Abstract
Background
MicroRNAs (miRNAs) are effective regulators of gene expression that play a pivotal role in the pathogenesis of colorectal cancer (CRC) and various other cancers. The high prevalence of aberrant miRNA expression in CRC suggests that they can be used as biomarkers and anticancer molecules for therapeutic purposes. There is evidence that microRNA-299-5p (miR-299-5p) is associated with vital cell processes (e.g. epithelial-mesenchymal transition, proliferation, and tumorigenicity) and its improper expression with tumorigenesis in many types of human cancer. This prospective study investigated the contribution of miR-299-5p to CRC tumorigenesis.
Methods
The real-time reverse transcription-polymerase chain reaction was used to examine miR-299-5p expression levels prospectively in 40 sample pairs of CRC tissue and adjacent noncancerous tissue (>2 cm from cancer tissue). The ability of miR-299-5p to function as a tumor marker was also examined.
Results
The expression levels of miR-299-5p were significantly downregulated in the group of CRC samples compared with matched noncancerous tissue samples. No significant relationship was found between miR-299-5p expression levels and clinicopathological features. Receiver operating characteristic analysis gave an area under the curve of 71% for miR-299-5p with 68% sensitivity and 78% specificity (P=0.001).
Conclusion
The miRNA miR-299-5p may be considered as a tumor marker in CRC and could be of assistance as a potential predictive biomarker in the diagnosis of this cancer.
Keywords: Colorectal cancer, biomarker, microRNA, miR-299-5p
Introduction
Colorectal cancer (CRC) is considered to be the third most common cause of cancer-related death [1], accounting for 10% of the worldwide cancer incidence and mortality [2]. More specifically, it is recognized as the second and third most frequently diagnosed type of cancer in females and males, respectively [3]. It has been one of the prevalent cancers in western populations [4]; a low rate of this type of cancer has been observed in Asia, Africa, and South America [5]. Unfortunately, in developing countries and most Asian countries, its yearly incidence is expected to increase over the next two decades [6]. Recently conducted studies in an Iranian context are indicative of a rapidly increasing incidence of this type of cancer [7]. In this regard, a remarkable incidence of cancer occurring in the gastrointestinal tract has been discerned in East Azerbaijan, which is located in the north west of Iran and has a recently reported population of 3,724,620, comprising 50.5% males (1,882,031) and 49.5% females (1,842,589) [8]. Thus, knowing more about the biology of CRC could be very useful for designing effective prognostic, diagnostic, and therapeutic plans to help decrease the impact of this disease [9].
MicroRNAs (miRNAs) are small, non-coding RNA molecules (about 18-25 nucleotides in length) first discovered in the early 1990s in Caenorhabditis elegans [10]. They are the largest family of noncoding RNAs and are believed to regulate up to one third of all genes [11]. There is increasing evidence that miRNAs can function as tumor suppressors or oncogenes [12]. They can be used as biomarkers in diagnosing the progression of several cancers [13]. Several studies have revealed that the micro-RNA expression patterns found in CRC show frequent abnormalities in this malignancy [14].
MiR-299-5p is located in the imprinted Dlk1-Dio3 region on chromosome 14q32.31 [13]. The aforementioned cluster resides within the region of a parentally imprinted chromosome [15] and has been reported to be downregulated in ovarian cancer [16]. It is also involved in gastrointestinal stromal tumors and adenocarcinoma [17], melanoma [18], ependymoma [19], neuroblastoma [20], and osteosarcoma [21]. The importance of miR-299 in CRC and its variations have been reported previously [22]. This area is a great developmental region; it accompanies several phenotypes associated with the changes in the dosage of the genes within this region in humans and mice [23].
Patients and methods
Study population
Between November 2014 and June 2015, a total of 40 CRC samples and normal adjacent tissues were collected from 40 patients diagnosed with CRC following colonoscopy and sigmoidoscopy at Imam Reza Hospital (Tabriz, Iran), the first affiliated hospital of Tabriz University of Medical Sciences. None of the patients had undergone chemotherapy or radiotherapy, or any adjuvant therapy.
A section of the resected specimen at the farthest tumor distance was used to obtain the non-tumor counterparts (>2 cm from tumor). All study members were born in East Azerbaijan, Iran. The Research Ethics Committee of Imam Reza Hospital approved the present study, carried out in accordance with our institutional protocol; informed consent was obtained from all patients. Resected specimens were analyzed by standard histopathological examination and colorectal adenocarcinoma was selected for investigation.
Sample preparation and RNA isolation
In this study, all the tissue samples were freshly frozen at -80°C until RNA extraction. Total RNA was isolated using the TRIzol reagent (Invitrogen Carlsbad, CA) which was applied according to the manufacturer’s instructions. Briefly, tissue samples were homogenized with 1 mL TRIzol LS and incubated for 5 min at room temperature. Next, 200 μL chloroform was added, shaken vigorously for 15 sec, and incubated for 2-15 min at room temperature. Centrifugation was then applied at 12,000 rpm for 15 min at 4°C. After the aqueous phase had been transferred into a new Eppendorf tube, 500 μL of 100% isopropanol was added. The mixture was stored at 20°C overnight, followed by a 13,000 rpm centrifugation for 10 min at 4°C, pelleting the nucleic acid. After removal of the supernatant, 1 mL of 75% ethanol was added to wash the RNA pellet. The sample was then centrifuged at 7500 rpm for 5 min at 4°C, after which 25 μL RNase-free water was added to the RNA pellet, which was finally incubated in a water bath at 55-60°C for 10-15 min. PicoDrop 2000 (Bob Batty International, UK) was used to quantify the concentration of isolated RNA. The extracted RNAs were stored at -80°C until cDNA synthesis. Subsequently, a 10 μL DNase I treatment reaction (Fermentas, Canada) was performed to deteriorate any DNA contaminations in the extracted RNAs.
Reverse transcription and quantitative real-time polymerase chain reaction (PCR)
To perform reverse transcription, 120 ng of total RNA in a final volume of 10 μL reaction mixture was utilized. The 10 μL RT reaction mixture was incubated at 37°C for 60 min and at 85°C for 5 sec; then the mixture was held at 4°C using the Prime Script(R)miRNA cDNA Synthesis Kit (ParsGenome, Iran). Finally, 90 μL of the RNase-free water was added to dilute the RT product.
SYBR® Green was used to perform the real-time PCR. MiR-299-5p and 5s rRNA (as a control RNA) primers were also purchased from ParsGenome. All PCR reactions, including non-template controls, were run in triplicate using a Rotor-Gene Q - QIAGEN Real-time PCR Detection System. REST2009 Software was employed to analyze the raw data obtained. Analyses of all the samples were performed in triplicate. The cycle number in which the fluorescence passed the fixed threshold was regarded as the definition of the threshold cycle (Ct). One control sample without any templates was added to each experiment. Polyacrylamide gel electrophoresis confirmed the ultimate real-time PCR products.
Normalization and data analysis
To apply normalization, the housekeeping reference gene, 5srRNA, and adjacent non-tumor tissue were used as a calibrator to present more accurate quantification of mRNA levels. The relative expression of miR-299-5p was analyzed using a randomization test with the Relative Expression Software Tool (REST) 2009 (http://gene-quantification.com/rest-2009.html) and REST was using the 2-ΔΔCt method to examine the miR-299-5p expression levels in the CRC tissues relative to their matched non-tumor counterparts. The following step was the determination of the Ct of fluorescence for each of the samples. The ΔCt values (ΔCt = Ct miR-299-5p – Ct 5s) were indicative of the presence of differences in the expression levels between miR-299-5P and 5s rRNA. Moreover, ΔΔCt revealed differences between cancer tissue and the matched control in terms of the ΔCt values (ΔΔCt = ΔCt cancer – ΔCt control). In addition, the 2-ΔΔCt value (fold value) was determined. It was found that when the fold value was <1, a fold change <1 in the expression was defined as decreased expression.
Statistical analysis
SPSS 18.0 software (Chicago, IL, USA) was used to analyze the obtained results, with a two-sided P-value <0.05 considered as the criterion for statistical significance. Receiver operating characteristic (ROC) analysis was used to evaluate the specificity and sensitivity of miR-299-5p expression levels in discriminating between CRC and normal tissue. The sensitivity and specificity at various cutoff values were calculated using Sigma Plot 12.5. P-values <0.05 were regarded as indicative of a statistically significant result.
Results
Expression levels of miR-299-5p in CRC and normal tissues
To compare the level of miR-299-5p expression in CRC in relation to normal tissue, the researchers entered the Ct values of all the samples into the REST 2009 software. The results of the randomization test displayed that miR-299-5p expression in tumor samples decreased 5.4 times more than in normal tissue (Fig. 1).
Figure 1.
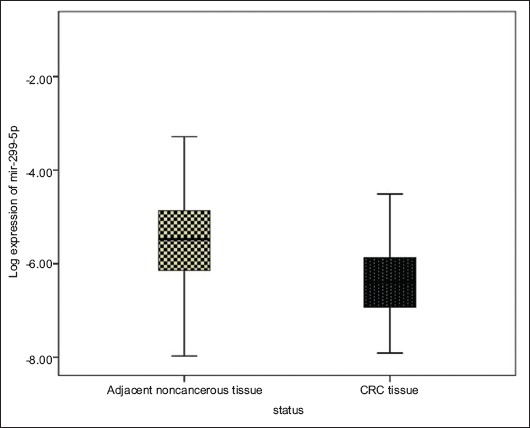
Figure 1 Disparate expression of miR-299-5p in tumor samples, showing a significantly lower value (4.7 times to 5.4) compared to normal samples.
CRC, colorectal cancer.
The association between the expression of miR-299-5p and clinicopathological features
As Table 1 shows, as regards clinicopathological characteristics, no significant relationship was detected between miR-299-5p expression and sex (P=0.837), age (P=0.839), tumor location (P=0.580), or histological grade (P=0.705).
Table 1.
The relationship between clinicopathological features and miR-299-5p expression levels in tissue samples with colorectal cancer

Ability of miR-299-5p to function as a CRC tumor marker
In the ROC analysis, the level of expression of miR-299-5p yielded an area under the curve of 71%, with 68% sensitivity and 78% specificity for the identification of CRC tumor samples (P=0.001). Consequently, miR-299-5p expression can be considered as a tumor marker (Fig. 2).
Figure 2.

Receiver operating characteristic (ROC) analysis of miR-299-5p as a biomarker in the detection of colorectal cancer. The ROC curve was generated from 40 points with a cutoff value set by SigmaPlot software. The area under the curve was 71% for miR-299-5p, with 68% sensitivity and 78% specificity
Discussion
It is crucial to improve the recognition of molecular factors that have prognostic and predictive significance in CRC [24]. MicroRNAs have an evident role in the initiation and progression of CRC. However, future research will have to precisely reveal the potential role of microRNA-based classifiers and therapeutics in medicine. A good number of studies have confirmed the potential of miRNAs as predictive/prognostic classifiers for early cancer detection [2]. There are many reports on the effects of miRNAs on oncogenes and tumor suppressor genes [25]. Downregulation of miRNAs is generally observed in human cancers [26]. It should be noted that many miRs have been previously studied in CRC [22,27,28].
The DLK1-DIO3 genomic locus present on chromosome 14q32.31 is a parentally imprinted region [13]; this region is one of the largest miRNA-containing clusters of the human genome and contains 54 miRNAs [13]. Most imprinted genes play key roles in placental growth and have been implicated in embryonic and developmental abnormalities in the mouse and human [29]. MiRNAs within the DLK1-DIO3 genomic region have been shown to be involved in disease pathogenesis, mainly of cancers. Multiple miRNAs from this cluster potentially target the factor of the Polycomb repressive complex 2 (PRC2), which plays a significant role in the prevention of PRC2 formation; hence, the aforementioned region could remain active in full pluripotent stem cells [13]. MiR-299-5p was found to be downregulated in oral squamous cell carcinoma cell lines [30] and prostate cancer cells [31]. Another study showed that miR-299-5p was upregulated in myelodysplastic syndrome [32]. MiR-299-5p was downregulated in metastatic breast cancer patients [33] and high risk neuroblastoma [34]. MiR-299-5p was downregulated in patients with t (11q23)/MLL-rearrangement and upregulated in t (15:17) cytogenetic subtypes [35]. Another study on hepatoma cells showed that hsa-miR-299-5p targeted osteopontin (OPN), which is known to be essential for enhancing proliferation and tumorigenicity [36]. Gain- and loss-of-function experiments revealed that miR-299-5p plays a role in the regulation of hematopoietic progenitor; it may therefore act as miRNA in the differentiation of megakaryoblasts [37]. Research on A549 human non-small cell lung cancer cells indicated that expression of miR-299-5p was upregulated [38].
One study revealed that miR-299-5p regulates OPN, and that increasing the expression of OPN via decreased levels of miR-299-5p plays an important role in enhancing proliferation, tumorigenicity, and the ability to display vasculogenic mimicry of the spheroid formation of breast cancer cells [39]. According to previous research, OPN is a multifunctional cytokine that influences several signaling pathways that are associated with cell proliferation, survival, drug resistance, invasion, stem-like behavior, apoptosis, and metastatic potential [40]. Past reports reveal that estrogen-related receptor α regulates OPN expression in human CRC. OPN is a direct target of estrogen-related receptor α, the induction of which could clearly contribute to CRC pathogenesis [41]. OPN in CRC may be a clinical prognostic marker significantly associated with prognosis [42]. OPN appears to regulate motility through interaction with CD44. An increase in OPN results in an increase in the expression of vascular endothelial growth factor [40]. There are mutual interactions between TAM receptors and CD44-positive CRC cancer cells via OPN/CD44 [43].
In conclusion, the present study revealed that miR-299-5p was significantly downregulated in CRC samples compared to their normal counterparts. However, the reason for the misregulation of this miRNA is unclear, though a loss of heterozygosity or epigenetic silencing could be responsible. Based on the above findings, miR-299-5p regulates OPN, which is a critical gene in several signaling pathways, especially in cancer. It is clear that more studies on the relationship between miR-299-5p and OPN in CRC may yield better results regarding the therapeutic targets.
The capability of miR-299-5p expression level to function as a tumor marker to distinguish CRC from normal tissue was also determined in the current study, suggesting that this miRNA has a high sensitivity and specificity; therefore, it can be regarded as a tumor marker in diagnosing CRC. However, to determine a more detailed association between these mRNAs and clinicopathological features (e.g. age, sex, tumor grade, and tumor location), other studies with larger sample sizes need to be conducted.
Summary Box.
What is already known:
Colorectal cancer (CRC) is considered as the third most common cause of cancer-related deaths, accounting for 10% of worldwide cancer incidence and mortality
The yearly incidence of CRC is expected to increase over the next two decades in developing countries, especially in Asia
Recent studies in Iran suggest a rapid increase in the incidence of CRC
What the new findings are:
MiR-299-5p was significantly downregulated in CRC samples compared to their normal counterparts
MiR-299-5p expression level may be regarded as a tumor marker for distinguishing CRC from normal tissue
As a potential predictive biomarker, miR-299-5p can help diagnose CRC
Acknowledgments
We would like to thank the patients, staff, and nurses in the Endoscopy and the Pathology departments of Tabriz Imam Reza hospital who kindly helped us conduct this study.
Biography
University of Tabriz, Tabriz, Iran
Footnotes
Conflict of interest: None
References
- 1.Ansari R, Mahdavinia M, Sadjadi A, et al. Incidence and age distribution of colorectal cancer in Iran: results of a population-based cancer registry. Cancer Lett. 2006;240:143–147. doi: 10.1016/j.canlet.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J. 2012;18:244–252. doi: 10.1097/PPO.0b013e318258b78f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rezaianzadeh A, Safarpour AR, Marzban M, Mohaghegh A. A systematic review over the incidence of colorectal cancer in Iran. Ann Colorectal Res. 2015;3:e25724. [Google Scholar]
- 4.Sung JJ, Lau JY, Goh KL, Leung WK Asia Pacific Working Group on Colorectal Cancer. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6:871–876. doi: 10.1016/S1470-2045(05)70422-8. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 6.Dolatkhah R, Somi MH, Bonyadi MJ, et al. Colorectal cancer in Iran: molecular epidemiology and screening strategies. J Cancer Epidemiol. 2015;2015:643020. doi: 10.1155/2015/643020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moghimi-Dehkordi B, Safaee A, Zali MR. Prognostic factors in 1,138 Iranian colorectal cancer patients. Int J Colorectal Dis. 2008;23:683–688. doi: 10.1007/s00384-008-0463-7. [DOI] [PubMed] [Google Scholar]
- 8.Somi MH, Golzari M, Farhang S, et al. Gastrointestinal cancer incidence in East Azerbaijan, Iran: update on 5 year incidence and trends. Asian Pac J Cancer Prev. 2014;15:3945–3949. doi: 10.7314/apjcp.2014.15.9.3945. [DOI] [PubMed] [Google Scholar]
- 9.Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2016 doi: 10.1136/gutjnl-2015-310912. gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Chang KH, Miller N, Kheirelseid EA, et al. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26:1415–1422. doi: 10.1007/s00384-011-1279-4. [DOI] [PubMed] [Google Scholar]
- 12.Faltejskova P, Svoboda M, Srutova K, et al. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16:2655–2666. doi: 10.1111/j.1582-4934.2012.01579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benetatos L, Hatzimichael E, Londin E, et al. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70:795–814. doi: 10.1007/s00018-012-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SP, Youngson N, Takada S, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 16.Stadtfeld M, Apostolou E, Akutsu H, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller F, von Heydebreck A, Zhang JD, et al. Localization- and mutation-dependent microRNA (miRNA) expression signatures in gastrointestinal stromal tumours (GISTs), with a cluster of co-expressed miRNAs located at 14q32.31. J Pathol. 2010;220:71–86. doi: 10.1002/path.2610. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa FF, Bischof JM, Vanin EF, et al. Identification of microRNAs as potential prognostic markers in ependymoma. PLoS One. 2011;6:e25114. doi: 10.1371/journal.pone.0025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gattolliat CH, Thomas L, Ciafrè SA, et al. Expression of miR-487b and miR-410 encoded by 14q32.31 locus is a prognostic marker in neuroblastoma. Br J Cancer. 2011;105:1352–1361. doi: 10.1038/bjc.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thayanithy V, Sarver AL, Kartha RV, et al. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50:171–181. doi: 10.1016/j.bone.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D’Angelo E, Fassan M, Maretto I, et al. Serum miR-125b is a non-invasive predictive biomarker of the pre-operative chemoradiotherapy responsiveness in patients with rectal adenocarcinoma. Oncotarget. 2016;7:28647–28657. doi: 10.18632/oncotarget.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Alimov A, Sundelin B, Wang N, Larsson C, Bergerheim U. Loss of 14q31-q32.2 in renal cell carcinoma is associated with high malignancy grade and poor survival. Int J Oncol. 2004;25:179–185. [PubMed] [Google Scholar]
- 25.Wang Y, Chen M, Tao Z, Hua Q, Chen S, Xiao B. Identification of predictive biomarkers for early diagnosis of larynx carcinoma based on microRNA expression data. Cancer Genet. 2013;206:340–346. doi: 10.1016/j.cancergen.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Perilli L, Vicentini C, Agostini M, et al. Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget. 2014;5:6611–6619. doi: 10.18632/oncotarget.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orang AV, Safaralizadeh R, Hosseinpour Feizi MA, Somi MH. Diagnostic and prognostic value of miR-205 in colorectal cancer. Asian Pac J Cancer Prev. 2014;15:4033–4037. doi: 10.7314/apjcp.2014.15.9.4033. [DOI] [PubMed] [Google Scholar]
- 29.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaillé J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 31.Formosa A, Markert E, Lena A, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32. 31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 32.Dostalova Merkerova M, Krejcik Z, Votavova H, et al. Distinctive microRNA expression profiles in CD34+bone marrow cells from patients with myelodysplastic syndrome. Eur J Hum Genet. 2011;19:313–319. doi: 10.1038/ejhg.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145–155. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mei H, Lin ZY, Tong QS. The roles of microRNAs in neuroblastoma. World J Pediatr. 2014;10:10–16. doi: 10.1007/s12519-014-0448-2. [DOI] [PubMed] [Google Scholar]
- 35.Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin JC, Jin XL, Zhang X, Piao YS, Liu SP. Effect of OSW-1 on microRNA expression profiles of hepatoma cells and functions of novel microRNAs. Mol Med Rep. 2013;7:1831–1837. doi: 10.3892/mmr.2013.1428. [DOI] [PubMed] [Google Scholar]
- 37.Tenedini E, Roncaglia E, Ferrari F, et al. Integrated analysis of microRNA and mRNA expression profiles in physiological myelopoiesis: role of hsa-mir-299-5p in CD34+progenitor cells commitment. Cell Death Dis. 2010;1:e28. doi: 10.1038/cddis.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bae S, Lee EM, Cha HJ, et al. Resveratrol alters microRNA expression profiles in A549 human non-small cell lung cancer cells. Mol Cells. 2011;32:243–249. doi: 10.1007/s10059-011-1037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shevde LA, Metge BJ, Mitra A, et al. Spheroid-forming subpopulation of breast cancer cells demonstrates vasculogenic mimicry via hsa-miR-299-5p regulated de novo expression of osteopontin. J Cell Mol Med. 2010;14:1693–1706. doi: 10.1111/j.1582-4934.2009.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irby R, McCarthy S, Yeatman T. Osteopontin regulates multiple functions contributing to human colon cancer development and progression. Clinical Exp Metastasis. 2004;21:515–523. doi: 10.1007/s10585-004-2873-4. [DOI] [PubMed] [Google Scholar]
- 41.Boudjadi S, Bernatchez G, Beaulieu JF, Carrier JC. Control of the human osteopontin promoter by ERRαin colorectal cancer. Am J Pathol. 2013;183:266–276. doi: 10.1016/j.ajpath.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 42.Likui W, Hong W, Shuwen Z. Clinical significance of the upregulated osteopontin mRNA expression in human colorectal cancer. J Gastrointest Surg. 2010;14:74–81. doi: 10.1007/s11605-009-1035-z. [DOI] [PubMed] [Google Scholar]
- 43.Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


