ABSTRACT
Proteolytic maturation involving cleavage of one nonstructural and six structural precursor proteins including pVIII by adenovirus protease is an important aspect of the adenovirus life cycle. The pVIII encoded by bovine adenovirus 3 (BAdV-3) is a protein of 216 amino acids and contains two potential protease cleavage sites. Here, we report that BAdV-3 pVIII is cleaved by adenovirus protease at both potential consensus protease cleavage sites. Usage of at least one cleavage site appears essential for the production of progeny BAdV-3 virions as glycine-to-alanine mutation of both protease cleavage sites appears lethal for the production of progeny virions. However, mutation of a single protease cleavage site of BAdV-3 pVIII significantly affects the efficient production of infectious progeny virions. Further analysis revealed no significant defect in endosome escape, genome replication, capsid formation, and virus assembly. Interestingly, cleavage of pVIII at both potential cleavage sites appears essential for the production of stable BAdV-3 virions as BAdV-3 expressing pVIII containing a glycine-to-alanine mutation of either of the potential cleavage sites is thermolabile, and this mutation leads to the production of noninfectious virions.
IMPORTANCE Here, we demonstrated that the BAdV-3 adenovirus protease cleaves BAdV-3 pVIII at both potential protease cleavage sites. Although cleavage of pVIII at one of the two adenoviral protease cleavage sites is required for the production of progeny virions, the mutation of a single cleavage site of pVIII affects the efficient production of infectious progeny virions. Further analysis indicated that the mutation of a single protease cleavage site (glycine to alanine) of pVIII produces thermolabile virions, which leads to the production of noninfectious virions with disrupted capsids. We thus provide evidence about the requirement of proteolytic cleavage of pVIII for production of infectious progeny virions. We feel that our study has significantly advanced the understanding of the requirement of adenovirus protease cleavage of pVIII.
KEYWORDS: adenovirus protease, BAdV-3, proteolytic cleavage, thermolabile, pVIII
INTRODUCTION
Adenoviruses are complex, large, nonenveloped viruses that have a linear double-stranded DNA genome (1). The in-depth molecular characterization of human adenoviruses and their properties, such as ease of genome manipulation, broad tropism, rapid growth to high titers in tissue culture, and transgene carrying capacity of up to 30 kb, have made adenoviruses a popular tool for gene transfer into mammalian cells (2, 3). However, although adenoviruses have been extensively studied for more than 5 decades, some aspects of adenovirus biology, such as virus assembly and final virus maturation, are not fully understood. Moreover, localization of some of the minor capsid proteins and their role(s) in the adenovirus life cycle are not clear yet.
As with many other viruses and bacteriophages, proteolytic maturation is a key step in the life cycle of adenoviruses. The key player in adenovirus maturation is adenovirus protease, which recognizes and cleaves at two consensus cleavage motifs, (M/I/L)XGX-G and (M/I/L)XGG-X (where X is any amino acid) (4). Adenoviral proteins that are substrates for adenovirus protease include three minor capsid proteins (pIIIa, pVI, and pVIII) and three core proteins (pVII, pμ, and pTP) (5–7). Apart from these six structural proteins, two nonstructural proteins, the L1 52,000-/55,000-molecular weight protein(52/55K) (8) and 100K (9), have been recently reported to be substrates of adenoviral protease.
One of the substrates for adenoviral protease is pVIII. The pVIII is a minor capsid protein that is present on the inner surface of the capsid and connects the core protein with the capsid (10). As pVIII is one of the least characterized adenoviral proteins, very little is known about the function of pVIII in the adenovirus life cycle. Earlier work suggested that the human adenovirus 5 (HAdV-5) pVIII encoded by the L4 region is 227 amino acids (aa) long (11) and is present in precursor form in immature virus particles and in a cleaved form in mature virions (12–14). Crystallographic studies could identify amino acids 31 to 90 of fragment 1 (consisting of aa 1 to 111) and amino acids 163 to 215 of fragment 2 (aa 158 to 227) of HAdV-5 pVIII in mature virions (15).
Recently, we have reported that bovine adenovirus 3 (BAdV-3) pVIII, a protein of 216 amino acids encoded by the L6 region (16), is localized to the nucleus of the infected cells by binding of pVIII (amino acids 52 to 72) with importin α-3 (17). The pVIII is expressed as 24-kDa (precursor form) and 8-kDa (cleaved form) proteins in BAdV-3-infected cells. Moreover, while the precursor form (24 kDa) could be detected in empty capsids, the cleaved form (8 kDa) could be detected in mature capsids (17), suggesting that BAdV-3 pVIII appears to be cleaved at the potential cleavage site (indicated by the arrow) 143LGGG↓S147 (17).
Although proteolytic maturation involving cleavage of some structural and nonstructural precursor proteins is an important aspect of the adenovirus life cycle (18, 19), it is not known what role each individual cleavage event plays in determining adenovirus particle stability and infectivity (18, 19). The present study was initiated to determine the role of proteolytic cleavage of pVIII in virus assembly, stability, and infection. Here, we report that BAdV-3 pVIII is cleaved by the adenovirus protease at both potential consensus protease cleavage sites. The cleavage at one consensus protease cleavage site of pVIII appears essential for the production of infectious progeny virus. Moreover, cleavage of pVIII at both potential protease cleavage sites (PPCS) is a major contributing factor in determining the stability of adenovirus particles.
RESULTS
Analysis of the pVIII protein sequences for potential protease cleavage sites.
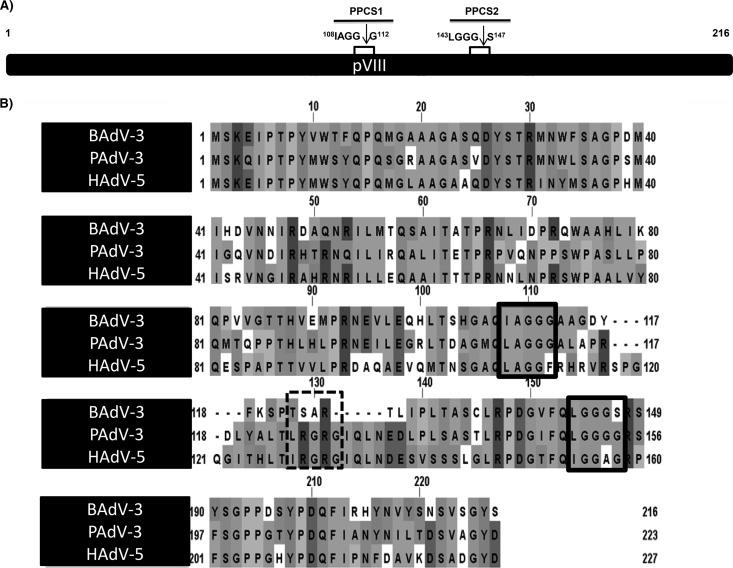
The adenovirus protease recognizes and cleaves at two consensus cleavage motifs, (M/I/L)XGX-G and (M/I/L)XGG-X (where X is any amino acid) (4), where glycine residues appear essential for the cleavage of substrate proteins (20, 21). Earlier, analysis of the amino acid sequence of BAdV-3 pVIII (17) revealed two potential adenoviral protease cleavage sites (Fig. 1A), which are conserved with potential protease cleavage sites in pVIII encoded by human adenovirus 5 (HAdV-5) or porcine adenovirus 3 (PAdV-3) (Fig. 1B). In contrast, analysis of pVIII encoded by HAdV-5 and PAdV-3 revealed the presence of three potential protease cleavage sites (Fig. 1B).
FIG 1.
Consensus protease cleavage sites of pVIII. (A) Schematic diagram of BAdV-3 pVIII showing the two potential protease cleavage sites (PPCS), PPCS1 (amino acids 108 to 112) and PPCS2 (amino acids 143 to 147). Arrows depict the site of cleavage. (B) Conservation of PPCS in pVIII protein. The PPCS conserved among pVIII of BAdV-3, PAdV-3, and HAdV-5 are indicated by boxed in solid lines, and PPCS conserved among PAdV-3 and HAdV-5 only are boxed in dotted lines. Sequences were downloaded from UniProt (BAdV-3, UniProt accession number O92788; PADV-3, Q83453; HAdV-5, P24936) and aligned with T-Coffee (42). The figure was created using JalView (43).
Analysis of cleavage of BAdV-3 pVIII at a potential protease cleavage site(s).
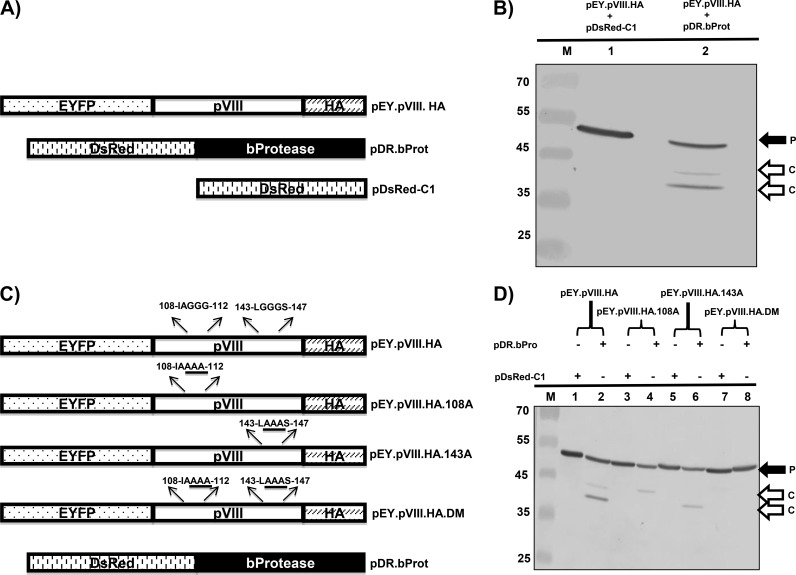
To determine whether pVIII is cleaved at one or both potential protease cleavage sites, we constructed plasmid pEY.pVIII.HA (expressing enhanced yellow fluorescent protein [EYFP] and hemagglutinin [HA]-tagged pVIII) (Fig. 2A). The 293T cells were cotransfected with plasmid pEY.pVIII.HA and pDsRed-C1 DNAs or plasmid pEY.pVIII.HA and pDR.bProt (expressing DsRed-tagged BAdV-3 protease) DNAs. At 48 h posttransfection, the cells were collected and lysed, and the lysates were analyzed by Western blotting using anti-green fluorescent protein (GFP) serum. As seen in Fig. 2B, anti-GFP serum detected a protein of 52 kDa (corresponding to the EYFP-pVIII-HA fusion protein) in the cells cotransfected with plasmid pEY.pVIII.HA and pDsRed-C1 DNAs. In contrast, anti-GFP serum detected proteins of 52 kDa, 40 kDa (corresponding to the cleavage product of the EYFP-pVIII fusion protein consisting of EYFP [26 kDa] and amino acids 1 to 111 of pVIII), and 43 kDa (corresponding to the cleavage product of the EYFP-pVIII fusion protein consisting of EYFP and amino acids 1 to 146 of pVIII) in the cells cotransfected with plasmid pEY.pVIII.HA and pDR.bProt DNAs, suggesting that pVIII fragment sizes are consistent with cleavage at the potential sites detected by bioinformatics.
FIG 2.
Cleavage of BAdV-3 pVIII protein in transfected cells. (A) Schematic diagram of plasmid DNAs. The names of the plasmids are depicted on the right of diagrams. The EYFP (enhanced yellow fluorescent protein)-pVIII (BAdV-3), bProtease (BAdV-3), and DsRed DNA plasmids are depicted. (B) Cleavage of BAdV-3 pVIII in transfected cells. Proteins from the lysates of 293T cells cotransfected with the indicated plasmid DNAs were separated by 12% SDS-PAGE, transferred to nitrocellulose membrane, and analyzed by Western blotting using anti-GFP antibody. P, uncleaved EY.pVIII.HA fusion protein; C, cleaved product. The molecular mass markers (lane M) in kilodaltons are shown on the left. (C) Schematic diagram of plasmid DNAs showing potential protease cleavage sites. The origin of the DNA is depicted. The amino acid sequences of potential protease cleavage sites are depicted above the diagrams, and glycine-to-alanine substitutions are underlined. (D) Cleavage of BAdV-3 pVIII mutant proteins in transfected cells. Proteins from the lysates of 293T cells cotransfected with the indicated plasmid DNAs were separated by 12% SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by Western blotting using anti-GFP antibody. The molecular mass markers (M) in kilodaltons are shown on the left. P, uncleaved fusion protein; C, cleaved product.
To further characterize the cleavage of pVIII by adenoviral protease, we constructed three additional plasmids (pEY.pVIII.HA.108A, pEY.pVIII.HA.143A, and pEY.pVIII.HA.DM) (Fig. 2C). Plasmid pEY.pVIII.HA.108A contains substitutions of alanines for glycines at amino acid positions 110, 111, and 112 of BAdV-3 pVIII, plasmid pEY.pVIII.HA.143A contain substitutions of alanines for glycines at positions 144, 145, and 146 of BAdV-3 pVIII, and plasmid pEY.pVIII.HA.DM contains substitutions of alanines for glycines at positions 110, 111, 112, 145, and 146 of BAdV-3 pVIII. Identity of these plasmid DNAs was confirmed by DNA sequencing. 293T cells were cotransfected with each of these plasmid DNAs with either plasmid pDsRed-C1 DNA or plasmid pDR.bProt DNA. After 48 h posttransfection, the cells were collected and lysed, and the lysates were analyzed by Western blotting using anti-GFP serum. As seen in Fig. 2D, anti-GFP serum detected a protein of 52 kDa in the lysates of the cells cotransfected with plasmid pEY.pVIII.HA and pDsRed-C1 (lane 1), pEY.pVIII.HA.108A and pDsRed-C1 (lane 3), pEY.pVIII.HA.143A and pDsRed-C1 (lane 5), pEY.pVIII.HA.DM and pDsRed-C1, and pEY.pVIII.HA.DM and pDR.bProt (lane 7) DNAs, suggesting that pVIII is not cleaved in the absence of protease. As seen earlier, anti-GFP serum detected proteins of 52 kDa, 43 kDa, and 40 kDa in the lysates of the cells cotransfected with plasmid pEY.pVIII.HA and pDR.bProt DNAs (lane 2). However, anti-GFP serum detected proteins of 52 kDa and 43 kDa in the lysates of the cells cotransfected with plasmid pEY.pVIII.HA.108A and pDR.bProt DNAs (lane 4). Similarly, anti-GFP serum detected two proteins of 52 kDa and 40 kDa in the lysates of the cells cotransfected with plasmid pEY.pVIII.HA.143A and pDR.bProt DNAs (lane 6). In contrast, anti-GFP serum detected only a protein of 52 kDa in the lysates of the cells cotransfected with plasmid pEY.pVIII.HA.DM and pDR.bProt DNAs (lane 8), indicating that protease is unable to cleave pVIII if potential cleavage sites are mutated. These results confirm the fact that the identified potential protease cleavage sites are indeed the actual protease cleavage sites of pVIII.
Cleavage of BAdV-3 pVIII by protease encoded by other mastadenoviruses.
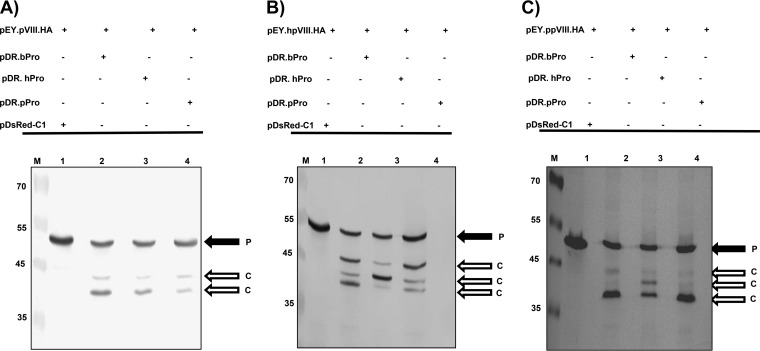
To determine if BAdV-3 pVIII protein can be cleaved by proteases encoded by other mastadenoviruses, the 293T cells were cotransfected with plasmid pEY.pVIII.HA DNA and plasmid pDsRed-C1, plasmid pDR.bProt (expressing BAdV-3 protease), plasmid pDR.hProt (expressing human adenovirus 5 [HAdV-5] protease), or plasmid pDR.pProt (expressing porcine adenovirus 3 [PAdV-3] protease)DNA. At 48 h posttransfection, the cells were collected and lysed, and the lysates were analyzed by Western blotting using anti-GFP serum. As seen in Fig. 3A, anti-GFP serum detected a protein of 52 kDa (uncleaved EYFP-pVIII-HA fusion protein) in the cells cotransfected with plasmid pEY.pVIII.HA and pDsRed-C1 DNAs (lane 1). In contrast, anti-GFP serum detected proteins of 52 kDa (uncleaved EYFP-pVIII-HA fusion protein), 43 kDa (corresponding to the cleavage product of the EYFP-pVIII fusion protein consisting of EYFP and amino acids 1 to 146 of pVIII), and 40 kDa (corresponding to the cleavage product of the EYFP-pVIII fusion protein consisting of EYFP and amino acids 1 to 111 of pVIII) in the cells cotransfected with plasmid pEY.pVIII.HA DNA and either pDR.bProt (lane 2), pDR.hProt (lane 3), or pDR.pProt (lane 4) DNA, indicating that BAdV-3 pVIII is cleaved at both potential cleavage sites by proteases encoded by HAdV-5 and PAdV-3.
FIG 3.
Cleavage of pVIII by different adenoviral proteases in transfected cells. Proteins from the lysates of 293T cells cotransfected with the indicated plasmid DNAs were separated by 12% SDS-PAGE, transferred to nitrocellulose membrane, and analyzed by Western blotting using anti-GFP antibody. The cleavage of pVIII of BAdV-3 pVIII (A), HAdV-5 (B), and PAdV-3 (C) by adenoviral proteases is represented. The molecular mass markers (lanes M) are shown (in kilodaltons) on the left of each panel. P, uncleaved fusion protein; C, cleaved product.
Cleavage of pVIII protein encoded by other mastadenoviruses.
Analysis of the amino acid sequence of pVIII of HAdV-5 and PAdV-3 revealed three potential protease cleavage sites. To analyze the cleavage of the pVIII proteins of HAdV-5 and PAdV-3, we constructed plasmids expressing the pVIII protein of HAdV-5 (pEY.hpVIII.HA) or PAdV-3 (pEY.ppVIII.HA) as an EYFP fusion protein. The 293T cells were cotransfected with plasmid pEY.hpVIII.HA DNA and plasmid pDsRed-C1 (control), plasmid pDR.bProt (expressing BAdV-3 protease), plasmid pDR.hProt (expressing HAdV-5 protease), or plasmid pDR.pProt (expressing PAdV-3 protease) DNA. At 48 h posttransfection, the cells were collected and lysed, and the cell lysates were analyzed by Western blotting using anti-GFP serum. As seen in Fig. 3B, anti-GFP serum detected a protein of 52 kDa (uncleaved EYFP-hpVIII-HA fusion protein) in the cells cotransfected with plasmids pEY.hpVIII.HA and pDsRed-C1 DNAs (lane 1). In contrast, proteins of 52 kDa, 40 kDa (amino acids 1 to 111 of the cleavage product of the EYFP-hpVIII fusion protein), 42 kDa (amino acid 1 to 131 of the cleavage product of the EYFP-hpVIII fusion protein), and 45 kDa (amino acids 1 to 157 of the cleavage product of the EYFP-hpVIII fusion protein) were detected in lysates of the cells cotransfected with plasmid pEY.hpVIII.HA DNA and pDR.bProt (lane 2), pDR.hProt (lane 3), or pDR.pProt (lane 4) DNA, indicating that HAdV-5 pVIII can be cleaved at all three potential cleavage sites by a protease encoded by HAdV-5, BAdV-3, or PAdV-3 (Fig. 3B, lane 3).
Similarly, anti-GFP serum detected a protein of 53 kDa (uncleaved EYFP-ppVIII-HA fusion protein) in the cells cotransfected with plasmid pEY.ppVIII.HA and pDsRed-C1 DNAs (Fig. 3C, lane 1). In contrast, proteins of 53 kDa, 40 kDa (corresponding to the cleavage product of the EYFP-pVIII fusion protein consisting of EYFP and amino acids 1 to 111 of pVIII), 42 kDa (corresponding to the cleavage product of the EYFP-pVIII fusion protein consisting of EYFP and amino acids 1 to 131 of pVIII), and 45 kDa (corresponding to the cleavage product of the EYFP-pVIII fusion protein consisting of EYFP and amino acids 1 to 157 of pVIII) were detected in the lysates of the cells cotransfected with plasmid pEY.ppVIII.HA DNA and either plasmid pDR.bProt (lane 2), plasmid pDR.hProt (lane 3), or plasmid pDR.pProt (lane 4) DNA, indicating that PAdV-3 pVIII can also be cleaved at all three potential cleavage sites by proteases encoded by HAdV-5, BAdV-3, or PAdV-3 (Fig. 3C).
Construction of BAdV-3 expressing pVIII protein with an altered protease cleavage site(s).
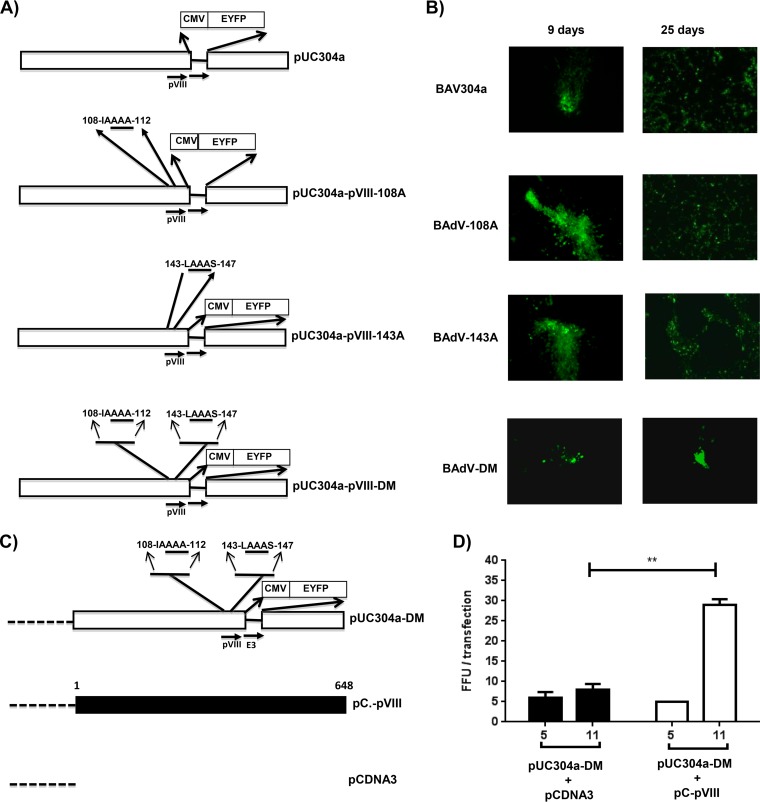
To determine the role of cleavage of pVIII protein in BAdV-3 replication, we decided to generate recombinant BAdV-3 expressing a protease cleavage site(s) mutant pVIII. To achieve this, we constructed three different plasmids containing a mutant full-length genomic clone of BAdV-3 (Fig. 4A). The plasmid pUC304a-pVIII-108A contained a BAdV-3 pVIII gene with substitutions of alanines for glycines at amino acids 110, 111, and 112 in potential protease cleavage site I of the protein. The plasmid pUC304a-pVIII-143A contained a BAdV-3 pVIII gene with substitutions of alanines for glycines at amino acids 144, 145, and 146 in potential protease cleavage site II of the protein. Similarly, plasmid pUC304a-pVIII-DM contained a BAdV-3 pVIII gene with substitutions of alanines for glycines at amino acids 110, 111, 112, 144, 145, and 146 in potential protease cleavage site I and site II of the protein.
FIG 4.
Isolation and characterization of mutant BAdV-3. (A). Schematic representation of plasmids. The BAdV-3 sequence is represented by an open box. The thin line represents the deleted E3 region (44). The alanines substituted for glycines are underlined. The numbers represent the amino acids of BAdV-3 pVIII; the human cytomegalovirus (CMV) immediate early promoter and EYFP (enhanced yellow fluorescent protein) are also indicated. Horizontal arrows indicate the direction of transcription. (B) Direct fluorescence. A monolayer of VIDO DT1 cells (22) was transfected with 4 to 6 μg of plasmid DNA of pUC304a (BAV304a), pUC304a-pVIII-108A (BAdV-108A), pUC304a-pVIII-143A (BAdV-143A), or pUC304a-pVIII-DM (BAdV-DM) and visualized at the indicated times posttransfection for the expression of EYFP and development of cytopathic effects using a TCS SP5 (Leica) fluorescence microscope. An increase in green fluorescence indicates that virus production could be seen after the 9th day posttransfection. (C). Schematic representation of plasmid pUC304a-DM. The dotted line represents the plasmid sequence. The BAdV-3 sequence is represented by an open box. The thin line represents the deleted E3 region (44). The alanines substituted for glycines are underlined. The numbers represent the amino acids of BAdV-3 pVIII; the human cytomegalovirus (CMV) immediate early promoter and EYFP (enhanced yellow fluorescent protein) are also shown. Horizontal arrows indicate the direction of transcription. The filled box represents the nucleotide sequence of BAdV-3 pVIII. The dotted line represents the nucleotide sequence of plasmid DNA. (D) Complementation assay. The VIDO DT1 cells (22) were cotransfected with the indicated plasmids, and fluorescent focus forming units were counted at the indicated day posttransfection (x axis). Values represent averages from two independent replicates, and error bars indicate the standard deviations. Statistical differences among the groups were calculated using an unpaired t test. **, P < 0.01.
To isolate mutant BAdV-3s, VIDO DT1 cells (cotton rat lung [CRL] cells expressing I-SceI protein) (22) were transfected with individual plasmid (pUC304a-pVIII-108A, pUC304a-pVIII-143A, or pUC304a-pVIII-DM) DNAs using Lipofectamine 2000 reagent and observed daily for the development of cytopathic effects (CPE). The cells transfected with either plasmid pUC304a-pVIII-108A or pUC304a-pVIII-143A DNA showed an increase in green fluorescence on the 9th day posttransfection, indicating virus production (Fig. 4B). The cells showing cytopathic effects were harvested at 25 days posttransfection, freeze-thawed three times, and used to purify virus (23). The virus was amplified in MDBK cells and purified using a cesium chloride density gradient. The purified viruses were lysed in 0.1% SDS, and absorbance at 260 nm was measured to calculate the physical particle yield.
The recombinant viruses were named BAdV-108A(pUC304a-pVIII-108A), with a ratio of particles to fluorescent focus units (FFU) of 425:1, and BAdV-143A(pUC304a-pVIII-143A) with a particle-to-FFU ratio of 645:1. The particle-to-FFU ratio of BAV304a (BAdV-3 containing the EYFP gene under control of the human cytomegalovirus immediate early promoter inserted in the E3 deletion BAdV-3 genome) was 65:1.
However, though fluorescent focus-forming units could be observed after day 9 posttransfection in VIDO DT1 cells transfected with plasmid pUC304a-pVIII-DM DNA, these fluorescent foci gradually disappeared without showing any observable cytopathic effect (Fig. 4B).
BAdV-3 pVIII complements the defect of BAV.pVIIIDM.
In order to determine if cleavage of pVIII using at least one of the two potential protease consensus sites is essential for viral replication, we performed a complementation assay. VIDO DT1 cells in six-well plates were cotransfected with plasmid DNAs (pUC304a-pVIII-DM and pC-pVIII DNAs or pUC304a-pVIII-DM and pCDNA3 DNAs) (Fig. 4C). The transfected cells were observed daily under a fluorescence microscope for the appearance of fluorescent focus-forming units. As seen in Fig. 4D, cotransfection of VIDO DT1 cells with plasmid pUC304a-pVIII-DM and pCDNA3 DNAs did not produce any progeny virus, as indicated by the absence of any increase in the number of FFU from day 5 to day 11 posttransfection. In contrast, cotransfection of VIDO DT1 cells with plasmid pUC304a-pVIII-DM and pC-pVIII DNAs produced progeny virus, as indicated by the significant increase in the number of FFU from day 5 to day 11 posttransfection (Fig. 4D).
Virus growth.
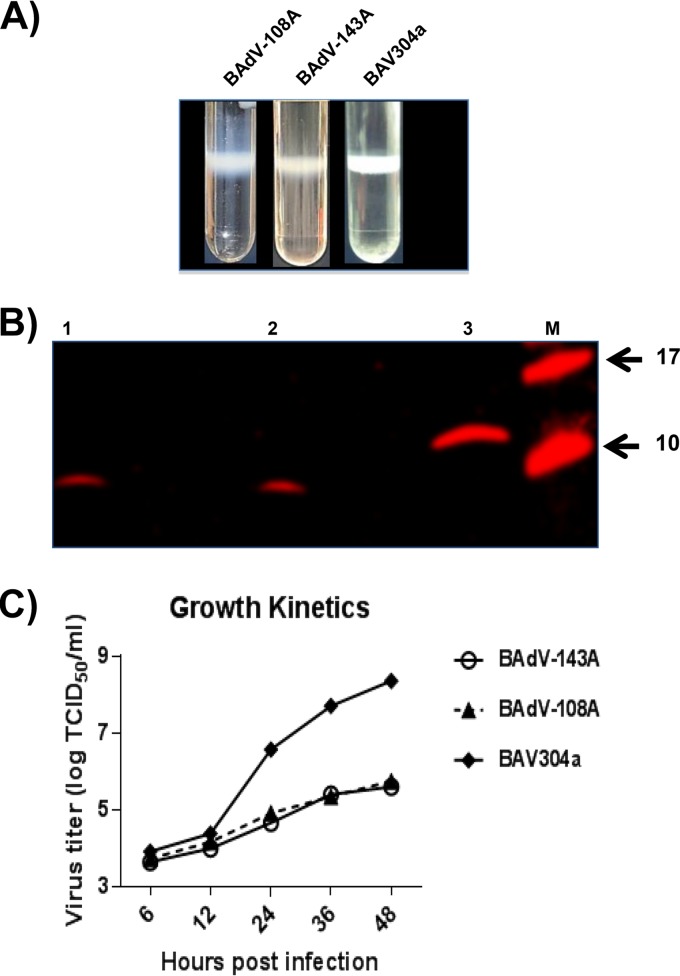
The BAV304a, BAdV-108A, and BAdV-143A viruses were propagated in MDBK cells and purified by double CsCl density gradients (Fig. 5A). To determine the incorporation of pVIII in purified virions, proteins from purified virions were separated by 4 to 20% gradient SDS-PAGE gels, transferred to nitrocellulose, and probed by Western blotting using anti-pVIIIb serum. As expected (Fig. 5B), anti-pVIIIb serum detected a protein of 8 kDa in purified BAV304a (lane 1) and BAdV-108A (lane 2) and a protein of 12 kDa in purified BAdV-143A (lane 3).
FIG 5.
Virus growth. (A) CsCl purification. Virus was purified from infected cell lysates by CsCl density gradient purification. Mature virion bands after double density gradient centrifugation are shown (B). Western blotting. Proteins from purified BAV304a, BAdV-108A, or BAdV-143A were separated on 4 to 20% gradient SDS-PAGE gels, transferred to nitrocellulose, and probed by Western blotting using anti-pVIIIb serum (17). (C) Virus titer. The infected MDBK cells were harvested at the indicated times postinfection and freeze-thawed, and virus was titrated on MDBK cells.
To determine if the mutation of protease cleavage sites of pVIII affects BAdV-3 replication, we compared the growth kinetics of BAV304a, BAdV-108A, and BAdV-143A. The monolayers of MDBK cells in 24-well plates were infected with CsCl2-purified BAV304a, BAdV-108A, or BAdV-143A at a multiplicity of infection (MOI) of 1. At different times postinfection (6, 12, 24, 36, and 48 h), the infected cells were collected and freeze-thawed three times, and virus in the cell lysate was titrated by 50% tissue culture infective dose (TCID50) in MDBK cells (24). As seen in Fig. 5C, BAV304a grew to a titer of 108.4 TCID50s/ml. However, BAdV-108A and BAdV-143A could only grow to titers of 105.75 and 105.6 TCID50s/ml, respectively.
Virus infectivity and genome replication.
To determine the infectivity of mutant viruses, GFP expression was used as an indicator to determine the virus infectivity in a single-round infection assay. As seen in Fig. 6A, there was no significant difference in the number of GFP-expressing cells when MDBK cells infected with equal amounts of infectious virus were analyzed at 18 h postinfection (hpi). The particle-to-FFU ratio was 8- to 12-fold higher for BAdV-108A and BAdV-143A than for BAV304a.
FIG 6.
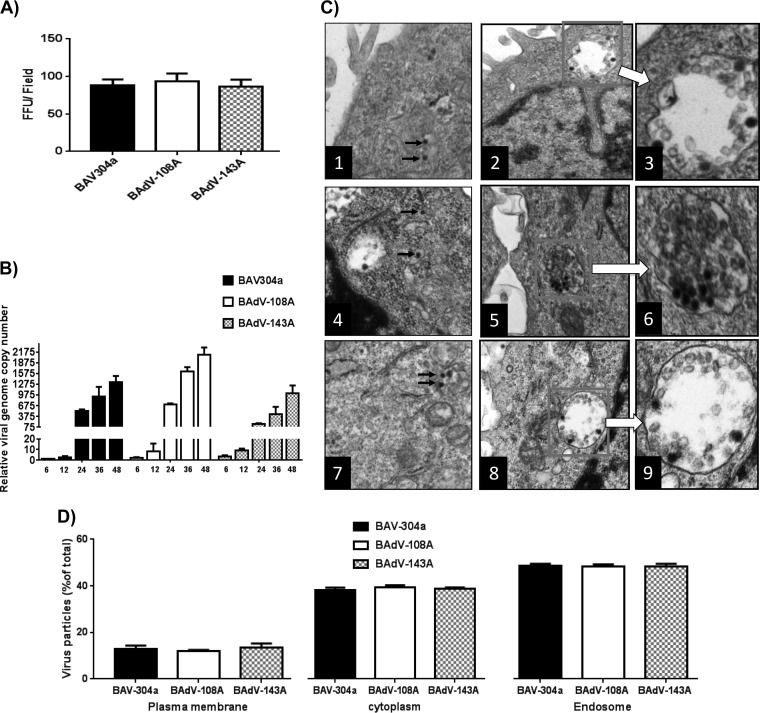
Analysis of mutant BAdV-3 viruses. (A) Virus infectivity. The MDBK cells were infected with equivalent amounts of infectious particles of the indicated viruses in triplicate. The infected cells were visualized for the expression of GFP at 18 h postinfection by a TCS SP5 (Leica) fluorescence microscope. (B) Viral genome replication. The MDBK cells were infected with equivalent amounts of infectious particles of the indicated virus in triplicate. At indicated times postinfection, the infected cells were collected, and genomic DNA was isolated. The viral genome copy number was determined by quantitative PCR and divided by actin copy number for normalization. For comparison, the normalized genome copy number values for each virus at each time point were compared to the value of wild-type virus at 6 hpi. (C) Subcellular distribution of BAdV-3. Monolayers of MDBK cells (1 × 106 cells/well) were incubated with 1.4 × 107 purified virions at 4°C. After 1 h of incubation, the cells were incubated at 37°C for 30 min. Finally, the cells were processed and visualized by transmission electron microscopy: frames 1 to 3, BAV304a; frames 4 to 6, BAdV-108A; frames 7 to 9, BAdV-143A. Boxed areas are enlarged, as indicated. (D) For each virus, 10 cells were selected randomly, and virus particles in endosomes, cytoplasm, and at plasma membrane were counted. Error bars indicate standard errors of the means of three independent experiments.
To determine if the alteration of protease cleavage of pVIII modulates the replication of mutant viruses, we determined the mutant viral DNA replication by quantitative real-time PCR as described in Materials and Methods. As seen in Fig. 6B, during the early phase of infection (6 hpi and 12 hpi) virus genome replication levels appeared similar in different viruses. However, during the late phase of infection (24 hpi and 36 hpi) BAdV-108A replicated marginally better than BAV304a or BAdV-143A. Similarly, BAV304a replicated marginally better than BAdV-143A.
To determine if the alteration of protease cleavage of pVIII modulates the release of the virus from endosomes, MDBK cells infected with equal amounts of individual infectious viruses were analyzed by transmission electron microscopy (TEM) after 90 min of infection. As seen in Fig. 6C, no significant difference could be observed in the number of virus particles in the endosome or cytosol between BAV304a, BAdV-108A, and BAdV-143A virus particles at 90 min postinfection.
Thermostability of virus.
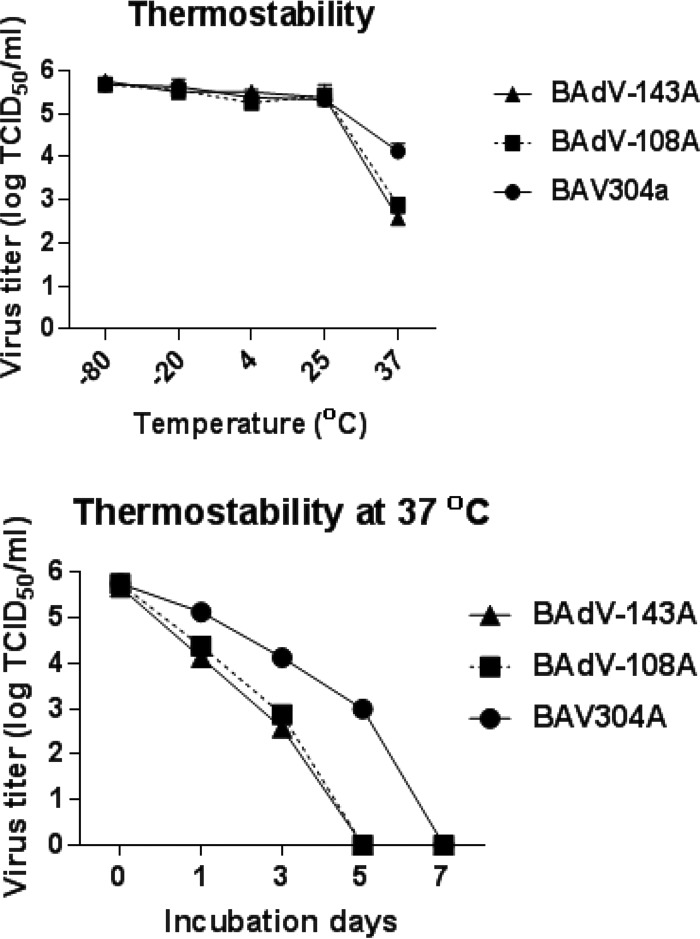
To determine if mutation of the protease cleavage site of the pVIII protein alters the stability of mutant BAdV-3, we compared the thermostabilities of BAV304a, BAdV-108A, and BAdV-143A. About 105 purified virus particles were incubated at different temperatures (−80°C, −20°C, 4°C, 25°C, and 37°C) for 3 days in phosphate-buffered saline (PBS) containing 10% glycerol. Finally, the infectivity of viable virus was determined by TCID50 assay. As seen in Fig. 7A, there was not much decrease in virus titers when viruses were incubated at −80°C, −20°C, 4°C, and 25°C for 3 days. However, the virus titers decreased significantly when viruses were incubated at 37°C for 3 days. The drop in the titer was greater for BAdV-108A and BAdV-143A than for BAV304a (Fig. 7A). To further assess the thermostability, 105 purified virus particles were incubated at 37°C for 0, 1, 3, 5, and 7 days in PBS containing 10% glycerol before the remaining infectivity was measured by TCID50 assay. As seen in Fig. 7B, when viruses were incubated at 37°C, more rapid loss of infectivity was observed for BAdV-108A and BAdV-143A. While BAdV-108A and BAdV-143A lost infectivity within 5 days, BAV304a lost infectivity only at 7 days postincubation at 37°C.
FIG 7.
Thermostability of mutant virions. (A) Purified virions (105 TCID50) grown in MDBK cells were incubated at various temperatures for 3 days in PBS containing 10% glycerol, and the residual viral infectivity was determined by titration on MDBK cells. (B) Purified virions (105 TCID50) grown in MDBK cells were incubated at 37°C for the indicated periods of time in PBS containing 10% glycerol, and the residual viral infectivity was determined by titration on MDBK cells.
Analysis of protein expression in infected cells.
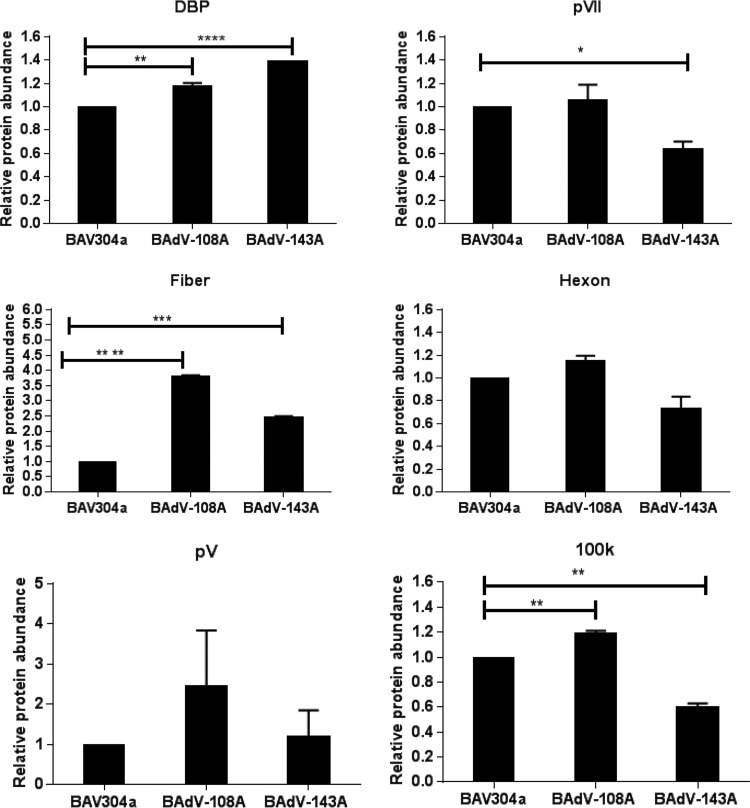
To analyze if there is any difference in expression levels of viral proteins, MDBK cells were infected with BAV304a and BAdV-108A or BAdV-143A. After 24 h postinfection, the cells were collected and lysed, and the lysates were analyzed by Western blotting using protein-specific rabbit antiserum. The blots were analyzed by using an Odyssey Infrared Imaging System. As seen in Fig. 8, no appreciable decrease could be detected in the expression of viral proteins in BAdV-108A-infected cells compared to levels in BAV304a-infected cells. Interestingly, expression levels of all tested proteins appeared increased in BAdV-108A-infected cells. In contrast, although expression levels of an early viral protein (DNA binding protein [DBP]) and late protein fiber appear increased in BAdV-143A-infected cells compared to levels in BAV304a-infected cells, the expression levels of some of the viral late proteins (pVII, 100K, and hexon) appear reduced in BAdV-143A-infected cells.
FIG 8.
Analysis of viral protein expression in infected cells. Proteins from the lysates of MDBK cells were separated by 4 to 20% gradient SDS-PAGE gels, transferred to nitrocellulose membranes, and probed by Western blotting using anti-DNA binding protein (DBP) (36), anti-pVII (37), anti-V (38), anti-hexon (39), and anti-100K (9) serum followed by Alexa Fluor 680-conjugated goat anti-rabbit antibody (Invitrogen). The results were analyzed by using an Odyssey Infrared Imaging System. Values represent averages from two independent replicates, and error bars indicate standard deviations. All data were analyzed using GraphPad Prism, version 6 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical differences among the groups were calculated using an unpaired t test, and significance is indicated as follows: *, P < 0.05; **, P < 0.01, ***, P < 0.001; ****, P < 0.0001.
Analysis of protein incorporation in viral particles.
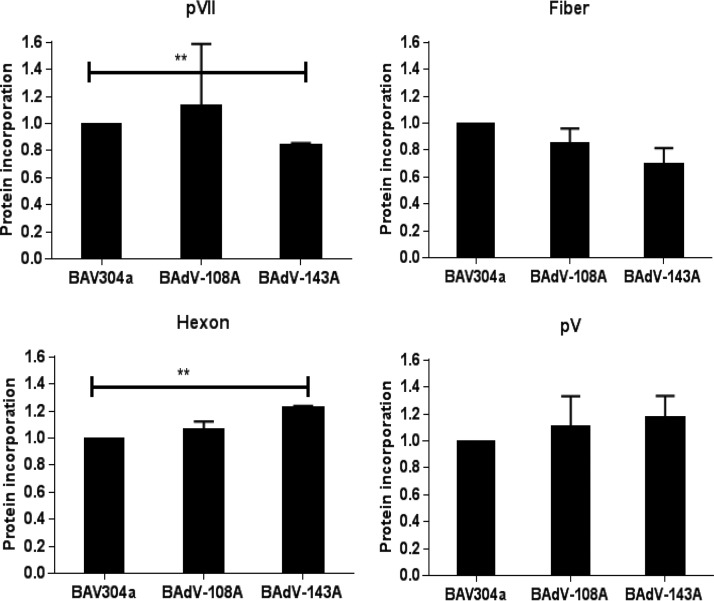
To analyze the incorporation of viral proteins in the progeny virions, the proteins from purified virions were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed in Western blotting using viral protein-specific serum. The blots were analyzed by using an Odyssey Infrared Imaging System. As seen in Fig. 9, compared to BAV304a, no significant reduction could be observed in the incorporation of the viral proteins, except pVII, in purified BAdV-108A or BAdV-143A virions.
FIG 9.
Analysis of viral protein incorporation in purified virions. Proteins from virions of BAV304a, BAdV-108A, or BAdV-143A subjected to two round of CsCl purification were separated by 4 to 20% gradient SDS-PAGE gels, transferred to nitrocellulose, and probed by Western blotting using protein-specific antiserum. All data were analyzed using GraphPad Prism, version 6 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical differences among the groups were calculated using an unpaired t test. **, P < 0.01.
Transmission electron microscopy.
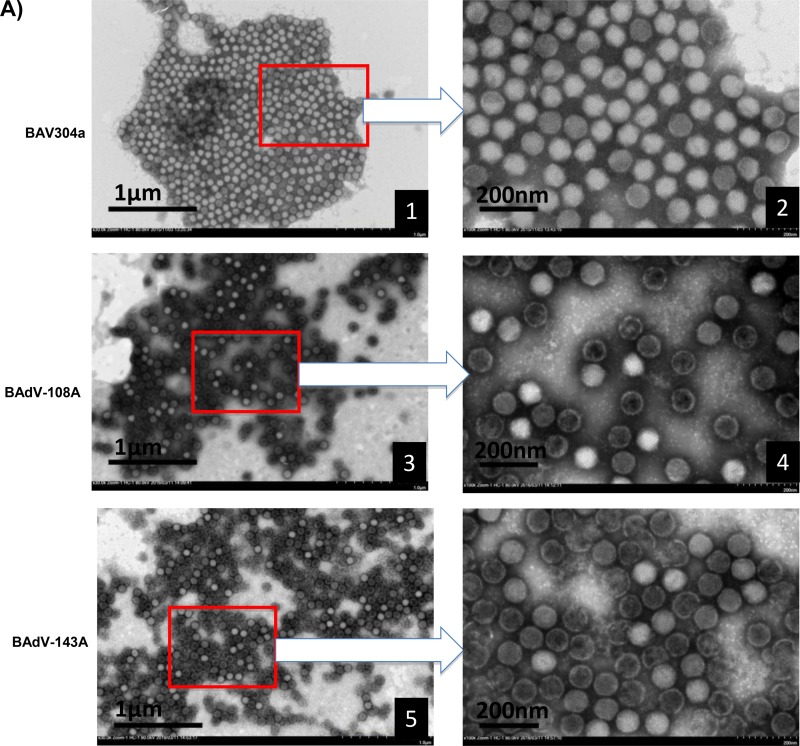
To assess whether mutation of a single protease cleavage site of the pVIII protein has any effect on virus structure, BAV304a, BAdV-108A, or BAdV-143A was propagated in MDBK cells and purified by CsCl density gradient (Fig. 5A) as described in Materials and Methods. The virus particles were then observed by negative-staining electron microscopy. As seen in Fig. 10A, the BAV304a particles (frames 1 and 2) had typical icosahedral structures, and most of them were intact. However, unlike the BAV304a particles, virus particles of BAdV-108A (frames 3 and 4) or BAdV-143A (frames 5 and 6) appeared more rounded, with disrupted capsids.
FIG 10.
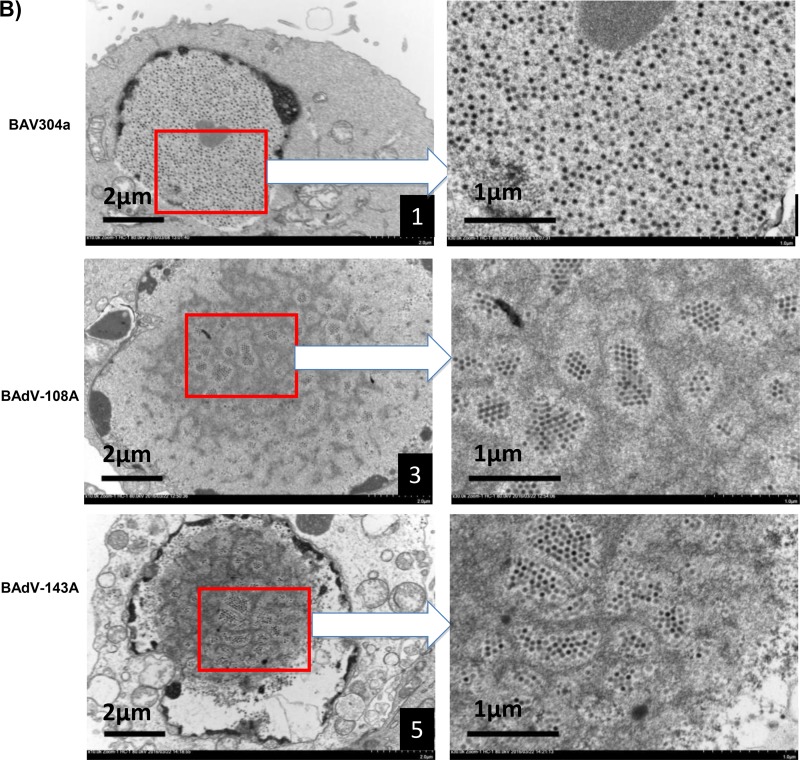
Electron microscopic analysis. (A) Purified BAV304a, BAdV-108A, or BAdV-143A is shown, as indicated, at a magnification of ×30,000. The arrows indicate enlargements of selected boxed regions of each virus (magnification, ×1,000,000). (B) MDBK cells infected with BAV304a, BAdV-108A, or BAdV-143A, as indicated, are shown at a magnification of ×10,000. The arrows indicate the enlargements of selected boxed regions at a magnification of ×30,000.
To further assess if the mutation of a single protease cleavage site of the pVIII protein affects the formation of BAdV-3 particles, transmission electron microscopy was performed on MDBK cells infected with BAV304a, BAdV-108A, or BAdV-143A. The infected cells were harvested at 48 h postinfection and examined by transmission electron microscopy. As seen in Fig. 10B, BAV304a particles were uniformly distributed and loosely arranged (frames 1 and 2). However, BAdV-108A (frames 3 and 4) and BAdV-143A (frames 5 and 6) particles were clustered together in groups and appeared tightly organized in rows.
DISCUSSION
The final step in adenovirus morphogenesis involves the maturation of immature virus particles by proteolytic cleavage of three minor capsid proteins, three core proteins, and one nonstructural protein (8, 18). The major player in this complex process is a 23-kDa adenovirus protease, which recognizes consensus motifs (M/I/L)XGX-G and (M/I/L)XGG-X in precursor proteins (4). Two cofactors, viral DNA and a C-terminal 11-amino-acid peptide of adenovirus protein pVI, are required for the optimal activation of protease (18). In addition, the cellular protein actin can act as a cofactor for adenovirus proteinase (AVP) (25). Much of the knowledge about the requirement of proteolytic cleavage of some precursor proteins for maturation of adenovirus has come from studies involving a temperature-sensitive mutant of HAdV-C2 (5). However, the importance of cleavage of each precursor protein in determining the infectivity of progeny virus is not clear (18). A recent study has demonstrated the importance of N-terminal cleavage of pVI in cell entry and capsid assembly (19). Thus, a better understanding of this complex process of proteolytic maturation of adenovirus is required. Here, we have analyzed the role of cleavage of pVIII in BAdV-3 virus assembly, stability, and infection. To our knowledge, this is the first study to investigate the role of proteolytic cleavage of pVIII in the adenoviral life cycle.
Analysis of BAdV-3 pVIII amino acid sequence revealed two potential protease cleavage sites (108IAGG-G112 and 143LGGG-S147). Our results suggest that the BAdV-3 protease cleaves BAdV-3 pVIII at both potential protease cleavage sites, a process which appears specific as mutation of protease cleavage sites abrogated the cleavage of pVIII. Moreover, cleavage at both potential sites does not appear to be sequential since mitigating cleavage at the 108IAGG-G112 motif does not block cleavage at the 143LGGG-S147 motif and vice versa. An earlier report has suggested the usage of only one potential protease cleavage site (143LGGG-S147) of pVIII (17). The difference in this previous report could be due to use of a different approach. Similarly, analysis of HAdV-5 pVIII revealed three potential adenoviral protease cleavage sites (amino acids 111, 131, and 157). Our results suggest that HAdV-5 protease can cleave pVIII at all three potential protease cleavage sites. Earlier studies have suggested the usage of only two potential cleavage sites, at amino acid 111 and amino acid 157, of HAdV-5 pVIII (12, 26–28). Although maturation of progeny adenovirus particles requires cleavage of adenovirus proteins, including pVIII, to occur in the nucleus of virus-infected cells (8, 18), the cleavage of adenovirus proteins (9) including pVIII (this study) appears to occur in the cytoplasm of the transfected cells.
Analysis of the amino acid sequences revealed a high degree of homology between proteases encoded by BAdV-3, HAdV-2, and PAdV-3 (16). Moreover, the conservation of residues involved in the catalytic activity among proteases encoded by BAdV-3, HAdV-5, and PAdV-3 (16) suggested that the mechanism of cleavage of proteins harboring the potential cleavage site(s) might be conserved in mastadenoviruses. Interestingly, detection of cleavage of pVIII encoded by BAdV-3, HAdV-5, or PAdV-3 by an individual protease encoded by BAdV-3, HAdV-5, or PAdV-3 at potential protease cleavage sites provides evidence that the mechanism of cleavage of proteins appears conserved in the mastadenoviruses.
However, we detected differences in the efficiencies of usage of protease cleavage sites between the HAdV-5, BAdV-3, and PAdV-3 proteases. This could be due to a difference in the type of potential protease cleavage site (Fig. 1, GX-G type and GG-X type). Earlier reports have suggested that efficiency of cleavage by adenovirus protease is dependent upon the amino acid sequence at the cleavage site. The cleavage site sequence that conforms to the GX-G type is cleaved more efficiently than that of the GG-X type (29). Moreover, the four amino acids on either side of the scissile bond determine the specificity of adenovirus protease-mediated cleavage (4, 21). Our results confirm these findings as BAdV-3 pVIII was cleaved more efficiently at protease potential cleavage site 1 (PPCS1; GX-G type) than at PPCS2 (GG-X type). Interestingly, there appears to be some difference in cleavage site preferences between the HAdV-5, BAdV-3, and PAdV-3 proteases as each of them cleaved different HAdV-5 pVIII cleavage sites with different efficiencies.
Although abrogation of protease cleavage at either potential cleavage site (PPCS1 or PPCS2) affects the efficient production of infectious progeny virions, the protease cleavage at individual sites does not appear to be essential for the production of progeny BAdV-3 virus. However, alteration of both protease cleavage sites (PPCS1, amino acids 110 to 112; PPCS2, amino acids 146 to 148) of BAdV-3 pVIII by replacement of glycines with alanines obviated the production of progeny virus, suggesting that the absence of protease cleavage of pVIII is lethal for the production of progeny BAdV-3. These results are consistent with the suggestion that the function of each protease cleavage site of BAdV-3 pVIII in viral replication may be redundant.
The cleavage of a few precursor proteins including pVIII has been postulated to be required for the production of infectious progeny virions (30). Absence of cleavage of these precursor proteins appears to make virions noninfectious as this partially alters the uncoating and release of virions from the endosome (31–33). In our experiments, mutant viruses produced significantly fewer infectious virus particles than BAV304a when cells were infected with equal numbers of infectious virus particles. The defect could be in different steps of virus gene expression (receptor binding to virus assembly) including endosome escape. Several lines of evidence suggest that capsid formation and virus assembly are occurring in mutant viruses. First, our quantitative TEM analysis suggests that the absence of cleavage at individual protease cleavage sites of BAdV-3 pVIII does not appear to impair the release of partially uncoated BAdV-3 from the endosome(s). Second, there is no significant difference in genome replication rates of mutant virus genomes. Third, alteration of individual protease cleavage sites does not significantly affect the incorporation of proteins in mutant virus capsids. Finally, capsid formation and virus assembly appeared to occur as progeny virions produced in infected cells banded at the CsCl gradient density, which is consistent with the formation of mature virions.
Earlier structural studies have revealed that mature HAdV-5 virions contain N-terminal (112 residues) and C-terminal (70 residues) proteolytic fragments of pVIII which interact with the bases of hexons (15). While the ternary complex of V-VI-VIII stabilizes the peripentonal hexons, the binary complex VIII-VIB contributes to the stabilization of a group of nine hexons (GONs) (15), providing help in maintaining the integrity of the capsids. Thus, a significant decrease in the production of infectious BAdV-108A and BAdV-143A viruses could be due to the production of less stable virions. Indeed, our results confirm the role of pVIII in stabilization of virion capsid. First, the altered proteolytic processing of the pVIII of BAdV-108A or BAdV-143A resulted in the production of thermolabile virions with fragile capsids, which may be disrupted by CsCl/TEM, leading to the production of significant numbers of broken capsids. Second, cleavage of pVIII at both potential cleavage sites appears essential for maintaining the integrity and stability of BAdV-3 virions. However, it is not clear whether virion capsid instability is due to loss of direct interactions between pVIII and other capsid proteins (15, 17) or to inefficient packaging of virion DNA (34).
In conclusion, we have demonstrated that inactivation of both protease cleavage sites of pVIII is detrimental for progeny virus production. Moreover, single pVIII protease cleavage site mutants assemble virus particles. However, these progeny virus particles appear thermolabile and may lead to the production of noninfectious virions with broken capsids.
MATERIALS AND METHODS
Cell lines and viruses.
Madin-Darby bovine kidney (MDBK) cells (24), cotton rat lung (CRL) cells (35), and VIDO DT1 cells (cotton rat lung cells expressing endonuclease I-SceI) (22) were grown in minimum essential medium (MEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS). 293T cells (ATCC CRL-3216) were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS. BAV304a (BAdV-3 containing the EYFP gene under control of the human cytomegalovirus immediate early promoter inserted in the E3-deleted region of the BAdV-3 genome [22]) and mutant BAdV-3 viruses were propagated in MDBK cells in MEM supplemented with 2% fetal bovine serum.
Antibodies.
Polyclonal anti-pVIIIb (pVIII peptide, residues 187 to 216) serum recognizes a protein of 24 kDa and 8 kDa in BAdV-3-infected cells (17). Production and characterization of serum recognizing DNA binding protein (DBP) (36), pVII (37), polypeptide V (38), hexon (39), and 100K (9) of BAdV-3 have been described previously. Anti-GFP antibody (Cell Signaling), Alexa Fluor 680-conjugated goat anti-rabbit antibody (Invitrogen), anti-β-actin monoclonal antibody (Sigma-Aldrich), IRDye 800-conjugated goat anti-mouse antibody (Rockland), and alkaline phosphatase (AP)-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) were purchased.
Plasmid construction.
Construction of plasmids pDR.bProt, pDR.hProt, and pDR.pProt has been described earlier (9). Plasmid pUC304a contains the BAV304a genome flanked by I-SceI endonuclease recognition sites (22). The other plasmids were constructed using standard DNA manipulation techniques (data not shown). The information will be provided on request.
Western blotting.
Proteins from purified virus, virus-infected cell lysates, or plasmid DNA transfected cell lysates were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), transferred to polyvinylidene difluoride (PVDF) or nitrocellulose membrane (Bio-Rad), and probed by Western blotting using protein-specific antiserum and Alexa Fluor 680- or IRDye 800-conjugated antibodies or alkaline phosphatase (AP)-conjugated goat anti-rabbit IgG (Sigma).The membranes probed with fluorophore-conjugated secondary antibody were scanned and analyzed by an Odyssey CLx Imaging System (Li-Cor).
Isolation of mutant BAdV-3.
Monolayers of VIDO DT1 cells (22) in six-well plates were transfected with 4 to 6 μg of individual plasmid DNA using Lipofectamine 2000 reagent (Invitrogen). At 4 h posttransfection, the medium was replaced with fresh MEM containing 2% FBS. The cells were observed daily for appearance of any cytopathic effects (CPE). On appearance of CPE, the cells were harvested and freeze-thawed three times before being used for reinfection of fresh MDBK cells
CsCl gradient centrifugation.
The CsCl gradient centrifugation was carried out as described earlier (23). Briefly, confluent monolayers of MDBK cells were infected with wild-type or recombinant virus. The cells showing more than 80% cells CPE were collected, pelleted by centrifugation, and resuspended in 5 ml of medium. The cell lysate was freeze-thawed five times and subjected to CsCl density gradient centrifugation at 35,000 rpm for 1 h at 4°C. The band containing virus was collected and subjected to a second round of CsCl density gradient centrifugation at 35,000 rpm for 16 h at 4°C. The virus was collected, dialyzed against three changes of dialysis buffer to remove traces of cesium chloride, and stored in small aliquots at −80°C.
Virus growth kinetics.
MDBK cells in 24-well plates were infected with either BAV304a, BAdV-108A, or BAdV-143A at an MOI of 1. Infected cells were then collected at different time points (0, 6, 12, 24, 36, and 48 h) postinfection and freeze-thawed three times, and virus in cell lysate was titrated by TCID50 assay in MDBK cells (24).
Virus thermostability assay.
The virus thermostability assay was performed as described earlier (40). Briefly, about 105 purified virus particles were incubated at different temperatures (−80°C, −20°C, 4°C, 25°C, and 37°C) for 3 days in PBS containing 10% glycerol. Finally, the infectivity of viable virus was determined by TCID50 assay. To further assess the thermostability, 105 purified virus particles were incubated at different temperatures (−80°C, 4°C, and 37°C) for 0, 1, 3, and 7 days in PBS containing 10% glycerol, and a TCID50 assay was performed to determine the remaining infectivity.
Transmission electron microscopy.
To analyze CsCl gradient-purified BAdV-3 virions by TEM, 300-mesh grids (catalog number G300; Ted Pella) that were previously covered with 0.25% Formvar, followed by carbon coating (Denton Vacuum, Inc.), were suspended for 2 min onto a 20-μl droplet containing CsCl gradient-purified virus particles and fixed with 5% electron microscopy-grade glutaraldehyde (catalogue number 16316-10; Electron Microscopy Sciences). After three washes with water, the grids were suspended on a droplet of 0.5% phosphotungstic acid (J. B. EM Services, Inc. Dorval, QC, Canada) for 1 min. The excess solution was removed, and the grids were allowed to dry. Finally, the grids with negatively stained virus particles were examined on an HT7700 transmission electron microscope (Hitachi, Inc.) at 80 kV.
To analyze BAdV-3-infected cells by TEM, the monolayers of MDBK cells were infected with BAV304a, BAdV-108A, or BAdV-143A. At 48 h postinfection, the cells were harvested and fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer at 4°C. Following fixation, cells were pelleted in 1% low-melting-point agarose, postfixed at room temperature in 1% OsO4 in 0.1 M sodium cacodylate buffer, and stained en bloc with saturated uranyl acetate in 70% ethanol. Dehydration was carried out with a graded ethanol series and finalized in propylene oxide (catalog number 20401; Electron Microscopy Sciences). Dehydrated cells were infiltrated with Epon 812/Araldite embedding medium by gradually increasing concentrations in propylene oxide and then polymerized in molds with pure Epon/Araldite at 60°C for 24 h. Finally, the pellet was sectioned with a Reichert Ultracut ultramicrotome, and each section was stained with Reynolds lead citrate and 2% uranyl acetate and viewed on an HT7700 transmission electron microscope (Hitachi, Inc.) at 80 kV.
Fluorescent focus assay.
The monolayers of MDBK cells grown in 12-well plates were infected with individual viruses (at an MOI of 2). After 18 h, GFP-positive cells/foci were counted for each well using a fluorescence microscope. The average number of GFP-positive cells was quantified as the number of fluorescent focus units (FFU) per field.
TEM analysis of subcellular distribution of purified viruses.
The monolayers of MDBK cells in one well of a six-well plate were incubated with individual viruses at an MOI of 10 at 4°C. After 1 h of incubation, the cells were washed before being incubated again at 37°C. After incubation for 30 min, the cells were then processed for TEM analysis as described earlier (24).
Viral genome replication.
Viral genome replication was measured as described previously (41), with minor modifications. Briefly, MDBK cells were infected with BAV304a, BAdV-108A, or BAdV-143A and harvested at the times postinfection indicated on Fig. 6B. The infected cells were pelleted, washed once with phosphate-buffered saline (PBS), and then resuspended in NP-40 buffer (150 mM NaCl, 10 mM Tris [pH 7.5],1.5 mM MgCl2, 0.6% NP-40). After the cells were incubated on ice for 10 min, the nuclei were then pelleted by centrifugation at 2,000 × g at 4°C for 5 min. The pelleted nuclei were resuspended in 200 μl of PBS and used for DNA extraction using a DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions. Finally, the DNA was resuspended in double-distilled water and subjected to quantitative PCR using a Bio-Rad real-time PCR system. The two sets of primers used for quantitative PCR were the primer pair Q adeno Fwd and Q adeno Rev and the primer pair Actin Fwd and Actin Rev (Table 1). Genome replication was calculated as the value of viral genome copy number divided by actin copy number. For comparison, the value of each virus was normalized to that of BAV304a at 6 hpi and is shown as relative viral genome copy number.
TABLE 1.
List of primers
| Primer name | Primer sequence |
|---|---|
| Q adeno Fwd | CAGGTGCCAGTCAAGATTAC |
| Q adeno Rev | ATGGCCGACTGAGTCATAAG |
| Actin Fwd | CTAGGCACCAGGGCGTAATG |
| Actin Rev | CCACACGGAGCTCGTTGTAG |
Statistical analysis.
All data were analyzed using GraphPad Prism, version 6 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical differences among the groups were calculated using an unpaired t test. P values are indicated in the figure legends.
ACKNOWLEDGMENTS
We thank members of the Adenovirus Vectored Vaccines Laboratory for their help and suggestions and staff of the animal care unit for production of antiserum.
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to S.T.
Footnotes
This article is VIDO article 786.
REFERENCES
- 1.Berk AJ. 2007. Adenoviridae: the viruses and their replication, p 2355–2395. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Campos SK, Barry MA. 2007. Current advances and future challenges in adenoviral vector biology and targeting. Curr Gene Ther 7:189–204. doi: 10.2174/156652307780859062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatsis N, Ertl HC. 2004. Adenoviruses as vaccine vectors. Mol Ther 10:616–629. doi: 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster A, Russell S, Talbot P, Russell Wt, Kemp G. 1989. Characterization of the adenovirus proteinase: substrate specificity. J Gen Virol 70:3225–3234. doi: 10.1099/0022-1317-70-12-3225. [DOI] [PubMed] [Google Scholar]
- 5.Weber J. 1976. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J Virol 17:462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangel WF, Toledo DL, Brown MT, Martin JH, McGrath WJ. 1996. Characterization of three components of human adenovirus proteinase activity in vitro. J Biol Chem 271:536–543. doi: 10.1074/jbc.271.1.536. [DOI] [PubMed] [Google Scholar]
- 7.Challberg MD, Kelly TJ Jr. 1981. Processing of the adenovirus terminal protein. J Virol 38:272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez-Berna AJ, Mangel WF, McGrath WJ, Graziano V, Flint J, San Martin C. 2014. Processing of the l1 52/55k protein by the adenovirus protease: a new substrate and new insights into virion maturation. J Virol 88:1513–1524. doi: 10.1128/JVI.02884-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makadiya N, Gaba A, Tikoo SK. 2015. Cleavage of bovine adenovirus type 3 non-structural 100K protein by protease is required for nuclear localization in infected cells but is not essential for virus replication. J Gen Virol 96:2749–2763. doi: 10.1099/vir.0.000205. [DOI] [PubMed] [Google Scholar]
- 10.Rohn K, Prusas C, Monreal G, Hess M. 1997. Identification and characterization of penton base and pVIII protein of egg drop syndrome virus. Virus Res 47:59–65. doi: 10.1016/S0168-1702(96)01407-4. [DOI] [PubMed] [Google Scholar]
- 11.Chroboczek J, Bieber F, Jacrot B. 1992. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology 186:280–285. doi: 10.1016/0042-6822(92)90082-Z. [DOI] [PubMed] [Google Scholar]
- 12.Chelius D, Hühmer AF, Shieh CH, Lehmberg E, Traina JA, Slattery TK, Pungor E. 2002. Analysis of the adenovirus type 5 proteome by liquid chromatography and tandem mass spectrometry methods. J Proteome Res 1:501–513. doi: 10.1021/pr025528c. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi E, Cohen SL, Tsai P, Sweeney JA. 2006. Quantitation of adenovirus type 5 empty capsids. Anal Biochem 349:208–217. doi: 10.1016/j.ab.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy VS, Nemerow GR. 2014. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci U S A 111:11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy PS, Idamakanti N, Zakhartchouk AN, Baxi MK, Lee JB, Pyne C, Babiuk LA, Tikoo SK. 1998. Nucleotide sequence, genome organization, and transcription map of bovine adenovirus type 3. J Virol 72:1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayalew LE, Gaba A, Kumar P, Tikoo SK. 2014. Conserved regions of bovine adenovirus-3 pVIII contain functional domains involved in nuclear localization and packaging in mature infectious virions. J Gen Virol 95:1743–1754. doi: 10.1099/vir.0.065763-0. [DOI] [PubMed] [Google Scholar]
- 18.Mangel WF, San Martín C. 2014. Structure, function and dynamics in adenovirus maturation. Viruses 6:4536–4570. doi: 10.3390/v6114536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyer CL, Besser ES, Nemerow GR. 2015. A single maturation cleavage site in adenovirus impacts cell entry and capsid assembly. J Virol 90:521–532. doi: 10.1128/JVI.02014-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruzindana-Umunyana A, Sircar S, Weber JM. 2000. The effect of mutant peptide cofactors on adenovirus protease activity and virus infection. Virology 270:173–179. doi: 10.1006/viro.2000.0253. [DOI] [PubMed] [Google Scholar]
- 21.Ruzindana-Umunyana A, Imbeault L, Weber JM. 2002. Substrate specificity of adenovirus protease. Virus Res 89:41–52. doi: 10.1016/S0168-1702(02)00111-9. [DOI] [PubMed] [Google Scholar]
- 22.Du E, Tikoo SK. 2010. Efficient replication and generation of recombinant bovine adenovirus-3 in nonbovine cotton rat lung cells expressing I-SceI endonuclease. J Gene Med 12:840–847. doi: 10.1002/jgm.1505. [DOI] [PubMed] [Google Scholar]
- 23.Tollefson AE, Kuppuswamy M, Shashkova EV, Doronin K, Wold WSM. 2007. Preparation and titration of CsCl-banded adenovirus stocks, p 223–235. In Wold WSM, Tollefson AE (ed), Adenovirus methods and protocols. Adenoviruses, Ad vectors, quantitation, and animal models, 2nd ed, vol 1 Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 24.Kulshreshtha V, Babiuk LA, Tikoo SK. 2004. Role of bovine adenovirus-3 33K protein in viral replication. Virology 323:59–69. doi: 10.1016/j.virol.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 25.Brown MT, McBride KM, Baniecki ML, Reich NC, Marriott G, Mangel WF. 2002. Actin can act as a cofactor for viral proteinase in the cleavage of the cytoskeleton. J Biol Chem 277:46298. doi: 10.1074/jbc.M202988200. [DOI] [PubMed] [Google Scholar]
- 26.Lehmberg E, Traina JA, Chakel JA, Chang R, Parkman M, McCaman MT, Murakami PK, Lahidji V, Nelson JW, Hancock WS. 1999. Reversed-phase high-performance liquid chromatographic assay for the adenovirus type 5 proteome. J Chromatogr B Biomed Sci Appl 732:411–423. doi: 10.1016/S0378-4347(99)00316-3. [DOI] [PubMed] [Google Scholar]
- 27.Blanche F, Monegier B, Faucher D, Duchesne M, Audhuy F, Barbot A, Bouvier S, Daude G, Dubois H, Guillemin T. 2001. Polypeptide composition of an adenovirus type 5 used in cancer gene therapy. J Chromatogr A 921:39–48. doi: 10.1016/S0021-9673(01)00896-2. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Vellekamp G, Chen G, Mirza UA, Wylie D, Twarowska B, Tang JT, Porter FW, Wang S, Nagabhushan TL. 2003. Proteomic study of recombinant adenovirus 5 encoding human p53 by matrix-assisted laser desorption/ionization mass spectrometry in combination with database search. Int J Mass Spectrom 226:55–69. doi: 10.1016/S1387-3806(02)00975-2. [DOI] [Google Scholar]
- 29.Diouri M, Keyvani-Amineh H, Geoghegan KF, Weber JM. 1996. Cleavage efficiency by adenovirus protease is site-dependent. J Biol Chem 271:32511–32514. doi: 10.1074/jbc.271.51.32511. [DOI] [PubMed] [Google Scholar]
- 30.Greber UF, Webster P, Weber J, Helenius A. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J 15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 31.Gastaldelli M, Imelli N, Boucke K, Amstutz B, Meier O, Greber UF. 2008. Infectious adenovirus type 2 transport through early but not late endosomes. Traffic 9:2265–2278. doi: 10.1111/j.1600-0854.2008.00835.x. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Berna AJ, Ortega-Esteban A, Menendez-Conejero R, Winkler DC, Menendez M, Steven AC, Flint SJ, de Pablo PJ, San Martin C. 2012. The role of capsid maturation on adenovirus priming for sequential uncoating. J Biol Chem 287:31582–31595. doi: 10.1074/jbc.M112.389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotten M, Weber JM. 1995. The adenovirus protease is required for virus entry into host cells. Virology 213:494–502. doi: 10.1006/viro.1995.0022. [DOI] [PubMed] [Google Scholar]
- 34.Smith AC, Poulin KL, Parks RJ. 2009. DNA genome size affects the stability of the adenovirus virions. J Virol 83:2025–2028. doi: 10.1128/JVI.01644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papp Z, Middleton DM, Mittal SK, Babiuk LA, Baca-Estrada ME. 1997. Mucosal immunization with recombinant adenoviruses: induction of immunity and protection of cotton rats against respiratory bovine herpesvirus type 1 infection. J Gen Virol 78:2933–2943. doi: 10.1099/0022-1317-78-11-2933. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y, Pyne C, Tikoo SK. 2001. Determination of bovine adenovirus-3 titer based on immunohistochemical detection of DNA binding protein in infected cells. J Virol Methods 94:147–153. doi: 10.1016/S0166-0934(01)00286-5. [DOI] [PubMed] [Google Scholar]
- 37.Anand SK, Gaba A, Singh J, Tikoo SK. 2014. Bovine adenovirus 3 core protein precursor pVII localizes to mitochondria, and modulates ATP synthesis, mitochondrial Ca2+ and mitochondrial membrane potential. J Gen Virol 95:442–452. doi: 10.1099/vir.0.057059-0. [DOI] [PubMed] [Google Scholar]
- 38.Kulshreshtha V, Tikoo SK. 2008. Interaction of bovine adenovirus-3 33K protein with other viral proteins. Virology 381:29–35. doi: 10.1016/j.virol.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 39.Patel AK, Tikoo SK. 2006. 293T cells expressing simian virus 40 T antigen are semi-permissive to bovine adenovirus type 3 infection. J Gen Virol 87:817–821. doi: 10.1099/vir.0.81305-0. [DOI] [PubMed] [Google Scholar]
- 40.Zhao X. 2016. The role of bovine adenovirus-3 protein v (pv) in virus replication. PhD thesis University of Saskatchewan, Saskatoon, SK, Canada. [Google Scholar]
- 41.Wu K, Orozco D, Hearing P. 2012. The adenovirus L4-22K protein is multifunctional and is an integral component of crucial aspects of infection. J Virol 86:10474–10483. doi: 10.1128/JVI.01463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 43.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zakhartchouk AN, Reddy PS, Baxi M, Baca-Estrada ME, Mehtali M, Babiuk LA, Tikoo SK. 1998. Construction and characterization of E3-deleted bovine adenovirus type 3 expressing full-length and truncated form of bovine herpesvirus type 1 glycoprotein gD. Virology 250:220–229. doi: 10.1006/viro.1998.9351. [DOI] [PubMed] [Google Scholar]