ABSTRACT
The HIV-1 envelope (Env) trimer is a target for vaccine design as well as a conformational machine that facilitates virus entry by transitioning between prefusion-closed, CD4-bound, and coreceptor-bound conformations by transitioning into a postfusion state. Vaccine designers have sought to restrict the conformation of the HIV-1 Env trimer to its prefusion-closed state as this state is recognized by most broadly neutralizing, but not nonneutralizing, antibodies. We previously identified a disulfide bond, I201C-A433C (DS), which stabilizes Env in the vaccine-desired prefusion-closed state. When placed into the context of BG505 SOSIP.664, a soluble Env trimer mimic developed by Sanders, Moore, and colleagues, the engineered DS-SOSIP trimer showed reduced conformational triggering by CD4. Here, we further stabilize DS-SOSIP through a combination of structure-based design and 96-well-based expression and antigenic assessment. From 103 designs, we identified one, named DS-SOSIP.4mut, with four additional mutations at the interface of potentially mobile domains of the prefusion-closed structure. We also determined the crystal structures of DS-SOSIP.4mut at 4.1-Å resolution and of an additional DS-SOSIP.6mut variant at 4.3-Å resolution, and these confirmed the formation of engineered disulfide bonds. Notably, DS-SOSIP.4mut elicited a higher ratio of tier 2 autologous titers versus tier 1 V3-sensitive titers than BG505 SOSIP.664. DS-SOSIP.4mut also showed reduced recognition of CD4 and increased thermostability. The improved antigenicity, thermostability, and immunogenicity of DS-SOSIP.4mut suggest utility as an immunogen or a serologic probe; moreover, the specific four alterations identified here, M154, M300, M302, and L320 (4mut), can also be transferred to other HIV-1 Env trimers of interest to improve their properties.
IMPORTANCE One approach to elicit broadly neutralizing antibodies against HIV-1 is to stabilize the structurally flexible HIV-1 envelope (Env) trimer in a conformation that displays predominantly broadly neutralizing epitopes and few to no nonneutralizing epitopes. The prefusion-closed conformation of HIV-1 Env has been identified as one such preferred conformation, and a current leading vaccine candidate is the BG505 DS-SOSIP variant, comprising two disulfides and an Ile-to-Pro mutation of Env from strain BG505. Here, we introduced additional mutations to further stabilize BG505 DS-SOSIP in the vaccine-preferred prefusion-closed conformation. In guinea pigs, our best mutant, DS-SOSIP.4mut, elicited a significantly higher ratio of autologous versus V3-directed neutralizing antibody responses than the SOSIP-stabilized form. We also observed an improvement in thermostability and a reduction in CD4 affinity. With improved antigenicity, stability, and immunogenicity, DS-SOSIP.4mut-stabilized trimers may have utility as HIV-1 immunogens or in other antigen-specific contexts, such as with B-cell probes.
KEYWORDS: HIV-1, immunogen design, protein stabilization
INTRODUCTION
An effective human immunodeficiency virus type 1 (HIV-1) vaccine has long been sought as a means to contend with the AIDS pandemic (1). Specifically, the HIV-1 envelope glycoprotein (Env) trimer is the sole viral antigen on the surface of the HIV-1 virion and is responsible for mediating HIV-1 attachment and entry into CD4+ T cells. A soluble version of the Env trimer has been sought to serve as the basis for a B cell-based subunit vaccine against HIV-1 (2). As the Env trimer is conformationally flexible (3), an Env trimer-based immunogen stabilized in a conformation that presents broadly neutralizing antibody (bNAb) epitopes and few to no nonneutralizing antibody epitopes has been sought (4). Ideally, such a conformationally fixed Env immunogen should have high thermostability and should remain in the desired antigenic state, even in the presence of CD4.
Significant efforts have been made to obtain HIV-1 Env trimers with desired antigenicity. Sanders, Moore, and colleagues developed a soluble, cleaved Env trimer, SOSIP.664, from the BG505 strain, which possesses multiple epitopes for bNAbs and few epitopes for nonneutralizing antibodies (4–7). We along with others determined structures of BG505 SOSIP.664, both ligand free (8) and complexed with bNAbs (9–18), and these show BG505 SOSIP.664 to prefer the antigenically desired prefusion-closed conformation (8–13, 15). Furthermore, BG505 SOSIP.664 reactivity toward nonneutralizing antibodies, including CD4-induced (CD4i) and V3 nonneutralizing or weakly neutralizing antibodies, could be further reduced by negative-selection purification (8, 19). However, BG505 SOSIP.664 gp140 undergoes conformational change in the presence of soluble CD4 (sCD4), which can result in the display of nonneutralizing or weakly neutralizing CD4i and V3 epitopes (8). We along with others have succeeded in further stabilizing BG505 SOSIP.664 in the prefusion-closed conformation (8, 20). The stabilized version of BG505 SOSIP.664 we developed, DS-SOSIP (8), incorporates an additional engineered disulfide bond (I201C-A443C), which locks the Env trimer in the prefusion-closed state and impedes binding of CD4i or V3 nonneutralizing or weakly neutralizing antibodies, even in the presence of sCD4 (8). However, DS-SOSIP retains the ability to bind a single CD4 molecule per trimer (8, 67).
In this study, we investigated whether further stabilization of DS-SOSIP in its prefusion-closed conformation could yield improved immunogenicity. We designed 103 variants of DS-SOSIP and screened for variants that displayed the desired antigenicity and suitable expression by using an enzyme-linked immunosorbent assay (ELISA) coupled to 96-well transient expression. We characterized four candidates in detail. Three of the four showed increased melting temperatures, and all four showed improved antigenicity and reduced sCD4 affinity (to as low as ∼0.5 μM) while retaining the ability to bind CD4-binding site-directed antibodies. We solved the crystal structures of two variants, DS-SOSIP.4mut (incorporating mutations L154M, N300M, N302M, and T320L [4mut]) and DS-SOSIP.6mut (incorporating 4mut mutations as well as Y177W and I420M), at nominal resolutions of 4.1 Å and 4.3 Å, respectively. These structures provided atomic details of the 201C-433C disulfide bond as well as of the designed hydrophobic interactions. Notably, guinea pig immunization studies showed DS-SOSIP.4mut to have an increased ratio of autologous tier-2 to V3-sensitive tier-1 neutralizing titer compared to that of BG505 SOSIP.664.
RESULTS
Design and antigenic assessment of 103 DS-SOSIP variants.
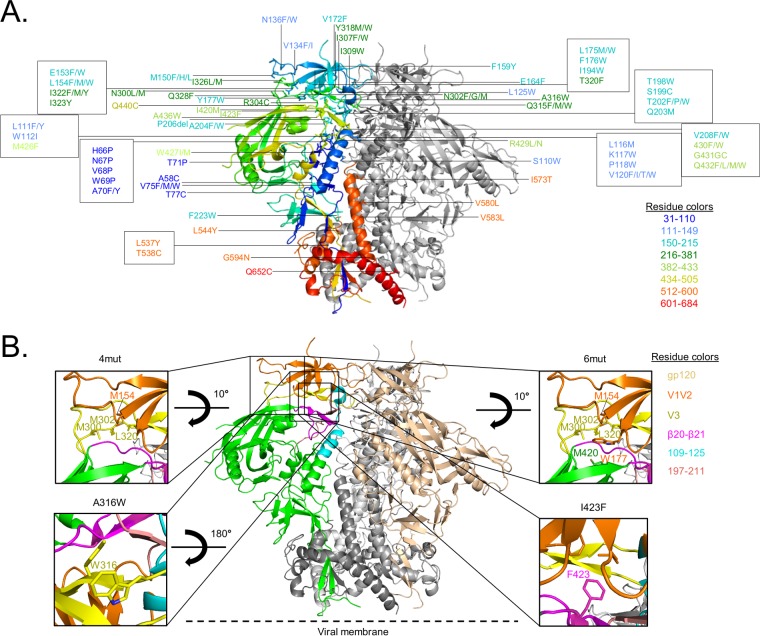
We employed two strategies to stabilize DS-SOSIP in the vaccine-preferred prefusion-closed conformation, resulting in a total of 103 DS-SOSIP variants (Fig. 1A). One strategy was to introduce mutations into DS-SOSIP that were predicted to form hydrophobic interactions at the interfaces of the mobile apex region based on computational structural modeling (see Materials and Methods) (see Table S1 in the supplemental material). A second strategy was to introduce mutations into DS-SOSIP that we identified previously in the BG505 SOSIP.664 context (8) to yield increased reactivity to the antibodies CAP256-VRC26.09 and PGT145 (21, 22), which recognized the trimer apex, as reactivity to these antibodies suggested increased presentation of the prefusion-closed state (Table S2). The antigenicity of each design was assessed after each construct was transiently expressed with 293T cells in a 96-well plate format, and the antigenicity of supernatants was assessed by ELISA (see Materials and Methods) (Table S3). We used the sum of the bNAbs PGT145 and CAP256-VRC26.09 reactivities divided by the sum of weakly neutralizing antibody reactivity (for the V3-directed antibodies 447-52D and 3074 [23, 24] in the presence of CD4) to rank each design. Based on this scoring metric, we selected four designs: DS-SOSIP.4mut (L154M/N300M/N302M/T320L), DS-SOSIP.6mut (L154M/N300M/N302M/T320L/Y177W/I420M), DS-SOSIP.I423F, and DS-SOSIP.A316W for further characterization (Fig. 1B). The mutations in the DS-SOSIP.4mut design (L154M/N300M/N302M/T320L) were predicted to form hydrophobic interactions at the interface between the first and second variable (V1V2) regions and the third variable (V3) loop region interface. The two mutations (Y177W/I420M) that were introduced in addition to the four mutations in the DS-SOSIP.6mut design were designed to strengthen interaction between V1V2 and the mobile β20-β21 hairpin. The I423F mutation was also predicted to strengthen the interaction between V1V2 and β20-β21. Finally, the A316W mutation was predicted to stabilize V3 in its prefusion conformation through an aromatic interaction with Y318. Coincidentally, A316W was reported in a prior study (31) to facilitate the stabilization of BG505 SOSIP.664 in the prefusion-closed conformation and to reduce immunogenicity toward the V3 loop.
FIG 1.
Structure-based stabilization of DS-SOSIP into its prefusion-closed conformation. (A) Amino acid mutational positions from 103 DS-SOSIP variants evaluated antigenically and mapped onto the structure of BG505 SOSIP.664 (PDB accession number 4TVP), with one protomer shown in rainbow colors (residues colored coded as defined in the legend) and the other two in gray ribbon representation. (B) Crystal structure of BG505 SOSIP.664 (PDB accession number 4TVP) shown in gray, with one gp120 protomer shown in green and another shown in salmon (central image). The gp120 domains are color coded as defined in the legend on the figure. Structural models of DS-SOSIP variants with the most favorable antigenic profiles (Table S3) are shown in the insets; for each variant, stabilizing mutations relative to DS-SOSIP are modeled in stick representation and labeled.
DS-SOSIP variants showed improved antigenicity for the prefusion-closed state.
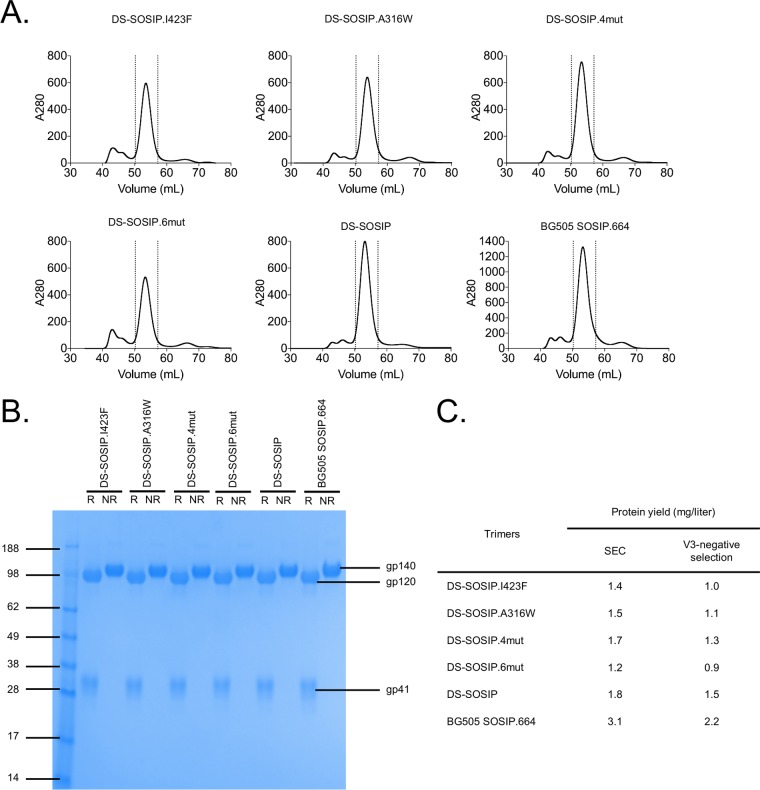
We expressed and purified the four DS-SOSIP variants, identified from the initial antigenic assessment of the 103 designs. Size exclusion chromatography (SEC) profiles of these variants were similar to that of DS-SOSIP (Fig. 2A), and all four showed expression yields (0.9 to 1.3 mg/liter) comparable to the yield of DS-SOSIP (1.5 mg/liter) (Fig. 2B). SDS-PAGE analysis indicated all four DS-SOSIP variants to be fully cleaved and to run as separate gp120 and gp41 subunits under reducing conditions (Fig. 2C).
FIG 2.
Properties of stabilized HIV-1 Env trimers. (A) Size exclusion chromatography (SEC) profiles of stabilized DS-SOSIP variants prior to V3 negative selection. (B) SDS-PAGE of stabilized DS-SOSIP variants prior to V3 negative selection. R, reducing conditions; NR, nonreducing conditions. (C) Expression yield of stabilized DS-SOSIP variants after SEC or SEC with V3 negative selection.
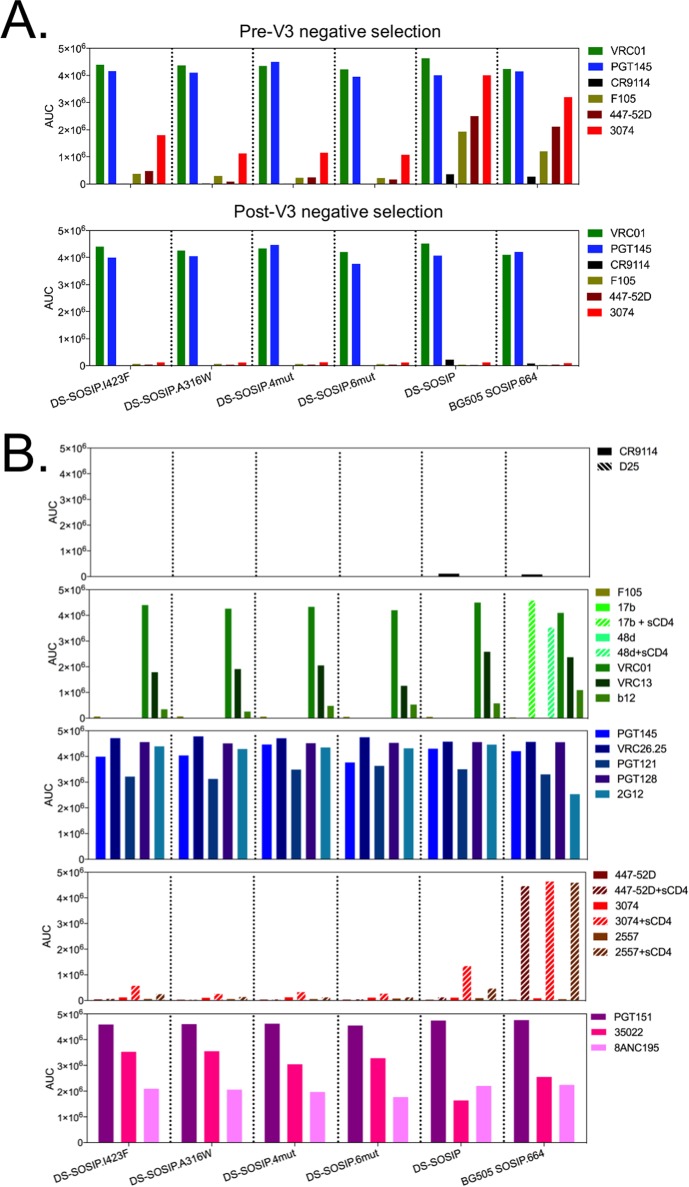
To define the antigenicity of these top four DS-SOSIP variants, we evaluated their binding affinities to a panel of 17 HIV-1 antibodies by a Meso Scale Discovery (MSD) Multi-Array (see Materials and Methods). Prior to negative selection with V3-directed antibodies, we observed all four DS-SOSIP variants to exhibit lower binding affinities toward V3-directed antibodies 447-52D and 3074 than DS-SOSIP (Fig. 3A). After negative selection with V3-directed antibodies, the four DS-SOSIP variants displayed even lower V3-directed antibody binding, even in the presence of CD4 (Fig. 3B). The four DS-SOSIP variants also showed higher affinity toward 35O22, a gp120-gp41 interface antibody, than DS-SOSIP.
FIG 3.
Antigenicity of stabilized Env trimer determined by Meso Scale Discovery (MSD). (A) Antigenicity of stabilized DS-SOSIP prior to V3 negative selection (top) or after V3 negative selection (bottom), assessed with broadly neutralizing antibodies (VRC01 and PGT145), weakly or nonneutralizing antibodies (F105, 447-52D, and 3074), and a negative control antibody (CR9114, an influenza virus antibody with no recognition of HIV-1 Env). AUC, area under the concentration-time curve. (B) Antigenicity of stabilized DS-SOSIP variants after V3 negative selection assessed on a panel of CD4-induced antibodies (17b and 48d, with and without soluble CD4), CD4-binding site antibodies (VRC01, VRC13, and b12), V2 apex-directed antibodies (PGT145 and CAP256-VRC26.25), glycan-V3 antibodies (PGT121, PGT128, and 2G12), weakly neutralizing V3-directed antibodies (447-52D, 3074, and 2557, with and without soluble CD4), and gp41-gp120 interface antibodies (PGT151, 35O22, and 8ANC195).
To determine if V3 antigenicity of stabilized DS-SOSIP variants changed over time, we performed a time course study by evaluating the antigenicity of the trimer variants after 0, 4, and 6 weeks of incubation at 37°C using biolayer interferometry (Fig. S1). We observed reactivity toward antibody 3074 to increase substantially for BG505 SOSIP.664 and DS-SOSIP, but not for the stabilized DS-SOSIP variants, after 4 weeks of incubation at 37°C.
DS-SOSIP variants formed prefusion-closed trimers.
To determine whether the introduction of the designed mutations altered structural conformation, we performed negative-stain electron microscopy (EM) on the purified DS-SOSIP variants (Fig. 4) after SEC and V3 negative selection. Similar to DS-SOSIP, all four DS-SOSIP variants uniformly formed trimers in the closed prefusion conformation without monomers and aggregates. In particular, visual inspection of the two-dimensional (2D) classes revealed compact propeller-like shapes typical of the closed prefusion conformation. There were no classes corresponding to the open conformation or nonnative trimers as defined by Pugach et al. (25).
FIG 4.
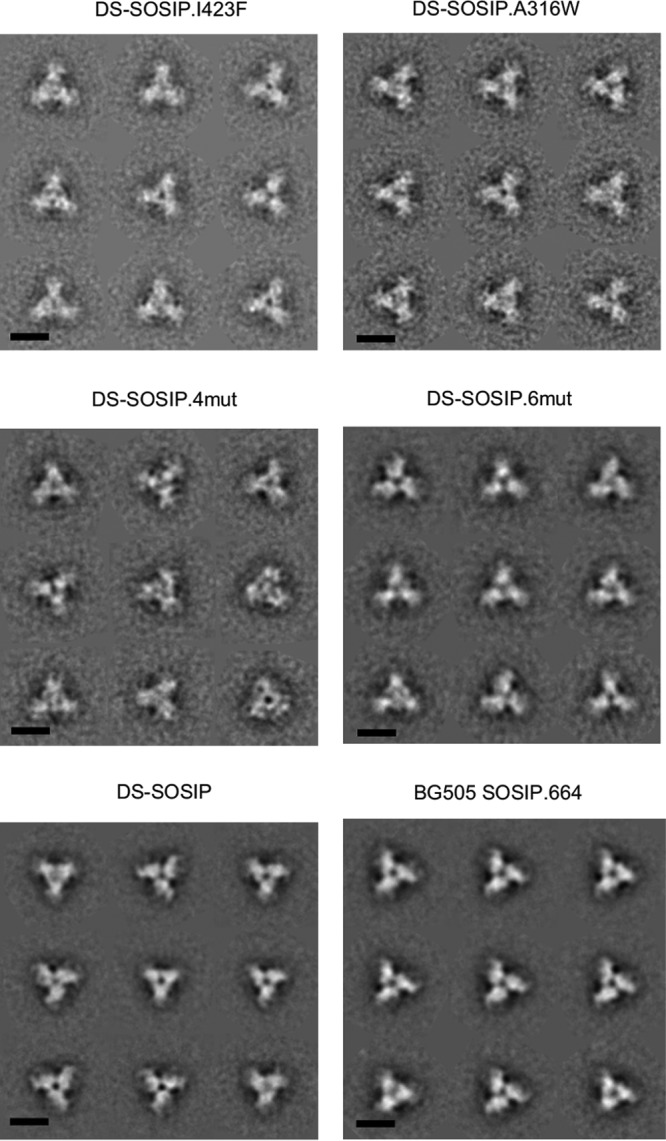
Negative-stain electron microscopy of stabilized Env trimers. Reference-free classification and averaging produced symmetrical propeller-like classes typical of the prefusion-closed conformation of the HIV-1 Env trimer, indicative of high homogeneity and correct folding/assembly of the proteins. Scale bar, 10 nm.
CD4 binding of DS-SOSIP variants.
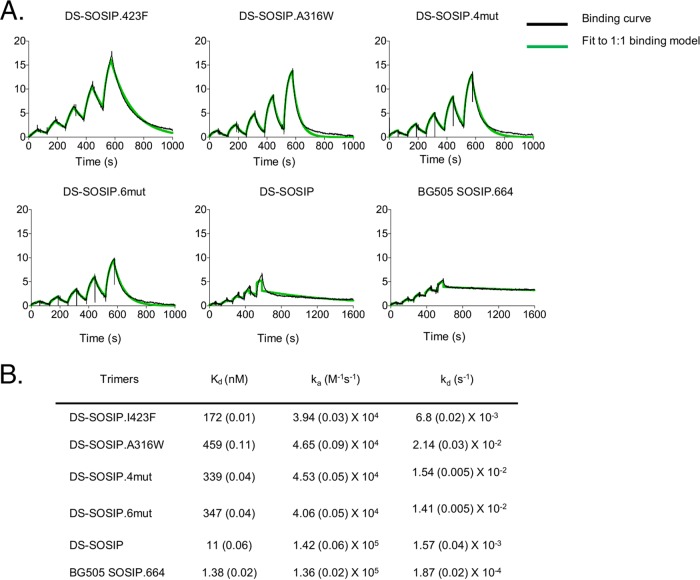
To determine whether the DS-SOSIP variants demonstrated a difference in CD4 binding, we measured the binding of sCD4 to the DS-SOSIP variants using surface plasmon resonance (SPR) (Fig. 5). All four DS-SOSIP variants showed substantially lower sCD4-binding affinities (the equilibrium dissociation constant, Kd, was reduced by 15- to 40-fold) than the binding affinity of DS-SOSIP (Kd = 11 nM), which was already reduced substantially from that of SOSIP.664 (Kd = 1.4 nM).
FIG 5.
Binding of soluble CD4 to stabilized HIV-1 Env trimers as measured by SPR with single-cycle kinetics. (A) SPR curves. For BG505 SOSIP.664 and DS-SOSIP, the concentrations of CD4 injected were 180, 90, 45, 22.5, and 11.25 nM. For the DS-SOSIP mutants, the concentrations of CD4 injected were 500, 250, 125, 62.5, and 31.25 nM. (B) Dissociation constant (Kd), on-rate constant (ka), and off-rate constant (kd) values. Values in parentheses report standard errors from fitting data to a 1:1 Langmuir binding model.
Thermostability of DS-SOSIP variants.
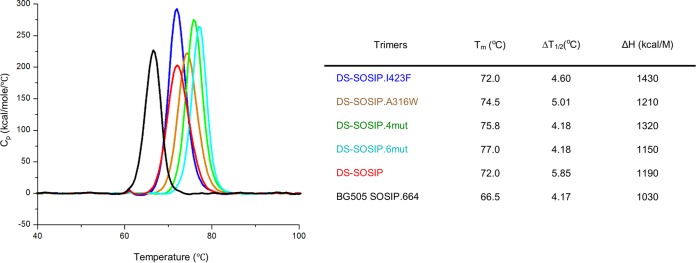
To examine whether the designed DS-SOSIP variants displayed a difference in thermostability, we evaluated the thermostability of the DS-SOSIP variants by differential scanning calorimetry (DSC) (Fig. 6). DS-SOSIP.6mut and DS-SOSIP.4mut showed approximately 5°C and 4°C increases in melting temperature (Tm) (to 77.0°C and 75.8°C, respectively) relative to that of DS-SOSIP (72.0°C). DS-SOSIP.A316W showed a 2°C increase in Tm (74.5°C), while DS-SOSIP.I423F showed no increase in Tm (72.0°C) relative to that of DS-SOSIP.
FIG 6.
Thermostability of stabilized HIV-1 Env trimers assessed by differential scanning calorimetry (DSC). At left are raw data from DSC shown in solid lines for the Env trimers identified by color according to the scheme shown on the right. Values for Tm, ΔT1/2 (width at half peak height), and enthalpy of unfolding from DSC (ΔH) are also shown. Cp, heat capacity at constant pressure.
We observed a positive correlation between CD4 Kd and Tm values (r = 0.804, P = 0.054) for the trimer immunogens. In terms of heat absorption, DS-SOSIP.I423F and DS-SOSIP.4mut had the highest enthalpy values of unfolding at 1,430 and 1,320 kcal/M, respectively, representing a substantial increase compared to the enthalpy values of unfolding for DS-SOSIP and BG505 SOSIP.664, which are 1,190 and 1,030 kcal/M, respectively.
Structure determination of stabilized DS-SOSIP variants.
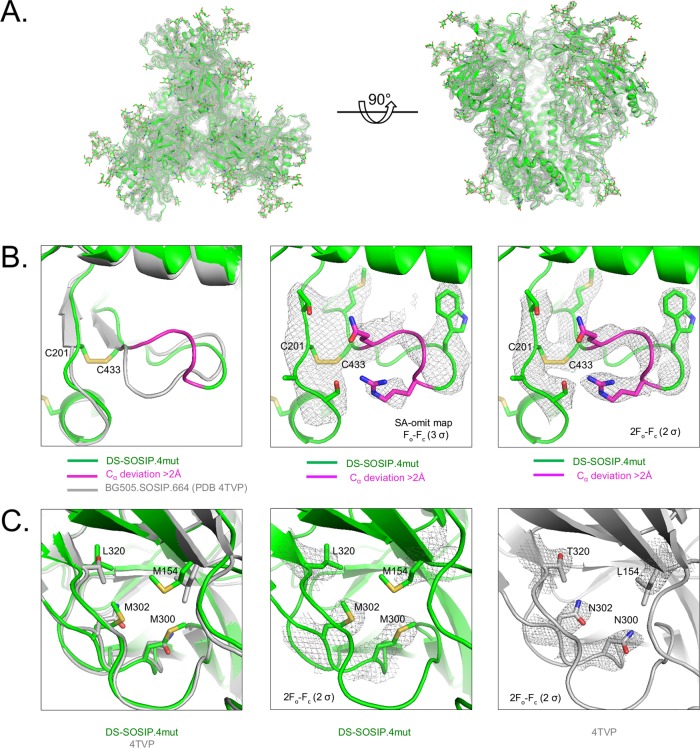
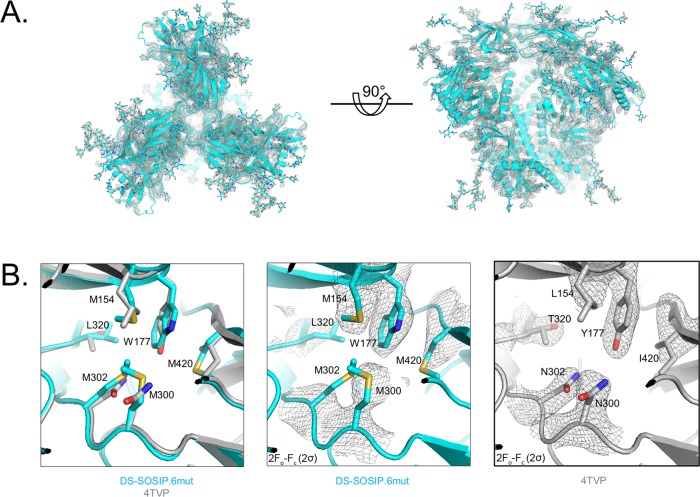
We determined crystal structures of DS-SOSIP.4mut and DS-SOSIP.6mut, the two variants with the highest Tm values, in complex with antibodies PGT122 and 35O22 (Fig. 7 and 8; Table S4). The Cα root mean square deviation (RMSD) between the DS-SOSIP.4mut and DS-SOSIP.6mut structures was 0.76 Å, and the Cα RMSDs of DS-SOSIP.4mut and DS-SOSIP.6mut versus BG505 SOSIP.664 (PDB accession number 4TVP) were 0.95 and 0.79 Å, respectively. The two structures revealed atomic-level detail of the engineered disulfide bond in DS-SOSIP (201C-443C). In both structures, the conformation of the β21-strand appeared to differ from the conformation observed in the prefusion-closed BG505 SOSIP.664 structure (e.g., PDB accession number 4TVP), possibly due to the introduction of the DS disulfide bond proximal to β21 (Fig. 7B). Notably, none of the hydrophobic mutations in 4mut or 6mut appeared to alter backbone conformation although these hydrophobic mutations did form hydrophobic interactions, as anticipated from the computational design (Fig. 7C and 8B). Despite the 4-Å resolution, a number of the bulkier side chains could be clearly observed (Fig. 7C and 8B).
FIG 7.
Crystal structure of DS-SOSIP.4mut. (A) Crystal structure of DS-SOSIP.4mut Env trimer, as extracted from the crystallized ternary complex with antibodies 35O22 and PGT122 (11), with amino acids displayed in ribbon representation and glycans in stick representation. The 2Fo − Fc (observed and calculated structure factor amplitudes, respectively) electron density is displayed as a gray mesh at 1σ. (B) Structural details of the engineered disulfide in DS-SOSIP (I201C/A433C, shown in sticks). The crystal structure of BG505 SOSIP.664 (PDB accession number 4TVP) is shown in gray. DS-SOSIP.4mut residues with Cα deviations greater than 2 Å from BG505 SOSIP.664 are shown in magenta. Simulated annealing (SA)-omit and 2Fo − Fc electron density map around the disulfide bond region are also shown as gray mesh in the middle and the right panels, respectively. (C) Structural details of designed hydrophobic side chains (shown in sticks) for DS-SOSIP.4mut; in the left panel, the crystal structure of BG505 SOSIP.664 (4TVP) is provided in gray for comparison. The 2Fo − Fc electron density map of the four designed residues in the DS-SOSIP.4mut and BG505 SOSIP.664 (4TVP) structures are shown in the middle and right panels, respectively, as gray mesh.
FIG 8.
Crystal structure of DS-SOSIP.6mut. (A) Crystal structure of DS-SOSIP.6mut Env trimer, as extracted from the crystallized ternary complex with antibodies 35O22 and PGT122 (11), with amino acids displayed in ribbon representation and glycans in stick representation. The 2Fo − Fc electron density at 1σ is displayed as a gray mesh. (B) Structural details of designed hydrophobic side chains (shown in sticks) for DS-SOSIP.6mut; in the left panel, the crystal structure of BG505 SOSIP.664 (PDB accession number 4TVP) is provided in gray for comparison. The 2Fo − Fc electron density map of the four designed residues in the DS-SOSIP.6mut and BG505 SOSIP.664 (4TVP) structures are shown in the middle and right panels, respectively, as gray mesh.
Immunogenicity of stabilized DS-SOSIP variants.
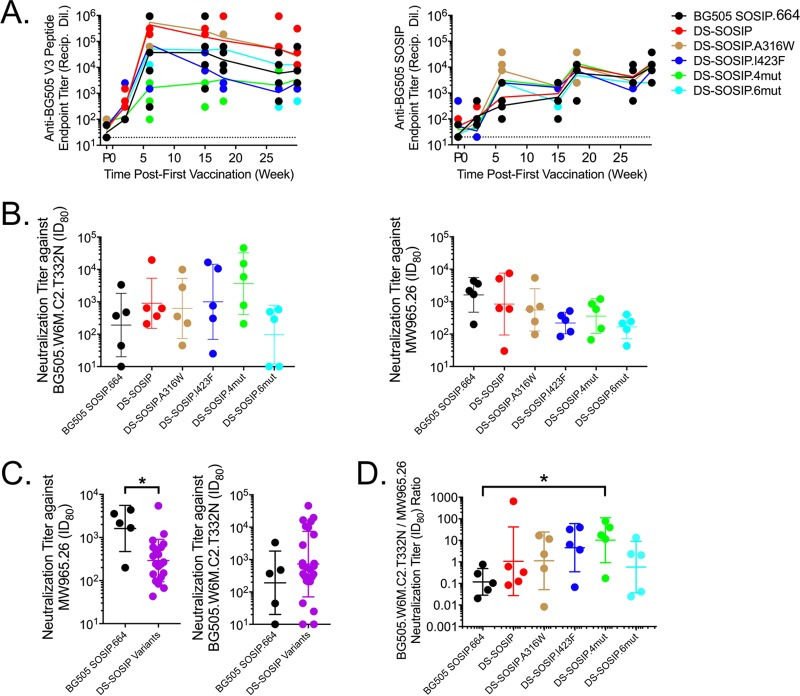
To measure the immunogenicities of the stabilized DS-SOSIP variants, we immunized guinea pigs at weeks 0, 4, 16, and 28 using V3 negatively selected trimers and assessed sera at weeks −1 (1 week prior to first injection), 2, 6, 15, 18, 27, and 30. All DS-SOSIP variants elicited trimer-specific responses comparable to those of BG505 SOSIP.664, while DS-SOSIP.I423F and DS-SOSIP.4mut elicited lower anti-V3 responses (Fig. 9A). Elicitation of neutralization against heterologous tier-2 strains was not observed for any of the trimers tested in this study (Table S5). Furthermore, each DS-SOSIP variant trended toward a decreased V3-sensitive tier-1 (MW965.26) response and, with the exception of DS-SOSIP.6mut, increased autologous (BG505.W6M.C2.T332N) neutralization activity at week 30 (Fig. 9B and Table S5). Specifically, the neutralization titer (80 percent inhibitory dilution [ID80] at week 30) elicited by DS-SOSIP.4mut was 19-fold higher than the neutralization titer elicited by BG505 SOSIP.664 (P = 0.095) and 4-fold higher than that elicited by DS-SOSIP. Comparison of the pooled neutralization results of the DS-SOSIP variants with those elicited by BG505 SOSIP.664 revealed reduced MW965.26 neutralizing activity (P = 0.024) (Fig. 9C). The ratio of BG505 neutralizing titer to MW965.26 neutralization titer for DS-SOSIP.4mut was significantly greater than the ratio of BG505 SOSIP.664 compared group-wise, with the remaining DS-SOSIP variants trending toward greater ratios (Fig. 9D) than BG505 SOSIP.664. With a geometric mean ratio of 0.1, BG505 SOSIP.664 induced a relatively high titer of tier-1 MW965.26 neutralizing activity. The ratio for DS-SOSIP.4mut was as high as 10.2, indicating the autologous neutralizing activity to be 10-fold higher than that of the tier-1 MW965.26. Such differences in immunogenicity are reflected in both reduced tier-1 neutralizing activity and increased autologous neutralizing activity for DS-SOSIP.4mut compared to the activity of BG505 SOSIP.664 (Fig. 9B). In addition, we observed the enthalpy of unfolding of the immunogens (Fig. 6) to have a high degree of positive correlation with both BG505 neutralizing titer (ρ = 0.829, P = 0.058) and the ratio of BG505 neutralizing titer to MW965.26 neutralizing titer (ρ = 0.943, P = 0.017) (Fig. 10).
FIG 9.
Immunogenicity of stabilized HIV-1 Env trimers. (A) Longitudinal V3 peptide-specific (left) and native trimer-specific (right) serum immunogenicity. Recip Dil, reciprocal dilution. (B) Week 30 serum neutralization (ID80) of autologous (left) and V3-sensitive tier-1 (right) pseudoviruses. (C) DS-SOSIP and its variants elicited reduced V3-sensitive tier-1 neutralization (left) while maintaining autologous neutralization compared to neutralizing activity of BG505 SOSIP.664 (*, P < 0.05, by a two-tailed Mann-Whitney test). (D) Autologous- to V3-sensitive tier-1 neutralization ratio of DS-SOSIP.4mut is greater than that of BG505 SOSIP.664 (*, P < 0.05, by Kruskal-Wallis test with a post hoc Dunn's multiple-comparison test).
FIG 10.
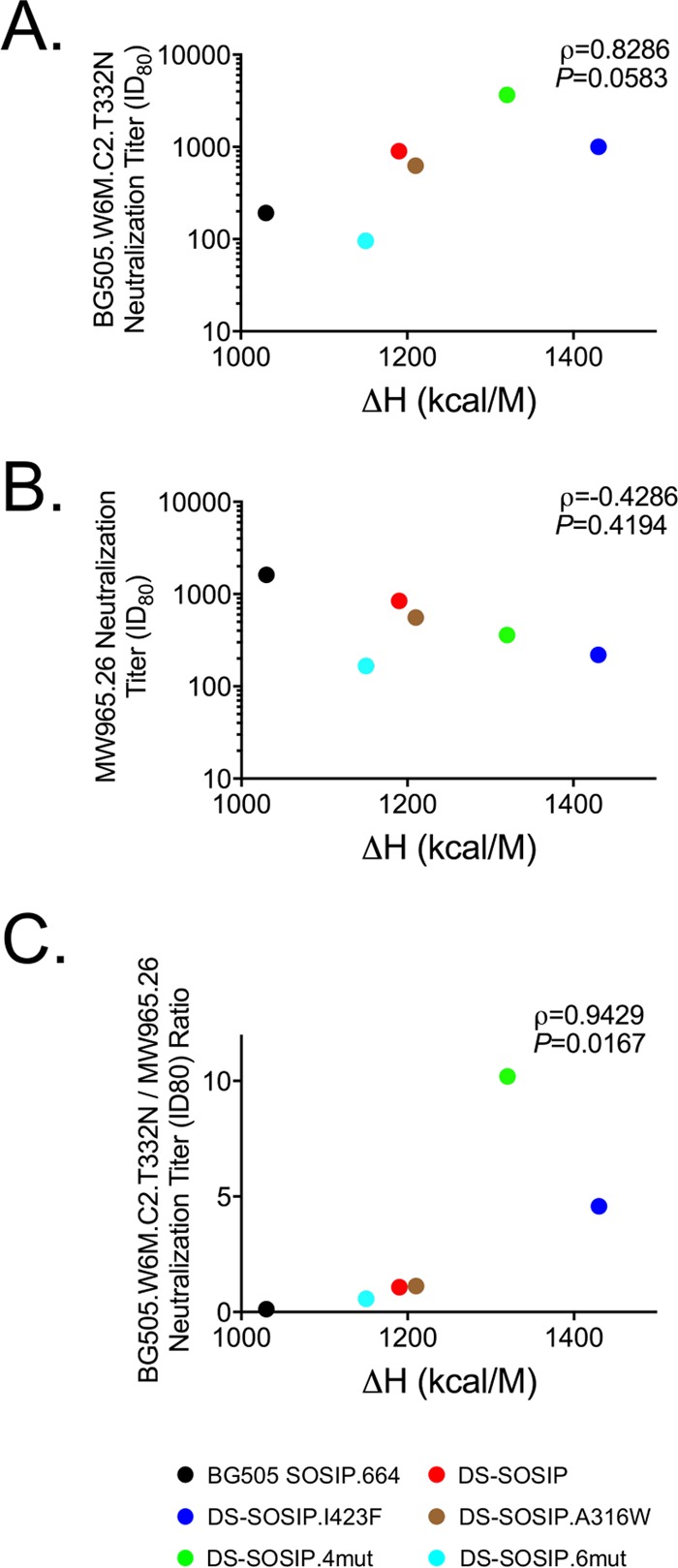
Correlation between immunogenicity and thermostability. (A) Spearman correlation between autologous neutralization (ID80 from week 30, group geometric means) and enthalpy of unfolding. (B) Spearman correlation between V3-sensitive tier-1 neutralization and enthalpy of unfolding. (C) Spearman correlation between autologous-to-V3-sensitive tier-1 neutralization ratio and enthalpy of unfolding.
DISCUSSION
The HIV-1 Env trimer evades the humoral immune response through three main mechanisms: genetic variation (26, 27), conformational flexibility (3, 28), and glycan masking (29). In terms of conformational flexibility, the HIV-1 Env trimer can exist in a number of conformations, related to its essential role in virus cell entry as a type 1 fusion machine, which uses structural rearrangements to fuse viral and target cell membranes (30). The conformational states accessed by HIV-1 Env trimer include the prefusion-closed state, CD4- and coreceptor-bound conformations, and intermediate conformations as well as a postfusion state. Of these, the prefusion-closed conformation presents mostly broadly neutralizing epitopes and few nonneutralizing or weakly neutralizing epitopes, and it thus has been the focus of vaccine design. Here, we used structure-based design to further stabilize DS-SOSIP in the prefusion-closed conformation.
The further stabilized DS-SOSIP trimers showed substantial reduction of weakly neutralizing antigenicity prior to V3 negative selection, indicating reduction of spontaneous V3 transition. The results suggest that these trimers may be better suited for genetic immunization, where the antigenic quality of the immunogen without purification may be crucial, although an additional stabilization effort will be needed to remove fully V3 reactivity. For the V3 negatively selected trimers, the further stabilized DS-SOSIP trimers also displayed reduction of V3 antigenicity in the presence of CD4 compared to that of DS-SOSIP and BG505 SOSIP.664 without CD4.
In addition to a reduction of weakly neutralizing V3 antigenicity, we found further stabilization of the DS-SOSIP variant of the HIV-1 Env trimer in the prefusion-closed conformation to reduce the affinity to CD4 to submicromolar levels and to increase immunogenicity for autologous neutralizing antibodies while reducing the immunogenicity for nonneutralizing and weakly neutralizing antibodies. Thus, stabilizing the Env trimer in the prefusion-closed conformation appears to prevent the HIV-1 Env trimer from transitioning to other intermediates beyond the prefusion-closed state (including CD4-bound conformations). The reduced affinity of these trimers to CD4 suggests that these trimers will be less likely to interfere with or to localize to CD4+ T cells when they are used as immunogens, an advantage when these immunogens are used to immunize humans.
In our guinea pig immunization study, we observed DS-SOSIP.4mut to have a higher ratio of autologous tier-2 to V3-sensitive tier-1 neutralizing titer than BG505 SOSIP.664 (P = 0.032), which is due to both an increase in autologous tier-2 and a decrease in V3-sensitive tier-1 neutralizing titers. A similar, significantly higher ratio for further stabilized SOSIP.664 trimer was observed by de Taeye et al. in rabbits (31) and as a result of PGT145 complexation in guinea pigs (32). Of note, we observed a 19-fold increase of autologous neutralization with DS-SOSIP.4mut compared to that with BG505 SOSIP.664 (P = 0.095) using five animals in each group; a larger group size would be needed to further determine if the observed increase in elicitation of autologous neutralization titer for DS-SOSIP.4mut is significant. In addition, we used the MW965.26 strain to evaluate the anti-V3 response as most of the MW965.26 neutralization was V3 mediated (32); we observed a reduction of V3 antigenicity to translate into a reduction of anti-V3 serum response in immunized guinea pigs for the stabilized DS-SOSIP variants (4.5-fold reduction for DS-SOSIP.4mut; P = 0.056).
Despite having similar antigenicity, thermostability, and structural conformation characteristics as DS-SOSIP.4mut, DS-SOSIP.6mut elicited lower autologous neutralization than DS-SOSIP.4mut and all other trimer immunogens tested in this study. While intriguing, the lower DS-SOSIP.6mut autologous neutralization did not achieve statistical significance relative to that of the other study groups; further investigation is required to determine if Y177W and I420M, two additional mutations in DS-SOSIP.6mut at the V1V2/β20-β21 interface, do indeed reduce the elicitation of autologous neutralization in a statistically significant way.
In summary, we have developed DS-SOSIP.4mut, a BG505 SOSIP.664 variant that is stabilized in the prefusion-closed conformation with improved antigenicity, thermostability, and immunogenicity. Importantly, DS-SOSIP.4mut displayed submicromolar affinity for CD4; while a number of other stabilized forms of Env have been reported (31–36), none of these prior studies report low CD4 affinity while maintaining recognition of CD4-binding site antibodies. The four stabilizing mutations (M154, M300, M302, and L320) we identified in DS-SOSIP.4mut can be added to immunogen candidates to improve their various properties. In addition, highly stabilized trimers may have utility as serological probes to isolate antibodies that target the prefusion-closed conformation of Env.
MATERIALS AND METHODS
Computational structure-based design to stabilize DS-SOSIP.
Six sets of residues at the interface of V1V2/V3 or V1V2/β20-β21 were selected manually for redesign based on residue properties and the interface cavity upon manual inspection of the BG505 SOSIP.664 trimer structure (PDB accession number 4TVP) (see Table S1 in the supplemental material). For each set of residues, Osprey, version 1.0 (37), was used to enumerate and evaluate the lowest energy for all mutation combinations, where each residue was allowed to mutate to any hydrophobic residue (phenylalanine, tyrosine, tryptophan, valine, leucine, isoleucine, methionine, alanine, glycine, or histidine). Two to four mutation combinations were selected for each set based on the energy scores. A number of designs were also developed by combining mutations from multiple sets (Table S1).
Antigenic analysis of DS-SOSIP variants in a 96-well microplate by ELISA.
Antigenicities of the following antibodies were assayed for DS-SOSIP variants: VRC01, CAP256-VRC26.09, PGT145, PGT122, F105, 447-52D (with and without the presence of CD4), 3074 (with and without the presence of CD4), and 17b in the presence of CD4. D7324 antibody-coated 96-well ELISA plates were prepared by incubating 2 μg/ml of affinity-purified sheep anti-HIV-1 gp120 antibody (D7324) (catalog number D7324; Aalto Bio Reagents, Ireland) in 100 μl of phosphate-buffered saline (PBS) in a 96-well flat-bottom Nunc-Immuno Plate (Thermo Scientific, IL) overnight at 4°C, followed by the removal of the coating solution and incubation of the wells with 200 μl of 2% (wt/vol) dry milk in PBS overnight at 4°C. Then, the wells were washed five times with PBS–0.05% Tween 20. Thirty microliters of supernatant expressed in each well of the 96-well microplate was incubated with 70 μl of PBS in each well of a D7324 antibody-coated 96-well ELISA plate for 1 h at 60°C, after which the wells were washed five times with PBS–0.05% Tween 20. One hundred microliters of anti-specific-epitope primary antibody at a concentration of 10 μg/ml in PBS with 0.2% (wt/vol) dry milk and 0.2% Tween 20 was added to each well and incubated for 1 h at room temperature (RT), after which the wells were washed five times with PBS–0.05% Tween 20. One hundred microliters of horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories, Inc., PA) at 1:10,000 in PBS with 1.0% (wt/vol) dry milk and 0.2% Tween 20 was added to each well and incubated for 30 min at RT, after which the wells were washed five times with PBS–0.05% Tween 20. The wells were developed with tetramethylbenzidine (TMB) at RT for 10 min, after which the reaction was stopped with 180 mM HCl. The readout was measured at a wavelength of 450 nm. All samples were read in duplicate.
Protein expression and purification.
The various BG505 DS-SOSIP trimer mutants were produced in 293 FreeStyle cells, as described previously (4). Briefly, 600 μg of BG505 DS-SOSIP trimer construct was cotransfected with 150 μg of furin plasmid DNA into 1 liter of cells. After 6 days, the transfected supernatants were harvested, filtered, and loaded over a VRC01 affinity column. After being washed with phosphate-buffered saline (PBS), the bound proteins were eluted with 3 M MgCl2 and 30 mM Tris at a pH of 7.0. The eluate was concentrated to 2 to 3 ml using an Amicon Ultracel-50K instrument (Millipore) and applied to a Superdex 200 16/600 gel filtration column (GE Healthcare) equilibrated in PBS. The peak corresponding to trimeric HIV-1 Env was identified, pooled, and subjected to negative selection with 447-52D (PDB accession number 4M1D) (23) and V3 cocktail columns to remove aberrant trimer species (8). The V3 cocktail column contains six V3-directed antibodies: 1006-15D, 2219, 2557, 2558, 3074, and 50.1 (PDB accession numbers 3MLW [38], 2B0S [39], 3MLS [38], 3UJI [40], 3MLX [38], and 1GGI [41], respectively).
Antigenic analysis of DS-SOSIP variants by MSD-electrochemiluminescence immunoassay (ECLIA).
Standard 96-well bare Multi-Array Meso Scale Discovery (MSD) plates (catalog number L15XA-3) were coated with a panel of HIV neutralizing antibodies (VRC01 [42], b12 [43], VRC13 [44], PGT121 [21], PGT128 [21], 2G12 [45], PGT145 [21], CAP256-VRC26.25 [46], 35O22 [47], 8ANC195 [48], and PGT151 [49]), nonneutralizing or weakly neutralizing monoclonal antibodies (F105 [50], 17b [51] with sCD4, 48D [52] with sCD4, 447-52D [23] with sCD4, 3074 [24] with sCD4, and 2557 [38] with sCD4), and noncognate antibodies (anti-influenza virus antibody CR9114 [53] and anti-respiratory syncytial virus [RSV] antibody D25 [54]) in duplicate (30 μl/well) at a concentration of 4 μg/ml diluted in 1× PBS by incubation overnight at 4°C. The following day, the plates were washed (wash buffer, 0.05% Tween 20–1× PBS) and blocked with 150 μl of blocking buffer (5%, wt/vol) (MSD blocker A, catalog number R93BA-4; MSD) by incubation for 1 h on a vibrational shaker (Titramax 100, catalog number 544-11200-00; Heidolph Instruments) at 650 rpm. All incubations were performed at room temperature except the coating step. During the incubation, BG505 DS-SOSIP trimers were titrated in serial 2-fold dilutions starting at a concentration of 5 μg/ml of the trimer in the assay diluent (1% [wt/vol] MSD blocker A–0.05% Tween 20). For soluble CD4 (sCD4) induction, the trimer was combined with sCD4 at a constant molar concentration of 1 μM before being added to the MSD plate. After the incubation with blocking buffer was complete, the plates were washed, and the diluted trimer was transferred (25 μl/well) to the MSD plates and incubated for 2 h on the vibrational shaker at 650 rpm. After the 2-h incubation with trimer, the plates were washed again, and 2G12 antibody labeled with Sulfo-Tag (catalog number R91AO-1; MSD) at a conjugation ratio of 1:15 (2G12/Sulfo-Tag), diluted in assay diluent at 2 μg/ml, was added to the plates (25 μl/well) and incubated for 1 h on a vibrational shaker at 650 rpm. The plates were washed and read using 1× read buffer (catalog number R92TC-1, 4× Read Buffer T; MSD) on an MSD Sector Imager 2400.
Negative-stain electron microscopy.
The samples were diluted to approximately 0.02 mg/ml, adsorbed to a freshly glow-discharged carbon film grid for 15 s, and stained with 0.7% uranyl formate at a pH of 5. Images were collected semiautomatically using SerialEM (55) on an FEI Tecnai T20 microscope operating at 200 kV and equipped with a 2,000- by 2,000-pixel (px) Eagle charge-coupled device (CCD) camera. The pixel size was 0.22 nm/px. Particles were selected using the swarm mode in e2boxer from the EMAN2 software package (56). Reference-free 2D class averages were obtained using EMAN2.
Surface plasmon resonance analysis.
Binding affinities and kinetics of soluble CD4 (sCD4) to various HIV-1 DS-SOSIP trimers were assessed by single-cycle kinetics analysis using surface plasmon resonance on a Biacore T-200 instrument (GE Healthcare) at 25°C with HBS-EP+ buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P-20). First, 2G12 antibody was immobilized on flow cells of a CM5 chip at ∼2,000 response units. Next, 200 nM trimer was captured onto the sample flow cell at a flow rate of 5 μl/min for 120 s. Finally, sCD4 at five concentrations (180 nM, 90 nM, 45 nM, 22.5 nM, and 11.25 nM for BG505 SOSIP.664 and BG505 DS-SOSIP; 500 nM, 250 nM, 125 nM, 62.5 nM, and 31.25 nM for optimized DS-SOSIP trimers) was injected incrementally in a single cycle, starting from the lowest concentration at a flow rate of 50 μl/min for 60s, which was followed by a dissociation phase of 30 min. Blank sensorgrams were obtained by injection of the same volume of HBS-EP+ buffer in place of sCD4. Sensorgrams of the concentration series were corrected with corresponding blank curves and fitted globally with Biacore T200 evaluation software with a 1:1 Langmuir model of binding.
Differential scanning calorimetry.
A high-precision differential scanning VP-DSC microcalorimeter (GE Healthcare/MicroCal) was employed to measure the heat capacity of the trimers. In brief, samples were diluted to 0.3 mg/ml with PBS. Thermal denaturation scans were performed from 30°C to 110°C at a rate of 1°C/min.
Expression and purification of BG505 DS-SOSIP trimer mutants for crystallization.
BG505 DS-SOSIP trimer mutant constructs were cotransfected with furin in HEK293S GnTI−/− cells using 600 μg of plasmid DNA and 150 μg of furin. The supernatants were harvested 7 days after transfection and run over a VRC01 affinity column. Following a PBS wash, the proteins were eluted using 3 M MgCl2, pH 7.4. The eluate was concentrated using a Centricon-70 and directly purified through Superdex 200 size exclusion chromatography in 5 mM HEPES, pH 7.5, 150 mM NaCl, and 0.02% azide. The trimeric peak was isolated, concentrated, and used directly or flash frozen in liquid nitrogen and kept at −80°C until further use.
Fragment antigen-binding (Fab) fragment expression and purification.
PGT122 and 35O22 IgG were expressed as previously described. Heavy-chain plasmids with a human rhinovirus 3C protease (HRV3C) cleavage site in the hinge region were cotransfected with corresponding light-chain plasmids in the HEK293-Expi cell line using TrueFect-Max transfection reagent (United Biosystems) according to the manufacturer's protocol. Cultures were fed with fresh medium 4 h posttransfection and with enriched medium containing valproic acid (4 mM final concentration) 24 h after transfection. Cultures were then incubated at 33°C for 5 more days, and supernatants were harvested and passed though a protein A column. After PBS wash and low-pH elution, eluate was collected. Fab fragments were obtained by using HRV3C cleavage, and Fab fragments were purified over a Superdex 200 column (GE) in 5 mM HEPES, pH 7.5, 150 mM NaCl, and 0.02% azide.
Complex preparation.
PGT122 and 35O22 Fab fragments were added to a solution of purified BG505 DS-SOSIP trimer mutants in a 2-fold molar excess for 2 h at RT. The ternary complex was then purified over gel filtration, and fractions of complex were pooled, concentrated down to 6 mg/ml, and used immediately for crystal screening or flash frozen in liquid nitrogen and kept at −80°C until further use.
Crystal screening.
The ternary complexes were screened for crystallization using 572 conditions from Hampton, Wizard, and Precipitant Synergy screens using a Cartesian Honeybee crystallization robot as described previously (57) and a Mosquito Crystal robot. Crystals were observed and were manually reproduced. The complex crystal of DS-SOSIP.4mut with PGT122 and 35O22 Fab fragments grew in 10.5% polyethylene glycol (PEG) 4000 (P4K), 0.2 M AmSO4, 0.1 M sodium acetate (NaOAc), pH 4.6. The crystals were cryoprotected in 20% glycerol. Crystals of DS-SOSIP.6mut in complex with PGT122 and 35O22 Fab fragments grew in 13.2% PEG 400, 6.6% PEG 8000, and 0.1 M. sodium acetate/acetic acid, pH 4.5, and were cryoprotected in 15% 2R-3R-butanediol. Data were collected at a wavelength of 1.00 Å at the Southeast Regional Collaborative Access Team (SER-CAT) beamline ID-22 (Advanced Photon Source, Argonne National Laboratory).
X-ray data collection, structure solution, and model building.
Diffraction data were processed with the HKL2000 suite (58). The overall resolution was determined as the highest resolution for which the completeness was greater than 50% and the I/σI was greater than 2.0. Thus, for DS-SOSIP.4mut and DS-SOSIP.6mut ternary complexes, the overall resolution was 4.1 Å and 4.3 Å, respectively. The DS-SOSIP.4mut and DS-SOSIP.6mut ternary complex crystal diffraction data were then assessed for anisotropy through use of the Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/), which indicates the resolution at which F/sigma drops below 3.0 along a, b, and c axes; for the two lattices, these were 4.9 Å, 4.9 Å, and 3.4 Å and 5.5 Å, 5.5 Å, and 3.5 Å, respectively. Because we sought to use as much of the data as possible for refinement, we used the overall resolution described above to define the resolution of the a and b axes, with the resolution limit for the c axis defined by the Diffraction Anisotropy Server.
We note that other investigators have determined overall resolutions by the following equation: Reseff = (high resolution)(completeness)(−1/3) (59), where Reseff is effective resolution. For DS-SOSIP.4mut ternary complex, the untruncated data had a completeness of 61.0%, and the overall resolution is calculated as Reseff = (3.4)(0.610)(−1/3) = 4.0 Å; the truncated data had a completeness of 58% and an effective resolution Reseff = (3.4)(0.58)(−1/3) = 4.1 Å. For DS-SOSIP.6mut ternary complex, the untruncated data had a completeness of 63.2%, and the overall resolution is calculated as Reseff = (3.5)(0.632)(−1/3) = 4.1 Å; the truncated data had a completeness of 51% and an effective resolution Reseff = (3.5)(0.51)(−1/3) = 4.4 Å. These alternatively defined resolutions are 0.1 and 0.2 Å, respectively, higher than our 50% completeness, 2 I/sigma-defined overall resolutions.
We also report the CC1/2 value (Pearson correlation coefficient between intensities averaged in two subsets of data) of the highest resolution shell (0.345 at 3.4 Å for 4mut; 0.299 at 3.5 Å for 6mut); CC1/2 values of >0.15 are reported to be significant (60).
Structure solution was obtained by molecular replacement with Phaser using the BG505 SOSIP.664 ternary complex structure (PDB accession number 4TVP) (11) as a search model. Refinement was carried out with Phenix (61). Model building was carried out with Coot (62). The Ramachandran plot was determined by MolProbity (63). Data collection and refinement statistics are shown in Table S4.
Immunogenicity in guinea pig.
Two-month-old Hartley guinea pigs were purchased from Charles River and immunized intramuscularly at the hind leg muscles as previously described (32). Injections consisted of 25 μg of immunogen formulated in 400 μl of PBS with 80 μl of Adjuplex (Sigma). The immunizations were administered as two separate injections of 200 μl into each quadriceps muscle. Immunizations were performed at weeks 0, 4, 16, and 28. Blood draws for immune assessment included a prebleed week −1 sample and blood draws performed 2 weeks after each immunization. All animal experiments were reviewed and approved by the Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH, and all animals were housed and cared for in accordance with local, state, federal, and institute policies in an American Association for Accreditation of Laboratory Animal Care-accredited facility at the NIH. Collected sera were heat inactivated for 1 h at 56°C before being analyzed.
Anti-BG505 V3 peptide ELISA.
Ninety-six-well plates (Costar High Binding Half-Area; Corning, Kennebunk, ME) were coated overnight at 4°C with 50 μl/well of 2 μg/ml BG505 V3 peptide (TRPNNNTRKSIRIGPGQAFYATG; Bio-Synthesis, Inc., Lewisville, TX). Between subsequent steps, plates were washed five times with PBS–0.05% Tween (PBS-T) and incubated at 37°C for 50 min, except as otherwise noted. After being coated, plates were blocked with 100 μl/well of blocking buffer (B3T; 150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 3.3% fetal bovine serum [FBS], 2% bovine albumin, 0.07% Tween 20, 0.02% thimerosal). Next, 50 μl/well serially diluted (5-fold; starting dilution, 1:20) guinea pig serum in B3T buffer was added. Afterward, goat anti-guinea pig IgG antibody conjugated to horseradish peroxidase (KPL, Gaithersburg, MD) diluted 1:10,000 in B3T buffer was added at 50 μl/well for 50 to 65 min. Plates were developed with tetramethylbenzidine (TMB) substrate (SureBlue; KPL, Gaithersburg, MD) for 10 min before the addition of 1 N sulfuric acid (Fisher Chemical, Fair Lawn, NJ), without washing, to stop the reaction. Plates were read at 450 nm (SpectraMax using SoftMax Pro, version 5, software; Molecular Devices, Sunnyvale, CA), and the optical densities (OD) were analyzed following subtraction of the nonspecific horseradish peroxidase background activity. The endpoint titer was defined as the reciprocal of the greatest dilution with an OD value above 0.1 (10 times average raw plate background) or, when this dilution's OD was greater than 0.2, the reciprocal of the midpoint between this dilution and the subsequent dilution.
Anti-BG505 SOSIP.664 ELISA.
Ninety-six-well plates (Costar High Binding Half-Area; Corning, Kennebunk, ME) were coated overnight at 4°C with 50 μl/well of 2 μg/ml D7324 monoclonal antibody (Aalto Bio Reagents, Dublin, Ireland). Between subsequent steps, plates were washed five times with PBS-T (PBS plus 0.05% Tween) and incubated at room temperature for 60 min, except as otherwise noted. After being coated, plates were blocked with 100 μl/well of blocking buffer (5% skim milk in PBS), followed by trimer capture with 50 μl/well of 0.5 μg/ml D7324-tagged BG505 SOSIP.664 in 10% FBS-PBS for 240 to 248 min. Following trimer addition, subsequent procedures mimicked the anti-BG505 V3 peptide ELISA, except that the dilutions were made in 0.2% Tween-PBS instead of B3T buffer, the endpoint titer was defined at an OD value of 0.1 (2 times average raw plate background), and the diluted guinea pig serum was incubated for 60 min.
Neutralization assays.
Single-round-of-replication Env pseudoviruses were prepared, titers were determined, and the pseudoviruses were used to infect TZM-bl target cells as described previously (64, 65). Neutralization curves were fit by nonlinear regression using a 5-parameter Hill slope equation as previously described (66). The 50% and 80% inhibitory dilutions (ID50 and ID80) were reported as the serum concentrations required to inhibit infection by 50% and 80%, respectively.
Accession number(s).
Atomic coordinates and structure factors of the DS-SOSIP.4mut and DS-SOSIP.6mut X-ray crystal structures have been deposited with the Protein Data Bank (PDB) under accession numbers 5UTY and 5UTF, respectively. In these PDB deposits, we provide the overall resolution in the title of the PDB entries as follows: “Crystal Structure of a Stabilized DS-SOSIP.4mut BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, N300M, N302M, T320L in Complex with Human Antibodies PGT122 and 35O22 at 4.1 Å” and “Crystal Structure of a Stabilized DS-SOSIP.6mut BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, Y177W, N300M, N302M, T320L, I420M in Complex with Human Antibodies PGT122 and 35O22 at 4.3 Å.” We have also added remarks to the PDB headers clarifying how the overall resolutions were determined.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Stuckey for assistance with figures and the Structural Biology Section, Structural Bioinformatics Core, Humoral Immunity Section, and the BSL-3 Core at the NIH Vaccine Research Center for helpful discussions and comments on the manuscript. We thank Dennis Burton and Mark Feinberg for antibody PGT122 and Mark Connors for antibody 35O22 used in structural analysis.
Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. Leidos Biomedical Research, Inc., provided support in the form of a salary for author Y.T. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02268-16.
REFERENCES
- 1.McElrath MJ, Haynes BF. 2010. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity 33:542–554. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Earl PL, Broder CC, Long D, Lee SA, Peterson J, Chakrabarti S, Doms RW, Moss B. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol 68:3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 4.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders RW, Wilson IA, Moore JP. 2016. HIV's Achilles' heel. Sci Am 315:50–55. doi: 10.1038/scientificamerican1216-50. [DOI] [PubMed] [Google Scholar]
- 8.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schon A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, McDermott AB, Mascola JR, Kwong PD. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. 2015. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell 162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong L, Torrents de la Pena A, Deller MC, Garces F, Sliepen K, Hua Y, Stanfield RL, Sanders RW, Wilson IA. 2015. Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer. Acta Crystallogr D Biol Crystallogr 71:2099–2108. doi: 10.1107/S1399004715013917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, de Val N, Lyumkis D, Ward AB. 2015. Model building and refinement of a natively glycosylated HIV-1 Env protein by high-resolution cryoelectron microscopy. Structure 23:1943–1951. doi: 10.1016/j.str.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garces F, Lee JH, de Val N, Torrents de la Pena A, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity 43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. 2016. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gristick HB, von Boehmer L, West AP Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, Bjorkman PJ. 2016. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol 23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jardine JG, Sok D, Julien JP, Briney B, Sarkar A, Liang CH, Scherer EA, Henry Dunand CJ, Adachi Y, Diwanji D, Hsueh J, Jones M, Kalyuzhniy O, Kubitz M, Spencer S, Pauthner M, Saye-Francisco KL, Sesterhenn F, Wilson PC, Galloway DM, Stanfield RL, Wilson IA, Burton DR, Schief WR. 2016. Minimally mutated HIV-1 broadly neutralizing antibodies to guide reductionist vaccine design. PLoS Pathog 12:e1005815. doi: 10.1371/journal.ppat.1005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. 2015. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog 11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Taeye SW, Moore JP, Sanders RW. 2016. HIV-1 envelope trimer design and immunization strategies to induce broadly neutralizing antibodies. Trends Immunol 37:221–232. doi: 10.1016/j.it.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, NISC Comparative Sequencing Program, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killikelly A, Zhang HT, Spurrier B, Williams C, Gorny MK, Zolla-Pazner S, Kong XP. 2013. Thermodynamic signatures of the antigen binding site of mAb 447-52D targeting the third variable region of HIV-1 gp120. Biochemistry 52:6249–6257. doi: 10.1021/bi400645e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Konings FA, Nadas A, Anyangwe CA, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. 2006. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol 80:6865–6872. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, Julien JP, Burton DR, Wilson IA, Sanders RW, Klasse PJ, Ward AB, Moore JP. 2015. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol 89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC, Wongstaal F. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 27.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 28.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 30.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 31.de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. 2015. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng C, Pancera M, Bossert A, Schmidt SD, Chen RE, Chen X, Druz A, Narpala S, Doria-Rose NA, McDermott AB, Kwong PD, Mascola JR. 2015. Immunogenicity of a prefusion HIV-1 envelope trimer in complex with a quaternary-structure-specific antibody. J Virol 90:2740–2755. doi: 10.1128/JVI.02380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, Briney B, Schiffner T, Garces F, Freund NT, Gitlin AD, Menis S, Georgeson E, Kubitz M, Adachi Y, Jones M, Mutafyan AA, Yun DS, Mayer CT, Ward AB, Burton DR, Wilson IA, Irvine DJ, Nussenzweig MC, Schief WR. 2016. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. 2015. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, Schon A, Van Galen J, Chuang GY, Gorman J, Harned A, Pancera M, Stewart-Jones GB, Cheng C, Freire E, McDermott AB, Mascola JR, Kwong PD. 2015. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol 89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, Sok D, Zhou B, Burton DR, Ward AB, Wilson IA, Zhu J. 2016. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun 7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CY, Georgiev I, Anderson AC, Donald BR. 2009. Computational structure-based redesign of enzyme activity. Proc Natl Acad Sci U S A 106:3764–3769. doi: 10.1073/pnas.0900266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong XP. 2010. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat Struct Mol Biol 17:955–961. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- 39.Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. 2006. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol 80:6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorny MK, Sampson J, Li H, Jiang X, Totrov M, Wang XH, Williams C, O'Neal T, Volsky B, Li L, Cardozo T, Nyambi P, Zolla-Pazner S, Kong XP. 2011. Human anti-V3 HIV-1 monoclonal antibodies encoded by the VH5-51/VL lambda genes define a conserved antigenic structure. PLoS One 6:e27780. doi: 10.1371/journal.pone.0027780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rini JM, Stanfield RL, Stura EA, Salinas PA, Profy AT, Wilson IA. 1993. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc Natl Acad Sci U S A 90:6325–6329. doi: 10.1073/pnas.90.13.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O'Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng HP, Yang AS, Program NCS, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD. 2015. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 46.Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, Pancera M, Cale EM, Ernandes MJ, Louder MK, Asokan M, Bailer RT, Druz A, Fraschilla IR, Garrett NJ, Jarosinski M, Lynch RM, McKee K, O'Dell S, Pegu A, Schmidt SD, Staupe RP, Sutton MS, Wang K, Wibmer CK, Haynes BF, Abdool-Karim S, Shapiro L, Kwong PD, Moore PL, Morris L, Mascola JR. 2015. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol 90:76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 49.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, McBride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 57.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.Weiss MS. 2001. Global indicators of X-ray data quality. J Appl Crystallogr 34:130–135. doi: 10.1107/S0021889800018227. [DOI] [Google Scholar]
- 60.Karplus PA, Diederichs K. 2012. Linking crystallographic model and data quality. Science 336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Davis IW, Murray LW, Richardson JS, Richardson DC. 2004. MOLPROBITY: structure validation and all-atom contact analysis for Nucleic acids and their complexes. Nucleic Acids Res 32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 65.Shu Y, Winfrey S, Yang ZY, Xu L, Rao SS, Srivastava I, Barnett SW, Nabel GJ, Mascola JR. 2007. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine 25:1398–1408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for neutralizing antibody assessment. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Q, Acharya P, Dolan MA, Zhang P, Guzzo C, Lu J, Kwon A, Gururani D, Miao H, Bylund T, Chuang GY, Druz A, Zhou T, Rice WJ, Wigge C, Carragher B, Potter CS, Kwong PD, Lusso P. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat Struct Mol Biol, in press. doi: 10.1038/nsmb.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.