ABSTRACT
Nipah virus (NiV), a paramyxovirus in the genus Henipavirus, has a mortality rate in humans of approximately 75%. While several studies have begun our understanding of NiV particle formation, the mechanism of this process remains to be fully elucidated. For many paramyxoviruses, M proteins drive viral assembly and egress; however, some paramyxoviral glycoproteins have been reported as important or essential in budding. For NiV the matrix protein (M), the fusion glycoprotein (F) and, to a much lesser extent, the attachment glycoprotein (G) autonomously induce the formation of virus-like particles (VLPs). However, functional interactions between these proteins during assembly and egress remain to be fully understood. Moreover, if the F-driven formation of VLPs occurs through interactions with host cell machinery, the cytoplasmic tail (CT) of F is a likely interactive domain. Therefore, we analyzed NiV F CT deletion and alanine mutants and report that several but not all regions of the F CT are necessary for efficient VLP formation. Two of these regions contain YXXØ or dityrosine motifs previously shown to interact with cellular machinery involved in F endocytosis and transport. Importantly, our results showed that F-driven, M-driven, and M/F-driven viral particle formation enhanced the recruitment of G into VLPs. By identifying key motifs, specific residues, and functional viral protein interactions important for VLP formation, we improve our understanding of the viral assembly/egress process and point to potential interactions with host cell machinery.
IMPORTANCE Henipaviruses can cause deadly infections of medical, veterinary, and agricultural importance. With recent discoveries of new henipa-like viruses, understanding the mechanisms by which these viruses reproduce is paramount. We have focused this study on identifying the functional interactions of three Nipah virus proteins during viral assembly and particularly on the role of one of these proteins, the fusion glycoprotein, in the incorporation of other viral proteins into viral particles. By identifying several regions in the fusion glycoprotein that drive viral assembly, we further our understanding of how these viruses assemble and egress from infected cells. The results presented will likely be useful toward designing treatments targeting this aspect of the viral life cycle and for the production of new viral particle-based vaccines.
KEYWORDS: paramyxovirus, Paramyxoviridae, viral assembly, budding, Nipah virus, cytoplasmic tail, fusion protein, matrix, attachment, glycoprotein
INTRODUCTION
The Paramyxoviridae family includes a wide array of medically relevant pathogens, including but not limited to measles (MeV), mumps, and parainfluenza viruses. The deadliest members of this family belong to the genus Henipavirus, with two of its members, Nipah virus (NiV) and Hendra virus causing encephalitis, respiratory disease, and approximately 40 to 75% mortality rates in humans (1). Moreover, new henipa-like viruses have been discovered recently, such as Mojiang virus, which is suspected for three deaths in China (2), and close to 20 new bat viral species (3). Further, henipaviruses (HNVs) are zoonotic and can infect many mammalian orders (4). During its first outbreak in 1998 and 1999, NiV caused over 100 human deaths and prompted the culling of over one million Malaysian pigs (5, 6). There are no approved human treatments or vaccines for NiV infections, leading to its classification as a biosafety level 4 (BSL-4) agent and a potential threat to international security (7). Furthermore, the World Health Organization has recently listed Nipah virus as one of the top pathogens most likely to cause major epidemics in the future (8).
Paramyxoviruses carry negative-sense, single-stranded RNA genomes 15 to 19 kb in size. While the exact number of protein products made from the genome varies from 6 to 10 among viral species, all paramyxoviruses produce a matrix (M), fusion (F), and attachment glycoprotein (G in the case of HNVs) (9). For HNVs, the F and G glycoproteins are both necessary for viral entry (viral-cell membrane fusion) and are responsible for cell-cell fusion events (syncytia) between infected and naive cells (10, 11). On the other hand, the M protein is generally the main driver in paramyxoviral assembly and budding (12). F and G operate together to mediate viral entry and syncytium formation. G recognizes the host cell receptor ephrinB2 or ephrinB3, triggering F to induce virus-cell or cell-cell membrane fusion (13–18). Although the presence of NiV M has been shown to affect the transport of F and G in polarized epithelia, the mechanism(s) of these effects remains to be elucidated (19). The glycoproteins F and G have also been shown to affect the transport of each other; however, these functional interactions appear to be dependent on cell type (19–21). While NiV M has important roles in protein transport and, ultimately, particle formation, attempts so far to show physical interactions of M with either of the glycoproteins have failed.
The mechanisms of viral particle formation vary considerably between paramyxoviruses (12). M is generally considered essential for these processes, since it acts to incorporate other viral components and create an infectious virion. In support of this central role, M proteins of most paramyxoviruses have the ability to form virus-like particles (VLPs) without any other viral factors present (22–29). Despite assembly and budding for most paramyxoviruses being driven largely by the M protein, other proteins, including the glycoproteins, have been shown to support and in some cases be required for these processes. For example, the particle formation of mumps virus is considerably enhanced by the expression of the fusion but not the hemagglutinin-neuraminidase (HN) attachment glycoprotein (30). Similarly, the fusion but not the attachment glycoprotein of Sendai virus can significantly enhance particle formation (29). Interestingly, for parainfluenza virus 5, either F or HN but not both are required for viral budding (31). This is in contrast to some paramyxoviruses, where the F protein likely does not have an important, or at least a driving function in budding, as is the case for MeV and Newcastle disease virus (24, 26, 27). Previous findings for NiV suggest that F, G, and M can each induce VLP formation when expressed alone, albeit at different efficiencies (25, 32). Specifically, one study showed that while M and F exhibited high abilities to bud autonomously, G did so with much lower efficiency (25). Although it is known that these proteins can induce particle formation, there is very limited knowledge on how NiV F, G, and M functionally interact during viral assembly and whether they influence incorporation of each other into viral particles (25).
In support of an important role for NiV F during particle formation, one study using a new flow virometric tool suggested that incorporation of G into VLPs was considerably more efficient when F in addition to M was expressed (33). Overall, the involvement of F with G and M during particle assembly remains unclear and the functions and mechanism(s) for the autonomous budding of F is unknown. In this study, we quantitatively show that either F or M can enhance the incorporation of G into VLPs severalfold more efficiently than G can do autonomously. In addition, we demonstrate that the ability of NiV F to form VLPs is principally dependent on three regions of its cytoplasmic tail, including a polybasic motif and two tyrosine motifs known to be important for F trafficking or surface expression (20, 21, 34–36). Some of these functions may be dependent on interactions with clathrin adaptor protein (AP) complexes (21, 37).
RESULTS
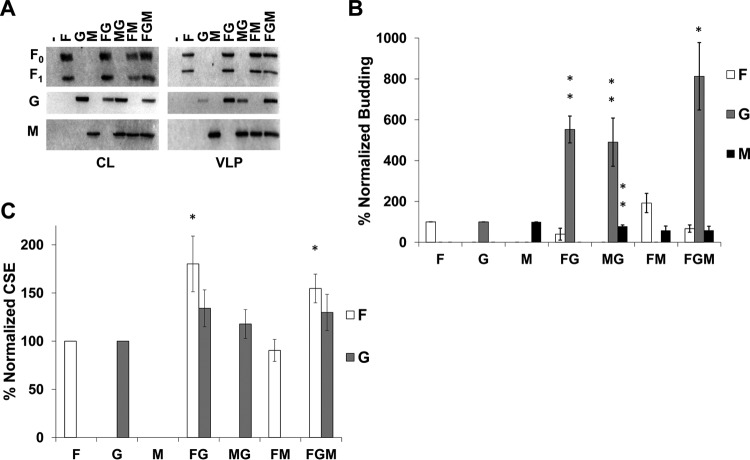
NiV F and M support incorporation of NiV G into VLPs.
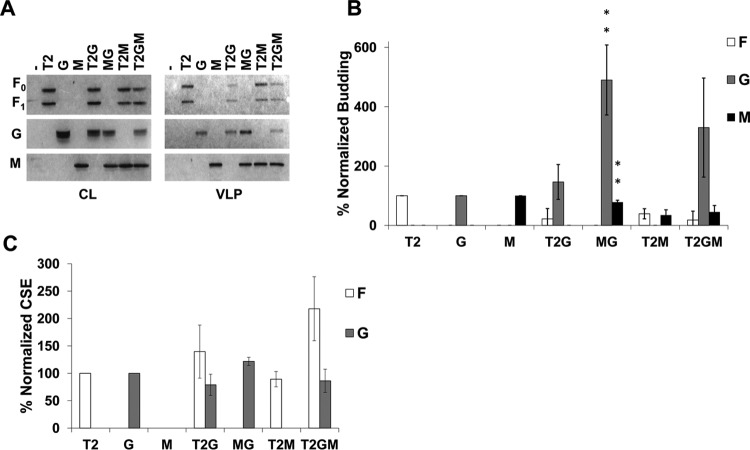
By expressing the NiV F, G, and M proteins in different combinations, we first aimed to understand the roles and relative importance of each of these proteins during assembly and incorporation of one another into viral particles. All combinations of F, G, and M (4:1:1 ratio of their expression plasmids, respectively) were assessed for levels of whole-cell expression by Western blotting and at the cellular surface (for F or G) by flow cytometry. In addition, levels of each protein incorporated into VLPs were determined by sucrose cushion ultracentrifugation of cleared supernatants, followed by Western blotting. In agreement with a previous study, when expressed in HEK293T cells, F and M efficiently formed VLPs autonomously, whereas G did so less efficiently (Fig. 1A) (25). In addition, both F and M showed the ability to incorporate G into VLPs roughly five times more efficiently than when G was expressed alone (Fig. 1B; both are P < 0.01). Interestingly, coexpression of F, G, and M led to a >8-fold increase in G incorporation (P < 0.05) while maintaining a nonsignificant difference in F or M incorporation (Fig. 1B). This finding supports cooperation between F and M in the incorporation of NiV G. We also report that, at least in the case of coexpression of M and G, the incorporation of M was reduced by about 20% (Fig. 1B; P < 0.01) compared to the single expression of M, suggesting that there may be some cost of incorporating G into viral particles. Possible explanations for this observation include competition for M binding between G and host cellular factors that support particle egress or that G can alter M trafficking to assembly sites. Although cellular surface expression (CSE) data showed that the levels of F CSE are higher when F was coexpressed with G (with or without M present) (Fig. 1C; P < 0.05), this did not increase the incorporation of F into VLPs (Fig. 1B).
FIG 1.
NiV F and M support incorporation of NiV G into VLPs. (A) Cell lysates (CL) and viruslike particles (VLPs) were isolated from HEK293T cells transfected for 24 h with the corresponding combinations of NiV F, G, and M. Transfections were done with F:G:M ratios of 4:1:1, with pcDNA3.1 as an empty vector. All samples were run through SDS-PAGE and Western blotting. For NiV F, the uncleaved form is designated F0, and part of the cleaved form is designated F1. (B) Densitometric analysis was used to quantify all bands and budding indices were calculated by dividing the band intensity of each VLP band by their corresponding band in CL. This was normalized to the ratio of individually expressed F, G, or M and is shown as a percentage of that ratio. (C) Flow cytometric analysis was used to assess cellular surface expression of F and G and these values were normalized to corresponding single expression values after removal of background. Error bars designate values for standard errors of the mean. Three or more independent experiments were used for each ratio, and the statistical significance was evaluated with one-sample t tests. Statistical significance is indicated by asterisks: *, P < 0.05; and **, P < 0.01.
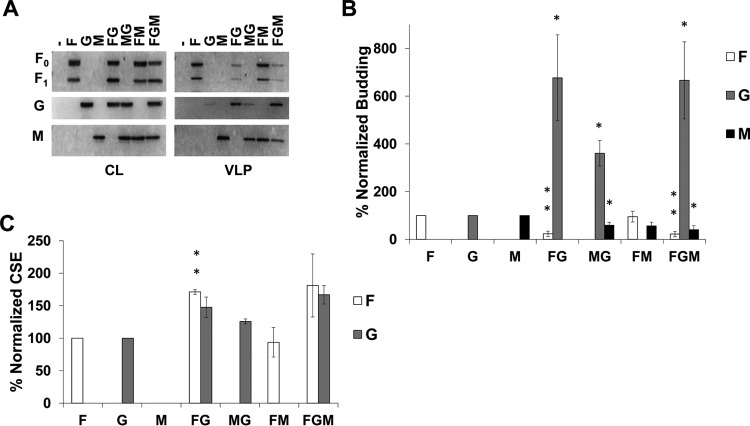
NiV G can reduce incorporation of F and M into VLPs.
We have shown that coexpression of NiV F or M with G significantly increased the incorporation efficiency of NiV G into VLPs (Fig. 1). Since it also appeared that the expression of G reduced the incorporation of F and/or M into VLPs, we next sought to determine whether increasing the levels of NiV G expression would more clearly reduce incorporation of F and M into VLPs. We report that transfection of F, G, and M expression plasmids at a 4:2:1 ratio instead of 4:1:1 (Fig. 1) (while maintaining a total of 3 μg of DNA per well) significantly altered the levels of F and M in VLPs relative to their individual expression (Fig. 2A and B). Specifically, we show that F incorporation was reduced by almost 80% when coexpressed with G (Fig. 2B; P < 0.01). Importantly, this was not due to a decrease in F CSE when G is coexpressed. In fact, in the presence of G, NiV F cell surface expression maintained the ∼70% increase as seen with the 4:1:1 ratio (Fig. 2C; P < 0.01). Interestingly, we saw the same reduction of F in VLPs even when M was expressed in addition to G, indicating that M could not rescue the incorporation of F into VLPs (Fig. 2B; P < 0.01).
FIG 2.
NiV G can reduce incorporation of F and M into VLPs. The same experiments and analyses were completed as in Fig. 1 except the F:G:M ration was changed to 4:2:1. Error bars designate values for standard errors of the mean. Three or more independent experiments were used for each ratio, and the statistical significance was evaluated with one-sample t tests. Statistical significance is indicated by asterisks: *, P < 0.05; and **, P < 0.01.
In addition, M incorporation was significantly reduced by expression of G. Specifically, M levels in VLPs decreased by about 40% (P < 0.05) and 60% (P < 0.05) when coexpressed with G or both F and G, respectively (Fig. 2B). These findings, in conjunction with those of Fig. 1, support a mechanism of F, G, and M budding that requires a delicate balance among the relative expression levels of each of these viral proteins.
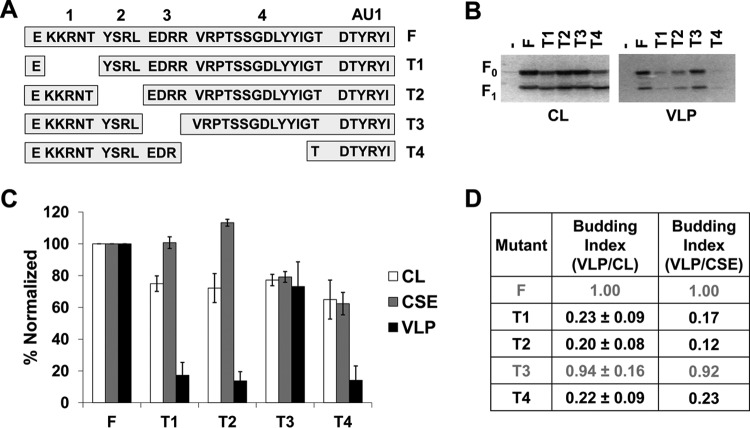
Specific regions of the NiV F cytoplasmic tail drive its ability to form VLPs.
The functions of paramyxoviral glycoproteins during assembly and budding are usually associated with motifs in the cytoplasmic tails (CTs) of these proteins (38). We have shown an important role for NiV F during particle formation (Fig. 1) and now look to elaborate on the mechanism of this behavior by identifying what drives the budding capabilities of F. The NiV F CT has been previously studied in the context of viral entry and membrane fusion; thus, we have assessed the budding capacities of previously studied mutants harboring deletions of regions in the CT (Fig. 3A) (35). Based on total expression and corresponding presence in VLPs (Fig. 3B and C), we calculated budding indexes for these deletion mutants as the ratio of normalized VLP signals to normalized cell lysate signals, so that the index for wild-type (WT) F would be 1.00. Interestingly, the T1, T2, and T4 mutants exhibited budding indices of 0.23 (P < 0.01), 0.20 (P < 0.01), and 0.22 (P < 0.01), respectively (Fig. 3D). This was in direct contrast to the budding index of the T3 deletion mutant, which was not significantly different from that of WT F (Fig. 3D). In addition, we report that the T1 and T2 mutants exhibited levels of CSE that appeared to be higher than would be expected from their total cell expression (CL versus CSE values, Fig. 3C). Thus, relative budding indexes were also calculated based on expression of each mutant with their corresponding levels of CSE (Fig. 3D). The marked decrease in budding by deleting regions T1, T2, or T4 suggest that these regions are functionally important for the budding ability of NiV F.
FIG 3.
Specific regions of the NiV F cytoplasmic tail drive its ability to form VLPs. (A) Schematic designating regions of the NiV F CT and showing the sequence for four deletion mutants made. (B) HEK293T cells were transfected for 24 h with an empty vector, wild-type NiV F, or one of the four CT mutants. Both cell lysates and VLPs were harvested and subjected SDS-PAGE, followed by Western blot analysis. (C) Densitometry was used to quantify band intensity for cell lysates and VLPs. In addition, flow cytometric analyses were used to evaluate the levels of cell-surface expression (CSE) for each of the mutants relative to the wild-type F. (D) As in Fig. 1, the cell lysate and VLP band intensities were used to calculate budding indices, and all were normalized to wild-type F. A similar index is also shown using values for VLP intensity and corresponding CSE values. Mutants exhibiting budding at levels significantly below those of wild-type F have their values in black. Error bars designate values for standard errors of the mean. Three or more independent experiments were used for each ratio, and the statistical significance was evaluated with one-sample t tests.
A region including a membrane-proximal polybasic cluster mildly modulates the budding of F.
Polybasic motifs, such as that in KKRNT of the NiV F CT, are sometimes associated with host cellular factors such as members of the actin cytoskeleton (39). To test which residues of this region may be responsible for the phenotype associated with deletion of the T1 region, we used alanine mutation screening and tested these mutants for any budding phenotypes (Fig. 4A). For both total expression and CSE, all five single-alanine mutants in T1 exhibited levels roughly consistent with those of wild-type F (Fig. 4B and C). In contrast to the substantial decrease in particle formation seen with deleting all of T1 (Fig. 3), single alanine mutations did not show any significant residues that indicate responsibility for this phenotype. While not at, but approaching, statistical significance, the N4A and T5A mutants appeared interesting as potential contributing factors in the T1 deletion budding phenotype (Fig. 4B and C). To further test this, an N4A/T5A double mutant was generated; however, this mutant also failed to produce the budding phenotype seen when the entire T1 region was deleted (Fig. 4B, C, and D). These findings support the possibilities that several residues of this region act functionally together or that this region is important in distancing other motifs in the NiV F CT from the membrane.
FIG 4.
A region including a membrane-proximal polybasic cluster mildly modulates budding of F. (A) Schematic for the cytoplasmic tail of NiV F designating the region being alanine screened. (B) HEK293T cells were transfected for 24 h with an empty vector, wild-type NiV F, one of the five alanine mutants of region T1, or the NT double mutant. Both cell lysates and VLPs were harvested and run in SDS-PAGE followed by Western blot analysis. (C) Densitometry was used to quantify band intensity for cell lysates and VLPs. In addition, flow cytometric analyses were used to evaluate the levels of CSE for each of the mutants relative to the wild-type F. (D) The cell lysate and VLP band intensities were used to calculate budding indices and all were normalized to wild-type F. A similar index is also shown using values for VLP intensity and corresponding CSE values. Error bars designate values for standard errors of the mean. Three or more independent experiments were used for each ratio, and the statistical significance was evaluated with one-sample t tests.
A tyrosine motif in the NiV F cytoplasmic tail modulates NiV F budding.
In other viruses such as HIV-1 and HIV-2, YXXØ motifs such as the T2 region have been linked to viral protein sorting as well as particle assembly in some cases (40–43). Interestingly, a YXXØ motif in the NiV M protein has been suggested to influence M trafficking and budding (22). In addition, studies have identified this motif as contributing to the endocytosis event leading to NiV F maturation into a cleaved, fusogenic form (34, 44). Despite the role of this motif in supporting endocytosis of F, several studies have suggested that mutation of this region is not necessary for cleavage to occur for NiV F (35, 36). Other motifs such as the polybasic cluster may also be important for endocytosis (36). Also, one such study demonstrated a much more significant reduction of processing when the YXXØ motif was deleted in Hendra virus F than seen for NiV F (36). Region T2 (YSRL) has also been shown to be involved in sorting of NiV F based on identified interactions with clathrin adaptor AP proteins (21). Since deletion of T2 also led to a marked drop in budding, single residue analysis was conducted to understand the importance of each for appearance of this phenotype (Fig. 5A). Alanine scanning yielded that mutation of the tyrosine (73% decrease; P < 0.05) and leucine (67% decrease; P < 0.001) residues, but not the serine and arginine residues, led to significant decreases in budding (Fig. 5D). These findings strongly support the importance of the YXXØ motif itself as a driving factor for budding. Further, based on values for cell surface expression, the budding defects were not caused by a failure to transport F to the plasma membrane. While the mechanism for the YXXØ motif to modulate budding is unknown, it is likely related to interactions with host cellular machinery that either support trafficking to specific sites of assembly or drive the process of viral budding directly.
FIG 5.
A tyrosine motif in the NiV F cytoplasmic tail modulates NiV F budding. (A) Schematic for the cytoplasmic tail of NiV F designating the region being alanine screened. (B) HEK293T cells were transfected for 24 h with an empty vector, wild-type NiV F, or one of the four alanine mutants of region T2. Both cell lysates and VLPs were harvested and run in SDS-PAGE followed by Western blot analysis. (C) Densitometry was used to quantify band intensity for cell lysates and VLPs. In addition, flow cytometric analyses were used to evaluate the levels of CSE for each of the mutants relative to the WT. (D) The cell lysate and VLP band intensities were used to calculate budding indices and all were normalized to wild-type F. A similar index is also shown using values for VLP intensity and corresponding CSE values. Mutants exhibiting budding at levels significantly below WT have their values in black. Error bars designate values for standards error of the mean. Three or more independent experiments were used for each ratio, and the statistical significance was evaluated with one-sample t tests.
A dityrosine trafficking motif drives NiV F budding and likely the T4 deletion phenotype.
We hypothesized that the budding phenotype observed for the largest NiV F CT deletion mutant, T4, would be most likely dependent on a couple of motifs. Considering our mass spectrometric data indicating phosphorylation of two adjacent serine residues in T4 (SS), and a dityrosine (YY) motif known to be important in NiV F trafficking in some cell types (20, 21), double-alanine mutants were generated for each of these motifs (Fig. 6A). While the SS mutant exhibited roughly WT levels of total and surface expression in addition to efficient formation of VLPs, mutation of both tyrosine residues nearly abrogated budding completely (95% reduction; P < 0.0001; Fig. 6A, B, and C). This effect was not due to failure to express on the cellular surface, as levels of CSE were close to wild-type CSE levels. Interestingly, a mild increase in the total expression level was observed for this mutant (P < 0.05; Fig. 6B and C).
FIG 6.
A dityrosine trafficking motif drives NiV F budding and likely the T4 deletion phenotype. (A) Schematic for the cytoplasmic tail of NiV F designating both double-alanine mutations. (B) HEK293T cells were transfected for 24 h with an empty vector, wild-type NiV F, or one of the two double-alanine mutants. Both cell lysates and VLPs were harvested and run in SDS-PAGE followed by Western blot analysis. (C) Densitometry was used to quantify band intensity for cell lysates and VLPs. In addition, flow cytometric analyses were used to evaluate the levels of CSE for each of the mutants relative to the WT. (D) The cell lysate and VLP band intensities were used to calculate budding indices and all were normalized to WT F. A similar index is also shown using values for VLP intensity and corresponding CSE values. Mutants exhibiting budding at levels significantly below wild-type F have their values in black. Error bars designate values for standard errors of the mean. Three or more independent experiments were used for each ratio, and the statistical significance was evaluated with Student t tests.
The budding capabilities of NiV F are important for G incorporation into VLPs.
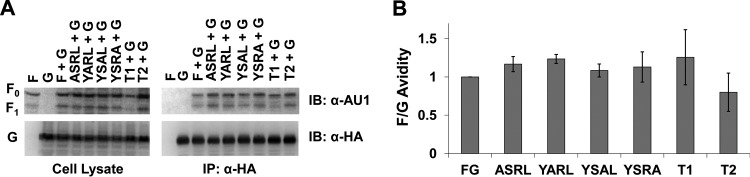
To verify the importance of autonomous NiV F budding in the context of F, G, and M incorporation into viral particles, we cotransfected the T2 deletion mutant of F with combinations of the other structural proteins, as shown for Fig. 1. Based on better incorporation of each protein, the 4:1:1 DNA expression plasmid ratio was selected. As would be expected if F drove G incorporation into VLPs, the T2G combination did not show statistically significant increases in G levels beyond that of single G expression (Fig. 7A and B). Analysis with coimmunoprecipitation of G with T2 supports that this behavior is not due to any change in F:G avidities caused by the T2 deletion (Fig. 8A and B). Further, we observed that while G appears to be incorporated about three times more in the T2GM combination than for G alone (Fig. 7A and B), the difference is not statistically significant (P > 0.2). None of these effects appeared to be caused by changes in cell surface expression (Fig. 7C). Together, these and the above findings highlight a crucial role of NiV F, driven by regions and residues of its CT, in the presence or absence of M expression during the incorporation of G into viral particles.
FIG 7.
The budding capabilities of NiV F are important for G incorporation into VLPs. The same experiments and analyses were completed as in Fig. 1 except the T2 mutant was used in place of the wild type. Error bars designate values for standard errors of the mean. Three or more independent experiments were used for each ratio, and the statistical significance was evaluated with one-sample t tests. Statistical significance is designated by asterisks: *, P < 0.05; and **, P < 0.01.
FIG 8.
Mutation of the NiV F CT does not alter F-G avidity. HEK293T cells were transfected for G and different mutants of NiV F at an F:G ratio of 4:1. Cells were harvested and lysed. Part of the lysate was immunoprecipitated for NiV G. (A) Both the cell lysates and eluates were run through SDS-PAGE and Western blotted for F and G. (B) bands were quantified with blot densitometry, and the results for two experiments were averaged. Avidity was determined from the levels of (IP F)/(IP G * CL F). The standard error for each set is shown.
DISCUSSION
Previous work has suggested that NiV M, as for many of the paramyxoviruses, is the key player in viral assembly and budding (12, 23, 24, 26, 27, 29). As with other paramyxoviruses, studies for NiV have so far suggested that NiV M does not require F or G to form VLPs (22, 25). While our findings support this conclusion, prior studies have focused primarily on M as the indicator for VLP formation, whereas we have increased the scope to the incorporation of F, G, and M in all possible combinations. In doing so, we have increased the knowledge on the functional roles for these three structural proteins and advanced toward the understanding of viral particle formation. Our results show that both F and M can drive incorporation of G into viral particles several times more effectively than G alone (Fig. 1). While F and M significantly affected G incorporation, we report that F and M did not appear to significantly affect incorporation of each other (Fig. 1 and 2). Interestingly, coexpression of the F T2 deletion mutant with M did not result in any significant recovery of F budding (Fig. 7). This may suggest that F-M interactions are limited and/or that the mechanisms of particle egress driven by F or M differ significantly.
For several paramyxoviruses, including parainfluenza virus 5 and mumps virus, viral budding is driven by proteins of the endosomal sorting complex required for transport (ESCRT) system (30, 31). ESCRT proteins have also been shown to be important in viral particle formation for many enveloped viruses (45). Still, in cases such as influenza virus and vesicular stomatitis virus, viral assembly and budding are thought to follow ESCRT-independent pathways (46–49). Importantly, initial experiments with NiV supported that this virus also utilized ESCRT-independent methods of budding. Specifically, when NiV M was previously coexpressed with dominant negative Vps4, the ATPase responsible for driving ESCRT function, no significant decrease in budding was found (50). Such findings are not unprecedented for viruses somewhat related to NiV. Specifically, human respiratory syncytial virus can bud independently of the ESCRT system in a mechanism driven by Rab11 family interacting protein 2 (51). Importantly, a recent study revealed interactions between the NiV C protein and Tsg101, an ESCRT factor. These interactions increase M budding efficiency. The same study also demonstrated that live NiV titers decreased when the ESCRT machinery was inhibited (52). While the exact mechanism of NiV F budding is not known, one possibility is that regions of the cytoplasmic tail interact with ESCRT proteins to drive viral particle assembly and/or egress from the cell. A likely binding partner of NiV F is the ESCRT-associated protein, AIP-1/Alix. Several viruses, including some poxviruses, arenaviruses, and lentiviruses utilize YXXL motifs similar to that in the NiV F CT to interact with Alix/AIP1 and recruit ESCRT machinery during egress (53–55). Further experiments would be needed to explore this possibility.
The ability for a paramyxovirus protein other than M to bud autonomously has been seen in the cases of Sendai and measles virus F proteins but is rather rare among paramyxoviruses (26, 28, 29). We have also shown that autonomous budding behaviors of F are at least partially dependent on regions of its cytoplasmic tail (Fig. 3). These regions include a polybasic motif, an YXXL motif, and a dityrosine motif (Fig. 4, 5, and 6). All of these regions have been previously implicated in modulating biological functions of NiV F such as trafficking, endocytosis, and/or fusogenicity (21, 34, 35, 56). The envelope glycoproteins of both HIV-1 and -2 have cytoplasmic tails that contain YXXØ motifs important for proper trafficking (40–43). For HiV-2, proper trafficking has been shown to be dependent on interactions between the YXXØ motif and AP-2 (42). Further, this tyrosine motif has also been shown to be a significant factor driving HIV-2 viral particle formation (42, 43). Although we report that T2 and T4 contain specific motifs that may be responsible for modulation of budding by those regions, we failed to identify a residue in the T1 region that confers a strong budding phenotype (Fig. 4). One possibility, supported by decreased budding after deletion of the whole region, is that T1 is important for spacing of the YXXØ motif directly following it. Other studies with transmembrane proteins have shown that specific trafficking patterns can depend not only on the presence of a YXXØ motif in the CT but also by the distance of the motif from the membrane. Lysosomal targeting, in particular, is characterized by a YXXØ motif six to nine residues from the membrane, which is interesting as the YXXØ motif in the NiV F CT starts at seven residues after the end of the transmembrane domain (57, 58).
The ties between the motifs we have identified as modulators of budding and lysosomal signaling are strengthened by the identification of the dityrosine motif in region T4 as basically necessary for F budding. We found that mutation of the dityrosine motif to alanines led to the nearly complete loss of VLP production. Importantly, diaromatic motifs similar to YY have been shown to be signals for lysosomal avoidance (58). While the underlying mechanism of budding remains unknown for NiV F, it may be that the interactions between this protein and trafficking factors, such as AP complexes (21, 34), indirectly or directly drive particle formation.
The observations concerning incorporation of NiV F, G, and M are most similar to the patterns of budding that have been reported for Sendai virus. Specifically, the fusion, but not the attachment glycoprotein, assists in budding (28, 29). Our findings that the substantial increase of G incorporation when G is coexpressed with F and/or M is lost when the T2 mutant is used in place of WT F, supports the role of F as significantly supportive during viral particle formation. While we have shown the importance of the ability for F to drive budding during incorporation of itself and of G, it remains clear from previous studies that M is essential for the efficient production of infectious virions (59). Importantly, we must keep in mind that in the context of simultaneous expression of the M, F, and G proteins, VLPs assembled and budded will include M-only, F-only, G-only, MF, MG, FG, and MFG VLPs. In this mixture of VLP populations, it remains to be seen what proportions of VLPs incorporate each of these proteins best. Although studies concerning the analysis of multiple populations are technically complicated, continually advancing techniques such as flow virometry may make these analyses more feasible, and we are currently enhancing this technique to accomplish these studies (33).
Although we have made strides in understanding the interactions between NiV F, G, and M in the process of particle formation, several questions remain. The importance of the F CT regions and residues we have identified as crucial for F budding should be tested in the context of the live virus under BSL-4 conditions. Our findings also support the importance of cellular factors in F budding and highlight several possible interactions of interest. Further studies could be conducted to identify which cellular factors are responsible for these assembly/budding phenotypes and help identify possible targets for treatments. While the mechanisms of NiV and closely related viruses remain to be completely understood, our identification of NiV F as a viral factor with importance not only in entry but also in egress may support future studies of assembly and VLP-based vaccine development.
MATERIALS AND METHODS
Antibodies.
Mouse AU1 and HA monoclonal antibodies were purchased from BioLegend. For flow cytometric analyses, rabbit antiserum 835 was used to detect NiV F (60). Rabbit AU1 and HA antibodies were obtained from Bethyl Laboratories and used to detect NiV F and G, respectively, during Western blot analysis. Mouse antibodies against the FLAG epitope on NiV M were purchased from Thermo Scientific Pierce.
Cell culture.
HEK293T (ATCC) cells were cultured in Dulbecco modified Eagle medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS) and 1% PenStrep (Life Technologies). All experiments were conducted using HEK293T cells.
Plasmids.
Codon-optimized sequences for NiV F and G were inserted into pcDNA3.1 expression vectors and tagged with AU1 (C terminal) and hemagglutinin (HA; C terminal), respectively, as published previously (61). Codon-optimized NiV M was inserted into a pCMV-3×-FLAG vector with an N-terminal FLAG tags as described previously (62). NiV F and all mutants were produced as described previously (35).
Cell surface expression.
HEK293T cells were transfected in six-well plates with 3 μg of total DNA per well between 65 and 85% confluence using TurboFect (Thermo Fisher). After 24 h, cells were harvested with 10 mM EDTA and then collected by centrifugation at 350 × g for 30 s at 4°C. The pellet was resuspended with Dulbecco phosphate-buffered saline supplemented with 1% FBS. After staining of NiV F with rabbit polyclonal antibody 835 (1:250 dilution) and NiV G with mouse monoclonal antibody against HA (BioLegend; 1:200 dilution) for 1 h, three washes were completed at 4°C and 800 × g for 5 min each. Afterward, goat anti-mouse and goat anti-rabbit secondary antibodies (Life Technologies) were applied at 1:2,000 dilution for 30 min, followed by two more washes and a fixation in 0.5% paraformaldehyde. After fixation, the cells were processed using a Millipore Guavasoft flow cytometer 8HT; 10,000 live cells were analyzed per sample.
Western blot analysis.
SDS–10% polyacrylamide gels were run and transferred onto polyvinylidene difluoride (PVDF) membranes. Odyssey blocking buffer (LiCor Biosciences) was used to block membranes overnight. Primary and secondary antibodies were incubated for 1 h and 45 min, respectively. Washing with 0.2% Tween 20 in PBS was completed after each antibody incubation. After membrane blotting, imaging and densitometric analyses were completed on a Bio-Rad ChemiDoc MP imaging system. When necessary, blot stripping was done with NewBlot PVDF stripping buffer according to manufacturer suggestions.
Coimmunoprecipitation of F and G.
HEK293T cells were seeded into six-well plates, and each well was transfected with F and/or G at a 4:1 ratio (a total of 2 μg of DNA per well) with Lipofectamine 2000 for 24 h. To ensure plenty of protein, three wells were combined for each sample. After transfection, the cells were lysed in lysis buffer (Miltenyi Biotec). Immunoprecipitations were completed according to manufacturer specification (Miltenyi Biotec) with the exception that all washes were completed with lysis buffer containing 150 mM NaCl, 1% Triton X-100, and 50 mM Tris HCl (pH 8.0) provided containing added the 1× protease inhibitor cOmplete ULTRA (Roche). Coimmunoprecipitation eluates and cell lysates were subjected to SDS–10% PAGE and analyzed as described above.
Virus-like particle detection.
Cells were transfected with TurboFect (Thermo Fisher) at 65 to 85% confluence in six-well plates. Then, 3 μg of total DNA was used per well as described for cell surface expression. After 24 h, the medium was collected and spun down at 376 × g for 10 min to pellet cellular debris. The supernatant medium was then loaded over 2 ml of 20% sucrose in NTE buffer and spun at 110,000 × g for 90 min at 4°C. After sucrose cushion ultracentrifugation, the pellets were resuspended in loading dye with 2% β-mercaptoethanol and stored until loaded into a gel for SDS-PAGE.
ACKNOWLEDGMENTS
This study was supported by NIH/NIAID grant AI109022 to H.C.A. Graduate students G.P.J., E.M.C., and V.O. were supported by NIH/NIGMS training grant T32GM008336.
REFERENCES
- 1.Luby SP, Hossain MJ, Gurley ES, Ahmed B-N, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA, Kenah E, Ksiazek TG, Rahman M. 2009. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001-2007. Emerg Infect Dis 15:1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, Sun L, Zhu Y, Zhou H, Jin Q. 2014. Novel henipa-like virus, Mojiang paramyxovirus, in rats, China, 2012. Emerg Infect Dis 20:1064–1066. doi: 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Rasche A, Yordanov S, Seebens A, Oppong S, Sarkodie YA, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stöcker A, Carneiro AJB, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EKV, Kruppa T, Franke CR, Kallies R, Yandoko ERN, Herrler G, Reusken C, Hassanin A, Krüger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. 2012. Bats host major mammalian paramyxoviruses. Nat Commun 3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chua KB. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 5.Enserink M. 1999. Epidemiology: new virus fingered in Malaysian epidemic. Science 284:407–410. doi: 10.1126/science.284.5413.407. [DOI] [PubMed] [Google Scholar]
- 6.Parashar UD, Sunn LM, Ong F, Mounts AW, Arif MT, Ksiazek TG, Kamaluddin MA, Mustafa AN, Kaur H, Ding LM, Othman G, Radzi HM, Kitsutani PT, Stockton PC, Arokiasamy J, Gary HE Jr, Anderson LJ. 2000. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998-1999 outbreak of severe encephalitis in Malaysia. J Infect Dis 181:1755–1759. doi: 10.1086/315457. [DOI] [PubMed] [Google Scholar]
- 7.Aguilar HC, Lee B. 2011. Emerging paramyxoviruses: molecular mechanisms and antiviral strategies. Expert Rev Mol Med 13:e6. doi: 10.1017/S1462399410001754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. 2015. Blueprint for R&D preparedness and response to public health emergencies due to highly infectious pathogens. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 9.Samal SK. (ed). 2011. The biology of paramyxoviruses. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 10.Wong KT, Shieh W-J, Kumar S, Norain K, Abdullah W, Guarner J, Goldsmith CS, Chua KB, Lam SK, Tan CT, Goh KJ, Chong HT, Jusoh R, Rollin PE, Ksiazek TG, Zaki SR; Nipah Virus Pathology Working Group. 2002. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol 161:2153–2167. doi: 10.1016/S0002-9440(10)64493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguilar HC, Iorio RM. 2012. Henipavirus membrane fusion and viral entry, p 79–94. In Lee B, Rota PA (ed), Henipavirus. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 12.Harrison MS, Sakaguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol 42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilar HC, Henderson BA, Zamora JL, Johnston GP. 2016. Paramyxovirus glycoproteins and the membrane fusion process. Curr Clin Microbiol Rep 3:142–154. doi: 10.1007/s40588-016-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negrete OA, Wolf MC, Aguilar HC, Enterlein S, Wang W, Mühlberger E, Su SV, Bertolotti-Ciarlet A, Flick R, Lee B. 2006. Two key residues in EphrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog 2:e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, Lee B. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401–405. [DOI] [PubMed] [Google Scholar]
- 16.Aguilar HC, Aspericueta V, Robinson LR, Aanensen KE, Lee B. 2010. A quantitative and kinetic fusion protein-triggering assay can discern distinct steps in the Nipah virus membrane fusion cascade. J Virol 84:8033–8041. doi: 10.1128/JVI.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, Aguilar HC. 2013. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog 9:e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang L-F, Eaton BT, Broder CC. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A 102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamp B, Dietzel E, Kolesnikova L, Sauerhering L, Erbar S, Weingartl H, Maisner A. 2013. Nipah virus entry and egress from polarized epithelial cells. J Virol 87:3143–3154. doi: 10.1128/JVI.02696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erbar S, Maisner A. 2010. Nipah virus infection and glycoprotein targeting in endothelial cells. Virol J 7:305. doi: 10.1186/1743-422X-7-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattera R, Farías GG, Mardones GA, Bonifacino JS. 2014. Co-assembly of viral envelope glycoproteins regulates their polarized sorting in neurons. PLoS Pathog 10:e1004107. doi: 10.1371/journal.ppat.1004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciancanelli MJ, Basler CF. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol 80:12070–12078. doi: 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coronel EC, Murti KG, Takimoto T, Portner A. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J Virol 73:7035–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. 2006. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol 80:11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patch JR, Crameri G, Wang L-F, Eaton BT, Broder CC. 2007. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol J 4:1. doi: 10.1186/1743-422X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pohl C, Duprex WP, Krohne G, Rima BK, Schneider-Schaulies S. 2007. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J Gen Virol 88:1243–1250. doi: 10.1099/vir.0.82578-0. [DOI] [PubMed] [Google Scholar]
- 27.Runkler N, Pohl C, Schneider-Schaulies S, Klenk H-D, Maisner A. 2007. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell Microbiol 9:1203–1214. doi: 10.1111/j.1462-5822.2006.00860.x. [DOI] [PubMed] [Google Scholar]
- 28.Sugahara F, Uchiyama T, Watanabe H, Shimazu Y, Kuwayama M, Fujii Y, Kiyotani K, Adachi A, Kohno N, Yoshida T, Sakaguchi T. 2004. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology 325:1–10. doi: 10.1016/j.virol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J Virol 75:11384–11391. doi: 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. 2009. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J Virol 83:7261–7272. doi: 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt AP, Leser GP, Waning DL, Lamb RA. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J Virol 76:3952–3964. doi: 10.1128/JVI.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walpita P, Barr J, Sherman M, Basler CF, Wang L. 2011. Vaccine potential of Nipah virus-like particles. PLoS One 6:e18437. doi: 10.1371/journal.pone.0018437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landowski M, Dabundo J, Liu Q, Nicola AV, Aguilar HC. 2014. Nipah virion entry kinetics, composition, and conformational changes determined by enzymatic virus-like particles and new flow virometry tools. J Virol 88:14197–14206. doi: 10.1128/JVI.01632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt C, Eickmann M, Diederich S, Moll M, Maisner A. 2005. Endocytosis of the Nipah virus glycoproteins. J Virol 79:3865–3872. doi: 10.1128/JVI.79.6.3865-3872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguilar HC, Matreyek KA, Choi DY, Filone CM, Young S, Lee B. 2007. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol 81:4520–4532. doi: 10.1128/JVI.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khetawat D, Broder CC. 2010. A functional henipavirus envelope glycoprotein pseudotyped lentivirus assay system. Virol J 7:312. doi: 10.1186/1743-422X-7-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weise C, Erbar S, Lamp B, Vogt C, Diederich S, Maisner A. 2010. Tyrosine residues in the cytoplasmic domains affect sorting and fusion activity of the Nipah virus glycoproteins in polarized epithelial cells. J Virol 84:7634–7641. doi: 10.1128/JVI.02576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Najjar F, Schmitt A, Dutch R. 2014. Paramyxovirus glycoprotein incorporation, assembly and budding: a three-way dance for infectious particle production. Viruses 6:3019–3054. doi: 10.3390/v6083019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S, Tsukita S. 1998. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol 140:885–895. doi: 10.1083/jcb.140.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deschambeault J, Lalonde JP, Cervantes-Acosta G, Lodge R, Cohen EA, Lemay G. 1999. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol 73:5010–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lodge R. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J 16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noble B, Abada P, Nunez-Iglesias J, Cannon PM. 2006. Recruitment of the adaptor protein 2 complex by the human immunodeficiency virus type 2 envelope protein is necessary for high levels of virus release. J Virol 80:2924–2932. doi: 10.1128/JVI.80.6.2924-2932.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abada P, Noble B, Cannon PM. 2005. Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production. J Virol 79:3627–3638. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diederich S, Sauerhering L, Weis M, Altmeppen H, Schaschke N, Reinheckel T, Erbar S, Maisner A. 2012. Activation of the Nipah virus fusion protein in MDCK cells is mediated by cathepsin B within the endosome-recycling compartment. J Virol 86:3736–3745. doi: 10.1128/JVI.06628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen BJ, Leser GP, Morita E, Lamb RA. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol 81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irie T, Licata JM, McGettigan JP, Schnell MJ, Harty RN. 2004. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J Virol 78:2657–2665. doi: 10.1128/JVI.78.6.2657-2665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patch JR, Han Z, McCarthy SE, Yan L, Wang L-F, Harty RN, Broder CC. 2008. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol J 5:137. doi: 10.1186/1743-422X-5-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Utley TJ, Ducharme NA, Varthakavi V, Shepherd BE, Santangelo PJ, Lindquist ME, Goldenring JR, Crowe JE. 2008. Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc Natl Acad Sci U S A 105:10209–10214. doi: 10.1073/pnas.0712144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park A, Yun T, Vigant F, Pernet O, Won ST, Dawes BE, Bartkowski W, Freiberg AN, Lee B. 2016. Nipah virus C protein recruits Tsg101 to promote the efficient release of virus in an ESCRT-dependent pathway. PLoS Pathog 12:e1005659. doi: 10.1371/journal.ppat.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honeychurch KM, Yang G, Jordan R, Hruby DE. 2007. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J Virol 81:7310–7315. doi: 10.1128/JVI.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shtanko O, Watanabe S, Jasenosky LD, Watanabe T, Kawaoka Y. 2011. ALIX/AIP1 is required for NP incorporation into Mopeia virus Z-induced virus-like particles. J Virol 85:3631–3641. doi: 10.1128/JVI.01984-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bieniasz PD. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 56.Weis M, Maisner A. 2015. Nipah virus fusion protein: Importance of the cytoplasmic tail for endosomal trafficking and bioactivity. Eur J Cell Biol 94:316–322. doi: 10.1016/j.ejcb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohrer J. 1996. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J Cell Biol 132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 59.Dietzel E, Kolesnikova L, Sawatsky B, Heiner A, Weis M, Kobinger GP, Becker S, von Messling V, Maisner A. 2016. Nipah virus matrix protein influences fusogenicity and is essential for particle infectivity and stability. J Virol 90:2514–2522. doi: 10.1128/JVI.02920-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeBuysscher BL, Scott D, Marzi A, Prescott J, Feldmann H. 2014. Single-dose live-attenuated Nipah virus vaccines confer complete protection by eliciting antibodies directed against surface glycoproteins. Vaccine 32:2637–2644. doi: 10.1016/j.vaccine.2014.02.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Bertolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA, Su SV, Wolf MC, Kohatsu L, Baum LG, Lee B. 2006. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 80:4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang YE, Park A, Lake M, Pentecost M, Torres B, Yun TE, Wolf MC, Holbrook MR, Freiberg AN, Lee B. 2010. Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog 6:e1001186. doi: 10.1371/journal.ppat.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]