ABSTRACT
Biotechnological production of cis,cis-muconic acid from renewable feedstocks is an environmentally sustainable alternative to conventional, petroleum-based methods. Even though a heterologous production pathway for cis,cis-muconic acid has already been established in the host organism Saccharomyces cerevisiae, the generation of industrially relevant amounts of cis,cis-muconic acid is hampered by the low activity of the bacterial protocatechuic acid (PCA) decarboxylase AroY isomeric subunit Ciso (AroY-Ciso), leading to secretion of large amounts of the intermediate PCA into the medium. In the present study, we show that the activity of AroY-Ciso in S. cerevisiae strongly depends on the strain background. We could demonstrate that the strain dependency is caused by the presence or absence of an intact genomic copy of PAD1, which encodes a mitochondrial enzyme responsible for the biosynthesis of a prenylated form of the cofactor flavin mononucleotide (prFMN). The inactivity of AroY-Ciso in strain CEN.PK2-1 could be overcome by plasmid-borne expression of Pad1 or its bacterial homologue AroY subunit B (AroY-B). Our data reveal that the two enzymes perform the same function in decarboxylation of PCA by AroY-Ciso, although coexpression of Pad1 led to higher decarboxylase activity. Conversely, AroY-B can replace Pad1 in its function in decarboxylation of phenylacrylic acids by ferulic acid decarboxylase Fdc1. Targeting of the majority of AroY-B to mitochondria by fusion to a heterologous mitochondrial targeting signal did not improve decarboxylase activity of AroY-Ciso, suggesting that mitochondrial localization has no major impact on cofactor biosynthesis.
IMPORTANCE In Saccharomyces cerevisiae, the decarboxylation of protocatechuic acid (PCA) to catechol is the bottleneck reaction in the heterologous biosynthetic pathway for production of cis,cis-muconic acid, a valuable precursor for the production of bulk chemicals. In our work, we demonstrate the importance of the strain background for the activity of a bacterial PCA decarboxylase in S. cerevisiae. Inactivity of the decarboxylase is due to a nonsense mutation in a gene encoding a mitochondrial enzyme involved in the biosynthesis of a cofactor required for decarboxylase function. Our study reveals functional interchangeability of Pad1 and a bacterial homologue, irrespective of their intracellular localization. Our results open up new possibilities to improve muconic acid production by engineering cofactor supply. Furthermore, the results have important implications for the choice of the production strain.
KEYWORDS: biotechnology, muconic acid, phenylacrylic acid decarboxylase, prenylated flavin mononucleotide, protocatechuic acid decarboxylase, Saccharomyces cerevisiae, styrene
INTRODUCTION
The unsaturated dicarboxylic acid cis,cis-muconic acid is a valuable precursor for the production of bulk chemicals such as adipic acid, terephthalic acid, and trimellitic acid (1), which have wide uses in chemical industry. For example, adipic acid is used as a building block to produce nylon, polyurethanes, as a reactant to form plasticizers, lubricant components, polyester polyols, and food ingredients. In industrial processes, adipic acid is mainly produced from benzene, which is reduced to cyclohexane, subsequently oxidized to cyclohexanone and cyclohexanol, and further converted to adipic acid in the presence of HNO3 (2). The release of the greenhouse gas N2O as a byproduct and the dependence on nonrenewable fossil fuels cause major environmental concerns. Therefore, biotechnological processes for the production of cis,cis-muconic acid from renewable feedstocks are regarded as attractive alternatives to conventional methods.
A heterologous biosynthetic pathway for production of cis,cis-muconic acid from glucose, initially established in Escherichia coli (3, 4), consists of three enzymatic steps. First, an intermediate of the shikimic acid pathway, 3-dehydroshikimate (3-DHS), is converted to protocatechuic acid (PCA) by 3-dehydroshikimate dehydratase AroZ. In the subsequent step, PCA is decarboxylated to catechol by PCA decarboxylase AroY. Finally, the ring-opening reaction of catechol is catalyzed by catechol dioxygenase CatA, and the product cis,cis-muconic acid is formed.
Due to its acid tolerance and resistance to microbial contaminations, Saccharomyces cerevisiae makes an ideal host organism for production of cis,cis-muconic acid, and thus the pathway has been successfully implemented in S. cerevisiae by several groups (5–8). Several approaches have been applied to improve the yield of the pathway. These approaches include deletion or inactivation of the E-domain of the pentafunctional yeast enzyme Aro1 responsible for the shikimate 5-dehydrogenase function of the enzyme (5, 7), leading to accumulation of the precursor 3-DHS. Overexpression of a feedback-resistant mutant of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase ARO4 improved entry of precursors into the shikimic acid pathway (6). Increasing the flux into the nonoxidative pentose phosphate pathway by deletion of glucose-6-phosphate dehydrogenase ZWF1 and overexpression of transketolase TKL1 (6, 8) resulted in elevated levels of shikimic acid pathway precursor erythrose-4-phosphate, allowing for further increases in muconic acid production. Even though these modifications permit the production of high titers, the decarboxylation of PCA to catechol in the heterologous pathway remains a major bottleneck.
The bacterial decarboxylases utilized for catalysis of this step belong to a family of bacterial nonoxidative, reversible hydroxyarylic acid decarboxylases/phenol carboxylases (9). Members of this family are usually encoded by genes organized in a cluster consisting of B, C, and D genes. The D gene is not present in the cluster in all microorganisms. In Klebsiella pneumoniae, two homologues of the C gene (aroY subunit C gene [aroY-C]) exist. One of the homologues (accession no. AAY57855) is organized in a BCD cluster and has been demonstrated to be highly oxygen-sensitive (5), whereas the other homologue (isomeric aroY-C [aroY-Ciso], AB479384) has been successfully utilized for cis,cis-muconic acid production in S. cerevisiae (5–8) and E. coli (3, 4, 10, 11). Contradictory results exist about the necessity to coexpress B (aroY-B; AAY57854) and D (aroY-D; AAY57856) genes to obtain decarboxylase activity in S. cerevisiae. Although in strain BY4741, AroY-Ciso and its close homologue ECL_01944 from Enterobacter cloacae required no expression of additional subunits for their activity (6, 7), AroY-Ciso and a homologue from Acetobacter tropicalis were inactive in the CEN.PK2 background (5, 12), and activity could only be achieved upon coexpression of AroY-B and -D (5).
These contradictory results about the activity of AroY-Ciso homologues in different genetic backgrounds prompted us to investigate the function of AroY-B and -D. We noticed that AroY-B is closely related to E. coli UbiX and S. cerevisiae Pad1, which have been demonstrated to be responsible for the synthesis of a prenylated form of flavin mononucleotide (prFMN) required for the function of the decarboxylases UbiD and Fdc1, respectively (13). In S. cerevisiae, Pad1 is localized in mitochondria, whereas Fdc1 is found in the cytosol (14). Both genes have been shown to be involved in detoxification of phenylacrylic acids (15, 16). Recent genetic analyses have revealed that PAD1 and FDC1 genes are interrupted by nonsense mutations in many industrial yeast strains (17). Our findings demonstrate that the functionality of AroY-Ciso or Fdc1 is dependent on the presence of an intact copy of PAD1 or coexpression of aroY-B, thereby providing evidence that AroY-B is functionally equivalent to Pad1.
RESULTS
Activity of AroY-Ciso in strains of S. cerevisiae is dependent on the presence of Pad1.
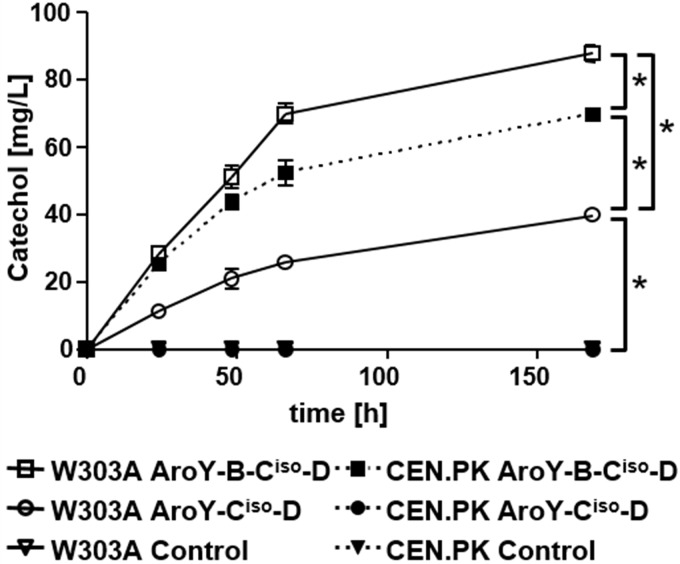
To investigate whether the absence of intact PAD1 in some strains is responsible for the lack of activity of AroY-Ciso, we compared the activity of the protein in two strains. While strain W303A contains an intact copy of PAD1 in its genome, the PAD1 open reading frame (ORF) in CEN.PK2-1C is interrupted by a nonsense mutation at nucleotide 294 (T294A) compared to the reference strain S288C (ORF YDR538W [Saccharomyces genome database, www.yeastgenome.org/]). We expressed AroY-Ciso and D in the presence or absence of subunit B from a multicopy vector in both strains and monitored the conversion of PCA to catechol for a period of 168 h (Fig. 1). Surprisingly, in strain W303A, AroY-Ciso catalyzed the conversion of PCA to catechol in the absence of AroY-B, reaching levels of 40 mg/liter after 168 h. In contrast, in CEN.PK2-1C, no formation of catechol was detectable. Overexpression of AroY-B enhanced the activity of AroY-Ciso in both strains, albeit not to the same levels (88 mg/liter in W303A and 70 mg/liter in CEN.PK2-1C). Thus, we can conclude that the inactivity of AroY-Ciso in CEN.PK2-1C is most likely caused by the absence of active Pad1 and that overexpression of AroY-B can complement this absence in strain CEN.PK2-1. Interestingly, even upon coexpression of AroY-B, the level of cofactor still seems to be limiting for enzymatic activity of AroY-Ciso in CEN.PK2-1C, since the level in W303A is higher.
FIG 1.
Activity of AroY-Ciso in S. cerevisiae strains CEN.PK2-1C and W303A. Cells transformed with expression plasmids encoding AroY-Ciso and AroY-D, or, in addition, AroY-B were cultivated in SC medium (2% glucose), which was supplemented with 5 mM protocatechuic acid (PCA). Catechol formation was measured by HPLC analysis of the culture supernatant after 0, 24, 48, 66, and 168 h. Error bars representing the standard deviations of three independent biological replicates are depicted in the graph, but they are masked by the size of the symbols at some time points. CEN.PK2-1C cells expressing AroY-Ciso and AroY-D, as well as cells transformed with empty plasmid (Control), were not able to convert protocatechuic acid to catechol, and thus the corresponding curves overlap with the x axis. An unpaired Student t test was used to determine the statistical significance of the difference between samples at 168 h, as indicated by connecting lines. Significant differences (P < 0.05) are indicated by asterisks.
Pad1 can replace AroY-B in its function in decarboxylation of protocatechuic acid.
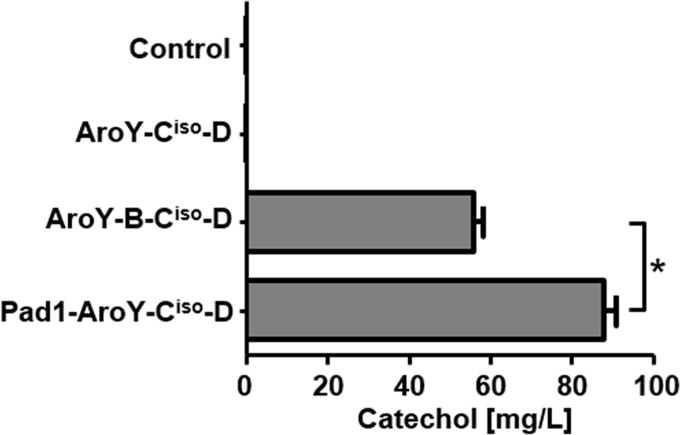
A sequence comparison of AroY-B and Pad1 using Clustal Omega (18) reveals 53% identity between the two proteins (see Fig. S1 in the supplemental material). To further verify our hypothesis that Pad1 and AroY-B perform the same function, providing prFMN as a cofactor for AroY-Ciso, we exchanged aroY-B for PAD1 in our multicopy vector and compared the influence of both enzymes on the activity of AroY-Ciso in CEN.PK2-1C (Fig. 2). Although upon coexpression of aroY-Ciso with aroY-B and -D, catechol at 56 mg/liter could be produced from PCA, coexpression of PAD1 instead of aroY-B resulted in a titer of 88 mg/liter catechol after 144 h. Hence, we could demonstrate that Pad1 not only can replace AroY-B in its function but also proves to be even more efficient in stimulating the decarboxylase activity of AroY-Ciso.
FIG 2.
Functional interchangeability of AroY-B and Pad1. Yeast cells (strain CEN.PK2-1C) were transformed with expression vectors encoding AroY-Ciso and -D alone or in combination with AroY-B or Pad1. As a control, empty plasmid was transformed. Cells were subsequently grown in SC medium (2% glucose), which was supplemented with 5 mM PCA. The production of catechol was measured by HPLC after 144 h. Error bars represent the standard deviations of three independent biological replicates. An unpaired Student t test was used to determine the statistical significance of the difference between samples AroY-B-Ciso-D and Pad1-AroY-Ciso-D, as indicated by connecting lines. A significant difference (P < 0.05) is indicated by an asterisk.
Influence of intracellular localization of Pad1 and AroY-B on their activity.
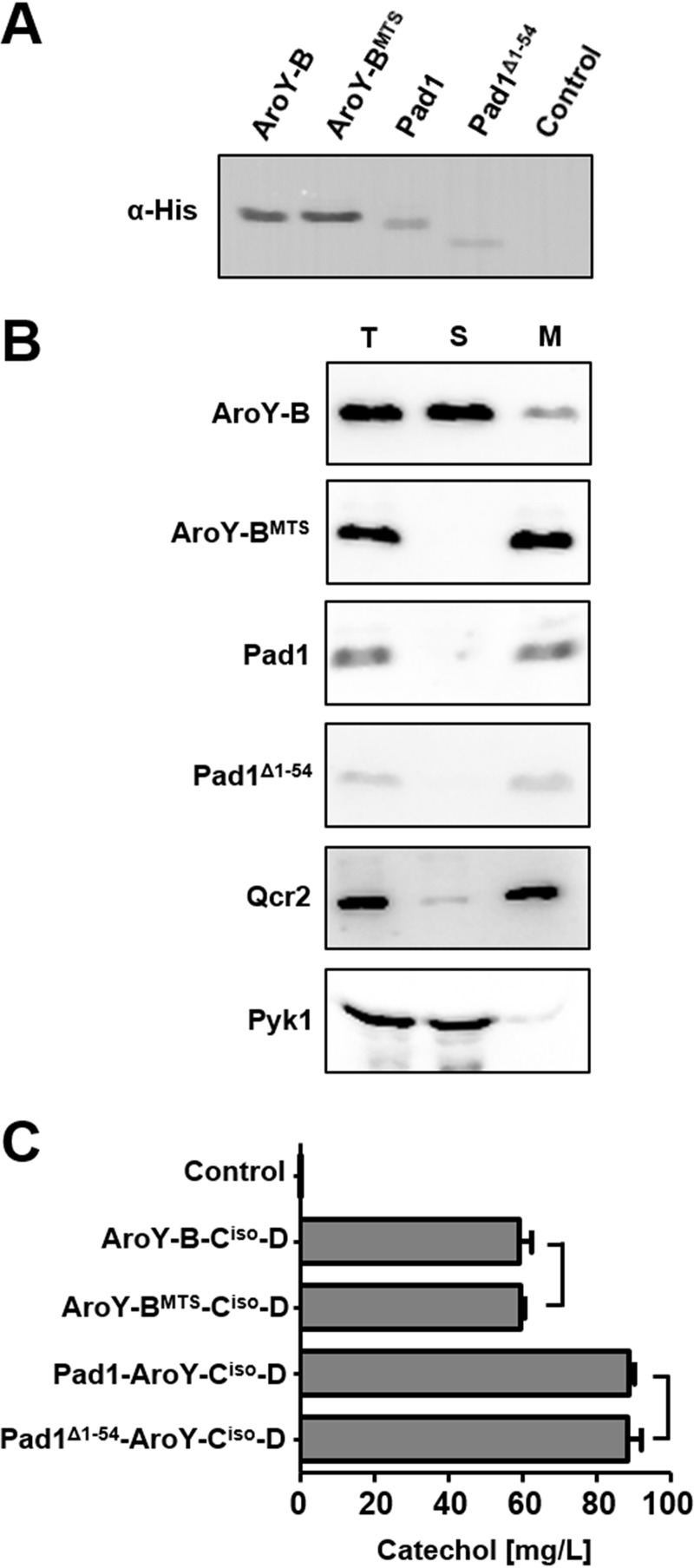
Pad1 was shown to be localized to mitochondria in yeast (14), whereas the only yeast enzyme known to depend on its function, Fdc1, is localized in the cytosol. Likewise, AroY-B, due to its bacterial origin, is most likely localized in the cytosol when expressed in yeast. Thus, prFMN generated by Pad1 has to be transported from mitochondria to the cytosol to fulfill its function. To test whether the intracellular localization of Pad1 and AroY-B has an impact on their ability to stimulate the activity of AroY-C, we attempted to relocate Pad1 to the cytosol and, vice versa, AroY-B to mitochondria. Pad1 is predicted to possess an N-terminal mitochondrial targeting signal (MTS) of 65 amino acid residues length (MitoProt [https://ihg.gsf.de/ihg/mitoprot.html]) but attempts to relocate Pad1 to the cytosol by removing 58 or 57 amino acid residues of its targeting signal resulted in an entire loss of activity (19). A sequence alignment of Pad1, AroY-B, and UbiX reveals that Pad1 possesses an N-terminal extension of 54 amino acid residues compared to its prokaryotic homologues (see Fig. S1 in the supplemental material). Thus, we presumed that the whole conserved part might be essential to retain activity and generated constructs for expression of Pad1 with an N-terminal deletion of 54 amino acids (Pad1Δ1−54). For targeting of AroY-B to mitochondria, the cleavable MTS of Neurospora crassa F0 ATP synthase subunit 9 (20) was added to the N terminus (AroY-BMTS). To study the expression of the newly generated variants in comparison to their unmodified counterparts, the proteins were provided with a C-terminal His tag and analyzed by SDS-PAGE and immunoblotting (Fig. 3A). Even though 69 amino acid residues were added to the N terminus of the protein, AroY-BMTS and AroY-B were detected at the same height by immunoblot analysis, indicating that the MTS of the protein is cleaved off after import into mitochondria. Interestingly, Pad1Δ1−54 migrated faster on the SDS-gel than Pad1, suggesting that either the N-terminal MTS of Pad1 is even shorter than 54 amino acid residues or that Pad1 does not contain a canonical, cleavable N-terminal MTS.
FIG 3.
Influence of intracellular localization of Pad1 and AroY subunit B on their activity. (A) Immunoblot analysis of the expression of Pad1, an N-terminal truncation of Pad1 (Pad1Δ1−54), AroY-B, and AroY-B fused to the mitochondrial targeting signal of N. crassa ATPase subunit 9 (AroY-BMTS) in CEN.PK2-1C. Cells were grown in SC medium containing 2% glucose prior to the preparation of cell extracts. All proteins were provided with a C-terminal His tag. Cell extracts were subjected to SDS-PAGE and immunoblotting, and His-tagged proteins were detected with antibodies against the His tag. (B) Analysis of intracellular localization. Cell extracts containing Pad1, Pad1Δ1−54, AroY-B, or AroY-BMTS with a C-terminal His tag (T) were fractionated into a cytosolic (S) or crude mitochondrial fraction (M) by differential centrifugation and subsequently analyzed by SDS-PAGE and immunoblotting. Enrichment of soluble, cytosolic proteins in fraction S and mitochondrial proteins in fraction M was controlled using antibodies against cytosolic Pyk1 (pyruvate kinase 1) or the inner mitochondrial membrane protein subunit 2 of ubiquinol cytochrome c reductase (Complex III; Qcr2), respectively. (C) Bioconversion of PCA to catechol in yeast cells coexpressing AroY-Ciso and -D either with Pad1, Pad1Δ1−54, Aro Y-B, and AroY-BMTS. Cultures were grown in SC medium (2% glucose) supplemented with 5 mM PCA. The conversion of PCA to catechol was determined after 144 h of cultivation by HPLC analysis of the culture supernatant. Error bars represent the standard deviations of three independent biological replicates. The statistical difference between the samples indicated by connecting lines was determined using an unpaired Student t test. Differences were shown to be nonsignificant (P > 0.05).
To answer this question, the intracellular localization of the proteins was determined by cell fractionation and subsequent immunoblotting (Fig. 3B). As expected, AroY-B could be localized mainly in the soluble, cytosolic fraction. A minor amount of the protein was detected in the mitochondrion-enriched fraction. Pad1 was found predominantly in the mitochondrion-enriched fraction. AroY-BMTS was successfully targeted to mitochondria and was detectable mainly in the mitochondrion-enriched fraction. Surprisingly, even though the first 54 amino acid residues of Pad1 were deleted, the protein was still detected in the mitochondrial fraction, indicating that the targeting is mediated by a noncanonical signal.
In the next step, we tested if the altered localization of AroY-B leads to a change in activity. For this aim, the mitochondrial and cytosolic variants were coexpressed with AroY-Ciso and AroY-D, and the impact on decarboxylation activity of AroY-Ciso was determined by measuring conversion of PCA to catechol in vivo in a feeding experiment (Fig. 3C). Surprisingly, upon the coexpression of AroY-B or AroY-BMTS with AroY-Ciso and -D, similar levels of catechol (59 and 60 mg/liter, respectively) were generated after prolonged incubation with 5 mM PCA in glucose containing medium. Deletion of the first 54 amino acid residues of Pad1 did not alter its activity. Coexpression of both Pad1 and Pad1Δ1−54 led to the production of 89 mg/liter catechol. Thus, we could show that an enrichment of AroY-B inside mitochondria did not improve its ability to synthesize the cofactor required for activity of AroY-Ciso. Since the production of catechol was higher upon the coexpression of Pad1, it appears that the cofactor availability is limiting the activity of AroY-Ciso.
Pad1 and AroY-B support the function of Fdc1 in detoxification of trans-cinnamic acid.
In natural environments, Pad1 provides prFMN to allow for Fdc1 function in detoxification of phenylacrylic acids present in plant cell walls, such as ferulic, coumaric, or cinnamic acid, to more volatile compounds (15). In biotechnology, this reaction is relevant for decarboxylation of cinnamic acid to styrene, a building block required in plastics production (21, 22). To test whether AroY-B can replace Pad1 in its function in providing the cofactor required for decarboxylase activity of Fdc1, we analyzed the impact of the coexpression of both enzymes on the detoxification of trans-cinnamic acid. For this aim, yeast cells of the strain CEN.PK2-1C were transformed with expression vectors encoding Fdc1 alone or Fdc1, together with Pad1 or AroY-B. AroY-Ciso was coexpressed with AroY-B and -D to determine whether AroY-Ciso is able to act as a decarboxylase on trans-cinnamic acid as well (Fig. 4A). Transformed cells were spotted onto plates containing increasing amounts of trans-cinnamic acid. After incubation for 4 days, pronounced differences in growth became visible. On plates containing 100 mg/liter trans-cinnamic acid, the coexpression of Pad1 and Fdc1, as well as of AroY-B and Fdc1, strongly supported growth, whereas the expression of Fdc1 alone resulted only in weak growth, comparable to that observed for the empty vector control. Likewise, the expression of AroY-Ciso, together with AroY-B and -D, resulted in only weak growth. On plates containing 200 mg/liter trans-cinnamic acid, the growth of cells expressing Fdc1 alone or Aro-Y-Ciso, -B, and -D and of cells transformed with empty vector was almost completely suppressed, whereas the growth of cells expressing AroY-B or Pad1, together with Fdc1, was still possible. Thus, AroY-B is functionally equivalent to Pad1. The combination of Fdc1 with AroY-B provided the cells with even more resistance against trans-cinnamic acid than did the combination with Pad1. This is surprising, since in our previous experiments, Pad1 seemed to be more effective in converting FMN to its prenylated form, even though measured only indirectly (Fig. 2). Therefore, we wondered whether the overexpression of Pad1 causes a general impairment in growth. Since a balanced flavin homeostasis inside mitochondria is important for maintaining respiratory function, we speculated that a stronger depletion of the FMN pool by prenylation would impair mitochondrial function, especially when cells are grown on nonfermentable carbon sources such as ethanol or glycerol. To test this hypothesis, we analyzed growth of cells transformed with plasmids overexpressing Pad1, AroY-B, or empty vector on plates containing 2% glucose or containing 2% ethanol and 2% glycerol (Fig. 4B). When grown on glucose as a carbon source, transformants expressing Pad1 exhibited a slight growth retardation compared to cells transformed with empty plasmid or AroY-B, resulting in a smaller colony size. On glycerol and ethanol containing agar, the growth difference became even more pronounced. Growth was most severely impaired when cells overexpressed Pad1. Hence, a general impairment in growth, and not a lower activity of Pad1, could at least partially account for the slower growth of cells overexpressing Pad1 together with Fdc1 in the detoxification experiments with trans-cinnamic acid.
FIG 4.
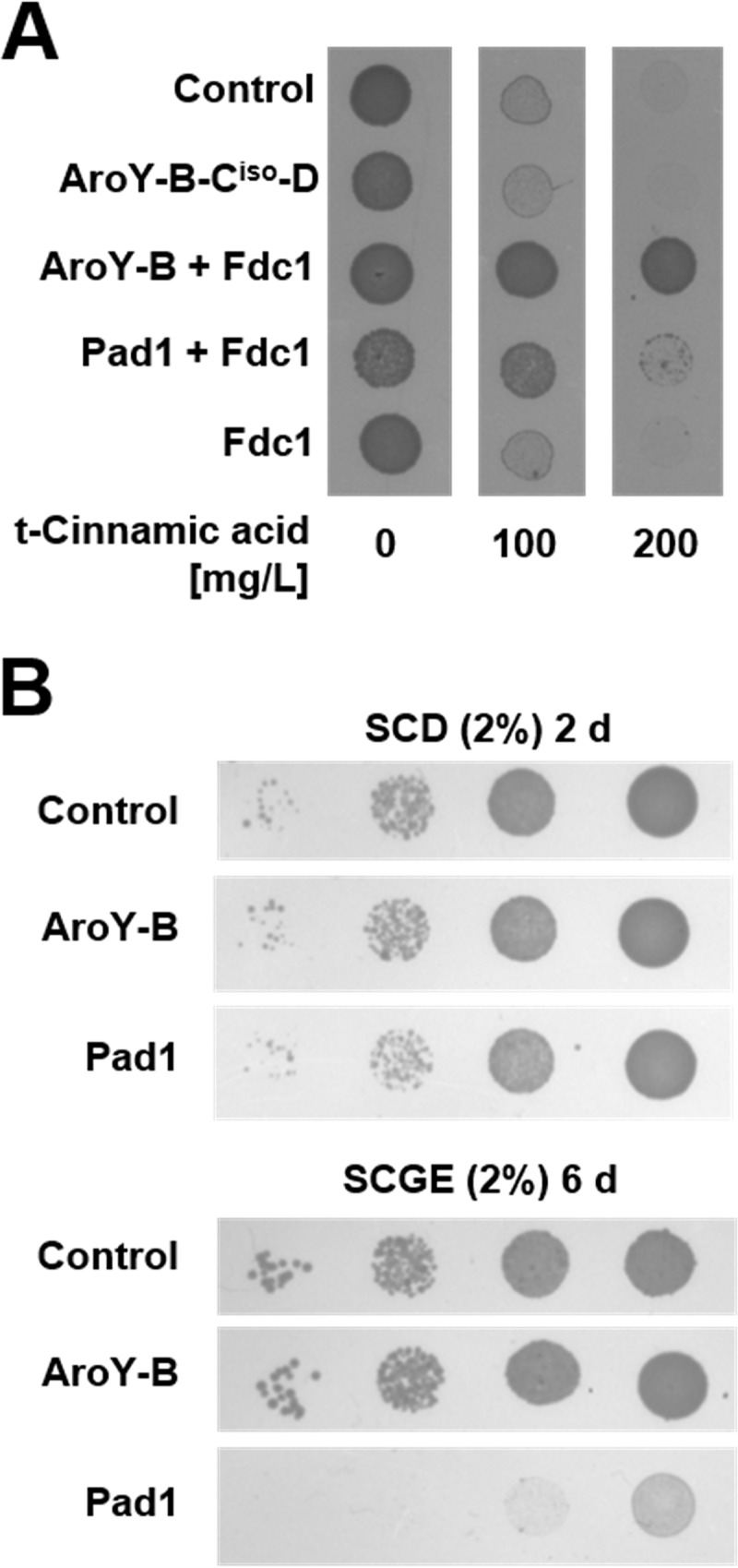
Replacement of Pad1 by AroY-B in detoxification of trans-cinnamic acid by Fdc1. (A) CEN.PK2-1C cells expressing Fdc1 alone, Pad1, or AroY-B together with Fdc1 or AroY-Ciso together with AroY-B and -D were spotted onto agar plates (SC, 2% glucose) supplemented with various concentrations of trans-cinnamic acid, as indicated. Cells transformed with empty vector were included as a control. The results were documented after 4 days of incubation at 30°C. (B) Growth phenotype of Pad1-expressing cells on different carbon sources. Cells expressing Pad1 or AroY-B with a C-terminal His tag were spotted onto agar plates with SC medium containing 2% glucose (SCD) or containing 2% glycerol and 2% ethanol (SCGE). Plates were incubated for 2 days (SCD plates) or 6 days (SCGE plates) at 30°C prior to documentation.
AroY-D is dispensable for activity of AroY-Ciso.
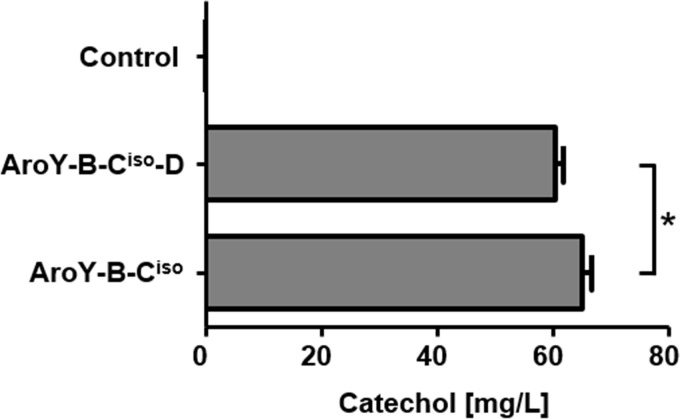
In S. cerevisiae, AroY-Ciso was so far either expressed alone (6) or in combination with AroY-B and -D (5, 8), but the contribution of AroY-D expression has never been studied systematically. In E. coli, it was demonstrated that AroY-B stimulates the activity of AroY-Ciso, but the effect of additional coexpression of Aro-D was dependent on plasmid design, and no conclusive statement could be made about its functional relevance (23). Similar contradictory results have been obtained with the AroY-D homologue EcdD from E. cloacae (24). In the results for a feeding experiment (Fig. 1), we already demonstrated that AroY-D alone is not sufficient to improve AroY-Ciso activity, since there is an absolute requirement for AroY-B (or Pad1). In our next experiment, we investigated whether the additional coexpression of AroY-D has any stimulatory effect on AroY-Ciso in the presence of AroY-B (Fig. 5). We determined that cells expressing AroY-Ciso with or without AroY-D showed a similar capability to convert PCA to catechol, which was even slightly improved in the absence of AroY-D (60 mg/liter with AroY-D versus 65 mg/liter without AroY-D after 144 h). Thus, AroY-D is dispensable for activity of AroY-C in S. cerevisiae.
FIG 5.
Dispensability of AroY-D for decarboxylation of protocatechuic acid. Yeast cells (strain CEN.PK2-1C) expressing AroY-Ciso and AroY-B with or without AroY-D were cultivated in SC medium (2% glucose) supplemented with 5 mM PCA. The conversion of PCA to catechol was determined by HPLC analysis of the culture supernatant after 144 h. Error bars represent the standard deviations of three independent biological replicates. An unpaired Student t test was used to determine the statistical significance of the difference between samples AroY-B-Ciso-D and AroY-B-Ciso, as indicated by connecting lines. The significant difference (P < 0.05) is indicated by an asterisk.
DISCUSSION
In S. cerevisiae, PAD1 and FDC1 encode two enzymes responsible for the detoxification of antimicrobial phenylacrylic acids present in plant tissue, such as cinnamic, ferulic, or coumaric acid (16). The product formed by decarboxylation of ferulic acid, 4-vinylguaiacol, is a compound with smokelike characteristics that is considered an off-flavor in most beers and wines. For this reason, many loss-of-function mutations were acquired in PAD1 or FDC1 in strains used for wine or beer production during their domestication (17, 25). In a variety of other yeast species (e.g., Kluyveromyces marxianus or Yarrowia lipolytica), no Pad1 homologues can be found and, consistently, these species are not able to decarboxylate phenylacrylic acids (26). However, in most laboratory strains of S. cerevisiae, intact copies of FDC1 and PAD1 are present (25). In this respect, laboratory strain CEN.PK2-1C is an exception, as PAD1 is interrupted by a nonsense mutation, leading to the production of a nonfunctional protein. Even though an insertion is present in FDC1 of CEN.PK2-1C, which is defined as an intron in the Saccharomyces genome database (www.yeastgenome.org/), the gene seems to encode an active protein (19). In biotechnology, both genes have gained attention for their use in a biosynthetic pathway for the production of styrene. In our study, we demonstrate that the lack of active Pad1 in CEN.PK2-1C is not only relevant for the decarboxylation of phenylacrylic acids by Fdc1 but also for the activity of a bacterial, heterologous enzyme, the PCA decarboxylase AroY-Ciso from K. pneumoniae. We found that AroY-Ciso could only be actively expressed in a strain with an intact copy of PAD1, W303A, whereas the protein is inactive in CEN.PK2-1C (Fig. 1). The lack of intact PAD1 in CEN.PK2-1C could be complemented by expression of an intact copy of PAD1 from S288C or of the homologous gene aroY-B from K. pneumoniae (Fig. 1 and 2), revealing that both Pad1 and AroY-B perform the same function. Vice versa, the functional identity of the two proteins was demonstrated in an experiment were the ability of Fdc1 to detoxify trans-cinnamic acid was tested (Fig. 4). Both Pad1 and AroY-B could compensate the lack of functional Pad1 in CEN.PK2-1C. In the light of our results, previous, inconsistent reports about the activity of AroY-Ciso and other bacterial homologues in different strain backgrounds (5–7, 12) become explainable. Furthermore, our data explain why an active pathway could not be implemented in another industrially relevant yeast strain, Kluyveromyces lactis (12), since its genome does not contain a PAD1 gene. Thus, it is important to consider the influence of the strain background when a production strain is chosen for integration of the pathway. The function of aroY-D remains enigmatic. In our experiments, coexpression of the protein provided no benefit for the activity of AroY-Ciso (Fig. 5), but even a slight disadvantage. The gene encodes a protein of only 78 amino acid residues, and in bacteria no conclusive results about is function could be obtained. In E. coli, depending on the experimental design, coexpression of AroY-D had inhibitory effects on the production of catechol as well (23).
Recently, it has been demonstrated that Fdc1 and its bacterial homologue UbiD perform the actual decarboxylase function, whereas Pad1 and its bacterial homologue UbiX are responsible for the prenylation of flavin mononucleotide (FMN), leading to the generation of a cofactor required for decarboxylase function (13, 27, 28). In contrast to Fdc1/Pad1, UbiD/UbiX is involved in the biosynthesis of ubiquinone in E. coli. Synthesis of FMN from riboflavin is catalyzed by a flavokinase, which in S. cerevisiae is encoded by the gene FMN1 (29). In subcellular fractionations, the enzyme has been found in mitochondrial and microsomal fractions, presumably providing a supply of the cofactor both in mitochondria and the cytosol. Furthermore, FMN can be exported from mitochondria by the transporter Flx1 (30). The reason for mitochondrial localization of Pad1 in S. cerevisiae remains a matter of speculation. Our results presented in Fig. 2 suggest that the mitochondrial localization might provide an advantage for the activity of the protein, since mitochondrial Pad1 seems to produce more cofactor for the decarboxylation reaction than its bacterial homologue AroY-B, even though its expression is lower (Fig. 3A). One could speculate that a higher concentration of FMN inside mitochondria could allow for a better efficiency of the reaction, or that mitochondria provide a better folding environment for Pad1 and potentially also for AroY-B. Unfortunately, it was not possible to create a functional, cytoplasmic form of Pad1 (Fig. 3B and C). More experiments are necessary to identify the exact targeting determinants of the protein. However, when AroY-B is targeted to mitochondria by fusion to a mitochondrial targeting signal (Fig. 3B), the function of AroY-Ciso is not improved (Fig. 3C), arguing against an influence of a higher FMN concentration or of the mitochondrial folding environment, which is more related to the bacterial one. Even though the presence of small amounts of the cytoplasmic control Pyk1 in the mitochondrial fraction indicates a minor carryover of soluble proteins to the mitochondrial fraction, it cannot be entirely excluded that the minor amount of AroY-B found in the mitochondrial fraction might be responsible for the activity of AroY-Ciso.
Interestingly, when mitochondrial functionality is assessed by growth on nonfermentable carbon sources, overexpression of Pad1 causes an impairment in growth, whereas AroY-B does not (Fig. 4B). This phenomenon could be explained by the apparently different activities of the two proteins (Fig. 2). It has to be considered that the activity of the two proteins was determined indirectly by their influence on the activity of AroY-Ciso. Production of prFMN by Pad1 might be much higher, but this might not become obvious when the cofactor demand of AroY-Ciso is already saturated. The high activity might lead to a depletion of FMN and, consequently, of flavin adenine dinucleotide (FAD) and thus impair respiratory function. However, it is possible that Pad1 performs a second function inside mitochondria or that the growth deficiency is caused by an aggregation of Pad1 due to its overexpression.
In our work, we have investigated a new example of a decarboxylase belonging to the UbiD family which depends on the presence of a functional FMN prenyltransferase for its activity. Sequence comparison of AroY-Ciso to other enzymes of this family with an already described prFMN binding capability, Fdc1 from S. cerevisiae and Aspergillus niger and UbiD from E. coli (13, 31) using Clustal Omega (18) reveals a relatively low sequence conservation between AroY-Ciso and these enzymes (28% identity to E. coli UbiD, 25% identity to A. niger Fdc1, and 23% to S. cerevisiae Fdc1) (see Fig. S2 in the supplemental material). However, important residues involved in interaction with the cofactor and its maturation are conserved. Based on the presence of this conserved residues and its organization in the BCD cluster, it is likely that the other homologue from K. pneumoniae, AroY-C, requires prFMN for its activity as well. Further studies have to reveal if other, unrelated enzymes with the ability to bind prFMN exist. Summarizing, our data provide new insights into the function of AroY-B and disclose new opportunities for improvement of the decarboxylation reaction in cis,cis-muconic acid production.
MATERIALS AND METHODS
Yeast strains and growth media.
All experiments were performed in S. cerevisiae strains CEN.PK2-1C (MATa leu2-3,112 ura3-52 trp1-289 his3-1 MAL2-8c SUC2; Euroscarf) or W303A (MATa ura3-1 trp1-Δ2 leu2-3,112 his3-11 ade2-1 can1-100). Competent cells were prepared and transformed as described previously (32). Transformed cells were cultivated at 30°C on a rotary shaker (180 rpm) in synthetic complete (SC) medium supplemented with 2% (wt/vol) glucose, whereby amino acids and nucleobases were added as required for the selection of the respective plasmids.
Plasmid construction.
Plasmids were generated by homologous recombination in the S. cerevisiae strain CEN.PK2-1C as described previously (33). Relevant information about plasmids, including promoters and terminators, are given in Table 1. The sequences of the primers used to generate plasmids are listed in Table 2.
TABLE 1.
Plasmids used in this study
| Plasmid | Regulatory elements, markers, and/or genes | GenBank accession no. | Source or reference(s) |
|---|---|---|---|
| p426-MET25 | 2μ; MET25p; CYC1t; URA3 | 34, 38 | |
| p426-aroY-B-C-D | MET25p; aroY subunit B; CYC1t; PGK1p; aroY subunit C isoform; PGK1t; TPI1p; aroY subunit D; TAL1t; all subunits from K. pneumoniae codon optimized (DNA2.0 optimized) | AAY57854, AB479384, AAY57856 | This study |
| p426-aroY-PAD1-C-D | MET25p; PAD1 from strain S288C; CYC1t; PGK1p; aroY subunit C isoform; PGK1t; TPI1p; aroY subunit D; TAL1t | NP_010827, AB479384, AAY57856 | This study |
| p426-aroY-BMTS-C-D | MET25p; aroY subunit B with mitochondrial targeting signal of ATPase subunit 9 (amino acids 1 to 69); CYC1t; PGK1p; aroY subunit C isoform; PGK1t; TPI1p; aroY subunit D; TAL1t | CAA24230, AAY57854, AB479384, AAY57856 | This study |
| p426-aroY-PAD1Δ1–54-C-D | MET25p; PAD1 with an N-terminal deletion of amino acid residues 1 to 54; CYC1t; PGK1p; aroY subunit C isoform; PGK1t; TPI1p; aroY subunit D; TAL1t | NP_010827, AB479384, AAY57856 | This study |
| p426-AroY-B-C | MET25p; aroY subunit B; CYC1t; PGK1p; aroY subunit C isoform; PGK1t | AAY57854, AB479384 | This study |
| p426-FDC1 | TPI1p; FDC1 from strain S288C; TPI1t | NP_010828 | This study |
| p426-aroY-B-FDC1 | MET25p; aroY subunit B; CYC1t; TPI1p; FDC1; TPI1t | AAY57854, NP_010828 | This study |
| p426-PAD1-FDC1 | MET25p; PAD1; CYC1t; TPI1p; FDC1; TPI1t | NP_010827, NP_010828 | This study |
| p426-PAD1-His | MET25p; PAD1; CYC1t | NP_010827 | This study |
| p426-PAD1Δ1–54-His | MET25p; PAD1 with an N-terminal deletion of amino acid residues 1 to 54; CYC1t | NP_010827 | This study |
| p426-aroY-B-His | MET25p; aroY subunit B; CYC1t | AAY57854 | This study |
| p426-aroY-BMTS-His | MET25p; aroY subunit B with mitochondrial targeting signal of ATPase subunit 9; CYC1t | CAA24230, AAY57854 | This study |
TABLE 2.
Primers used in this study
| Plasmid | Primer sequence (5′-3′) | Binding sites and overhang |
|---|---|---|
| p426-aroY-B-C-Da | AATTCTATTACCCCCATCCATACTCTAGAAATGAAACTGATAATCGGGATGAC | aroY-B (ovMET25p) f |
| AATGTAAGCGTGACATAACTAATTACATGATTATTCGATTTCCTGAGCGAATTG | aroY-B (ovCYC1t) r | |
| TCATGTAATTAGTTATGTCACGCTTAC | CYC1t f | |
| GTTTTTTCAGTTTTGTTCTTTTTGCAAACAGTCGACGGTACCGGCCGCAAATTAAAG | CYC1t (ovPGK1p) r | |
| TGTTTGCAAAAAGAACAAAACTG | PGK1p f | |
| GCGTAATACGACTCACTATAGGGCGAATTGGGATCCGACGTTGATTTAAGGTGGTTC | tTAL1t (ovp426) r | |
| p426-PAD1-aroY-C-D | AATTCTATTACCCCCATCCATACTCTAGAAATGCTCCTATTTCCAAGAAGAAC | PAD1 (ovMET25p) f |
| AATGTAAGCGTGACATAACTAATTACATGATTACTTGCTTTTTATTCCTTCCCAAC | PAD1 (ovCYC1t) r | |
| See above | CYC1t f | |
| See above | tTAL1t (ovp426) r | |
| p426-aroY-BMTS-C-D | AATTCTATTACCCCCATCCATACTCTAGAAATGGCTAGTACCAGAGTTTTAG | MTS (ovMET25p) f |
| GGAGGAGTAAGCTCTCTTTTGGAAGGCTTG | MTS (ovAroY-B) r | |
| AAGCCTTCCAAAAGAGAGCTTACTCCTCCAAACTGATAATCGGGATGACC | aroY-B (ovMTS) f | |
| See above | aroY-B (ovCYC1t) r | |
| See above | CYC1t f | |
| See above | tTAL1t (ovp426) r | |
| p426-aroY-PAD1Δ1–54-C-D | AATTCTATTACCCCCATCCATACTCTAGAAATGAAGAGAATTGTTGTCGCAATTAC | PAD1 (ovMET25p) f |
| See above | PAD1 (ovCYC1t) r | |
| See above | CYC1t f | |
| See above | tTAL1t (ovp426) r | |
| p426-aroY-B-C | Derived from p426-aroY-B-C-D by removal of aroY-D expression cassette using BamHI | |
| p426-FDC1 | ATTAACCCTCACTAAAGGGAACAAAAGCTGGCTAGCATTTAAACTGTGAGGACCTTAATAC | TPI1p (ovp426) f |
| CTCTAAATTCTAAAGCTGGATTTAGCTTCCTCATTTTTAGTTTATGTATGTGTTTTTTG | TPI1p (ovFdc1) r | |
| CTATAACTACAAAAAACACATACATAAACTAAAAATGAGGAAGCTAAATCCAGC | FDC1 (ovTPI1p) f | |
| GAAAAGAAGATAATATTTTTATATAATTATATTAATCTTATTTATATCCGTACCTTTTCC | FDC1 (ovTPI1t) r | |
| TTAATATAATTATATAAAAATATTATCTTCTTTTC | TPI1t f | |
| GCGTAATACGACTCACTATAGGGCGAATTGGGATCCTGAGTAACCCATATAGAGATCGTAC | TPI1t (ovp426) r | |
| p426-aroY-B-FDC1 | See above | aroY-B (ovMET25p) f |
| GGTACCGGCCGCAAATTAAA | CYC1t r | |
| ACGCTCGAAGGCTTTAATTTGCGGCCGGTACCGTCGACATTTAAACTGTGAGGACCTTAATAC | TPI1p (ovCYC1t) | |
| See above | TPI1p (ovFdc1) r | |
| See above | TPI1t (ovp426) r | |
| p426-PAD1-FDC1 | AATTCTATTACCCCCATCCATACTCTAGAAATGCTCCTATTTCCAAGAAGAAC | PAD1 (ovMET25p) f |
| See above | CYC1t r | |
| See above | TPI1p (ovCYC1t) | |
| See above | TPI1t (ovp426) r | |
| p426-MET25-PAD1-His | AATTCTATTACCCCCATCCATACTCTAGAAATGCTCCTATTTCCAAGAAGAAC | PAD1 (ovMET25p) f |
| AATGTAAGCGTGACATAACTAATTACATGACTAGTGATGGTGATGGTGATGCTTGCTTTTTATTCCTTCCCAAC | PAD1-His (ovCYC1t) r | |
| p426-MET25-PAD1Δ1–54-His | AATTCTATTACCCCCATCCATACTCTAGAAATGAAGAGAATTGTTGTCGCAATTAC | PAD1 (ovMET25p) f |
| See above | PAD1-His (ovCYC1t) r | |
| p426-MET25-aroY-B-His | See above | aroY-B (ovMET25p) f |
| AATGTAAGCGTGACATAACTAATTACATGACTAGTGATGGTGATGGTGATGTTCGATTTCCTGAGCGAATTG | aroY-B-His (ovCYC1t) r | |
| p426-aroY-BMTS-His | See above | MTS (ovMET25p) f |
| See above | aroY-B-His (ovCYC1t) r |
The expression cassette PGK1p-aroY-Ciso-PGK1t-TPI1p-aroY-D-TAL1t is derived from Weber et al. (5).
Biotransformation experiments.
Precultures (50 ml) were grown overnight into the exponential phase in selective SC media with 2% (wt/vol) glucose and 2 mM methionine to repress the expression of plasmid-borne, heterologous genes under the control of the MET25 promoter (34). Cells were washed with sterile water and inoculated to an optical density at 600 nm (OD600) of 0.8 in selective SC medium with 2% (wt/vol) glucose but without methionine to allow for derepression of the MET25 promoter. PCA was added to a concentration of 5 mM. Biotransformation experiments were performed in three independent biological triplicates. Samples for metabolite analyses were taken at different time points for a time period of up to 168 h.
HPLC analysis of metabolites.
For high-pressure liquid chromatography (HPLC) analysis of metabolites, samples were removed from the culture and centrifuged for 5 min at 12,000 × g to separate culture supernatant and cells. To precipitate proteins in the culture supernatant, 5-sulfosalicylic acid was added to a final concentration of 5% (wt/vol). After removal of precipitated proteins by centrifugation for 5 min at 12,000 × g, samples were subjected to HPLC (Dionex Ultimate3000; Thermo Fisher Scientific GmbH, Dreieich, Germany) using a Nucleogel Sugar 810H exchange column (Macherey-Nagel GmbH & Co, Düren, Germany). Metabolites were eluted with 5 mM H2SO4 as a mobile phase and a flow rate of 0.6 ml/min at 65°C. For the detection of glucose, a Shodex RI-101 refractive index detector was used. For the detection of PCA and catechol, a UV detector was used at 220 nm. Data were collected and analyzed with the Chromeleon software (v6.80). Processing and statistical analysis of the data was performed using GraphPad Prism. Using a two-tailed, unpaired Student t test, differences between sample means were considered statistically significant if the P value was <0.05.
Preparation of cell extracts and isolation of mitochondria.
Cell extracts for direct loading of total protein were prepared as described previously (35). Mitochondria were isolated as described earlier (36), with minor modifications. Briefly, cultures of CEN.PK2-1C transformed with expression plasmids encoding Pad1, Pad1Δ1−54, AroY-B, or AroY-BMTS were grown for 48 h at 30°C in SC medium containing 2% glucose and no methionine. A total of 100 OD600 units of each culture were collected by centrifugation, subjected to zymolyase 20 T treatment, and washed in ice cold homogenization buffer (10 mM Tris/HCl [pH 7.4], 0.6 M sorbitol, 1 mM EDTA, 0.2% [wt/vol] bovine serum albumin). Spheroplasts were resuspended in 8 ml of ice-cold homogenization buffer and disrupted in a Potter-Elvehjem homogenizer. Unbroken cells, nuclei, and large cellular debris were removed by centrifugation for 5 min at 1,500 × g at 4°C, followed by centrifugation of the supernatant for 5 min at 3,000 × g at 4°C. The resulting extract was separated into cytosol and a crude mitochondrial fraction by centrifugation (15 min at 12,000 × g at 4°C). Protein extract corresponding to 0.6 OD600 units was loaded onto each lane.
Protein samples were separated by SDS-PAGE on 12.5% polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The membranes were subsequently stained with Penta His antibody (Qiagen, Hilden, Germany). As a control, mitochondrial inner membrane protein Qcr2 (subunit 2 of ubiquinol cytochrome c reductase [Complex III]) (37) and the cytoplasmic protein Pyk1 (pyruvate kinase 1; kindly provided by Jeremy Thorner, University of California, Berkeley, CA) were detected. As secondary antibodies, goat anti-mouse IgG (Penta His) or goat anti-rabbit (Qcr2 and Pyk1) peroxidase-conjugated antibodies were used (Sigma-Aldrich, Taufkirchen, Germany). Western blots were developed using the SuperSignal West Dura extended duration substrate (Thermo Fisher Scientific GmbH). The signals were documented using the Vilber Fusion SL3 Xpress system (VWR Life Science Competence Center, Erlangen, Germany).
Supplementary Material
ACKNOWLEDGMENTS
We thank Ilka Wittig, Goethe University, Frankfurt, Germany, for antibodies against Qcr2. Antibodies against Pyk1 were kindly provided by Jeremy Thorner, University of California at Berkeley, Berkeley, CA.
Financial support was provided by the German Federal Ministry of Education and Research following a decision of the German Bundestag (grant 031A542 [E.B.]) and by the European Commission under the 7th Framework Programme, Marie-Curie ITN YeastCell (grant 606795 [E.B.]).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03472-16.
REFERENCES
- 1.Xie NZ, Liang H, Huang RB, Xu P. 2014. Biotechnological production of muconic acid: current status and future prospects. Biotechnol Adv 32:615–622. doi: 10.1016/j.biotechadv.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Polen T, Spelberg M, Bott M. 2013. Toward biotechnological production of adipic acid and precursors from biorenewables. J Biotechnol 167:75–84. doi: 10.1016/j.jbiotec.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Draths KM, Frost JW. 1994. Environmentally compatible synthesis of adipic acid from d-glucose. J Am Chem Soc 116:399–400. doi: 10.1021/ja00080a057. [DOI] [Google Scholar]
- 4.Niu W, Draths KM, Frost JW. 2002. Benzene-free synthesis of adipic acid. Biotechnol Prog 18:201–211. doi: 10.1021/bp010179x. [DOI] [PubMed] [Google Scholar]
- 5.Weber C, Brückner C, Weinreb S, Lehr C, Essl C, Boles E. 2012. Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl Environ Microbiol 78:8421–8430. doi: 10.1128/AEM.01983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curran KA, Leavitt JM, Karim AS, Alper HS. 2013. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab Eng 15:55–66. doi: 10.1016/j.ymben.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Suastegui M, Matthiesen JE, Carraher JM, Hernandez N, Rodriguez Quiroz N, Okerlund A, Cochran EW, Shao Z, Tessonnier JP. 2016. Combining metabolic engineering and electrocatalysis: application to the production of polyamides from sugar. Angew Chem Int Ed Engl 55:2368–2373. doi: 10.1002/anie.201509653. [DOI] [PubMed] [Google Scholar]
- 8.Skjoedt ML, Snoek T, Kildegaard KR, Arsovska D, Eichenberger M, Goedecke TJ, Rajkumar AS, Zhang J, Kristensen M, Lehka BJ, Siedler S, Borodina I, Jensen MK, Keasling JD. 2016. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat Chem Biol 12:951–958. doi: 10.1038/nchembio.2177. [DOI] [PubMed] [Google Scholar]
- 9.Lupa B, Lyon D, Gibbs MD, Reeves RA, Wiegel J. 2005. Distribution of genes encoding the microbial non-oxidative reversible hydroxyarylic acid decarboxylases/phenol carboxylases. Genomics 86:342–351. doi: 10.1016/j.ygeno.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Li Z, Pereira B, Stephanopoulos G. 2015. Engineering E. coli-E. coli cocultures for production of muconic acid from glycerol. Microb Cell Fact 14:134. doi: 10.1186/s12934-015-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Pereira B, Li Z, Stephanopoulos G. 2015. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci U S A 112:8266–8271. doi: 10.1073/pnas.1506781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horwitz AA, Walter JM, Schubert MG, Kung SH, Hawkins K, Platt DM, Hernday AD, Mahatdejkul-Meadows T, Szeto W, Chandran SS, Newman JD. 2015. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst 1:88–96. doi: 10.1016/j.cels.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Payne KA, White MD, Fisher K, Khara B, Bailey SS, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Beveridge R, Barran P, Rigby SE, Scrutton NS, Hay S, Leys D. 2015. New cofactor supports alpha, beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature 522:497–501. doi: 10.1038/nature14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 15.Mukai N, Masaki K, Fujii T, Kawamukai M, Iefuji H. 2010. PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae. J Biosci Bioeng 109:564–569. doi: 10.1016/j.jbiosc.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Clausen M, Lamb CJ, Megnet R, Doerner PW. 1994. PAD1 encodes phenylacrylic acid decarboxylase which confers resistance to cinnamic acid in Saccharomyces cerevisiae. Gene 142:107–112. [DOI] [PubMed] [Google Scholar]
- 17.Gallone B, Steensels J, Prahl T, Soriaga L, Saels V, Herrera-Malaver B, Merlevede A, Roncoroni M, Voordeckers K, Miraglia L, Teiling C, Steffy B, Taylor M, Schwartz A, Richardson T, White C, Baele G, Maere S, Verstrepen K. 2016. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166:1397–1410. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard P, Viljanen K, Penttila M. 2015. Overexpression of PAD1 and FDC1 results in significant cinnamic acid decarboxylase activity in Saccharomyces cerevisiae. AMB Express 5:12. doi: 10.1186/s13568-015-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benisch F, Boles E. 2014. The bacterial Entner-Doudoroff pathway does not replace glycolysis in Saccharomyces cerevisiae due to the lack of activity of iron-sulfur cluster enzyme 6-phosphogluconate dehydratase. J Biotechnol 171:45–55. doi: 10.1016/j.jbiotec.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 21.McKenna R, Thompson B, Pugh S, Nielsen DR. 2014. Rational and combinatorial approaches to engineering styrene production by Saccharomyces cerevisiae. Microb Cell Fact 13:123. doi: 10.1186/s12934-014-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna R, Nielsen DR. 2011. Styrene biosynthesis from glucose by engineered Escherichia coli. Metab Eng 13:544–554. doi: 10.1016/j.ymben.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Sonoki T, Morooka M, Sakamoto K, Otsuka Y, Nakamura M, Jellison J, Goodell B. 2014. Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J Biotechnol 192(Pt A):71–77. doi: 10.1016/j.jbiotec.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 24.Johnson CW, Salvachua D, Khanna P, Smith H, Peterson DJ, Beckham GT. 2016. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metabolic Eng Commun 3:111–119. doi: 10.1016/j.meteno.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukai N, Masaki K, Fujii T, Iefuji H. 2014. Single nucleotide polymorphisms of PAD1 and FDC1 show a positive relationship with ferulic acid decarboxylation ability among industrial yeasts used in alcoholic beverage production. J Biosci Bioeng 118:50–55. doi: 10.1016/j.jbiosc.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Stratford M, Plumridge A, Archer DB. 2007. Decarboxylation of sorbic acid by spoilage yeasts is associated with the PAD1 gene. Appl Environ Microbiol 73:6534–6542. doi: 10.1128/AEM.01246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin F, Ferguson KL, Boyer DR, Lin XN, Marsh EN. 2015. Isofunctional enzymes PAD1 and UbiX catalyze formation of a novel cofactor required by ferulic acid decarboxylase and 4-hydroxy-3-polyprenylbenzoic acid decarboxylase. ACS Chem Biol 10:1137–1144. doi: 10.1021/cb5008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White MD, Payne KA, Fisher K, Marshall SA, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Rigby SE, Scrutton NS, Hay S, Leys D. 2015. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 522:502–506. doi: 10.1038/nature14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos MA, Jimenez A, Revuelta JL. 2000. Molecular characterization of FMN1, the structural gene for the monofunctional flavokinase of Saccharomyces cerevisiae. J Biol Chem 275:28618–28624. doi: 10.1074/jbc.M004621200. [DOI] [PubMed] [Google Scholar]
- 30.Tzagoloff A, Jang J, Glerum DM, Wu M. 1996. FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J Biol Chem 271:7392–7397. doi: 10.1074/jbc.271.13.7392. [DOI] [PubMed] [Google Scholar]
- 31.Marshall SA, Fisher K, Ni Cheallaigh A, White MD, Payne KA, Parker DA, Rigby SE, Leys D. 2017. Oxidative maturation and structural characterization of prenylated-FMN binding by UbiD, a decarboxylase involved in bacterial ubiquinone biosynthesis. J Biol Chem pii:jbc.M116.762732. doi: 10.1074/jbc.M116.762732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz RD, Schiestl RH. 2007. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:1–4. doi: 10.1038/nprot.2007.17. [DOI] [PubMed] [Google Scholar]
- 33.Oldenburg KR, Vo KT, Michaelis S, Paddon C. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mumberg D, Müller R, Funk M. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang T, Lei J, Yang H, Xu K, Wang R, Zhang Z. 2011. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 28:795–798. doi: 10.1002/yea.1905. [DOI] [PubMed] [Google Scholar]
- 36.Gregg C, Kyryakov P, Titorenko VI. 2009. Purification of mitochondria from yeast cells. J Vis Exp 30:1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrozzo R, Wittig I, Santorelli FM, Bertini E, Hofmann S, Brandt U, Schagger H. 2006. Subcomplexes of human ATP synthase mark mitochondrial biosynthesis disorders. Ann Neurol 59:265–275. doi: 10.1002/ana.20729. [DOI] [PubMed] [Google Scholar]
- 38.Hamacher T, Becker J, Gardonyi M, Hahn-Hagerdal B, Boles E. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.