ABSTRACT
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) replication in human cells is restricted at early postentry steps by host inhibitory factors. We previously described and characterized an early-phase restriction of HIV-1 and -2 replication in human cell lines, primary macrophages, and peripheral blood mononuclear cells. The restriction was termed lentiviral restriction 2 (Lv2). The viral determinants of Lv2 susceptibility mapped to the HIV-2 envelope (Env) and capsid (CA). We subsequently reported a whole-genome small interfering RNA screening for factors involved in HIV that identified RNA-associated early-stage antiviral factor (REAF). Using HIV-2 chimeras of susceptible and nonsusceptible viruses, we show here that REAF is a major component of the previously described Lv2 restriction. Further studies of the viral CA demonstrate that the CA mutation I73V (previously called I207V), a potent determinant for HIV-2, is a weak determinant of susceptibility for HIV-1. More potent CA determinants for HIV-1 REAF restriction were identified at P38A, N74D, G89V, and G94D. These results firmly establish that in HIV-1, CA is a strong determinant of susceptibility to Lv2/REAF. Similar to HIV-2, HIV-1 Env can rescue sensitive CAs from restriction. We conclude that REAF is a major component of the previously described Lv2 restriction.
IMPORTANCE Measures taken by the host cell to combat infection drive the evolution of pathogens to counteract or sidestep them. The study of such virus-host conflicts can point to possible weaknesses in the arsenal of viruses and may lead to the rational design of antiviral agents. Here we describe our discovery that the host restriction factor REAF fulfills the same criteria previously used to describe lentiviral restriction (Lv2). We show that, like the HIV-2 CA, the CA of HIV-1 is a strong determinant of Lv2/REAF susceptibility. We illustrate how HIV counteracts Lv2/REAF by using an envelope with alternative routes of entry into cells.
KEYWORDS: Lv2, REAF, antiviral
INTRODUCTION
Infection of cells by human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) is initiated by binding of the viral envelope (Env) to CD4. Conformational changes in the viral Env expose a site that can interact with a chemokine receptor, either CXCR4 or CCR5, expressed at the cell surface of CD4+ T cells and primary macrophages (1, 2). Viruses in general can enter cells through different routes, either directly at the plasma membrane (PM) or through a number of endocytic pathways (3). Influenza virus is a prototypical virus that enters cells through an endocytic route and requires the acid environment of the late endosome to trigger its fusion to and entry into cells. Since the mechanism of HIV fusion is pH independent (4), it has been widely assumed that HIV fuses at the PM (5–7). pH-independent endocytic entry has recently been observed (8–15) and is thus a possible mechanism of HIV entry; this, however, remains a topic of considerable controversy (16, 17). Regardless of the route, once HIV fuses at the PM, the conical core is released into the cytoplasm. The viral genomic RNA is reverse transcribed by the virus-encoded RNA/DNA-dependent reverse transcriptase (RT), resulting in virus-encoded RNA-DNA and single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) intermediates. The RNase H activity of RT degrades the RNA from these hybrids, resulting in ssDNA from which the second DNA strand is synthesized (18, 19). Once reverse transcription is complete, the proviral dsDNA is processed for integration into the host cell genome.
HIV must overcome many cellular barriers to its replication as it journeys to the nucleus to integrate into the host genome (20, 21). Interferon-induced transmembrane proteins can inhibit virus-cell membrane fusion (22), and the process of reverse transcription itself is also vulnerable. Immediately after initiation, members of the apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like family of restriction factors induce deoxycytidine to deoxyuridine mutations in the nascent DNA (23). Further disabling reverse transcription, SAMHD1 depletes the deoxynucleoside triphosphate substrates required (24). RNA-associated early-stage antiviral factor (REAF) was reported to inhibit HIV and SIV replication during reverse transcription (25). REAF is intrinsically expressed and provides an initial line of defense against HIV and SIV infection. It associates with reverse transcripts, either ssDNA or RNA-DNA hybrids; however, the precise mechanism of its action is not yet understood. A more recently described restriction factor, MX2/MXB, inhibits replication at a later stage, suppressing nuclear import and provirus formation (26–28). Integration is inhibited by the TRIM28 (KAP1)/SETDB1 complex (29). Once the provirus is integrated, the late phase of the replication cycle begins with the production of viral proteins (30). A PM-located restriction factor, tetherin/BST2/CD317, prevents viruses from leaving the cell at the late budding stage of the life cycle (31).
The first lentiviral restriction factor 1 (Lv1) effective against HIV-1 was identified as rhesus TRIM5α (32, 33). Lv1/TRIM5α is species specific and active against HIV-1 in nonhuman primate cells. TRIM5α forms a lattice around the capsid (CA), resulting in premature disassembly of the conical core (34). It is not known if Lv2 is species specific. It inhibits HIV-1 and -2 during reverse transcription, and susceptibility is determined by the viral CA (35). Lv2 differs from Lv1 in that Env is an additional determinant of restriction. Approximately half of HIV-1 and HIV-2 strains are susceptible to Lv2 (35). Lv3 is a postentry restriction to infection of simian MAGI cells by HIV-1 and, similar to Lv2, is dependent on fusion events at the cell membrane (36). Lv4 restricts the nuclear entry of SIV isolates in human cells (37). A recently described restriction to HIV-1 induced by Toll-like receptor agonist TLR7/8 in human monocytes is termed Lv5 (38). So far, the identities of Lv3, Lv4, and Lv5 are unknown. Here we describe the identification of REAF as a potent component of Lv2.
To identify components of Lv2, we designed a whole-genome small interfering RNA (siRNA) screening method (20). HeLa-CD4 cells were transfected with an siRNA library targeting 19,121 human genes and then challenged with an HIV-189.6 (molecular clone restricted [MCR]) pseudovirus (20). One factor identified was RPRD2 (here called REAF), which we now show fulfills the characteristics used to define Lv2 (35, 38).
RESULTS
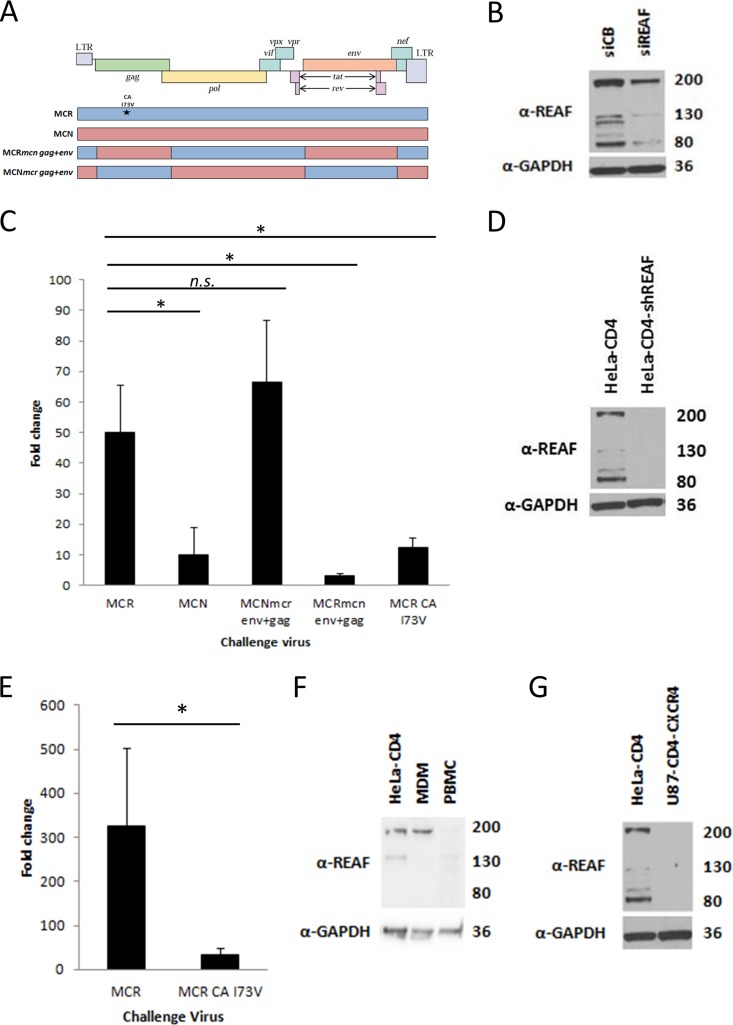
The HIV-2 molecular determinants of Lv2 restriction were previously mapped by using two HIV-2 molecular clones of isolates derived from the same patient, HIV-2MCR and HIV-2MCN, which are differentially sensitive to Lv2. A gene swapping approach between the viruses identified the gag and env genes as critical determinants of Lv2 restriction (39). These chimeric viruses (shown schematically in Fig. 1A) were tested to determine if they had the same pattern of susceptibility to REAF.
FIG 1.
REAF is an important component of Lv2. (A) A schematic representation of the HIV-2 molecular clones, chimeric viruses, and SDM (*) used to map the determinants of REAF restriction. LTR, long terminal repeat. (B) WB analysis of HeLa-CD4 cell lysate following REAF siRNA knockdown compared with the nontargeting control (siCB). GAPDH was added as a loading control. (C) Titration of constructs on HeLa-CD4 cells transiently transfected with siREAF showing fold changes compared to cells transfected with the siCB control (compared with HIV-2MCR: HIV-2MCN, P = 0.004; HIV-2MCNmcr env+gag, no significant difference; HIV-2MCRmcn env+gag, P < 0.001; HIV-2MCR CA I73V, P = 0.003). (D) WB of REAF knockdown in HeLa-CD4-shREAF cells compared to HeLa-CD4 cells. GAPDH was added as a loading control. (E) Fold changes in HeLa-CD4-shREAF cells infected with HIV-2MCR and HIV-2MCR CA I73V confirm the Lv2 phenotype in the stable knockdown cells (P = 0.016). (F) WB of REAF levels in MDMs and PBMCs compared to those in HeLa-CD4 cells. GAPDH was added as a loading control. (G) WB of REAF levels in U87-CD4-CXCR4 cells compared to those in HeLa-CD4 cells. GAPDH was added as a loading control. Gel run with that shown in panel D. The values to the right of panels B, D, F, and G are approximate molecular sizes in kilodaltons.
HeLa-CD4 cells were knocked down for REAF by using specific siRNA (siREAF) or nontargeting control siRNA (cyclophilin B, siCB) (Fig. 1B). Figure 1C shows viral rescue in HeLa-CD4 cells following treatment with siREAF and compared with cells treated with siCB. Repeat experiments consistently show, as expected for a virus highly sensitive to Lv2 (39), that HIV-2MCR is potently rescued in comparison to HIV-2MCN (50-fold versus 10-fold; P = 0.004). When env and gag from restricted HIV-2MCR were inserted in place of relatively insensitive HIV-2MCN env and gag (HIV-2MCNmcr env+gag), greater sensitivity of this virus to REAF was observed (66-fold). In the reciprocal experiment, where env and gag from HIV-2MCN replaced the HIV-2MCR genes (HIV-2MCRmcn env+gag), the resulting chimera was only rescued 3-fold (P < 0.001). These results for susceptibility to REAF are consistent with the Lv2 phenotype previously described (39). A single point mutation in HIV-2MCR CA at position 73 is known to be a critical determinant of Lv2 restriction (previously labeled position 207). Figure 1C shows that HIV-2MCR CA I73V is rescued only 12-fold from REAF restriction compared to 50-fold for wild-type HIV-2MCR (P = 0.003).
To confirm these results and for further experiments, we generated HeLa-CD4 cell lines permanently expressing shRNA specific for REAF mRNA. Western blot (WB) analysis shows that the HeLa-CD4-shREAF cells expressed much less REAF protein than the parental HeLa-CD4 cells did (Fig. 1D). The phenotype of REAF knockdown in this cell line was confirmed by using HIV-2MCR and HIV-2MCR CA I73V. HIV-2MCR was restricted 326-fold, compared to the 33-fold restriction of HIV-2MCR CA I73V (Fig. 1E; P = 0.016).
We previously reported that Lv2 was active in HeLa-CD4 cells, human primary peripheral blood mononuclear cells (PBMCs), and monocyte-derived macrophages (MDMs) but not in U87-CD4-CXCR4 cells (39). REAF mRNA (data not shown) and protein are present in MDMs and at much lower levels in PBMCs (Fig. 1F), while WB analysis shows it to be barely detectable in U87-CD4-CXCR4 cells (Fig. 1G).
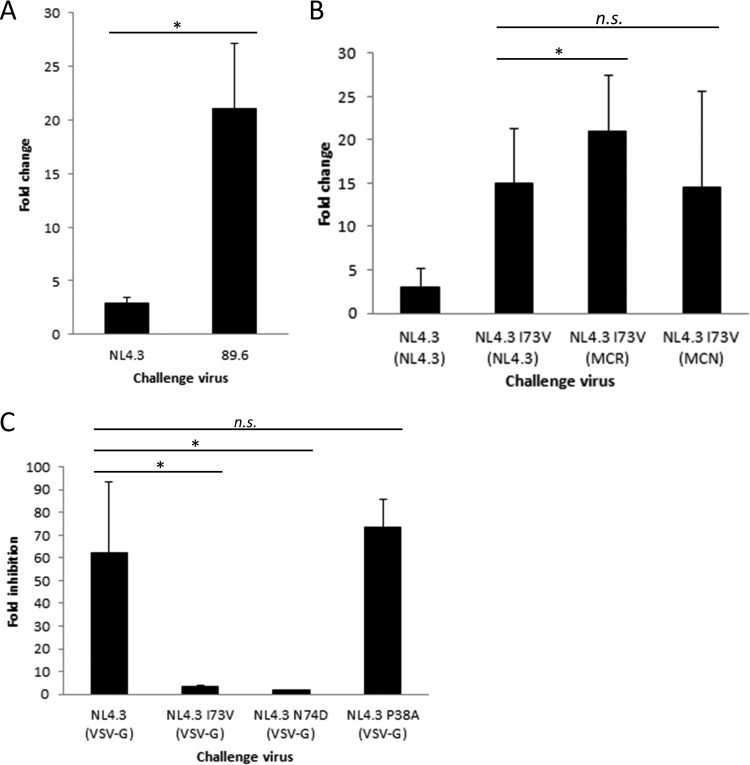
As previously reported for Lv2 (39) and further demonstrated here, the HIV-2 determinants for REAF are Env and CA (specifically, amino acid 73). We sought to identify the determinants of REAF restriction for HIV-1. Figure 2A shows that, compared to HIV-189.6, HIV-1NL4.3 is more resistant to REAF restriction (3-fold versus 21-fold; P < 0.001). We used HIV-1NL4.3 to further establish if the equivalent HIV-2 CA mutation 73 plays a role in Lv2/REAF restriction in HIV-1. Using site-directed mutagenesis (SDM), we generated HIV-1NL4.3 CA I73V. Both wild-type and mutant CAs were pseudotyped with HIV-2MCR Env and/or HIV-1NL4.3 Env and tested for their susceptibility to REAF by using the HeLa-CD4-shREAF cell line. Figure 2B shows that wild-type HIV-1NL4.3 CA is only weakly susceptible to REAF when pseudotyped with HIV-1NL4.3 Env. However, when the CA is mutated (HIV-1NL4.3 CA I73V [NL4.3]), the restriction is more potent but still relatively weak compared to that of HIV-2 (15-fold, compare to Fig. 1E for HIV-2). CA I73V was further restricted when pseudotyped with HIV-2MCR Env (21-fold, P = 0.03) but not when pseudotyped with HIV-2MCN Env (16-fold, no significant difference). Thus, CA amino acid 73 and Env are determinants of Lv2/REAF restriction for both HIV-1 and HIV-2.
FIG 2.
The HIV-1 CA determinants of REAF-associated restriction are the same as those for HIV-2 and are in the CPSF6 binding pocket. (A) A comparison of the susceptibility of HIV-1NL4.3 and HIV-189.6 following transient knockdown of REAF by siRNA (*, P < 0.001). (B) Mutation of HIV-1NL4.3 CA (HIV-1NL4.3 CA I73V [MCR] or HIV-1NL4.3 CA I73V [NL4.3]) renders it susceptible to restriction in HeLa-CD4-shREAF cells (*, P = 0.03; n.s., not significant). (C) Fold inhibition of HIV-1NL4.3 (VSV-G) with I73V and N74D mutant CAs (*, P < 0.001; n.s., not significant) in the presence of mutant CPSF6-358 compared to the vector alone.
The CA amino acid at position 73 lies in the binding domain of the cleavage- and polyadenylation-specific factor 6 (CPSF6) protein (40). This is of particular interest, as the adjacent CA mutation (N74D) has been shown to affect the sensitivity of HIV-1 to the depletion of nuclear pore proteins RanBP2, Nup153, and TNPO3 (41, 42).
N74D is an HIV-1 escape mutant that was generated by the passage of HIV-1NL4.3 in cells expressing artificially mutated CPSF6-358, which perturbs HIV-1 nuclear entry (42). CPSF6 is a pre-mRNA processing protein that shuttles between the nucleus and the cytoplasm (43). The mutant form CPSF6-358 lacks a C-terminal nucleus-targeting arginine/serine-rich domain and so is confined to the cytoplasm and restricts HIV-1 before nuclear entry (42, 44, 45).
We tested whether the Lv2/REAF HIV-2 CA determinant I73V was similar to N74D with respect to resistance to CPSF6-358. Another mutant CA with a P38A mutation, which is located outside the CPSF6 CA binding region, was included as a negative control (46). HeLa-CD4 target cells permanently expressing mutant CPSF6-358 (HeLa EKVΔCPSF6-358) or the vector alone (HeLa EKV) (40, 47) were challenged with pseudotypes carrying a mutant or wild-type CA. Figure 2C shows that, as expected, infection of the pseudotypes with wild-type and P38A mutant CA-producing virus was inhibited (62- and 73-fold, respectively). However, both CA mutants HIV-1NL4.3 CA I73V (vesicular stomatitis virus G protein [VSV-G]) and HIV-1NL4.3 CA N74D (VSV-G) were resistant to CPSF6-358 (3.3- and 1.6-fold; both P < 0.001).
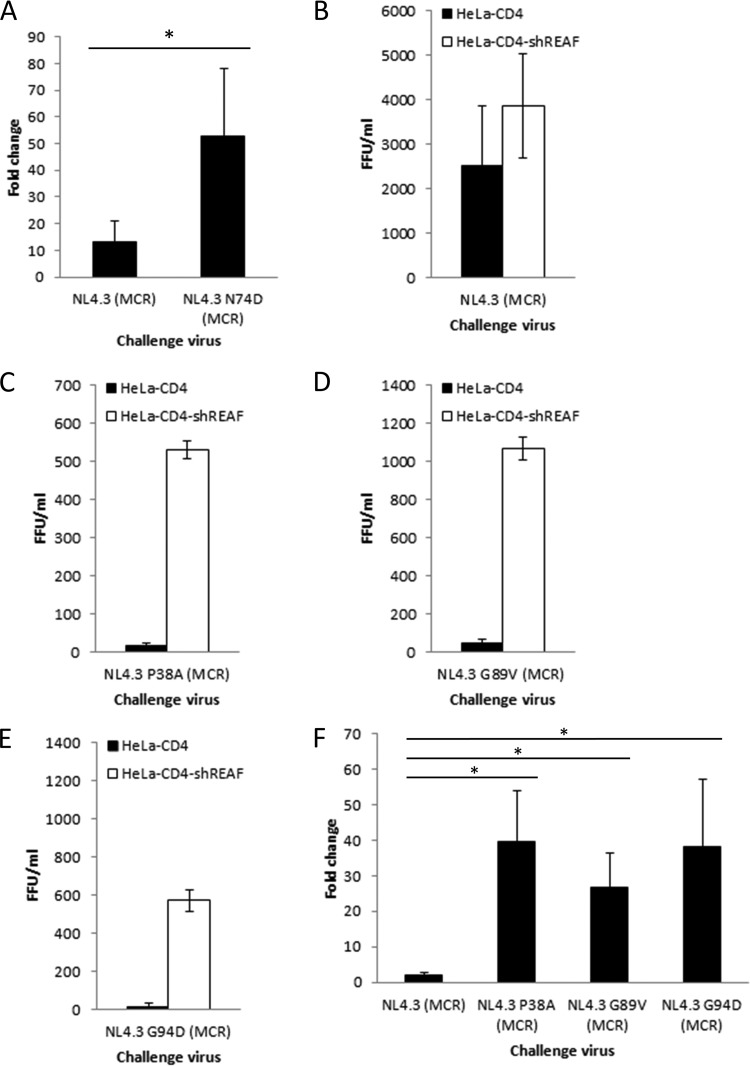
Given this similarity in resistance to CPSF6-358, we also sought to determine whether, similar to I73V, N74D is more susceptible than the wild-type virus to REAF restriction. HeLa-CD4 and HeLa-CD4-shREAF cells were challenged by pseudotypes with either wild-type or mutated CA. Figure 3A shows that HIV-1NL4.3 (MCR) was rescued 13-fold. Surprisingly, HIV-1NL4.3 CA N74D (MCR) is even more restricted and was rescued 53-fold (P < 0.001), suggesting that N74D mutant CA is a more potent determinant of REAF restriction in HIV-1.
FIG 3.
Mutant CAs are sensitive to REAF restriction. (A) Infection of HeLa-CD4-shREAF cells with HIV-1NL4.3 CA N74D (MCR) renders it susceptible to REAF compared to the viral pseudotype with wild-type CA (HIV-1NL4.3 [MCR]) (P < 0.001). (B to E) Titers of HIV-1NL4.3 (MCR), HIV-1NL4.3 CA P38A (MCR), HIV-1NL4.3 CA G89V (MCR), and HIV-1NL4.3 CA G94D (MCR) following a challenge of HeLa-CD4 and HeLa-CD4-shREAF cells. (F) Fold changes in HIV-1NL4.3 CA P38A (MCR), HIV-1NL4.3 CA G89V (MCR), and HIV-1NL4.3 CA G94D (MCR) on HeLa-CD4-shREAF cells compared to wild-type CA (HIV-1NL4.3 [MCR]) show that they are also susceptible to REAF (all P < 0.001 [*]).
As well as being less sensitive to CPSF6-358, N74D mutant CA has a more stable conical core, which results in delayed disassembly and reverse transcription (48). It is thought that optimal disassembly of the conical core is required for successful infection, as mutations interfering with core stability often result in a disturbance of reverse transcription kinetics (46, 49–51). We previously reported that REAF was transiently downmodulated shortly after infection (25). We hypothesized that unstable CAs will prematurely disassemble and expose reverse transcripts to REAF. The corollary of this is that CAs that disassemble too late will miss the time window during which REAF is absent. To test the hypothesis that CA stability is a determinant of REAF/Lv2 restriction, we investigated the REAF susceptibility of mutant CAs with various levels of stability. P38A mutant CA, in contrast to N74D mutant CA, is highly unstable (46, 52), while G94D mutant CA is unaffected (48). The G89V mutation in the cyclophilin binding loop was chosen because it has previously been shown to affect sensitivity to host restriction factors (46, 47, 50, 51). P38A mutant CA is also distinct from N74D mutant CA in that it is sensitive to CPSF6-358 (Fig. 2C). Figure 3C to E show the infectivity of all three mutant CAs. HIV-1NL4.3 CA P38A (MCR), HIV-1NL4.3 CA G89V (MCR), and HIV-1NL4.3 CA G94D were severely compromised on HeLa-CD4 cells compared to wild-type CA (titer of <500 focus-forming units [FFU]/ml; Fig. 3B). However, their replication was rescued in the absence of REAF, similar to that of HIV-1NL4.3 CA N74D (MCR) (Fig. 3F; all P < 0.001).
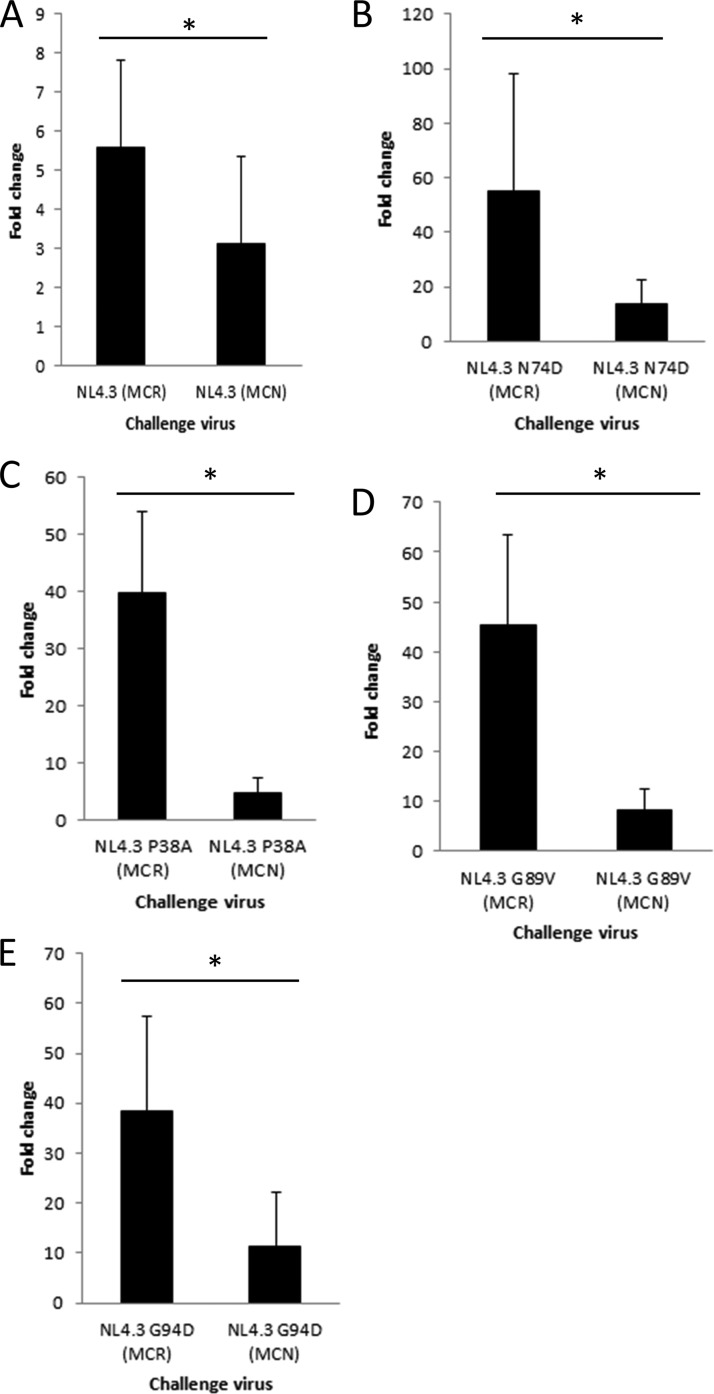
We showed above that, in addition to CA, HIV-1 Env also confers sensitivity to REAF (Fig. 2B). We previously determined that HIV-2MCN Env has the ability to overcome Lv2 (35, 39). If REAF is Lv2, HIV-2MCN Env would overcome REAF restriction. HeLa-CD4-shREAF cells were challenged with HIV-1NL4.3 with the wild-type CA pseudotyped with HIV-2MCN Env. As expected, the wild-type HIV-1NL4.3 CA was only slightly restricted and the HIV-2MCN Env could reduce this to a small but significant degree (5.6- to 3.1-fold; P = 0.007) (Fig. 4A). In contrast, the HIV-1NL4.3 strains carrying REAF-sensitive CA with the N74D, P38A, G89V, and G94V mutations were potently rescued with the HIV-2MCN Env (Fig. 4B to E, all P values < 0.001), further confirming that REAF and Lv2 are similarly rescued by viral Env.
FIG 4.
Viral Env determines susceptibility to REAF-associated restriction. (A) Infection of HeLa-CD4-shREAF cells with HIV-1NL4.3 (MCN) decreases sensitivity to REAF-associated restriction compared to HIV-1NL4.3 (MCR) (P = 0.007). Comparison of HIV-2MCR and HIV-2MCN Envs on pseudotypes carrying N74D (P < 0.001) (B), P38A (P < 0.001) (C), G89V (P < 0.001) (D), and G94D (P = 0.001) (E) mutant CAs showing that HIV-2MCN Env makes these viruses less sensitive to REAF. *, P < 0.001.
DISCUSSION
Here we show that REAF is a major component of the previously described restriction Lv2 (35, 53, 54). HIV chimeric viruses and mutants that delineate susceptible and resistant clones demonstrate that Lv2 and REAF are indistinguishable. Both Lv2 and REAF restriction activities are molecularly determined by viral Env and CA. The amino acid at position 73 in the HIV-2 CA was a crucial determinant of Lv2 and confirmed here for REAF (39). The equivalent amino acid at position 73 also affects HIV-1 susceptibility to REAF. Although the effects of I73V are statistically significant, they are much weaker than those of HIV-2. Indeed, the same amino acid substitution rendered HIV-2MCN less susceptible but HIV-1NL4.3 more susceptible to REAF. This led us to the hypothesis that the overall structure or stability of the CA, rather than precise molecular interactions, is critical. It has been proposed that disassembly too early prior to localization at the nuclear pore would result in exposure to restriction factors and premature termination of reverse transcription (19, 55–58). Indeed, it has been proposed that HIV-1 disassembly involves a regulated collapse of the conical core that protects viral reverse transcription complexes (59). Also, it has been shown that disassembly occurs within an hour of fusion and is facilitated by reverse transcription (60–62). We have observed that cellular REAF is reduced within 1 h of a viral challenge, but importantly, levels are rapidly replenished an hour later (25). REAF associates with viral nucleic acid and restricts replication during reverse transcription (25). We therefore suggested that the temporary reduction of the REAF protein level allows viruses to reverse transcribe in the absence of REAF-associated activity. This model is in keeping with the notion that CA stability is a determinant of REAF-associated restriction. We further tested the hypothesis that CA stability is a determinant of REAF susceptibility by using already well-characterized mutants with different CA stabilities. Compared to the I73V mutant CA (20-fold), more potent effects (>50-fold) were observed with the P38A (unstable), N74D (hyperstable), and G94D (unaffected) mutant CAs (48). Since all of these CA mutations result in greater susceptibility to REAF-associated activity despite having divergent CA stabilities, we cannot conclude that CA stability is a major determinant of susceptibility to REAF. Regardless, we show that the CA is a strong determinant of REAF restriction in HIV-1. Given the multifunctional role of CA in HIV-1 replication (63), understanding the role of CA in susceptibility to REAF-associated activity may shed light on more specific interactions of CA with host cell factors required for efficient infectivity of HIV.
It is highly controversial whether or not infectious HIV conical cores enter the cytoplasm after fusion at the PM or through an endocytic route (8–17, 35, 53). Our previous observation of Lv2 suggests that although either route is possible, infection is more successful when the virus fuses at the PM and avoids an endocytic entry pathway (35, 53). Our results here confirm that, like Lv2, the choice of entry route, as determined by the viral Env, is a determinant of susceptibility to REAF-associated activity. HIV-1NL4.3 can be rendered sensitive to REAF if pseudotyped with HIV-2MCR, while the HIV-2MCN Env cannot. These previously characterized HIV-2 Envs fuse either at the PM (MCN) or via an endocytic route (MCR) (35, 38). Furthermore, all of the CA mutants that were rendered highly sensitive to REAF-associated activity were protected when pseudotyped with an MCN Env.
We propose that, similar to Lv2, REAF may be more active against viruses attempting to access the cytoplasm via an endosome. Therefore, fusion at the PM is a more efficient replication pathway. However, we cannot eliminate the possibility that some viruses will bypass REAF-associated activity regardless of the entry route, for example, if they have a conical core without the sensitivity-conferring mutations described here. Future studies that address the roles of Env and CA in determining REAF-associated restriction will shed light on these host cell interactions in the early life cycle of HIV.
MATERIALS AND METHODS
Cells.
Buffy coats from seronegative donors were obtained from the National Blood Service (Brentwood, United Kingdom). The donors were anonymous, and patient consent was not required. PBMCs were prepared by density gradient centrifugation (Lymphoprep; Axis-Shield). MDMs were isolated from PBMCs with CD14+ MACS Microbeads (Miltenyi Biotec) and left to differentiate for 5 days in RPMI 1640–10% fetal calf serum–15 ng/ml granulocyte macrophage colony-stimulating factor (Peprotech). HEK 293T, HeLa-CD4, U87-CD4-CXCR4, HeLa EKV, and HeLa EKVΔCPSF6-358 cells and their optimal culture conditions have been described previously (39, 47, 64, 65).
Preparation of REAF knockdown cells.
The pSUPER RNA interference system (pSUPER.retro.puro; Oligoengine) was used for the expression of short hairpin RNA (shRNA) in mammalian cells (66, 67). For REAF knockdown, pSUPER.retro.puro(shREAF) was generated by digestion with BglII and HindIII, annealing of the specific primers, and ligation. The primers used were shREAF-BglII (5′ GATCCCCCACGTAAGCCCTCAGATGATTCAAGAGATCATCTGAGGGCTTACGTGTTTTTA 3′ [the shRNA target sequences are underlined]) and shREAF-HindIII (5′ AGCTTAAAAACACGTAAGCCCTCAGATGATCTCTTGAATCATCTGAGGGCTTACGTGGGG 3′).
The vector was either transfected directly into HeLa-CD4 cells for transient knockdown or used to generate stable knockdown cell lines. Briefly, retroviruses were produced by cotransfecting pSUPER.retro.puro(shREAF) with an HIV-1 gag-pol expression vector (p8.91) (68) and pMDG VSV-G Env into HEK 293T cells. Supernatant containing virus was harvested after 48 h and used to transduce HeLa-CD4 cells under puromycin selection. REAF silencing in transient and stably knocked down cells was confirmed by WB analysis.
Plasmids and virus production.
The infectious molecular clone for HIV-189.6 was obtained from the Centre for AIDS Research (National Institute for Biological Standards and Control [NIBSC], United Kingdom). Infectious full-length and chimeric HIV clones were prepared by polyethylenimine (PEI; Polysciences) or Lipofectamine 2000 (Invitrogen) transfection of HEK 293T cells. The virus named in parentheses for each pseudotype denotes the Env used.
Production of CA mutant viruses.
HIV-1 CA mutants were generated by SDM of the HIV-1NL4.3-derived viral clone pBR-NL43-IRES-eGFP (69) with further modification to introduce stop codons in the first and third codons of the Env coding sequence. HIV-1 pseudovirus particles were produced by PEI transfection of HEK 293T cells by using a 1:1 molar ratio of viral plasmid to MCR/MCN/VSV-G/NL4.3 Env expression plasmid.
WB analysis.
Proteins separated by SDS-PAGE were detected with the primary rabbit polyclonal antibody against REAF (Eurogentec) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Abcam), a loading control, followed by a horseradish peroxidase-conjugated donkey anti-rabbit antibody (GE Healthcare). Protein was visualized with an enhanced-chemiluminescence (ECL) kit (GE Healthcare).
siRNA transfection and infection with replication-competent virus.
HeLa-CD4 cells were seeded at 2.5 × 104 cells/well in 24-well plates. siRNA transfection (30 nM) was performed with HiPerfect (Qiagen) according to the manufacturer's instructions with the following sequences: siREAF, (5′ CACGTAAGCCCTCAGATGATA 3′); siCB, (5′ ACAGCAAATTCCATCGTGT 3′). At 72 h after siRNA transfection, cells were challenged with virus for up to 5 h. Infection was assessed for up to 48 h by intracellular p24 staining.
In situ immunostaining for p24 antigen.
Infected cells were fixed with cold (−20°C) methanol-acetone (1:1), washed with phosphate-buffered saline, and then immunostained for p24 with mouse anti-HIV-1 p24 monoclonal antibodies EVA365 and EVA366 (1:50; NIBSC, United Kingdom) as previously described (70). Infected-cell foci stained blue (regarded as foci of infection) and were quantitated in FFU per milliliter by light microscopy.
Statistical analysis.
The results presented here are derived from a minimum of three independent experiments performed in duplicate at a minimum. Differences between two treatments were tested for statistical significance by using unpaired two-tailed t tests. An asterisk denotes a P value of <0.05, and n.s. indicates no significant difference.
ACKNOWLEDGMENTS
The monoclonal antibodies to p24 (EVA365 and EVA366) were provided by the EU Programme EVA Centre for AIDS Reagents, NIBSC, United Kingdom (AVIP contract no. LSHP-CT-2004-503487). We thank Laura Hilditch and Greg Towers for providing HeLa cells expressing CPSF6-358. Thanks to Uta von Schwedler and Wesley Sundquist for the very kind donation of HIV-1-NL4.3 CA mutants. We would like to remember Uta, who is no longer with us to see the publication of this paper.
Á.M., K.M.M., and R.D.S. designed the experiments. Á.M., K.M., E.O.S., C.E.J., J.D.D., C.P., and R.D.S. performed the experiments. Á.M., K.M.M., and R.D.S. analyzed the data. Á.M. and K.M.M. wrote the manuscript.
This work was supported by The Rosetrees Trust (M275 and M275-F1) and a Queen Mary University of London, Life Sciences Institute Ph.D. studentship (C.E.J.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Clapham PR, McKnight A. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J Gen Virol 83:1809–1829. doi: 10.1099/0022-1317-83-8-1809. [DOI] [PubMed] [Google Scholar]
- 2.Doms RW, Trono D. 2000. The plasma membrane as a combat zone in the HIV battlefield. Genes Dev 14:2677–2688. doi: 10.1101/gad.833300. [DOI] [PubMed] [Google Scholar]
- 3.Marsh M, Helenius A. 2006. Virus entry: open sesame. Cell 124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClure MO, Sommerfelt MA, Marsh M, Weiss RA. 1990. The pH independence of mammalian retrovirus infection. J Gen Virol 71(Pt 4):767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 5.Maddon PJ, McDougal JS, Clapham PR, Dalgleish AG, Jamal S, Weiss RA, Axel R. 1988. HIV infection does not require endocytosis of its receptor, CD4. Cell 54:865–874. doi: 10.1016/S0092-8674(88)91241-X. [DOI] [PubMed] [Google Scholar]
- 6.Pelchen-Matthews A, Clapham P, Marsh M. 1995. Role of CD4 endocytosis in human immunodeficiency virus infection. J Virol 69:8164–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein BS, Gowda SD, Lifson JD, Penhallow RC, Bensch KG, Engleman EG. 1987. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell 49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- 8.Vidricaire G, Tremblay MJ. 2007. A clathrin, caveolae, and dynamin-independent endocytic pathway requiring free membrane cholesterol drives HIV-1 internalization and infection in polarized trophoblastic cells. J Mol Biol 368:1267–1283. doi: 10.1016/j.jmb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 9.de la Vega M, Marin M, Kondo N, Miyauchi K, Kim Y, Epand RF, Epand RM, Melikyan GB. 2011. Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 8:99. doi: 10.1186/1742-4690-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter GC, Bernstone L, Baskaran D, James W. 2011. HIV-1 infects macrophages by exploiting an endocytic route dependent on dynamin, Rac1 and Pak1. Virology 409:234–250. doi: 10.1016/j.virol.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Clotet-Codina I, Bosch B, Senserrich J, Fernández-Figueras MT, Peña R, Ballana E, Bofill M, Clotet B, Esté JA. 2009. HIV endocytosis after dendritic cell to T cell viral transfer leads to productive virus infection. Antiviral Res 83:94–98. doi: 10.1016/j.antiviral.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Daecke J, Fackler OT, Dittmar MT, Kräusslich HG. 2005. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J Virol 79:1581–1594. doi: 10.1128/JVI.79.3.1581-1594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maréchal V, Prevost MC, Petit C, Perret E, Heard JM, Schwartz O. 2001. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J Virol 75:11166–11177. doi: 10.1128/JVI.75.22.11166-11177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sloan RD, Kuhl BD, Mesplède T, Münch J, Donahue DA, Wainberg MA. 2013. Productive entry of HIV-1 during cell-to-cell transmission via dynamin-dependent endocytosis. J Virol 87:8110–8123. doi: 10.1128/JVI.00815-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herold N, Müller B, Kräusslich HG. 2015. Reply to “Can HIV-1 Entry Sites Be Deduced by Comparing Bulk Endocytosis to Functional Readouts for Viral Fusion? ” J Virol 89:2986–2987. doi: 10.1128/JVI.03376-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marin M, Melikyan GB. 2015. Can HIV-1 entry sites be deduced by comparing bulk endocytosis to functional readouts for viral fusion? J Virol 89:2985. doi: 10.1128/JVI.03352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterlin BM, Trono D. 2003. Hide, shield and strike back: how HIV-infected cells avoid immune eradication. Nat Rev Immunol 3:97–107. doi: 10.1038/nri998. [DOI] [PubMed] [Google Scholar]
- 19.Campbell EM, Hope TJ. 2015. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13:471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Oliveira NM, Cheney KM, Pade C, Dreja H, Bergin AM, Borgdorff V, Beach DH, Bishop CL, Dittmar MT, McKnight A. 2011. A whole genome screen for HIV restriction factors. Retrovirology 8:94. doi: 10.1186/1742-4690-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. 2011. The IFITM proteins inhibit HIV-1 infection. J Virol 85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop KN, Holmes RK, Sheehy AM, Malim MH. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 24.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marno KM, Ogunkolade BW, Pade C, Oliveira NM, O'Sullivan E, McKnight Á. 2014. Novel restriction factor RNA-associated early-stage anti-viral factor (REAF) inhibits human and simian immunodeficiency viruses. Retrovirology 11:3. doi: 10.1186/1742-4690-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, Schaller T, Hué S, Barclay WS, Schulz R, Malim MH. 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C. 2013. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. 2011. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9:484–495. doi: 10.1016/j.chom.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Goff SP. 2007. Host factors exploited by retroviruses. Nat Rev Microbiol 5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 31.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 32.Towers G, Bock M, Martin S, Takeuchi Y, Stoye JP, Danos O. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci U S A 97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci U S A 99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pertel T, Hausmann S, Morger D, Züger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grütter MG, Luban J. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchant D, Neil SJ, Aubin K, Schmitz C, McKnight A. 2005. An envelope-determined, pH-independent endocytic route of viral entry determines the susceptibility of human immunodeficiency virus type 1 (HIV-1) and HIV-2 to Lv2 restriction. J Virol 79:9410–9418. doi: 10.1128/JVI.79.15.9410-9418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pineda MJ, Orton BR, Overbaugh J. 2007. A TRIM5alpha-independent post-entry restriction to HIV-1 infection of macaque cells that is dependent on the path of entry. Virology 363:310–318. doi: 10.1016/j.virol.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizzato M, Neagu M, Pertel T, Firrito C, Ziglio S, Zufferey M, Berthoux L, Luban J. 2013. Lv4, an activity that restricts nuclear entry of SIVMAC/SIVSM in human blood cells. Retrovirology 10(Suppl 1):O28. doi: 10.1186/1742-4690-10-S1-O28. [DOI] [Google Scholar]
- 38.Hofmann H, Vanwalscappel B, Bloch N, Landau NR. 2016. TLR7/8 agonist induces a post-entry SAMHD1-independent block to HIV-1 infection of monocytes. Retrovirology 13:83. doi: 10.1186/s12977-016-0316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz C, Marchant D, Neil SJ, Aubin K, Reuter S, Dittmar MT, McKnight A. 2004. Lv2, a novel postentry restriction, is mediated by both capsid and envelope. J Virol 78:2006–2016. doi: 10.1128/JVI.78.4.2006-2016.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AJ, Fletcher AJ, Schaller T, Elliott T, Lee K, KewalRamani VN, Chin JW, Towers GJ, James LC. 2012. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog 8:e1002896. doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hué S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruepp MD, Aringhieri C, Vivarelli S, Cardinale S, Paro S, Schümperli D, Barabino SM. 2009. Mammalian pre-mRNA 3′ end processing factor CF I m 68 functions in mRNA export. Mol Biol Cell 20:5211–5223. doi: 10.1091/mbc.E09-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem 279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 45.Rüegsegger U, Blank D, Keller W. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell 1:243–253. doi: 10.1016/S1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 46.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, Jacques DA, Selwood DL, James LC, Noursadeghi M, Towers GJ. 2013. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hulme AE, Kelley Z, Okocha EA, Hope TJ. 2015. Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. J Virol 89:643–651. doi: 10.1128/JVI.03043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzon T, Leschonsky B, Bieler K, Paulus C, Schröder J, Wolf H, Wagner R. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294–307. doi: 10.1006/viro.1999.0178. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Kar AK, Sodroski J. 2009. Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. J Virol 83:10951–10962. doi: 10.1128/JVI.00682-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah VB, Shi J, Hout DR, Oztop I, Krishnan L, Ahn J, Shotwell MS, Engelman A, Aiken C. 2013. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J Virol 87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol 77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrison IP, McKnight A. 2011. Cellular entry via an actin and clathrin-dependent route is required for Lv2 restriction of HIV-2. Virology 415:47–55. doi: 10.1016/j.virol.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Reuter S, Kaumanns P, Buschhorn SB, Dittmar MT. 2005. Role of HIV-2 envelope in Lv2-mediated restriction. Virology 332:347–358. doi: 10.1016/j.virol.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 55.Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, El Marjou A, Lacabaratz C, Lelièvre JD, Manel N. 2013. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Landau NR. 2014. The innate immune response to HIV-1: to sense or not to sense. DNA Cell Biol 33:271–274. doi: 10.1089/dna.2014.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Towers GJ, Noursadeghi M. 2014. Interactions between HIV-1 and the cell-autonomous innate immune system. Cell Host Microbe 16:10–18. doi: 10.1016/j.chom.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fassati A, Goff SP. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A 108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kutluay SB, Perez-Caballero D, Bieniasz PD. 2013. Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLoS Pathog 9:e1003214. doi: 10.1371/journal.ppat.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y, Fricke T, Diaz-Griffero F. 2013. Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J Virol 87:683–687. doi: 10.1128/JVI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD. 2013. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog 9:e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng HK, Unutmaz D, KewalRamani VN, Littman DR. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 65.Salahuddin SZ, Markham PD, Wong-Staal F, Franchini G, Kalyanaraman VS, Gallo RC. 1983. Restricted expression of human T-cell leukemia–lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology 129:51–64. doi: 10.1016/0042-6822(83)90395-1. [DOI] [PubMed] [Google Scholar]
- 66.Brummelkamp TR, Bernards R, Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 67.Brummelkamp TR, Bernards R, Agami R. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243–247. doi: 10.1016/S1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 68.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 69.Schindler M, Münch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Müller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, Bailes E, Roques P, Sodora DL, Silvestri G, Sharp PM, Hahn BH, Kirchhoff F. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 70.McKnight A, Clapham PR, Weiss RA. 1994. HIV-2 and SIV infection of nonprimate cell lines expressing human CD4: restrictions to replication at distinct stages. Virology 201:8–18. doi: 10.1006/viro.1994.1260. [DOI] [PubMed] [Google Scholar]