Abstract
Identification of in vivo direct RNA targets for RNA binding proteins (RBPs) provides critical insight into their regulatory activities and mechanisms. Recently, we described a methodology for enhanced crosslinking and immunoprecipitation followed by high-throughput sequencing (eCLIP-seq) using antibodies against endogenous RNA binding proteins. However, in many cases it is desirable to profile targets of an RNA binding protein for which an immunoprecipitation-grade antibody is lacking. Here we describe a scalable method for using CRISPR/Cas9-mediated homologous recombination to insert a peptide tag into the endogenous RNA binding protein locus. Further, we show that TAG-eCLIP performed using tag-specific antibodies can yield the same robust binding profiles after proper control normalization as eCLIP with antibodies against endogenous proteins. Finally, we note that antibodies against commonly used tags can immunoprecipitate significant amounts of antibody-specific RNA, emphasizing the need for paired controls alongside each experiment for normalization. TAG-eCLIP enables eCLIP profiling of new native proteins where no suitable antibody exists, expanding the RBP-RNA landscape.
Keywords: CLIP-seq, eCLIP-seq, RNA binding protein, CRISPR/Cas9, protein tagging
1.1 Introduction
Believed previously to be a mere intermediary between DNA and protein, RNA is becoming increasingly appreciated as subject to a variety of post-transcriptional processing steps prior to translation (1). Analogous to transcription factors and histones that interact with DNA, transcribed RNA is associated with RNA binding proteins (RBPs) which have numerous regulatory functions. These RBPs transport RNAs from the nucleus and throughout the cell, carry out splicing, regulate stabilization, degradation, and translation of RNAs, and form ribonucleoprotein complexes with non-coding RNAs to confer regulatory activity (1). Recent work indicates that there are likely over a thousand RBPs encoded in the human genome that play a wide range of developmental roles, and mutation or dysfunction of numerous RBPs have been linked to a wide variety of defects including neurodegenerative and autoimmune diseases (1–4).
For an RBP of interest, identifying its binding sites in vivo is a critical step towards understanding its functions at the molecular and physiological level. The development of microarray and high-throughput sequencing technologies rapidly led to the development of RNA Immunoprecipitation (RIP) and Crosslinking and Immunoprecipitation (CLIP) methods to profile RNA binding protein target sites transcriptome-wide (5). Initial RIP methods focused on profiling RBP targets at the transcript level, by pulling down an RBP and its bound RNA for quantification by microarray (6). Building upon this work, CLIP utilizes crosslinking (typically with UV irradiation) to covalently couple the RBP to its RNA targets. With this irreversible and stable linkage, CLIP allows stringent wash conditions and an RNA fragmentation step to bring target identification from the kilobase transcript-level to clusters that are less than a hundred bases in length (5). Further work improved crosslinking efficiency through incorporation of the photoactivatable nucleoside analog 4-thiouridine into RNAs during transcription in living cells (PAR-CLIP) (7), and iCLIP described altered library preparation steps to improve efficiency and enable identification of binding sites with single-nucleotide resolution (8). Recently, we developed an enhanced CLIP (eCLIP) method that builds upon these methods by dramatically improving the efficiency of converting immunoprecipitated RNA into an adapter-ligated and amplified sequencing library, enabling the incorporation of paired input samples to improve signal-to-noise in identifying true binding sites above common artifacts. The robust success of eCLIP enabled profiling of over one hundred RNA binding proteins in K562 and HepG2 cells, and has proven successful in a variety of other cell-types and tissues (9).
However, one major limitation for all RIP and CLIP methods is that they require antibodies for immunoprecipitation. Thus, to profile the targets of an RBP under study, one must first screen through expensive antibodies, oftentimes with irregular success and high levels of background. In many other cases no suitable commercially available antibody yet exists for the RBP of interest, thus requiring custom generation at high cost. To help address this concern, we recently performed a large-scale effort to identify antibodies that could successfully immunoprecipitate RBPs in K562 cells, identifying antibodies for 365 RBPs (10). Although this was highly successful, hundreds of RBPs remain without antibodies suitable for immunoprecipitation. Additionally, the concern that each antibody may have its own individual off-target or background interactions would be alleviated if all experiments were performed using the same antibody.
One common solution to the lack of suitable antibodies is to utilize peptide tags which already have high-quality, immunoprecipitation-grade antibodies. Most commonly, the protein of interest, flanked by either N- or C-terminal tags is exogenously expressed and the tag is used to immunoprecipitate the protein of interest along with its interactors (11). Numerous such tags exist, including the well-characterized V5 and FLAG tags, which have proven successful in a variety of experimental regimes (12, 13). However, over-expression of various DNA- or RNA-binding proteins has sometimes revealed amplified binding to the same targets and other times led to interactions with ectopic or low-affinity sites, complicating interpretation of large-scale over-expression experiments (14, 15).
The recent development of CRISPR technologies has made it possible to rapidly and successfully insert these tags into endogenous gene loci (16–18), which enables profiling of RBPs within their normal regulatory context. A recent method to perform endogenous tagging followed by ChIP-seq (CETCh-seq) demonstrated successful use of the CRISPR-Cas9 system to introduce a 3xFLAG tag at the 3′ end of transcription factors (19). Specifically, ChIP-seq using the FLAG tag yielded substantially similar binding site identification to parallel experiments performed with antibodies targeting native proteins, confirming this approach as a general scheme for profiling DNA binding proteins lacking antibodies.
Here, we describe a scalable methodology for performing and validating CRISPR-mediated tag insertion into RNA binding protein loci. Using two tags (V5 and FLAG), we show that TAG-eCLIP yields the same high-quality target identification as eCLIP with native antibodies. Furthermore, we characterize common non-specific background identified by anti-V5 and anti-FLAG antibodies in wild-type cells, which indicates that such TAG-eCLIP experiments require proper controls for robust analysis. These methods provide further improvements to simpler, more cost-effective RBP target identification in cases where high-quality antibodies do not currently exist.
1.2 Methods
1.2.1 Cloning of CRISPR/Cas9 sgRNA vectors
The 100nt sequence centered on the annotated stop codon was obtained for each desired transcript. sgRNA sequences targeting the 3′ end of the RBP of interest were identified using the Zhang lab CRISPR design tool (available at http://crispr.mit.edu). The sgRNA sequences that were closest to the stop codon, but had maximal score (minimal predicted off-targets), were selected. Two methods were tested for different RBPs: using a single double-strand nuclease Cas9 (pX330-U6-Chimeric_BB-CBh-hSpCas9; Addgene plasmid # 42230, pSpCas9(BB)-2A-GFP (PX458); Addgene plasmid # 48138 and pSpCas9(BB)-2A-Puro (PX459); Addgene plasmid # 62988 were a gift from Feng Zhang), or using a pair of single-strand nickase mutant Cas9 vectors (pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A); Addgene plasmid # 42335 was a gift from Feng Zhang). For nickase experiments, the pair of sgRNAs that flanked the stop codon with the highest combined score (fewest predicted off-targets) was chosen (Fig. 1B). Cloning was performed by gel extraction of the BbsI-cut backbone, and ligation with phosphorylated oligonucleotides, as previously described (16).
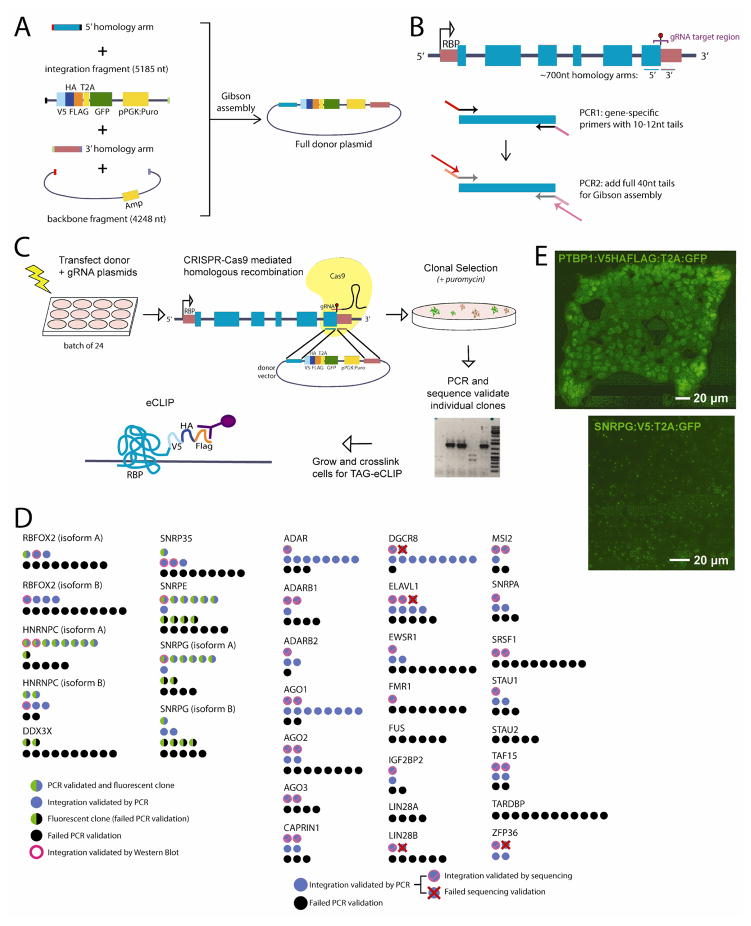
Figure 1. Generation of peptide-tagged RBP lines.
(A) Strategy to generate donor vectors for homologous recombination. Colored boxes indicate sequence features; small colored boxes at fragment ends indicate 40nt homology regions for Gibson assembly. (B) Strategy for two-stage PCR amplification of 5′ and 3′ donor arms flanking the annotated stop codon for an RBP. (C) Strategy for transfection, selection, and validation of proper integration of peptide tags. We found that this strategy could be performed in batches of 24 RBPs. (D) Success rate for validation for 29 RBPs. For the first batch, circles indicate the number of clones that did not validate by PCR (black), validated by PCR (blue) and showed GFP fluorescence (green) or both (green and blue), and confirmed by Western Blot (pink boundary). For the second batch, a subset of PCR validated clones (blue) were sequenced and either validated (pink checkmark) or did not validate (red X). (E) Fluorescence microscopy of validated lines for (left) PTBP1 and (right) SNRPG. As T2A cleavage is ~60–80% efficient, 20–40% of target protein is translationally fused with GFP, enabling visualization of sub-cellular localization.
1.2.2 Cloning of homology-directed repair (HDR) donor vectors
For chosen RBPs, the ~800nt regions immediately upstream (5′ homology arm) and downstream (3′ homology arm) of the stop codon were computationally identified. The forward primer for the 5′ arm and reverse primer for the 3′ arm were selected using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) to be ~700nt away from the stop codon. This 700-800nt homology arm size was chosen based on standard recommendations in the field (https://www.addgene.org/crispr/zhang/faq/). The reverse primer for the 5′ arm and forward primer for the 3′ arm were selected by starting at the base flanking the stop codon, and taking the smallest region (20–28 nt long) with a melting temperature > 57°C. Homology tails for Gibson assembly were added in two PCR steps. First, a short extension was added to the 5′ end of the gene-specific primers as follows (see Supplemental Table 1 for gene-specific primers used):
| PCR_5_F: | CGACGGCCAGTG - gene-specific primer |
| PCR_5_R: | GGCTTACCGAATTC - gene-specific primer (starts at base before stop codon) |
| PCR_3_F: | CTAGATCGGATCC - gene-specific primer (starts at base after stop codon) |
| PCR_3_R: | GCATGCAGTCGA - gene-specific primer |
The first PCR amplification was performed using Phusion polymerase (NEB) on human genomic DNA (gDNA) with 38 cycles of amplification, with 2% DMSO added to aid amplification. After agarose gel extraction (Qiagen) of the specific product, a second PCR was performed (NEB Q5; 6 cycles of amplification at 45°C followed by 6 cycles at 62°C) using the following primers to add full homology tails:
| 2ndPCR_5_L | GGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTG |
| 2ndPCR_5_R | CGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCGAATTC |
| 2ndPCR_3_L | TATCACGTAAGTAGAACATGAAATAACCTAGATCGGATCC |
| 2ndPCR_3_R | CTGCCTTGGGAAAAGCGCCTCCCCTACCCGCATGCAGTCGA |
This second PCR product was prepared for Gibson assembly by PCR cleanup kit (Qiagen).
To enable efficient screening of properly integrated clones, the HR130 backbone was obtained (System Biosciences). To insert either V5 or V5:HA:FLAG upstream of the T2A site at the EcoRI site, phosphorylated oligonucleotides were annealed and ligated with EcoRI-digested backbone, with insert sequences as below (brackets indicate flanking EcoRI overhang regions):
V5 [GAATTC]GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG[AAATTC]
V5:HA:FLAG [GAATTC]GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGAAATTCTACCCATACGATGTTCCAGATTACGCTGACTACAAAGACGATGACGACAAG[AAATTC]
To clone donor vectors, two backbone regions were obtained by restriction digestion of the HR130 backbone: a ~4200 nt backbone product from EcoRI+EcoRV+SalI (E/E/S) digestion containing origin of replication, bacterial resistance, and a diphtheria toxin cassette for negative -selection against random integrations, and a ~5500 nt ‘insert’ product from EcoRI + BamHI (E/B) digestion that contains the tags, GFP, and Puromycin positive selection marker. The two homology arm PCR products (5′ arm and 3′ arm) and digested backbone products (E/E/S and E/B) were added to a Gibson assembly reaction in 2:2:1:1 molar ratio, assembled at 50°C for one hour, and transformed and screened using standard methods. Proper homology arms were validated by sequencing with primers in the GFP cassette (CCACCAGCTCGAACTCCAC; for 5′ arm) and core insulator (GGGCTGTCCCTGATATCAAAC; for 3′ arm). The annotated plasmid map is available at https://benchling.com/s/scSbNllF and is provided as Supplemental Data 1.
1.2.3 Transfection and clonal selection
Donor vectors were linearized with a unique backbone-cutting restriction enzyme (one of AhdI, NdeI, PciI, or ScaI, depending on the homology arm sequence) and purified by phenol-chloroform extraction. HEK293T cells were seeded into 12-well plates with 200,000 cells in 1 mL standard media (DMEM + 10% FBS), grown for 24 hours, and then transfected using Lipofectamine 2000 with 750 ng of donor and either 750 ng of sgRNA plasmid (double-strand cutting version) or 500 ng of each of two sgRNA plasmids (dual nickase version). After 36 to 48 hours, cells in each well were seeded into a 10 cm plate in the presence of 1 μg/mL puromycin to select for integrants. Once sufficiently grown (typically after ~4–5 days), colonies derived from single cells were isolated into individual wells of 96-well plates, and grown for ~2 weeks until they reached sufficient density for PCR screening. GFP-positive colonies were preferentially selected, followed by the selection of random other colonies.
Genomic DNA was released from clonal isolates in 96-well format by detachment of cells (TrypLE, Thermo Fisher), transferring 1/3–1/2 of cells into 96-well PCR plates, and addition of 50 μl QuickExtract solution (Epicentre). Cells were pipette-mixed, incubated at 65°C for 6 minutes, mixed, and incubated at 95°C for 2 minutes. A portion of the lysates was used for PCR validation using a reverse primer in the 5′ end of GFP (CCACCAGCTCGAACTCCAC), and a gene-specific primer outside of the 5′ homology arm region used in the donor vector. Presence of a single PCR product of the correct size (typically ~800–1000nt) indicated potential successful integration. PCR products were then purified (Qiagen) and submitted for Sanger sequencing to confirm absence of insertions or deletions in the 3′ end of the RBP of interest, or western blot using anti-tag antibody was performed to validate presence of the expected RBP:tag fusion protein.
1.2.4 TAG-eCLIP experimental methods
eCLIP experiments were performed as previously described in a detailed standard operating procedure (9). Briefly, 107 cells were UV-crosslinked (254 nm, 400 mJ/cm2), lysed in 1 mL of 4°C eCLIP lysis buffer (50 mM TrisHCl pH 7.4, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 1:200 Protease Inhibitor Cocktail III (EMD Millipore)), incubated at 37°C for 5 min with 40 U of RNase I (Ambion) and 4 U Turbo DNase (Ambion), treated with 11 μL Murine RNase inhibitor (NEB), and clarified by centrifugation (4°C, 15kg for 15 min). Immunoprecipitation was performed at 4°C overnight using sheep anti-rabbit or anti-mouse IgG Dynabeads (ThermoFisher) precoupled with primary antibodies as follows: V5 (A190-120A lot 006, Bethyl), FLAG (F1804 lot SLBQ6349V, Sigma), HNRNPC (RN052PW lot 001, MBL), RBFOX2 (A300-864A lot 002, Bethyl), FMR1 (RN016P lot 001, MBL), LIN28B (A303-588A lot 001, Bethyl), DGCR8 (A302-468A lot 001, Bethyl), TAF15 (A300-307A lot 001, Bethyl), EWSR1 (A300-417A lot 001, Bethyl), and IGF2BP2 (RN008P lot 001, MBL).
After incubation, immunoprecipitation samples were magnetically separated and washed twice in high salt wash buffer (50 mM Tris-HCl pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, and 0.5% sodium deoxycholate) and twice in wash buffer (20 mM Tris HCl pH 7.4, 10 mM MgCl2, 0.2% Tween-20). Next, remaining 5′ phosphates were removed with FastAP (ThermoFisher) and 3′ phosphates of RNA fragments generated by RNase I digestion were removed with T4 PNK (NEB) at low pH in the absence of ATP. 3′ adapters were then ligated to RNA fragments with T4 RNA Ligase I (NEB), using optimized reaction conditions including 18% PEG 8000 and 0.3% DMSO. Adapters used for eCLIP and input libraries were as previously described (9). After one additional wash with high salt wash buffer and two with wash buffer, samples were run on 4–12% NuPAGE Novex Bis-Tris protein gels (ThermoFisher) and transferred to either PVDF (for chemiluminescent imaging) or nitrocellulose (for RNA extraction) membranes. For TAG-eCLIP experiments, all western blot imaging to validate successful immunoprecipitation was done using primary antibody against the native protein (including immunoprecipitations with anti-tag antibodies).
A range from protein size to 75 kDa above protein size was isolated, incubated first for 20 minutes at 37°C with 200 μL PK buffer (160 μL of 100 mM TrisHCl, pH 7.4, 50 mM NaCl, 10 mM EDTA plus 40 μL Proteinase K (NEB P8107S)), followed by 20 minutes at 37°C with 200 μL PK-Urea buffer (160 μL of 100 mM TrisHCl, pH 7.4, 50 mM NaCl, 10 mM EDTA, 7M Urea plus 40 μL Proteinase K (NEB P8107S), after which RNA was isolated using phenol-chloroform extraction followed by RNA Clean & Concentrator column cleanup (Zymo). RNA was reverse transcribed with AffinityScript (Agilent), treated with ExoSap-IT (Affymetrix) to remove excess oligonucleotides, and a DNA adapter was ligated to the 3′ end using T4 RNA Ligase I (NEB) in optimized reaction conditions including 22% PEG 8000. Libraries were PCR amplified with Q5 master mix (NEB) for 6–18 cycles (chosen by performing qPCR on the pre-amplified library). The 175–300nt fragment was size-selected by agarose gel electrophoresis and gel extracted (MinElute Gel Extraction, Qiagen). Libraries were quantitated and validated by Tapestation (Agilent), and sequenced on the Illumina HiSeq 4000 platform.
1.2.5 Processing of TAG-eCLIP sequencing data
Sequencing reads obtained for all datasets were processed as previously described, including adapter trimming, discarding of reads mapping to repetitive elements, identifying reads uniquely mapping to the human genome (hg19), removing PCR duplicate reads, initial cluster identification with the CLIPper algorithm, and normalization against paired size-matched inputs (9). Where described, tag-derived datasets were then additionally compared against tag immunoprecipitation in wild-type cells to identify tag-derived artifacts.
To compare read density within peak regions, one eCLIP dataset was first selected as a ‘pivot’ dataset. All CLIPper-identified clusters (regardless of enrichment above size-matched input) were then considered in both the pivot and comparison dataset (and their respective inputs) to determine fold-enrichment in each dataset. Correlation was determined across all clusters, with significance calculated by converting the Pearson’s correlation to p-value using a standard Student’s t distribution transformation in MATLAB.
To compare enrichment for TAG-eCLIP peaks relative to tag-only eCLIP in wild-type cells, the number of reads overlapping each CLIPper-identified cluster was counted for TAG-eCLIP IP and input, as well as tag-only IP and input samples, and two comparisons were performed: TAG-eCLIP IP versus input, and TAG-eCLIP IP versus tag-only IP. The lesser of the fold-enrichment and greater of p-values were taken as final enrichment values for analysis.
1.2.6 Data and code availability
eCLIP and TAG-eCLIP datasets have been deposited at the Gene Expression Omnibus (accession number GSE88722). Additional antibody validation information for RBP antibodies is available at the ENCODE portal (http://www.encodeproject.org). eCLIP data processing and analysis pipeline code has been publicly released (https://github.com/gpratt/gatk/releases/tag/2.3.2), and was previously described in additional detail (9).
1.3 Results & Discussion
1.3.1 Large-scale generation of donor and sgRNA plasmids for integration of peptide tags using CRISPR/Cas9
Enhanced crosslinking and immunoprecipitation followed by high-throughput sequencing (eCLIP-seq) enabled robust profiling of RNA binding proteins by utilizing antibodies against endogenous proteins to specifically pull down and isolate RBP-bound RNA. To address the limitation of requiring antibodies against each RBP of interest, we took advantage of peptide tags. There are multiple commonly used tags in molecular biology of various sizes and affinities, many of which have been shown to immunoprecipitate DNA and RNA binding proteins successfully. In order to minimize the impact the tag protein would have on endogenous RBP activity, we selected three small but widely used tags for further consideration: V5, FLAG, and HA. Our first validation experiments used only V5; later, we integrated all three tags together to enable side-by-side comparison of their immunoprecipitation success in the eCLIP protocol. These tags were followed by a fluorescent transcriptional reporter separated by a protein cleavage signal (T2A:GFP), enabling visual confirmation of successful integration (Fig. 1A). In addition, to aid in selection of recombined clones, our donor vector contained a puromycin resistance cassette. This resistance marker is under a separate PGK promoter, so that the knock-in line could be generated regardless of the endogenous expression level of the RBP. This consideration is particularly important for generating tagged lines for proteins with stress-, differentiation-, or other condition-specific expression.
Although there are now multiple options for incorporating a peptide tag into a protein of interest, we chose the CRISPR/Cas9 system as it enabled efficient integration of a tag into the endogenous gene locus, maintaining proper transcriptional regulation of the tagged peptide. Each desired endogenous knock-in requires two custom reagents with the CRISPR/Cas9 system: an sgRNA targeting Cas9 cleavage proximal to the target loci of interest, and a homology repair ‘donor’ containing the desired integration sequence flanked by ~700 nt 5′ and 3′ regions of homology (Fig. 1B). sgRNA targeting constructs were cloned using standard Gibson assembly as previously described (16). For each RBP, we determined whether we could design a pair of suitable guides flanking the stop codon. If so, we generated a pair of sgRNA vectors using the Cas9n (D10A) nickase-variant; if not, we generated a single sgRNA vector using the standard Cas9 double-strand cutter. We note that in later experiments we did not observe significant differences in efficiency or off-target integration between these two variants, suggesting that the single standard Cas9 vector is likely sufficient for future experiments.
Cloning of donor vectors and in particular the isolation of homology arm regions with tails for Gibson assembly of donor vectors involves trade-offs between cost and efficiency, particularly at large scale. For individual experiments, the pair of homology arms can now be ordered as synthesized gene products from a number of commercial sources; however, this was cost-prohibitive at large scale. Thus, we designed a PCR-based strategy to minimize the need for long (and expensive) synthetic DNA fragments. First, we designed PCR primers to amplify the desired ~700nt homology arms, with the 5′ arm ending at the base upstream of the stop codon and the 3′ arm beginning at the base downstream of the stop codon (Fig. 1B). We added short 10–12nt tails onto these first PCR primers, and amplified these regions off of genomic DNA using standard high-fidelity polymerase. Next, we performed a second round of PCR to add ~40nt tails to these PCR products for assembly with restriction digested backbone in a 4-way Gibson assembly reaction. We found this procedure to be highly efficient; out of 56 RBPs attempted, we were able to amplify homology arms for 54 (96%), most with one standard set of PCR conditions. After assembly we obtained sequence validated properly assembled vectors typically selecting only 2–4 clones for each, confirming the high efficiency and fidelity of this approach.
1.3.2 Cell line generation and validation
We selected 32 RBP isoforms (29 RBPs, plus 3 isoforms each for RBFOX2, HNRNPC, and SNRPG) for further experiments. We initially chose to linearize donor plasmids before transfection to reduce the time needed for antibiotic selection following transfection, although in later experiments we did not observe a benefit to this additional step. We found that the most efficient method of performing these experiments at scale was to transfect HEK293T cells in 12-well format, grow for 36–48 hours (to enable Cas9-mediated homology repair), and then split each well into a 10 cm plate with media supplemented with puromycin (Fig. 1C). This typically yielded < 50 puromycin-resistant colonies, which after an additional 4–5 days of growth were of sufficient size and sufficiently dispersed around the plate to be suitable for manual single-colony isolation into 96-well plates. We typically selected up to 12 puromycin-resistant clones for further validation, and found that this typically yielded at least one PCR validated clone (Fig. 1D). If robust GFP fluorescence was observed, we found that prioritizing GFP-positive clones could increase validation efficiency; however, in the more frequent case of low or no visible GFP signal due to low expression of single-copy integrations, colonies were chosen at random. Next, we used PCR screening to validate that the donor region had correctly integrated into the desired RBP loci. Out of 208 colonies isolated, 95 (46%) validated by PCR, confirming the high efficiency of obtaining properly targeted integrations using this approach (Fig. 1D). We note that the success rate was highly variable for different RBPs; while 10 out of 11 (91%) of clones for DGCR8 and 10 out of 12 (83%) for AGO1 validated by PCR, we were unable to validate any of 12 TARDBP or 6 FUS clones (Fig. 1D). To confirm in-frame integration into the proper loci, we further performed either western blot with anti-tag antibody or Sanger sequencing across the RBP:TAG:GFP junction. We observed that the majority of tested PCR-positive clones validated using these methods. The low fraction (typically <1%) of Puromycin-resistant cells suggests an extremely low probability that these lines contain a second non-target integration, which should be tested for stem cell models or disease studies.
It is possible to use the GFP visualization as secondary validation, as the fluorescent expression pattern is dependent on successful integration. As previously reported, we observed that the T2A self-cleavage site is only ~60–80% efficient, leading to 20–40% translational read-through expression of a full protein-GFP fusion (20). This allowed for visualization of subcellular localization consistent with previously studies for a number of RBPs, including PTBP1 (nuclear) and SNRPG (nuclear foci), enabling additional confirmation of proper RBP tagging and expression (Fig. 1E). These translational fusions were not typically observed in immunoprecipitation during CLIP, possibly due to inaccessibility of the tag (data not shown).
1.3.3 Validation of successful eCLIP with peptide tags (TAG-eCLIP)
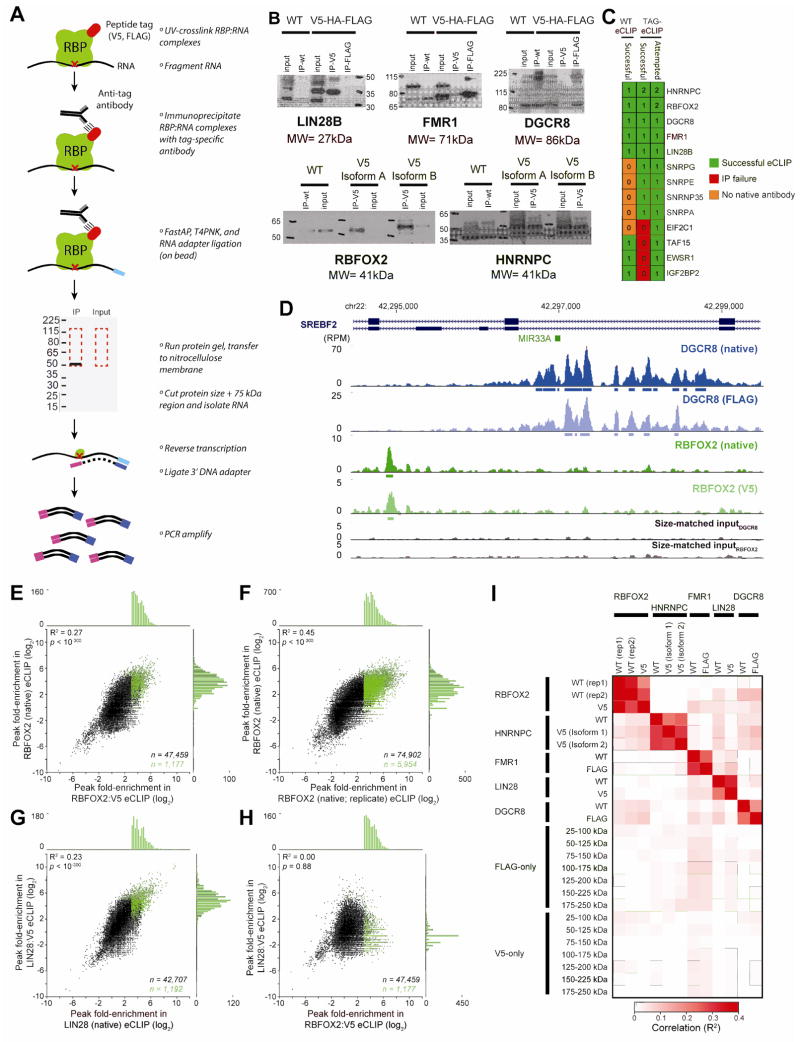
Next, we performed eCLIP experiments to test whether peptide-tagged RBPs were suitable for eCLIP, using our standard eCLIP methodology (Fig. 2A). We selected 15 tagged lines for 13 RBPs (including two annotated stop codons each for RBFOX2 and HNRNPC). Using anti-V5 and anti-FLAG antibodies, we observed successful immunoprecipitation for at least one of V5 or FLAG antibody in 11 out of 15 cases (Fig. 2B–C). Interestingly, we often observed highly variable immunoprecipitation between anti-V5 and anti-FLAG antibody despite the tags located proximal to each other, indicating that local protein context can significantly alter tag accessibility (Fig. 2B). We additionally noted that immunoprecipitation of HNRNPC:V5 with anti-V5 antibody co-immunoprecipitated wild-type HNRNPC, likely reflecting the known oligomerization of HNRNPC into tetramers (21).
Figure 2.
TAG-eCLIP compared to eCLIP of native proteins. (A) Schematic of TAG-eCLIP method (left), with individual steps detailed (right). (B) Western blots performed after the immunoprecipitation stage for native antibody eCLIP in wild-type (WT) cells, or anti-V5 or anti-FLAG antibody in endogenously tagged RBP lines, with protein size markers indicated in kDa. For RBFOX2 and HNRNPC, two annotated stop codons were targeted for independent tagging. (C) Numbers indicate the total number of wild-type RBP tagged lines in which eCLIP was attempted or successful in generating libraries. (D) Tracks show read density (in reads per million; RPM) for DGCR8 (native eCLIP and FLAG TAG-eCLIP) and RBFOX2 (native eCLIP and V5 TAG-eCLIP), with boxes underneath indicating peaks significantly enriched above size-matched input (below). (E–H) Points indicate fold-enrichment in eCLIP relative to size-matched input for peak regions called in one dataset (x-axis) in (E) RBFOX2 native eCLIP versus V5 TAG-eCLIP, (F) RBFOX2 native eCLIP biological replicates, (G) LIN28 native eCLIP versus V5 TAG-eCLIP, and (H) LIN28 V5 TAG-eCLIP versus RBFOX2 V5 TAG-eCLIP. All CLIPper-identified clusters are shown in black, with significantly enriched peaks relative to size-matched input indicated in green (with number of peaks indicated). Histograms above and to the right indicate the number of significantly enriched peaks in the indicated bin. Correlation is calculated across all points, with p-value calculated in MATLAB. (I) Heatmap reflects correlation as calculated in (E–H) for all pairwise comparisons. Each point reflects correlation in eCLIP fold-enrichment relative to input for clusters identified in the dataset on the x-axis. See Figure 3 for FLAG-only and V5-only eCLIP.
Next, we asked whether eCLIP with peptide tags (TAG-eCLIP) could recapitulate results obtained using native antibody immunoprecipitation of wild-type cells. We were able to obtain a commercially available antibody previously validated to immunoprecipitate the endogenous RBP for 8 out of the 11 tagged lines described above (10). In all eight cases, we were able to successfully immunoprecipitate the wild-type protein in HEK293T cells, albeit sometimes with altered efficiency relative to the tagged protein (Fig. 2B–C). We discarded three factors (TAF15, EWSR1, and IGF2BP2) for which immunoprecipitation was successful for wild-type but not tagged peptide, leaving five factors (HNRNPC, RBFOX2, DGCR8, FMR1, and LIN28B) with eCLIP libraries generated for both wild-type and tagged protein. Manual inspection of significantly enriched regions indicated highly similar signal between native protein and TAG-eCLIP (Fig. 2D). To consider reproducibility we first compared the number of significantly enriched peaks identified using both methods, using previously described cutoffs for significantly enriched peaks in order to limit analysis to a set of high-confidence peaks (9). RBFOX2 V5 TAG-eCLIP identified 1,177 peaks significantly enriched above paired input, on part with 1,911 identified in wild-type eCLIP. 5,977 CLIPper-identified clusters were depleted in CLIP relative to input, similar to false positive rates previously shown in eCLIP of RBFOX2 (9). Next, to quantitatively measure reproducibility we considered the correlation in peak-level read density relative to size-matched inputs across datasets. We observed that RBFOX2 showed significant correlation (R2 = 0.27; p < 10−300) between V5-tagged and native eCLIP (Fig. 2E). Although significant, this correlation was decreased from that observed for biological replicates of native RBFOX2 (R2 = 0.45; p < 10−300) (Fig. 2F), potentially due to TAG-eCLIP profiling only the subset of isoforms which share the tagged stop codon. Similarly, LIN28 also showed significant correlation between native and V5-TAG-eCLIP (R2 = 0.23; p < 10−300) (Fig. 2G). In contrast, V5-TAG-eCLIP of RBFOX2 and LIN28 showed little correlation (R2 = 0.00; p = 0.88) (Fig. 2H). Comparing all five RBPs, we observed this same pattern of high correlation between native and tagged RBP, with little correlation across RBPs regardless of tagged or wild-type status (Fig. 2I). Thus, these results confirm that TAG-eCLIP yields substantially similar results to native protein pulldown in wild-type cells.
1.3.4 Tag-specific concerns and normalization methods for TAG-eCLIP
These results confirmed that the replacement of native antibodies with peptide tags can generally yield high-quality eCLIP data for the RBPs profiled. Next, we asked whether there were tag-specific artifacts due to anti-tag antibody recognition of native peptides. We performed eCLIP with anti-FLAG, anti-V5, and rabbit IgG isotype control in wild-type K562 cells, size-selecting seven different 75 kDa size windows at the nitrocellulose membrane step (25–100, 50–125, 75–150, 100–175, 125–200, 150–225, and 175–250 kDa) (Fig. 3A). To compare library yield across experiments, for each library we used the measured library concentration and number of PCR cycles performed to calculate an extrapolated Ct (eCT), defined as the number of PCR cycles required to obtain 100 femtomoles of library (assuming 2-fold amplification with each cycle) (9). In 7 out of 7 V5 and 4 out of 7 FLAG size ranges in K562, we observed more than 2-fold greater library yield relative to IgG-only, with a 5.20-fold median increase (Fig. 3B). These results confirmed that both anti-V5 and anti-FLAG antibodies immunoprecipitate significant amounts of RNA even in wild-type cells.
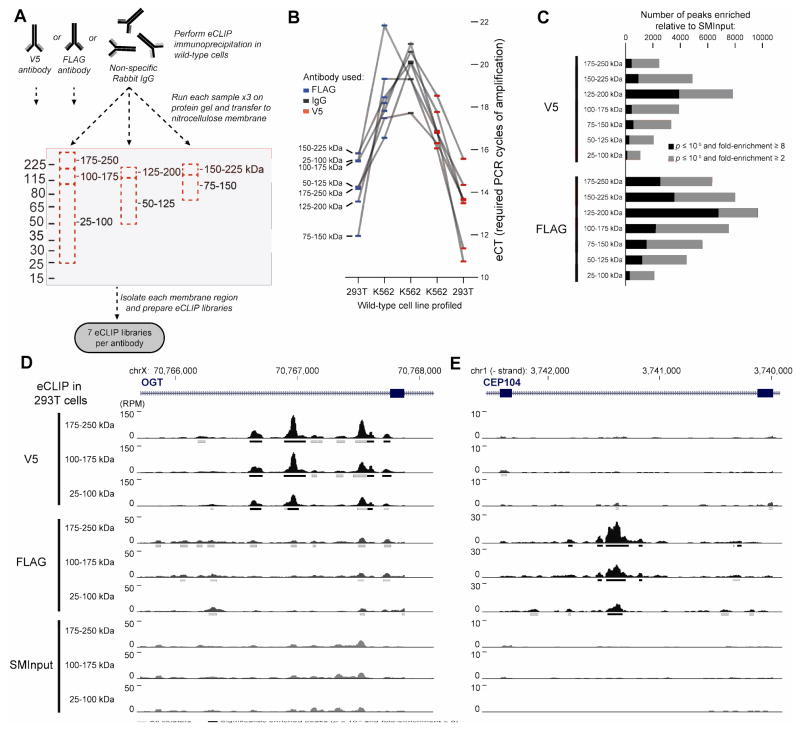
Figure 3.
Tag-only eCLIP in wild-type cells identifies tag-specific artifacts. (A) Experimental framework for testing tag-specific artifacts. Anti-V5, anti-FLAG, or isotype control was used for eCLIP in wild-type cells (K562 and 293T), along with a paired size-matched input. For each experiment, samples were run on 3 lanes of a protein gel and transferred to nitrocellulose membranes as in standard eCLIP. Seven size ranges (each 75 kDa) were isolated, and libraries prepared for each. (B) Boxes indicate library yield for anti-V5 (red), anti-FLAG (blue), and isotype control (black) antibodies in wild-type cells as indicated. Library yield is quantified as extrapolated CT (eCT), calculated by taking the number of PCR cycles performed and normalizing to a final yield of 100 femtomoles. Lines connect libraries from the indicated protein size range. (C) Bars indicate the number of peaks identified for V5 and FLAG experiments in 293T at two significance thresholds. (D–E) Genome browser tracks indicate read density (represented as Reads Per Million; RPM) for three example size ranges of V5, FLAG, and size-matched input. Clusters identified by CLIPper and significantly enriched peaks relative to input are indicated as bars underneath the read density track.
Next, to provide a point of comparison for the HEK293T TAG-eCLIP experiments, we repeated the V5 and FLAG pulldown in wild-type 293T cells. Surprisingly, these experiments showed even greater yield in HEK293T than K562 (Fig. 3B), indicating that the abundance of this tag-specific background can vary dramatically across cell types. After sequencing and standard eCLIP data analysis, we observed that anti-V5 and anti-FLAG antibodies each yielded antibody-specific significantly enriched peaks that were not observed in either the other antibody or the paired size-matched input (Fig. 3C). The number of significantly enriched peaks ranged from 125 (V5 25–100 kDa) to over 6,700 (FLAG 125–200), with maximum signal for both observed in the 125–200 kDa size range (Fig. 3C). Anti-FLAG antibody yielded an average of 3.6-fold more peaks than anti-V5 antibody for the same size range (Fig. 3C), possibly reflecting previously characterized anti-FLAG antibody interactions with abundant RBPs including EEF1A1, EIF4B, and SF3B3 among others (22). Visual inspection of individual binding sites indicated that these regions show similar profiles to true binding sites for standard RBPs profiled by eCLIP (Fig. 3D–E).
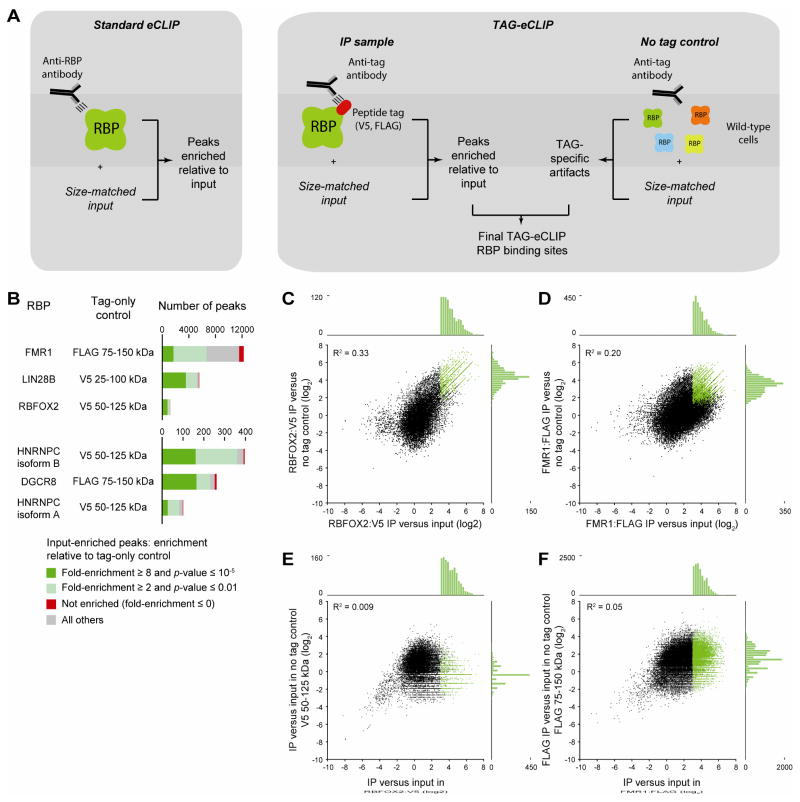
Thus, for TAG-eCLIP experiments, we highly recommend that this additional control (tag-specific antibody pulldown in wild-type cells, or ‘tag-only eCLIP’) be performed in parallel, with an additional analysis step of normalization against both the size-matched input as well as the non-RBP control to remove these non-specific peaks (Fig. 4A). To test the degree to which this would alter the list of observed binding sites, we performed such normalization on the six TAG-eCLIP datasets described above. We observed that many input-enriched peaks were similarly significantly enriched relative to size-matched tag-only eCLIP performed in wild-type cells, ranging from 1,701 out of 12,244 for FMR1 (13.9%) to 810 out of 1,117 (68.8%) for RBFOX2 (Fig. 4B). Directly comparing peak-level fold-enrichment, we observed that normalizing a RBP:tag eCLIP experiment against either its paired size-matched input or the size-matched tag-only IP in wild-type cells yielded generally similar results (Fig. 4C–D). In contrast, there was little correlation observed between RBP:tag IP and tag-only IP for both RBFOX2 (R2 = 0.009; p < 10−90) and FMR1 (R2 = 0.05; p < 10−300), though these were statistically significant over the large number of peaks queried. However, there were a small number of peaks that showed significant enrichment in both, indicating potential tag-dependent, antibody-specific false positives (Fig. 4E–F). The number of peaks with lower values of enrichment over input in TAG-eCLIP relative to tag-only eCLIP in wild-type cells varied among datasets, ranging from 1 out of 1,177 (0.0009%) for RBFOX2 to 671 out of 12,244 (0.05%) for FMR1 (Fig. 4B). Thus, the degree of false-positive signal due to anti-tag antibody-specific false positives can vary dramatically across different datasets and RBPs profiled.
Figure 4.
Normalization of TAG-eCLIP with wild-type control. (A) Schematic for identifying true binding sites for (left) standard eCLIP using RBP-specific antibodies, and (right) TAG-eCLIP using anti-tag antibodies. (B) Considering peaks significantly enriched in TAG-eCLIP over size-matched input, bars indicate the number of peaks significantly enriched versus size-matched tag-only eCLIP in wild-type cells (dark green), enriched with lower stringency (light green), depleted (red), or others (grey). (C–D) Scatter plot indicates fold-enrichment (log2) in tag antibody immunoprecipitation sample (IP) relative to size-matched input (x-axis) and in IP relative to “no tag control” (wild-type cells not expressing the tagged RBP immunoprecipitated with the same anti-tag antibody) (y-axis), for all clusters identified in (C) RBFOX2:V5 TAG-eCLIP or (D) FMR1:FLAG TAG-eCLIP. Attached histograms indicate the number of clusters in each bin. (E–F) Scatter plot indicates fold-enrichment in IP relative to size-matched input for all clusters identified in (E) RBFOX2:V5 TAG-eCLIP or (F) FMR1:FLAG TAG-eCLIP (x-axis), compared to enrichment in no tag control IP versus paired size-matched input (y-axis).
1.4 Conclusions
eCLIP provided a dramatic improvement in robustness and success in profiling RNA binding protein targets in vivo using antibodies derived against native proteins (9). To further assist such efforts, here we have presented an experimental strategy for parallelizable tagging of multiple RBPs. Standardized and cost-efficient tagging strategies will enable large-scale profiling of RBP targets with the same antibody across experiments, making it possible to profile the ever-expanding list of RNA binding proteins with consistent experimental conditions that will reduce antibody-specific background artifacts. As we have previously shown that a single set of experimental conditions can be used for RBPs that bind less than one kilobase RNAs (histone RNAs, RN7SK) as well as those binding unspliced pre-mRNAs of hundreds of kilobases in length (9), these results suggest that TAG-eCLIP experiments can generally be performed without significant factor-specific optimization. However, we note that lysis, RNA fragmentation, and protection from endogenous RNases and proteinases must still be optimized for each sample type of interest, as these can vary widely across different cell lines and tissue types.
Here, we show that for proteins where immunoprecipitation-grade antibodies are not available, integration of peptide tags into the endogenous gene loci followed by TAG-eCLIP provides a highly successful alternative strategy that, in all five tested cases, recapitulates binding patterns observed with antibodies targeting native proteins. In this work we describe a method for integration of C-terminal tags, as the Puromycin resistance cassette simplifies selection of rare integration events but integration at the 5′ end would disrupt endogenous transcription. However, improvements in performing seamless tag integration (using either Cre-mediated recombination or smaller tags lacking the resistance cassette) would enable N-terminal tagging, which may be essential for some RBPs for which C-terminal tags alter native protein structure or activity. We further note the presence of tag-specific background signal when anti-tag peptides are used in wild-type cells, indicating that paired control experiments in which anti-tag antibody is used in wild-type cells is an essential control to such TAG-eCLIP experiments. These results should aid in the design and implementation of eCLIP experiments, particularly for poorly characterized RBPs, in the same way that validation of peptide tag usage for DNA binding proteins provided a boost to the study of transcription factors (19).
Supplementary Material
Highlights.
Peptide tags can be inserted into RNA binding protein loci with CRISPR/Cas9
Streamlined process enables large-scale tagging efforts with high validation rate
TAG-eCLIP of peptide-tagged proteins recapitulates RNA targets of native proteins
Different peptide tags have distinct artifacts and require paired controls
Acknowledgments
The authors would like to thank members of the Yeo lab for insightful discussions and critical reading of the manuscript, particularly S. Aigner. This work was supported by grants from the National Institute of Health [HG004659, HG007005 and NS075449 to G.W.Y.]. E.L.V.N. is a Merck Fellow of the Damon Runyon Cancer Research Foundation [DRG-2172-13]. G.A.P. is supported by the National Science Foundation Graduate Research Fellowship. G.W.Y. is an Alfred P. Sloan Research Fellow.
Abbreviations
- RBP
RNA binding protein
- eCLIP-seq
enhanced crosslinking and immunoprecipitation followed by high-throughput sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nature reviews Genetics. 2014;15(12):829–45. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandziulis RJ, Swanson MS, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes & development. 1989;3(4):431–7. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 3.Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends in neurosciences. 2015;38(4):226–36. doi: 10.1016/j.tins.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Advances in experimental medicine and biology. 2014;825:1–55. doi: 10.1007/978-1-4939-1221-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–5. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 6.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14085–90. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17(7):909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nature methods. 2016;13(6):508–14. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sundararaman B, Zhan L, Blue SM, Stanton R, Elkins K, Olson S, et al. Resources for the Comprehensive Discovery of Functional RNA Elements. Molecular cell. 2016;61(6):903–13. doi: 10.1016/j.molcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munro S, Pelham HR. Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70. The EMBO journal. 1984;3(13):3087–93. doi: 10.1002/j.1460-2075.1984.tb02263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanke T, Young DF, Doyle C, Jones I, Randall RE. Attachment of an oligopeptide epitope to the C-terminus of recombinant SIV gp160 facilitates the construction of SMAA complexes while preserving CD4 binding. Journal of virological methods. 1995;53(1):149–56. doi: 10.1016/0166-0934(95)00003-d. [DOI] [PubMed] [Google Scholar]
- 13.Einhauer A, Jungbauer A. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. Journal of biochemical and biophysical methods. 2001;49(1–3):455–65. doi: 10.1016/s0165-022x(01)00213-5. [DOI] [PubMed] [Google Scholar]
- 14.Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schubeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528(7583):575–9. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 15.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8(11):2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savic D, Partridge EC, Newberry KM, Smith SB, Meadows SK, Roberts BS, et al. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome research. 2015;25(10):1581–9. doi: 10.1101/gr.193540.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one. 2011;6(4):e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnett SF, Friedman DL, LeStourgeon WM. The C proteins of HeLa 40S nuclear ribonucleoprotein particles exist as anisotropic tetramers of (C1)3 C2. Molecular and cellular biology. 1989;9(2):492–8. doi: 10.1128/mcb.9.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen GI, Gingras AC. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42(3):298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
eCLIP and TAG-eCLIP datasets have been deposited at the Gene Expression Omnibus (accession number GSE88722). Additional antibody validation information for RBP antibodies is available at the ENCODE portal (http://www.encodeproject.org). eCLIP data processing and analysis pipeline code has been publicly released (https://github.com/gpratt/gatk/releases/tag/2.3.2), and was previously described in additional detail (9).