ABSTRACT
To our knowledge, fecal microbiota collection methods have not been evaluated in low- and middle-income countries. Therefore, we evaluated five different fecal sample collection methods for technical reproducibility, stability, and accuracy within the Health Effects of Arsenic Longitudinal Study (HEALS) in Bangladesh. Fifty participants from the HEALS provided fecal samples in the clinic which were aliquoted into no solution, 95% ethanol, RNAlater, postdevelopment fecal occult blood test (FOBT) cards, and fecal immunochemical test (FIT) tubes. Half of the aliquots were frozen immediately at −80°C (day 0) and the remaining samples were left at ambient temperature for 96 h and then frozen (day 4). Intraclass correlation coefficients (ICC) were calculated for the relative abundances of the top three phyla, for two alpha diversity measures, and for four beta diversity measures. The duplicate samples had relatively high ICCs for technical reproducibility at day 0 and day 4 (range, 0.79 to 0.99). The FOBT card and samples preserved in RNAlater and 95% ethanol had the highest ICCs for stability over 4 days. The FIT tube had lower stability measures overall. In comparison to the “gold standard” method using immediately frozen fecal samples with no solution, the ICCs for many of the microbial metrics were low, but the rank order appeared to be preserved as seen by the Spearman correlation. The FOBT cards, 95% ethanol, and RNAlater were effective fecal preservatives. These fecal collection methods are optimal for future cohort studies, particularly in low- and middle-income countries.
IMPORTANCE The collection of fecal samples in prospective cohort studies is essential to provide the opportunity to study the effect of the human microbiota on numerous health conditions. However, these collection methods have not been adequately tested in low- and middle-income countries. We present estimates of technical reproducibility, stability at ambient temperature for 4 days, and accuracy comparing a “gold standard” for fecal samples in no solution, 95% ethanol, RNAlater, postdevelopment fecal occult blood test cards, and fecal immunochemical test tubes in a study conducted in Bangladesh. Fecal occult blood test cards and fecal samples stored in 95% ethanol or RNAlater adequately preserve fecal samples in this setting. Therefore, new studies in low- and middle-income countries should include collection of fecal samples using fecal occult blood test cards, 95% ethanol, or RNAlater for prospective cohort studies.
KEYWORDS: feces, low/middle-income countries, microbiota, sampling methods, stability
INTRODUCTION
The human microbiome has garnered increasing scientific interest over the past few years. There is a growing appreciation that microbiota, particularly the fecal microbiota, is associated with specific health conditions, such as diabetes (1), obesity (2), inflammatory bowel disease (3), and cancer (4). However, much of this research has been cross-sectional, since there are very few existing cohorts from which fecal samples were collected, although there are many large cohorts for which fecal samples are currently being collected and new collections are planned for other prospective studies.
It has become clear that numerous factors may impact measurements of the microbiota, such as the laboratory handling and bioinformatic processing of the data (5). The impact of the type of fecal sample collection method on microbial data has been considered in a number of previous studies (6–23). However, the majority of these studies were conducted in North American or European populations near academic or clinical laboratories and typically included only a small number of methods. Since the microbiota appears to be distinct by geographic location (24), it is unknown whether the identified fecal sample collection methods are adequate in populations with differing microbial compositions. In addition, it is not clear if certain sample collection methods would be more feasible under field conditions, particularly in low- and middle-income countries. Therefore, we tested the technical reproducibility of five different fecal sample collection methods (no solution, 95% ethanol, RNAlater, postdevelopment fecal occult blood test [FOBT] cards, and fecal immunochemical test [FIT] tubes), the impact of delayed freezing on these collection methods, and the accuracy of each method compared with that of a “gold standard” within the Health Effects of Arsenic Longitudinal Study (HEALS) in Bangladesh.
(Part of this work was presented at the International Human Microbiome Consortium Congress in Houston, Texas, 9 to 11 November 2016.)
RESULTS
Descriptive characteristics.
There was a higher proportion of female (60.0%) than male participants in this sample, for which ages ranged from 25 to 50 years with a mean age of 37.6 (standard deviation [SD], 6.9) years. Participants had a mean body mass index (BMI) of 19.5 (SD, 2.6), and 38.0% had smoked cigarettes or bidi (unprocessed tobacco wrapped in leaves) (Table 1). Overall, it appeared that interindividual difference explained the majority of microbial variability for all beta diversity estimates, but the differences by sample collection method were still apparent, particularly for the weighted UniFrac (see Fig. S2 in the supplemental material). For alpha diversity measured using observed operational taxonomic units (OTUs), the FOBT cards and samples in RNAlater had a slightly increased alpha diversity, while the FIT tubes and samples preserved in 95% ethanol had a slightly decreased alpha diversity compared with that of the no-solution day 0 samples. For the Shannon index, only the FOBT cards had a slightly increased alpha diversity, while the FIT tubes and samples preserved in RNAlater and 95% ethanol had a lower diversity compared with that of the no-solution day 0 samples (see Fig. S3A and B). At the phylum level, there were differences by collection method where the no-solution day 0 samples had a lower relative abundance of Bacteroidetes and a higher relative abundance of Firmicutes than the other methods. In general, the freezing timepoint did not appear to change the relative abundances within each collection method (Fig. S4A and B).
TABLE 1.
Descriptive statistics of study participantsa
| Parameter | Value |
|---|---|
| Age (mean [SD]) (year) | 37.6 (6.9) |
| Sex (n [%]) | |
| Male | 20 (40.0) |
| Female | 30 (60.0) |
| BMI (mean [SD]) (kg/m2) | 19.5 (2.6) |
| Ever smoked cigarettes or bidi (n [%]) | |
| Yes | 19 (38.0) |
| No | 31 (62.0) |
Includes 50 participants of the HEALS (Health Effects of Arsenic Longitudinal Study). BMI, body mass index.
Technical reproducibility.
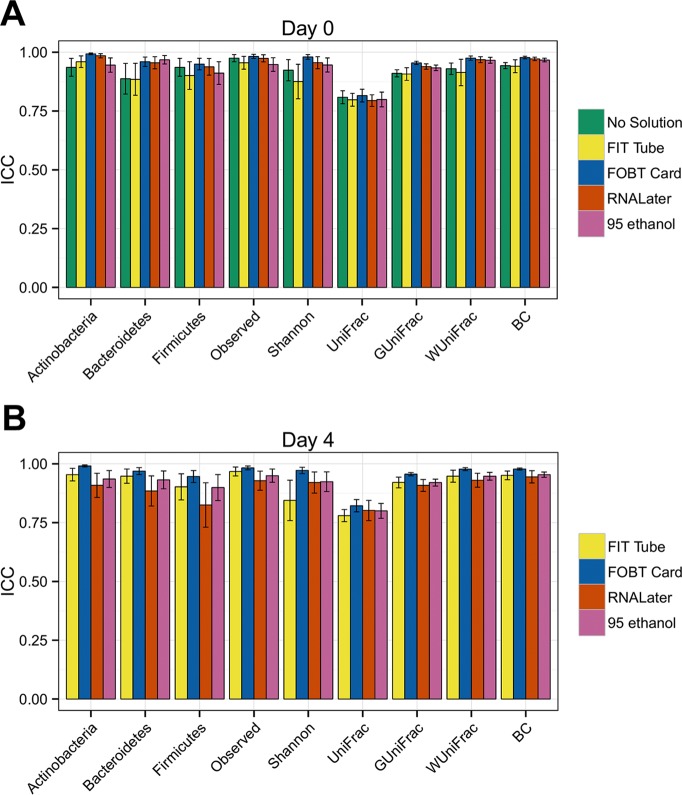
The duplicate samples had relatively high intraclass correlation coefficients (ICCs) for technical reproducibility at day 0 and day 4, with almost all ICCs greater than 0.80. For example, for day 0 samples, the ICCs for the weighted UniFrac distance were 0.93 (95% confidence interval [CI], 0.91 to 0.95) for no solution, 0.91 (95% CI, 0.86 to 0.97) for the FIT tube, 0.98 (95% CI, 0.97 to 0.98) for the FOBT card, 0.97 (95% CI, 0.96 to 0.98) for RNAlater, and 0.97 (95% CI, 0.95 to 0.98) for 95% ethanol (Fig. 1; see also Table S2A and B). In general, the ICCs were high for the relative abundances of the top genera for day 0 (see Fig. S5A and Table S3A) and day 4 (Fig. S5B and Table S3B) samples.
FIG 1.
Mean technical reproducibility and 95% CIs from day 0 replicates (A) and day 4 replicates (B) for the relative abundance of three phyla, two alpha diversity metrics, and four beta diversity metrics by fecal sample collection method using intraclass correlation coefficients. For day 0, samples from 38, 36, 46, 42, and 47 individuals were included for no solution, FIT tube, FOBT card, RNAlater, and 95% ethanol samples, respectively. For day 4, samples from 41, 50, 43, and 46 individuals were included for FIT tube, FOBT card, RNAlater, and 95% ethanol samples, respectively. BC, Bray-Curtis distance.
Stability at ambient temperature.
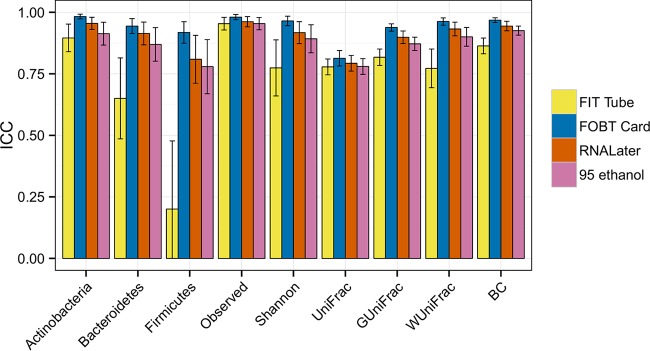
The fecal collection methods, particularly the FOBT card and the preservation of samples in RNAlater and 95% ethanol, had high ICCs for stability. For the relative abundance of Actinobacteria, the ICCs ranged from 0.90 (95% CI, 0.84 to 0.95) for the FIT tube to 0.98 (95% CI, 0.97 to 0.99) for the FOBT card. However, for the relative abundances of the other two phyla, Bacteroidetes and Firmicutes, the ICCs for the FIT tube were lower with ICCs of 0.65 (95% CI, 0.49 to 0.82) and 0.20 (95% CI, 0.00 to 0.48), respectively. The ICCs for alpha and beta diversity measures were all high (Fig. 2; see also Table S4). At the genus level, the ICCs were generally high except for Blautia and Faecalibacterium (both from the Firmicutes phylum) for the FIT tube (see Fig. S6 and Table S5).
FIG 2.
Mean stability and 95% CIs by fecal sample collection methods (i.e., day 4 fecal samples compared with day 0 fecal samples) for the relative abundance of three phyla, two alpha diversity metrics, and four beta diversity metrics using intraclass correlation coefficients. Samples from 47, 50, 48, and 49 individuals were included for the FIT tube, FOBT card, RNAlater, and 95% ethanol samples, respectively.
Accuracy compared to the gold standard.
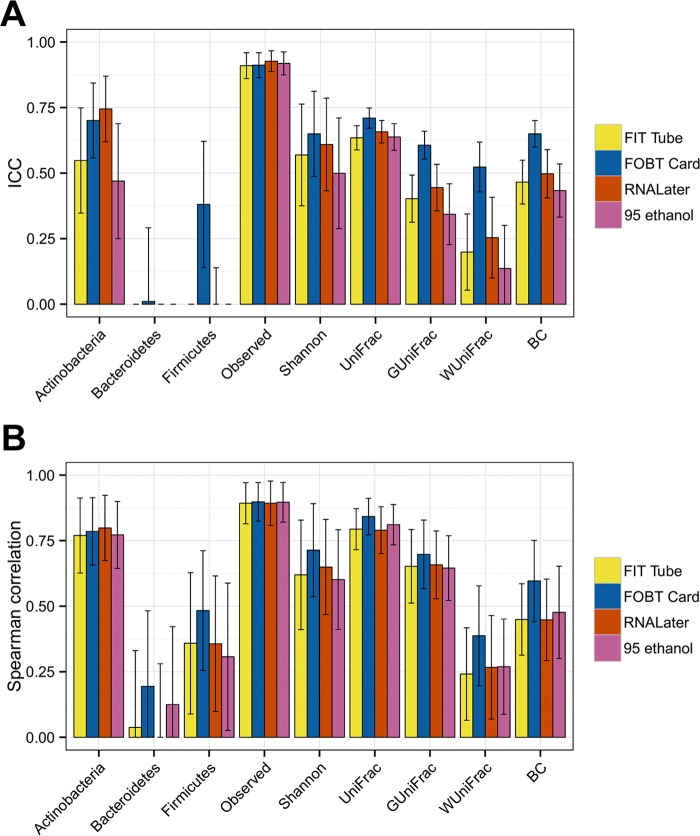
In comparison to the gold standard method in which fecal samples are immediately frozen with no solution, the ICCs for many of the microbial metrics were low, although the FOBT cards tended to have the highest ICCs for most measures. For instance, for weighted UniFrac, the ICCs for the FIT tube, RNAlater, and 95% ethanol were all less than 0.30, while the ICC for the FOBT card was 0.52 (95% CI, 0.43 to 0.62). Unweighted UniFrac, which does not take into account relative abundances, had ICCs higher than 0.60 for each of the methods (Fig. 3 top; see also Table S6A). The values from Spearman correlations for the same comparisons were higher. For the Shannon index, the Spearman correlations ranged from 0.60 (95% CI, 0.41 to 0.79) for 95% ethanol to 0.71 (95% CI, 0.52 to 0.89) for the FOBT card in comparison to the gold standard (Fig. 3 bottom; see also Table S6B). At the genus level, the ICCs were low for some genera, including Prevotella, Blautia, and Sutterella (see Fig. S7A and Table S7A), but these were strengthened with the Spearman correlation (see Fig. S7B and Table S7B).
FIG 3.
Mean accuracy and 95% CIs from day 0 fecal samples compared with the gold standard no solution sample frozen immediately. Intraclass correlation coefficients (A) and Spearman correlations (B) were calculated for the relative abundance of three phyla, two alpha diversity metrics, and four beta diversity metrics by sample collection method. Samples from 47, 49, 49, and 49 individuals were included for the FIT tube, FOBT card, RNAlater, and 95% ethanol samples, respectively.
DISCUSSION
In this study, nested within the HEALS cohort in Bangladesh, we found that 32.5% of the eligible cohort study participants provided in-clinic fecal samples. The response rate may be increased in the future by using in-home collection. For the different collection methods, the primary source of variability was intersubject variability, although some variability was related to the collection method, particularly for weighted UniFrac. The ICCs for the technical replicates were high (>0.80) for both the day 0 and day 4 samples, which suggests that a single aliquot from a sample is a good representation of the sample. For stability, the FOBT card and samples stored in RNAlater or 95% ethanol had high ICCs (>0.75), while the FIT tubes had lower ICCs for a number of metrics, which suggests that the FIT tubes were not as stable at room temperature in Bangladesh. When the collection methods were compared with the gold standard of immediately freezing fecal samples with no solution, the ICCs were generally low for measures that incorporated relative abundance values but higher for observed OTUs and unweighted UniFrac, which only takes into account the presence or absence of an OTU. By using the Spearman correlation, which takes into account the rank order of the data, the accuracy was increased. However, most of the metrics remained low, which suggests that each collection method creates different microbial compositions compared with that of the gold standard fecal sample. This suggests that it may be difficult to combine results using different collection methods, and so future studies, particularly those considering geographic differences in the microbiota, should collect samples using at least one similar method for comparability.
In general, our findings agree with those from many of the previous studies considering the impact of the collection method and short-term storage at room temperature in predominantly Western populations (6–23). In general, FOBT cards or Whatman FTA cards, which have feces smeared on the card itself that then dries, have been found to be relatively stable at room temperature and similar to immediately frozen samples without preservatives (6, 10, 15, 21, 22). Fecal samples stored in 95% ethanol (12, 21) and RNAlater (6, 11, 12, 15, 19, 22) appear to be adequately preserved in most studies, although some studies found some decreased diversity, DNA quality, or stability in samples stored in RNAlater (9, 10, 13, 19, 21). We and another group previously observed that FIT tubes, which store a small amount of feces from a probe in a buffer solution, stabilize fecal samples (22, 23), but we were unable to replicate this finding in this study in Bangladesh.
This study has some limitations. For the accuracy calculations, we compared each of the collection methods to fecal samples frozen immediately without preservative, the gold standard. It has recently been shown that fecal samples immediately frozen without preservative have some microbial composition changes (21), and so this sample may not represent the ideal gold standard of an immediately extracted sample that has not been frozen. However, in a large epidemiological study, it is almost impossible to immediately extract thousands of samples, and since DNA extraction is a potential source of variability in microbial data (5), we did not include an immediately extracted sample in this study. The lower accuracy measures may also limit the pooling of microbial data between different geographical regions if each study utilizes a unique collection method, as each collection may have unique microbial artifacts. However, if all studies use the same collection method, this issue will be minimized. We also had a relatively low proportion of eligible participants involved in the fecal collection. It is unknown whether cohort participants who refused to provide a fecal specimen had different microbial compositions that might have affected our measurements, although this is unlikely. In addition, the fecal samples were shipped from Bangladesh to the National Cancer Institute and to the University of California, San Diego, and may have experienced some thawing in transit, which might have been related to some of our observed decreases in technical reproducibility, stability, and accuracy compared with those from our previous studies (6, 22). We also only investigated 16S rRNA gene sequencing, and the stability and accuracy may differ for other technologies, such as whole-genome shotgun metagenomics and metabolomics.
This study also has a number of strengths. This is the first study, to our knowledge, that has evaluated fecal collection methods in a low- to middle-income country. Although this study was conducted in Bangladesh, these findings may also extend to other field-based settings, such as rural areas in developed/Western nations. It was also conducted within the context of a larger cohort study to help evaluate the uptake of fecal collections in this population. We had participants provide fecal samples in the clinic to eliminate any differences related to time in transport or home freezing.
The collection of fecal samples in large cohort studies is vital for investigating prospective associations between the fecal microbiota and health conditions, such as cancer. Given the fecal microbial differences detected by geographic location (24), likely related to unique lifestyles and exposures, identifying fecal collection methods that can successfully be used in low- and middle-income countries is essential. Here, we found that FOBT cards, 95% ethanol, and RNAlater successfully stabilized fecal samples at room temperature and were generally able to rank the participants' microbial characteristics compared with those of an immediately frozen, no-solution fecal sample. These three methods appear to be optimal for future cohort studies, particularly in low- and middle-income countries; however, it is vital that all comparisons for one study are made using one selected method.
MATERIALS AND METHODS
Study participants.
The HEALS study has been previously described in detail (25). In brief, HEALS is a prospective cohort study which recruited participants from Araihazar, Bangladesh, from October 2000 to May 2002. Participants had to be married, be a resident in the study area for at least 5 years, and have a local well as a primary water source. A total of 11,746 participants were recruited. Since the baseline data were collected, participants have been followed up approximately every 2 years.
For this microbiota study, we aimed to collect fecal samples from HEALS participants living in the 6 nearby villages surrounding the clinic in 2015. A trained village health worker visited participants' homes to ensure eligibility using a structured questionnaire and to set up appointments to visit the study clinic for recruitment. To be eligible, the participants had to be free from any major illness. The village health worker visited 310 participants, and 154 fulfilled the eligibility requirements. Of the 154 eligible participants, 71 (46.1%) agreed to visit the clinic for the study and 50 (32.5%) participants visited the clinic and completed all of the study procedures. This study was approved by the institutional review board (IRB) at the University of Chicago.
Fecal specimen collection.
When participants arrived at the clinic, a senior research officer obtained informed consent and administered a questionnaire regarding tobacco use, alcohol consumption, recent antibiotic exposure, colon health, and demographics. An empty ThermoFisher Scientific vial (Waltham, MA, USA) was provided to each participant for fecal collection. The participant collected the feces and study personnel delivered it to the laboratory for processing.
The fecal specimen was mixed using a spatula, and aliquots for the different collection methods were generated in a random order. For each participant, 10 aliquots of feces, 2 triple-slide FOBT cards, and 2 FIT tubes were created. Approximately 1 to 2 g of feces, representing a full scoop of feces, was placed in a Sarstedt feces tube (Numbrecht, Germany) containing no solution (2 aliquots), 2.5 ml of RNAlater (Ambion, Austin, TX; 4 aliquots), or 2.5 ml of 95% ethanol (Sigma-Aldrich, St. Louis, MO; 4 aliquots). Two triple-slide Hemoccult II Elite Dispensapak Plus test kits for FOBT (Beckman Coulter, Brea, CA), a guaiac-based test, were smeared thinly with feces and the flaps were closed. Two FIT tubes (Polymedco, Inc., Cortlandt Manor, New York) were created by dipping the FIT probes into the fecal specimen and the tubes were shaken. Two aliquots from each FIT tube were created and stored in cryovials.
As seen in Table 2, two replicates from the no solution, 95% ethanol, RNAlater, and FIT cryovials, and one FOBT card were frozen immediately at −80°C (day 0). Two replicates from the 95% ethanol, RNAlater, and FIT cryovials, and one FOBT card were left at ambient temperature for 96 h and then frozen at −80°C (day 4).
TABLE 2.
Collection methods for HEALS fecal samples and number of aliquots used for analysis of the microbiota
| Collection method | No. of aliquots frozen on: |
|
|---|---|---|
| Day 0 | Day 4 | |
| No solution | 2 | 0 |
| RNAlater | 2 | 2 |
| 95% ethanol | 2 | 2 |
| FOBT carda | 1 | 1 |
| FIT tube | 2 | 2 |
Triple-slide (3-window) card developed with peroxide on day 0 or day 4. Three windows were used per card, and each window was considered a separate aliquot.
DNA extraction and sequencing.
The samples were shipped on dry ice from Bangladesh to the National Cancer Institute, and the samples were then shipped to the University of California, San Diego. The samples were thawed at 4°C and kept on ice while being plated for DNA extraction. From the no solution, 95% ethanol, and RNAlater aliquots, the fecal material was pulled out and swabbed using a wooden swab (Puritan cotton tipped applicators; Puritan Medical Products) as described in previous studies (6, 21, 22). A dry swab was rubbed vigorously on the window of the FOBT cards and a swab was dipped into each FIT aliquot. The swabs were then used for the DNA extraction.
DNA was extracted from each of the samples using the Mo Bio PowerMag soil DNA isolation kit as described on the Earth Microbiome Project website, which incorporates a bead-beating step (http://www.earthmicrobiome.org/emp-standard-protocols/dna-extraction-protocol/). PCR amplification and sequencing were performed as described previously (22). Briefly, barcoded 515f/806r primers were used for PCR amplification of the V4 region of the 16S rRNA gene. Barcoded amplicons were pooled in equal concentrations and were sequenced using the Illumina HiSeq. After removing singletons and reads with read errors, the average coverage was approximately 102,000 reads per sample.
Bioinformatic data processing.
Bioinformatic processing of the data was conducted as described previously (22). In brief, the reads were demultiplexed and quality filtered using QIIME 1.9 (27) with -q 3. OTUs were then obtained using deblur (https://github.com/biocore/deblur; our unpublished data). Deblur is a greedy denoising algorithm that removes all reads that are candidate PCR/read errors using a maximal error probability per hamming distance. Deblur was run on the forward reads using default parameters with -negate (negative filtering mode which removes PhiX and adapter sequences), -t 150 (read trimming to 150 bp), and -min-reads 10 (final removal of OTUs with less than 10 reads total in all samples). A summary of read counts before and after using Deblur is shown in Table S1 in the supplemental material. The output of Deblur is a biom table with each OTU being a specific sequence present in the original sample. Therefore, OTUs from Deblur do not have a similarity threshold and are sensitive to a single nucleotide difference. Taxonomy was then assigned using the QIIME command assign_taxonomy.py with the -m rdp option using both Greengenes database version 13.8 (28) and RDP classifier 2.2 (29). A phylogenetic tree for the samples was built using make_phylogeny.py in QIIME.
After rarefaction to 10,000 reads per sample, alpha diversity measures (observed OTUs and the Shannon diversity index) were calculated using the R phyloseq package (30), the Bray-Curtis distance was calculated using the R vegan package, and the additional beta diversity measures (unweighted UniFrac, generalized UniFrac, and weighted UniFrac) were calculated using the R GUniFrac package (31). Unweighted UniFrac is based on the presence and absence of OTUs, while weighted UniFrac is calculated from the relative abundance of the OTUs. Generalized UniFrac was developed as the midpoint between the unweighted and weighted UniFrac distances (6).
Statistical analysis.
Due to the handling of a large number of samples, there is a possibility for sample mislabeling or swapping, in addition to potential cross-sample contamination. Therefore, we removed outliers in the microbial data on the basis of the unweighted UniFrac distance. This outlier index for a given sample A from subject S is defined as the ratio between the average distance from sample A to the other samples from subject S and the median within-subject S distance. When a sample had an outlier index larger than 1.4, this sample was checked using a principal-coordinate analysis (PCoA) plot and genus-level bar plot. Samples that showed obvious deviations were removed. Using this method, we excluded four no-solution samples, two 95% ethanol samples, and one FOBT card and FIT tube each for day 0 samples, in addition to three FIT tubes, one FOBT card, and one sample each for 95% ethanol and RNAlater for day 4 samples. Figure S1 in the supplemental material demonstrates the strong clustering by subject regardless of collection method or freezing timepoint.
To evaluate the percentages of microbial variability due to subject, sample collection type, and day of freezing, we calculated a distance-based coefficient of determination, R2, from beta diversity estimates (i.e., unweighted UniFrac, generalized UniFrac, weighted UniFrac, and Bray-Curtis distances). Then, we calculated the alpha diversity (i.e., observed OTUs and the Shannon index) by collection method and freezing timepoint. We determined the phylum-level relative abundance for individual samples and the relative abundance at the phylum level for each collection method and freezing timepoint.
We calculated intraclass correlation coefficients (ICCs) from a mixed-effects model to evaluate the technical reproducibility (i.e., comparability of replicates), stability at ambient temperature, and accuracy of the different fecal collection methods compared with the gold standard fecal sample frozen immediately with no solution using the method described previously (6). For accuracy, if replicates of the gold standard were available, these values were averaged. The ICCs were calculated based on the square root of the relative abundances of the three most dominant phyla (Actinobacteria, Bacteroidetes, and Firmicutes), two alpha diversity metrics (observed OTUs and the Shannon index), and four beta diversity metrics (unweighted UniFrac, generalized UniFrac, weighted UniFrac, and Bray-Curtis distances). To use all of the information from the beta diversity distance matrix, we computed a distance-based ICC for beta diversity, which uses the within-subject squared distances and between-subjects squared differences to determine the biological and technical variance (see Methods in the supplemental material). For technical reproducibility and stability, we calculated 95% confidence intervals (95% CI) using the R “ICC” package (CI = “Smith”). For the beta diversity (i.e., distance-based) ICCs, we calculated the 95% CIs using 1,000 bootstrap samples. All of these 95% CIs were averaged over 100 random samplings of replicates. For accuracy, the Spearman correlation, a nonparametric measure of rank, was also calculated to determine whether the rank order compared with the gold standard was preserved. The 95% CIs for accuracy were calculated using the same method as for the beta diversity ICCs. We also conducted a supplemental analysis of technical reproducibility, stability at ambient temperature, and accuracy compared with the gold standard at the genus level. All statistical analyses were conducted using R (3.1.2).
Accession number(s).
The sequencing data are available at the European Nucleotide Archive under accession number ERP020918.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge James Gaffney, Greg Humphrey, and Tara Schwartz (Department of Pediatrics, University of California, San Diego) for their technical assistance in this study.
This work was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health and by grants from the National Institutes of Health (1R01CA179243 to N.C., P42ES10349 and R01CA107431 to H.A.), the Center for Individualized Medicine at the Mayo Clinic (to J.C.), and the Howard Hughes Medical Institute and the Sloan Foundation (to R.K.).
R. Knight is chief science officer at Biota Technology, Inc., and a consultant/advisory board member for J&J/Janssen, CommenSe, Inc., and Prometheus Therapeutics & Diagnostics and has received honoraria from the Speakers Bureau of the American Academy of Anti-Aging Medicine and Zurich Insurance Company. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00361-17.
REFERENCES
- 1.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Krogh Pedersen H, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Igor Costea P, Kultima JR, Li J, Jorgensen T, Levenez F, Dore J, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O. 2015. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE. 2010. Obesity and the human microbiome. Curr Opin Gastrenterol 26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 3.Kostic AD, Xavier RJ, Gevers D. 2014. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogtmann E, Goedert JJ. 2016. Epidemiologic studies of the human microbiome and cancer. Br J Cancer 114:237–242. doi: 10.1038/bjc.2015.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. 2015. The microbiome quality control project: baseline study design and future directions. Genome Biol 16:276. doi: 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha R, Chen J, Amir A, Vogtmann E, Shi J, Inman KS, Flores R, Sampson J, Knight R, Chia N. 2016. Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiol Biomarkers Prev 25:407–416. doi: 10.1158/1055-9965.EPI-15-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J, Azpiroz F, Roca J, Guarner F, Manichanh C. 2012. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol 12:158. doi: 10.1186/1471-2180-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll IM, Ringel-Kulka T, Siddle JP, Klaenhammer TR, Ringel Y. 2012. Characterization of the fecal microbiota using high-throughput sequencing reveals a stable microbial community during storage. PLoS One 7:e46953. doi: 10.1371/journal.pone.0046953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo JM, Leong LE, Rogers GB. 2015. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep 5:16350. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominianni C, Wu J, Hayes RB, Ahn J. 2014. Comparison of methods for fecal microbiome biospecimen collection. BMC Microbiol 14:103. doi: 10.1186/1471-2180-14-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores R, Shi J, Yu G, Ma B, Ravel J, Goedert JJ, Sinha R. 2015. Collection media and delayed freezing effects on microbial composition of human stool. Microbiome 3:33. doi: 10.1186/s40168-015-0092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, Izard J, Garrett WS, Chan AT, Huttenhower C. 2014. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A 111:E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorzelak MA, Gill SK, Tasnim N, Ahmadi-Vand Z, Jay M, Gibson DL. 2015. Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS One 10:e0134802. doi: 10.1371/journal.pone.0134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. 2010. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol Lett 307:80–86. doi: 10.1111/j.1574-6968.2010.01965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ, Matherly LH, Majumdar AP, Kato I. 2008. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Methods 72:124–132. doi: 10.1016/j.mimet.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Ott SJ, Musfeldt M, Timmis KN, Hampe J, Wenderoth DF, Schreiber S. 2004. In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn Microbiol Infect Dis 50:237–245. doi: 10.1016/j.diagmicrobio.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Roesch LF, Casella G, Simell O, Krischer J, Wasserfall CH, Schatz D, Atkinson MA, Neu J, Triplett EW. 2009. Influence of fecal sample storage on bacterial community diversity. Open Microbiol J 3:40–46. doi: 10.2174/1874285800903010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedjo DI, Jonkers DM, Savelkoul PH, Masclee AA, van Best N, Pierik MJ, Penders J. 2015. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One 10:e0126685. doi: 10.1371/journal.pone.0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voigt AY, Costea PI, Kultima JR, Li SS, Zeller G, Sunagawa S, Bork P. 2015. Temporal and technical variability of human gut metagenomes. Genome Biol 16:73. doi: 10.1186/s13059-015-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, Li H, Bushman FD. 2010. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol 10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, Knight R. 2016. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems 1:e00021-16. doi: 10.1128/mSystems.00021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogtmann E, Chen J, Amir A, Shi J, Abnet CC, Nelson H, Knight R, Chia N, Sinha R. 2017. Comparison of collection methods for fecal samples in microbiome studies. Am J Epidemiol 185:115–123. doi: 10.1093/aje/kww177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baxter NT, Koumpouras CC, Rogers MA, Ruffin MT IV, Schloss PD. 2016. DNA from fecal immunochemical test can replace stool for detection of colonic lesions using a microbiota-based model. Microbiome 4:59. doi: 10.1186/s40168-016-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahsan H, Chen Y, Parvez F, Argos M, Hussain AI, Momotaj H, Levy D, van Geen A, Howe G, Graziano J. 2006. Health Effects of Arsenic Longitudinal Study (HEALS): description of a multidisciplinary epidemiologic investigation. J Expo Sci Environ Epidemiol 16:191–205. doi: 10.1038/sj.jea.7500449. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD, Li H. 2012. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28:2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.