Abstract
Stressful environmental exposures incurred early in development can affect postnatal metabolic health and susceptibility to non-communicable diseases in adulthood, although the molecular mechanisms by which this occurs have yet to be elucidated. Here we use a mouse model to investigate how assorted in vitro exposures restricted exclusively to the preimplantation period affect transcription both acutely in embryos and long-term in subsequent offspring adult tissues, to determine if reliable transcriptional markers of in vitro stress are present at specific developmental time points and throughout development. Each in vitro fertilization or embryo culture environment led to a specific and unique blastocyst transcriptional profile, but we identified a common 18-gene and 9-pathway signature of preimplantation embryo manipulation that was present in all in vitro embryos irrespective of culture condition or method of fertilization. This fingerprint did not persist throughout development and there was no clear transcriptional cohesion between adult IVF offspring tissues or compared to their preceding embryos, indicating a tissue-specific impact of in vitro stress on gene expression. However, the transcriptional changes present in each IVF tissue were targeted by the same upstream transcriptional regulators, which provide insight as to how acute transcriptional responses to stressful environmental exposures might be preserved throughout development to influence adult gene expression.
Keywords: developmental origins of health and disease, assisted reproductive technologies, in vitro fertilization, blastocyst, embryo culture, microarray, gene expression
INTRODUCTION
There is robust epidemiological and animal evidence that exposure to different environmental conditions during early development affects postnatal growth, metabolism, and disease susceptibility in adulthood. As dictated by the Developmental Origins of Health and Disease (DOHaD) hypothesis, poor or suboptimal developmental experiences—including nutritional, oxidative, or in vitro stress—can predispose chronic diseases, including hypertension and components of the metabolic syndrome (Gillman 2005).
Arguably the most pressing question in the DOHaD field is identifying how the molecular changes occurring in a developing organism secondary to adverse environmental or nutritional exposures alter growth and metabolic trajectories across the life course. Interestingly, many models of DOHaD exhibit common long-term outcomes, including glucose intolerance, hypertension and vascular dysfunction, irrespective of both the form of developmental stress and the timing of exposure (Jungheim, et al. 2010, Simmons, et al. 2001, Watkins, et al. 2011). This suggests that common mechanisms may be involved in both sensing and transducing environment stimuli, leading to a reprogrammed epigenetic, transcriptional and/or metabolic state that could potentiate an increased susceptibility for metabolic disease throughout postnatal life. As a result, understanding the mechanisms by which the developing embryo and fetus assimilate environmental input into a programmed metabolic response is relevant across multiple DOHaD fields.
Appropriate to this discussion is the evidence that preimplantation development is an important period of environmental sensitivity, and that stresses incurred within this window can affect glucose metabolism, β-cell function, blood pressure and fat deposition in adulthood (Donjacour, et al. 2014, Fernandez-Gonzalez, et al. 2004, Kwong, et al. 2000, Rexhaj, et al. 2013, Watkins, et al. 2010). The preimplantation period is a particularly advantageous model for investigating DOHaD-related questions because exposures are limited to a time frame of 4-5 days and embryos exhibit cellular uniformity (only 2 cell types are formed after 4 days in culture: the inner cell mass and trophectoderm)—thereby providing an opportunity to connect precise variations in exposure to outcome. In addition, embryo manipulation holds wide clinical significance due to the routine use of Assisted Reproductive Technologies (ART) such as in vitro fertilization (IVF), which has resulted in the birth of over 5 million children as of 2012 (ICMART 2012).
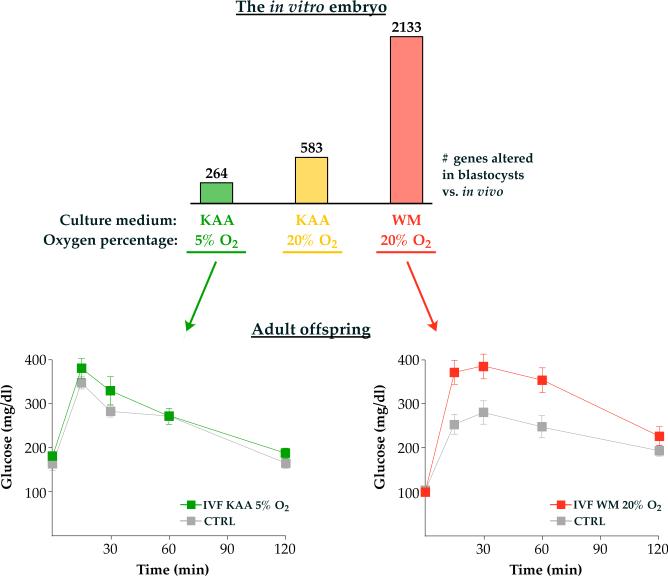
It is also clear that increasing the severity of a particular stressor leads to progressively worse phenotypes in adulthood. For example, mouse IVF and embryo culture performed under clinically optimized conditions affect the expression of nearly 300 genes in blastocysts but do not alter adult glucose tolerance. Conversely increasing embryo culture stress with a higher oxygen tension (20% O2) or an inferior culture medium exacerbates the transcriptional changes (over 2000 misexpressed genes in blastocysts) and results in significant glucose intolerance in adulthood (Figure 1) (Donjacour, et al. 2014, Feuer, et al. 2014).
Figure 1. Embryos with more severe changes in gene expression after in vitro manipulation will manifest abnormal glucose tolerance as adults.
Different culture media and oxygen tensions in combination have distinct effects on blastocyst gene expression and adult glucose tolerance. The number indicates statistically different gene expression compared to blastocysts conceived naturally and flushed from the uterus (control). In general, suboptimal conditions such as Whitten's medium (WM) or higher oxygen concentration (20% Ox) will more severely affect embryo transcription and result in worse offspring glucose phenotypes (red panel). Conversely, optimized conditions such as K simplex optimized medium (KAA) and physiologic oxygen concentration (5% O2) have a lesser impact on blastocyst transcription with normal offspring glucose tolerance (green panel). IVF, in vitro fertilization.
Over the past 15 years the Rinaudo laboratory has performed microarray studies in mice to examine how the many different components of in vitro embryo manipulation (culture medium composition, oxygen tension, method of fertilization) have influenced transcriptional profiles in both blastocysts and adult offspring tissues (Feuer, et al. 2014, Giritharan, et al. 2012, Giritharan, et al. 2010, Giritharan, et al. 2007, Rinaudo and Schultz 2004, Rinaudo, et al. 2006). These experiments have been conducted by several scientists in multiple locations, using various microarray platforms and analysis software. Further, there have been significant advancements in our knowledge of the genome over the past decade. This manuscript provides a systematic analysis and integration of the gene expression changes present in 1) blastocysts exposed to assorted culture conditions and methods of fertilization, and 2) selected adult tissues from offspring generated by in vitro fertilization. The ultimate goal of this study was to compare the transcriptional profiles to identify common signatures of in vitro embryo manipulation, and to determine if a relationship exists between the acute transcriptional response to in vitro stress and subsequent adult gene expression.
METHODS
Animals
All animals were maintained according to institutional regulations and NIH guidelines, under a constant 12-hour light/dark cycle with ad libitum access to water and standard chow (PicoLab® #5058). The strains used in this study included vasectomized CD-1 males, B6D2F1/J males, CF-1 females, and C57Bl/6J males and females. Beginning at 24 weeks, the postnatal IVF and control inbred C57Bl/6J cohorts were placed on a high fat diet (Research Diets, Inc. #D12492) until time of sacrifice at 29 weeks. The postnatal IVF and control outbred CF1 × B6D2F1 cohorts were maintained on a standard chow diet (20% protein, 9% fat, LabDiet) until sacrifice at 40 weeks.
Embryo generation, culture and collection
A detailed methodology of the embryo collection and transfer techniques has been published and may be found in (Feuer, et al. 2014, Giritharan, et al. 2012, Giritharan, et al. 2010, Giritharan, et al. 2007, Rinaudo and Schultz 2004, Rinaudo, et al. 2006). Briefly, CF-1 females aged 6-8 weeks were injected with 5 IU PMSG followed 46-68 hours later by 5 IU hCG to induce superovulation.
For in vitro culture (IVC) experiments, superovulated dams were mated overnight post-hCG injection with B6D2F1/J males. The next morning, fertilized zygotes were flushed from ampullae, washed and cultured to the blastocyst stage at 37°C under Ovoil™ (Vitrolife, #10029) with 5% CO2 in a modular humidified chamber. At the two-cell stage, all unfertilized oocytes were removed. Culture conditions included either Whitten's medium [WM (Whitten 1971); made in house] or potassium simplex optimization medium supplemented with amino acids [KAA (Ho, et al. 1995); Millipore, MR-106-D], with 5% or 20% oxygen (4 possible conditions).
For in vitro fertilization (IVF) experiments, 13-15 hours following hCG administration the cumulous-oocyte complexes were isolated from ampullae and incubated 4-6 hours in HTF medium (Millipore, MR-070-D) with capacitated (1 hour) cauda epididymal sperm from B6D2F1/J males. Fertilized zygotes were washed and cultured to the blastocyst stage as described above in WM and 20% oxygen. A second IVF cohort was generated using C57Bl/6J animals cultured under KAA and 5% oxygen conditions.
For intracytoplasmic sperm injection (ICSI) experiments, oocytes were obtained the morning after superovulation as described for IVF. Cauda epididymides were isolated from B6D2F1/J males aged 10-11 weeks, punctured gently with 30-G needles and incubated 10 min in a microcentrifuge tube at 37°C in pre-warmed 1ml EGTA Tris-HCl buffered solution (10mM Tris-HCl pH 8.2, 50mM EGTA and 50mM NaCl). ICSI was performed using the top 800μl sperm suspension, and oocytes containing the first polar body were microinjected using a Piezo drill (PMM Controller, Prime Tech) as described (Li, et al. 2009). Fertilized zygotes were washed and cultured to the blastocyst stage in WM and humidified air.
To obtain late-cavitating blastocysts of similar morphology, embryos were harvested at different time points: after 96-100 hr (IVC), 106-110 hr (IVF), or 112 hr (ICSI) and snap frozen for microarray experiments. The in vivo-derived control group was represented by blastocysts isolated from naturally mated superovulated dams 96 hrs post-hCG injection (CF-1 x B6D2F1/J, or C57Bl/6J, as indicated in the text). At all times, the experimental groups were compared to the same in vivo control breeds. All embryo generation experiments were performed ≥4 times.
We have previously shown that IVF and in vivo embryos derived by these protocols contain a similar number of inner cell mass (ICM) cells (12.8 ± 0.4 vs. 13.8 ± 0.5, not significant) (Giritharan, et al. 2012). ICM was isolated from CF-1 × B6D2F1 control and IVF blastocysts cultured in WM and 20% oxygen by immunosurgery (n=3 times per treatment group). Briefly, trophectoderm cells were lysed by incubating embryos 30 min in WM containing 20μg/ml anti-mouse rabbit antibodies (Sigma) and 30 min in WM with 5μg/ml rabbit complement (Invitrogen) at 37°C. ICM samples were cleaned of destroyed TE via repeated pipetting using a glass pipette with a 30-40μm diameter under a dissecting microscope. Upon collection, ICM samples were immediately transferred to cell lysis buffer provided in the PicoPure RNA isolation kit (Molecular Devices) and frozen at −80°C.
Embryo transfer
For postimplantation cohorts, pseudopregnancy was induced by mating naturally cycling CF-1 females to vasectomized CD-1 males, confirmed by the presence of a copulation plug the next morning (considered day 0.5). Late-cavitating blastocysts were transferred to the uterine horns of recipients on day 2.5 of pseudopregnancy. For control experiments, superovulated dams were mated overnight; embryonic day 3.5 blastocysts (96 hrs after hCG administration) were flushed from the uterine horns and transferred immediately to the uterine horns of CF-1 recipients. This experimental strategy controls for litter size and any effects of superovulation or the embryo transfer procedure, both of which can influence imprinted gene expression (Fortier, et al. 2008, Rivera, et al. 2008). Resulting pups were not cross-fostered, as this procedure may alter adult phenotypes and imprinted gene expression (Hager, et al. 2009, Matthews, et al. 2011).
Microarray preparation
Comprehensive descriptions of RNA extraction, amplification, fragmentation and hybridization to Affymetrix GeneChips for each microarray experiment may be found in (Feuer, et al. 2014, Giritharan, et al. 2012, Giritharan, et al. 2010, Giritharan, et al. 2007, Rinaudo and Schultz 2004, Rinaudo, et al. 2006). Table 1 outlines the individual microarrays presented or re-analyzed in this study.
Table 1.
Summary of the microarray experiments used in this study.
| Experiment | Fertilization | Culture | Tissue | Platform | Location | Publication | |
|---|---|---|---|---|---|---|---|
| Effect of embryo culture | Natural (IVC) | KAA 5% O2 | CF1 × B6D2F1/J Blastocsyt | Affymetrix MOE 430A (22,690 probes) | University of Pennsylvania | (Rinaudo and Schultz 2004, Rinaudo, et al. 2006) | |
| WM 5% O2 | |||||||
| KAA 20% O2 | |||||||
| WM 20% O2 | |||||||
| Effect of fertilization | Natural (IVC) | WM 20% O2 | CF1 × B6D2F1/J Blastocsyt | Affymetrix MOE 430.2 (45,101 probes) | University of California, San Francisco | (Giritharan, et al. 2010, Giritharan, et al. 2007) | |
| IVF | |||||||
| ICSI | |||||||
| Long-term effects: adult tissues | IVF | KAA 5% O2 | C57Bl/6J 29 week female | Liver | Affymetrix Mouse 1.0 ST array (>770,000 probes) | University of California, San Francisco | (Feuer, et al. 2014) |
| Gonadal fat | |||||||
| Skeletal muscle | |||||||
| Pancreatic islets | |||||||
| Long-term effects: adult tissues | IVF | WM 20% O2 | CF1 × B6D2F1/J 40 week male | Heart | Affymetrix Mouse 1.0 ST array (>770,000 probes) | University of California, San Francisco | Unpublished |
| Cell type specificity | IVF | WM 20% O2 | CF1 × B6D2F1/J inner cell mass (ICM) | Affymetrix MOE 430.2 (45,101 probes) | University of California, San Francisco | (Giritharan, et al. 2012) | |
For embryo studies, total RNA was extracted either with Trizol containing 2ml Pellet Paint (Novagen) or a PicoPure RNA Isolation kit (Arcturus) from 3-4 pooled embryo replicates comprised of 80 blastocysts (IVC), 10 blastocysts (IVF or ICSI), or 40 ICM samples. RNA samples were submitted for preparation and hybridization to the University of Pennsylvania Microarray Facility (IVC) or the Genomic Core Facility at the University of California, San Francisco (IVF, ICSI, ICM).
In adult offspring, microarray analysis was performed on two postnatal cohorts: 1) 29 week old C57Bl/6J female gonadal fat, liver, skeletal muscle and pancreatic islets derived from IVF – KAA 5% oxygen or naturally (n=3 per condition, with each animal providing the 4 tissues to minimize variation), and 2) 40 week old CF1 × B6D2F1/J male cardiac tissue derived from IVF – WM 20% oxygen or naturally (n=3 per condition). Animals contributing to these analyses were selected from at least two separate litters of 5-10 pups per condition. Total RNA was extracted and purified from previously frozen tissues using the RNeasy mini kit (Qiagen), then submitted to the Gladstone Genomics Core Facility at the University of California, San Francisco for labeling, hybridization and scanning of the microarrays. Whole frozen islets were also sent to the core facility for RNA isolation and amplification prior to microarray processing.
Microarray data analysis
All microarray data were analyzed with GeneSpring GX 13.1 software (Agilent Technologies). Scanned arrays were uploaded into GeneSpring for background adjustment, summarization, log transformation and baseline transformation. Samples were interpreted by culture medium, oxygen, method of fertilization and cell-type, via individual microarray platform. In addition to β-actin, Gapdh signal is used as an internal quality control in both Affymetrix MOE 430A and 430.2 arrays, and its intensity level can lead to the inclusion versus exclusion of specific samples as outliers in a dataset during a quality control analysis. We recently discovered that Gapdh is altered in IVF blastocysts compared to naturally-derived control blastocysts (data not shown), suggesting that previous analysis of these microarrays may have led to the incorrect inclusion/exclusion of data. As a result, these experiments were re-analyzed excluding Gapdh signal as a quality control influence. The Principal Component Analysis (PCA) algorithm in GeneSpring was applied to all specimens grouped by individual ART condition (including culture medium, oxygen percentage, and method of fertilization) using all genes and expressed sequence tags, separated by microarray platform, to evaluate for patterns in gene expression and underlying cluster structures. Fold-changes were calculated based on the normalized scores instead of the raw expression data. To identify differentiated genes between two groups (e.g. ART versus in vivo), an un-paired t-test with a significance threshold of p-value <0.05 was applied to compare ART to control samples. The Effect-of-Culture and Cell-Type-Specificity experiments were analyzed using a 30% cutoff with Benjamini-Hochberg correction; the Effect-of-Fertilization experiments were analyzed using a 50% cutoff with Benjamini-Hochberg correction; and the Long-Term adult tissue data were analyzed using a 30% cutoff without correction.
Gene functional analysis and pathway analysis
Post-processing of the resulting gene lists was conducted using Ingenuity Pathway Analysis (IPA, June 2015 release), a structured knowledge repository that identifies biologically significant relationships and networks based on previously characterized functional associations of genes (http://ingenuity.com). The analysis includes canonical pathways overrepresented in gene lists, how genes and pathways integrate into broader biological networks, as well as predicted upstream regulators responsible for the cascade of gene expression changes. Only data experimentally observed in animal tissues were considered, and fold-change thresholds were set such that the number of entities contributing to each analysis remained between the recommended 100-2000.
Heatmaps were generated using GENE-E software developed by the Broad Institute (available at http://www.broadinstitute.org/cancer/software/GENE-E/).
Microarray validation by real-time quantitative RT-qPCR
To validate the microarray data, blastocysts derived in vivo or cultured from the zygotic stage (IVC, WM with 20% oxygen or KAA with 5% oxygen) were collected as described above, and real-time quantitative RT-qPCR was conducted on 3-4 independent biological replicates containing 10 or more pooled blastocysts. Total RNA was extracted using the PicoPure RNA Isolation Kit (Applied Biosystems) and reverse transcribed to cDNA using an iScript cDNA synthesis kit (Bio-Rad Laboratories). Quantification of gene transcripts was performed in duplicate with SyBr Green PCR Supermix using 0.2 embryo equivalents of cDNA from each treatment group per reaction. Amounts of Arrdc4 (Forward: 5’-CCC TTA TTG ACT CCT ATG CT and reverse: 3’-CTT CTC CGT TAC AGT AGC CC); Slc7a3 (Forward: 5’-TTC TGG CCG AGT TGT CTA TGT TTG and reverse: 3’-AGT GCG GTT CTG TGG CTG TCT C (Bloise, et al. 2012));Socs3 (Forward: 5’-GCA GGA GAG CGG ATT CTA CT and reverse: 3’-ACG CTC AAC GTG AAG AAG TG); and Fgf4 (Forward: 5’-CCG GTG CAG CGA GGC GTG GT and reverse: 3’-GGA AGG AAG TGG GTG ACC TTC AT (Rappolee, et al. 1994)) transcripts were normalized to levels of histone 2A (H2A) transcript (Forward: 5’-ACA TGG CGG CGG TGC TGG AGT A and reverse: 3-‘CGG GAT GCG CGT CTT CTT GTT). H2A was selected as a reference because it is stably and reliably expressed across preimplantation embryo development (Jeong, et al. 2005, Kuijk, et al. 2007).
Primers were designed using PerlPrimer software (http://perlprimer.sourceforge.net/) unless otherwise indicated. ART blastocyst transcriptional results are reported as normalized to gene expression in vivo.
RESULTS
More severe preimplantation manipulation increases levels of blastocyst transcriptional alteration
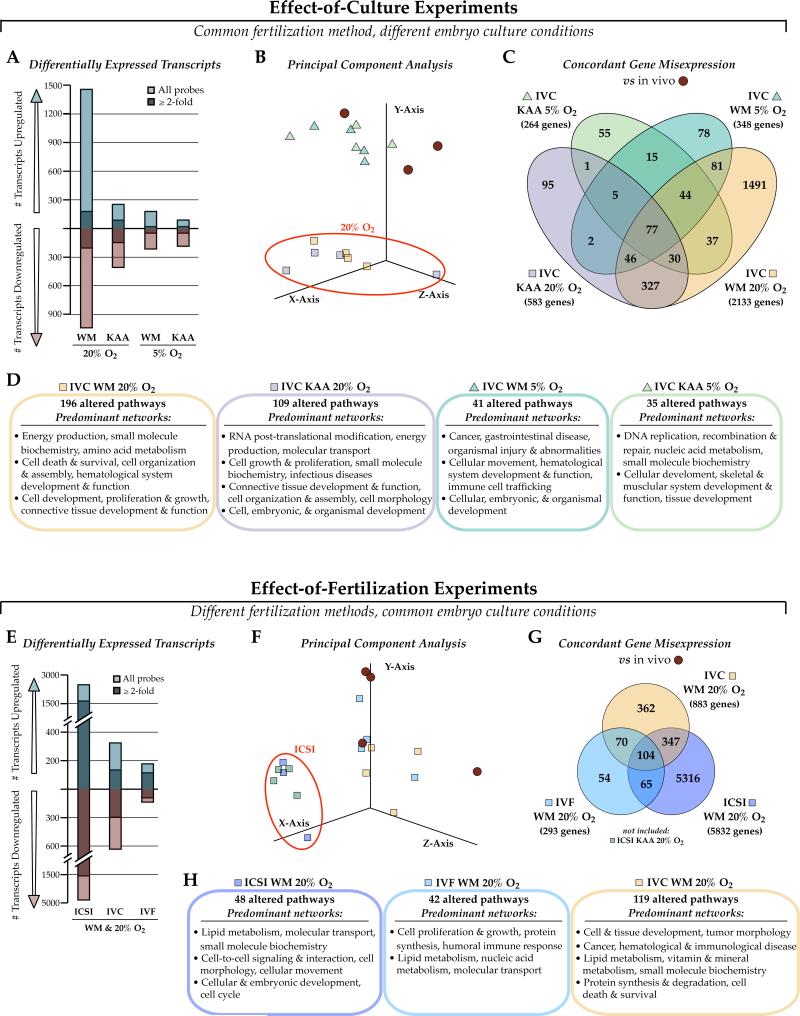
We examined the impact of in vitro manipulation on blastocyst gene expression by testing either the specific effects of different preimplantation embryo culture conditions, or by varying the method of fertilization and maintaining the culture conditions constant (Figure 2). First, superovulated and naturally fertilized zygotes were cultured in either WM or KAA, and 5% or 20% oxygen (4 conditions total) and compared to in vivo-generated blastocysts from superovulated dams (flushed blastocyst control). Both WM and higher oxygen tensions are considered stressful; conversely KAA and 5% oxygen are optimal for mouse embryo culture and represent current IVF clinical practices (Chronopoulou and Harper 2015, Schwarzer, et al. 2012). This gradation in stress was reflected in the number of transcripts altered by each culture condition, with WM and higher oxygen levels increasingly perturbing blastocyst gene expression (Figure 2A). Principal component analysis revealed a marked effect of oxygen on transcription, significantly more pronounced than the relative influence culture medium composition (Figure 2B, red circle). Each condition begat a unique blastocyst transcriptome, with gene expression perturbations associated with distinct pathways and gene networks (Figure 2C, 2D). 77 genes were commonly misexpressed in all four in vitro culture (IVC) conditions, and were enriched for stress response pathways, cell cycle control, cancer and pluripotency signaling (Supplemental Figure 1, Supplemental File 1).
Figure 2. Effect of different embryo culture conditions and types of fertilization on blastocyst gene expression.
Microarrays were performed on blastocysts either generated naturally and cultured in WM or KAA with 5% or 20% oxygen (Effect-of-Culture experiments, A-D), or produced by IVF, ICSI or naturally and cultured in WM and 20% oxygen (Effect-of-Fertilization experiments, E-H), with naturally-derived blastocysts as controls. A) Number of transcripts altered after each culture condition shows a strong impact of suboptimal WM and high oxygen on transcription. B) Principal Component Analysis (PCA) indicates that oxygen concentration has a more robust effect on gene expression than culture medium composition. C) Venn diagram of concordant gene misexpression across the 4 culture conditions. D) Ingenuity Pathway Analysis (IPA) highlighting the predominant networks (network score ≥30) associated with the gene expression changes. E-H) Same as A-D for the Effect-of-Fertilization experiments. E) There was a severe effect of fertilization by ICSI on blastocyst gene expression, which F) contributed significantly to PCA clustering. G) Venn diagram comparing the concordance of gene misexpression after each fertilization type, with H) top IPA networks associated with the gene lists. WM Whitten's medium; KAA potassium simplex optimization medium with amino acids; IVC in vitro culture, IVF in vitro fertilization, ICSI intracytoplasmic sperm injection.
We next evaluated the impact of different fertilization techniques by comparing blastocyst transcriptomes between embryos fertilized via ICSI, IVF, or naturally (in vitro culture only, IVC) and cultured from zygote to blastocyst in WM and 20% oxygen, versus in vivo. ICSI—a more mechanically intrusive procedure—had a dramatic effect on transcription, altering the expression of over 7,000 transcripts corresponding to 5,832 genes (Figure 2E to 2G). Unexpectedly, the IVC condition had a more prominent effect on gene expression than IVF. As with the effect-of-culture experiments, each fertilization method had a separate impact on blastocyst gene expression, with altered gene lists associated with unique pathways and networks (Figure 2H and Supplemental File 2). 104 genes showed concordant misexpression in each fertilization condition, and were associated with a wide range of cellular processes including cytoskeletal dynamics, metabolite biosynthesis, the cell cycle and growth (Supplemental Figure 2).
Blastocysts exhibit a common gene signature of in vitro embryo manipulation
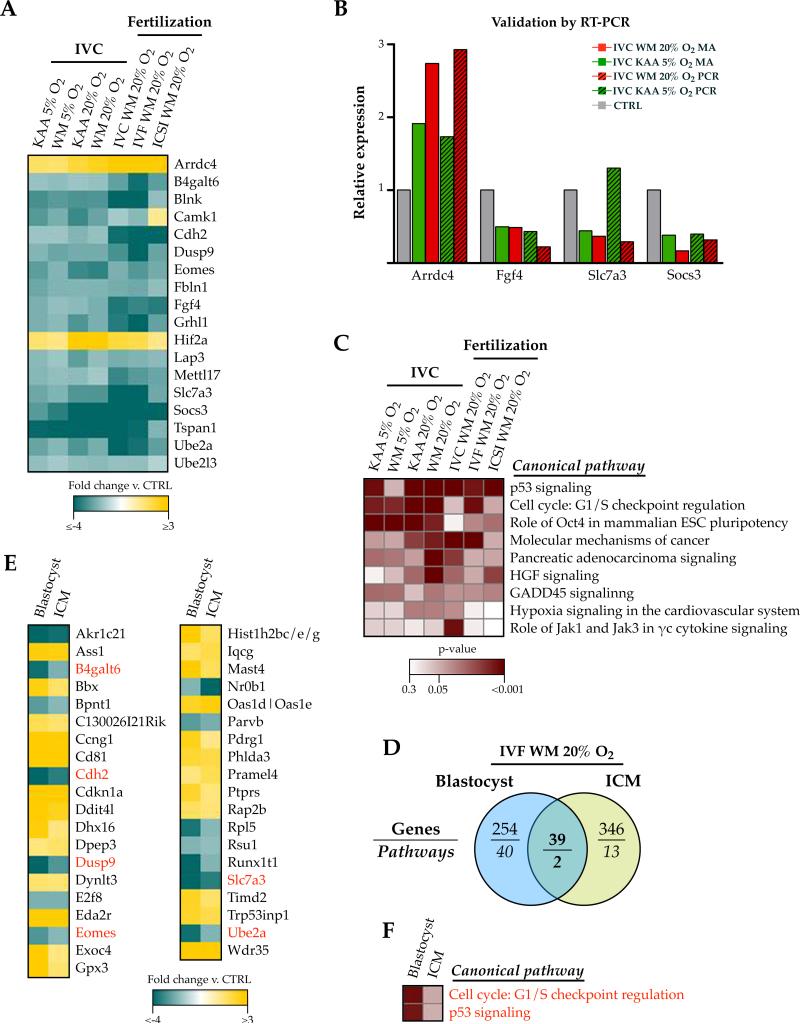
Following the observation that each fertilization method or culture condition resulted in unique gene expression changes, we searched for evidence of a ubiquitous transcriptomic fingerprint indicative of a shared effect of embryo manipulation compared to in vivo blastocyst gene expression. We identified a list of 18 genes concordantly misexpressed in all in vitro conditions (Figure 3A). Importantly, all of the genes were similarly increased or decreased compared to in vivo embryos except for Camk1 (increased in the ICSI microarray only), suggesting a common effect of in vitro manipulation on blastocyst gene expression. We chose 4 genes (Arrdc4, Fgf4, Slc7a3, Socs3) for validation in naturally derived blastocysts cultured from the zygotic stage (IVC) in either KAA-5% oxygen or WM-20% oxygen, which confirmed the microarray data for all conditions except for Slc7a3 expression in IVC KAA-5% O2 blastocysts (Figure 3B). Further, 9 pathways associated with growth, cancer, pluripotency and response to stress (Figure 3C) were collectively enriched in the in vitro manipulated embryos.
Figure 3. A common, but cell type-specific, fingerprint of embryo manipulation in vitro.
A) A core list of 18 genes were altered by in vitro embryo manipulation, regardless of the culture condition or the method of fertilization. B) Real time qPCR verification of the microarray data. Four of the 18 genes were selected for analysis in IVC blastocysts cultured in KAA-5% O2 (green) or WM-20% O2 (red), and the graph shows their expression relative to controls (normalized to 1) from both the microarray (MA) and PCR data. C) 9 pathways were collective enriched in association with the individual microarray experiments evaluating the impacts of fertilization conditions and culture conditions on blastocyst transcription. D-F) A cell type-specific effect of in vitro fertilization. D) Venn diagram showing the differential effects of IVF-WM and 20% oxygen culture conditions on blastocyst vs. inner cell mass (ICM) gene expression and pathway association. E) Only 39 genes and F ) two pathways were concordantly affected in both blastocysts and ICM, in spite of similar conditions of fertilization and culture in vitro. Genes and pathways also present in the ‘Common Fingerprint’ are highlighted in red. Legends (fold changes and p-value) are applicable to all heatmaps in the figure.
The impact of IVF on gene expression is cell type-specific
Because blastocysts are comprised of both trophectoderm and the inner cell mass (ICM), we further probed our embryo data by comparing the blastocyst IVF WM and 20% oxygen transcriptome to blastocyst ICM derived from the same conditions (Supplemental File 3). Of the 293 genes and 42 pathways affected in the blastocysts, only 39 genes and 2 pathways were similarly affected in the ICM (Figure 3D), confirming that controlling for different cell types is highly relevant when analyzing the impact of in vitro embryo manipulation. Only 6 of the 39 genes, as well as both pathways (cell cycle: G1/S checkpoint regulation and p53 signaling) were also highlighted in the ‘Common Fingerprint’ of in vitro embryo manipulation (Figure 3E-3F, red text).
There is no common gene signature of in vitro embryo manipulation in adult tissues
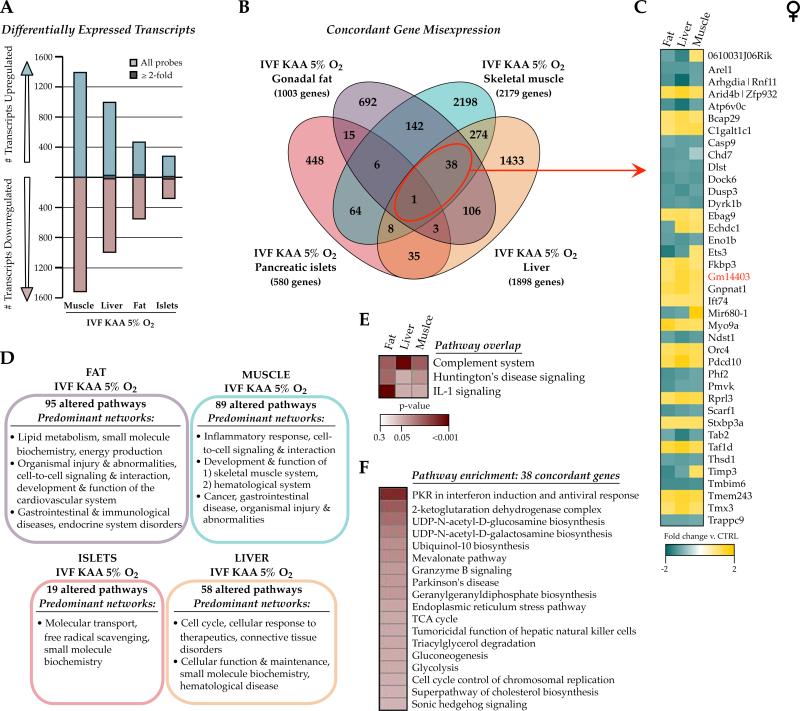
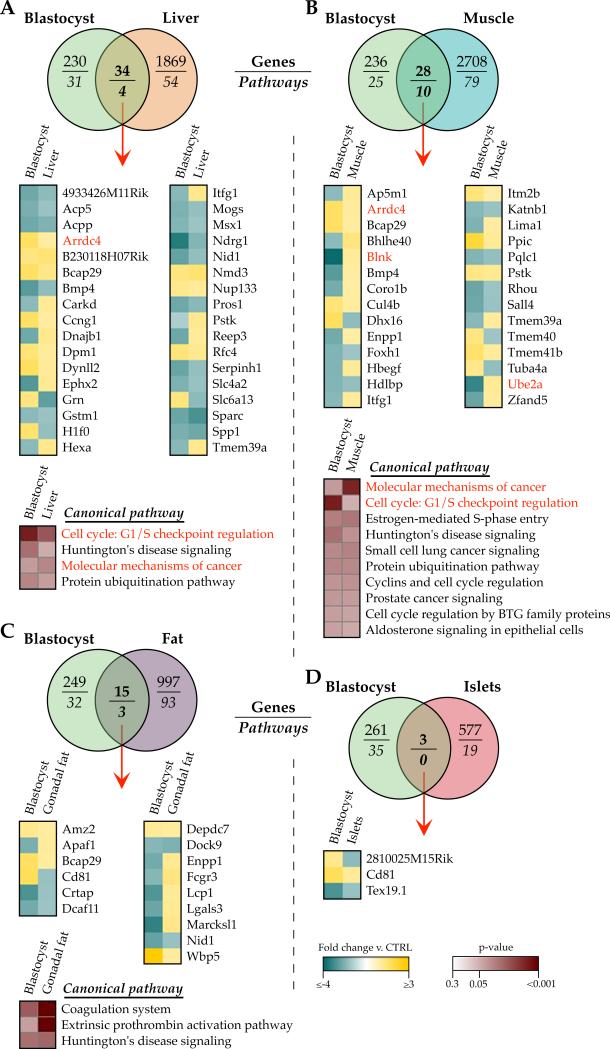
Previously we examined the long-term effects of IVF (KAA and 5% O2) on adult offspring liver, skeletal muscle, gonadal fat and pancreatic islet transcriptomes in an inbred C57Bl/6J mouse model and found no concordant changes or common biological themes (Feuer, et al. 2014). These particular tissues were selected for their involvement in either secretion of or response to insulin, as these IVF offspring show altered growth curves, pancreatic beta cell hyperinsulinemia, and are predisposed to glucose intolerance (Figure 1). The gene expression changes were subtle (>95% under 2-fold change), and re-analysis with updated software confirmed that only one gene, Gm14403, and no pathways were altered in all four IVF tissues compared to naturally-conceived controls (Figure 4 and Supplemental File 4), indicating a tissue- and/or cell type-specific impact of IVF on gene expression. This was corroborated by the fact that removal of the pancreatic islet data (a highly complex tissue with many cell types) led to modest overlap (38 genes and 3 pathways) between fat, liver and muscle IVF transcriptomes (Figure 4C, 4E). Interestingly, pathway analysis revealed that the 38 concordantly misexpressed genes were largely associated with glucose metabolic flux (through glycolysis, the TCA cycle and mitochondrial function, gluconeogenesis, and UDP-N-acetylglucosamine biosynthesis, Figure 4F).
Figure 4. Adult IVF transcriptional signatures are tissue-specific.
Microarray comparison between female 29wk gonadal fat, liver, skeletal muscle and pancreatic islets derived from IVF KAA 5% oxygen animals and in vivo flushed blastocyst controls. A) Number of transcripts altered in each IVF tissue. The majority of changes was modest and ≤2-fold different from controls. B) Venn diagram comparing the concordance of gene misexpression after IVF across the 4 tissues. C) Fold-change directionality of concordant IVF gene misexpression between fat, liver and muscle, with the one gene altered in all 4 tissues (Gm14403) indicated in red. D) Ingenuity Pathway Analysis (IPA) highlighting the predominant networks (network score >30) associated with the gene expression changes in each tissue, and E) the commonly altered pathways between IVF fat, liver and muscle gene lists. F) Top canonical pathways enriched in the 38 genes misexpressed in IVF fat, liver and muscle.
There is no obvious transcriptional link between in vitro embryos and their corresponding adult tissues
Given that in vitro embryo manipulation results in transcriptional changes in both embryos and adult tissues, we next inquired if the gene misexpression in adult tissues could be attributed to or explained by the transcriptional alterations in blastocysts. We compared the KAA and 5% oxygen microarray data between IVC embryos and their analogous adult IVF liver, skeletal muscle, gonadal fat, and pancreatic islets in order to determine if different tissues were vulnerable to retaining the in vitro embryo signature (Figure 5 and Supplemental File 5). There were minimal commonalities between the two time points, with fewer than 2% of the genes affected in each adult IVF tissue similarly altered in the blastocyst. Further, the shared gene misexpression between embryos and adult tissues was frequently altered in opposite directions. Pathway analysis demonstrated that although the gene misexpression was highly tissue-specific, some pathways were consistently enriched in both KAA – 5% O2 blastocysts and adult tissues, including cell cycle G1/S checkpoint regulation, molecular mechanisms of cancer, protein ubiquitination pathway, and Huntington's disease signaling.
Figure 5. No common in vitro signature of KAA and 5% oxygen between IVC blastocysts and IVF adult tissues.
Comparison of microarray data between IVC KAA 5% oxygen CF1 x B6D2F1/J blastocysts and IVF KAA 5% oxygen C57Bl/6J 29wk female A) liver, B) skeletal muscle, C) gonadal fat and D) pancreatic islets, versus in vivo-derived controls. Venn diagrams show overlap of gene misexpression and Ingenuity Pathway Analysis, with heatmaps depicting the fold-change directionality of concordant gene misexpression as well as pathway enrichment between blastocysts and corresponding adult tissues. Genes and pathways also present in the ‘Embryo Common Fingerprint’ of in vitro manipulation are highlighted in red. Legends are applicable to all heatmaps in the figure.
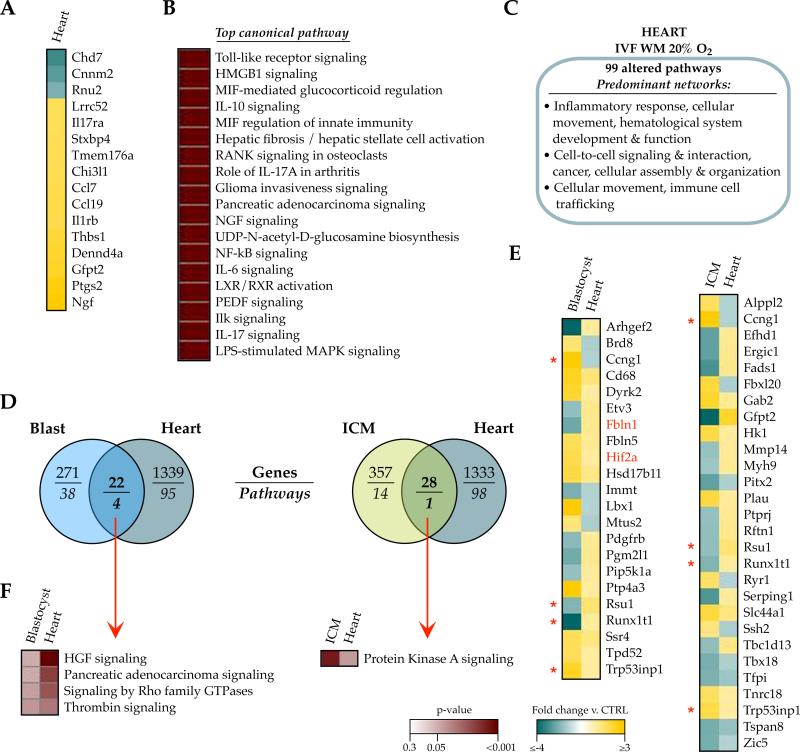
Although this initial comparison found negligible transcriptional linkage between in vitro embryos and adult tissues, the findings could have been influenced by differences in mouse strain and conception condition (CF-1 × B6D2F1/J IVC blastocysts, C57Bl/6J IVF adult tissues). We therefore expanded our analysis by investigating whether more stringent and homogenous conditions would reveal a relationship between the acute transcriptional response to in vitro stress and subsequent gene expression in adulthood. Microarray was performed on 40 week-old IVF and control cardiac tissue and compared to data from ICM or whole blastocysts derived from analogous conditions (outbred CF-1 × B6D2F1 tissues conceived by IVF with WM and 20% oxygen). We selected the heart because in addition to glucose intolerance, these mice exhibit cardiac left ventricular hypertrophy (Donjacour, et al. 2014), a condition that has also been reported in IVF/ICSI infants (Valenzuela-Alcaraz, et al. 2013). Relative to controls, 1443 transcripts corresponding to 1361 genes were altered in the IVF hearts, and only 16 genes exhibited fold-changes greater than ±2-fold (Figure 6A). 99 pathways were significantly enriched in the transcriptional profiles, with overrepresentation in the inflammatory response, hematological system development and function, cancer, cellular assembly and organization, as well as cell-to-cell signaling, interaction and movement (Figure 6B, 6C and Supplemental File 6).
Figure 6. No common IVF signature between embryos and adult tissues from homogenous conditions.
A-C) Misregulation in adult male 40wk cardiac tissue derived from IVF WM 20% oxygen mice. A) Of the 1361 genes significantly misexpressed in IVF heart compared to controls, only 16 showed a fold-change greater than ± 2-fold. B) Ingenuity Pathway Analysis of canonical pathways most significantly enriched (p<0.001) in the altered IVF heart genes and C) their associated networks (network score ≥25). D) Venn diagram indicating the low number of genes (top) and number of pathways (bottom) overlapping between the IVF WM 20% oxygen blastocyst (blue) or ICM (green) transcriptomes and corresponding adult heart (grey). E) The genes similarly misexpressed between blastocyst and heart, or ICM and heart, and their directionality of change. Red asterisks indicate genes commonly altered in both blastocyst and ICM, versus heart. F) Overlap of pathways associated with the altered genes between the embryo data and the heart. Legends are applicable to all heatmaps in the figure.
When we compared the IVF - WM 20% oxygen transcriptional profiles between the early embryo and adult heart, there again was minimal concordance in gene misexpression: 22 genes and 4 pathways were commonly altered in blastocysts and heart, whereas 28 genes and 1 pathway were shared between ICM and heart (Figure 6D-6F and Supplemental File 5). Only 4 genes (Ccng1, Rsu1 Runx1t1, Trp53inp1) were misexpressed in blastocysts, ICM, and heart tissue (Figure 4E, red asterisks). Surprisingly, more than half of the genes with shared misexpression were altered in opposite ways between the embryos and heart.
A set of common upstream regulators are predicted to drive the transcriptional changes in embryos and adult tissues
Ingenuity Pathway Analysis offers an upstream regulator prediction algorithm that evaluates gene lists and microarray fold-change to identify molecules (transcriptional regulators, growth factors, receptors, transporters, etc.) with direct actions on the differentially expressed genes. Based on fold-change directionality of the downstream target genes, IPA predicts whether upstream regulator inhibition or activation might govern the differences observed. Given the significant cell type-specificity observed in each IVF tissue transcriptome, we hypothesized that common regulators might control the gene expression changes, but based on tissue or cell type this would manifest in unique transcriptional signatures (Figure 7 and Supplemental Files 1-6).
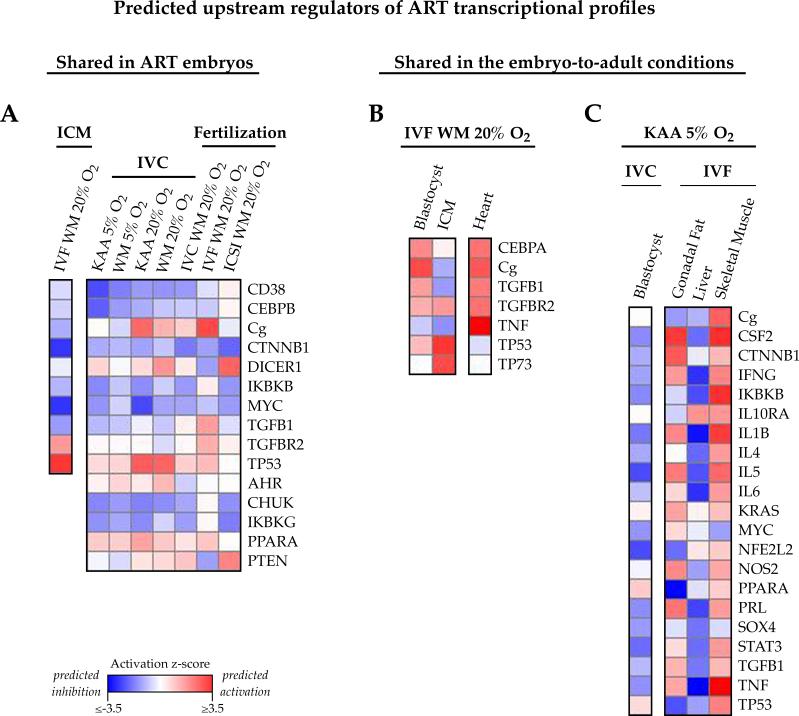
Figure 7. Common upstream regulators may govern the transcriptional changes present in in vitro embryos and adult tissues.
Ingenuity Pathway Analysis identifies regulators functioning upstream of the altered genes, and predicts whether these regulators are impaired (negative z-score, blue) or activated (positive z-score, red) based on fold-change misexpression. A) Shared upstream regulators of the ART embryo expression profiles. 15 molecules were predicted to function upstream of the transcriptional alterations present in the in vitro blastocyst, 10 of which were similarly highlighted in ICM. B, C) Upstream regulators present in the embryo-to-adult condition-specific comparisons. B) 7 regulators commonly target the altered genes in IVF WM 20% blastocysts, ICM and adult heart, and C) 21 regulators collectively mediate the gene expression changes observed in KAA 5% oxygen blastocysts and corresponding adult female fat, liver and muscle.
Analysis of the transcriptional alterations present in blastocysts after each condition of in vitro manipulation identified a core group of 15 regulators—comprised of kinases, enzymes, growth factors, phosphatases, ligand-dependent nuclear receptors and transcriptional regulators—that collectively function directly upstream of the altered genes, 10 of which additionally target the misregulated ICM genes (Figure 7A). The predicted activation or inhibition of these regulators was largely concordant across all of the in vitro embryo conditions.
Focusing on the embryo-to-adult comparisons (Figure 7B, 7C), there were 7 putative regulators of the genes altered in blastocysts, ICM and adult male heart derived from the IVF WM and 20% oxygen conditions, compared to in vivo controls (Figure 7B). Based on the directionality of the gene expression changes, the majority of these putative regulators were predictably activated (e.g. functionally increased) in blastocysts and heart, but inhibited in the ICM.
Regarding the KAA and 5% oxygen blastocysts and their corresponding adult female fat, liver, and muscle, 21 regulators operate upstream of the misexpressed genes at both time points (Figure 7C). Interestingly, these molecules showed predicted inhibition in blastocyst and liver profiles, but activation in fat and muscle, suggesting that differential mediation of gene expression changes by common regulators is tissue-specific. Supplemental Figure 3 summarizes the predicted upstream regulators shared by the ART embryo expression profiles and the condition-specific embryo versus adult comparisons.
DISCUSSION
This study used a mouse model to determine if there are common mechanisms involved in the preimplantation embryo's response to different in vitro exposures frequently used in assisted reproductive technologies (embryo culture conditions, method of fertilization), and if there exists a relationship between these acute transcriptional responses and gene expression in subsequent offspring adult tissues. This work has wide clinical relevance not only because well over 5 million children have been born from ART (ESHRE 2014), but because it remains unclear how environmentally-induced developmental reprogramming can predispose to abnormal metabolic phenotypes in adulthood. The main findings of the manuscript are that: 1) mouse blastocysts exhibit a common gene signature of in vitro embryo manipulation; 2) blastocyst transcriptional alterations are increased with greater preimplantation manipulation or stress; 3) the impact of embryo manipulation on gene expression is cell type-specific; 4) there is minimal overlap between IVF transcriptional profiles in different adult tissues; and 5) there is no obvious transcriptional link between in vitro embryos and their corresponding adult tissues derived from similar in vitro conditions. Finally, 6) we identified a set of common upstream regulators that could drive the transcriptional changes present in IVF embryos and adult tissues.
The first notable finding is that although each in vitro condition led to a specific and unique blastocyst transcriptional profile, we identified a common 18-gene and 9-pathway signature of preimplantation embryo manipulation that was present in all in vitro embryos irrespective of culture condition or method of fertilization. Coupled with the variation in time, location, scientist and microarray platform for these experiments, the emergence of a core fingerprint of in vitro embryo manipulation is particularly robust. Further, the 18 genes were altered in the same direction except for Camk1 expression. Camk1 operates within the calcium-triggered signaling cascade; a hallmark of mammalian fertilization is the presence of repetitive Ca2+ oscillations from the site of sperm penetration. The unique increase in Camk1 expression in ICSI blastocysts compared to other ART conditions might be explained by data showing that ICSI induces subtle changes in oocyte Ca2+ oscillatory patterns compared to conventional insemination (Miao, et al. 2012, Sato, et al. 1999).
In addition to several key transcriptional regulators with crucial roles in development and cellular differentiation (Eomes, Fgf4, Grhl1, Hif2α), several of the genes affected by in vitro manipulation reflect an increase in reactive oxygen species (ROS). Indeed, there is abundant evidence that embryo culture induces an increase in ROS (Cebral, et al. 2007, Goto, et al. 1993, Martin-Romero, et al. 2008). In our data set, oxidative stress is exemplified by changes in the hypoxia-inducible transcription factor Hif2α, the expression of genes involved in ubiquitination and protein turnover [Ube2a, Ube2l3, Lap3, Socs3, Arrdc4, (Shang and Taylor 2011)], and in particular Arrdc4 transcription.
Within this common fingerprint, Hif2α is a well-known transcriptional regulator of cellular and organismal responses to oxygen deprivation (Semenza 2016). Oxygen deprivation is sensed by mitochondria and converted to a reactive oxygen species-dependent signal subunit that results in HIF2α subunit stabilization (Chandel 2010). Although transcriptional induction of Hif2α subunit gene expression under hypoxic conditions is less widely reported, it has been demonstrated in hematopoietic stem cells that redox stress can trigger Hif2α expression to help modulate redox balance via the homeodomain transcription factor Meis1 (Kocabas, et al. 2012, Miller, et al. 2016). Along these lines, the arrestin family member Arrdc4 also plays roles in cell metabolism and is related to molecules such as Txnip, which can influence cellular redox status in response to glucose availability (Zhou and Chng 2013) and thus integrate cellular oxidative and metabolic states (Patwari and Lee 2012). Txnip expression is upregulated in IVF embryos (Feuer, et al. 2014) and other models have correlated this increase with impaired glucose tolerance (Parikh, et al. 2007), oxidative stress (Schulze, et al. 2004), apoptosis (Chen, et al. 2008), and diabetes pathogenesis (Shalev 2008). The induction of these transcripts suggests a perturbation of the metabolic and redox axes in early embryos conceived and cultured in vitro. In further support of this, GADD45 and p53 signaling—identified as one of the most significantly misregulated pathways in our dataset—are implicated in cellular and organismal responses to perturbations in cell metabolism or redox status (Salvador, et al. 2013, Zhuang, et al. 2012). Finally, Socs3 regulates cytokine signaling through a negative feedback loop in the Jak/Stat pathway and is also linked to insulin resistance (Jorgensen, et al. 2013, Palanivel, et al. 2012).
The remaining conserved transcripts were repressed in embryos following in vitro manipulation. Many of these, including Eomes, Fgf4, Socs3 and Dusp9 are implicated in placental development or are critical for trophoblast stem cell function (Maltepe and Fisher 2015). We have documented changes in mouse IVF placentae in prior studies (Bloise, et al. 2014, Bloise, et al. 2012, Delle Piane, et al. 2010), which suggests that dysregulation of critical placental genes is conserved across IVF techniques and could potentially contribute to alterations to placental development and function in this setting. Further, ART-induced remodeling of the placental landscape could contribute to the adult metabolic phenotypes observed in these mice. Another noteworthy gene in this common fingerprint is Ube2a, which catalyzes the monoubiqitination of histone H2B at Lys-120 to form H2BK120ub1; this marks epigenetic transcriptional activation, elongation by RNA pol II, telomeric silencing, and is a requirement for H3K4 me and H3K79me formation. Together these genes suggest a common fingerprint of in vitro embryo manipulation linking cellular differentiation, oxidative stress, glucose metabolism and insulin resistance with epigenetic changes.
The pathways with significant overrepresentation following any in vitro manipulation are also implicated in cellular development and differentiation, proliferation and the cell cycle (including cancer signaling), as well as stress response signaling via p53 and GADD45. Given that DOHaD-related phenotypes are often metabolic, it is notable that there was no enrichment for any particular metabolic pathways. However, many of the pathways with enrichment reveal interesting connections between the acute responses to in vitro embryo manipulation and long-term metabolic phenotypes. Oxygen homeostasis is important for cardiovascular system patterning during embryogenesis, including adaptive responses in signaling and redox homeostasis [reviewed in (Simon, et al. 2008)]; adult mice generated by IVF with WM - 20% O2 conditions exhibit cardiac hypertrophy (Donjacour, et al. 2014). GADD45 and p53 signaling reflect cell stress and DNA damage, with known roles in regulation of the cell cycle, senescence, survival and apoptosis. The link between GADD45 and apoptosis is significant, as GADD45 induction is known to downregulate pro-apoptotic JNK signaling and therefore suggests a pro-survival function (De Smaele, et al. 2001). Further GADD45 proteins promote active DNA demethylation and thus mediate gene activation, which is particularly important for cell differentiation and transcriptional regulation during development (Schafer 2013). Given GADD45 function in the stress response and role in age-related disorders such as insulin resistance (Moskalev, et al. 2012), alterations in this signaling pathway during an epigenetically vulnerable period might have lasting consequences for metabolic health.
This is highly relevant, as it is widely believed that epigenetic changes mediate developmental plasticity and contribute significantly to the programming of environmental signals (Gluckman, et al. 2011). Given that both DNA methylation and histone modifications are extensively remodeled in the preimplantation embryo (Reik 2007), and because culture conditions can affect chromatin marks (Doherty, et al. 2000), it is possible that IVF-induced changes in transcriptional and epigenetic regulation are responsible for propagating the adult DOHaD phenotypes.
Although each in vitro condition exerted a marked effect on blastocyst transcription, we additionally found that more severe transcriptional changes occurred with increased preimplantation stress. In particular, culture of embryos using atmospheric oxygen had a dramatic effect on gene expression (Figure 2A, 2B). This was compounded by the addition of a suboptimal culture medium (WM), whereas the relative influence of culture medium composition alone on transcriptional profiles was small in comparison. Further the Effect-of-Fertilization experiments (Figure 2E-2H) showed an outstanding effect of ICSI compared to IVC or IVF. The severity of the stress (oxygen, ICSI) was also reflected in the degree of fold change for the 18-gene signature of in vitro manipulation in embryos. Interestingly, embryo culture of naturally fertilized zygotes had a stronger impact on gene expression than in vitro fertilization and culture in the same conditions (Figure 2G, 883 versus 293 altered genes). This highlights the remarkable vulnerability of zygotes to the stresses incurred through the embryo isolation process, which may be related to the timing of zygotic genome activation. Indeed, transfer of mouse zygotes following in vitro exposure to different nutritional milieu during the pronuclear stage alters birth weight in a condition-specific manner (Banrezes, et al. 2011). A notable physiological feature that separates the IVC group is that these in vivo-generated embryos are exposed to seminal fluid, which can elicit unique gene expression changes (Schjenken and Robertson 2015). In addition to gene expression, protocol-specific effects have also been described for blastocyst cell number and lineage ratio, implantation efficiency, fetal and placental growth, postnatal growth and adult glucose metabolism (Kohda, et al. 2011, Schwarzer, et al. 2012, Scott, et al. 2010). Along these lines, it is likely that additional ART procedures such as in vitro or in vivo oocyte maturation can distinctly modify embryo, fetal and postnatal phenotypes. Moreover this would suggest that the recipient uterine environment could facilitate supplementary changes to transcriptional signatures of transferred embryos, further compounding the effects of ART techniques on gene expression. Overall, these findings are in agreement with the ’quiet embryo’ hypothesis (Leese, et al. 2008) and argue strongly for avoiding extensive embryo manipulation whenever possible.
Another key finding of this analysis is that significant cell type-transcriptional specificity of IVF is already observable at the blastocyst stage, where only 2 cell types are present. In fact, comparison of blastocyst vs. ICM profiles derived from the same in vitro conditions showed an overlap of only 39 genes, 6 of which are present in the common fingerprint. Such nominal concordance demonstrates that individual cell types are primed to differentially respond to environmental perturbation, and that pooling multiple cell types may obscure investigations into DOHaD mechanisms. The relevance of cell type is further supported by the lack of a collective IVF fingerprint across the adult tissues, and the evidence that pancreatic islets—a highly differentiated and complex tissue—exhibited fewer transcriptional and pathway changes following IVF than other tissues. Interestingly, the adult transcriptional changes in IVF offspring were minimal (over 95% of the gene misexpression was ≤2 fold-change different). This was true for both tissues derived from optimized IVF – KAA 5% oxygen conditions (islets, liver, fat and muscle) but also for cardiac tissue from suboptimal IVF – WM 20% O2 offspring. Other models of developmental environmental perturbation have similarly reported only subtle and tissue-distinct transcriptional profiles. For example, investigation of the transgenerational effects of gestational vinclozolin exposure demonstrated that F3 generation rats exhibited unique, tissue-specific changes to adult expression, with no common pathway overrepresentation (Skinner, et al. 2012). Another study showed gene expression changes in ICSI but not IVF neonates, and no transcriptional or phenotypic differences by 8 weeks of age (Kohda, et al. 2011). Given the lack of a common signature in adults, it is not surprising that the transcriptional changes observed in manipulated embryos did not persist through development.
The reasons for the tissue-specific variation and the lack of stability of the IVF-induced transcriptional changes throughout in utero and postnatal development are unknown. Because only 4-8 epiblast cells contribute to the adult body (Morris and Zernicka-Goetz 2012, Soriano and Jaenisch 1986), persistence and penetrance of early gene expression patterns into adulthood could depend on which particular founder cells contributed to organogenesis. Separately, the necessity of pooling blastocysts to assess gene expression could have captured embryos capable of adapting to a stressful in vitro environment but not necessarily able to develop beyond the blastocyst stage. This would introduce a strong transcriptional bias in embryos compared to postnatal tissues, although given the robust concordance in gene expression amongst embryos derived from different in vitro conditions, we believe this an unlikely possibility. It is also possible that some ART-induced changes are resolved during development. However, the presence of distinct adult phenotypes (e.g. the manifestation of glucose intolerance in mouse IVF offspring) makes this scenario unlikely. Another explanation is that the molecular changes present in blastocysts following preimplantation disturbance are differentially affected by organogenesis, growth factors, or sexually dimorphic signals occurring during later stages of development. As a result, cell physiology and metabolism within each developing tissue is altered in accordance with new, tissue-specific developmental cues. Fitting within this framework, we have previously investigated whether specific genes altered in IVF embryos exhibit tissue-specific maintenance of transcriptional and epigenetic changes throughout development. We showed that expression of the glucose-sensitive gene Txnip was significantly increased in IVF blastocysts, selectively increased in female IVF fat and muscle tissues but not liver, and that this dysregulation was associated with enriched H4 acetylation at the Txnip promoter (Feuer, et al. 2014). In the current study, Txnip is upregulated in all ART blastocysts except for the ICSI and the IVC – KAA 20% oxygen conditions. In another study we reported that glucocorticoid receptor (GR) expression is increased in IVF – KAA 5% O2 embryos and male offspring fat, but not liver or muscle (Simbulan, et al. 2015).
Finally, it is possible that the gene expression changes in adult IVF tissues occur secondary to an altered function of key upstream transcriptional regulators, which would not necessarily be identifiable in a microarray profile. Transcriptional regulators are able to control diverse processes with cell type- and temporal-specificity through combinatorial interplay with other transcriptional factors and modulators. In this manner, altering the function of a transcriptional regulator would exert a variety of different effects depending upon the cell type, which could explain the stark tissue specificity observed in the ART expression profiles. Several of the proposed upstream regulators are highly relevant to DOHaD phenotypes and provide grounds for directly testable hypotheses in future investigations. CEBPA/B play essential and redundant functions during early embryogenesis and placentation (Begay, et al. 2004), and CEBPA enhances iPS cell reprogramming efficiency (Di Stefano, et al. 2014). CEBPA is crucial for liver and lung development, adipocyte terminal differentiation, the establishment and maintenance of energy homeostasis, lipid storage and gluconeogenesis, all through combinatorial interactions with other transcription factors such as Myc or members of the PPAR family (Flodby, et al. 1996, Wang, et al. 1995, Wu, et al. 1999). Context-specific interactions might explain why some transcriptional regulators exhibited predicted activation in certain tissues and inhibition in others. Further, the Cepba locus encodes a functional RNA that sequesters DMNT1 to ensure robust Cebpa transcription though local inhibition of Cepba gene methylation, thus directly linking Cepba activity to site-specific genomic methylation (Di Ruscio, et al. 2013).
Separately, NFE2L2 (also known as NRF2) is involved in antioxidant defense mechanisms, and binds DNA antioxidant response elements (AREs) to regulate the adaptive response to oxidative stress [reviewed in (Ma 2013)]. NFE2L2 was predictedly inhibited in our ART datasets, which is particularly relevant because mice deficient in NFE2L2 exhibit increased sensitivity to oxidative stress. There is ample evidence in the literature that embryo culture induces an increase in reactive oxygen species (ROS) (Cebral, et al. 2007, Goto, et al. 1993, Martin-Romero, et al. 2008), and ROS induction is a central event precipitating diabetes pathogenesis (Nishikawa, et al. 2000, Sakai, et al. 2003). ROS can also impair telomerase activity, with telomere attrition being a predictive marker for cardiovascular dysfunction, metabolic syndrome and other age- and DOHaD-related pathologies (Hallows, et al. 2012). It is therefore possible that an in increase in ROS levels is the initial stimulus that reprograms the ART embryo, which employs transcription factors like NFE2L2, CEBPs (Manea, et al. 2014), and p53 to exert acute transcriptional responses and subsequent chromatin remodeling that persists throughout development, leading to different cell type-contextual expression patterns.
Among the potential caveats of this paper is the fact that the results presented are specific to data from one laboratory and one species. Whilst this strength enables direct comparisons between datasets, small technical discrepancies between different laboratories can affect results and subsequent conclusions. Similarly, choice of mouse strain(s) can influence gene expression signatures (Turk, et al. 2004). The mouse data provided may not apply to other species and thus should be confirmed in, for example, bovine (Smith, et al. 2009) or sheep (Wei, et al. 2016) models. Finally, given the progressive maturation of translation machinery in the early embryo, changes in RNA may not be reflected by changes in protein abundance (Seydoux, et al. 1996).
To conclude, we have provided a systematic, integrated analysis of the acute and long-term transcriptional effects of different mechanisms of in vitro embryo manipulation commonly used in assisted reproduction. The results of this study indicate that the expression of 18 genes may be used to evaluate ART embryo health and could be developed into a new tool for identifying embryos with the greatest implantation potential. Because fertilization and embryo culture practices vary worldwide, this list of 18 genes is valuable for its robustness and potential application across the entire ART field. This in vitro fingerprint does not persist throughout development, and there is no clear transcriptional cohesion between adult IVF offspring tissues compared to their preceding embryos. However, the unique transcriptional signatures present in each IVF tissue are collectively targeted by the same upstream transcriptional regulators, which provides insight into DOHaD mechanisms regarding how acute transcriptional responses to stressful environmental exposures might be preserved throughout development to influence adult gene expression and manifest metabolic phenotypes. Our results suggest that the activity of these regulators may be reliable markers of in vitro stress present at specific developmental points extending into adulthood.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by R01: HD 082039 and ASRM award to PFR. SKF was supported by the NIH training fellowship 5T32HD007263-32.
ABBREVIATIONS & DEFINITIONS
- IVC
in vitro culture
- IVF
in vitro fertilization
- ICSI
intracytoplasmic sperm injection
- KAA
potassium simplex optimized medium supplemented with amino acids
- WM
Whitten's medium
Footnotes
DECLARATION OF INTEREST
The authors have nothing to disclose.
REFERENCES
- Banrezes B, Sainte-Beuve T, Canon E, Schultz RM, Cancela J, Ozil JP. Adult body weight is programmed by a redox-regulated and energy-dependent process during the pronuclear stage in mouse. PLoS One. 2011;6:e29388. doi: 10.1371/journal.pone.0029388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begay V, Smink J, Leutz A. Essential requirement of CCAAT/enhancer binding proteins in embryogenesis. Mol Cell Biol. 2004;24:9744–9751. doi: 10.1128/MCB.24.22.9744-9751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloise E, Feuer SK, Rinaudo PF. Comparative intrauterine development and placental function of ART concepti: implications for human reproductive medicine and animal breeding. Hum Reprod Update. 2014 doi: 10.1093/humupd/dmu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloise E, Lin W, Liu X, Simbulan R, Kolahi KS, Petraglia F, Maltepe E, Donjacour A, Rinaudo P. Impaired placental nutrient transport in mice generated by in vitro fertilization. Endocrinology. 2012;153:3457–3467. doi: 10.1210/en.2011-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebral E, Carrasco I, Vantman D, Smith R. Preimplantation embryotoxicity after mouse embryo exposition to reactive oxygen species. Biocell. 2007;31:51–59. [PubMed] [Google Scholar]
- Chandel NS. Mitochondrial regulation of oxygen sensing. Adv Exp Med Biol. 2010;661:339–354. doi: 10.1007/978-1-60761-500-2_22. [DOI] [PubMed] [Google Scholar]
- Chen J, Saxena G, Mungrue IN, Lusis AJ, Shalev A. Thioredoxin-interacting protein: a critical link between glucose toxicity and beta-cell apoptosis. Diabetes. 2008;57:938–944. doi: 10.2337/db07-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronopoulou E, Harper JC. IVF culture media: past, present and future. Hum Reprod Update. 2015;21:39–55. doi: 10.1093/humupd/dmu040. [DOI] [PubMed] [Google Scholar]
- De Smaele E, Zazzeroni F, Papa S, Nguyen DU, Jin R, Jones J, Cong R, Franzoso G. Induction of gadd45beta by NF-kappaB downregulates proapoptotic JNK signalling. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- Delle Piane L, Lin W, Liu X, Donjacour A, Minasi P, Revelli A, Maltepe E, Rinaudo PF. Effect of the method of conception and embryo transfer procedure on mid-gestation placenta and fetal development in an IVF mouse model. Hum Reprod. 2010;25:2039–2046. doi: 10.1093/humrep/deq165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D'Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP, Lu J, Thieffry D, Beato M, Graf T. C/EBPalpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature. 2014;506:235–239. doi: 10.1038/nature12885. [DOI] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Donjacour A, Liu X, Lin W, Simbulan R, Rinaudo PF. In vitro fertilization affects growth and glucose metabolism in a sex-specific manner in an outbred mouse model. Biol Reprod. 2014;90:80. doi: 10.1095/biolreprod.113.113134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESHRE ART fact sheet. [Online.] 2014 posting date. [Google Scholar]
- Fernandez-Gonzalez R, Moreira P, Bilbao A, Jimenez A, Perez-Crespo M, Ramirez MA, Rodriguez F, Fonseca De, Pintado B, Gutierrez-Adan A. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc Natl Acad Sci U S A. 2004;101:5880–5885. doi: 10.1073/pnas.0308560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuer SK, Liu X, Donjacour A, Lin W, Simbulan RK, Giritharan G, Piane LD, Kolahi K, Ameri K, Maltepe E, Rinaudo PF. Use of a mouse in vitro fertilization model to understand the developmental origins of health and disease hypothesis. Endocrinology. 2014;155:1956–1969. doi: 10.1210/en.2013-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17:1653–1665. doi: 10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- Gillman MW. Developmental origins of health and disease. N Engl J Med. 2005;353:1848–1850. doi: 10.1056/NEJMe058187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritharan G, Delle Piane L, Donjacour A, Esteban FJ, Horcajadas JA, Maltepe E, Rinaudo P. In vitro culture of mouse embryos reduces differential gene expression between inner cell mass and trophectoderm. Reprod Sci. 2012;19:243–252. doi: 10.1177/1933719111428522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritharan G, Li MW, De Sebastiano F, Esteban FJ, Horcajadas JA, Lloyd KC, Donjacour A, Maltepe E, Rinaudo PF. Effect of ICSI on gene expression and development of mouse preimplantation embryos. Hum Reprod. 2010;25:3012–3024. doi: 10.1093/humrep/deq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritharan G, Talbi S, Donjacour A, Di Sebastiano F, Dobson AT, Rinaudo PF. Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction. 2007;134:63–72. doi: 10.1530/REP-06-0247. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Low FM. The role of developmental plasticity and epigenetics in human health. Birth Defects Res C Embryo Today. 2011;93:12–18. doi: 10.1002/bdrc.20198. [DOI] [PubMed] [Google Scholar]
- Goto Y, Noda Y, Mori T, Nakano M. Increased generation of reactive oxygen species in embryos cultured in vitro. Free Radic Biol Med. 1993;15:69–75. doi: 10.1016/0891-5849(93)90126-f. [DOI] [PubMed] [Google Scholar]
- Hager R, Cheverud JM, Wolf JB. Change in maternal environment induced by cross-fostering alters genetic and epigenetic effects on complex traits in mice. Proc Biol Sci. 2009;276:2949–2954. doi: 10.1098/rspb.2009.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows SE, Regnault TR, Betts DH. The long and short of it: the role of telomeres in fetal origins of adult disease. J Pregnancy. 20122012:638476. doi: 10.1155/2012/638476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Wigglesworth K, Eppig JJ, Schultz RM. Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol Reprod Dev. 1995;41:232–238. doi: 10.1002/mrd.1080410214. [DOI] [PubMed] [Google Scholar]
- ICMART International Committee Monitoring Assisted Reproductive Technology (ICMART) World Report. Preliminary 2008 Data. 2012 [Google Scholar]
- Jeong YJ, Choi HW, Shin HS, Cui XS, Kim NH, Gerton GL, Jun JH. Optimization of real time RT-PCR methods for the analysis of gene expression in mouse eggs and preimplantation embryos. Mol Reprod Dev. 2005;71:284–289. doi: 10.1002/mrd.20269. [DOI] [PubMed] [Google Scholar]
- Jorgensen SB, O'Neill HM, Sylow L, Honeyman J, Hewitt KA, Palanivel R, Fullerton MD, Oberg L, Balendran A, Galic S, Poel C van der, Trounce IA, Lynch GS, Schertzer JD, Steinberg GR. Deletion of skeletal muscle SOCS3 prevents insulin resistance in obesity. Diabetes. 2013;62:56–64. doi: 10.2337/db12-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungheim ES, Schoeller EL, Marquard KL, Louden ED, Schaffer JE, Moley KH. Diet-induced obesity model: abnormal oocytes and persistent growth abnormalities in the offspring. Endocrinology. 2010;151:4039–4046. doi: 10.1210/en.2010-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas F, Zheng J, Thet S, Copeland NG, Jenkins NA, DeBerardinis RJ, Zhang C, Sadek HA. Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood. 2012;120:4963–4972. doi: 10.1182/blood-2012-05-432260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda T, Ogonuki N, Inoue K, Furuse T, Kaneda H, Suzuki T, Kaneko-Ishino T, Wakayama T, Wakana S, Ogura A, Ishino F. Intracytoplasmic sperm injection induces transcriptome perturbation without any transgenerational effect. Biochem Biophys Res Commun. 2011;410:282–288. doi: 10.1016/j.bbrc.2011.05.133. [DOI] [PubMed] [Google Scholar]
- Kuijk EW, du Puy L, van Tol HT, Haagsman HP, Colenbrander B, Roelen BA. Validation of reference genes for quantitative RT-PCR studies in porcine oocytes and preimplantation embryos. BMC Dev Biol. 2007;7:58. doi: 10.1186/1471-213X-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WY, Wild AE, Roberts P, Willis AC, Fleming TP. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development. 2000;127:4195–4202. doi: 10.1242/dev.127.19.4195. [DOI] [PubMed] [Google Scholar]
- Leese HJ, Baumann CG, Brison DR, McEvoy TG, Sturmey RG. Metabolism of the viable mammalian embryo: quietness revisited. Mol Hum Reprod. 2008;14:667–672. doi: 10.1093/molehr/gan065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MW, Willis BJ, Griffey SM, Spearow JL, Lloyd KC. Assessment of three generations of mice derived by ICSI using freeze-dried sperm. Zygote. 2009;17:239–251. doi: 10.1017/S0967199409005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltepe E, Fisher SJ. Placenta: the forgotten organ. Annu Rev Cell Dev Biol. 2015;31:523–552. doi: 10.1146/annurev-cellbio-100814-125620. [DOI] [PubMed] [Google Scholar]
- Manea SA, Todirita A, Raicu M, Manea A. C/EBP transcription factors regulate NADPH oxidase in human aortic smooth muscle cells. J Cell Mol Med. 2014:181467–1477. doi: 10.1111/jcmm.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Romero FJ, Miguel-Lasobras EM, Dominguez-Arroyo JA, Gonzalez-Carrera E, Alvarez IS. Contribution of culture media to oxidative stress and its effect on human oocytes. Reprod Biomed Online. 2008;17:652–661. doi: 10.1016/s1472-6483(10)60312-4. [DOI] [PubMed] [Google Scholar]
- Matthews PA, Samuelsson AM, Seed P, Pombo J, Oben JA, Poston L, Taylor PD. Fostering in mice induces cardiovascular and metabolic dysfunction in adulthood. J Physiol. 2011;589:3969–3981. doi: 10.1113/jphysiol.2011.212324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao YL, Stein P, Jefferson WN, Padilla-Banks E, Williams CJ. Calcium influx-mediated signaling is required for complete mouse egg activation. Proc Natl Acad Sci U S A. 2012;109:4169–4174. doi: 10.1073/pnas.1112333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ME, Rosten P, Lemieux ME, Lai C, Humphries RK. Meis1 Is Required for Adult Mouse Erythropoiesis, Megakaryopoiesis and Hematopoietic Stem Cell Expansion. PLoS One. 2016;11:e0151584. doi: 10.1371/journal.pone.0151584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Zernicka-Goetz M. Formation of distinct cell types in the mouse blastocyst. Results Probl Cell Differ. 2012;55:203–217. doi: 10.1007/978-3-642-30406-4_11. [DOI] [PubMed] [Google Scholar]
- Moskalev AA, Smit-McBride Z, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Tacutu R, Fraifeld VE. Gadd45 proteins: relevance to aging, longevity and age-related pathologies. Ageing Res Rev. 2012;11:51–66. doi: 10.1016/j.arr.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Edelstein D, Du XL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, Giardino I, Brownlee M. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404:787–790. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- Palanivel R, Fullerton MD, Galic S, Honeyman J, Hewitt KA, Jorgensen SB, Steinberg GR. Reduced Socs3 expression in adipose tissue protects female mice against obesity-induced insulin resistance. Diabetologia. 2012;55:3083–3093. doi: 10.1007/s00125-012-2665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ, Jensen CB, Krook A, Bjornholm M, Tornqvist H, Zierath JR, Ridderstrale M, Altshuler D, Lee RT, Vaag A, Groop LC, Mootha VK. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P, Lee RT. An expanded family of arrestins regulate metabolism. Trends Endocrinol Metab. 2012;23:216–222. doi: 10.1016/j.tem.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappolee DA, Basilico C, Patel Y, Werb Z. Expression and function of FGF-4 in peri-implantation development in mouse embryos. Development. 1994;120:2259–2269. doi: 10.1242/dev.120.8.2259. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rexhaj E, Paoloni-Giacobino A, Rimoldi SF, Fuster DG, Anderegg M, Somm E, Bouillet E, Allemann Y, Sartori C, Scherrer U. Mice generated by in vitro fertilization exhibit vascular dysfunction and shortened life span. J Clin Invest. 2013;123:5052–5060. doi: 10.1172/JCI68943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo P, Schultz RM. Effects of embryo culture on global pattern of gene expression in preimplantation mouse embryos. Reproduction. 2004;128:301–311. doi: 10.1530/rep.1.00297. [DOI] [PubMed] [Google Scholar]
- Rinaudo PF, Giritharan G, Talbi S, Dobson AT, Schultz RM. Effects of oxygen tension on gene expression in preimplantation mouse embryos. Fertil Steril. 2006;86:1252–1265. 1265, e1251–1236. doi: 10.1016/j.fertnstert.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet. 2008;17:1–14. doi: 10.1093/hmg/ddm280. [DOI] [PubMed] [Google Scholar]
- Sakai K, Matsumoto K, Nishikawa T, Suefuji M, Nakamaru K, Hirashima Y, Kawashima J, Shirotani T, Ichinose K, Brownlee M, Araki E. Mitochondrial reactive oxygen species reduce insulin secretion by pancreatic beta-cells. Biochem Biophys Res Commun. 2003;300:216–222. doi: 10.1016/s0006-291x(02)02832-2. [DOI] [PubMed] [Google Scholar]
- Salvador JM, Brown-Clay JD, Fornace AJ., Jr. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol. 2013;793:1–19. doi: 10.1007/978-1-4614-8289-5_1. [DOI] [PubMed] [Google Scholar]
- Sato MS, Yoshitomo M, Mohri T, Miyazaki S. Spatiotemporal analysis of [Ca2+]i rises in mouse eggs after intracytoplasmic sperm injection (ICSI). Cell Calcium. 1999;26:49–58. doi: 10.1054/ceca.1999.0053. [DOI] [PubMed] [Google Scholar]
- Schafer A. Gadd45 proteins: key players of repair-mediated DNA demethylation. Adv Exp Med Biol. 2013;793:35–50. doi: 10.1007/978-1-4614-8289-5_3. [DOI] [PubMed] [Google Scholar]
- Schjenken JE, Robertson SA. Seminal Fluid Signalling in the Female Reproductive Tract: Implications for Reproductive Success and Offspring Health. Adv Exp Med Biol. 2015;868:127–158. doi: 10.1007/978-3-319-18881-2_6. [DOI] [PubMed] [Google Scholar]
- Schulze PC, Yoshioka J, Takahashi T, He Z, King GL, Lee RT. Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. J Biol Chem. 2004;279:30369–30374. doi: 10.1074/jbc.M400549200. [DOI] [PubMed] [Google Scholar]
- Schwarzer C, Esteves TC, Arauzo-Bravo MJ, Gac S Le, Nordhoff V, Schlatt S, Boiani M. ART culture conditions change the probability of mouse embryo gestation through defined cellular and molecular responses. Hum Reprod. 2012;27:2627–2640. doi: 10.1093/humrep/des223. [DOI] [PubMed] [Google Scholar]
- Scott KA, Yamazaki Y, Yamamoto M, Lin Y, Melhorn SJ, Krause EG, Woods SC, Yanagimachi R, Sakai RR, Tamashiro KL. Glucose parameters are altered in mouse offspring produced by assisted reproductive technologies and somatic cell nuclear transfer. Biol Reprod. 2010;83:220–227. doi: 10.1095/biolreprod.109.082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Dynamic regulation of stem cell specification and maintenance by hypoxia-inducible factors. Mol Aspects Med. 2016:47–48. 15–23. doi: 10.1016/j.mam.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Shalev A. Lack of TXNIP protects beta-cells against glucotoxicity. Biochem Soc Trans. 2008;36:963–965. doi: 10.1042/BST0360963. [DOI] [PubMed] [Google Scholar]
- Shang F, Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med. 2011;51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan RK, Liu X, Feuer SK, Maltepe E, Donjacour A, Rinaudo P. Adult male mice conceived by in vitro fertilization exhibit increased glucocorticoid receptor expression in fat tissue. J Dev Orig Health Dis. 2015:1–10. doi: 10.1017/S2040174415007825. [DOI] [PubMed] [Google Scholar]
- Simmons RA, Templeton LJ, Gertz SJ. Intrauterine growth retardation leads to the development of type 2 diabetes in the rat. Diabetes. 2001;50:2279–2286. doi: 10.2337/diabetes.50.10.2279. [DOI] [PubMed] [Google Scholar]
- Simon MC, Liu L, Barnhart BC, Young RM. Hypoxia-induced signaling in the cardiovascular system. Annu Rev Physiol. 2008;70:51–71. doi: 10.1146/annurev.physiol.70.113006.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Haque MM, Zhang B, Savenkova MI. Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions. Genome Biol. 2012;13:R91. doi: 10.1186/gb-2012-13-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SL, Everts RE, Sung LY, Du F, Page RL, Henderson B, Rodriguez-Zas SL, Nedambale TL, Renard JP, Lewin HA, Yang X, Tian XC. Gene expression profiling of single bovine embryos uncovers significant effects of in vitro maturation, fertilization and culture. Mol Reprod Dev. 2009;76:38–47. doi: 10.1002/mrd.20927. [DOI] [PubMed] [Google Scholar]
- Soriano P, Jaenisch R. Retroviruses as probes for mammalian development: allocation of cells to the somatic and germ cell lineages. Cell. 1986;46:19–29. doi: 10.1016/0092-8674(86)90856-1. [DOI] [PubMed] [Google Scholar]
- Turk R, t Hoen PA, Sterrenburg E, de Menezes RX, de Meijer EJ, Boer JM, van Ommen GJ, den Dunnen JT. Gene expression variation between mouse inbred strains. BMC Genomics. 2004;5:57. doi: 10.1186/1471-2164-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Alcaraz B, Crispi F, Bijnens B, Cruz-Lemini M, Creus M, Sitges M, Bartrons J, Civico S, Balasch J, Gratacos E. Assisted reproductive technologies are associated with cardiovascular remodeling in utero that persists postnatally. Circulation. 2013;128:1442–1450. doi: 10.1161/CIRCULATIONAHA.113.002428. [DOI] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- Watkins AJ, Lucas ES, Torrens C, Cleal JK, Green L, Osmond C, Eckert JJ, Gray WP, Hanson MA, Fleming TP. Maternal low-protein diet during mouse preimplantation development induces vascular dysfunction and altered reninangiotensin-system homeostasis in the offspring. Br J Nutr. 2010;103:1762–1770. doi: 10.1017/S0007114509993783. [DOI] [PubMed] [Google Scholar]
- Watkins AJ, Lucas ES, Wilkins A, Cagampang FR, Fleming TP. Maternal periconceptional and gestational low protein diet affects mouse offspring growth, cardiovascular and adipose phenotype at 1 year of age. PLoS One. 2011;6:e28745. doi: 10.1371/journal.pone.0028745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Xiaoling Z, Kai M, Rui W, Jing X, Min G, Zhonghong W, Jianhui T, Xinyu Z, Lei A. Characterization and comparative analyses of transcriptomes for in vivo and in vitro produced peri-implantation conceptuses and endometria from sheep. J Reprod Dev. 2016;62:279–287. doi: 10.1262/jrd.2015-064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten WK. Nutrient requirements for the culture of preimplantation embryos in vitro. Advances in Bioscience. 1971;6:129–141. [Google Scholar]
- Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chng WJ. Roles of thioredoxin binding protein (TXNIP) in oxidative stress, apoptosis and cancer. Mitochondrion. 2013;13:163–169. doi: 10.1016/j.mito.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Ma W, Lago CU, Hwang PM. Metabolic regulation of oxygen and redox homeostasis by p53: lessons from evolutionary biology? Free Radic Biol Med. 2012;53:1279–1285. doi: 10.1016/j.freeradbiomed.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.