Abstract
RNA-binding proteins (RBPs) participate in diverse cellular processes and have important roles in human development and disease. The human genome, and that of many other eukaryotes, encodes hundreds of RBPs that contain canonical sequence-specific RNA-binding domains (RBDs) as well as numerous other unconventional RNA binding proteins (ucRBPs). ucRBPs physically associate with RNA but lack common RBDs. The degree to which these proteins bind RNA, in a sequence specific manner, is unknown. Here, we provide a detailed description of both the laboratory and data processing methods for RNAcompete, a method we have previously used to analyze the RNA binding preferences of hundreds of RBD-containing RBPs, from diverse eukaryotes. We also determine the RNA-binding preferences for two human ucRBPs, NUDT21 and CNBP, and use this analysis to exemplify the RNAcompete pipeline. The results of our RNAcompete experiments are consistent with independent RNA-binding data for these proteins and demonstrate the utility of RNAcompete for analyzing the growing repertoire of ucRBPs.
Keywords: RNAcompete, RNA-binding protein, DNA microarray, binding site, NUDT21, CNBP
1. Introduction
Hundreds of thousands of annotated RBPs are encoded by the 742 sequenced eukaryotic genomes. The vast majority of these RBPs, including those from well-studied organisms, have unknown RNA-binding preferences. For example, in human, recent evidence indicates that there are approximately 1,200–1,500 human proteins that associate with RNA, representing approximately 6–8% of the annotated human proteome [1, 2]; however, less than half of human RBPs that contain canonical RBDs (see below) have an established RNA-binding motif [3, 4]. This poor characterization of RBP sequence-binding preferences presents a significant barrier in the analysis of post-transcriptional gene regulation.
Most known sequence-specific RBPs contain canonical RBDs that mediate RNA-binding through protein-RNA interactions [5]. The most common sequence-specific eukaryotic RBDs are the RNA recognition motif (RRM) (~246 in human), the Cys-Cys-Cys-His (CCCH-zf) type zinc finger domain (~60 in human), and the HNRNP K homology (KH) domain (~38 in human) [6]. Among these, the ~90 amino acid RRM domain is the most extensively studied [5]. Here, RNA recognition typically occurs on the β-sheet surface and is often mediated by three exposed aromatic residues [7]. Eukaryotic KH domains span ~70 amino acids and contain an RNA-binding cleft formed by a conserved GXXG motif flanked by two α-helices, a variable loop, and a β-strand [8]. Lastly, CCCH-zf domains are 12–30 amino acids long and bind RNA through stacking interactions, and hydrogen bonding between the protein backbone and Watson-Crick edges of bases [9]. Individual RBDs tend to bind RNA in the micromolar range [5]; however RNA-binding affinity and specificity is significantly increased by combinations of RBDs or by additional RNA contacting residues located in regions outside of the RBD [5].
Not all sequence-specific RBPs contain a canonical RBD. For example, several well-characterized proteins, including the pre-mRNA 3′ cleavage and polyadenylation specificity factor 5, NUDT21 (Nudix hydrolase domain and N-terminal region: [10]), histone stem-loop binding protein, SLBP (SLBP domain: [11, 12]), and Drosophila brain tumor protein, BRAT (NHL domain: [13, 14]), use unconventional RBDs—indicated in parentheses—for sequence-specific RNA recognition. In a genome-wide context, proteomic analyses of proteins UV cross-linked to mRNA in human HeLa, HEK293, and HuH7 cell lines have identified over 800 that associate with mRNA, but do not contain canonical RBDs [2, 15, 16]. We refer to these potentially sequence-specific RBPs as unconventional RBPs (ucRBPs). A similar analysis in Drosophila identified ~300 ucRBPs, one of which (CG3800) was shown by CLIP-seq to bind RNA containing specific 5-mer sequences enriched for G and A residues [17]. It was not shown, however, whether CG3800 binds to these sequences autonomously.
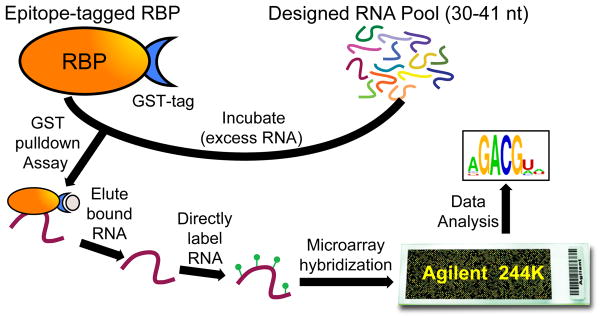
RNAcompete is an in vitro method (see Figure 1) that we developed [18] and have applied to hundreds of RBPs [3]. It recapitulates RNA-binding motifs for a diverse set of well-studied RBPs previously identified in vitro (e.g. SELEX experiments: Systematic Evolution of Ligands by Exponential Enrichment [19]) and in vivo (e.g. CLIP experiments: UV Cross-Linking and Immuno-Precipitation [20]). In RNAcompete experiments, purified epitope-tagged RBPs select RNA sequences from a designed (non-randomized) RNA pool. Bound RNAs are identified using microarray hybridizations and analyzed computationally to determine RBP-specific 7-mer RNA-binding profiles. Several in vitro methods have been described since the inception of RNAcompete, including RNA Bind-n-Seq (RNBS) [21], SEQRS [22], RNA-MaP [23], HiTS-RAP [24], and RNA-MITOMI [25]. Of these, only RNA Bind-n-seq has been used in a large-scale study (unpublished ENCODE online data). The RNAcompete methodology has several attractive features including: i) no antibodies are required; ii) no iterative selection or library preparation is necessary; iii) does not require RBP-specific optimizations; iv) is relatively inexpensive at scale; v) is amenable to large-scale studies [3]; and, vi) has an established and validated uniform computational analysis pipeline.
Figure 1.
Schematic of the RNAcompete assay. A GST-tagged RBP (RBP is orange oval, GST-tag is blue crescent), is incubated with a 75-fold excess of a non-random, custom designed RNA pool (multicoloured lines). RNA selectively bound (purple line) to an RBP during a GST-pulldown assay (GST bead is represented as a beige oval) is eluted, directly labeled with either Cy3 or Cy5 (green circles), and hybridized to a custom Agilent 244K microarray. Microarray data is analyzed computationally to generate RNA-binding motifs represented as logos.
In this report, we present a detailed protocol for the experimental and computational components of the RNAcompete system. We also highlight the utility of RNAcompete via analysis of the RNA-binding preferences of two human ucRBPs, NUDT21 and CNBP.
2. Materials
2.1 Materials
| Name | Manufacturer |
|---|---|
| 3M NaOAc pH 5.2 | BioShop (SAA333.100) |
| 6× Orange Loading Dye | Thermo Scientific (R0631) |
| Acrylamide/Bis 30% (29:1) | Bio-Rad (161-0156) |
| Agarose | Thermo Scientific (16500100 ) |
| Agilent 244K Microarray | Agilent (AMADID# 024519) |
| Ampicillin | BioShop (AMP201.25) |
| AscI | NEB (R0558S) |
| Axon GenePix 4000B Microarray Scanner | Molecular Devices |
| β-mercaptoethanol (BME) | Sigma (M7522) |
| Bio-Rad Mini-Protean Gel Electrophoresis System | Bio-Rad (165-3301) |
| Bromophenol Blue (BB) | BioShop (BRO777.10) |
| BSA Standard (2 mg/mL) | Pierce (23209) |
| BspQI | NEB (R0712L) |
| C41 E.coli competent cells | Lucigen Corporation (60442-1) |
| Coomassie Brilliant Blue | BioShop (CBB555.25) |
| dNTPs | Thermo Scientific (R0181) |
| Dithiothreitol (DTT) | BioShop (DTT001.5) |
| Ethylenediaminetetraacetic Acid (EDTA) | Bioshop (EDT111.100) |
| Ethidium Bromide | BioShop (ETB444.10) |
| Ethyl Alcohol (Ethanol) | Commercial Alcohols (P016EAAN) |
| FirstChoice Human Total RNA Survey Panel | Ambion (AM6000) |
| Formamide | BioShop (FOR001.500) |
| Gasket Slide | Agilent (G2534-60003) |
| Glutathione, reduced | BioShop (GTH001.5) |
| Glycerol | BioShop (GLY001.1) |
| Glycogen | Thermo Scientific (R0551) |
| GST-Sepharose 4B beads | GE Healthcare (17-0756-05) |
| N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid (HEPES) | BioShop (HEP001.500) |
| High Performance Glutathione Sepharose | GE Healthcare (17-5279-01) |
| Hybridization Incubator (Model 1000) | Robbins Scientific |
| Illustra G-25 Spin Columns (50 purifications) | GE Healthcare (27-5325-01) |
| Incubator Shaker | New Brunswick Scientific (Innova 4300) |
| Isopropyl-β-D-thiogalactopyranoside (IPTG) | BioShop (IPT001.10) |
| Klenow Fragment (3′-5′ exo-) | NEB (M0212S) |
| LabQuake Shaker | Barnstead Thermolyne (4152110) |
| LB Broth, Lennox | BioShop (LBL405.1) |
| LB-Agar | Bioshop (LBA408.1) |
| Lysozyme | BioShop (LYS702.1) |
| Megascript T7 Transcription Kit | Thermo Scientific (AM1334) |
| Methanol | BioShop (MET302.4) |
| Methyl Ethane Sulfonate (MES) | Sigma (M2933-100G) |
| Microarray Hybridization Chamber | Agilent (G2534A) |
| MisoNix 3000 Sonicator | Mandel |
| NaCl | BioShop (SOD002.205) |
| Nanodrop (ND-1000) | Thermo Scientific |
| Nuclease-free H2O | Thermo Scientific (10977023) |
| PageRuler Prestained Protein Ladder | Thermo Scientific (26616) |
| Phosphate-Buffered Saline (PBS) Buffer | BioShop (PBS408.500) |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1) | BioShop (PHE512.400) |
| Phenylmethylsulfonyl Fluoride (PMSF) | Sigma (P7626-5G) |
| Platinum Taq DNA Polymerase High Fidelity | Thermo Scientific (11304011) |
| Protein Assay Dye Reagent Concentrate | Bio-Rad (#5000006) |
| Qiaex II Gel Extraction Kit | Qiagen (20021) |
| QIAquick PCR Purification Kit | Qiagen (28104) |
| Refrigerated Microcentrifuge | Eppendorf (5415R) |
| Salmon Sperm DNA | Thermo Scientific (15632011) |
| SbfI | NEB (R3642S) |
| Sodium Dodecyl Sulphate (SDS) | Bioshop (SDS001.100) |
| SOC Media | Thermo Scientific (15544034) |
| Sodium N-lauroyl Sarcosine (SLS) | BioShop (SLS002.100) |
| Sorvall LYNX 4000 Superspeed Centrifuge | Thermo Scientific (75006580) |
| Spectra Max Plus 384 | Medical Devices |
| NaCl-NaH2PO4-EDTA (SSPE) Buffer | BioShop (SSP333.1) |
| Superscript II Reverse Transcriptase | Thermo Scientific (18064014) |
| T4 DNA Ligase | NEB (M0202T) |
| T4 DNA Polymerase | NEB (M0203S) |
| Tris-Acetate-EDTA (TAE) Buffer | BioShop (TAE222.1) |
| Tecan HS4800 Pro Hybridization Workstation | Tecan |
| Tris base | BioShop (TRS001.1) |
| Tris-HCl pH 7.5 | Life Technologies (15567027) |
| Triton X-100 | Sigma (T9284) |
| ULS Labeling Kit for Agilent Arrays | Kreatech (EA-021) |
2.2 Buffers
| Buffer | Components |
|---|---|
| 5× SDS Loading Buffer | 350 mM Tris-HCl pH 6.8, 10% SDS, 50% glycerol, 0.005% BB, 0.5M DTT |
| Agilent Hybridization Buffer | 1M NaCl, 0.5 % SLS, 50 mM MES pH 6.5, 50% formamide |
| Binding Buffer | 20 mM HEPES pH 7.5, 70 mM KCl, 10 mL NaCl, 2 mM DTT, 10% glycerol, BSA 100 ug/mL |
| Buffer C | 20 mM HEPES pH 7.5, 100 mM NaCl, 100 mM EDTA pH 8.0, 0.07% BME |
| Buffer D | 500 mM NaCl, 0.07% BME |
| Elution Buffer | 30mM reduced glutathione, 250 mM NaCl, 20% glycerol, 50 mM Tris-HCl pH 8.8, 0.07% BME |
| Hybe Wash Buffer #1 | 6 × SSPE, 0.005% SLS |
| Hybe Wash Buffer #2 | 0.06× SSPE |
| Hybridization Buffer | 10 mM Tris-HCl pH 7.5, 1M NaCl, 0.5% Triton X-100, 0.75 mM DTT |
| Klenow Fill-In Reaction Mix | 10 mM Tris-HCl pH 7.9, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 1 × NEB BSA, 0.1 mM dNTPs |
| Ligation Reaction Mix | 50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1 mM ATP, 10 mM DTT |
| Lysis Buffer | 20 mM HEPES pH7.5, 0.1 mM EDTA, 1 M NaCl, 10 mM BME, 1 mM PMSF, Triton X-100 |
| Oligo Annealing Buffer | 50 mM NaCl, 1 mM EDTA pH 8.0 |
| Pre-hybridization Buffer | 50 mM MES pH 6.5, 1 M NaCl, 0.5% SLS |
| STE Buffer | 10 mM Tris-HCl pH 7.5, 1% SDS, 5mM EDTA |
| Wash Buffer #1 | 6× SSPE/0.05% Triton X-100 |
| Wash Buffer #2 | 0.06 × SSPE |
| Wash Buffer #3 | 10 mM Tris-HCl pH 7.5, 50 mM NaCl, 10 mM MgCl2 |
3. RNAcompete Protocol
We have organized the procedures comprising the RNAcompete pipeline as follows: custom microarray design and synthesis (section 3.1); DNA pool generation (section 3.2); RNA pool generation (section 3.3); cloning RBPs into E. coli expression vectors (section 3.4); purification of GST-tagged RBPs (section 3.5); RNA pulldown assay (section 3.6); microarray analysis (section 3.7); and, RNAcompete data analysis (section 3.8). The following sections detail the experimental and computational methods encompassed by RNAcompete. The experimental methods emphasize small-scale investigation (for simplicity) however it is important to note that the scale can be readily increased as previously demonstrated [3]. Buffers, reagents and other materials are listed in sections 2.1/2.2 unless otherwise noted. Also, note that we use Nuclease-free H2O for all experiments involving RNA; however, DEPC-H2O could also be used.
3.1 Custom Microarray Design and Synthesis
In principle, the experimental pipeline described below could be run with any set of RNAs (some considerations are discussed in section 3.8). However, all RNAcompete experiments to date have employed defined RNA pools that are generated from 244K Agilent custom DNA microarrays, with designs similar to those employed for universal Protein Binding Microarrays [26]. Our current design is based on a de Bruijn sequence of order 11 that was subsequently modified to minimize secondary structure in the designed sequences and minimize intramolecular RNA cross-hybridization. The final pool consists of 241,399 individual sequences up to 38 nucleotides in length – its construction is described in more detail in [3]. After these modifications, not every 11-mer is represented but each 9-mer is represented at least 16 times in the pool. Moreover, to facilitate internal data comparisons, the pool is split computationally into two sets, Set A and Set B. Each set contains at least 155 copies of all 7-mers except GCTCTTC and CGAGAAG which are removed because they correspond to the SapI/BspQI restriction site (see section 3.2.7). A ϕ2.5 bacteriophage T7 promoter initiating with an AGA or AGG sequence is added at the beginning of each probe sequence to enable RNA synthesis (see section 3.3) [27]. The microarray design is detailed in a Supplementary Information file from [3] and can be ordered from Agilent Technologies using AMADID# 024519.
Where microarray technologies are no longer available, one could order the RNAcompete library as a ssDNA pool from Agilent and replace the microarray readout with high-throughput sequencing designed for small RNAs [28, 29]. The sequencing counts could be processed and analyzed in the same way as scanner intensities (see section 3.8). Like RNAcompete, this modification would be an inexpensive alternative to RBNS [21] and SEQRS [22] because the defined RNA pool requires fewer reads to quantify and simplifies the analysis of the data.
The current RNAcompete RNA pool was designed to measure the ssRNA sequence binding preferences of RBPs. As such, this library was designed to be depleted of secondary structure. Nonetheless, we were able to recover sequence preferences for RBPs that bind hairpin loops (Vts1p, SNRPA) and for Vts1p the preferred 7-mers suggest a hairpin loop structure. Also, because some weak secondary structure remains in the pool, we had the statistical power to define some secondary structure preference for approximately 25% of RBPs (Supplementary Data 3 in Ray et al., Nature 2013). A full and quantitative assessment of secondary structure preference would require a complex starting pool to ensure a complete representation of possible secondary structures. Sequencing-based technologies with random starting pools might provide the required complexity [21, 22]; however, to date, they have only been used to define accessible bases for a handful of RBPs.
3.2 DNA Pool Generation
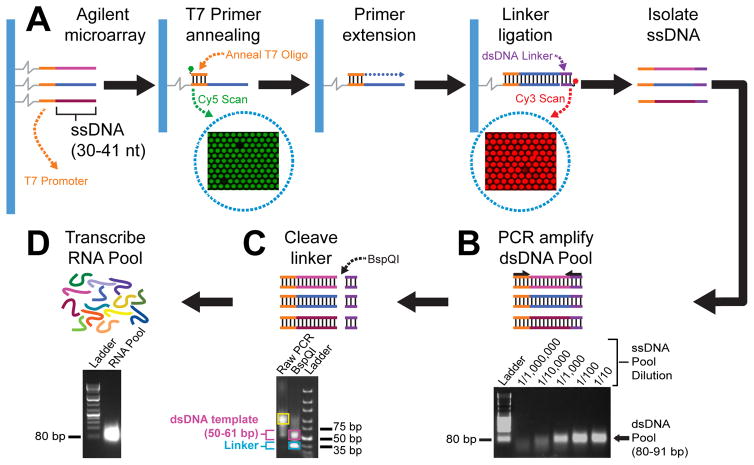
Prior to RNA pool generation, ssDNA probes on a custom microarray undergo several biochemical treatments to generate a corresponding DNA pool (Figure 2A). First, a Cy3-labeled T7 promoter oligonucleotide is annealed to complementary sequences on all microarray probes (verified by Cy3 fluorescence) and converted to dsDNA through primer extension using T4 DNA polymerase. Next, a Cy5-labeled dsDNA linker is ligated to these dsDNAs using T4 DNA ligase (verified by Cy5 fluorescence) and the sense strand, which is not covalently anchored to the microarray, is “stripped” using high heat and NaOH, and purified. The purified ssDNA pool is used as template during PCR for dsDNA pool generation and amplification using primers that anneal to the T7 promoter and linker sequences (Figure 2B). Lastly, the linker is removed using a Type IIS restriction enzyme, BspQI, and the dsDNA template is gel-purified (Figure 2C) prior to RNA pool synthesis (Figure 2D).
Figure 2.
Custom DNA and RNA pool generation from microarray. A) Agilent microarrays contain ssDNA probes that have variable designed sequences (multicoloured) and a T7 promoter sequence (orange lines). A Cy3 (green circle) labeled T7 promoter primer (orange line) is annealed to ssDNA microarray probes which are subsequently double-stranded by enzymatic primer extension using T4 DNA polymerase (blue dotted line). A dsDNA linker (purple lines), labeled with Cy5 (red circle), is ligated to the dsDNA microarray probes using T4 DNA ligase. Cy3 and Cy5 signals are detected on microarrays by scanning at 532 and 635 nm, respectively, to verify efficient T7 primer annealing and linker ligation. Scanned Cy3 and Cy5 microarray images are shown (enclosed by dotted blue circles). B) PCR amplification of the dsDNA pool using ssDNA pool purified from the microarray as template. ssDNA pool (PCR template) dilutions as well as bands corresponding to an 80 bp DNA ladder and a 80–91 bp dsDNA pool are indicated on the ethidium bromide stained agarose gel image. C) Linker cleavage is mediated by a Type IIS restriction enzymes, BspQI. Representative bands corresponding to undigested PCR amplified dsDNA pool (enclosed by yellow box), BspQI digested dsDNA template (enclosed by pink box), and cleaved dsDNA linker (enclosed by blue box) are indicated. D) The non-random RNA pool (multicoloured lines and high intensity band in the agarose gel image) is generated via T7 RNA polymerase mediated transcription reactions, using gel purified dsDNA templates (shown in panel 2C).
3.2.1 Annealing T7 promoter to microarray probes
Resuspend 8 nmoles of T7 promoter oligo 5′ end labelled with Cy3 (5′-Cy3-CTAATACGACTCACTATTAG; Integrated DNA Technologies) in 20 μL of H2O.
Heat for 2 minutes at 85°C, centrifuge briefly, and keep on ice.
Place Gasket Slide into Microarray Hybridization Chamber.
Add resuspended T7 oligo to 880 μL of Hybridization Buffer and pipette into Gasket Slide.
Slowly and carefully place Agilent 244K Microarray on (probe side down or “label to label”) Gasket Slide (to minimize introduction of air bubbles between the microarray and Gasket Slides), assemble Microarray Hybridization Chamber, and incubate for 4 hours at 30°C in a Microarray Hybridization Incubator with rotation (setting 8).
Remove microarray from Microarray Hybridization Chamber and Gasket Slide and place in 50 mL conical centrifuge tube containing 45 mL of Wash Buffer #1 and invert 5 times.
Transfer slide to a 50 mL conical centrifuge tube containing Wash Buffer #2 and invert 5 times.
Remove microarray slide and air dry.
Scan the microarray for Cy3 signal using Axon Genepix 4000B Microarray Scanner or an alternative microarray scanner (400 PMT at a wavelength of 532 nm), to confirm primer hybridization. All probes excluding empty Agilent control spots should have a high intensity Cy3 signal.
3.2.2 Double-stranding microarray probes by primer extension
Place Gasket Slide into the Microarray Hybridization Chamber.
Assemble and pipette 900 μL of Klenow Fill-In Reaction Mix containing 50 Units of T4 DNA Polymerase and 36 Units of Klenow Fragment (3′-5′ exo-).
Place microarray slide onto Gasket Slide and assemble Microarray Hybridization Chamber (see section 3.2.1).
Incubate at 30°C for 30 minutes in a Microarray Hybridization Incubator with rotation (setting 8).
Remove microarray slide and wash in 45 mL of Wash Buffer #3 by inverting 5 times in a 50 mL conical tube.
Repeat wash and leave slide in Wash Buffer #3 until the linker ligation step (see section 3.2.4).
3.2.3 Generating dsDNA linker
Note: dsDNA linkers can be prepared in advance and stored at −20°C.
Combine 15 nmoles of Cy5 labeled oligo, and 15 nmoles of 5′-phosphate labeled and 3′-dideoxycytosine (ddC) oligo (5′-P-TGAAGAGCGAGCGGATACAG-ddC-3′, 5′-Cy5-CTGTATCCGCTCGCTCTTCA; the core SapI/BspQI restriction site is underlined, Integrated DNA Technologies [IDT]) in 50 μL of H2O with 50 μL Oligo Annealing Buffer.
Heat at 95°C for 2 minutes, 65°C for 10 minutes, 37°C for 10 minutes, room temperature for 10 minutes, centrifuge briefly and keep on ice.
3.2.4 Ligating dsDNA linker to microarray probes
Place Gasket Slide into the Microarray Hybridization Chamber.
Add dsDNA linker (100 μL) to 800 μL of Ligation Mix containing 18,000 Units of T4 DNA ligase, assemble Microarray Hybridization Chamber and incubate overnight in a Microarray Hybridization Incubator at room temperature with rotation (setting 8).
Remove microarray slide and wash twice in 50 mL conical centrifuge tube containing 45 mL of Wash Buffer #3.
Remove microarray slide and air dry.
Scan microarray for Cy5 signal using Axon Genepix 4000B Microarray Scanner, or an alternative microarray scanner, (600 PMT at a wavelength of 635 nm) to confirm that all spots excluding empty Agilent controls have a high intensity Cy5 signal.
3.2.5 Purifying ssDNA from microarray
Heat 1 mL of 20 mM NaOH solution, Gasket Slide, and Microarray Hybridization Chamber to 65°C in Microarray Hybridization Incubator.
For the following steps, work in the Microarray Hybridization Incubator (set at 65°C) to keep all materials and reagents warm.
Place Gasket Slide into Microarray Hybridization Chamber.
Pipette 900 μL of heated 20 mM NaOH solution to Gasket Slide and carefully place microarray slide (probe side down) onto Gasket Slide.
Assemble Microarray Hybridization Chamber and place into a Microarray Hybridization Incubator and incubate at 65°C for 20 minutes with rotation (setting 8).
Carefully remove microarray slide and pipette all liquid (which now contains stripped ssDNA pool from the microarray) to a new Eppendorf tube.
Scan both Cy3 and Cy5 using previous settings to ensure microarrays have been stripped. If significant signal is detected, repeat the stripping procedure and pool samples together.
Increase volume of ssDNA sample to 740 μL with H2O, transfer 180 μL to each of 4 Eppendorf tubes and, to each tube, add 20 μL 3M NaOAc (pH 5.2), 2 μL glycogen, and 500 μL 100% ethanol. Mix by inverting several times.
Precipitate DNA: Incubate samples for 15 minutes on dry ice, 1+ hours at −80°C, or overnight at −20°C, centrifuge at maximum speed for 30 minutes (4°C), wash pellet with 700 μL 75% ethanol, spin at maximum speed for 10 minutes, remove ethanol, centrifuge briefly and remove residual ethanol, let air dry for 2 minutes.
Resuspend ssDNA pellets with 40 μL with H2O.
3.2.6 Amplification of dsDNA pool
The stripped ssDNA pool is amplified by PCR using common primers and scaled as required. As a general working rule, approximately three PCR reactions are required for every RBP to be assayed. We amplify a large quantity of DNA pool, enough for hundreds of experiments, to limit pool variation between experiments. Furthermore, our computational analysis requires data from multiple experiments run using the same DNA pool in order to estimate oligo abundance in this pool.
Dilute DNA 20-fold by adding 2.5 μL of stripped ssDNA pool to 47.5 μL of H2O.
Assemble PCR reaction (scale as required): 61.5 μL H2O, 7.5 μL 10× Platinum Taq PCR Buffer, 1 μL 10 mM dNTPs, 2 μL 50 mM MgOAc, 0.5 μL Forward Primer (5′-CTAATACGACTCACTATTAG, 100 pmol/μL, IDT), 0.5 μL Reverse Primer (5′-CCAGTCAGCACTGTATCCGCTCGCTCTTCA, 100 pmol/μL, IDT), 1 μL of diluted ssDNA pool, and 1 μL Platinum Taq DNA Polymerase High Fidelity (5U/μL).
PCR conditions: heat sample at 94°C for 120 seconds; 30 cycles of 98°C for 30 seconds, 48°C for 30 seconds, and 68°C for 15 seconds; 68°C for 5 minutes.
Phenol-chloroform-isoamyl alcohol (PCI) treatment: Increase volume to 180 μL with H2O, add 20 μL 3M NaOAc pH 5.2, add 200 μL PCI and vortex vigorously for 10 seconds, centrifuge at maximum speed for 2 minutes, transfer 180 μL of aqueous layer into a new Eppendorf tube, add 500 μL of ethanol and precipitate DNA (see section 3.2.5 for details).
Resuspend in 15 μL H2O.
Add 3 μL of 6× Orange Loading Dye, load entire sample into a 3.0% agarose gel containing Ethidium Bromide (0.2 μg/mL), and separate DNA in 1× TAE Buffer (electrophorese samples at 120 Volts for 20 minutes).
Excise DNA band (80–91 bp), and purify using recommended instructions from Qiaex II Gel Extraction Kit (elute with 40 μL of H2O).
Note: if a large number of RBPs are to be analyzed, the dsDNA Pool can be re-amplified in 96-well plates, and purified at a larger scale.
3.2.7 Removing linker with BspQI and purification of dsDNA template
To 40 μL of eluted dsDNA pool add 5 μL of 10× NEB Buffer #4, 4 μL of H2O, and 1 μL of BspQI and incubate at 50°C for 4 hours.
Add an additional 1 μL of BspQI and incubate at 50°C for 4 hours.
Perform PCI/ethanol precipitation treatment (see sections 3.2.5 and 3.2.6) and resuspend pellet in 40 μL H2O.
Repeat BspQI digestion, PCI/ethanol precipitation, and resuspend pellet in 15 μL of H2O.
Add 3 μL of 6× Orange Loading Dye, load entire sample into a 4.0% agarose gel (containing 0.2 μg/mL Ethidium Bromide), and separate DNA using 1× TAE Buffer at 150 Volts for 70 minutes.
Excise DNA band (50–61 bp) and purify using recommended instructions from Qiaex II Gel Extraction Kit (elute with 20 μL of nuclease-free H2O).
Perform PCI/ethanol precipitation treatment (see sections 3.2.5 and 3.2.6) and resuspend dsDNA template pellet in 10 μL Nuclease-free H2O and quantify using a Nanodrop or alternative spectrophotometer.
3.3 RNA Pool Generation
RNA pool synthesis is mediated by T7 RNA polymerase using the dsDNA template (refer to Figure 2D). It is important to note that we use a bacteriophage ϕ2.5 T7 RNA polymerase promoter which, unlike the “classical” T7 promoter, initiates transcription with AGA or AGG sequences and has the advantage of generating homogeneous RNA 5′-ends [27].
Use 2.0 pmol (~40 ng) of purified, BspQI-digested dsDNA template in each 20 μL Megascript T7 Transcription reaction and incubate overnight at 37°C.
Add 1 μL of DNAse I (using Turbo DNase I from MegaScript T7 Transcription Kit) and incubate at 37°C for 2 hours.
Purify RNA pool twice using Illustra G-25 Spin Columns as recommended by manufacturer with the following exceptions: i) pack columns by centrifugation at 1,300 × g for 1 minute; and, ii) elute RNA by centrifugation at 1,090 × g for 4 minutes.
Perform PCI/ethanol precipitation as previously described (see section 3.2.5 and 3.2.6) and resuspend pellet using 40 μL of Nuclease-free H2O.
Quantify RNA levels using a Nanodrop or alternative spectrophotometer.
Typically, one transcription reaction yields enough RNA pool for one RNAcompete experiment.
3.4 Cloning RBPs into E.coli expression vectors
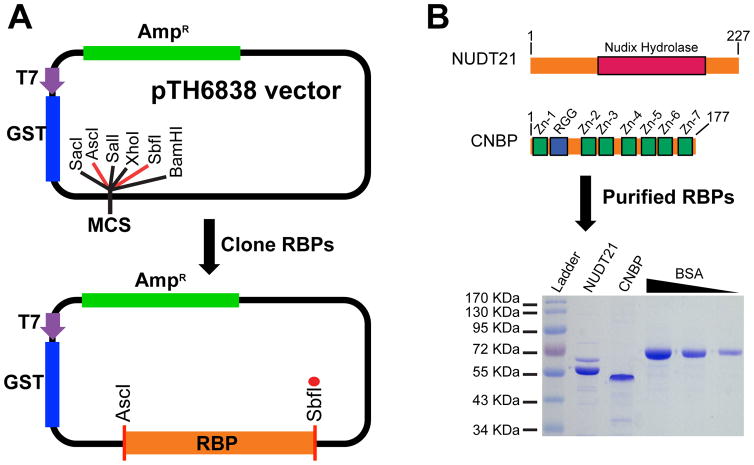
RBP inserts are typically cloned into the AscI and SbfI restriction sites in the multiple cloning site of a pTH6838 expression vector (Figure 3A) using standard cloning procedures. The vector map and sequence for pTH6838 can be found at http://hugheslab.ccbr.utoronto.ca/supplementary-data/RNAcompete_eukarya/ [3]. The full-length ucRBP inserts analyzed below were generated using a Superscript II Reverse Transcription Kit, FirstChoice Human Total RNA Survey Panel as template and gene-specific primers for NUDT21 (5′-TGTCACGGACCTACGGCGCGCCATGTCTGTGGTACCGCCCAATCGC, and TAGTGCACCACAATCCTGCAGGTGTAAATAAAATTGAACCTGCTCAACA; IDT) or CNBP (5′-TGTCACGGACCTACGGCGCGCCATGAGCAGCAATGAGTGCTTC, 5′-TAGTGCACCACAATCCTGCAGGCTGTAGCCTCAATTGTGCATTCC; IDT) following the manufacturer’s recommendations.
Figure 3.
Cloning and protein purification of human RBPs. A) Individual human RBP inserts (orange) are cloned into the pTH6838 multiple cloning site (MCS), typically using AscI and SbfI restriction sites. The red circle indicates a stop codon engineered immediately after the SbfI restriction site. The T7 promoter (purple) and coding sequences for GST (blue) and ampicillin resistance (green) are shown. B) Diagram of full-length NUDT21 and CNBP show the Nudix hydrolase domain (red rectangle) from NUDT21 as well as the RGG Box (blue rectangle) and seven CCHC zinc fingers (green rectangles) from CNBP (top panel). The first and last amino acid for NUDT21 and CNBP are also indicated. GST-tagged NUDT21 and CNBP RBPs expressed and affinity purified from E. coli cells are visualized on protein SDS-PAGE gels (bottom panel). Purified proteins for NUDT21 and CNBP as well as BSA standards (2.5 μg, 1.25 μg, and 0.62 μg) are labeled above the Coomassie blue stained SDS-PAGE protein gel image.
3.5 Purification of GST-tagged RBPs
For both small- and large-scale purification of GST-tagged RBPs, we have developed a streamlined protein purification protocol (sections 3.5.1 to 3.5.8) that operates at a >90% success rate. Specifically, a protein of expected size is produced from >90% of clones encoding RBPs of less than 600 amino acids. All protein work is performed on ice and Nuclease-free H2O is used for all washing and elution buffers. The results for our GST-tagged NUDT21 and CNBP protein purifications are shown in Figure 3B.
3.5.1 Transformation of E.Coli with RBP expression vector
Add 25–50 ng of plasmid DNA (e.g. pGEX or pTH6838 based RBP constructs) to 50 μL of E.coli C41 cells, gently mix, and incubate for 30 minutes on ice.
Incubate in a 42°C water bath for 45 seconds, transfer to ice for 10 minutes, add 500 μL of SOC media, and incubate at 37°C for 1 hour.
Centrifuge cells 1,500 × g for 3 minutes.
Remove all but ~50 μL of supernatant and resuspend cell pellet with remaining supernatant.
Plate cells onto LB-Agar plate containing 100 μg/mL Ampicillin and incubate for 16–18 hours at 37°C.
3.5.2 Expression of GST-tagged RBPs in E.coli
Inoculate 5 mL of LB Media containing Ampicillin (100 μg/μL) with a single colony and incubate for 16–18 hours in an Incubator Shaker (INNOVA 4300) at 37°C (220 RPM).
Transfer 5 mL pilot culture into a 500 mL Erlenmeyer flask containing 250mL of LB Media and Ampicillin (100 μg/μL), and grow at 37°C (220 RPM) until OD600nm reaches 0.6 to 0.8.
Add 250 μL of 1M IPTG and incubate at 16°C for 16–18 hours at 220 RPM.
Centrifuge cells at 2,755 × g for 40 minutes, remove supernatant and resuspend cells with 10 mL of Lysis Buffer and transfer to 50 mL conical centrifuge tubes. Optional: flash freeze cell suspension using liquid nitrogen and store at −80°C.
3.5.3 Lysing induced E.coli cells
If frozen: thaw E.coli cells on ice, add 25 mL of Lysis Buffer, 150 μL of 100 mg/mL Lysozyme Solution, and keep on ice for 15 minutes.
Place cell suspension into a 500 mL beaker filled with ice and sonicate (MisoNix 3000 Sonicator) setting the power output 8.5 – with 2 seconds on, 3 seconds off – and total time 2 minutes.
Centrifuge at 22,600 × g for 15 minutes at 4°C (Sorvall LYNX 4000 Superspeed Centrifuge), and transfer supernatant to new falcon tube, and keep samples on ice.
3.5.4 Preparation of GST Sepharose beads
Transfer 250 μL of High Performance Glutathione Sepharose beads to a 15 mL conical centrifuge tube.
Add 6 mL of cold PBS, invert several times, centrifuge 2,755 × g for 3 minutes at 4°C, and remove supernatant. Repeat.
Increase volume to 1 mL with Lysis Buffer and resuspend beads by pipetting.
3.5.5 Binding and washing of RBPs immobilized on GST Sepharose beads
Add 1 mL of PBS equilibrated GST beads to lysed cells, mix by inverting several times, and rotate at 4°C for 2 hours.
Centrifuge RBPs bound to GST beads at 2,755 × g for 3 minutes at 4°C and remove supernatant.
Add 6 mL of Buffer D, invert 5 times, centrifuge at 3,700 RPB for 3 minutes at 4°C, and discard the supernatant.
Repeat the above step two times using Buffer C instead of Buffer D.
3.5.6 Elution of RBPs from GST Sepharose beads
Add 300 μL of Elution Buffer to RBPs bound to GST beads, mix by pipetting up and down, and rotate at 4°C for 2 hours.
Centrifuge at 3,220 × g for 3 min, transfer supernatant to new Eppendorf tubes being careful to avoid transferring GST beads.
Flash freeze eluted GST-tagged RBP with liquid nitrogen and store at −80°C.
Save residual protein (above beads) for estimating concentration and analysis by SDS-PAGE.
3.5.7 Estimating RBP concentration by Bradford assay
Dilute Protein Assay Dye Reagent Concentrate (Bradford solution) 5-fold, add 4 μL of eluted RBP to 1 mL of dilute Bradford solution, incubate at room temperature for 5 minutes, and measure OD595nm values spectrophotometrically (Medical devices, Spectra Max Plus 384 or alternative device).
Dilute BSA Standard to 750 μg/mL, 500 μg/mL, 250 μg/mL, 125 μg/mL, and 62.5 μg/mL.
Measure OD595nm values spectrophotometrically and calculate purified RBP concentration using a standard curve.
3.5.8 Verification of RBP integrity and concentration
Transfer 2.5 μg of purified RBP (based on the Bradford assay estimate) and increase volume to 20 μL with water, add 5 μL of 5× SDS Loading Buffer, mix, heat at 95°C for 5 minutes, and load 10 μL into wells of a SDS-PAGE gel (stacking gel-4%, separation gel-10%)
Also, load appropriate amounts of BSA standards and 7.5 μL of PageRuler Prestained Protein Ladder.
Electrophorese samples at 190V for 35 minutes using a Bio-Rad Mini-Protean III system.
Stain gel with 1% Coomassie Blue solution for 40 minutes and destain with 100 mL of destaining solution.
Assess protein size and purity and adjust RBP concentration based on visual inspection of gel.
3.6 RNA pulldown assay
During RNAcompete assays, an RNA-binding pulldown experiment is used to capture RNA bound by an RBP of interest (Figure 1). Here, an epitope-tagged RBP is incubated with a 75-fold excess of RNA pool in the presence of a high affinity matrix. Iterative washing removes unbound RNA whereas bound RNAs are eluted, purified, directly labeled with either Cy3 or Cy5, and hybridized to Agilent custom 244K arrays. Note: as identical microarrays are used, a single microarray design is sufficient for both dsDNA pool generation and microarray hybridization experiments. Although our RNAcompete pipeline utilizes GST-tagged RBPs, in principle, other epitope tags could be used.
3.6.1 Preparation of GST-Sepharose beads
For each RNAcompete assay, prepare 10 μL of GST-Sepharose 4B beads. Increase scale as required.
Thoroughly mix GST beads and pipette 10 μL into an Eppendorf tube.
Add 100 μL PBS, invert 5X, centrifuge at 2,000 × g (4°C) for 1 minute. Repeat twice.
Remove supernatant (leave ~15 μL of liquid on beads to avoid sucking up beads)
Add 100 μL of Binding Buffer, invert 5x, spin at 2,000 × g (4°C) for 1 minute. Repeat once.
Resuspend beads, with Binding Buffer, to a final volume of 40 μL.
3.6.2 RNA-binding reaction
The values listed below should be used as a guideline as volumes will vary based on the concentration of the purified RBPs and RNA pool.
Pipette 930 μL of Binding Buffer into a new Eppendorf.
Add 40 μL of washed GST-Sepharose 4B beads, 20 pmoles of GST-tagged RBP adjusted to a volume of 20 μL with Binding Buffer, and 10 μL of treated RNA (18.2 ug).
Rotate at 4°C for 30 minutes.
Spin sample(s) at 2,000 g for 1 minute and remove supernatant.
To remove unbound RNA: add 500 μL of Binding Buffer, invert 5 times, centrifuge at 2,000 × g (4°C) for 1 minute, and remove supernatant.
Repeat washing steps 3 times.
3.6.3 Elution of Bound RNA
Add 200 μL of STE Buffer, heat at 95°C for 2 minutes, and centrifuge at maximum speed for 1 minute.
Transfer 180 μL of supernatant into a new Eppendorf tube.
Add 20 μL of 3M NaOAc pH 5.2, 200 μL of PCI, vortex vigorously for 10 seconds, and centrifuge at maximum speed for 2 minutes.
Transfer 180 μL of aqueous phase to a new Eppendorf tube.
-Add 2.0 μL of glycogen and 500 μL ethanol, invert 5 times to mix, precipitate RNA as described for DNA in section 3.2.6, and resuspend RNA pellet with 17.5 μL of H2O.
3.7 Microarray analysis
RBP-specific pulldown RNA are identified using microarray analysis and relative RNA-binding levels are quantified based on fluorescence intensity. To do this, we directly label pulldown RNA with either Cy3 or Cy5 fluorescent dyes, hybridize the labeled RNA to a custom designed Agilent 244K microarrays, and measure the fluorescence signal using an Axon Genepix 4000B Microarray Scanner. In principle, a variety of scanners and image analysis software can be used to extract data from microarray slides.
3.7.1 Labeling RNA with Fluorescent Dyes
Pulldown RNA is directly labeled with either Cy3 or Cy5 using Kreatech ULS Labeling Kit for Agilent Arrays. Note: it is important to keep dyes, reactions, and labeled RNA in the dark as Cy3 and Cy5 are light sensitive.
Assemble RNAcompete pulldown reaction: 17.5 μL Pulldown RNA (all), 0.5 μL Cy5 dye, 2 μL 10× Kreatech Labeling Buffer.
Assemble RNA pool reaction: 0.5 μL RNA pool (0.9 μg), 17 μL H2O, 0.5 μL Cy3 dye, 2 μL 10× Kreatech Labeling Buffer.
Incubate at 85°C for 15 minutes.
To each labeling reaction, add 160 μL H2O, 20 μL 3M NaOAc (pH 5.2), 500 μL ethanol, and 2 μL of glycogen.
Precipitate RNA as described for DNA in section 3.2.5 and resuspend RNA pellets with 8.0 μL of H2O.
3.7.2 Hybridizing labeled RNA to microarrays
For small-scale experiments, each microarray is hybridized with 2 samples—the first sample is Cy3-labeled reference RNA pool and the second is Cy5-labeled RNA from an RNAcompete pulldown experiment [18]. For large-scale experiments, microarrays can be hybridized with Cy3- and Cy5-labeled RNA from two RNAcompete pulldown experiments when one or more reference RNA pool hybridization(s) have already been performed.
-Assemble Hybridization Mixture: Add 8.0 μL Cy3 labeled RNA pool (0.9 μg), 8.0 μL Cy5 labeled RNA from an RNAcompete experiment, 105 μL of Agilent Hybridization Buffer.
Heat Salmon Sperm DNA at 98°C for 10 minutes, centrifuge briefly, and store on ice.
Heat hybridization sample mixtures at 65°C for 5 minutes and centrifuge at maximum speed for 1 minute.
Add 4 μL of denatured salmon sperm DNA to each hybridization sample mixture.
Using a Tecan HS4800 Pro Hybridization Workstation, wash Agilent 244K microarrays with Pre-hybridization Buffer for 30 seconds (at room temperature), load 120 μL of sample into the microarray sample loading slot, hybridize samples for 20 hours at 42°C, wash for 30 seconds in Hybe Wash Buffer #1, and 30 seconds in Hybe Wash Buffer #2.
If a Tecan or comparable microarray hybridization workstation is not available, Cy3- and Cy5-labeled RNA samples can be pipetted into a Gasket Slide after which an Agilent 244K Microarray is placed on the Gasket Slide (probe side down) and then into a Microarray Hybridization Chamber (see section 3.2.1), and samples are hybridized (using conditions stated above) in a Hybridization Incubator with rotation (setting 8). Subsequent washing steps should be performed manually in 50 mL conical centrifuge tubes.
3.7.3 Scanning microarray slides and extracting intensity data
Microarrays are placed probe side down in an Axon Genepix 4000B Microarray Scanner (or alternative), scanned at 5 micron resolution with PMT values in the range 400–560 for Cy3 and 480–600 for Cy5 (fluorescent intensities should be below saturation; see section 3.8.1), and saved as multi-channel TIFF images.
Fluorescence signals captured in TIFF images are quantified using Imagene software version 3.0 and aberrant spots are manually flagged as “poor quality”. Presumably, other image processing software with similar functions and settings would perform comparably.
The raw data is saved as a text file for computational analysis (see section 3.8).
3.8. RNAcompete Data Analysis
3.8.1. Data pre-processing and normalization
Our normalization procedure corrects for array-level variation in the distribution in probe intensities due to, for example, differences in laser power or background intensity. We use affine transforms (in other words, rescaling and subtracting an offset) when normalizing in order to preserve the linear relationship between the RNA oligo concentration in the pulldown and the normalized probe intensity. So long as this relationship remains linear, our assay stays quantitative.
First, when scanning the arrays, we set the laser power to minimize saturation while maintaining as high a dynamic range of the probe intensity distribution as possible. Saturation would destroy the desired linear relationship. After quantifying the probe intensities, we flag, through visual inspection, probes affected by spatial trends or image analysis artifacts. Flagged probe intensities are discarded and not used in any further analysis.
Next, we combine RNAcompete microarray data into batches which use the same initial RNA pool, because our probe-level normalization corrects for initial RNA pool concentration which can vary between batches. Each batch was represented as a matrix where rows correspond to probes and columns to pulldown intensities of each RBP profiled. Note that the red and green channels of the array are treated as separate “one colour” hybridizations.
Array-level normalization begins by applying an affine transformation to each column (i.e. array) separately so that the median and inter-quartile range (IQR) of each column was equal to the median of the column medians and the median of the column IQRs, respectively. This normalization is intended to make probe intensities comparable between arrays. Then, we perform a robust z-transform for each row in which we subtracted the row median from each element in the row, and then divide by a robust estimate of the standard deviation, which is set equal to 1.4826 times the median absolute deviation of the row. This normalization is intended to correct for probe-specific scaling differences and for differences in the initial concentration of the corresponding RNA. In our original version of RNAcompete [18], we attempted to measure RNA concentration in the initial pool; but we found that a reference-free approach (used in [3]) provided better validation rates on our positive controls and more reproducible 7-mers scores.
We perform a final array-specific normalization by doing a robust z-transform on the column. After this, the median normalized probe intensity for an RNA oligo is zero, and each probe’s score is expressed in terms of units of standard deviation above this level. Because of our reference-free approach, we recommend that the computational analyses detailed here be followed when “batches” (>30) of RNAcompete experiments that use the same RNA pool are available. If experiments are performed at a smaller scale, then each RNA pulldown should be paired with a measured of the pool on the same array, and the alternative data analysis pipeline reported in Ray et al., 2009 ([18]) should be used.
3.8.2. Calculation of 7-mer scores
For most analyses of RNAcompete data, we convert the initial probe data to 7-mer scores (typically Z-scores). Anecdotally, the highest-scoring 7-mers reflect the known binding sites for well-studied proteins. 7-mers are relatively specific (i.e. can discriminate individual sites within a typical mRNA) and are well represented in the RNAcompete pool. Each 7-mer appears in at least 155 sequences in each of Set A and B, in a variety of diverse sequence contexts. 7-mers scores are averaged over all of these contexts thus minimizing the impact of any single one.
To derive 7-mer scores, normalized probe intensities less than zero are set to zero, in order to reduce the impact of technical variation. 7-mer scores are then set equal to the trimmed mean of the intensities of all probes containing that 7-mer, where the trimming removes the highest and lowest 5% intensities. Then, a Z-score transformation on the 7-mer scores is performed to generate our final “Z-score” values.
3.8.3. Derivation of “top-10” 7-mer motifs
We initially evaluated several strategies for deriving motifs from the RNAcompete data, and concluded that a simple 7-mer alignment provides a surprisingly good overall outcome (outlined in [3]). First, the ten 7-mers with the highest Z-scores are aligned using pairwise gapless alignments (i.e., shifting the 7-mers by all possible offsets and counting the number of matching bases), and a pairwise alignment score for each pair of 7-mers is then calculated from the number of matching bases. An overall alignment score is then calculated for each 7-mer by summing the alignment scores to each other 7-mer and the 7-mer with the highest overall alignment score is selected as the seed 7-mer. Then, all other 7-mers were aligned to the seed 7-mer by maximizing their pairwise alignment scores.
Based on this alignment, a position Z-score matrix was created by summing the Z-scores of the 7-mers that contained the given base at that position. Gaps at the 3′ and 5′ ends are scored by dividing that 7-mer’s Z-score evenly between all four bases. Zeroes in the Z-score matrix are replaced by a pseudocount value of one. The matrix is then trimmed by removing positions for which the number of non-gap rows is ≤ 50% of the maximum number of non-gap rows across all positions. Finally, the Z-score matrix is converted into a PFM by dividing each element by the sum of Z-score for that position. Logos are generated using the LogoGenerator tool, which is part of the REDUCE suite (https://systemsbiology.columbia.edu/reduce-suite) [30]. Scripts for normalizing RNAcompete microarray data, calculating 7-mer Z-scores and motif generation are now publicly available (https://github.com/morrislab/RNAcompete). The computational pipeline described here can be adjusted using command line parameters to identify k-mers 4–8 nucleotides in length. K-mers longer than 8 nts are not represented in a sufficient number of probes to provide accurate estimates of their specificity.
3.8.4 Evaluating RNAcompete experiments
During RNAcompete experiments we can visually assess the extent of RNA binding during the Cy3/Cy5 labeling step by inspecting the colour of the precipitated RNA pellet and when scanning microarrays by measuring fluorescence of microarray spots. In addition, we have developed several criteria to identify successful RNAcompete experiments where we have correctly recovered the sequence specificity of an RBP:
Selectively: High Z-scores for some 7-mers – most successful experiments have maximum Z-scores > 5 – and a clear separation between these Z-scores and the distribution of other Z-scores.
-
Consistency between Set A and B: Consistency in the 7-mer Z-scores distributions including:
High, linear correlation between Set A and Set B 7-mer Z-scores,
Overlap in the 7-mers with the top 10 Z-scores from each set.
-
Interpretabilty: The presence of a clear, low entropy motif in the vast majority of the top 10 7-mers, this includes::
A clear and consistent low entropy motif among the top 10 7-mers from Set A and B and the top 10 7-mers from both Set A and B.
The absence of confounding “frequent flyer” 7-mers in the top 10 7-mers (e.g. homopolymers or simple repeats, which could reflect preferences for base content), or more complex motifs that are obtained from multiple unrelated protein in the same batch. We reason that these frequent flyers could reflect unanticipated biases in the experimental procedures. Poly-G is a common frequent flyer and the significance of this observation is not yet known.
In some cases, an RBP will bind two related motifs that differ by a gap [31]; these cases can be identified by manual inspection.
-
Reproducibility: If an RBP satisfies most, but not all, of the top three criteria, we will still deem it successful in some cases if either:
we perform another replicate and generate similar results
its implied sequence specificity closely matches that of highly related RBPs.
Currently, we apply these criteria through expert curation by group that includes the corresponding and first author(s) of the manuscripts. We curate the RNAcompete experiments in batches in order to help identify the frequent flier 7-mers [32]. Additionally, we use CISBP-RNA to survey new motifs by identifying identical or similar motifs corresponding to previously analyzed RBPs. This can be done by using "Motif Scan" in the "Tools" section of the CISBP-RNA database, http://cisbp-rna.ccbr.utoronto.ca [3].
3.8.5 Troubleshooting RNAcompete experiments
During ssDNA pool generation, if full coverage of Cy3 signal following dsDNA linker ligation is not observed (excluding Agilent control spots), the process should be performed using higher amounts of T4 DNA polymerase and T4 DNA ligase, to improve double-stranding and linker ligation reactions. Also, during the ligation step, rotation is highly recommended, to improve ligation efficiency.
If linker cleavage by BspQI is incomplete, it is generally helpful to re-purify the DNA pool using PCI extraction and ethanol precipitation (to remove trace contaminants that potentially inhibit restriction digestion) and re-digest with excess enzyme. This process can be repeated until the digestion of the DNA pool is essentially complete.
During RNA pool synthesis, if there is low RNA yield, it is important to increase or decrease the dsDNA template amounts and incubation times to maximize RNA yield. Supplementing the transcription reaction with additional T7 RNA polymerase can also significantly increase RNA levels.
If an RBP is not binding the RNA pool well (as observed by visual inspection of dye labeling or scanned microarray images), the binding conditions could be altered by changing buffer, temperature, or other variables and the new RNA pulldown yields can be assessed using spectrophotometric analysis.
If positive controls do not yield expected results, but the experiments otherwise appear to be working, then the data processing should be examined. In general, the normalization and QC procedures are more effective with larger batches of experiments done in parallel.
3.8.6 Evaluation of RNAcompete-derived CNBP motif in Drosophila CG3800 CLIP-Seq data
To evaluate the RNAcompete-derived motif for human CNBP, we analyzed CLIP-seq data for Drosophila ortholog CG3800 [17]. We used the pre-processed alignments of CG3800 and matched input for our analysis (see Methods section in [17]). Uniquely mapping reads were extracted from the BAM files using SAMtools [33]. These reads were then clustered using Piranha [34] and setting the bin size to be 100 nt. Replicates for CG3800 and input were independently combined by retaining only the overlapping cluster sites using BEDtools [35]. The input clusters were further filtered by removing regions that overlap with CG3800. Sequences for each cluster were extracted from the BDGP6 Drosophila melanogaster genome (Ensembl v81). Next, each sequence S was scanned for the human CNBP motif using the RNAcompete-derived position frequency matrix (PFM) and scored by log odds using the Bio.motifs module in Biopython [36]. The log odds score for a subsequence in S is defined as:
where pij is the probability of observing nucleotide j at position i of motif M, and qj is the background probability of nucleotide j. The background nucleotide frequencies were computed from the CG3800 and Input sequences. Each sequence was then scored by taking the sum of scores using LogSumExp (LSE):
where Lk is the log odds score at position k in sequence S, L* is the maximum log odds score in S, and W is the length of S subtracted by the length of M.
3.8.5 Evaluating the human CNBP motif against 5-mers enriched in CG3800 CLIP data
As a comparison, we also scanned for the occurrence of the top five 5-mers enriched in CG3800 CLIP (GAGGA, AGGAG, GGAGG, GAAGA, and AGAAG) as reported in [17]. We scored clusters by computing the frequency of that 5-mer (single 5-mer models) or of any of the 5-mers (combined 5-mer model).
Next, we evaluated the performance of the above motif models (RNAcompete motif; single 5-mers; and combined 5-mers) at predicting the CG3800 CLIP 100 nt clusters (positive set) compared to 100 nt clusters that appear only in the input (and do not overlap with CG3800 clusters) (negative set). For the RNAcompete motifs, we evaluated the ability of the LSE scores to distinguish positive and negative clusters. For the single and combined 5-mers, the motif frequencies were used. The R package ROCR [37] was used to produce Receiver Operating Characteristic (ROC) curves and compute the area under the curve of the ROC (AUC-ROC).
4 Results/Discussion
We used RNAcompete to examine the sequence preferences of human ucRBPs NUDT21 and CNBP. Both proteins were recently identified in proteomic mRNA-binding screens [15, 16], lack canonical RBDs and have other existing RNA-binding against which to compare [10, 17]. NUDT21 is 227 amino acid protein that binds UGUA sequences typically located 40–50 nucleotides upstream of cleavage and polyadenylation signals in pre-mRNA and is an essential component of the 3′ pre-mRNA cleavage and polyadenylation machinery [38]; thus, it serves as a positive control. The Nudix hydrolase domain and flanking N-terminal region form RNA-protein contacts crucial for the recognition of these UGUA sequences [10]. CNBP, the human ortholog of Drosophila CG3800 (74% similarity at the amino acid level), is a 177 amino acid protein that contains 7 CCHC-type zinc finger domains and an RGG box located between zinc fingers 1 and 2 [39]. This protein binds both single-stranded DNA and RNA and has been suggested to remodel and stabilize nucleic acid secondary structure [40]. Studies in CNBP have also reported transcription factor activity [40], linkage to myotonic dystrophy [41], and regulatory activity in cardiomyocytes differentiation through binding to a specific G-rich sequences in Braveheart long noncoding RNA [42].
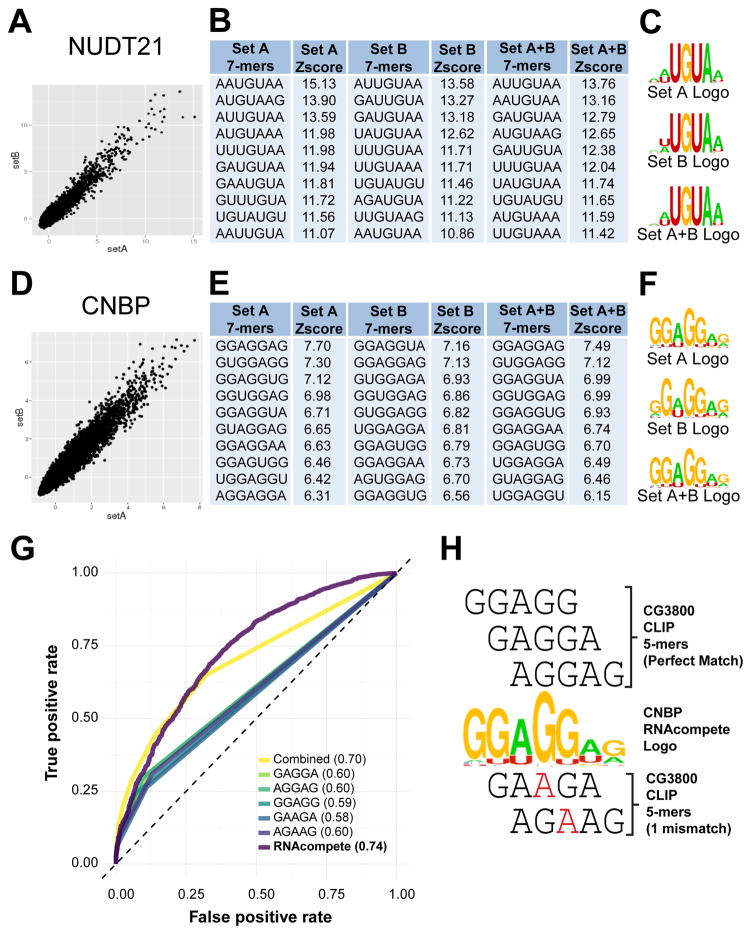
RNAcompete analysis of full-length GST-tagged NUDT21 and CNBP purified from E.coli revealed strong relative preference for individual 7-mers, quantified as Z-scores (Figures 4A and 4D displays the top 10 7-mers and Supplementary Table 1 contains the full 7-mer profiles for NUDT21 and CNBP). For both NUDT21 and CNBP, individual 7-mers (Figures 4B and 4E) and corresponding logos (Figures 4C and 4F) from RNA Sets A and B are highly similar, and therefore, we categorize these as “successful” experiments. Our RNAcompete-based data for NUDT21 demonstrates a clear preference for a core UGUA sequence, entirely consistent with its previously reported binding site in sequences located upstream of the cleavage and polyadenylation signal in the 3′-UTR of pre-mRNA [10, 43, 44]. The RNA-binding specificity of human CNBP has not been previously characterized. As noted above, however, in vivo CLIP-tags associated with the Drosophila CNBP ortholog, CG3800—which contains 6 instead of 7 CCHC-type zinc fingers [39]—are enriched for GGAGG, AGGAG, GAAGA, GAGGA, AGAAG 5-mers [17]. Our in vitro RNAcompete data for human CNBP identifies sequence preferences that contain or are highly related to the CG3800 5-mers listed above (the 5 top-scoring 7-mers are GGAGGAG, GUGGAGG, GGAGGUA, GGUGGAG, and GGAGGUG, Figure 4E). The high degree of overlap is illustrated in Figure 4H where it is clear that most CG3800 5-mers are contained in different registers within the CNBP RNAcompete-derived motif. Moreover, our human CNBP RNAcompete-derived motif displays good predictive capacity for CG3800 CLIP targets, outperforming both individual and combined 5-mers derived from the same dataset (Figure 4G).
Figure 4.
RNAcompete analysis of ucRBPs NUDT21 and CNBP. A) and D) Scatterplots show correlation between 7-mers Z-scores from Set A and Set B sequences. B) and E) Top ten 7-mers and corresponding Z-scores for Set A, Set B, and the average of Set A and Set B (Set A+B) are indicated. C) and F) Logos derived from the top 10 7-mers shown in panels B) and E). G) Analysis of true and false positive rates using AUC-ROC analysis for individual CG3800 CLIP-derived 5-mer, combined 5-mer (representing simultaneous analysis of all 5 CG3800 5-mers), and human RNAcompete PFM modes, in CG3800 CLIP data, are shown. The AUC-ROC values are indicated in parentheses. H) Manual alignment of RNAcompete-derived logo for human CNBP with CLIP-derived 5-mers for CG3800 are labeled, with nucleotides in red representing mismatches between RNAcompete and CLIP motifs.
In this study, we use the term “unconventional” to describe RBPs lacking a well-characterized RBD. Several terms have been previously used to describe these RBPs including "atypical" [45], "noncanonical" [46], “nonprofessional" [47], and "enigmatic" [48]. These terms suggest that ucRBPs are rare, or that their RNA-binding function is secondary, but recent observations suggests that ucRBPs are quite common. We also note that “atypical” and “noncanonical” have confusing abbreviations: atRBPs would denote RBPs from Arabidopsis thaliana, while ncRBPs would seem to bind noncoding RNAs. A more recent term, “EnigmRBPs” [2], does not capture instances where ucRBPs have established roles in RNA biology (e.g. Aconitase and NUDT21). As such, we propose that the term “ucRBPs” for proteins that directly bind RNA but lack a well-characterized RBD.
5. Conclusions
In this report, we present a detailed description of the experimental and computational methodologies encompassed by the RNAcompete system and show the value of RNAcompete for the analysis of ucRBPs. Given the expanding repertoire of both conventional RBPs and ucRBPs; large-scale characterization of RNA-binding specificity is critical. RNAcompete’s simplicity, scalability, and labour/cost-effectiveness make it an important tool for analyzing the RNA-binding preferences of proteins and facilitating the cellular and physiological characterization of RBP function.
Supplementary Material
Acknowledgments
We would like to thank Hans-Hermann Wessels and Markus Landthaler for providing the CG3800 CLIP-seq dataset alignment files. This work was supported by the National Institutes of Health (grant number R01HG008613) to TRH, QDM, Jack Greenblatt, and Ben Blencowe, and by the Canadian Institute for Health Research (MOP-125894) to QDM and TRH. KCH was partially supported by an Ontario Graduate Scholarship and a CIHR Frederick Banting and Charles Best Canada Graduate Scholarship
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nature reviews Genetics. 2014;15(12):829–45. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, Schwarzl T, Curk T, Foehr S, Huber W, Krijgsveld J, Hentze MW. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nature communications. 2015;6:10127. doi: 10.1038/ncomms10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–7. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic acids research. 2011;39(Database issue):D301–8. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nature reviews. Molecular cell biology. 2007;8(6):479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook KB, Hughes TR, Morris QD. High-throughput characterization of protein-RNA interactions. Briefings in functional genomics. 2015;14(1):74–89. doi: 10.1093/bfgp/elu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daubner GM, Clery A, Allain FH. RRM-RNA recognition: NMR or crystallography...and new findings. Curr Opin Struct Biol. 2013;23(1):100–8. doi: 10.1016/j.sbi.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic acids research. 2006;34(17):4943–59. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murn J, Teplova M, Zarnack K, Shi Y, Patel DJ. Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt. Nature structural & molecular biology. 2016;23(1):16–23. doi: 10.1038/nsmb.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Q, Gilmartin GM, Doublie S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(22):10062–7. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ZF, Whitfield ML, Ingledue TC, 3rd, Dominski Z, Marzluff WF. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes & development. 1996;10(23):3028–40. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 12.Tan D, Marzluff WF, Dominski Z, Tong L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science. 2013;339(6117):318–21. doi: 10.1126/science.1228705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laver JD, Li X, Ray D, Cook KB, Hahn NA, Nabeel-Shah S, Kekis M, Luo H, Marsolais AJ, Fung KY, Hughes TR, Westwood JT, Sidhu SS, Morris Q, Lipshitz HD, Smibert CA. Brain tumor is a sequence-specific RNA-binding protein that directs maternal mRNA clearance during the Drosophila maternal-to-zygotic transition. Genome Biol. 2015;16:94. doi: 10.1186/s13059-015-0659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, Hennig J, Cook KB, Morris Q, Hughes TR, Engelmann JC, Krahn MP, Meister G. The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain-Tumor-Mediated Gene Regulation. Cell reports. 2015;13(6):1206–20. doi: 10.1016/j.celrep.2015.09.068. [DOI] [PubMed] [Google Scholar]
- 15.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 16.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Molecular cell. 2012;46(5):674–90. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Wessels HH, Imami K, Baltz AG, Kolinski M, Beldovskaya A, Selbach M, Small S, Ohler U, Landthaler M. The mRNA-bound proteome of the early fly embryo. Genome research. 2016;26(7):1000–9. doi: 10.1101/gr.200386.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nature biotechnology. 2009;27(7):667–70. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 19.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 20.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–5. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 21.Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Molecular cell. 2014;54(5):887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell reports. 2012;1(5):570–81. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buenrostro JD, Araya CL, Chircus LM, Layton CJ, Chang HY, Snyder MP, Greenleaf WJ. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nature biotechnology. 2014;32(6):562–8. doi: 10.1038/nbt.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tome JM, Ozer A, Pagano JM, Gheba D, Schroth GP, Lis JT. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat Methods. 2014;11(6):683–8. doi: 10.1038/nmeth.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin L, Meier M, Lyons SM, Sit RV, Marzluff WF, Quake SR, Chang HY. Systematic reconstruction of RNA functional motifs with high-throughput microfluidics. Nat Methods. 2012;9(12):1192–4. doi: 10.1038/nmeth.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nature biotechnology. 2006;24(11):1429–35. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman TM, Wang G, Huang F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic acids research. 2004;32(1):e14. doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, Konig J, Ule J. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65(3):274–87. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roven C, Bussemaker HJ. REDUCE: An online tool for inferring cis-regulatory elements and transcriptional module activities from microarray data. Nucleic acids research. 2003;31(13):3487–90. doi: 10.1093/nar/gkg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith SA, Ray D, Cook KB, Mallory MJ, Hughes TR, Lynch KW. Paralogs hnRNP L and hnRNP LL exhibit overlapping but distinct RNA binding constraints. PloS one. 2013;8(11):e80701. doi: 10.1371/journal.pone.0080701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson IJ. Mouse genomics: making sense of the sequence. Curr Biol. 2001;11(8):R311–4. doi: 10.1016/s0960-9822(01)00172-5. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin RS. Genome Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LO, Smith AD. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28(23):3013–20. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–3. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–1. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 38.Hardy JG, Norbury CJ. Cleavage factor Im (CFIm) as a regulator of alternative polyadenylation. Biochem Soc Trans. 2016;44(4):1051–7. doi: 10.1042/BST20160078. [DOI] [PubMed] [Google Scholar]
- 39.Antonucci L, D’Amico D, Di Magno L, Coni S, Di Marcotullio L, Cardinali B, Gulino A, Ciapponi L, Canettieri G. CNBP regulates wing development in Drosophila melanogaster by promoting IRES-dependent translation of dMyc. Cell cycle. 2014;13(3):434–9. doi: 10.4161/cc.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armas P, Aguero TH, Borgognone M, Aybar MJ, Calcaterra NB. Dissecting CNBP, a zinc-finger protein required for neural crest development, in its structural and functional domains. Journal of molecular biology. 2008;382(4):1043–56. doi: 10.1016/j.jmb.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 41.Meyer A, Lannes B, Carapito R, Bahram S, Echaniz-Laguna A, Geny B, Sibilia J, Gottenberg JE. Eosinophilic myositis as first manifestation in a patient with type 2 myotonic dystrophy CCTG expansion mutation and rheumatoid arthritis. Neuromuscular disorders : NMD. 2015;25(2):149–52. doi: 10.1016/j.nmd.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 42.Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, Boyer LA. A G-Rich Motif in the lncRNA Braveheart Interacts with a Zinc-Finger Transcription Factor to Specify the Cardiovascular Lineage. Molecular cell. 2016 doi: 10.1016/j.molcel.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. Rna. 2005;11(10):1485–93. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Molecular cell. 2003;12(6):1467–76. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 45.Kenan DJ, Query CC, Keene JD. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16(6):214–20. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 46.Mili S, Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Molecular and cellular biology. 2003;23(14):4972–82. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley interdisciplinary reviews. RNA. 2010;1(2):266–86. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myllykoski M, Kursula P. Expression, purification, and initial characterization of different domains of recombinant mouse 2′,3′-cyclic nucleotide 3′-phosphodiesterase, an enigmatic enzyme from the myelin sheath. BMC research notes. 2010;3:12. doi: 10.1186/1756-0500-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.