ABSTRACT
Several virulence genes have been identified thus far in the herpes simplex virus 1 genome. It is also generally accepted that protein heterogeneity among virions further impacts viral fitness. However, linking this variability directly with infectivity has been challenging at the individual viral particle level. To address this issue, we resorted to flow cytometry (flow virometry), a powerful approach we recently employed to analyze individual viral particles, to identify which tegument proteins vary and directly address if such variability is biologically relevant. We found that the stoichiometry of the UL37, ICP0, and VP11/12 tegument proteins in virions is more stable than the VP16 and VP22 tegument proteins, which varied significantly among viral particles. Most interestingly, viruses sorted for their high VP16 or VP22 content yielded modest but reproducible increases in infectivity compared to their corresponding counterparts containing low VP16 or VP22 content. These findings were corroborated for VP16 in short interfering RNA experiments but proved intriguingly more complex for VP22. An analysis by quantitative Western blotting revealed substantial alterations of virion composition upon manipulation of individual tegument proteins and suggests that VP22 protein levels acted indirectly on viral fitness. These findings reaffirm the interdependence of the virion components and corroborate that viral fitness is influenced not only by the genome of viruses but also by the stoichiometry of proteins within each virion.
IMPORTANCE The ability of viruses to spread in animals has been mapped to several viral genes, but other factors are clearly involved, including virion heterogeneity. To directly probe whether the latter influences viral fitness, we analyzed the protein content of individual herpes simplex virus 1 particles using an innovative flow cytometry approach. The data confirm that some viral proteins are incorporated in more controlled amounts, while others vary substantially. Interestingly, this correlates with the VP16 trans-activating viral protein and indirectly with VP22, a second virion component whose modulation profoundly alters virion composition. This reaffirms that not only the presence but also the amount of specific tegument proteins is an important determinant of viral fitness.
KEYWORDS: FACS, HSV-1, VP16, VP22, flow cytometry, flow virometry, herpes simplex virus, heterogeneity, tegument, viral particle
INTRODUCTION
Over the years, virulence genes that modulate the ability of viruses to propagate have been identified. Given their parasitic nature, viruses are also largely influenced by their hosts. This relates, for example, to both the innate and acquired immune responses and the presence of specific cellular receptors that contribute to tropism. However, many reports suggest that protein heterogeneity among viral particles additionally impacts viral fitness, but this has been challenging to probe at the individual virion level. One important reason is that most methods typically evaluate the average composition of whole viral particle populations and rarely address individual particle content. On the other hand, techniques such as immunoelectron microscopy (immuno-EM) and fluorescence microscopy do evaluate particle-to-particle variation but not infectivity. Thus, no tool existed until now to directly evaluate the biological relevance of protein variability among individual virions. The present work attempts to tackle this issue.
Mature herpes simplex virus 1 (HSV-1) virions are composed of 4 distinct structural components, encompassing the double-stranded DNA viral genome, a capsid, a so-called tegument, and a host-derived lipid envelope. Altogether, at least 44 different viral proteins and potentially as many cellular proteins are incorporated in mature extracellular HSV-1 viruses (1). Interestingly, the bulk of these proteins are contained within the tegument, an intricate layer whose composition is dependent on multiple interactions among the various tegument proteins as well as with capsid and envelope proteins (2). As for other viruses, virulence varies within and among HSV-1 stocks, while viral preparations typically contain a vast array of heterogeneous entities dominated by the presence of noninfectious particles (3). These properties make HSV-1 an ideal candidate to study protein variability among viral particles and probe the impact of this variability on the ability of the virus to infect cells.
The HSV-1 tegument structurally bridges the viral envelope to the capsid. It is multifunctional and is implicated in capsid transport, targeting of the incoming capsids to the nuclear pores, egress of newly made viral particles, and acquisition of the final envelope (4, 5). Among the tegument proteins are trans-activating molecules that modulate the expression of other viral proteins. These include ICP0, ICP4, and VP16, which are all present in virions, presumably to jump start the infection cycle (6–8). Not surprisingly, depleting or altering these proteins genetically or by RNA interference (RNAi) leads to reduced viral yields (9–13). Thus, the tegument proteins are not merely structural but also are active participants in most aspects of the viral life cycle and are important determinants of viral fitness. It is very likely that, in addition to their qualitative presence, their relative amounts in a given viral particle also modulate the infection, but evaluating this relationship has been difficult.
While the stoichiometry of the components forming the highly structured HSV-1 capsid has been determined with good accuracy (14), only rough estimates prevail for the more loosely organized tegument layer. These last estimates were originally derived from semiquantitative Sypro red or Coomassie staining of protein gels and autoradiograms (15–17). More recently, tegument abundance has also been monitored by coincidence fluorescence spectroscopy and, in the case of pseudorabies herpesvirions (PRV), by fluorescence microscopy of green fluorescent protein (GFP)-tagged components (18, 19). Several pieces of evidence suggest that some tegument proteins have a tightly controlled stoichiometry, while others may be more flexible. For instance, overexpression of the abundant VP22 tegument leads to its greater incorporation in mature virions (20). This contrasts with UL37, whose quantity remains stable in virions even when exogenously overexpressed (21). Moreover, an analysis by quantitative mass spectrometry of deletion mutants of the related pseudovirus hinted that VP1/2 and VP13/14 are incorporated in virions in fixed amounts, while VP11/12, VP16, and VP22 are present in variable quantities (22). These findings are consistent with work by Smith and colleagues, who found some heterogeneity of specific tegument proteins among viral particles (19). However, none of the abovementioned studies addressed whether this heterogeneity is mirrored by corresponding changes in infectivity at the individual particle level. Interestingly, the addition of extraneous tegument proteins in the form of so-called L particles, which contain an envelope and tegument proteins but are devoid of viral capsid and genome, enhances the ability of transfected viral DNA to generate infectious particles (23), pointing to the importance of the relative amount of tegument proteins.
The goal of the present study was to quantitatively evaluate the variability of the HSV-1 tegument proteins among viral particles and to evaluate the biological relevance of that particle-to-particle variability. To this end, we resorted to flow cytometry (also referred to as flow virometry), a powerful method we recently employed to characterize and purify HSV-1 nuclear capsids (24). The present study shows that flow cytometry can also be used to study naturally occurring heterologous populations of mature extracellular HSV-1 virions. It further confirms that some tegument proteins are more stable in their stoichiometry from one viral particle to the next, while others, such as VP16 and VP22, are incorporated in greatly more flexible amounts. Interestingly, the sorting by flow cytometry of virions based on their VP16 or VP22 content translated into a modest but reproducible modulation of infectivity, with particles containing more VP16 or VP22 being slightly more infectious. Interestingly, RNA interference studies targeting VP16 corroborated these findings, while virion depletion of VP22 hinted at a more complex scenario. Quantitative Western blot analyses of the viral tegument layer thus indicated that the latter phenotype is multifactorial and indirectly related to VP22 protein levels. They also revealed that altering one tegument component, for instance, by changing its stoichiometry or by tagging it with GFP, can have profound implications for the overall composition of the virions. We ultimately confirmed at the individual viral particle level that the amount of individual tegument proteins in the virions, as opposed to merely their presence, is indeed a contributing factor that modulates infectivity of the viral particles. Thus, not only the presence but also the amount of specific proteins in mature virions constitutes a bona fide viral fitness factor.
RESULTS
Successful detection of mature HSV-1 enveloped virions by flow cytometry.
We previously showed that we can detect HSV-1 nonenveloped nuclear capsids by flow cytometry despite their being smaller than the theoretical resolution limit of 0.5 μm of most instruments (24). Since it is not possible to discern 100-μm from 200-μm particles, whether commercial beads or viruses or the background signal based on light scattering, we circumvented this issue in the past by GFP tagging nuclear capsids or by staining the viral genome with the nucleic acid Syto 13 dye, whose fluorescence is strongly stimulated when bound to nucleic acids. This not only allowed us to tell the capsids apart from the background but also proved to be an efficient tool to efficiently enrich for so-called C nuclear capsids, reaching 90% purity in a single purification step (24).
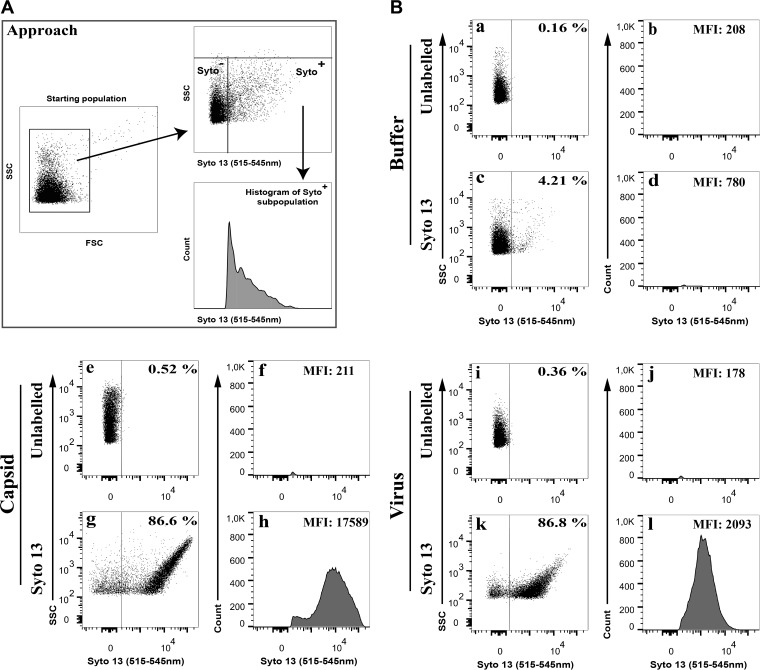
To evaluate if mature enveloped virions also could be monitored by flow cytometry, we labeled them with the membrane-permeable Syto 13 along with control nuclear capsids or buffer alone. Figure 1A depicts our analysis strategy. As previously reported (24), background particles present in the 0.22-μm-filtered phosphate-buffered saline (PBS) shield buffer were detected by light scattering but were otherwise devoid of any intrinsic fluorescence (Fig. 1Ba and b). Similarly, no significant fluorescent signal was detected when Syto 13 was added to buffer alone (4% of particles emitted light with a mean fluorescence intensity [MFI] of 780) (Fig. 1Bc and d). In contrast, the addition of Syto 13 to nuclear capsids labeled nearly 90% of the capsids and gave rise to a 23-fold increase in fluorescence, with an MFI of 17,589 (Fig. 1Be to h). Interestingly, addition of Syto 13 to extracellular virions was equally efficient (87% efficiency) but resulted in only a 3-fold increase in fluorescence, for an MFI of 2,093 (Fig. 1Bi to l). These results suggested that the viral envelope or density of the tegument layer partially perturbed the entry of Syto 13 in the viral particles and/or quenched its fluorescence. However, the signal was amply sufficient to detect, analyze, and sort enveloped HSV-1 virions by flow cytometry.
FIG 1.
Analysis of Syto-stained HSV-1 virions by FACS. Vero cells were infected with wild-type HSV-1 at an MOI of 5 for 18 hpi. Extracellular virions were stained with 1 μM Syto 13 and analyzed by flow cytometry. (A) Schematic description of the gating strategy. Note that aggregated particles were first excluded by gating them out in the SSC versus FSC plots and by specific flow cytometry parameters (see Materials and Methods). (B) The graphs on the left for each condition (dot plots) show the fluorescence profiles of all nonaggregated particles, while only Syto 13+ samples (fluorescence above the buffer control) were considered on the right (histograms). Percentages denote the amount of Syto 13-positive particles relative to the starting population, which includes inert particles inherently present in the FACS buffer (24). Meanwhile, the mean fluorescence signal (MFI) is only that of the Syto 13+ viral particles.
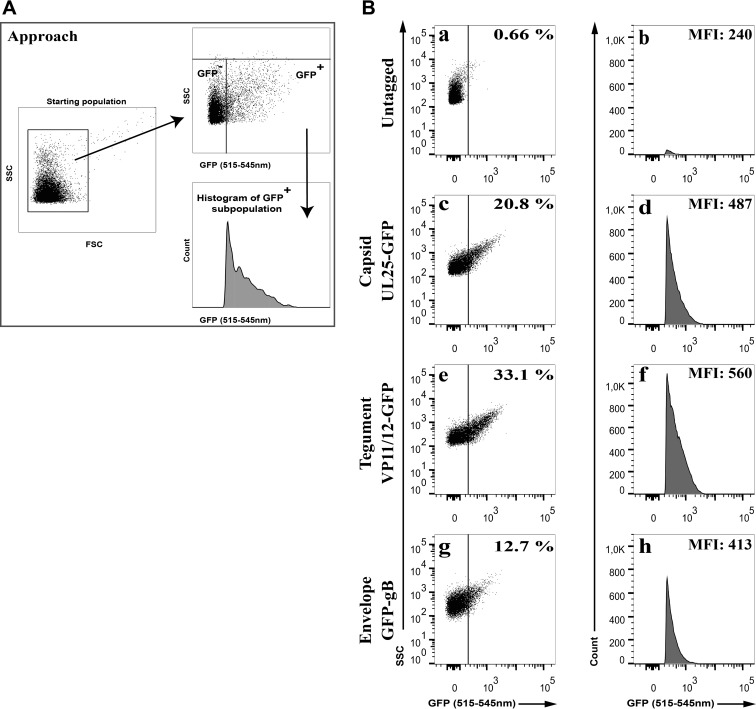
Since it was possible to detect Syto-stained virions by flow cytometry, we reasoned we could employ a different strategy to detect them by GFP tagging diverse virion components. As indicated in Fig. 2, extracellular mature virions encoding GFP-tagged capsids (UL25-GFP), tegument proteins (VP11/12-GFP), or envelope proteins (GFP-gB) all were readily detectable by flow cytometry over the background signal. Although this seemed less efficient than with Syto 13-stained virions, these findings paved the way for the analysis of the distinct tegument proteins present in extracellular virions.
FIG 2.
Analysis of GFP-tagged virions by FACS. Vero cells were infected with wild-type HSV-1 (untagged) or fluorescent recombinant viruses tagging the viral capsid (UL25-GFP), the tegument (VP11/12-GFP), or envelope (GFP-gB) at an MOI of 5 for 18 hpi. Virions were diluted and directly analyzed by flow cytometry. (A) Schematic description of the gating strategy. Virion aggregates were first removed by gating them out, and GFP+ particles were analyzed. (B) The proportion (%) and mean fluorescence levels (MFI) of the GFP particles are indicated in each graph (averages from 3 independent experiments). As described for Fig. 1, these percentages denote the amount of GFP-positive particles relative to the starting population, which includes inert particles inherently present in the FACS buffer (24). Meanwhile, the mean fluorescence signal (MFI) is only that of the GFP+ viral particles. Note the sharp edge on the left of the GFP histograms, a direct consequence of the gating.
Flow cytometry quantification of viral particles.
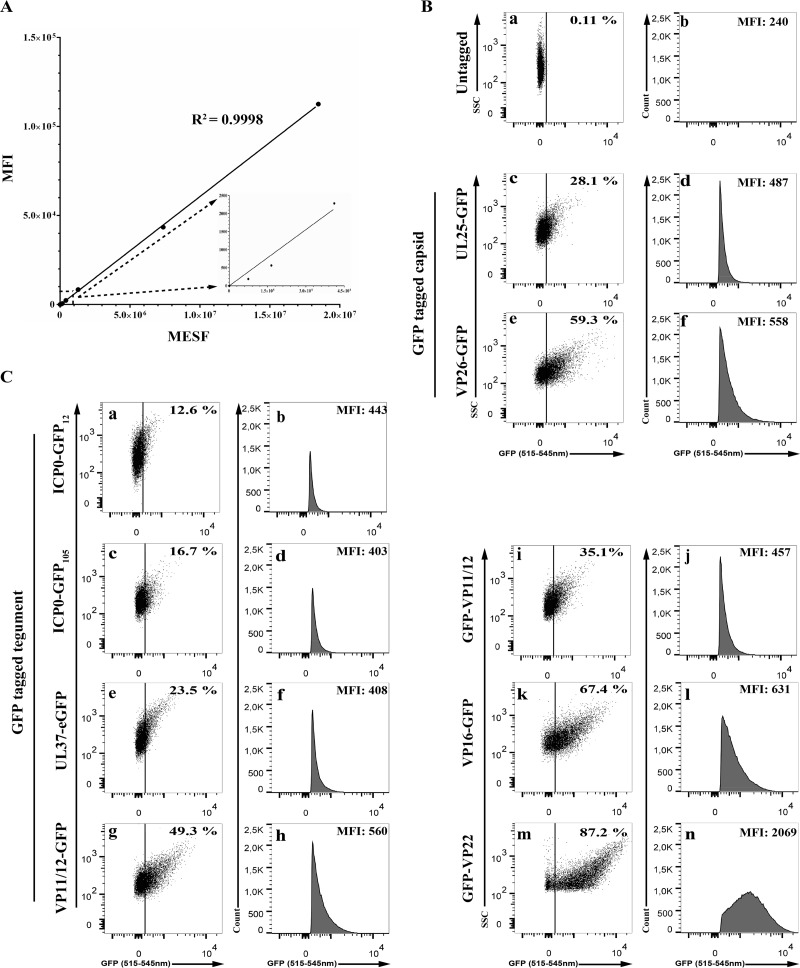
To define the usefulness of our flow cytometry approach, we first confirmed that the GFP signals could be quantitatively detected. Not surprisingly, commercials beads using known relative amounts of GFP signal lead to a very linear detection by the flow cytometer (Fig. 3A). This was largely expected, since these beads are commonly used to calibrate those instruments. Thus, we could use flow cytometry to measure GFP signals embedded in virions. We next probed a battery of recombinant viruses coding for various GFP-tagged tegument proteins that are incorporated in virions (Table 1 and Fig. 3C). We also used two capsid controls, i.e., the K26GFP virus, which tags the VP26 protein, as well as GS4677, which encodes UL25-GFP (Fig. 3B). Cells were infected with these recombinant viruses, and the extracellular virions were collected 18 h later and analyzed by fluorescence-activated cell sorting (FACS). In all cases, the virions were detected initially by light scattering, gating on the bulk of the particles using the same gating strategies that exclude large aggregates, as described for Fig. 1 and 2. All individual particles were analyzed subsequently in the fluorescence channel (Fig. 3B and C, dot blots) and then focusing on the GFP-positive particles (Fig. 3B and C, histograms). For all recombinant viruses, clear signals were detected above the control untagged wild-type virions. This was in full agreement with our previous report that particles as small as 100 μm could indeed be detected by flow cytometry (24). Note that UL25, which most recently has been estimated at 60 copies (25), also could be detected. We concluded that flow cytometry can be used to analyze individual virions tagged for various components, even when low-copy-number proteins are tagged.
FIG 3.
Flow cytometry detects all structural constituents of the virus. (A) Linear detection of AcGFP flow cytometer calibration beads analyzed by flow cytometry using a 488-nm laser line. Note the blow-up of the first few points. (B and C) Extracellular virions were purified from wild-type (i.e., nonfluorescent)-infected cells or from cells infected with fluorescent recombinant viruses and examined by flow cytometry as described for Fig. 1. The graphs on the left show total individual particles (i.e., excluding aggregates), while those on the right show the histogram profiles of the GFP-positive gated material for each virus. The MFI and the proportion (percentage) in each graph are averages from 4 independent experiments. Once again, these percentages denote the amount of GFP-positive particles relative to the starting population, which includes inert particles inherently present in the FACS buffer (24). Meanwhile, the mean fluorescence signal (MFI) is only that of the GFP+ viral particles. Once again, the sharp edge on the left of the GFP histograms is a direct consequence of the gating rather than a binomial distribution of the signal.
TABLE 1.
Viruses used in this study
| Virus | Strain | Tagged protein | Reference or source |
|---|---|---|---|
| HSV-1 GS4677 | F | UL25-GFP | Unpublished |
| HSV-1 GS3351 | F | GFP-VP11/12 | 42, 43 |
| HSV-1 GS2971 | F | GFP-gB | 43 |
| HSV-1 0+ GFP 12 | KOS | ICP0-GFP 12 | 44 |
| HSV-1 0+ GFP 105 | KOS | ICP0-GFP 105 | 44 |
| HSV-1 K26GFP | KOS | VP26GFP | 45 |
| HSV-1 K37eGFP | KOS | UL37eGFP | 46 |
| GHSV-UL46 | KOS | VP11/12-GFP | 47 |
| HSV-1 VP16-GFP | 17 | VP16-GFP | 48 |
| HSV-1 GFP VP22 | 17 | GFP-VP22 | 49 |
Individual viral particles vary in their tegument content.
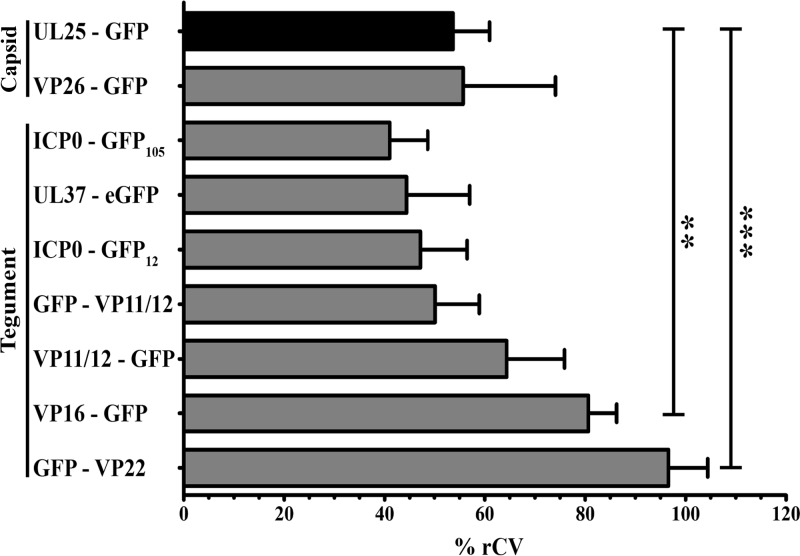
Having established a method to individually analyze large numbers of HSV-1 virions, we proceeded to compare the tegument content of extracellular particles using the aforementioned battery of GFP-tagged viruses. To this end, we characterized by flow cytometry 100,000 extracellular virions of each recombinant virus and calculated the variance of the GFP signals. Rather than employing the coefficient of variation of the mean (CV), we instead used that of the median (rCV), a more robust approach that naturally excludes outlying points that can strongly skew results. As a control, we evaluated the rCV of UL25-GFP-tagged capsid component, since this capsid component is believed to be invariant among the virions (25). Finally, we performed statistical comparisons between the rCV obtained for various samples with that for the UL25-GFP virus to detect tegument proteins that may vary significantly among viral particles. Figure 4 reveals that the capsid and tegument components VP26, ICP0, UL37, and VP11/12 did not significantly differ from UL25 in variance. In contrast, VP16 and VP22 were much more variable among viral particles (P < 0.01 and P < 0.001, respectively). Thus, while some tegument proteins are incorporated in similar amounts among viral particles, VP16 and VP22 vary considerably from particle to particle. This was consistent with their much broader distributions in the GFP profiles, as opposed to the tight distribution for UL25 and VP26 (Fig. 3A).
FIG 4.
Variability of the tegument among individual viral particles. Extracellular virions were purified from wild-type (i.e., nonfluorescent)-infected cells or from cells infected with the indicated fluorescent recombinant viruses and analyzed by flow cytometry. Tegument variability was measured by the coefficient of variability based on the medians (% rCV). Error bars represent the standard deviations from 4 independent experiments. Student t tests were performed to analyze the significance of the data (**, P < 0.01; ***, P < 0.001).
VP16 and VP22 levels in virions appear to correlate with infectivity.
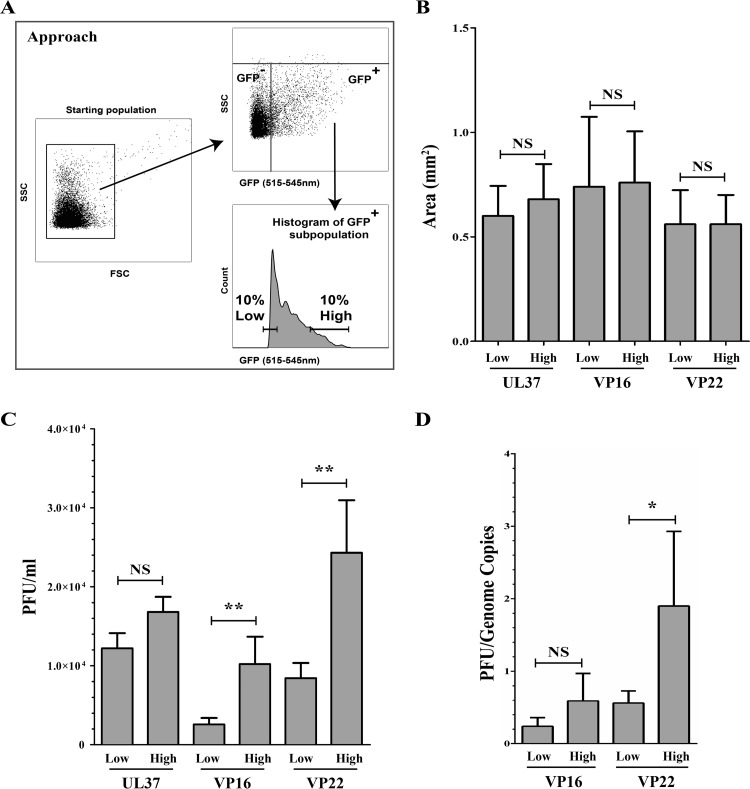
The most critical question was whether the higher variability in VP16 or VP22 content in virions had any biological relevance or simply reflected a reduced stringency in copy numbers. We therefore exploited the power of flow cytometry to sort viruses based on tegument abundance. To this end, we harvested viral populations at the extreme ends of the tegument distribution curves and got viral stocks incorporating either the lowest or top 10% amounts of VP16 or VP22 (Fig. 5A). We then measured their efficacy to infect Vero cells in plaque assays, initially using the same number of FACS events to infect cells. Under these conditions, no differences were noted in plaque size, indicating that cell-cell spread was not impaired (Fig. 5B). However, a modest but reproducible 2.5- to 4-fold difference in the number of PFU was found (Fig. 5C). In contrast, when the same experiment was done for the invariant UL37 tegument protein, selecting once again for viruses incorporating the lowest and top 10% levels of UL37, no differences in plaque size or infectivity were observed (Fig. 5B and C). As expected, virions with high VP22 content appeared to contain an increased proportion of infectious particles (Fig. 5D). Oddly, such difference was not statistically significant for VP16 but was reproducibly detected.
FIG 5.
VP16 and VP22 abundance correlate with infectivity. Extracellular virions were purified from cells infected with K37eGFP, VP16-GFP, or GFP-VP22 viruses and sorted by flow cytometry, as described for Fig. 1, for their low or high levels of UL37, VP16, or VP22, respectively. (A) Schematic description of the gating strategy applied for the sorting of viruses by FACS. Virion aggregates were first removed by gating them out. (B and C) Viral spread of the sorted viruses. The infectivity of the sorted virions was assessed by plaque assays on Vero cells, and plaque size (B) and abundance (C) were evaluated. (D) PFU/genome copy number ratios. The PFU/genome copy number ratio was obtained by dividing the PFU values by the corresponding values of genome copies measured by qPCR for the same viral samples. Error bars represent the standard deviations derived from 3 independent experiments and analyzed with the Student t test (NS, not significant, i.e., P > 0.05; **, P < 0.01).
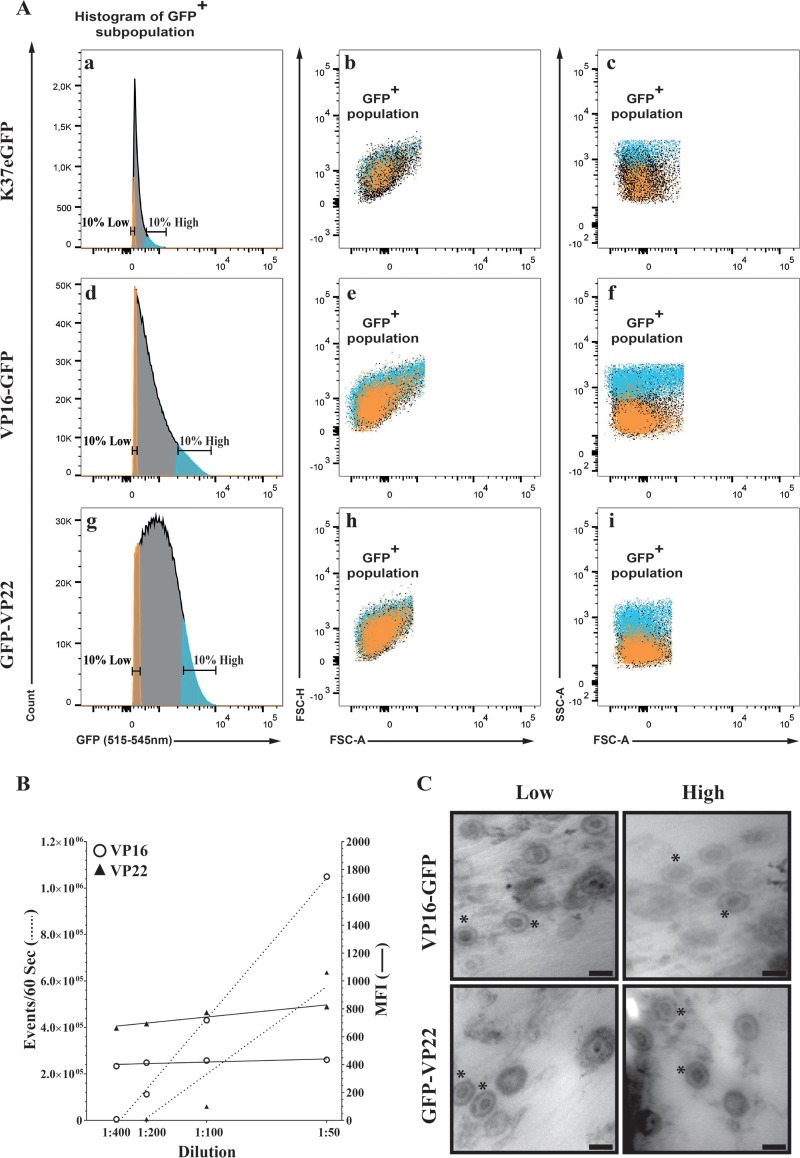
In an effort to better understand why virions containing different amounts of VP16 or VP22 differed, we considered the presence of doublets, aggregates, and coincidental events in the high-content-sorted fractions. Although sorting was performed under technical conditions that typically exclude such events (see Materials and Methods), we nonetheless used three independent approaches to rule them out. Given that forward scattering (FSC) is an indication of size while side scattering (SSC) is rather suggestive of sample granularity and internal complexity, we first compared the light-scattering profiles of the high/low fractions of VP16 and VP22 samples. Figure 6Aa, d, and g show the typical profiles of viral particles prior to their sorting into high- and low-GFP content. To evaluate the properties of these sorted viral particles, we examined their light-scattering profiles. Figure 6Ab, e, and h highlight the forward light scattering of these sorted particles, which is color coded for ease of analysis (orange for the lowest 10% GFP+ viral particles and blue for the highest 10% GFP+ viral particles, while black dots represent unselected particles not belonging to these two groups). Clearly, both GFP populations displayed similar forward-scattering properties and thus were likely of similar size. Interestingly, viral particles containing more VP16/VP22 exhibited an increased side-scattering profile, presumably reflecting a greater complexity than their low-content counterparts (Fig. 6Ac, f, and i). We conclude that viral particles containing high levels of tegument proteins are, by this criteria, of sizes similar to those of their low-content counterpart. We next performed a classical flow cytometry dilution analysis, which is based on the assumption that coincidental events would be detected at low sample dilution but would disappear when samples are highly diluted. In that scenario, the mean fluorescence intensities of the recorded events would be reduced by simply diluting the samples. Figure 6B shows that, as expected, diluting the viral stocks reduced the number of events recorded by the instrument during the fixed 60-s analysis but only marginally altered the fluorescence levels of each of those events. These results are once again consistent with the conclusion that the analysis of individual viral particles was achieved. Finally, sorted viruses were analyzed by electron microscopy. As shown in Fig. 6C, individual viral particles were readily detected with no evidence for viral aggregates. As expected, the bulk of these viruses shared the characteristics of mature virions with a DNA-containing capsid and an envelope. We conclude that the higher signals measured by FACS in the high-content fractions were unlikely to be attributed to aggregates, virion doublets, or coincidental events.
FIG 6.
Sorted viruses are single viral particles. (A) Extracellular virions were purified from cells infected with VP16-GFP or GFP-VP22 viruses, analyzed, and then sorted by flow cytometry as described for Fig. 5A for their low or high levels of VP16 or VP22 content, respectively. All images are only GFP+ particles, i.e., those already depleted of aggregates and nonfluorescent components. FSC-H and FSC-A are the height and area under the curve of the forward-scattering signal, respectively, while SSC-A measures the area under the side-scattering signal. Orange, particles with the lowest 10% GFP signal; blue, particles with the top 10% GFP signal; black, particles exhibiting an intermediate GFP signal and not subsequently analyzed. The linear edges around the dot blots reflect the gate used to sort the samples. (B) Extracellular virions purified from infected cells were first diluted 1:50, and then serial dilutions of 1:2 were prepared and analyzed by flow cytometry by continuous recording of events during a fixed time (60 s in these experiments). (C) Sorted viruses were concentrated on 0.1-μm filters, which were embedded and processed for electron microscopy (see Materials and Methods). For clarity, the presence of some capsids with their envelope is indicated by asterisks. Bars represent 100 nm.
Higher infectivity of sorted viral particles is not explained by L-particles.
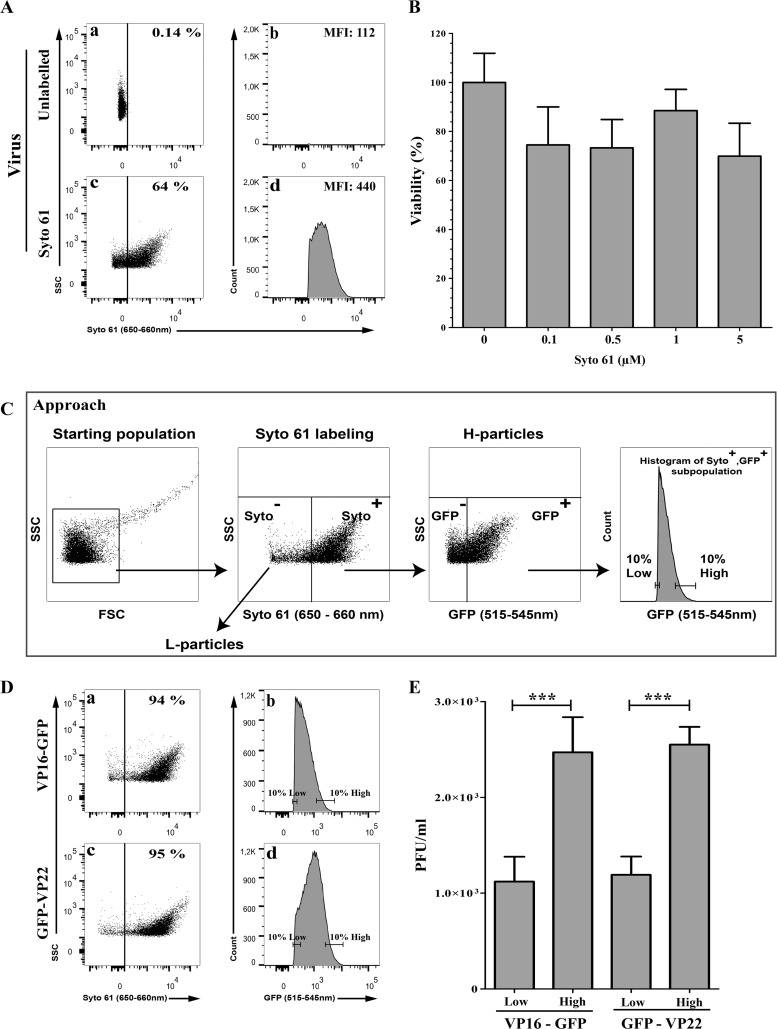
Typical HSV-1 preparations contain a proportion of so-called L-particles, noninfectious enveloped viral entities that are loaded with tegument proteins but lack a viral capsid or genome. It was therefore possible that the viral particles sorted as described above for their high tegument content proportionally contained fewer L-particles than their corresponding low-tegument counterpart, which would therefore be less infectious. To rule out this scenario, we performed dual labeling of the tegument and viral genome using the GFP-tagged viral strains and Syto dyes. Since GFP and Syto 13 share similar excitation and emission profiles, we screened a panel of red-emitting Syto dyes and identified Syto 61 as the best candidate (Fig. 7A and data not shown). Since we wanted to isolate intact virions, we first examined if the membrane-permeable Syto 61 perturbs the virus. We consequently compared the plaque-forming capacity of untreated or Syto 61-stained virions with various concentrations of the dye and found that it was well tolerated by the virions and had no impact on the ability of the viruses to propagate (Fig. 7B), as did Syto 13 (data not shown). It was therefore possible to label, with Syto 61, the viral genome of the above-described GFP recombinants and eliminate L-particles by first selecting Syto 61-positive events and then sorting GFP-positive viral particles (Fig. 7C, cartoon). Syto 61-positive viral particles sorted for their high content of VP16 or VP22 (Fig. 7D) again were significantly more infectious that their low-content VP16 or VP22 virion counterparts (Fig. 7E), reiterating the above-described findings and ruling out a significant implication of L-particles in the above-described results. We therefore concluded that particles sorted for their high VP16 or VP22 content seemed more infectious than particles containing smaller amounts of the same proteins.
FIG 7.
Difference in infectivity is not linked to L-particles. (A) Staining of virions with Syto 61. Vero cells were infected with untagged wild-type viruses at an MOI of 5 for 18 hpi. Purified extracellular virions were stained with 1 μM Syto 61 or mock treated and analyzed by flow cytometry. Aggregates were gated out and Syto 61 fluorescence evaluated. The percentages denote the amount of Syto 61+ particles relative to the starting population, which includes inert particles inherently present in the FACS buffer (24). Meanwhile, the mean fluorescence signal (MFI) is only that of the Syto 61+ viral particles. (B) Syto 61 does not affect the viability of the stained virions. Wild-type strain F extracellular virions were stained with 0 to 5 μM Syto 61 for 1 h at 4°C. The viability of the Syto 61-labeled virions was assessed by standard plaque assays. For comparison, the number of plaques obtained for the untreated sample was set at 100%. Error bars represent the standard deviations from 2 independent experiments. (C) Schematic description of the approach to deplete L-particles and enrich for heavy particles (H-particles). Once aggregates were gated out (leftmost image), VP16-GFP or GFP-VP22 Syto 61-positive events were selected to exclude L-particles (second image from the left). A second gating was then applied to select GFP-positive particles (third image from the left). The samples were finally sorted on the basis of their low or high levels of VP16 or VP22 (rightmost image). As before, all sorting took place under conditions where only single individual viral particles were present (see Materials and Methods). (D) Sorting of DNA-containing GFP-positive virions. High-content and low-content viral particles were sorted according to the scheme depicted in panel C. Percentages denote the amount of GFP+ particles, which only includes heavy particles with a potential contamination of a mere 5 to 6% by L-particles or H-particles that were not labeled with the Syto 61 dye. (E) Infectivity of the sorted virions. The infectivity of the Syto 61+/GFP+ sorted virions was assessed by plaque assay on Vero cells. Error bars represent the standard deviations from 2 independent experiments, and results were analyzed with the Student t test (***, P < 0.001).
Depletion of VP16 or VP22 in virions hints at a complex scenario.
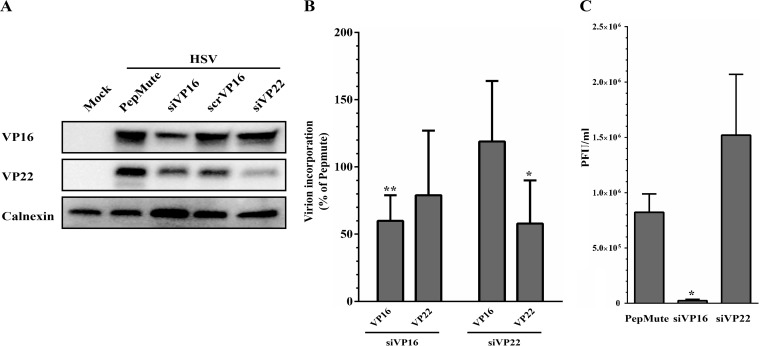
To orthogonally confirm our findings, we used an RNA interference strategy to lower the amounts of the proteins in the virions and assessed their infectivity, an approach we successfully used in the past for VP16 and several host proteins incorporated in virions (13, 26). As anticipated, short interfering RNA (siRNA) targeting VP16 or VP22 significantly reduced the level of expression of these proteins in infected cell lysates (Fig. 8A). Not surprisingly, this resulted in a concomitant depletion of the proteins in viral particles produced on siRNA-treated cells (Fig. 8B). Individual testing of the two siRNAs used to block VP22 expression also led to reduced levels of VP22, albeit with different efficacies, suggesting off-target effects were not an issue (data not shown). We then proceeded to evaluate the infectivity of these extracellular virions and found that the depletion of VP16 from the virus had a profound impact on its ability to form plaques (Fig. 8C), in full agreement with the flow cytometry data. Unexpectedly, VP22-depleted virions had the opposite effect on viral infectivity and exhibited an improved plaque-forming capacity. Thus, while the amount of VP16 in virions directly correlated with the infectivity of the viral particles, a more complex situation seemed to prevail for VP22.
FIG 8.
Analysis of VP16 and VP22 siRNA-depleted virions. 143B cells were transfected for 48 h using PepMute with a unique RNAi against VP16 or two pooled siRNAs targeting VP22. Cells were then infected with HSV-1 KOS at an MOI of 5 for 24 h. (A) Immunoblot of mock-treated or siRNA-transfected and HSV-1-infected cell lysate. Thirty micrograms of cell lysate was separated by SDS-PAGE, transferred to PVDF membrane, and probed with antibodies against VP16 and VP22. Calnexin was used as a loading control. (B) Impact on VP16/VP22 incorporation in virions. The amount of VP16 or VP22 in extracellular virions produced by cells transfected with the indicated siRNA was evaluated by Western blotting in 5 independent experiments, each normalized to the untreated sample (i.e., PepMute set at 100%). (C) Impact on infectivity. The infectivity of the VP16- or VP22-depleted extracellular virions was assessed in standard plaque assays. Error bars represent the standard deviations from 3 independent experiments (*, P < 0.05; **, P < 0.01).
The overall composition of the tegument can be altered by modulating single components.
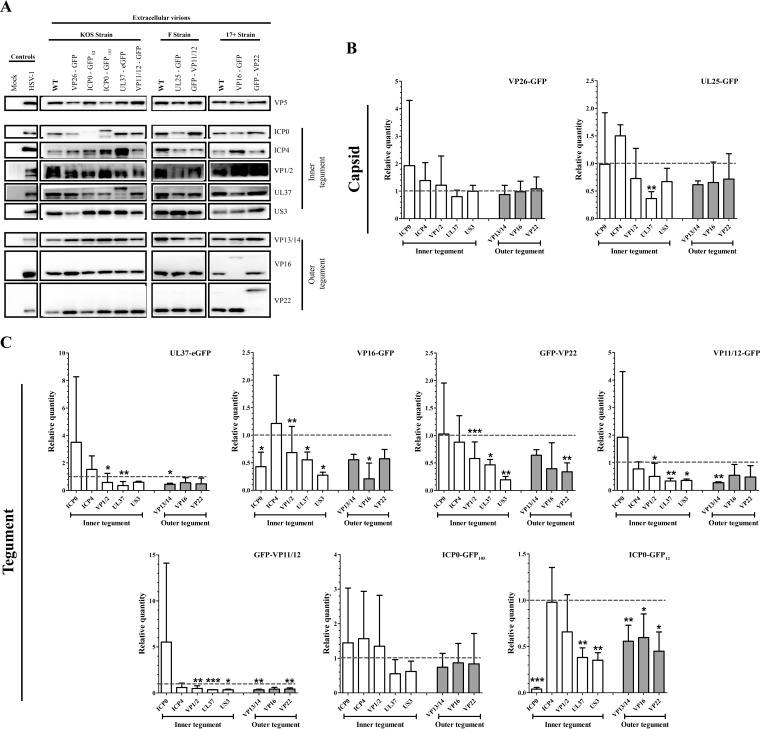
The tegument layer is the result of an intricate interconnectivity of its components, with the deletion of one tegument protein often impacting the incorporation of other viral proteins. We therefore asked whether tagging of the tegument proteins affected their level of incorporation as well as that of other virion components. This was achieved by quantitative Western blotting of all of the recombinant virions used in this study using a battery of antibodies (Fig. 9). To ensure equal numbers of viral particles were loaded, all values were normalized for VP5. These tegument/VP5 ratios were then compared to their corresponding wild-type strains (arbitrarily set at 1) to ensure the data were not biased by strain considerations. Interestingly, tagging of the capsid protein VP26 with GFP had no noticeable impact on its incorporation or any of the tegument proteins examined (Fig. 9). Similarly, tagging of UL25 seemed to incorporate slightly less UL37 but had an otherwise normal complement of other tegument proteins. In sharp contrast, tagging of most other tegument proteins drastically reduced their incorporation and that of multiple other virion components. Interestingly, while tagging of VP11/12 at either the carboxyl- or amino-terminal end yielded similar results. This was clearly not the case for ICP0, with the ICP0-GFP105 virus seemingly having a normal composition while the ICPO-GFP12 virus had a profoundly distinct virion content, suggesting a positional effect of the GFP moiety. Altogether, these results indicated that large tags such as GFP can impair the incorporation of the labeled proteins but can also alter the overall composition of viruses. We consequently conclude the observed impact of VP22 on infectivity was likely indirect and due to several factors subsequent to the overall change in virion composition.
FIG 9.
Interconnectivity of the virion components. (A) Western blot analyses. Extracellular virions indicated at the top of the blots were purified from infected cells and analyzed by Western blotting using antibodies indicated to the right of each blot. (B and C) Tegument quantification. The amounts of various tegument proteins present in virions tagged on the capsid (B) or the tegument (C) were determined by quantitative Western blotting. The data were normalized to VP5 to ensure even loading of the gels. These tegument/VP5 ratios were finally compared to those of the wild type for each strain, arbitrarily set at 1, to avoid biases among viral strains. Error bars represent the standard deviations from 3 independent experiments. Student t tests were performed to analyze the significance of the data (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Note that throughout this study, a ChemiDoc was used, not film, which is notoriously nonlinear.
DISCUSSION
The power of flow cytometry.
One limitation in the characterization of the heterogeneity of viral particles has been the limited tools to examine them individually, with the notable exception of labor-intensive electron microscopy and, more recently, fluorescence microscopy. The present study meaningfully contributes to these past findings in several ways. First, we probed by flow cytometry 100,000 particles, adding a significant level of power to such analyses. Second, we quantitatively looked at the protein content of individual viral particles, not entire viral populations, as is typically the case for Western blotting, biochemical assays, or mass spectrometry. Third, unlike the above-mentioned studies, we could directly correlate protein content changes to individual virion infectivity. Finally, the present work looks at a large picture by probing the impact of the modified protein on the overall composition of the virus. This, in principle, enables a much better view of the real consequence of manipulating virion content even if only altering one viral protein. Thus, our flow cytometry analysis of individual viral particles is clearly a very useful technical advancement but also leads to very relevant information.
Our previous (24) and present work show that it is possible to detect, analyze, and sort individual viral intermediates, including mature virions, despite their limited size below the theoretical detection limit of most FACS instruments. These viral particles can be labeled either with membrane-permeable Syto reagents, which emit fluorescence when bound to nucleic acid, or by genetically tagging various virion components with a fluorescent molecule. Interestingly, flow cytometry appears insensitive to the location of the tags in the virions, as DNA, capsid, tegument, and envelope proteins all could be detected. This is not surprising, since this methodology has been used to monitor both cell surface markers as well as internal molecules, either fluorescently labeled or through detection with antibodies following permeabilization of the cells. Consequently, the location of a given GFP-tagged protein in the virion, for instance, an inner or outer tegument protein, should have no bearing on its detection. These findings enabled us to individually and statistically probe the relative amounts of tegument proteins among very large numbers of viral particles. This unique advantage over existing methods is a very useful tool to explore the variability of virions in terms of their protein content, in addition to being a very efficient way to purify viral intermediates.
Variability of the tegument.
The HSV-1 tegument proteins UL37, ICP0, and VP11/12 did not statistically vary any more than the UL25 capsid proteins or VP26, for that matter. Nonetheless, the data suggest some level of variability. Whether this reflects real biological variability is not known. Intuitively, one expected no variation at all for VP26 if it is incorporated in a unique stoichiometry into the capsid. Thus, we were initially surprised by its rCV values. However, we find the same variability as Smith and colleagues (19) using fluorescence microscopy, who reported a variance of 0.48 while we got 0.53 (i.e., 53%). On the other hand, the variance of VP26 is clearly more restricted than that of VP22 and VP16 (see the tight bell curves for VP26 compared to others in Fig. 3Bd). The question remains: why this variation? Perhaps the stoichiometry of VP26 is not as fixed as expected, as evidenced by the lower incorporation of GFP tagged in the corresponding UL25 in PRV particles (27), or alternatively, that the technology we used (i.e., fluorescence) has limitations. Although unanticipated from past findings, this may nonetheless be significant. One case in point is the UL17/UL25 complex, which was first thought to specifically label C capsids but has since been found on other nuclear capsids, albeit at lower levels (28–30). Interestingly, levels of VP22 and, to a lesser extent, VP16 fluctuated significantly more among viral particles than the above-described proteins. This is in agreement with a quantitative mass spectrometry study of VP13/14, VP22, and US3 PRV deletion mutants, which hinted that VP1/2 and VP13/14 are incorporated in virions in fixed amounts, while VP11/12, VP16, and VP22 are present in variable quantities (22). This scenario is also consistent with work by Smith and colleagues, who found by fluorescence microscopy some heterogeneity of specific PRV tegument proteins among viral particles (19). Finally, our data are in line with various reports that indicate VP22 varies much more than other tegument proteins, with wildly ranging estimates of 450 to 2,640 copies per HSV-1 viral particle (15, 17, 18). Altogether, this reiterates the value of flow cytometry to quantitatively assess virion content.
The variability of some tegument proteins and not others is intriguing. Interestingly, tegument variability maps to abundant tegument proteins, but it is difficult to ascertain whether this is purely coincidental. A potential scenario may be that the outer tegument is spatially less stringent in the outskirts of the viral particles and/or is less organized there, allowing for more versatility in the copy number of proteins such as VP16 and VP22. This would contrast with inner tegument proteins, which interact with the highly structured viral capsid. This is consistent with the reported greater incorporation of the outer VP22 tegument protein in virions when overexpressed and the stable incorporation of the inner UL37 tegument protein under similar conditions (20, 21). Clearly, more spatial information on the arrangement of the tegument layer is required to fully assess this possibility. An alternative view is that some of the tegument variability is caused by unspecified mutations in some of the viral strains used in this study, particularly bacmid-derived strains. While most of them have been characterized (see references in Table 1), full sequencing data are not yet available. An additional aspect is tegument variability and overall protein composition of virions produced in various cell lines, which clearly add another layer of complexity to the present findings (31).
Interdependence of tegument components.
Tagging virions with GFP often impacts viral fitness to the point that some proteins cannot readily be tagged without consequences (32, 33). This implies a negative effect of the tag on the organization of the virions and/or function of the labeled proteins. The present study confirms this and shows that the GFP moiety can have consequences for other tegument components. Hence, while GFP labeling of VP26, UL25, or ICP0 at position 105 but not position 12 had minimal impacts on the composition of the viruses, the coupling of GFP to most tegument proteins clearly reduced their recruitment and limited the incorporation of several other viral proteins. Moreover, this does not even take into account untested viral tegument proteins as well as the many host proteins that are incorporated in mature viruses (1, 13). Thus, the impact of one molecule on other virion proteins may even be greater than that reported here. The low incorporation of GFP-tagged ICP0 might mean that its incorporation into virions is optional or that it is not rate limiting, and consequently little is required to initiate the infection.
Absolute values of individual tegument proteins.
The present study reports the relative amounts of tegument proteins, which has proved very useful to evaluate tegument variability among viral particles. Unfortunately, it has not been possible to define absolute copy numbers, although it was one of our original goals. Apparently, GFP fluorescence varies by as much as 10-fold depending on the size of the tagged proteins, with large proteins being comparatively less fluorescent (John Nolan, personal communication). For this reason, commercial beads with defined absolute GFP copy numbers cannot be used as standards to determine absolute tegument copy numbers. Consequently, it would not be possible to tag one viral component, for example, VP26, to evaluate the precise numbers of another virion component. However, as shown in Fig. 3B, it is readily possible to monitor abundance by measuring relative copy numbers of a given protein under different conditions, which is precisely what we did. It remains to be seen whether a different tagging strategy could circumvent this limitation.
Biological relevance of tegument heterogeneity.
Our flow cytometry and RNA interference findings show a clear correlation between VP16 abundance in virions and infectivity. One plausible explanation rests with the trans-activating properties of VP16, where presumably more of the protein in the incoming virions should stimulate the infection if it is rate limiting. This is consistent with the boosting impact that L-particles, which contain tegument proteins, have on the infectivity of transfected viral DNA (i.e., devoid of tegument) as well as their lack of stimulation when used in parallel with viruses containing a full set of tegument proteins, especially at high multiplicities of infection (MOIs) (23). Given the 73% reduced levels of VP16 in the VP16-GFP recombinant viruses compared to its wild-type counterpart, VP16 levels in this recombinant virus may indeed be suboptimal. Rather than being a caveat, this instead allowed us to appreciate its critical role in the incoming viruses early during the infection, particularly since it is otherwise a late protein. Alternatively, or perhaps additionally, this study may point to the contribution of other virion components, whose levels are indeed altered in the VP16-GFP viruses (this study), or the many host and viral proteins that recently have been shown to interact with VP16 (34). However, resolving this issue constitutes a significant challenge that will require multiparametric approaches. We also considered the possibility that high/low fractions had inherently different viral particle stability that could provide an alternative justification for our results. To actually measure viral particle stability may be challenging, given that it requires biophysics approaches that depend on large concentrations of material. Although we cannot formally rule out that low-VP22 or -VP16-content particles are less stable than their high-content counterparts, the data suggest it is the different amounts of these molecules in the virus, not their stability, that directly or indirectly impacts infectivity. First, both high and low populations of the control UL37eGFP virions were equally infectious (Fig. 5c), ruling out an indirect consequence of the sorting process or the flow cytometer itself. Second, a priori, there is no reason to believe that the VP16 and VP22 viral proteins would have the same structural impact on the virions or that less rather than more proteins should necessarily be more deleterious to virion stability. In agreement with this, VP16 and VP22 viral particles similarly impacted fitness, with both kinds of high-content particles being more infectious (Fig. 5c). Third, the PFU-to-genome-copy-number ratios are increased in high populations of both VP16 and VP22, strongly suggesting that these particles are indeed more infectious than their low-population counterparts (Fig. 5d). Finally, high- and low-population VP16 or VP22 particles have similar morphologies by EM with no obvious differences (Fig. 6c).
Regarding the HSV-1 VP22 tegument protein, our flow cytometry data initially suggested a direct link between infectivity and VP22 abundance. However, downregulating VP22 in virions by RNA interference resulted in a surprising increase in infectivity, in apparent contradiction with the above-described findings. The simplest interpretation is that VP22 abundance per se is not the main or sole contributing factor to viral fitness. In the present context, it is worth noting that although the levels of VP16 were statistically unaltered in the GFP-VP22 recombinant viruses, they nonetheless were reproducibly reduced. That VP22 interacts with the trans-activating domain of VP16 and has been postulated to modulate its function is also of particular interest (35). Moreover, VP22 has been implicated in the optimal localization and expression of several viral proteins or their incorporation into mature virions, including VP16 (36–38). Furthermore, it is possible that viruses sorted for their high levels of VP22 also contained more VP16, thereby promoting viral infectivity. To address the latter possibility, we attempted to compare by Western blotting the compositions of the flow cytometry-sorted VP16-GFP or GFP-VP22 with that of untagged virions. Unfortunately, this proves technically problematic, as we estimated that several days of continuous sorting were required to produce sufficient material, since this approach generates highly diluted samples and significant losses of material during sorting and in subsequent concentration steps (data not shown). We therefore cannot tell at this point if viral particles loaded with more VP22 were more infectious based on their VP16 content or activity and/or other proteins.
The minimal differences in infectivity reported here may seem unconvincing to some used to log differences. However, such small differences nonetheless may be significant. One appealing outcome of the present work is the potentially preferential infection of cells by the fittest viral particles. This would be consistent with reports indicating that both neuronal and nonneuronal cells are typically infected by only a few HSV-1 virions (39–41). Hence, a 2- to 3-fold advantage may be sufficient to preferentially promote the spread of the fittest virions in their hosts. However, an open question is how such a selection might operate, but resolving this issue may be challenging given the complex interconnectivity of the tegument proteins and the impact of GFP tags on the overall composition of virions. One other major difficulty is to evaluate the impact of tegument variability in vivo, i.e., in animal models, since every new round of infection will generate some diversity among the viral particles, even if we infect the animals with homogenous viral stocks. Thus, the infection of animals with FACS-sorted virions would quickly lead to a heterogeneous viral population, likely masking any phenotype linked to tegument amounts. Therefore, some ingenuity would be required to solve this difficult puzzle.
Concluding remarks.
Overall, the present study quantifies the generally accepted concept whereby viral fitness is dependent not merely on the genetic makeup of viruses but also on the precise levels of proteins that are incorporated into the viral particles. A second important aspect of this work is that modulating a single virion component can have profound impacts on the overall composition of a virus. Although the present study focuses on HSV-1, it likely applies to other members of the herpesvirus family, all complex entities containing a great number of components in their mature particles. Altogether, this opens up new research avenues to address the virulence of HSV-1 and potentially other viruses.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney Vero cells (ATCC CCL-81) were cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; HyClone), 2 mM l-glutamine (Life Technologies), and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). 143B cells (ATCC CRL-8303) were also supplemented with 15 mg/ml 5-bromo-2 deoxyuridine (BrdU; Sigma).
The GFP-tagged and corresponding wild-type HSV-1 viral stocks used in this study are summarized in Table 1. These include the parental strains KOS, F, and 17+, which were kindly provided by Beate Sodeik. The K26GFP and K37eGFP recombinants viruses, both in the strain KOS background and carrying a GFP tag on the capsid protein VP26 and the tegument protein UL37, respectively, were kindly provided by Prashant Desai (45, 46). The HSV-1 GS4677, GS3351, GS3330, and GS2971 recombinant viruses carrying a GFP tag on the capsid protein UL25, the tegument proteins VP11/12 and VP16, and the envelope glycoprotein B (gB), respectively (all strain F derived), were generously provided by Gregory Smith (42, 43). The 0+ GFP 12 and 0+ GFP 105 recombinant viruses, obtained from William Halford, are in the KOS background and carry a GFP tag on the tegument protein ICP0 between amino acids 11 to 12 and 104 to 105, respectively (44). The GHSV-UL46 recombinant virus tagging VP11/12 (strain KOS) was kindly provided by Jim Smiley (47), while the HSV-1 VP16-GFP recombinant virus (strain 17+) carrying a GFP tag on the aforementioned protein was obtained from Peter O'Hare (48). Finally, the HSV-1 GFP VP22 recombinant virus (strain 17+), carrying a GFP tag on the tegument protein VP22, was kindly provided by Gill Elliott (49). All viruses were amplified on BHK, 143B, or Vero cells, and titers were determined on Vero cells by plaque assay as previously described (50). Please note that, for clarity, all figures in this study refer to the tagged proteins rather than the official names of the viruses (Table 1).
Purification of extracellular virions.
Vero cells freshly passaged the day before were grown on 500-cm2 dishes until 80% confluence. Cells were mock treated or infected with wild-type or recombinant HSV-1 at a multiplicity of infection (MOI) of 5. At 18 h postinfection (hpi), the extracellular medium was harvested and clarified by centrifugation at 300 × g for 5 min at 4°C. The samples were then filtered through a 0.45-μm filter to eliminate intact cells and large cellular debris. Extracellular virions were subsequently pelleted by centrifugation at 20,000 × g for 1 h at 4°C in a Beckman SW32-Ti rotor. The viral pellets were resuspended in MNT buffer (30 mM morpholineethanesulfonic acid [MES], 100 mM NaCl, 20 mM Tris-HCl, pH 7.4) and treated for 30 min at 10°C with 500 U/ml of DNase I (Roche) and 2 mg/ml of RNase A (Invitrogen) to digest both cellular and nonencapsidated viral nucleic acids. The viral preparations were then stored at −80°C.
Antibodies.
Primary antibodies for Western blotting were graciously provided by various sources (and diluted) as follows. Anti-VP1/2 (1:300) was provided by R. Courtney (51), anti-UL37 (1:1,000) was provided by F. J. Jenkins (52), anti-US3 (1:4,000) was provided by B. Roizman (53), anti-VP16 (1:1,000) was provided by H. Browne (54), and anti-VP13/14 (1:5,000) and anti-VP22 (1:10,000) were provided by G. Elliot (55, 56). Commercial antibodies against VP5 (1:1,000; Cedarlane), ICP0 (1:5,000; Abcam), ICP4 (1:1,000; Abcam), and calnexin (1:1,000; Enzo Life Sciences) were also used. Horseradish peroxidase-coupled secondary antibodies (goat anti-mouse or anti-rabbit) were used at 1:10,000 and were purchased from Jackson ImmunoResearch.
Gel electrophoresis and quantitative Western blotting.
Five micrograms of extracellular virions was boiled for 10 min in loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 0.1% bromophenol blue, 10% glycerol, and 2% β-mercaptoethanol) and separated on 10% SDS-PAGE gels. Proteins were electrophoretically transferred from the gels to polyvinylidene difluoride (PVDF) membranes, and the membranes were incubated for 30 min in blocking buffer (10% nonfat dry milk, 13.7 mM NaCl, 0.27 mM KCl, 0.2 mM KH2PO4, 1 mM Na2HPO4, and 0.1% Tween 20). All primary antibodies were diluted in blocking buffer and added to blots for 1 to 2 h. Blots were then washed three times in blocking buffer and probed with secondary antibodies conjugated to horseradish peroxidase. The detection was done on a ChemiDoc MP system (Bio-Rad), which has a 4-orders-of-magnitude dynamic range, and Clarity Western ECL substrate (Bio-Rad). Images were acquired and analyzed with Image Lab software, version 5 (Bio-Rad). The volume intensity of each tegument band was divided by that of calnexin (in the case of cell lysates) or VP5 capsid protein (in the case of extracellular virions) for each sample to normalize for gel loading. These tegument/VP5 ratios were finally compared to that of the wild type of each strain, arbitrarily set at 1, to avoid biases among viral strains.
Syto 13 and Syto 61 fluorescent labeling of viral particles.
Five microliters of a diluted fraction of DNase/RNase-treated virions was incubated for 1 h at 4°C with 1 μM Syto 13 (green fluorescence) or Syto 61 (red fluorescence; Invitrogen), unless otherwise indicated. Note that either Syto signal is minimal as a free molecule but strongly stimulated when bound to nucleic acid. Consequently, unwashed and labeled virions were directly analyzed by flow cytometry (see below). To measure the impact of the Syto dyes on the viability of the virus, the stained samples were directly assessed in plaque assays as described below.
Flow cytometry analysis.
Flow cytometry was performed as previously described (24). Briefly, Syto 13- or 61-labeled particles and/or GFP recombinant viruses diluted 1:500 in 0.2-μm-filtered MNT were analyzed on a FACSAria II sorter (BD Biosciences) equipped with a 100-μm nozzle and 405-, 488-, and 633-nm lasers. The 100-μm nozzle, rather than a smaller one, was used to reduce the pressure and hence maximize the excitation time and signal strength (24). Analysis and sorting were performed in PBS at low pressure (23 lb/in2) and a flow rate between 1 and 3 for a maximum of 3,000 events/s to minimize coincidental events. For GFP and Syto 13, the samples were excited with a 488-nm laser coupled to an emission filter allowing the 515- to 545-nm wavelengths to go through. Syto 61 fluorescence was detected with the 633-nm laser coupled to a 660- to 670-nm bandpass. In all cases, a minimal threshold of 200 for the SSC channel was applied to remove the background signal and 100,000 particles were analyzed. They were then analyzed in the fluorescence channel, gating on GFP-tagged or Syto 13- and/or Syto 61-labeled virions as indicated. To ensure the approach only monitored single viral particles, as opposed to virion aggregates, several tools were used. First, as mentioned above, the samples all were prefiltered through a 0.45-μm filter. Second, the filtered virions were initially analyzed by light scattering, where FSC is a measure of size and SSC an indication of granularity and internal complexity. In all cases, a gate was applied on the bulk of the particles (>95%; see the corresponding figures), which excluded large aggregates. Third, a purity mask of 16 was applied upon sorting, meaning that each drop being scanned could not be sorted if a nontarget particle fell within the first or last 8/32 of the leading or trailing drop, thus effectively preventing doublets from being sorted and only allowing single-event sorting. The data were initially acquired with FACSDiva software, version 6.1.3 (BD Biosciences), and then processed with FlowJo, version 10.0.7r2 (TreeStar). Note that only fluorescent viral particles were considered for the histograms and the calculation of the mean fluorescence intensity (MFI). Upon sorting, the diluted virions were aliquoted and frozen at −80°C, and subsequently titers were determined on Vero cells as detailed below. The robust coefficient of variability (percent rCV) corresponds to the robust standard deviation divided by the median. Bilateral Student's t tests were used to determine any statistically significant differences between the samples.
AcGFP flow cytometer calibration beads.
Aequorea coerulescens GFP (AcGFP; no. 632594; Clontech) beads are a mixture of six distinct populations with a different amount of attached AcGFP molecules and, thus, a distinct fluorescent signature. The molecular equivalent of soluble fluorophore (MESF) per peak is obtained by correlating the MFI of each population with the amount of soluble AcGFP yielding the same fluorescence intensity. Twenty microliters of AcGFP calibration beads was resuspended in 1 ml of 1× flow cytometer calibration bead dilution buffer according to the manufacturer's instructions.
Plaque assays.
To assess viral particle infectivity, Vero cells were plated on 6-well plates and grown to confluence. The growth medium was removed, and 250-μl aliquots of 10-fold serial dilutions of virions in RPMI plus 0.1% bovine serum albumin (BSA) were placed on the cells in duplicates. After 1 h of adsorption to cells at 37°C, 2× DMEM mixed with 2% agarose (1:1) was added, and the cells were further incubated at 37°C. The medium-agarose mix typically was removed 72 hpi later, the cells were fixed with methanol, and the plaques were revealed with crystal violet as before (50).
qPCR analysis of sorted virions.
To determine genome copy numbers, 200 μl of FACS-sorted virus was first treated for 10 min at 37°C with 500 U/ml of DNase I (Roche) to digest nonencapsidated viral DNA. The encapsidated viral DNA was extracted using GenElute mammalian genomic DNA miniprep kits (Sigma-Aldrich) per the manufacturer's instructions. These samples were then analyzed by quantitative PCR using HSV-1 glycoprotein B (gB)-specific primers (forward, 5′ CCACGAGACCGACATGGAGC 3′; reverse, 5′ GTGTTCGGTGTGCGACCCCTC 3′). The quantitative PCR (qPCR) was performed with a LightCycler 480 (Roche) using the PerfeCTa SYBR green SuperMix (Quanta Biosciences). The data were analyzed using LightCycler I 480 software, version 1.5.
Electron microscopy.
For EM, sorted viruses first were fixed in 2% glutaraldehyde in 0.1 M phosphate buffer (PB; pH 7.3) for 1 h on ice. Viruses then were concentrated by passing them through a 13-mm Swinny stainless steel holder (EMD Millipore) containing a 0.1-μm Omnipore polytetrafluoroethylene hydrophobic membrane filter (EMD Millipore). The filter containing sorted viruses was washed with PB and postfixed in 1% OsO4 in PB for 1 h at 4°C. The filter was then rinsed, dehydrated in increased concentrations of ethanol, and embedded in Epon (57). Ultrathin sections of filter containing viruses were taken using a Reichert ultracut microtome and placed on naked nickel grids. The grids were then contrasted with 3% aqueous uranyl acetate (Canemco & Marivac). Samples were examined on a Philips CM100 transmission electron microscope, and digital micrographs were captured using an AMT XR80 charge-coupled-device digital camera.
Production of depleted virus using siRNA.
143B cells were seeded in 6-well plates at a concentration of 5 × 104 cells/well 24 h before transfection. Cells were transfected for 48 h using PepMute siRNA transfection reagent (SignaGen Laboratories) according to the manufacturer's instructions. RNA interference reagents (Dharmacon) used at 25 to 100 nM included a unique siRNA against VP16 (12, 13) and two distinct siRNAs against VP22. Cells then were either mock treated or infected with HSV-1 KOS at an MOI of 5. After the 1-h adsorption time, cells were washed with phosphate-buffered saline (PBS), and then complete DMEM without BrdU was added to the wells and infection was continued for another 24 h. The supernatant was then harvested and cleared from cell debris by centrifugation at 500 × g for 5 min at 4°C. Extracellular virions were then titrated on Vero cells and/or concentrated for 2 h at 18,000 × g, and the viral pellets were resuspended in MNT and analyzed by Western blotting.
ACKNOWLEDGMENTS
We acknowledge the significant contribution of Kerstin Radtke in the early stages of this project. We also thank John Nolan for judicious advice on the analysis of small particles by flow cytometry. We are particularly indebted to Danièle Gagné, who provided first-class advice and service at the flow cytometry platform of IRIC. Finally, we thank Beate Sodeik, Prashant Desai, Gregory A. Smith, William Halford, Jim Smiley, Steve Weinheimer, Peter O'Hare, Gill Elliott, Richard Courtney, Frank Jenkins, Bernard Roizman, and Helena Browne for providing viral strains and antibodies without which this work would not have been possible.
This study was funded by the Canadian Institutes of Health Research (grant MOP258030 to R.L.).
We have no conflicts of interest to report.
REFERENCES
- 1.Loret S, Guay G, Lippé R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol 82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mettenleiter TC. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet Microbiol 113:163–169. doi: 10.1016/j.vetmic.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 3.Taha MY, Brown SM, Clements GB. 1988. Neurovirulence of individual plaque stocks of herpes simplex virus type 2 strain HG 52. Arch Virol 103:15–25. doi: 10.1007/BF01319805. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 5.Smith G. 2012. Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol 66:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res 145:173–186. doi: 10.1016/j.virusres.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Loret S, Lippé R. 2012. Biochemical analysis of infected cell polypeptide (ICP)0, ICP4, UL7 and UL23 incorporated into extracellular herpes simplex virus type 1 virions. J Gen Virol 93:624–634. doi: 10.1099/vir.0.039776-0. [DOI] [PubMed] [Google Scholar]
- 8.Henaff D, Remillard-Labrosse G, Loret S, Lippé R. 2013. Analysis of the early steps of herpes simplex virus 1 capsid tegumentation. J Virol 87:4895–4906. doi: 10.1128/JVI.03292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preston CM. 1979. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol 29:275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacks WR, Schaffer PA. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol 61:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinheimer SP, Boyd BA, Durham SK, Resnick JL, O'Boyle DR II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J Virol 66:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YQ, Lai W, Li H, Li G. 2008. Inhibition of herpes simplex virus type 1 by small interfering RNA. Clin Exp Dermatol 33:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stegen C, Yakova Y, Henaff D, Nadjar J, Duron J, Lippé R. 2013. Analysis of virion-incorporated host proteins required for herpes simplex virus type 1 infection through a RNA interference screen. PLoS One 8:e53276. doi: 10.1371/journal.pone.0053276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baines J, Duffy C. 2006. Nucleocapsid assembly and envelopment of herpes simplex virus, p 175–204. In Sandri-Goldin RM. (ed), Alpha herpesviruses. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 15.Heine JW, Honess RW, Cassai E, Roizman B. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol 14:640–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roller RJ, Roizman B. 1992. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol 66:3624–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcomb WW, Jones LM, Dee A, Chaudhry F, Brown JC. 2012. Role of a reducing environment in disassembly of the herpesvirus tegument. Virology 431:71–79. doi: 10.1016/j.virol.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Clarke RW, Monnier N, Li H, Zhou D, Browne H, Klenerman D. 2007. Two-color fluorescence analysis of individual virions determines the distribution of the copy number of proteins in herpes simplex virus particles. Biophys J 93:1329–1337. doi: 10.1529/biophysj.107.106351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohannon KP, Jun Y, Gross SP, Smith GA. 2013. Differential protein partitioning within the herpesvirus tegument and envelope underlies a complex and variable virion architecture. Proc Natl Acad Sci U S A 110:E1613–E1620. doi: 10.1073/pnas.1221896110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leslie J, Rixon FJ, McLauchlan J. 1996. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology 220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 21.McLauchlan J. 1997. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J Gen Virol 78:189–194. doi: 10.1099/0022-1317-78-1-189. [DOI] [PubMed] [Google Scholar]
- 22.Michael K, Klupp BG, Mettenleiter TC, Karger A. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J Virol 80:1332–1339. doi: 10.1128/JVI.80.3.1332-1339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dargan DJ, Subak-Sharpe JH. 1997. The effect of herpes simplex virus type 1 L-particles on virus entry, replication, and the infectivity of naked herpesvirus DNA. Virology 239:378–388. doi: 10.1006/viro.1997.8893. [DOI] [PubMed] [Google Scholar]
- 24.Loret S, El Bilali N, Lippé R. 2012. Analysis of herpes simplex virus type I nuclear particles by flow cytometry. Cytometry A 81:950–959. [DOI] [PubMed] [Google Scholar]
- 25.Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol 397:575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khadivjam B, Stegen C, Hogue-Racine MA, El Bilali N, Döhner K, Sodeik B, Lippé R. 1 February 2017. The ATP-dependent RNA helicase DDX3X modulates herpes simplex virus type 1 gene expression. J Virol doi: 10.1128/JVI.02411-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohannon KP, Sollars PJ, Pickard GE, Smith GA. 2012. Fusion of a fluorescent protein to the pUL25 minor capsid protein of pseudorabies virus allows live-cell capsid imaging with negligible impact on infection. J Gen Virol 93:124–129. doi: 10.1099/vir.0.036145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A 108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol 85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang TY, Courtney RJ. 1995. Influence of the host cell on the association of ICP4 and ICP0 with herpes simplex virus type 1. Virology 211:209–217. doi: 10.1006/viro.1995.1393. [DOI] [PubMed] [Google Scholar]
- 32.Jambunathan N, Chouljenko D, Desai P, Charles AS, Subramanian R, Chouljenko VN, Kousoulas KG. 2014. Herpes simplex virus 1 protein UL37 interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment. J Virol 88:5927–5935. doi: 10.1128/JVI.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagel CH, Döhner K, Fathollahy M, Strive T, Borst EM, Messerle M, Sodeik B. 2008. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+). J Virol 82:3109–3124. doi: 10.1128/JVI.02124-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh HS, Knipe DM, Oh HS, Bryant KF, Nieland TJ, Mazumder A, Bagul M, Bathe M, Root DE, Knipe DM. 2015. Proteomic analysis of the herpes simplex virus 1 virion protein 16 transactivator protein in infected cells. Proteomics 5:e01086-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott G, Mouzakitis G, O'Hare P. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol 69:7932–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elliott G, Hafezi W, Whiteley A, Bernard E. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J Virol 79:9735–9745. doi: 10.1128/JVI.79.15.9735-9745.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duffy C, Lavail JH, Tauscher AN, Wills EG, Blaho JA, Baines JD. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J Virol 80:8664–8675. doi: 10.1128/JVI.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka M, Kato A, Satoh Y, Ide T, Sagou K, Kimura K, Hasegawa H, Kawaguchi Y. 2012. Herpes simplex virus 1 VP22 regulates translocation of multiple viral and cellular proteins and promotes neurovirulence. J Virol 86:5264–5277. doi: 10.1128/JVI.06913-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobiler O, Lipman Y, Therkelsen K, Daubechies I, Enquist LW. 2010. Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat Commun 1:146. doi: 10.1038/ncomms1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobiler O, Brodersen P, Taylor MP, Ludmir EB, Enquist LW. 2011. Herpesvirus replication compartments originate with single incoming viral genomes. mBio 2:e00278–11. doi: 10.1128/mBio.00278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor MP, Kobiler O, Enquist LW. 2012. Alphaherpesvirus axon-to-cell spread involves limited virion transmission. Proc Natl Acad Sci U S A 109:17046–17051. doi: 10.1073/pnas.1212926109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J Virol 84:1504–1512. doi: 10.1128/JVI.02029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antinone SE, Zaichick SV, Smith GA. 2010. Resolving the assembly state of herpes simplex virus during axon transport by live-cell imaging. J Virol 84:13019–13030. doi: 10.1128/JVI.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu M, Schmidt EE, Halford WP. 2010. ICP0 dismantles microtubule networks in herpes simplex virus-infected cells. PLoS One 5:e10975. doi: 10.1371/journal.pone.0010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai P, Person S. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol 72:7563–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desai P, Sexton GL, Huang E, Person S. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J Virol 82:11354–11361. doi: 10.1128/JVI.00956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willard M. 2002. Rapid directional translocations in virus replication. J Virol 76:5220–5232. doi: 10.1128/JVI.76.10.5220-5232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La Boissiere S, Izeta A, Malcomber S, O'Hare P. 2004. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J Virol 78:8002–8014. doi: 10.1128/JVI.78.15.8002-8014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elliott G, O'Hare P. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J Virol 73:4110–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turcotte S, Letellier J, Lippé R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol 79:8847–8860. doi: 10.1128/JVI.79.14.8847-8860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNabb DS, Courtney RJ. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221–232. doi: 10.1016/0042-6822(92)91208-C. [DOI] [PubMed] [Google Scholar]
- 52.Schmitz JB, Albright AG, Kinchington PR, Jenkins FJ. 1995. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology 206:1055–1065. doi: 10.1006/viro.1995.1028. [DOI] [PubMed] [Google Scholar]
- 53.Munger J, Chee AV, Roizman B. 2001. The U(S)3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J Virol 75:5491–5497. doi: 10.1128/JVI.75.12.5491-5497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean C, Buckmaster A, Hancock D, Buchan A, Fuller A, Minson A. 1982. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J Gen Virol 63:297–305. doi: 10.1099/0022-1317-63-2-297. [DOI] [PubMed] [Google Scholar]
- 55.Donnelly M, Verhagen J, Elliott G. 2007. RNA binding by the herpes simplex virus type 1 nucleocytoplasmic shuttling protein UL47 is mediated by an N-terminal arginine-rich domain that also functions as its nuclear localization signal. J Virol 81:2283–2296. doi: 10.1128/JVI.01677-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elliott G, O'Reilly D, O'Hare P. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 57.Luft JH. 1961. Improvements in epoxy resin embedding methods. J Biophys Biochem Cytol 9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]