peptide secretion by ABC transporter in Enterococcus faecalis, required for conjugation and biofilm formation.44
A typical ABC transporter has 4 functional domains or subunits including 2 nucleotide binding domains (NBD1 and NBD2) and 2 transmembrane domains (TMD1 and TMD2). NBDs and TMDs are held together with the help of a terminal coupling helix of the TMDs. In prokaryotes, these domains are synthesized from polycistronic mRNA and can be present as individual domains or in fused form of NBD-TMDs.45 For example, in E. coli, the TMDs and NBDs are fused as TMD-NBD-TMD-NBD in the exporter protein haemolysin B (HlyB).46 In eukaryotes, the transporter is synthesized from a monocistronic mRNA mostly as a single peptide chain containing all 4 domains, or as homodimeric or heterodimeric halves.41 The TMDs or the integral membrane protein subunits of ABC exporters differ in the number of transmembrane segments.47 Each of the TMDs or membrane-spanning domains (MSDs) typically contains 6–10 membrane-spanning α helices (transmembrane α helices) and together they form a transmembrane pore consisting of 12–20 α helices. This transmembrane pore determines the specificity of the transporter for substrates. Peptides from the surrounding aqueous environment bind to the TMDs when they are in outward-facing conformation and exposing the solute binding site to the exterior. Upon ATP hydrolysis in the cytoplasmic NBD domain, the TMDs change the conformation to inward-facing conformation and the substrate is released in the cytoplasm (Fig. 2).41
Figure 2.
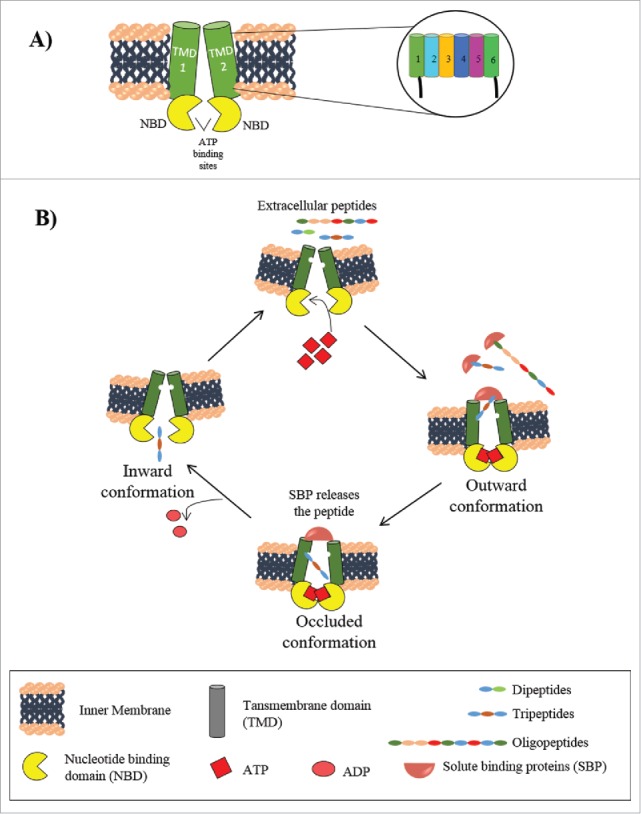
ABC peptide transporters. A. General structure of ABC transporters. A representation of single ABC transporter showing 2 transmembrane domains (TMD 1 and 2) and 2 nucleotide binding domains (NBDs) spanning the inner membrane. 6–10 transmembrane α helices constituting each TMDs are shown in enlarged view, showing a rough arrangement of the transmembrane domains. B. Mechanism of peptide transport by ABC transporters in Gram negative bacteria. The outward conformation allows the binding of 2 ATP molecules on the cytosolic face and SBP bound peptides on the extracellular face that leads to formation of the occluded conformation. ATP hydrolysis changes the conformation to the inward open state releasing the peptide in the cytoplasm.
Link of peptide transporters with virulence
Gram positive pathogens
Of the many ABC and PTR transporters known, only few ABC peptide transporters are known to affect pathogenesis in major Gram positive pathogens. To highlight the connection between nutritional peptide transporters and virulence, we have excluded the examples of bacterial pheromone peptides. Streptococcus sp. represents the group of opportunistic Gram positive pathogens that cause diseases like meningitis, pneumonia and pharyngitis. The significance of peptide transporters in virulence of various species of Streptococcus has been demonstrated in the past. The peptide permeases Ami (or AmiACDEF) and PlpA affect recognition of host cell receptor glycoconjugates GalNAcβ1-4Gal and GalNAcβ1-3Gal by Streptococcus pneumoniae.48 Genome wide Tn-seq screen revealed that S. pneumoniae requires its peptide transport system AmiACDEF for survival in human saliva, an essential strategy for transmission.49,50 Regulation of transcription of the adherence mediating cell surface polypeptide CshA by hexa-hepta peptide permease HppA in S. gordonii shows another example.51 The Gram positive cocci, S. agalactiae, responsible for causing major invasive infection in human newborns, requires 4 peptide permeases for attachment to the host cells and to express its virulence determinants.14 In S. pyogenes, Dpp transporter system controls the expression of cysteine protease SpeB, a virulence factor required for attachment to the host.52
The pathogen Staphylococcus aureus possesses one oligopeptide transporter system Opp and one di- as well as tripeptide permease namely DtpT.53 S. aureus was found to be attenuated in multiple animal models, after mutation in the oppC gene, a permease subunit of Opp peptide transporter assembly.54 In Corynebacterium pseudotuberculosis, which causes caseous lymphadenitis (CLA) in small ruminants, the peptide transporter system Opp is required for adhering to host cells.15 The oligopeptide permease systems, Opp and App, of the gastrointestinal pathogen Clostridium difficile, transport oligopeptides for nutrition. These transporters were found to inversely regulate sporulation, and thereby virulence, of C. difficile in hamster model. Unlike other sporulating Gram positive pathogens, C. difficile genome has not revealed the presence of any pheromone peptides in its genome. It is speculated that oligopeptides taken up as nutrients are responsible for this inverse regulation of virulence of C. difficile.55 In case of the facultative intracellular pathogen Listeria monocytogenes, the gene oppA is required for survival inside macrophages besides providing nutrition.56
Gram negative pathogens
Gram negative bacteria, like E. coli and Salmonella sp. can utilize small peptides for carbon and nitrogen sources,57 imported by transport systems like Dpp, Tpp and Opp. Dpp and Tpp are specific for dipeptides and hydrophobic tripeptides, respectively, whereas Opp can transport oligopeptides.30,58,59 While Dpp and Opp belong to ABC transporters, Tpp belongs to PTR family of transporters. Opp is the major peptide transporter in E. coli and S. Typhimurium, as strains with mutation in Opp were unable to transport peptides, whereas, strains with mutation in Dpp and Tpp, did not show any defect in transport of peptides.57 Surprisingly, there is poor information available on the role of peptide transporters in the virulence of Gram negative pathogens, where most of the knowledge comes from the studies carried out on S. Typhimurium (Table 2).
Table 2.
Peptide transporters in bacterial pathogens.
| Peptide substrates | Bacterial pathogen | Role in virulence | |
|---|---|---|---|
| PTR peptide transporters | |||
| Tpp | Tripeptides | Salmonella Typhimurium | Antimicrobial resistance58 |
| CstA | Dipeptides & Tripeptides | Salmonella Typhimurium | Virulence in Caenorhabditis elegans62 |
| YjiY | Dipeptides & Tripeptides | Salmonella Typhimurium | Induction of class III flagellar genes andvirulence in Balb/c mouse model13 |
| DtpT | Dipeptides & Tripeptides | Staphylococcus aureus | Unknown |
| ABC peptide transporters | |||
| Opp operon | Oligopeptides | Salmonella sp | Unknown |
| Mycobacterium tuberculosis | Glutathione import for methyl glyoxal detoxification, Induction of cytokine release and apoptosis in macrophages68 | ||
| Staphylococcus aureus | Bacteremia54 | ||
| Corynebacterium pseudotuberculosis | Adhesion to host cells15 | ||
| Clostridium difficile | Sporulation and virulence in hamster model55 | ||
| Listeria monocytogenes | Survival inside macrophages56 | ||
| Campylobacter jejuni | Growth inside host66 | ||
| Moraxella catarhhalis | Fitness inside host65 | ||
| Borrelia burgdorferi | Infectivity in mouse model70 | ||
| Dpp operon | Dipeptides | Salmonella Typhimurium | Chemotaxis30 |
| Streptococcus pyogenes | Expression of cysteine protease52 | ||
| Ami operon | Oligopeptides | Streptococcus pneumoniae | Survival inside human saliva and transmission49,50 |
| App operon | Oligopeptides | Clostridium difficile | Sporulation and virulence in hamster model55 |
| Yej operon | Oligopeptides | Salmonella Typhimurium | Antimicrobial resistance63 |
| Sap operon | Oligopeptides | Salmonella Typhimurium | Antimicrobial resistance64 |
| HppA | Hexapeptides | Streptococcus gordonii | Adhesion to host51 |
The enteropathogen Salmonella sp. colonizes the intestine of the host wherein the digestion of dietary proteins by host generates large amount of peptides. In fact, the PTR transporters are reported to be highly induced in the gut of Atlantic salmon.60 This gives the hint that bacteria residing in the gut may also express their counterpart PTR transporters to import peptides. In vitro, in peptide rich medium LB, the virulence associated genes encoded in the pathogenicity island SPI 1, are expressed only during the transition between exponential and stationary growth phases, i.e., late exponential phase.61 The stationary growth phase dependent expression of virulence factors is accompanied by decrease in amino acid biosynthesis.12 However, the concentration of amino acids is maintained in the stationary phase as well as during the later stage of infection inside host cell.12 This suggests parallel import of amino acids or peptides containing the required amino acids. Hence, peptide transporters of Salmonella may provide for the amino acids required for survival inside host and thereby play a role in the establishment of infection. The PTR transporters known in S. Typhimurium include tripeptide transporter (tpp)58 and carbon starvation genes i.e. cstA and yjiY.13 While CstA and YjiY are required for virulence of S. Typhimurium in Caenorhabditis elegans and mouse model respectively,13,62 tripeptide permease (TppB) is shown to be required for resistance against antimicrobial peptides.58 YjiY affects the transcription of flagellar class III genes, which explains the mechanism behind the reduced colonization of yjiY mutant in mouse model.13 However, the role of CstA in colonization of host is not addressed completely. The ABC peptide transporter system Dpp codes for proteins that can act as chemoreceptors and help the bacterium to move toward the source of peptides, showing peptide chemotaxis.30 Other ABC peptide transporter systems YejABEF and SapABCDF confer resistance against antimicrobial peptides.63,64 The probable mechanism behind this kind of resistance is that the antimicrobial peptides are transported inside cytoplasm, away from their targets and exposed to digestion inside the cytoplasm. The YejABEF operon also plays role in the virulence of S. Typhimurium in mouse model.63
The Gram negative diplococcus Moraxella catarhhalis causes otitis media in children and exacerbations of chronic obstructive pulmonary disease in adults. The oligopeptide transporter system Opp was found to be required for nutrition as well as fitness of M. catarhhalis in the host animal.65 Campylobacter jejuni, another Gram negative enteropathogen, requires peptide transporters for transport of peptides as a compensation for restricted growth in the host.66 Therefore, it is safe to say that peptide transporters in Gram negative pathogens hold importance in pathogenesis.
Other bacterial pathogens
Mycobacterium tuberculosis
As Mycobacteria cannot be categorized on the basis of Gram staining, it is discussed separately in this section. Mycobacterium tuberculosis poses biggest challenge in infectious diseases due to the emergence of multi drug resistant (MDR) and extensively drug resistant (XDR) strains causing tuberculosis. As a potential tool for delivery of therapeutic agents, transporters were identified across the genome of M. tuberculosis. Above 10% of a total of 171 transport systems were found to be peptide transporters in M. tuberculosis.17 The Opp system of peptide transport was shown to be required for chronic infection and expression of virulence associated surface lipids.67 The importance of peptide transporters in the pathogenesis of M. tuberculosis also gets emphasized by the finding that ABC peptide transporter system OppABCD is required for glutathione import in M. tuberculosis to regulate the detoxification of methyl glyoxal, cytokine release and apoptosis of macrophages.68
Borrelia burgdorferi
Borrelia burgdorferi is a spirochete that causes Lyme disease in human when bitten by the infested tick, Ixodes ricinus. The pathogen propagates through blood and lymphatics and causes persistent infection like M. tuberculosis. There is limited information available about the virulence factors in B. burgdorferi that help the pathogen to adapt to host environment. Transposon mutagenesis has been used to understand the biology and identify factors that contribute to virulence of B. burgdorferi.69 Such Tn-Seq data has revealed that the oligopeptide permease OppA2 is required for its infectivity in mouse model.70
Regulatory network of peptide transporters in pathogenic bacteria
There is limited information available about the environmental stimuli required for the expression of peptide transporters in pathogens. Commencement of nutrient starvation in stationary phase of growth is reported to induce carbon starvation (Cst) family of PTR peptide transporters, i.e., CstA and YjiY, via cAMP, in E. coli and Salmonella.13,71-73 In E. coli. the exponential phase specific global regulator protein CsrA negatively regulates the translation of CstA by binding to cstA mRNA.74 Nutrient starvation also induces the expression of YehU/YehT, the only 2 component system known to regulate cst genes, in E. coli as well as in Salmonella.72,75 TppB is transcriptionally regulated by leucine and anaerobiosis in Salmonella,76 whereas in E. coli, the tppB homolog ydgR is regulated by EnvZ/OmpR 2 component system,77 showing its involvement in stress response. The stationary phase specific sigma factor RpoS, which is known to regulate several virulence factors required in mammalian host, induces the expression of OppA2 in B. burgdorferi.70
The nutritional status is also required for the expression of ABC peptide transporters.78 For example, the amino acid leucine is known to induce the expression of OppA peptide transporter in E. coli.79 The exponential phase specific small RNA GcvB inhibits the binding of ribosome to the transcripts of ABC transporters OppA and DppA in S. Typhimurium.80 The expression of these peptide transporters may depend on the presence of specific substrate peptides. One example includes induction of dppA-E operon and repressed or unchanged expression of oppA1-F operon during growth of the fastidious organism S. agalactiae in media containing mixture of di- and oligopeptides.14 In case of S. pyogenes, repression of expression of peptide transporters dppA-E occur in free amino acid containing, di- and oligopeptide rich THY media.52
Presence of peptides is also shown to induce the expression of peptide transporters in E. coli.71,72 One of the mechanisms behind this regulation is addressed in an industrially important species Lactococcus lactis, which is commonly studied for peptide transport. L. lactis depends on its peptide transporters DtpT from the PTR family and Opp and Opt from the ABC transporter family, for growth during the manufacturing of various fermented dairy products.81 The PTR transporter DtpT in Lactococcus lactis transports di- and tripeptides and is shown to be expressed constitutively, except for heat shock and acidic stress,82 showing its essentiality in the growth of L. lactis. DtpT also causes repression of proteinase PrtP.83 OptS, on the other hand, in response to the import of peptides from milk, regulates the expression of another peptide transport system Opp.81 The mechanism behind is speculated to be the binding of peptides or amino acids generated from these peptides to certain transcription regulators.81
Clearly, the nutritional status of the bacteria and presence of peptide substrates or amino acids in the surroundings are important factors for the expression of peptide transporters. This gives an idea of the mode of function of peptide transporters in the context of virulence as well. For instance, the transcription of class III flagellar genes in Salmonella is shown to be dependent on the PTR peptide transporter YjiY13 (Table 2). However, the role of peptides in regulating transcription of virulence associated genes is not established. The host certainly provides a plethora of peptide substrates to the pathogen during infection. Once peptides are being imported, they may get degraded to generate amino acids. Also, antimicrobial peptides produced by the host, upon bacterial infection,84 can be digested by the pathogen generating amino acids.85 Besides serving as essential building blocks of cells, these amino acids may also play a more direct role in virulence. One recent example includes the transcriptional regulation of the virulence factor MgtC by the amino acid proline in Salmonella.86 The oligopeptide permeases of Bacillus cereus and B. thuringiensis, regulate the expression of a virulence regulon plcR, by transporting the pentapeptide PapR through Opp transport system.87,88 The first residue of this peptide determines strain specificity,88 suggesting that amino acid composition of the peptide substrates matters for virulence. However, this is an example of pheromone peptides which are different from the nutritional peptides in their origin and function. It is conceivable that peptide transporters affect pathogenesis probably by providing the regulatory amino acids. Therefore, similar phenomena should be displayed by amino acid transporters.
Several amino acid transporters are indeed reported to be related to pathogenesis. For instance, glutamine transporter GlnQ is required for the fitness of S. pneumoniae during infection.89 Mutation in GlnQ of group B streptococci, shows reduced attachment and invasion of the pathogen in vitro and reduced virulence in vivo.90 The transporter of the amino acid arginine, ArgT, is required for survival of S. Typhimurium inside host cells.91 Francisella tularensis depends on amino acid importers for adaptation to intracellular life.92 Transcriptome analysis shows that transporters of amino acids like glutamine and proline get upregulated in the uropathogenic E. coli during urinary tract infection.93 Therefore, there is certainly an overlap in the functions of peptide transporters and amino acid transporters. It is noteworthy that peptides provide a combination of amino acids and therefore may hold more importance during infection. Nevertheless, these transporters could also function completely independent of their peptide substrates to bring about virulence associated phenotypes. Therefore, a complex network of gene regulation connects peptide transporters with pathogenesis (Fig. 3).
Figure 3.
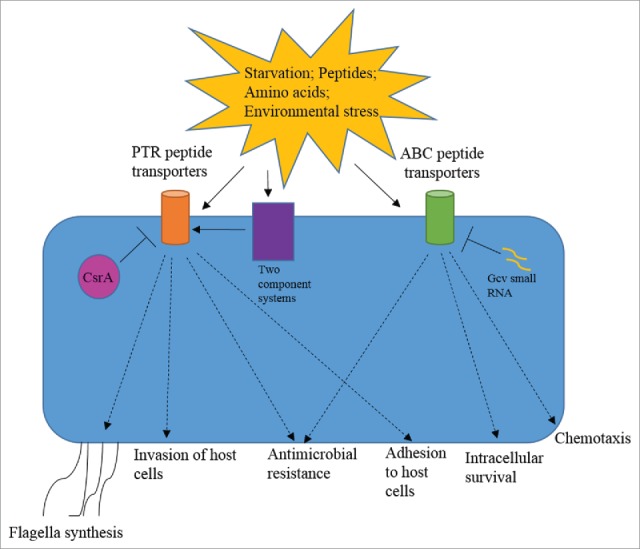
Interconnection of bacterial peptide transporters with pathogenesis. Nutritional starvation or presence of peptide substrates in the surroundings induces the expression of peptide transporters. Two component systems such as YehU/YehT and EnvZ/OmpR positively regulate the expression of few PTR transporters. Examples of negative regulation include inhibition of one of the PTR transporters (CstA) by CsrA and ABC transporters (Opp and Dpp) by small RNA Gcv. Major virulence associated phenotypes affected by peptide transporters include flagella synthesis, chemotaxis, adhesion to host cells, invasion of host cells, intracellular survival and antimicrobial resistance. The dashed arrows show contribution of a particular kind of peptide transporter in specific phenotype.
Characterization of unknown peptide transporters
Keeping the medical importance of peptide transporters in mind, there is strong requirement of identification and characterization of novel peptide transporters in pathogens. Genome of major bacterial pathogens may have several annotated peptide transporters which are yet to be characterized. Peptide transporters can be identified in the bacterial genome based on the similarity in the nucleotide sequence with previously characterized transporters in different bacterial species. Identification of transporters can be done with the help of Transporter Classification Database (TCDB; www.tcdb.org)17 along with prediction of transmembrane domains using programs like HMMTOP,94 TMHMM,95 MEMSAT396 and OCTOPUS.97 For the purpose of exploitation of these peptide transporters as drug delivery systems, it is necessary to have the knowledge of their peptide substrates. Structural studies of transporters in model pathogenic organisms would shed light on this. To establish the function of many unknown or putative peptide transporters, several direct and indirect peptide uptake assays have been designed in the past. Indirect methods include assessment of growth71 or expression of inducible enzymes in physiological auxotrophs in presence of candidate peptides.98 Phenotype microarray has turned out to be one of the most useful high throughput method for preliminary indirect assessment of function of peptide transporters.99 Direct methods like microscopic examination of fluorescently labeled peptides taken by bacterial cells,13 use of radiolabeled substrates68 and chromatography mediated analysis of the peptides present inside the cell100 would describe the function of the transporter more specifically. Use of proton ionophores38 or measuring alkalization as a result of proton translocation would be helpful to study PTR peptide transporters.26 Mammalian peptide transporters have been exogenously expressed in Xenopus oocytes to characterize their function and determine the nature of peptide substrates.60 Similar studies are carried out by expressing bacterial transporters in laboratory bacterial strains like E. coli BL21.38 Thus, with the help of these methodologies it is possible to characterize many unknown peptide transporters from the genome of pathogenic bacteria.
Conclusion and future perspectives
The research related to the link of peptide transport and pathogenesis is in its beginning phase, leaving us with several intriguing questions. For example, what are the environmental cues for the expression of peptide transporters by pathogens during infection? How do these transporters regulate expression of specific virulence associated genes and are these mechanisms conserved in all pathogens? How important is the sequence of peptide substrate for virulence? Interestingly, most of these transporters impact the step of adhesion of pathogen to the host, thereby its colonization, indicating toward a possible common modality of function. Although, the significance of peptide transporters in multiple steps of bacterial pathogenesis including adhesion, motility and biofilm formation, is reported, the mechanisms behind these phenotypes are never completely addressed. Also, there is less focus on identification and characterization of novel transporters of peptides in important pathogens. It is important to identify peptide transporters that hold maximum potential in delivering antimicrobial agents or serving as therapeutic targets. In conclusion, there is a wide avenue left for research regarding peptide transporters and their contribution in bacterial pathogenesis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol 2011; 19:341-8; PMID:21600774; http://dx.doi.org/ 10.1016/j.tim.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Poncet S, Milohanic E, Maze A, Nait Abdallah J, Ake F, Larribe M, Deghmane AE, Taha MK, Dozot M, De Bolle X, et al.. Correlations between carbon metabolism and virulence in bacteria. Contrib Microbiol 2009; 16:88-102; PMID:19494580; http://dx.doi.org/ 10.1159/000219374 [DOI] [PubMed] [Google Scholar]
- [3].Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol 2010; 8:401-12; PMID:20453875; http://dx.doi.org/ 10.1038/nrmicro2351 [DOI] [PubMed] [Google Scholar]
- [4].Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev 2009; 73:233-48; PMID:19487727; http://dx.doi.org/ 10.1128/MMBR.00005-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schoen C, Kischkies L, Elias J, Ampattu BJ. Metabolism and virulence in Neisseria meningitidis. Front Cell Infect Microbiol 2014; 4:114; PMID:25191646; http://dx.doi.org/ 10.3389/fcimb.2014.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ding Y, Liu X, Chen F, Di H, Xu B, Zhou L, Deng X, Wu M, Yang CG, Lan L. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci U S A 2014; 111:E4981-90; PMID:25368190; http://dx.doi.org/ 10.1073/pnas.1411077111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Heroven AK, Dersch P. Coregulation of host-adapted metabolism and virulence by pathogenic yersiniae. Front Cell Infect Microbiol 2014; 4:146; PMID:25368845; http://dx.doi.org/ 10.3389/fcimb.2014.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Steeb B, Claudi B, Burton NA, Tienz P, Schmidt A, Farhan H, Mazé A, Bumann D. Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog 2013; 9:e1003301; PMID:23633950; http://dx.doi.org/ 10.1371/journal.ppat.1003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bordbar A, Mo ML, Nakayasu ES, Schrimpe-Rutledge AC, Kim YM, Metz TO, Jones MB, Frank BC, Smith RD, Peterson SN, et al.. Model-driven multi-omic data analysis elucidates metabolic immunomodulators of macrophage activation. Mol Syst Biol 2012; 8:558; PMID:22735334; http://dx.doi.org/ 10.1038/msb.2012.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bowden SD, Rowley G, Hinton JC, Thompson A. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar typhimurium. Infect Immun 2009; 77:3117-26; PMID:19380470; http://dx.doi.org/ 10.1128/IAI.00093-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tchawa Yimga M, Leatham MP, Allen JH, Laux DC, Conway T, Cohen PS. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect Immun 2006; 74:1130-40; PMID:16428761; http://dx.doi.org/ 10.1128/IAI.74.2.1130-1140.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim YM, Schmidt BJ, Kidwai AS, Jones MB, Deatherage Kaiser BL, Brewer HM, Mitchell HD, Palsson BO, McDermott JE, Heffron F, et al.. Salmonella modulates metabolism during growth under conditions that induce expression of virulence genes. Mol Biosyst 2013; 9:1522-34; PMID:23559334; http://dx.doi.org/ 10.1039/c3mb25598k [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Garai P, Lahiri A, Ghosh D, Chatterjee J, Chakravortty D. Peptide utilizing carbon starvation gene yjiY is required for flagella mediated infection caused by Salmonella. Microbiology 2015; PMID:26497384 [DOI] [PubMed] [Google Scholar]
- [14].Samen U, Gottschalk B, Eikmanns BJ, Reinscheid DJ. Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J Bacteriol 2004; 186:1398-408; PMID:14973032; http://dx.doi.org/ 10.1128/JB.186.5.1398-1408.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Moraes PM, Seyffert N, Silva WM, Castro TL, Silva RF, Lima DD, Hirata R Jr, Silva A, Miyoshi A, Azevedo V. Characterization of the Opp peptide transporter of Corynebacterium pseudotuberculosis and its role in virulence and pathogenicity. BioMed research international 2014; 2014:489782; PMID:24895581; http://dx.doi.org/ 10.1155/2014/489782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Alix E, Blanc-Potard AB. Hydrophobic peptides: novel regulators within bacterial membrane. Molecular microbiology 2009; 72:5-11; PMID:19210615; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06626.x [DOI] [PubMed] [Google Scholar]
- [17].Youm J, Saier MH Jr. Comparative analyses of transport proteins encoded within the genomes of Mycobacterium tuberculosis and Mycobacterium leprae. Biochim Biophys Acta 2012; 1818:776-97; PMID:22179038; http://dx.doi.org/ 10.1016/j.bbamem.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pletzer D, Braun Y, Weingart H. Swarming motility is modulated by expression of the putative xenosiderophore transporter SppR-SppABCD in Pseudomonas aeruginosa PA14. Antonie Van Leeuwenhoek 2016; PMID:26995781 [DOI] [PubMed] [Google Scholar]
- [19].Garmory HS, Titball RW. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 2004; 72:6757-63; PMID:15557595; http://dx.doi.org/ 10.1128/IAI.72.12.6757-6763.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hassan M, Kjos M, Nes IF, Diep DB, Lotfipour F. Natural antimicrobial peptides from bacteria: characteristics and potential applications to fight against antibiotic resistance. J Appl Microbiol 2012; 113:723-36; PMID:22583565; http://dx.doi.org/ 10.1111/j.1365-2672.2012.05338.x [DOI] [PubMed] [Google Scholar]
- [21].Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 2010; 1:440-64; PMID:21178486; http://dx.doi.org/ 10.4161/viru.1.5.12983 [DOI] [PubMed] [Google Scholar]
- [22].Allaker RP, Ian Douglas CW. Non-conventional therapeutics for oral infections. Virulence 2015; 6:196-207; PMID:25668296; http://dx.doi.org/ 10.4161/21505594.2014.983783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Steiner HY, Naider F, Becker JM. The PTR family: a new group of peptide transporters. Molecular microbiology 1995; 16:825-34; PMID:7476181; http://dx.doi.org/ 10.1111/j.1365-2958.1995.tb02310.x [DOI] [PubMed] [Google Scholar]
- [24].Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol 1992; 8:67-113; PMID:1282354; http://dx.doi.org/ 10.1146/annurev.cb.08.110192.000435 [DOI] [PubMed] [Google Scholar]
- [25].Pletzer D, Braun Y, Dubiley S, Lafon C, Kohler T, Page MG, Mourez M, Severinov K, Weingart H. The pseudomonas aeruginosa PA14 ABC transporter NppA1A2BCD is required for uptake of peptidyl nucleoside antibiotics. J Bacteriol 2015; 197:2217-28; PMID:25917903; http://dx.doi.org/ 10.1128/JB.00234-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Prabhala BK, Aduri NG, Jensen JM, Ernst HA, Iram N, Rahman M, Mirza O. New insights into the substrate specificities of proton-coupled oligopeptide transporters from E. coli by a pH sensitive assay. FEBS letters 2014; 588:560-5; PMID:24440353; http://dx.doi.org/ 10.1016/j.febslet.2014.01.004 [DOI] [PubMed] [Google Scholar]
- [27].Newstead S. Molecular insights into proton coupled peptide transport in the PTR family of oligopeptide transporters. Biochimica et Biophysica Acta 2015; 1850:488-99; PMID:24859687; http://dx.doi.org/ 10.1016/j.bbagen.2014.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Paulsen IT, Skurray RA. The POT family of transport proteins. Trends Biochem Sci 1994; 19:404; PMID:7817396; http://dx.doi.org/ 10.1016/0968-0004(94)90087-6 [DOI] [PubMed] [Google Scholar]
- [29].Daniel H, Spanier B, Kottra G, Weitz D. From bacteria to man: archaic proton-dependent peptide transporters at work. Physiology (Bethesda) 2006; 21:93-102; PMID:16565475; http://dx.doi.org/ 10.1152/physiol.00054.2005 [DOI] [PubMed] [Google Scholar]
- [30].Abouhamad WN, Manson M, Gibson MM, Higgins CF. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol Microbiol 1991; 5:1035-47; PMID:1956284; http://dx.doi.org/ 10.1111/j.1365-2958.1991.tb01876.x [DOI] [PubMed] [Google Scholar]
- [31].Knutter I, Hartrodt B, Theis S, Foltz M, Rastetter M, Daniel H, Neubert K, Brandsch M. Analysis of the transport properties of side chain modified dipeptides at the mammalian peptide transporter PEPT1. Eur J Pharm Sci 2004; 21:61-7; PMID:14706812; http://dx.doi.org/ 10.1016/S0928-0987(03)00141-6 [DOI] [PubMed] [Google Scholar]
- [32].Biegel A, Knutter I, Hartrodt B, Gebauer S, Theis S, Luckner P, Kottra G, Rastetter M, Zebisch K, Thondorf I, et al.. The renal type H+/peptide symporter PEPT2: structure-affinity relationships. Amino Acids 2006; 31:137-56; PMID:16868651; http://dx.doi.org/ 10.1007/s00726-006-0331-0 [DOI] [PubMed] [Google Scholar]
- [33].Leibach FH, Ganapathy V. Peptide transporters in the intestine and the kidney. Annu Rev Nutr 1996; 16:99-119; PMID:8839921; http://dx.doi.org/ 10.1146/annurev.nu.16.070196.000531 [DOI] [PubMed] [Google Scholar]
- [34].Newstead S, Drew D, Cameron AD, Postis VL, Xia X, Fowler PW, Ingram JC, Carpenter EP, Sansom MS, McPherson MJ, et al.. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J 2011; 30:417-26; PMID:21131908; http://dx.doi.org/ 10.1038/emboj.2010.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Solcan N, Kwok J, Fowler PW, Cameron AD, Drew D, Iwata S, Newstead S. Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J 2012; 31:3411-21; PMID:22659829; http://dx.doi.org/ 10.1038/emboj.2012.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Doki S, Kato HE, Solcan N, Iwaki M, Koyama M, Hattori M, Iwase N, Tsukazaki T, Sugita Y, Kandori H. Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc Natl Acad Sci U S A 2013; 110:11343-8; PMID:23798427; http://dx.doi.org/ 10.1073/pnas.1301079110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fei YJ, Kanai Y, Nussberger S, Ganapathy V, Leibach FH, Romero MF, Singh SK, Boron WF, Hediger MA. Expression cloning of a mammalian proton-coupled oligopeptide transporter. Nature 1994; 368:563-6; PMID:8139693; http://dx.doi.org/ 10.1038/368563a0 [DOI] [PubMed] [Google Scholar]
- [38].Weitz D, Harder D, Casagrande F, Fotiadis D, Obrdlik P, Kelety B, Daniel H. Functional and structural characterization of a prokaryotic peptide transporter with features similar to mammalian PEPT1. J Biol Chem 2007; 282:2832-9; PMID:17158458; http://dx.doi.org/ 10.1074/jbc.M604866200 [DOI] [PubMed] [Google Scholar]
- [39].Kottra G, Stamfort A, Daniel H. PEPT1 as a paradigm for membrane carriers that mediate electrogenic bidirectional transport of anionic, cationic, and neutral substrates. J Biol Chem 2002; 277:32683-91; PMID:12082113; http://dx.doi.org/ 10.1074/jbc.M204192200 [DOI] [PubMed] [Google Scholar]
- [40].Chen XZ, Zhu T, Smith DE, Hediger MA. Stoichiometry and kinetics of the high-affinity H+-coupled peptide transporter PepT2. J Biol Chem 1999; 274:2773-9; PMID:9915809; http://dx.doi.org/ 10.1074/jbc.274.5.2773 [DOI] [PubMed] [Google Scholar]
- [41].Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep 2015; 7:14; PMID:25750732; http://dx.doi.org/ 10.12703/P7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Young J, Holland IB. ABC transporters: bacterial exporters-revisited five years on. Biochim Biophys Acta 1999; 1461:177-200; PMID:10581355; http://dx.doi.org/ 10.1016/S0005-2736(99)00158-3 [DOI] [PubMed] [Google Scholar]
- [43].Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 2002; 296:1091-8; PMID:12004122; http://dx.doi.org/ 10.1126/science.1071142 [DOI] [PubMed] [Google Scholar]
- [44].Varahan S, Harms N, Gilmore MS, Tomich JM, Hancock LE. An ABC transporter is required for secretion of peptide sex pheromones in Enterococcus faecalis. MBio 2014; 5:e01726-14; PMID:25249282; http://dx.doi.org/ 10.1128/mBio.01726-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Holland IB, Blight MA. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol 1999; 293:381-99; PMID:10529352; http://dx.doi.org/ 10.1006/jmbi.1999.2993 [DOI] [PubMed] [Google Scholar]
- [46].Schmitt L, Benabdelhak H, Blight MA, Holland IB, Stubbs MT. Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: identification of a variable region within ABC helical domains. J Mol Biol 2003; 330:333-42; PMID:12823972; http://dx.doi.org/ 10.1016/S0022-2836(03)00592-8 [DOI] [PubMed] [Google Scholar]
- [47].Khwaja M, Ma Q, Saier MH Jr. Topological analysis of integral membrane constituents of prokaryotic ABC efflux systems. Res Microbiol 2005; 156:270-7; PMID:15748994; http://dx.doi.org/ 10.1016/j.resmic.2004.07.010 [DOI] [PubMed] [Google Scholar]
- [48].Cundell DR, Pearce BJ, Sandros J, Naughton AM, Masure HR. Peptide permeases from Streptococcus pneumoniae affect adherence to eucaryotic cells. Infect Immun 1995; 63:2493-8; PMID:7790061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Verhagen LM, de Jonge MI, Burghout P, Schraa K, Spagnuolo L, Mennens S, Eleveld MJ, van der Gaast-de Jongh CE, Zomer A, Hermans PW, et al.. Genome-wide identification of genes essential for the survival of Streptococcus pneumoniae in human saliva. PloS one 2014; 9:e89541; PMID:24586856; http://dx.doi.org/ 10.1371/journal.pone.0089541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 2002; 45:1389-406; PMID:12207705 [PMC free article] [PubMed] [Google Scholar]
- [51].McNab R, Jenkinson HF. Altered adherence properties of a Streptococcus gordonii hppA (oligopeptide permease) mutant result from transcriptional effects on cshA adhesin gene expression. Microbiology 1998; 144(Pt 1):127-36; PMID:9467905; http://dx.doi.org/ 10.1099/00221287-144-1-127 [DOI] [PubMed] [Google Scholar]
- [52].Podbielski A, Leonard BA. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol Microbiol 1998; 28:1323-34; PMID:9680220; http://dx.doi.org/ 10.1046/j.1365-2958.1998.00898.x [DOI] [PubMed] [Google Scholar]
- [53].Hiron A, Borezee-Durant E, Piard JC, Juillard V. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J Bacteriol 2007; 189:5119-29; PMID:17496096; http://dx.doi.org/ 10.1128/JB.00274-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mei JM, Nourbakhsh F, Ford CW, Holden DW. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol 1997; 26:399-407; PMID:9383163; http://dx.doi.org/ 10.1046/j.1365-2958.1997.5911966.x [DOI] [PubMed] [Google Scholar]
- [55].Edwards AN, Nawrocki KL, McBride SM. Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect Immun 2014; 82:4276-91; PMID:25069979; http://dx.doi.org/ 10.1128/IAI.02323-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Borezee E, Pellegrini E, Berche P. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect Immun 2000; 68:7069-77; PMID:11083832; http://dx.doi.org/ 10.1128/IAI.68.12.7069-7077.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Goodell EW, Higgins CF. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J Bacteriol 1987; 169:3861-5; PMID:3301822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gibson MM, Price M, Higgins CF. Genetic characterization and molecular cloning of the tripeptide permease (tpp) genes of Salmonella typhimurium. J Bacteriol 1984; 160:122-30; PMID:6090406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hogarth BG, Higgins CF. Genetic organization of the oligopeptide permease (opp) locus of Salmonella typhimurium and Escherichia coli. J Bacteriol 1983; 153:1548-51; PMID:6338001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ronnestad I, Murashita K, Kottra G, Jordal AE, Narawane S, Jolly C, Daniel H, Verri T. Molecular cloning and functional expression of atlantic salmon peptide transporter 1 in Xenopus oocytes reveals efficient intestinal uptake of lysine-containing and other bioactive di- and tripeptides in teleost fish. J Nutr 2010; 140:893-900; PMID:20220205; http://dx.doi.org/ 10.3945/jn.109.118240 [DOI] [PubMed] [Google Scholar]
- [61].Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J Bacteriol 1999; 181:3433-7; PMID:10348855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tenor JL, McCormick BA, Ausubel FM, Aballay A. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr Biol 2004; 14:1018-24; PMID:15182677; http://dx.doi.org/ 10.1016/j.cub.2004.05.050 [DOI] [PubMed] [Google Scholar]
- [63].Eswarappa SM, Panguluri KK, Hensel M, Chakravortty D. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology 2008; 154:666-78; PMID:18227269; http://dx.doi.org/ 10.1099/mic.0.2007/011114-0 [DOI] [PubMed] [Google Scholar]
- [64].Parra-Lopez C, Baer MT, Groisman EA. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J 1993; 12:4053-62; PMID:8223423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jones MM, Johnson A, Koszelak-Rosenblum M, Kirkham C, Brauer AL, Malkowski MG, Murphy TF. Role of the oligopeptide permease ABC Transporter of Moraxella catarrhalis in nutrient acquisition and persistence in the respiratory tract. Infect Immun 2014; 82:4758-66; PMID:25156736; http://dx.doi.org/ 10.1128/IAI.02185-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Vorwerk H, Mohr J, Huber C, Wensel O, Schmidt-Hohagen K, Gripp E, Josenhans C, Schomburg D, Eisenreich W, Hofreuter D. Utilization of host-derived cysteine-containing peptides overcomes the restricted sulphur metabolism of Campylobacter jejuni. Mol Microbiol 2014; 93:1224-45; PMID:25074326 [DOI] [PubMed] [Google Scholar]
- [67].Flores-Valdez MA, Morris RP, Laval F, Daffe M, Schoolnik GK. Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J 2009; 23:4091-104; PMID:19671666; http://dx.doi.org/ 10.1096/fj.09-132407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dasgupta A, Sureka K, Mitra D, Saha B, Sanyal S, Das AK, Chakrabarti P, Jackson M, Gicquel B, Kundu M, et al.. An oligopeptide transporter of Mycobacterium tuberculosis regulates cytokine release and apoptosis of infected macrophages. PloS One 2010; 5:e12225; PMID:20808924; http://dx.doi.org/ 10.1371/journal.pone.0012225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lin T, Troy EB, Hu LT, Gao L, Norris SJ. Transposon mutagenesis as an approach to improved understanding of Borrelia pathogenesis and biology. Front Cell Infect Microbiol 2014; 4:63; PMID:24904839; http://dx.doi.org/ 10.3389/fcimb.2014.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Troy EB, Lin T, Gao L, Lazinski DW, Lundt M, Camilli A, Norris SJ, Hu LT. Global Tn-seq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol Microbiol 2016; PMID:27279039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Schultz JE, Matin A. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J Mol Biol 1991; 218:129-40; PMID:1848300; http://dx.doi.org/ 10.1016/0022-2836(91)90879-B [DOI] [PubMed] [Google Scholar]
- [72].Kraxenberger T, Fried L, Behr S, Jung K. First insights into the unexplored two-component system YehU/YehT in Escherichia coli. J Bacteriol 2012; 194:4272-84; PMID:22685278; http://dx.doi.org/ 10.1128/JB.00409-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Garai P, Gnanadhas DP, Chakravortty D. Salmonella enterica serovars Typhimurium and Typhi as model organisms: revealing paradigm of host-pathogen interactions. Virulence 2012; 3:377-88; PMID:22722237; http://dx.doi.org/ 10.4161/viru.21087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, Babitzke P. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol 2003; 185:4450-60; PMID:12867454; http://dx.doi.org/ 10.1128/JB.185.15.4450-4460.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wong VK, Pickard DJ, Barquist L, Sivaraman K, Page AJ, Hart PJ, Arends MJ, Holt KE, Kane L, Mottram LF, et al.. Characterization of the yehUT two-component regulatory system of Salmonella enterica Serovar Typhi and Typhimurium. PloS one 2013; 8:e84567; PMID:24386394; http://dx.doi.org/ 10.1371/journal.pone.0084567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Jamieson DJ, Higgins CF. Anaerobic and leucine-dependent expression of a peptide transport gene in Salmonella typhimurium. J Bacteriol 1984; 160:131-6; PMID:6434517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Goh EB, Siino DF, Igo MM. The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes. J Bacteriol 2004; 186:4019-24; PMID:15175316; http://dx.doi.org/ 10.1128/JB.186.12.4019-4024.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Detmers FJ, Lanfermeijer FC, Poolman B. Peptides and ATP binding cassette peptide transporters. Res Microbiol 2001; 152:245-58; PMID:11421272; http://dx.doi.org/ 10.1016/S0923-2508(01)01196-2 [DOI] [PubMed] [Google Scholar]
- [79].Andrews JC, Blevins TC, Short SA. Regulation of peptide transport in Escherichia coli: induction of the trp-linked operon encoding the oligopeptide permease. Journal of bacteriology 1986; 165:428-33; PMID:3511033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev 2007; 21:2804-17; PMID:17974919; http://dx.doi.org/ 10.1101/gad.447207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lamarque M, Aubel D, Piard JC, Gilbert C, Juillard V, Atlan D. The peptide transport system Opt is involved in both nutrition and environmental sensing during growth of Lactococcus lactis in milk. Microbiology 2011; 157:1612-9; PMID:21393368; http://dx.doi.org/ 10.1099/mic.0.048173-0 [DOI] [PubMed] [Google Scholar]
- [82].Xie Y, Chou LS, Cutler A, Weimer B. DNA macroarray profiling of lactococcus lactis subsp. lactis IL1403 gene expression during environmental stresses. Appl Environ Microbiol 2004; 70:6738-47; PMID:15528540; http://dx.doi.org/ 10.1128/AEM.70.11.6738-6747.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Marugg JD, Meijer W, van Kranenburg R, Laverman P, Bruinenberg PG, de Vos WM. Medium-dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. J Bacteriol 1995; 177:2982-9; PMID:7768792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Roy S, Marla S, Praneetha DC. Recognition of corynebacterium pseudodiphtheriticum by Toll-like receptors and up-regulation of antimicrobial peptides in human corneal epithelial cells. Virulence 2015; 6:716-21; PMID:26125127; http://dx.doi.org/ 10.1080/21505594.2015.1066063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 2005; 3:238-50; PMID:15703760; http://dx.doi.org/ 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- [86].Lee EJ, Choi J, Groisman EA. Control of a Salmonella virulence operon by proline-charged tRNA(Pro). Proc Natl Acad Sci U S A 2014; 111:3140-5; PMID:24516160; http://dx.doi.org/ 10.1073/pnas.1316209111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gominet M, Slamti L, Gilois N, Rose M, Lereclus D. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol Microbiol 2001; 40:963-75; PMID:11401703; http://dx.doi.org/ 10.1046/j.1365-2958.2001.02440.x [DOI] [PubMed] [Google Scholar]
- [88].Slamti L, Lereclus D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J 2002; 21:4550-9; PMID:12198157; http://dx.doi.org/ 10.1093/emboj/cdf450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hartel T, Klein M, Koedel U, Rohde M, Petruschka L, Hammerschmidt S. Impact of glutamine transporters on pneumococcal fitness under infection-related conditions. Infect Immun 2011; 79:44-58; PMID:21078855; http://dx.doi.org/ 10.1128/IAI.00855-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tamura GS, Nittayajarn A, Schoentag DL. A glutamine transport gene, glnQ, is required for fibronectin adherence and virulence of group B streptococci. Infect Immun 2002; 70:2877-85; PMID:12010975; http://dx.doi.org/ 10.1128/IAI.70.6.2877-2885.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Das P, Lahiri A, Lahiri A, Sen M, Iyer N, Kapoor N, Balaji KN, Chakravortty D. Cationic amino acid transporters and Salmonella Typhimurium ArgT collectively regulate arginine availability towards intracellular Salmonella growth. PloS one 2010; 5:e15466; PMID:21151933; http://dx.doi.org/ 10.1371/journal.pone.0015466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gesbert G, Ramond E, Tros F, Dairou J, Frapy E, Barel M, Charbit A. Importance of branched-chain amino acid utilization in Francisella intracellular adaptation. Infect Immun 2015; 83:173-83; PMID:25332124; http://dx.doi.org/ 10.1128/IAI.02579-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun 2004; 72:6373-81; PMID:15501767; http://dx.doi.org/ 10.1128/IAI.72.11.6373-6381.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 1998; 283:489-506; PMID:9769220; http://dx.doi.org/ 10.1006/jmbi.1998.2107 [DOI] [PubMed] [Google Scholar]
- [95].Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567-80; PMID:11152613; http://dx.doi.org/ 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- [96].Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 2007; 23:538-44; PMID:17237066; http://dx.doi.org/ 10.1093/bioinformatics/btl677 [DOI] [PubMed] [Google Scholar]
- [97].Viklund H, Elofsson A. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 2008; 24:1662-8; PMID:18474507; http://dx.doi.org/ 10.1093/bioinformatics/btn221 [DOI] [PubMed] [Google Scholar]
- [98].Cascieri T Jr., Mallette MF. New method for study of peptide transport in bacteria. Appl Microbiol 1974; 27:457-63; PMID:4596381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res 2001; 11:1246-55; PMID:11435407; http://dx.doi.org/ 10.1101/gr.186501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Charbonnel P, Lamarque M, Piard JC, Gilbert C, Juillard V, Atlan D. Diversity of oligopeptide transport specificity in Lactococcus lactis species. A tool to unravel the role of OppA in uptake specificity. J Biol Chem 2003; 278:14832-40; PMID:12590143; http://dx.doi.org/ 10.1074/jbc.M212454200 [DOI] [PubMed] [Google Scholar]


