Abstract
Purpose
Ionizing radiation (IR) is a ubiquitous environmental stressor with genotoxic and epigenotoxic capabilities. Terrestrial IR, predominantly a low-linear energy transfer (LET) radiation, is being widely utilized in medicine, as well as in multiple industrial applications. Additionally, an interest in understanding the effects of high-LET irradiation is emerging due to the potential of exposure during space missions and the growing utilization of LET radiation in medicine.
Conclusions
In this review, we summarize the current knowledge of the effects of IR on DNA methylation, a key epigenetic mechanism regulating the expression of genetic information. We discuss global, repetitive elements and gene-specific DNA methylation in light of exposure to high and low doses of high- or low-LET IR, fractionated IR exposure, and bystander effects. Finally, we describe the mechanisms of IR-induced alterations to DNA methylation and discuss ways in which that understanding can be applied clinically, including utilization of DNA methylation as a predictor of response to radiotherapy and in the manipulation of DNA methylation patterns for tumor radiosensitization.
Keywords: Epigenetics, ionizing radiation, DNA methylation, radiotherapy, space radiation, transposable elements
Introduction
Ionizing radiation (IR) is a ubiquitous environmental factor and a clinically important diagnostic and treatment modality. Approximately 50% of all cancer patients receive radiotherapy and over 70 million CT scans are performed annually in the US alone (Brenner, 2010; Moding et al., 2013). Exposures can also occur in occupational settings or as a consequence of nuclear accidents such as Chornobyl and Fukushima that have had devastating effects on the lives of millions of people.
Aside from the generally accepted potential to damage DNA, it is now evident that IR is also a potent epigenotoxic agent. Among epigenetic parameters, DNA methylation has arguably received the most attention in the context of radiation biology. DNA methylation is a key epigenetic mechanism in the regulation of the proper expression of genetic information. It is a covalent addition of a methyl group to the 5th position of carbon, facilitated by a complex interplay of DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) along with the protein ubiquitin-like with PHD and RING finger domains 1 (UHRF1), and methyl-CpG-binding proteins. In mammals, DNA methylation is found almost exclusively at cytosine residues located in the context of cytosine and guanine base sequences (CpG), approximately 70–90% of which are methylated. The majority of these methylated sites are located in either short (< 4 Kb) regions of DNA that contain large numbers of CpGs or in long domains of predominantly repetitive DNA elements (Jones, 2012).
In eukaryotes, more than half of the genes and a variety of repetitive elements (REs) contain CpG-rich regions, also known as CpG islands (CGIs). DNA methylation at CGIs that are located within the transcription start sites (TSS) has been attributed to transcriptional silencing, but it remains unclear whether this reflects repression or a lack of activation (Baubec & Schübeler, 2014). Strikingly, DNA methylation within gene bodies does not block transcription, and may stimulate elongation and splicing, or it may prevent aberrant transcription initiation from an alternative TSS (Portela & Esteller, 2010; Jones, 2012; Ehrlich & Ehrlich, 2014).
Alterations in DNA methylation may result in changes in gene expression or reactivation of REs, and may lead to genomic instability and the development of pathological states. Indeed, aberrant patterns of DNA methylation have been documented in a spectrum of human diseases (Portela & Esteller, 2010). Numerous environmental stressors, independent of their mode of action, have been shown to affect DNA methylation (Feil & Fraga, 2011; Koturbash et al., 2011a; Cortessis et al., 2012). Furthermore, interplay between DNA methylation and the environment is increasingly recognized as an important step in the response to environmental stimuli and the onset of disease (Aguilera et al., 2010; Stein, 2012). In this review we will discuss the current knowledge regarding the effects of IR on DNA methylation, their mechanisms, as well as laboratory and clinical applications.
Ionizing radiation and global DNA methylation
The first interest in the effects of IR on DNA methylation dates back to 1972, when a 15% increase in DNA methylation was reported in E.coli 15T-(555-7) after x-irradiation (Whitfield & Billen, 1972). The levels of 5-methylcytosine (5-mC) were then evaluated in the bone marrow and thymus of Wistar and outbred rats after exposure to 6.5 Gy and 7 Gy 60Co γ radiation, respectively (Rakova, 1979) (Table 1). Although the results of these studies were inconclusive, they conveyed three important points: 1) radiation can cause changes in global DNA methylation; 2) the extent of radiation-induced alterations in DNA methylation is tissue-dependent; and 3) radiation-induced alterations in DNA methylation may differ between experimental models and even among strains of the same species.
Table 1.
Effects of ionizing radiation on global DNA methylation.
| Model System/Exposure | Effect | Methodology | Reference |
|---|---|---|---|
| Escherichia coli 15T- irradiated with x-rays | DNA hypermethylation | L-methyl-3H methionine | (Whitfield and Billen 1972) |
| Wistar (6.5 Gy of 60Co) and outbred rats (7 Gy of 60Co); Thymus and bone marrow | Dynamic changes in DNA methylation with patterns of DNA hyper- and hypo-methylation; Lack of significant changes at the day 10 after exposure | 5-methylcytosine chromatography | (Rakova 1979) |
| Four cell lines (CHO, V79, HeLa, and C-1300) irradiated with 0–10 Gy 60Co gamma rays. 24, 48, and 72h post-exposure | DNA hypomethylation in all cell lines and time points, mostly dose-dependent | HPLC determination of 5-methylcytosine content | (Kalinich et al. 1989) |
| C57BL/6NJcl mice exposed to 4–10 Gy X-rays whole body radiation. 8, 24, 48, and 72h post-irradiation | DNA hypomethylation in the liver but not in the brain or spleen | HPLC determination of 5-methylcytosine content | (Tawa et al. 1998) |
| Male and female C57/Bl mice irradiated with 0–5 Gy x-rays, or exposed to a total of 5 Gy in a single versus fractionated (over 10 days) dose. | Dose-dependent hypermethylation was observed in female liver and spleen and in male spleen 6h after irradiation. 4 weeks after irradiation, hypermethylation was detected in male lung. For the fractionated dose, male and female lung showed hypermethylation 4 weeks after exposure. | Cytosine extension assay | (Pogribny et al. 2004) |
| Male and female C57/Bl mice irradiated with a total of 5 Gy x-rays in a single versus fractionated (over 10 days) dose. | DNA hypermethylation in male liver after a fractionated dose, no change in females or in muscle tissue | Cytosine extension assay | (Kovalchuk et al. 2004) |
| Male and female C57/Bl mice irradiated with a total of 5 Gy x-rays in a single versus fractionated (over 10 days) dose. | Hypomethylation in the thymus after acute and chronic exposure, male and female after 6h, and in male and female acute and male chronic exposure after 4 weeks. In the muscle, hypomethylation was observed in males and females 6h after acute exposure only. | Cytosine extension assay | (Koturbash et al. 2005) |
| CBA/H and C57BL/6 mice exposed to a single 3 Gy dose of x-rays, 10–14 days after exposure | Hypomethylation in the bone marrow of CBA/H animals but not C57BL/6 | HPLC determination of 5-methylcytosine content | (Giotopoulos et al. 2006) |
| Human fibroblast and bronchial epithelial cells exposed to 0.1, 1, and 4 Gy 137Cs | No significant differences 7 days after exposure | Methylated-CpG island recovery assay, microarray, and COBRA | (Lahtz et al. 2012) |
| Human-hamster hybrid cell line GM10115 exposed to 0.1 and 1 Gy of 56Fe or 0.5 and 2 Gy x-rays | Hypermethylation after 0.5 Gy X-rays and hypomethylation after 1 Gy 56Fe | Methylation-sensitive arbitrarily primed PCR | (Aypar et al. 2011) |
| Male BALB/c mice acutely exposed to 0.5 Gy x-rays or chronically exposed to a fractionated dose over 10 days | Genomic hypomethylation in blood 2h after exposure | HPLC determination of 5-methylcytosine content | (Wang et al. 2014) |
| Male and female C57BL/6 mice exposed acutely to a single dose of 0.5 Gy x-rays or to a fractionated equivalent dose over 10 days | Hypomethylation in the thymus 4h after acute and chronic exposure | Cytosine extension assay | (Pogrlbny et al. 2005) |
Similarly, early studies investigated the effects of IR doses on DNA methylation. Kalinich and colleagues used a dose range of 0.5 to 10 Gy of 60Co γ radiation to evaluate the epigenetic response in four cell lines – Chinese Hamster Ovary (CHO), Chinese hamster lung fibroblasts (V79A03), human HeLa (S-3) cells, and mouse neuroblastoma cells (C-1300N1E-115). All four cell lines exhibited significant dose-dependent decreases in 5-mC at 24, 48, and 72 h after irradiation, indicating a loss of global DNA methylation (Kalinich et al., 1989). Tawa and colleagues also showed that for C57BL/6NJcl mice, in vivo exposure to 4-10 Gy of x-rays resulted in a decrease in 5-mC content in the liver, but not in the spleen or brain of the irradiated mice (Tawa et al., 1998). These results validated the earlier findings suggesting tissues specificity in the level of change in DNA methylation in response to IR exposure. Conversely, it was somewhat unexpected that the liver, an organ relatively resistant to radiation-induced cell killing, exhibited a loss of DNA methylation, while the spleen, a relatively sensitive organ that belongs to the hematopoietic system, did not. Furthermore, this study was not able to reproduce the in vitro findings of Kalinich, as no changes in DNA methylation were detected in CHO and m5S/1m cell lines. The authors did not provide explanations for this discrepancy, but one might posit that phenotype variability in different clones for each cell line used in the two studies could play a significant role.
Subsequent in vivo studies with x-rays have confirmed radiation-induced loss of global DNA methylation in the liver. These studies also showed that exposures to doses higher than 1 Gy generally resulted in the loss of global DNA methylation in hematopoietic tissues including thymus, spleen, and bone marrow, as well as other target organs for radiation-induced carcinogenesis, such as the mammary gland, but not in the muscle and lung (Kovalchuk et al., 2004; Pogribny et al., 2004; Koturbash et al., 2005; Giotopoulos et al., 2006; Loree et al., 2006).
Ionizing radiation and gene-specific DNA methylation
While IR-induced changes in global methylation are important and may alter chromatin structure in critical ways, early studies regarding DNA methylation have not determined whether changes in DNA methylation occur uniformly throughout the genome, or whether particular genomic loci are more sensitive to changes in DNA methylation than others. DNA methylation profiles within specific genes can affect their transcriptional patterns and can be altered by exogenous stressors. Given that DNA hypermethylation-induced silencing of tumor-suppressor genes and hypomethylation-induced activation of oncogenes have been described in virtually all human cancers and are considered driving mechanisms of carcinogenesis (Portela & Esteller, 2010; Heyn & Esteller, 2012; Jones, 2012; Johnson et al., 2015; Nüsgen et al., 2015), the potential for IR-induced changes in gene-specific DNA methylation that affect changes in expression are critically important.
Early studies demonstrated significant DNA hypermethylation of cyclin-dependent kinase 2A, CDKN2A, (p16INK4A) (z=2.844, P=0.005), and O6-methylguanine-DNA methyltransferase (MGMT) (z=3.034, P=0.002) genes in the sputum of uranium miners (Su et al., 2006). (Table 2). These same tumor-suppressor genes are frequently found to be hypermethylated and inactivated during carcinogenesis in general, and lung cancer in particular (Kontic et al., 2012; Nikolaidis et al., 2012). Further, the level of hypermethylation of these genes significantly correlated with the cumulative doses of radon among the miners (cumulative exposure dose range 12±6 – 294±132; z=3.859, P=0.0001). Subsequent studies found 3.5 fold higher levels of p16INK4A hypermethylation in lung adenocarcinomas from plutonium-exposed workers at the Russian nuclear enterprise MAYAK as compared to non-IR worker controls (C.I. 1.5, 8.5; P=0.001) (Belinsky et al., 2004). Again, the increased probability for gene-specific methylation approximated a 4-fold increase in relative risk for adenocarcinoma in workers exposed to plutonium. Hypermethylation of p16INK4A and its corresponding transcriptional silencing was also reported in the murine model of radiation-induced thymic lymphoma when compared to normal thymus tissue (Song et al., 2014).
Table 2.
Effects of ionizing radiation on gene-specific DNA methylation.
| Model System/Exposure | Gene | Effect | Methodology | Reference |
|---|---|---|---|---|
| Male and female C57/Bl mice irradiated with a total of 5 Gyof x-rays in a single versus fractionated (over 10 days) dose. | p16INK4 and MGMT promoter | Hypermethylation of p16INK4 promoter in liver, more prominent in acutely exposed than in chronically exposed, and in males than in females. No change in muscle tissue or in MGMT promoter. | Bisulfite sequencing | (Kovalchuk et al. 2004) |
| C3H/HeN male mice exposed to 0.1, 0.3, or 1 Gy 56Fe and analyzed 1–120 days after exposure | DAPK1, EVL, 14.3.3, p16 INK4, MGMT, IGFBP3 | Hypermethylation at 1 and 30 days after exposure, and hypomethylation 7 and 120 days after exposure in the lung. No changes were observed in liver. | Bisulfite conversion, pyrosequencing | (Lima et al. 2014) |
| AG01522D and RKO cells irradiated with 0.1 and 1 Gy x-Ray, proton, or 56Fe ions | p16 INK4 and MGMT | No change in promoter methylation | COBRA | (Goetz et al. 2011) |
| Human-hamster hybrid cell line GM10115 exposed to 0.1 and 1 Gy of 56Fe or 0.5 and 2 Gy x-rays | NFκB, TSLC1, CDH1 | No change in promoter methylation | Methylation-specific PCR assay, bisulfite sequencing | (Aypar et al. 2011) |
| Male BALB/c mice acutely exposed to 0.5 Gy x-rays or chronically exposed to a fractionated dose over 10 days | Rad23b, Tdg, Ccnd1, Ddit3, Llg11, Rasl11a, Tbx2, and Scl6a15 | Hypermethylation in gene promoters after chronic exposure | Quantitative PCR on MeDIP-enriched DNA | (Wang et al. 2014) |
| Lung adenocarcinoma from workers from the MAYAK nuclear enterprise (γ rays) | Gata5, PAX5β, H-cadherin | Increase in Gata5 promoter methylation in tumors from workers compared to tumors from controls, no difference for PAX5β and H-cadherin | Methylation-specific PCR assay | (Lyon et al. 2007) |
| Male BALB/c mice exposed to whole body radiation split into four weekly 1.75 Gy doses | p16 INK4 | Hypermethylation in thymic tissue 6 months after exposure at 2 CpG islands in the p16 promoter | COBRA | (Song et al. 2014) |
| Adenocarcinoma from workers from the MAYAK nuclear enterprise (γ rays) | p16 INK4, MGMT, DAP-K, and RASSF1A | Hypermethylation in the promoter region of p16 and hypomethylation of RASSF1A in tumors from workers compared to tumors from controls | Methylation-specific PCR assay | (Belinsky et al. 2004) |
| Workers at a uranium mine | p16INK4 and O6-MGMT | Hypermethylation in p16INK4 and O6-MGMT proportional to the cumulative doses of radon among miners | Methylation-specific PCR assay | (Su et al. 2006) |
| MDA-MB-231 cells irradiated with X-rays (2 and 6 Gy). Endpoints at 1, 2, 4, 8, 24, 48, and 72 h post-irradiation | Whole genome | 1h post-irradiation with 2 Gy, RB1 was hypomethylated and IGF1R and KRas were hypomethylated. Fifteen genes were differentially methylated at all-time points post-2 Gy, and 23 genes were differentially methylated at all time-points post-6 Gy (direction of the change is not provided). GO terms associated point to changes in the cell cycle. | Bisulfite conversion and methylation microarray | (Antwih et al. 2013) |
Additional epigenetic evaluation of the MAYAK workers with lung adenocarcinomas demonstrated hypermethylation of GATA5, a transcription regulatory protein needed for proper development and maintenance of cellular differentiation (Lyon et al., 2007). Analysis of the promoter methylation of a panel of 5 genes performed within this study has also shown that at least one of the genes was hypermethylated in 93% of adenocarcinomas from MAYAK workers, while hypermethylation of at least one gene was observed in only 66% of non-workers’ tumors.
Several recent studies have evaluated the levels of gene-specific DNA methylation using more modern and high throughput microarrays that, given the stochastic effects of IR, allow for evaluation of gene-specific DNA methylation on the global scale. Lahtz et al. reported a lack of detectable changes in gene-specific methylation in normal human diploid fibroblasts and bronchial epithelial cells 1 week after exposure to 0.1 to 10 Gy doses of 137Cs γ-rays. (Lahtz et al., 2012). As was measured by the methylated-CpG island recovery assay (MIRA), only a small number of peaks characterizing differential DNA methylation were detected in irradiated cells when compared to controls. This may be explained by the possible dynamic nature of DNA methylation as has been observed in some studies; for instance, Antwih et al. reported dynamic changes in gene-specific DNA methylation in human breast cancer MDA-MB-231 cells at 1–72 h after 2 and 6 Gy of x-ray irradiation (320 kV at 0.86 Gy/min) (Antwih et al., 2013). It is worth mentioning that the differentially methylated genes in MDA-MB-231 cells induced by exposure were enriched in gene ontology categories relating to the cell cycle, DNA repair, and apoptosis, suggesting the possible role of DNA methylation in the cellular response to irradiation. Another possible explanation for the lack of the radiation-induced changes in gene-specific DNA methylation (despite overall alterations in global DNA methylation) is that these changes may stem primarily from the repetitive elements,.
Ionizing radiation-induced changes in the DNA methylation of repetitive elements
While genes comprise 1–2% of the typical mammalian genome, repetitive elements contribute to a significantly larger proportion. Recent advances in computational biology indicate that repetitive elements may comprise up to two thirds of mammalian genomes (de Koning et al., 2011). Among them, the Long Interspersed Nucleotide Element 1 (LINE-1) and Alu elements are of particular interest, since together they cover approximately 30% of the genomes and are heavily methylated in order to prevent their aberrant transcription (Figure 1). LINE-1 and Alu belong to a family of non-long terminal repeat retrotransposons (non-LTR) that propagate through the genome via an RNA intermediate in a ‘copy-and-paste’ mechanism. Alterations in the DNA methylation status of these repetitive elements often lead to their reactivation and retrotransposition, and are documented in human cancers as well as in response to environmental stressors (Miousse et al., 2015; Miousse & Koturbash, 2015). The Alu element is the only active Short Interspersed Nucleotide Element (SINE) in humans, while there are four distinct SINEs in mice (B1, B2, ID, and B4). The mouse B1 and human Alu SINE are unique in being derived from 7SL RNA and are believed to have a common genetic origin. A notable feature of LINEs and SINEs is the contrast in genomic distribution of the elements. LINEs are strongly biased towards (A+T)-rich genomic regions, while SINEs are strongly biased towards (G+C)-rich genomic regions, so that paired analysis can allow a more genome-wide evaluation of DNA methylation (Mouse Genome Sequencing Consortium et al., 2002).
Figure 1.
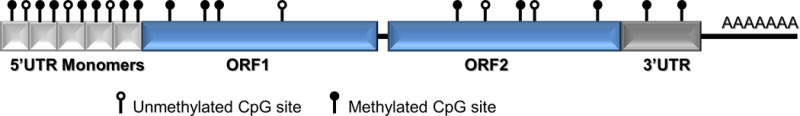
Schematic representation of the LINE-1 element. Mammalian LINE-1 elements is comprised of the heavily methylated 5′-Untranslated Region (5′-UTR), less rich on CpG sites two open reading frames ORF1 and ORF2, and a 3′-UTR that ends with a Poly-A tale.
Studies have demonstrated that alterations in global DNA methylation, at both early and late post-exposure time-points, may stem primarily from these transposable elements. Loss of LINE-1 DNA methylation was reported in the human-hamster hybrid cell line (GM10115) shortly after exposure to 2 Gy of x-rays, however the results were less consistent in other cell lines (250 kV peak, 13 mA, 165-mm copper at a 2.4 Gy/min dose rate) (Aypar et al., 2011). Hypomethylation of LINE-1 was detected in the rat spleen tissue 7 months after cranial irradiation to 20 Gy of x-rays (90 kV, 5 mA at a 3 Gy/min dose rate) and was associated with LINE-1 reactivation (Koturbash et al., 2007) (Table 3).
Table 3.
Effects of ionizing radiation on DNA methylation of repetitive elements.
| Model System/Exposure | Element | Effect | Methodology | Reference |
|---|---|---|---|---|
| C57BL/6 mice irradiated with 10 mGy of x-ray; Peripheral blood, spleen, and liver |
LINE-1, SINE B1, IAP | No persistent alterations in methylation at 297 or 420 days post-exposure | Bisulfite conversion, high resolution melt (HRM) Pyrosequencing |
(Newman et al. 2014a) |
| C57BL/6 and CBA mice irradiated with 1 Gy of x-ray; Peripheral blood, spleen, and liver |
LINE-1, SINE B1, IAP | Strain-, tissue-, sex-, and time-dependent alterations in DNA methylation. Hypomethylation of LINE-1 in C57BL/6 mice, while CBA was characterized by hypermethylation of LINE-1 in the spleen tissue shortly after exposure. Methylation of SINE B1 and IAP was affected to a lesser extent | Bisulfite conversion, high resolution melt (HRM) Pyrosequencing |
(Newman et al. 2014b) |
| Spleen tissue 7 months post localized hippocampal irradiation with two doses of 10 Gy x-rays | LINE-1 | Persistent global and LINE-1-associated hypomethylation after exposure has been observed in the spleen bystander tissue | COBRA | (Koturbash et al. 2007) |
| C57BL/6 mice, 56Fe 0.1–0.4Gy Bone marrow |
LINE-1, SINE B1 | Dose-dependent hypermethylation in hematopoietic stem and progenitor cells at 4 weeks after exposure, followed by DNA hypomethylation and LINE-1/SINE B1 reactivation at 22 weeks | Methylation-sensitive enzymatic digestion followed by qRT PCR | (Miousse et al. 2014) |
| C57BL/6 mice 56Fe 0.1–0.4Gy; Lung |
LINE-1, SINE B1, Charlie, Mariner, major and minor satellites | Dose-dependent hypermethylation and transcriptional silencing were observed 22 weeks after exposure | Methylation-sensitive enzymatic digestion followed by qRT PCR | (Nzabarushimana et al. 2014) |
| C3H/HeN mice irradiated with 10 and 100 cGy 56Fe; Liver and Lung |
LINE-1 | Hypomethylation in TF promoter types 7 days after exposure, and hypermethylation 30 days after exposure in the lung. No differences in A1 type promoters or in liver. | Bisulfite conversion, pyrosequencing | (Lima et al. 2014) |
| AG01522D and RKO cells irradiated with 0.1 and 1 Gy x-ray, proton, or 56Fe ions | LINE-1 and Alu | Hypomethylation at 16–20 population doublings after irradiation with proton and iron, and for x-rays there was hypomethylation in RKO and hypermethylation in AG01522D after 1 Gy | COBRA | (Goetz et al. 2011) |
| Human-hamster hybrid cell line GM10115 exposed to 0.1 and 1 Gy of 56Fe or 0.5 and 2 Gy x-rays | LINE-1 and Alu | LINE-1 hypomethylation of at the highest dose of 56Fe and X-rays, hypermethylation at 0.5 Gy x-rays Alu hypomethylation of at the highest dose of 56Fe and x-rays | COBRA | (Aypar et al. 2011) |
| C57BL/6 male mice 56Fe (0.1 Gy), protons (0.5 Gy) Heart |
LINE-1, ERV2, SINE-B1, major and minor satellites | LINE-1 DNA hypomethylation at day 7 after irradiation; LINE-1, ERV2, SINE B1, and major satellites DNA hypermethylation at day 90. | Pyrosequencing, Methylation-sensitive enzymatic digestion followed by qRT PCR | (Koturbash et al. 2016) |
| C57BL/6 male mice 56Fe (0.5 Gy), protons (0.1 Gy), or 56Fe (0.5 Gy)+protons (0.1 Gy) Lung |
LINE-1 | DNA hypermethylation of selective LINE-1 (L1Md_Gf, L1MdTf_III, L1MdF_V) elements detected in the lung 4 weeks after irradiation | Methylation-sensitive enzymatic digestion followed qPCR, Methylated DNA Immunoprecipitation (MeDIP) followed by qRT PCR | (Prior et al. 2016) |
Although early studies reported loss of global and RE-associated DNA methylation after irradiation, accumulative evidence indicates that these changes are rather dose-, cell/tissue-, time-, and radiation quality-dependent. For instance, DNA hypomethylation of LINE-1 was reported in RKO human colorectal carcinoma cells after exposure to 1 Gy of x-rays (250 kV peak, 13 mA at a 2.4 Gy/min dose rate); however the same dose of radiation led to hypermethylation of LINE-1 in AG01522D primary human diploid skin fibroblasts in the same study (Goetz et al., 2011). Furthermore, no significant changes in DNA methylation of Alu elements were detected in either of the cell lines in this study, suggesting that changes in DNA methylation of different repetitive elements are not necessarily unidirectional. This may be evidence of the differences of AT or CG rich regions of the genome in which LINE and Alu elements are found, respectively.
Irradiation of C57BL/6 mice with 10 mGy of x-rays (100–140 kVp at 13.9 mGy/min dose-rate) did not result in any persistent alterations in the DNA methylation of LINE-1, SINE B1 (non-autonomous retrotransposon that corresponds to human Alu elements), and Intracisternal A Particle (IAP) elements in peripheral blood, spleen, and liver tissues (Newman et al., 2014a). At the same time, exposure to a dose of 1 Gy of x-rays was characterized by the strain-, tissue-, sex-, and time-dependent alterations in methylation of the abovementioned repetitive elements (Newman et al., 2014b). Interestingly, loss of DNA methylation associated with LINE-1 was observed in the relatively radioresistant C57BL/6 strain, while the radiosensitive CBA and BALB/c strains showed a DNA hypermethylation response in the spleen tissue shortly after exposure (Newman et al., 2014b).
Fractionated exposure to ionizing radiation and DNA methylation
A limited number of studies have evaluated the effects of fractionated exposure to IR exposure on DNA methylation, although it is of particular interest given that clinical radiotherapy is usually delivered in fractionated doses. Furthermore, occupational exposures occur within the lower doses range over the protracted time-course.
Fractionated exposure to 0.5 Gy of x-rays, delivered in 10 consecutive days at a rate of 0.05 Gy/day, has led to a loss of global DNA methylation in the thymus tissue of male but not female C57BL/6 mice (Pogribny et al., 2005). Although acute exposure to 0.5 Gy of x-rays was also characterized by DNA hypomethylation, the extent of the observed DNA hypomethylation as a result of fractionated exposure was 2.5-fold higher. Fractionated exposure also resulted in substantially decreased protein levels of DNA methyltransferases Dnmt1 and Dnmt3b, as well as the methyl-binding protein MeCP2, suggesting a possible mechanism for the observed DNA hypomethylation.
A recent study performed by Wang and colleagues confirmed the loss of global DNA methylation using the same exposure regimen (0.05 Gy/day for 10 consecutive days) in male BALB/c mice and decreased mRNA and protein levels of Dnmt1 and MeCP2 (Wang et al., 2014). They further expanded their analysis on gene-specific methylation and identified 811 specific regions of hypermethylation in the animals that received fractionated exposure. Among them, DNA hypermethylation in Rad23b and Ddit3 genes was also associated with their transcriptional silencing and was detectable for at least 1 month after irradiation.
Another study looked at the effects of higher, radiotherapy-relevant fractions of 2 Gy with cumulative doses of 10 and 20 Gy on DNA methylation in human MCF7 breast cancer cells (Kuhmann et al., 2011). Cells were irradiated with 137Cs at a dose rate of 50 cGy/min and harvested at 48–72 h and up to 24 days after the last exposure. A subset of genes was found differentially methylated in response to radiation treatment using the methyl-CpG immunoprecipitation approach. At the same time, only a very small fraction of these changes were confirmed by the quantitative MassARRAY technique. Among them, significant hypermethylation of the Forkhead box C1 (FOXC1) and Trafficking protein particle complex 9 (TRAPPC9) genes was detected; however these changes in DNA methylation did not result in changes in the expression of these genes.
DNA methylation and ionizing radiation-induced bystander effect
The bystander effect is a phenomenon in which irradiated cells can induce genomic instability in unirradiated cells and has significant implications for diagnostic radiological procedures and radiotherapy (Sedelnikova et al., 2007). It currently remains unknown what are the specific factors that are involved in the transmission of the signal from the irradiated cell to the non-irradiated one, but it has been suggested that these factors are small, transient and, perhaps, not even chemical by nature (Mothersill & Seymour, 2012). It is, however, without a doubt that epigenetic alterations can be triggered by the bystander signal(s) and may play a significant role in the development and manifestation of the IR effects in non-irradiated cells and tissues.
Aberrant DNA methylation has been detected both in cell/tissue cultures and in vivo in distant bystander tissue shortly after irradiation and this effect can persist for a long time after exposure. In vitro studies reported the loss of genomic DNA methylation in both irradiated and bystander regions of the EpiAirway (Air-100) normal human three-dimensional tissue system 48 h after exposure (microbeam of 7.0 MeV 4He; LET 100–2000 keV/μm with a LET of 100 keV/μm of 3.2 Gy) (Sedelnikova et al., 2007). DNA hypomethylation exhibited a persistent nature and was detectable until the end of the experiment (7 days after irradiation). Similarly, cranial exposure to 1 Gy of x-rays resulted in global DNA hypomethylation in the murine spleen tissue 6 h after irradiation (Koturbash et al., 2008). This effect was sex-specific (observed in male mice only), but was diminished in male mice after orchiectomy and became detectable in female mice after ovariectomy (Koturbash et al., 2008). The long-term bystander effect was documented in the rat spleen 7 months after hippocampal irradiation to 20 Gy of x-rays (90 kV, 5 mA at 3 Gy/min) and was characterized by the loss of global and LINE-1 DNA methylation and subsequent reactivation of the latter (Koturbash et al., 2007).
Effects of high-linear energy transfer (LET) ionizing radiation on DNA methylation
High-LET IR is prevalent in the space environment, but it has increasingly been utilized in the clinic, due to the possibility of delivering a higher dose to a confined volume, thus potentially increasing the effects on the tumor tissue while decreasing the normal tissue toxicity (Durante & Cucinotta, 2008; Durante, 2014; Tommasino & Durante, 2015). Indeed, about forty proton and heavy ion centers dedicated to cancer therapy currently function worldwide and it has been predicted that particle therapy will become a major arm of cancer radiotherapy in the near future (Girdhani et al., 2013).
This introduction of high-LET radiation into clinical practice, as well as the necessity of understanding the effects of IR exposure during the space missions, has triggered the investigation of biological and molecular mechanisms of response to high-LET radiation. Exposure to the main sources of high-LET radiation – protons and heavy ions – results in more complex clustered and irreparable DNA damage, leading to greater relative biological effectiveness in cell death (Blakely & Kronenberg, 1998).
Accumulating evidence convincingly demonstrates that high-LET radiation possesses a distinct epigenotoxic potential as well. Exposures to low mean absorbed doses of protons (150 MeV/n, LET 0.55 keV/μm; dose rate 2 cGy/min) or 56Fe ions (56Fe+26, 1 GeV/n, LET 150 keV/μm; 10 cGy/min) result in the loss of DNA methylation within the LINE-1 and Alu elements in AG01522D and RKO cells at 16–20 population doublings after irradiation (Goetz, Morgan and Baulch 2011). Similar effects were reported in another study, where the exposure of GM10115 cells to 1 Gy of 56Fe (56Fe+26, 1GeV/amu, 150keV/μm, dose rate 2 cGy/min) led to DNA hypomethylation in both LINE-1 and Alu elements (Aypar et al., 2011). At the same time, exposure to 0.1 Gy of 56Fe did not affect the DNA methylation status of these retrotransposons.
A number of in vivo studies using the mouse model report global genomic and repetitive elements-associated DNA hypermethylation. Exposure to low absorbed mean doses of 56Fe (56Fe+26, 600 MeV/nucleon; dose-range 0.1 – 0.4 Gy) led to dose-dependent global DNA hypermethylation in the lungs of C57BL/6 mice at 22 weeks after exposure (Nzabarushimana et al. 2014). As has been reported, an increase in 5-mC content was not associated with increased DNA methylation in a panel of tumor-suppressor genes frequently hypermethylated and inactivated in lung cancer. At the same time, a number of repetitive elements, including retrotransposons LINE-1 and SINE B1, transposons Charlie and Mariner, as well major and minor DNA satellites, were found hypermethylated in a dose-dependent manner (Nzabarushimana et al., 2014). Importantly, this hypermethylation was associated with further transcriptional silencing of these repetitive elements. Hypermethylation of specific families of LINE-1 elements in the mouse lung and heart 1–4 months after exposure to high-LET radiation was also reported in a number of other recent studies (Lima et al., 2014; Koturbash et al., 2016; Prior et al., 2016). Another recent study documented induction of lung tumors in mice exposed to 56Fe only at doses below 0.4 Gy, suggesting that DNA hypermethylation and silencing of repetitive elements may have a protective effect against heavy ion-induced lung carcinogenesis (Christofidou-Solomidou et al., 2015).
Of particular interest are the epigenetic effects of high-LET radiation on the hematopoietic system because exposure to 56Fe and 28Si, similar to γ-rays, is associated with increased rates of acute myeloid leukemia (AML) in mice (Weil et al., 2009, 2014). Alterations in the gene-specific and repetitive elements-associated DNA methylation in AML patients are well-documented (Gebhard et al., 2006; Kroeger et al., 2008; Bujko et al., 2014). Exposure to 56Fe ion radiation (56Fe+26, 600 MeV/nucleon; dose-range 0.1 – 0.4 Gy) selectively targeted global and two transposable elements – LINE-1 and SINE B1 – DNA methylation in the less differentiated hematopoietic stem and progenitor cells, while only subtle effects were observed in mononuclear cells (Miousse et al., 2014). Importantly, changes in DNA methylation after exposure to the lowest known leukemogenic dose of 56Fe (0.4 Gy;(Weil et al., 2014)) persisted in the murine bone marrow for at least 22 weeks after exposure, while no detectable DNA damage, accumulation of reactive oxygen species, or cellular senescence were observed. These findings suggest that epigenetic alterations are not simply the passengers in radiation-induced AML, but may play a significant role in its development and progression.
What are the mechanisms of the radiation-induced effects on DNA methylation?
Despite significant progress in radiation epigenetics that has been achieved in the past decade, the mechanisms of radiation-induced changes in DNA methylation, mostly global genomic hypomethylation and hypermethylation of tumor-suppressor genes, remain largely unknown.
Effects of IR on DNA methyltransferases are among the most plausible scenarios. It has been shown that exposure to 60Co γ radiation decreases the nuclear maintenance and de novo methyltransferase activity (Kalinich et al., 1989). A number of studies also report decreases in DNA methyltransferases’ levels of mRNA (Miousse et al., 2014) and protein (Antwih et al., 2013) as a result of exposure. Furthermore, IR has been shown to affect several microRNAs that specifically target DNA methyltransferases, such as the miR-29 family, miR-141, and miR-152, which may lead to DNA methyltransferases’ mRNA degradation and subsequent alterations in DNA methylation (Koturbash et al., 2011b; Wang et al., 2011).
Another proposed mechanism associated with changes in DNA methylation is dependent on DNA damage and the repair status of irradiated cells. DNA methyltransferases and UHRF1 may be reassigned from copying the methylation patterns during replication to the repair of radiation-induced DNA damage (Kontic et al., 2012). This mechanism, however, may be considered only for the organs comprised of rapidly dividing cells and at early time-points – hours to days after induction of DNA damage. For instance, it has been shown that the decrease in the global DNA methylation 6 h after irradiation correlates with the accumulation of DNA strand breaks (r2>0.9) and the increase in recombination activity (r2>0.9) (Koturbash et al., 2006). Furthermore, it has been shown that DNA polymerases involved in DNA repair and recombination incorporate cytosine but not methylcytosine during repair synthesis (Pogribny et al., 2005).
Other less explored mechanisms consider alterations in the one-carbon metabolism pathways (Figure 2), which is essential for the synthesis of S-adenosylmethionine (SAM), the donor of methyl groups for DNA methylation (Batra et al., 2004; Batra & Mishra, 2007; Batra & Verma, 2014; Koturbash et al., 2016); interference of DNA damage with the ability of DNA methyltransferases to methylate DNA (Panayiotidis et al., 2004); and, in some cases, radiation-induced proliferation, when the function of DNA methyltransferases is not sufficient to sustain the methylation patterns in rapidly dividing cells (Koturbash et al., 2008).
Figure 2.
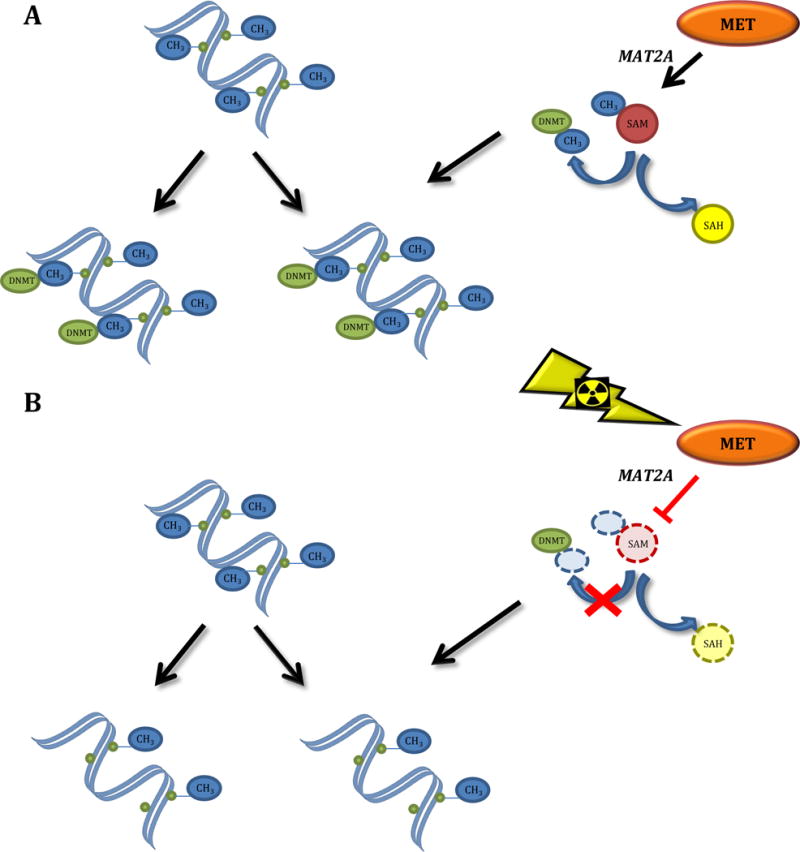
Alterations in the methionine cycle as a mechanism of ionizing radiation-induced DNA hypomethylation. (A) Under normal conditions, methionine (MET) is being converted into S-adenosylmethionine (SAM) by the methionine adenosyl transferase 2A (MAT2A). SAM donates its methyl group (CH3), which is further used by the DNA methyltransferase DNMT-1 (DNMT) for post-replicative maintenance of DNA methylation. (B) Ionizing radiation may affect synthesis of methionine and/or inhibit methionine biotransformation to SAM, thus exhausting the internal resources for donors of methyl groups, resulting in hemimethylated DNA after replication.
Clinical applications: Gene- and LINE-1- specific DNA methylation as a predictor of response to radiotherapy
Accumulating evidence demonstrates that the DNA methylation landscape may influence the tissue response to IR. This knowledge may be further utilized in order to modulate the normal and cancerous tissue response to IR, as well as development of predictive biomarkers of tumor recurrence and overall prognosis.
A microarray analysis of the human radiosensitive H460 and radioresistant H1299 non-small cell lung cancer (NSCLC) cell lines revealed 1091 differentially methylated genes (Kim et al., 2010). Of these genes, 747 were hypermethylated in radioresistant H1299 cells, including SERPINB5 and S100A6. These genes were also significantly down-regulated in H1299 cells compared to H460 cells. Furthermore, small interfering RNA-mediated down-regulation of SERPINB5 and S100A6 has led to the development of radioresistance in H460 cells, providing clear evidence of the role of DNA methylation in the regulation of the cancer cell response to radiotherapy.
The methylation status of MGMT, a gene that encodes for a DNA repair enzyme responsible for removing the alkyl groups from guanine, has been proven to serve as a prognostic marker in glioblastoma (Esteller et al., 2000). Hypermethylation of MGMT was associated with the regression of tumors and prolonged disease-free survival in response to the treatment with alkylating agent carmustine. This phenomena is linked to the hypermethylation-driven silencing of MGMT and, therefore, decreasing its inhibitory effect on alkylating agents in tumor cell killing. Similarly, after administration of temozolomide, concomitant with and adjuvant to radiotherapy, hypermethylation of MGMT promoter was strongly associated with better survival (Niyazi et al., 2012). Another study demonstrated that even in patients who received radiotherapy following tumor resection without chemotherapy as a component of adjuvant treatment, the methylation of the MGMT promoter correlated with the improved response to radiotherapy and overall patient survival (Rivera et al., 2010).
Hypomethylation of LINE-1 elements has been observed in virtually all human cancers and is associated with a poorer prognosis in the vast majority of them (Miousse & Koturbash, 2015). Some studies report that the hypomethylated status of LINE-1 is associated with a poor prognosis and overall survival after chemo- and radiotherapy. In a cohort of sixty-nine patients with esophageal squamous cell carcinoma at the Kumamoto University Hospital (Japan), among which 4 patients received radiotherapy and 24 patients received chemoradiotherapy, LINE-1 hypomethylation was significantly associated with a poorer disease-free survival (log-rank P=0.0008; HR: 2.32, 95% CI: 1.38–3.84, P=0.0017; multivariate HR: 1.81, 95% CI: 1.06–3.05, P=0.031) and cancer-specific survival (log-rank P=0.0020; univariate HR: 2.21, 95 CI: 1.33–3.60, P=0.0026; multivariate HR: 1.87, 95% CI: 1.12–3.08, P=0.018) (Iwagami et al., 2013).
Targeting DNA methylation by DNA demethylating agents
A number of DNMT inhibitors, such as nucleoside analogs 5-Azacytidine (5-aza), 5-Aza-2′-deoxycytidine (decitabine; DAC) and zebularine, are successfully used in the treatment of hematological malignancies and are being tested in some solid cancer (Ricketts et al., 2013; Momparler et al., 2014; Nervi et al., 2015). During the S-phase of DNA replication, they incorporate into DNA in place of cytosine and trap DNA methyltransferases resulting in further degradation of the latter (Song et al., 2011). This leads to the hypomethylation of the newly synthesized DNA. Furthermore, it has been shown that while decitabine and zebularine can incorporate into DNA only, 5-Azacytidine may incorporate into RNA as well.
Accumulating evidence indicates that DNMT inhibitors can potentiate the effects of other anti-cancer drugs, i.e., carboplatin (Qin et al., 2015). The results from recent in vitro clonogenic studies suggest that DNMT inhibitors may also modulate the cancer cell response to IR. For instance, nucleoside analogs increased the sensitivity to radiotherapy in glioblastoma (Dote et al., 2005), gastric (Qiu et al., 2009), colorectal (Hofstetter et al., 2010), head and neck (Brieger et al., 2012), and nasopharyngeal (Jiang et al., 2014) cancer cell lines. At the same time, the effect of DNMT inhibitors-induced radiosensitization was not uniform, and in some cell lines it was little to negligible (Qiu et al., 2009; Kim et al., 2012). Knowledge of the in vivo effects remains sparse and limited to several studies, reporting a significant tumor growth delay in the case of the U251 glioblastoma (Dote et al., 2005) and nasopharyngeal carcinoma (Jiang et al., 2014) xenografts.
It has to be also taken into consideration that the long-term effects of DNMT inhibitors on both normal and cancerous cells remain largely unknown. The loss of DNA methylation within the promoters of tumor-suppressor genes with their further reactivation is certainly beneficial in the treatment of cancer. At the same time, current nucleoside analogs do not possess selective DNA demethylating abilities, resulting in concomitant hypomethylation of repetitive elements and may subsequently lead to their aberrant transcription, retrotransposition, and development of genomic instability. Loss of methyl groups within the gene bodies may also result in spurious transcription from the exposed alternative transcription start sites (Portela & Esteller, 2010). Additionally, administration of nucleoside analogs has been also associated with increased mutation frequencies and decreased fertility (Gravina et al., 2010).
Current limitations and future prospects
The majority of our knowledge on the effects of IR on DNA methylation derives from in vitro and in vivo experimental systems, while our understanding of these effects in the normal tissue and tumors in humans is limited. Thus, the translational relevance of these studies should be interpreted with caution. Another considerable limitation is that in the majority of the in vivo rodent studies, only male animals were utilized, while sex differences in response to IR, including the effects of DNA methylation have been documented (Koturbash et al., 2008).
While the exposures to terrestrial IR at doses of 1 Gy and above are characterized by the loss of global DNA methylation and seem to stem primarily from repetitive elements, the effects of exposure at doses below 1 Gy, which are generally characterized by non-linear responses, remain to be a challenge. Some studies indicate that assessment of DNA methylation within specific genomic loci, i.e., particular families and sub-families of repetitive elements, may aid in identification of the DNA methylation signatures of exposure (Prior et al., 2016). In these regards, recent developments in high-throughput approaches and robust analytical platforms promise to further expand our understanding of the effects of low dose exposures. The certain degree of inconsistency, however, needs to be taken into consideration, as principal differences in analytical tools for assessment of DNA methylation – from PCR-based gene-specific methylation analysis to whole genome methylation array – exist.
The phenomenon of IR-induced alterations in DNA methylation is undeniable; however, our understanding on the functional outcomes of these alterations is very limited. Only a small fraction of studies perform simultaneous evaluation of DNA methylation and gene expression analyses in order to investigate the role of IR-induced changes in DNA methylation on the expression of genetic information. It is also known that loss of DNA methylation may result in the development of genomic instability, i.e., via activation of transposable elements, however the exact mechanisms remain unknown. LINE-1 hypomethylation and, associated with it, LINE-1 reactivation and retrotransposition, are well-described events and are suggested as potentially causative events in carcinogenesis (Lee et al., 2012). However, no studies, to our knowledge, have concomitantly addressed the DNA methylation and LINE-1 retrotransposition as a consequence of exposure to IR.
In recent years, other forms of DNA methylation, including 5-hydroxymethylcytosine (5hmC), have also been recognized (Ye & Li, 2014). A stable base, 5hmC is an oxidation product of 5mC that is actively converted by the TET1 enzyme (Ten-eleven translocation methyl cytosine dioxygenase; Fig. 1). The TET family of enzymes can also subsequently convert 5hmC to 5-formylcytosine and 5-carboxylcytosine in active demethylation processes. Rediscovery of DNA hydroxymethylation as an important step in the DNA demethylation process and further recognition of 5-hmC as an independent, not necessarily transient epigenetic mark, promises to bring new insights into the field of radiation epigenetics and beyond. Understanding the effects of IR exposure on DNA hydroxymethylation is in its infancy, but the pioneer studies demonstrated that 5-hmC is clearly affected by IR and may have substantial input on the regulation of gene expression and overall cellular and tissue response to exposure (Jangiam et al., 2015; Rithidech et al., 2015).
Epigenetic modifications are not limited to DNA only; histone proteins that form the structural unit of nucleosome can undergo those modifications as well. To date, about a dozen of histone modifications are being discovered and characterized, of which histone methylation, acetylation, and phosphorylation received the most attention. Phosphorylation of Ser 139 on histone H2AX is a critical step in the initiation of the radiation-induced damage repair and occurs within minutes after exposure (Sedelnikova et al., 2007). Histone methylation has been demonstrated to be affected by IR as well. For instance, histone marks that are responsible for the formation of transcriptionally silent heterochromatin structure – histone H3 lysine 9 (H3K9) and histone H4 lysine 20 (H4K20) trimethylation – are negatively regulated after exposure to both low- and high-dose IR (Pogribny et al., 2005; Koturbash et al., 2007). This relaxed chromatin structure may allow for easier access of repair complexes to the sites of DNA damage. More details on histone modifications in radiation biology and radiotherapy can be found in excellent reviews published elsewhere (Ma et al., 2011; Smits et al., 2014).
Of particular interest is the possibility of dietary modulation of the DNA methylation patterns. It is known that dietary intake of folates/folic acid, methionine and choline – the key players in the one-carbon metabolism pathway – affects the levels of DNA methylation in in vivo models (Wilson et al., 1984; Shivapurkar et al., 1986). The central role of one-carbon metabolism in cancer has long been recognized and has led to the development of methotrexate, an antifolate agent, for chemotherapy. In these regards, utilization of methionine, which serves as a precursor of S-adenosylmethionine – a major donor of methyl groups – and is also needed for the synthesis of glutathione, is another promising avenue.
It has been suggested that methionine supplementation may mitigate the effects of IR by maintaining the normal patterns of DNA methylation and the synthesis of glutathione needed for balanced cellular redox status. For instance, administration of the methionine-supplemented diet to male Swiss mice for 2 weeks before exposure to 2, 4, or 6 Gy of 60Co (dose rate of 50 cGy/min) maintained high levels of DNMT and methionine synthase activity, as well as genomic DNA methylation in comparison to mice fed a normal control diet (Batra et al., 2010). It has to be noted, however, that analysis was done only shortly after irradiation (24–48 h) and further studies dedicated to the investigation of the long-term effects are clearly needed. It also has to be taken into consideration that methionine is a difficult amino acid to digest, as well as being an important metabolite in microorganisms’ metabolism (Hondorp & Matthews, 2006). Therefore, methionine dietary overload may cause rotting in the intestine and boost microbe growth in the damaged gastrointestinal tract, possibly leading to inflammation, bacterial translocation and development of the IR-induced gastrointestinal/hematopoietic syndrome.
On the other hand, tumors were shown to be particularly sensitive to methionine deficiency that is critical for their metabolism. Restriction of methionine consumption by tumor cells results in their cell cycle arrest at the G2/S stage that makes them particularly sensitive to chemotherapeutic agents (Cellarier et al., 2003; Durando et al., 2008; Agrawal et al., 2012; Cavuoto & Fenech, 2012). In vitro, in vivo, and studies involving cancer patients demonstrated that methionine deprivation significantly potentiates the effects exerted by chemotherapeutic agents (Kokkinakis et al., 2006; Pavillard et al., 2006; Guénin et al., 2009; Durando et al., 2010; Strekalova et al., 2015). The exact mechanisms of this effect are not well understood, but the alterations in tumor DNA methylation caused by methionine deprivation are among those that were proposed (Cavuoto & Fenech, 2012). To our knowledge, this remarkable feature has not been investigated yet in regards to radiotherapy but may potentially provide a promising avenue in radiation oncology.
Acknowledgments
Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 1P20GM109005; NIH/UAMS Clinical and Translational Science Award UL1TR000039 and KL2TR000063, the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000, and the National Space Biomedical Research Institute through the National Aeronautics and Space Administration NCC 9-58, grant# RE03701. The authors are thankful to Dr. Janet Baulch for crticial reading and to Christopher Fettes for editing this manuscript.
Abbreviations
- CHO
Chinese Hamster Ovary
- CDKN2A
Cyclin Dependent Kinase 2A
- CI
Confidence Interval
- DNMT1
DNA Methyltransferase 1
- Gy
Gray
- HR
Hazard Ration
- IAP
Intracysternal A Particle
- IR
ionizing radiation
- LET
Linear Energy Transfer
- LINE-1
Long Interspersed Nucleotide Element 1
- MeV
Megaelectron Volt
- MGMT
O6-methylguanine-DNA methyltransferase
- MIRA
methylated-CpG island recovery assay
- ORF
Open Reading Frame
- SINE
Short Interspersed Nucleotide Element 1
- TE
transposable elements
- UHRF1
Ubiquitin-like with PHD and RING Finger Domains 1
- UTR
Untranslated Region
Footnotes
Disclosure statement: The authors report no conflicts of interest.
References
- Agrawal V, Alpini SEJ, Stone EM, Frenkel EP, Frankel AE. Targeting methionine auxotrophy in cancer: discovery & exploration. Expert Opin Biol Th. 2012 Jan;12:53–61. doi: 10.1517/14712598.2012.636349. [DOI] [PubMed] [Google Scholar]
- Aguilera O, Fernández AF, Muñoz A, Fraga MF. Epigenetics and environment: a complex relationship. J Appl Physiol. 2010 Jul;109:243–251. doi: 10.1152/japplphysiol.00068.2010. [DOI] [PubMed] [Google Scholar]
- Antwih DA, Gabbara KM, Lancaster WD, Ruden DM, Zielske SP. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics-Us. 2013 Aug 1;8:839–848. doi: 10.4161/epi.25498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aypar U, Morgan WF, Baulch JE. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat Res-Fund Mol M. 2011 Feb 10;707:24–33. doi: 10.1016/j.mrfmmm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Batra V, Kesavan V, Mishra KP. Modulation of enzymes involved in folate dependent one-carbon metabolism by gamma-radiation stress in mice. J Radiat Res. 2004 Dec;45:527–533. doi: 10.1269/jrr.45.527. [DOI] [PubMed] [Google Scholar]
- Batra V, Mishra KP. Modulation of DNA methyltransferase profile by methyl donor starvation followed by gamma irradiation. Mol Cell Biochem. 2007 Jan;294:181–187. doi: 10.1007/s11010-006-9258-8. [DOI] [PubMed] [Google Scholar]
- Batra V, Sridhar S, Devasagayam TPA. Enhanced one-carbon flux towards DNA methylation: Effect of dietary methyl supplements against gamma-radiation-induced epigenetic modifications. Chem-Biol Interact. 2010 Feb 12;183:425–433. doi: 10.1016/j.cbi.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Batra V, Verma P. Dietary L-methionine supplementation mitigates gamma-radiation induced global DNA hypomethylation: Enhanced metabolic flux towards S-adenosyl-L-methionine (SAM) biosynthesis increases genomic methylation potential. Food Chem Toxicol. 2014 Jul;69:46–54. doi: 10.1016/j.fct.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Baubec T, Schübeler D. Genomic patterns and context specific interpretation of DNA methylation. Curr Opin Genet Dev. 2014 Apr;25:85–92. doi: 10.1016/j.gde.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Klinge DM, Liechty KC, March TH, Kang T, Gilliland FD, Sotnic N, Adamova G, Rusinova G, Telnov V. Plutonium targets the p16 gene for inactivation by promoter hypermethylation in human lung adenocarcinoma. Carcinogenesis. 2004 Jun;25:1063–1067. doi: 10.1093/carcin/bgh096. [DOI] [PubMed] [Google Scholar]
- Blakely EA, Kronenberg A. Heavy-ion radiobiology: New approaches to delineate mechanisms underlying enhanced biological effectiveness. Radiat Res. 1998 Nov;150:S126–S145. [PubMed] [Google Scholar]
- Brenner DJ. Slowing the Increase in the Population Dose Resulting from CT Scans. Radiat Res. 2010 Dec;174:809–815. doi: 10.1667/RR1859.1. [DOI] [PubMed] [Google Scholar]
- Brieger J, Mann SA, Pongsapich W, Koutsimpelas D, Fruth K, Mann WJ. Pharmacological genome demethylation increases radiosensitivity of head and neck squamous carcinoma cells. Int J Mol Med. 2012 Mar;29:505–509. doi: 10.3892/ijmm.2011.843. [DOI] [PubMed] [Google Scholar]
- Bujko M, Musialik E, Olbromski R, Przestrzelska M, Libura M, Pastwinska A, Juszczynski P, Zwierzchowski L, Baranowski P, Siedlecki JA. Repetitive genomic elements and overall DNA methylation changes in acute myeloid and childhood B-cell lymphoblastic leukemia patients. Int J Hematol. 2014 Jul;100:79–87. doi: 10.1007/s12185-014-1592-0. [DOI] [PubMed] [Google Scholar]
- Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev. 2012 Oct;38:726–736. doi: 10.1016/j.ctrv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Cellarier E, Durando X, Vasson MP, Farges MC, Demiden A, Maurizis JC, Madelmont JC, Chollet P. Methionine dependency and cancer treatment. Cancer Treat Rev. 2003 Dec;29:489–499. doi: 10.1016/s0305-7372(03)00118-x. [DOI] [PubMed] [Google Scholar]
- Christofidou-Solomidou M, Pietrofesa RA, Arguiri E, Schweitzer KS, Berdyshev EV, Mccarthy M, Corbitt A, Alwood JS, Yu Y, Globus RK, et al. Space radiation-associated lung injury in a murine model. Am J Physiol Lung Cell Mol Physiol. 2015;308:L416–L428. doi: 10.1152/ajplung.00260.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Haile RW, Laird PW. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012 Oct;131:1565–1589. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning APJ, Gu WJ, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. Plos Genet. 2011 Dec;7 doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dote H, Cerna D, Burgan WE, Carter DJ, Cerra MA, Hollingshead MG, Camphausen K, Tofilon PJ. Enhancement of in vitro and in vivo tumor cell radiosensitivity by the DNA methylation inhibitor zebularine. Clin Cancer Res. 2005 Jun 15;11:4571–4579. doi: 10.1158/1078-0432.CCR-05-0050. [DOI] [PubMed] [Google Scholar]
- Durando X, Farges MC, Buc E, Abrial C, Petorin-Lesens C, Gillet B, Vasson MP, Pezet D, Chollet P, Thivat E. Dietary methionine restriction with FOLFOX regimen as first line therapy of metastatic colorectal cancer: a feasibility study. Oncology-Basel. 2010;78:205–209. doi: 10.1159/000313700. [DOI] [PubMed] [Google Scholar]
- Durando X, Thivat E, Gimbergues P, Cellarier E, Abrial C, Dib M, Tacca O, Chollet P. Methionine dependency of cancer cells: a new therapeutic approach? B Cancer. 2008 Jan;95:69–76. doi: 10.1684/bdc.2008.0550. [DOI] [PubMed] [Google Scholar]
- Durante M. New challenges in high-energy particle radiobiology. Brit J Radiol. 2014 Mar;:87. doi: 10.1259/bjr.20130626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante M, Cucinotta FA. Heavy ion carcinogenesis and human space exploration. Nat Rev Cancer. 2008 Jun;8:465–472. doi: 10.1038/nrc2391. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Ehrlich KC. DNA cytosine methylation and hydroxymethylation at the borders. Epigenomics-Uk. 2014;6:563–566. doi: 10.2217/epi.14.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012 Feb;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Gebhard C, Schwarzfischer L, Pham TH, Schilling E, Klug M, Andreesen R, Rehli M. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006 Jun 15;66:6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- Giotopoulos G, McCormick C, Cole C, Zanker A, Jawad M, Brown R, Plumb M. DNA methylation during mouse hemopoietic differentiation and radiation-induced leukemia. Exp Hematol. 2006 Nov;34:1462–1470. doi: 10.1016/j.exphem.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Girdhani S, Sachs R, Hlatky L. Biological effects of proton radiation: what we know and don’t know. Radiat Res. 2013 Mar;179:257–272. doi: 10.1667/RR2839.1. [DOI] [PubMed] [Google Scholar]
- Goetz W, Morgan MNM, Baulch JE. The effect of radiation quality on genomic DNA methylation profiles in irradiated human cell lines. Radiat Res. 2011 May;175:575–587. doi: 10.1667/RR2390.1. [DOI] [PubMed] [Google Scholar]
- Gravina GL, Festuccia C, Marampon F, Popov VM, Pestell RG, Zani BM, Tombolini V. Biological rationale for the use of DNA methyltransferase inhibitors as new strategy for modulation of tumor response to chemotherapy and radiation. Mol Cancer. 2010 Nov;25:9. doi: 10.1186/1476-4598-9-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénin S, Morvan D, Thivat E, Stepien G, Demidem A. Combined methionine deprivation and chloroethylnitrosourea have time-dependent therapeutic synergy on melanoma tumors that NMR spectroscopy-based metabolomics explains by methionine and phospholipid metabolism reprogramming. Nutr Cancer. 2009;61:518–529. doi: 10.1080/01635580902803727. [DOI] [PubMed] [Google Scholar]
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012 Oct;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- Hofstetter B, Niemierko A, Forrer C, Benhattar J, Albertini V, Pruschy M, Bosman FT, Catapano CV, Ciernik IF. Impact of genomic methylation on radiation sensitivity of colorectal carcinoma. Int J Radiat Oncol. 2010 Apr 1;76:1512–1519. doi: 10.1016/j.ijrobp.2009.10.037. [DOI] [PubMed] [Google Scholar]
- Hondorp ER, Matthews RG. Methionine. EcoSal Plus. 2006;2 doi: 10.1128/ecosalplus.3.6.1.7. [DOI] [PubMed] [Google Scholar]
- Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, Ishimoto T, Iwatsuki M, Sakamaki K, Ohashi Y, Baba H. LINE-1 Hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg. 2013 Mar;257:449–455. doi: 10.1097/SLA.0b013e31826d8602. [DOI] [PubMed] [Google Scholar]
- Jangiam W, Tungjai M, Rithidech KN. Induction of chronic oxidative stress, chronic inflammation and aberrant patterns of DNA methylation in the liver of titanium-exposed CBA/CaJ mice. Int J Radiat Biol. 2015 May;91:389–398. doi: 10.3109/09553002.2015.1001882. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li YQ, Liu N, Sun Y, He QM, Jiang N, Xu YF, Chen L, Ma J. 5-Azacytidine enhances the radiosensitivity of CNE2 and SUNE1 cells in vitro and in vivo possibly by altering DNA methylation. PLoS ONE. 2014 Apr;1:9. doi: 10.1371/journal.pone.0093273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KC, Koestler DC, Fleischer T, Chen PP, Jenson EG, Marotti JD, Onega T, Kristensen VN, Christensen BC. DNA methylation in ductal carcinoma in situ related with future development of invasive breast cancer. Clin Epigenetics. 2015 Jul;25:7. doi: 10.1186/s13148-015-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012 Jul;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kalinich JF, Catravas GN, Snyder SL. The effect of gamma-radiation on DNA methylation. Radiat Res. 1989 Feb;117:185–197. [PubMed] [Google Scholar]
- Kim EH, Park AK, Dong SM, Ahn JH, Park WY. Global analysis of CpG methylation reveals epigenetic control of the radiosensitivity in lung cancer cell lines. Oncogene. 2010 Aug;29:4725–4731. doi: 10.1038/onc.2010.223. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim JH, Chie EK, Young PD, Kim IA, Kim IH. DNMT (DNA methyltransferase) inhibitors radiosensitize human cancer cells by suppressing DNA repair activity. Radiat Oncol. 2012 Mar;20:7. doi: 10.1186/1748-717X-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinakis DM, Brickner AG, Kirkwood JM, Liu XY, Goldwasser JE, Kastrama A, Sander C, Bocangel D, Chada S. Mitotic arrest, apoptosis, and sensitization to chemotherapy of melanomas by methionine deprivation stress. Mol Cancer Res. 2006 Aug;4:575–589. doi: 10.1158/1541-7786.MCR-05-0240. [DOI] [PubMed] [Google Scholar]
- Kontic M, Stojsic J, Jovanovic D, Bunjevacki V, Ognjanovic S, Kuriger J, Puumala S, Nelson HH. Aberrant promoter methylation of CDH13 and MGMT genes is associated with clinicopathologic characteristics of primary non-small-cell lung carcinoma. Clin Lung Cancer. 2012 Jul;13:297–303. doi: 10.1016/j.cllc.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Beland FA, Pogribny IP. Role of epigenetic events in chemical carcinogenesis-a justification for incorporating epigenetic evaluations in cancer risk assessment. Toxicol Mech Meth. 2011a May;21:289–297. doi: 10.3109/15376516.2011.557881. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Boyko A, Rodriguez-Juarez R, McDonald RJ, Tryndyak VP, Kovalchuk I, Pogribny IP, Kovalchuk O. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis. 2007 Aug;28:1831–1838. doi: 10.1093/carcin/bgm053. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Kutanzi K, Hendrickson K, Rodriguez-Juarez R, Kogosov D, Kovalchuk O. Radiation-induced bystander effects in vivo are sex specific. Mutat Res-Fund Mol M. 2008 Jul 3;642:28–36. doi: 10.1016/j.mrfmmm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Loree J, Kutanzi K, Koganow C, Pogribny I, Kovalchuk O. In vivo bystander effect: Cranial X-irradiation leads to elevated DNA damage, altered cellular proliferation and apoptosis, and increased p53 levels in shielded spleen. Int J Radiat Oncol. 2008 Feb 1;70:554–562. doi: 10.1016/j.ijrobp.2007.09.039. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Miousse IR, Sridharan V, Nzabarushimana E, Skinner CM, Melnyk SB, Pavliv O, Hauer-Jensen M, Nelson GA, Boerma M. Radiation-induced changes in DNA methylation of repetitive elements in the mouse heart. Mutat Res-Fund Mol M. 2016 May;787:43–53. doi: 10.1016/j.mrfmmm.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue – A possible mechanism contributing to radiation carcinogenesis? Biochem Biophys Res Commun. 2005 Nov 18;337:526–533. doi: 10.1016/j.bbrc.2005.09.084. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Rugo RE, Hendricks CA, Loree J, Thibault B, Kutanzi K, Pogribny I, Yanch JC, Engelward BP, Kovalchuk O. Irradiation induces DNA damage and modulates epigenetic effectors in distant bystander tissue in vivo. Oncogene. 2006 Jul 20;25:4267–4275. doi: 10.1038/sj.onc.1209467. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Zemp F, Kolb B, Kovalchuk O. Sex-specific radiation-induced microRNAome responses in the hippocampus, cerebellum and frontal cortex in a mouse model. Mutat Res-Gen Tox En. 2011b Jun 17;722:114–118. doi: 10.1016/j.mrgentox.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Kovalchuk O, Burke P, Besplug J, Slovack M, Filkowski J, Pogribny I. Methylation changes in muscle and liver tissues of male and female mice exposed to acute and chronic low-dose X-ray-irradiation. Mutat Res-Fund Mol M. 2004 Apr 14;548:75–84. doi: 10.1016/j.mrfmmm.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Kroeger H, Jelinek J, Estécio MRH, He R, Kondo K, Chung W, Zhang L, Shen LL, Kantarjian HM, Bueso-Ramos CE, et al. Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse. Blood. 2008 Aug 15;112:1366–1373. doi: 10.1182/blood-2007-11-126227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann C, Weichenhan D, Rehli M, Plass C, Schmezer P, Popanda O. DNA methylation changes in cells regrowing after fractioned ionizing radiation. Radiother Oncol. 2011 Oct;101:116–121. doi: 10.1016/j.radonc.2011.05.048. [DOI] [PubMed] [Google Scholar]
- Lahtz C, Bates SE, Jiang Y, Li AX, Wu XW, Hahn MA, Pfeifer GP. Gamma irradiation does not induce detectable changes in DNA methylation directly following exposure of human cells. PLoS ONE. 2012 Sep;14:7. doi: 10.1371/journal.pone.0044858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang LX, Gokcumen O, Haseley P, Luquette LJ, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012 Aug 24;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima F, Ding DC, Goetz W, Yang AJ, Baulch JE. High LET Fe-56 ion irradiation induces tissue-specific changes in DNA methylation in the mouse. Environ Mol Mutagen. 2014 Apr;55:266–277. doi: 10.1002/em.21832. [DOI] [PubMed] [Google Scholar]
- Loree J, Koturbash I, Kutanzi K, Baker M, Pogribny I, Kovalchuk O. Radiation-induced molecular changes in rat mammary tissue: Possible implications for radiation-induced carcinogenesis. Int J Radiat Biol. 2006 Nov;82:805–815. doi: 10.1080/09553000600960027. [DOI] [PubMed] [Google Scholar]
- Lyon CM, Klinge DM, Liechty KC, Gentry FD, March TH, Kang T, Gilliland FD, Adamova G, Rusinova G, Telnov V, et al. Radiation-induced lung adenocarcinoma is associated with increased frequency of genes, inactivated by promoter hypermethylation. Radiat Res. 2007 Oct;168:409–414. doi: 10.1667/RR0825.1. [DOI] [PubMed] [Google Scholar]
- Liu Ma S, Jiao X, Yang B, Liu YX. Low-dose radiation-induced responses: focusing on epigenetic regulation. Int J Radiat Biol. 2010 Jul;86(7):517–28. doi: 10.3109/09553001003734592. [DOI] [PubMed] [Google Scholar]
- Miousse IR, Chalbot MCG, Lumen A, Ferguson A, Kavouras IG, Koturbash I. Response of transposable elements to environmental stressors. Mutat Res-Rev Mutat. 2015 Jul-Sep;765:19–39. doi: 10.1016/j.mrrev.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Koturbash I. The fine LINE: Methylation drawing the cancer landscape. BioMed Res Int. 2015;2015:131547. doi: 10.1155/2015/131547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Shao LJ, Chang JH, Feng W, Wang YY, Allen AR, Turner J, Stewart B, Raber J, Zhou DH, et al. Exposure to low-dose Fe-56-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat Res. 2014 Jul;182:92–101. doi: 10.1667/RR13580.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moding EJ, Kastan MB, Kirsch DG. Strategies for optimizing the response of cancer and normal tissues to radiation. Nat Rev Drug Discov. 2013 Jul;12:526–542. doi: 10.1038/nrd4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momparler RL, Côté S, Momparler LF, Idaghdour Y. Epigenetic therapy of acute myeloid leukemia using 5-aza-2′-deoxycytidine (decitabine) in combination with inhibitors of histone methylation and deacetylation. Clin Epigenetics. 2014 Oct;1:6. doi: 10.1186/1868-7083-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothersill C, Seymour C. Are epigenetic mechanisms involved in radiation-induced bystander effects? Frontiers in genetics. 2012;3:74. doi: 10.3389/fgene.2012.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002 Dec 5;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nervi C, De Marinis E, Codacci-Pisanelli G. Epigenetic treatment of solid tumours: a review of clinical trials. Clin Epigenetics. 2015 Dec 10;7:127. doi: 10.1186/s13148-015-0157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MR, Sykes PJ, Blyth BJ, Bezak E, Lawrence MD, Morel KL, Ormsby RJ. The methylation of DNA repeat elements is sex-dependent and temporally different in response to X radiation in radiosensitive and radioresistant mouse strains. Radiat Res. 2014b Jan;181:65–75. doi: 10.1667/RR13460.1. [DOI] [PubMed] [Google Scholar]
- Newman MR, Sykes PJ, Blyth BJ, Bezak E, Lawrence MD, Morel KL, Ormsby RJ. A single whole-body low dose X-irradiation does not affect L1, B1 and IAP repeat element DNA methylation longitudinally. Plos One. 2014a Mar 27;9:e93016. doi: 10.1371/journal.pone.0093016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis G, Raji OY, Markopoulou S, Gosney JR, Bryan J, Warburton C, Walshaw M, Sheard J, Field JK, Liloglou T. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res. 2012 Nov 15;72:5692–5701. doi: 10.1158/0008-5472.CAN-12-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyazi M, Schnell O, Suchorska B, Schwarz SB, Ganswindt U, Geisler J, Bartenstein P, Kreth FW, Tonn JC, Eigenbrod S, et al. FET-PET assessed recurrence pattern after radio-chemotherapy in newly diagnosed patients with glioblastoma is influenced by MGMT methylation status. Radiother Oncol. 2012 Jul;104:78–82. doi: 10.1016/j.radonc.2012.04.022. [DOI] [PubMed] [Google Scholar]
- Nüsgen N, Goering W, Dauksa A, Biswas A, Jamil MA, Dimitriou I, Sharma A, Singer H, Fimmers R, Frohlich H, et al. Inter-locus as well as intra-locus heterogeneity in LINE-1 promoter methylation in common human cancers suggests selective demethylation pressure at specific CpGs. Clin Epigenetics. 2015 Mar 1;7:17. doi: 10.1186/s13148-015-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzabarushimana E, Miousse IR, Shao LJ, Chang JH, Allen AR, Turner J, Stewart B, Raber J, Koturbash I. Long-term epigenetic effects of exposure to low doses of Fe-56 in the mouse lung. J Radiat Res. 2014 Jul;55:823–828. doi: 10.1093/jrr/rru010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayiotidis MI, Rancourt RC, Allen CB, Riddle SR, Schneider BK, Ahmad S, White CW. Hyperoxia-induced DNA damage causes decreased DNA methylation in human lung epithelial-like A549 cells. Antioxid Redox Sign. 2004 Feb;6:129–136. doi: 10.1089/152308604771978435. [DOI] [PubMed] [Google Scholar]
- Pavillard V, Nicolaou A, Double JA, Phillips RM. Methionine dependence of tumours: A biochemical strategy for optimizing paclitaxel chemosensitivity in vitro. Biochem Pharmacol. 2006 Mar 14;71:772–778. doi: 10.1016/j.bcp.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun. 2004 Aug 6;320:1253–1261. doi: 10.1016/j.bbrc.2004.06.081. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Koturbash I, Tryndyak V, Hudson D, Stevenson SML, Sedelnikova O, Bonner W, Kovalchuk O. Fractionated low-dose radiation exposure leads to accumulation of DNA damage and profound alterations in DNA and histone methylation in the murine thymus. Mol Cancer Res. 2005 Oct;3:553–561. doi: 10.1158/1541-7786.MCR-05-0074. [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010 Oct;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Prior S, Miousse IR, Nzabarushimana E, Pathak R, Skinner C, Kutanzi KR, Allen AR, Raber J, Tackett AJ, Hauer-Jensen M, et al. Densely ionizing radiation affects DNA methylation of selective LINE-1 elements. Environmental Research. 2016;150:470–481. doi: 10.1016/j.envres.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin TC, Si JL, Raynal NJM, Wang XD, Gharibyan V, Ahmed S, Hu X, Jin CL, Lu Y, Shu JM, et al. Epigenetic synergy between decitabine and platinum derivatives. Clin Epigenetics. 2015 Sep;11:7. doi: 10.1186/s13148-015-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Yashiro M, Shinto O, Matsuzaki T, Hirakawa K. DNA methyltransferase inhibitor 5-aza-CdR enhances the radiosensitivity of gastric cancer cells. Cancer Sci. 2009 Jan;100:181–188. doi: 10.1111/j.1349-7006.2008.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakova I. Methylation of newly synthesized DNA in rat bone marrow and thymus after irradiation. Radiobiologiia. 1979;19:413. Russian. [PubMed] [Google Scholar]
- Ricketts CJ, Morris MR, Gentle D, Shuib S, Brown M, Clarke N, Wei WB, Nathan P, Latif F, Maher ER. Methylation profiling and evaluation of demethylating therapy in renal cell carcinoma. Clin Epigenetics. 2013 Sep 13;5:16. doi: 10.1186/1868-7083-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rithidech KN, Honikel LM, Reungpathanaphong P, Tungjai M, Jangiam W, Whorton EB. Late-occurring chromosome aberrations and global DNA methylation in hematopoietic stem/progenitor cells of CBA/CaJ mice exposed to silicon (Si-28) ions. Mutat Res-Fund Mol M. 2015 Nov;781:22–31. doi: 10.1016/j.mrfmmm.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, Aldape KD. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma (vol 12, pg 116, 2010) Neuro-Oncology. 2010 Jun;12:617–617. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OA, Nakamura A, Kovalchuk O, Koturbash I, Mitchell SA, Marino SA, Brenner DJ, Bonner WM. DNA double-strand breaks form in bystander cells after microbeam irradiation of three-dimensional human tissue models. Cancer Res. 2007 May 1;67:4295–4302. doi: 10.1158/0008-5472.CAN-06-4442. [DOI] [PubMed] [Google Scholar]
- Shivapurkar N, Wilson MJ, Hoover KL, Mikol YB, Creasia D, Poirier LA. Hepatic DNA methylation and liver-tumor formation in male C3h mice fed methionine-deficient and choline-deficient diets. J Natl Cancer I. 1986 Jul;77:213–217. [PubMed] [Google Scholar]
- Smits KM, Melotte V, Niessen HE, Dubois L, Oberije C, Troost EG, Starmans MH, Boutros PC, Vooijs M, van Engeland M, Lambin P. Epigenetics in radiotherapy: where are we heading? Radiother Oncol. 2014 May;111(2):168–77. doi: 10.1016/j.radonc.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Song SH, Han SW, Bang YJ. Epigenetic-based therapies in cancer progress to date. Drugs. 2011;71:2391–2403. doi: 10.2165/11596690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Song WG, Liu YZ, Liu Y, Zhang C, Yuan B, Zhang LB, Sun SL. Increased P16 DNA methylation in mouse thymic lymphoma induced by irradiation. PLoS ONE. 2014 Apr 18;9:e93850. doi: 10.1371/journal.pone.0093850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RA. Epigenetics and environmental exposures. J Epidemiol Community Health. 2012 Jan;66:8–13. doi: 10.1136/jech.2010.130690. [DOI] [PubMed] [Google Scholar]
- Strekalova E, Malin D, Good DM, Cryns VL. Methionine deprivation induces a targetable vulnerability in triple-negative breast cancer cells by enhancing TRAIL receptor-2 expression. Clin Cancer Res. 2015 Jun 15;21:2780–2791. doi: 10.1158/1078-0432.CCR-14-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Su SB, Zhang YL, Yang W, Shen LJ, Cao YP, Tong YJ. Aberrant promoter methylation of p16 (INK4a) and O-6-methylguanine-DNA methyltransferase genes in workers at a Chinese uranium mine. J Occup Health. 2006 Jul;48:261–266. doi: 10.1539/joh.48.261. [DOI] [PubMed] [Google Scholar]
- Tawa R, Kimura Y, Komura J, Miyamura Y, Kurishita A, Sasaki MS, Sakurai H, Ono T. Effects of X-ray irradiation on genomic DNA methylation levels in mouse tissues. J Radiat Res. 1998 Dec;39:271–278. doi: 10.1269/jrr.39.271. [DOI] [PubMed] [Google Scholar]
- Tommasino F, Durante M. Proton radiobiology. Cancers. 2015;7:353–381. doi: 10.3390/cancers7010353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, Zhang YW, Xu K, Mao XB, Xue LJ, Liu XB, Yu HJ, Chen LB, Chu XY. Genome-wide screen of DNA methylation changes induced by low dose X-Ray radiation in mice. PLoS ONE. 2014 Mar;10:9. doi: 10.1371/journal.pone.0090804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Scheiber MN, Neumann C, Calin GA, Zhou DH. MicroRNA regulation of ionizing radiation-induced premature senescence. Int J Radiat Oncol. 2011 Nov 1;81:839–848. doi: 10.1016/j.ijrobp.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil MM, Bedford JS, Bielefeldt-Ohmann H, Ray FA, Genik PC, Ehrhart EJ, Fallgren CM, Hailu F, Battaglia CLR, Charles B, et al. Incidence of acute myeloid leukemia and hepatocellular carcinoma in mice irradiated with 1 GeV/nucleon Fe-56 ions. Radiat Res. 2009 Aug;172:213–219. doi: 10.1667/RR1648.1. [DOI] [PubMed] [Google Scholar]
- Weil MM, Ray FA, Genik PC, Yu YJ, McCarthy M, Fallgren CM, Ullrich RL. Effects of Si-28 ions, Fe-56 ions, and protons on the induction of murine acute myeloid leukemia and hepatocellular carcinoma. Plos One. 2014 Aug 15;9:e104819. doi: 10.1371/journal.pone.0104819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield Billen D. In vivo methylation of Escherichia coli DNA following ultraviolet and X-irradiation. J Mol Biol. 1972;63:363–372. doi: 10.1016/0022-2836(72)90433-0. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Shivapurkar N, Poirier LA. Hypomethylation of hepatic nuclear-DNA in rats fed with a carcinogenic methyl-deficient diet. Biochem J. 1984;218:987–990. doi: 10.1042/bj2180987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Li LJ. 5-hydroxymethylcytosine: a new insight into epigenetics in cancer. Cancer Biol Ther. 2014 Jan 1;15:10–15. doi: 10.4161/cbt.27144. [DOI] [PMC free article] [PubMed] [Google Scholar]


