ABSTRACT
Persistent immune activation during chronic human immunodeficiency virus type 1 (HIV-1) infection facilitates immune dysfunction and thereby fuels disease progression. The translocation of bacterial derivatives into blood and the hyperinflammatory responsiveness of monocytes have been considered important causative factors for persistent immune activation. Whether microRNAs (miRNAs) are involved in regulating monocyte-mediated inflammatory responses during chronic HIV-1 infection remains elusive. In this study, we show that miR-126-5p functions as a positive regulator of monocyte-mediated inflammatory responses. Significantly increased miRNA miR-126-5p and decreased cylindromatosis (CYLD) were observed in primary monocytes from chronic HIV-1 patients. Inhibition of miR-126-5p in monocytes from chronic HIV-1 patients attenuated the responsiveness of these cells to lipopolysaccharide (LPS) stimulation. Gain-of-function assays confirmed that miR-126-5p could downregulate CYLD, which in turn caused an upregulation of phosphorylation of JNK protein (pJNK) and enhanced inflammatory responses of monocytes to LPS stimulation. Overall, miR-126-5p upregulates the responsiveness of monocytes to LPS stimulation in chronic HIV-1 infection, and the suppression of miR-126-5p and the promotion of CYLD expression in primary monocytes may represent a practical immune intervention strategy to contain persistent inflammation in chronic HIV-1 infection.
IMPORTANCE Monocyte-mediated hyperinflammatory responses during chronic HIV-1 infection are important causative factors driving AIDS progression; however, the underlying mechanism has not been fully addressed. We demonstrated that miR-126-5p, one of the most upregulated miRNAs during chronic HIV-1 infection, could enhance the inflammatory responses of monocytes to LPS by suppressing the inhibitory protein CYLD and thereby unleashing the expression of pJNK in the LPS/Toll-like receptor 4/mitogen-activated protein kinase pathway. This observation reveals a new mechanism for HIV-1 pathogenesis, which could be targeted by immune intervention.
KEYWORDS: CYLD, HIV-1, inflammatory responses, miR-126-5p, monocytes
INTRODUCTION
AIDS progression is fueled by chronic immune hyperactivation mediated by inflammatory cytokines (1–5). Monocyte activation and inflammation are commonly found in chronic human immunodeficiency virus type 1 (HIV-1) patients (6–11). The translocation of bacterial derivatives from the compromised gastrointestinal (GI) tract into the blood observed in chronic HIV-infected subjects (1, 5, 12) and the hyperresponsiveness of monocytes to lipopolysaccharide (LPS) have been considered important causative factors for persistent immune activation (6, 13). Preexposure of monocytes to Toll-like receptor 7/8 (TLR7/8) ligands derived from HIV-1 single-stranded RNA (ssRNA) (14, 15) and upregulated TLR2 expression on monocytes (16) could enhance the inflammatory responsiveness of these cells. In addition, skewed differentiation of monocytes into proinflammatory CD16+ monocytes and tumor necrosis factor alpha (TNF-α)-producing M-DC8+ CD14+ CD16++ monocytes during chronic HIV-1 infection have been observed in the presence of HIV-1 viremia compared to that observed in HIV-1 seronegative subjects (7, 9, 17). Effective antiretroviral therapy (ART) successfully suppresses HIV-1 replication but fails to normalize monocyte activation/inflammation (3, 18–21). Our previous report shows that interferon-stimulated gene lymphocyte antigen complex 6, locus E (LY6E), in monocytes regulates the CD14/TLR4 pathway but inadequately restrains the hyperactivation of monocytes during chronic HIV-1 infection (13), indicating the presence of additional mechanisms to promote the inflammatory responses of monocytes.
MicroRNAs (miRNAs) are ∼22-nucleotide (nt)-long small RNAs that are important regulators of mRNA turnover and translation. The importance of miRNAs in HIV-1 infection, particularly in interpreting the replication restriction of HIV-1 in primary monocytes and resting primary CD4+ T lymphocytes, has been well established (22–29). miRNAs, such as miR-21 (30), miR-146a (31), and miR-155 (32), are important regulators in the TLR4 pathway and exert their influence by targeting multiple sites in the signaling cascade, but little is known about the role of miRNAs in regulating the immune response during chronic HIV-1 infection in monocytes, an important source of inflammatory cytokines. We hypothesized that miRNAs play a critical role in regulating monocyte-mediated inflammatory responses and enhanced immune activation, thereby facilitating disease progression.
In the current study, we used small-RNA sequencing-based miRNA profiling to explore the miRNAs that are differentially expressed in healthy donors and HIV-1 patients who are naive to ART treatment and found that miR-126-5p was one of the most significantly upregulated miRNAs in monocytes derived from chronic HIV-1-infected patients. Inhibition of miR-126-5p expression in monocytes from chronic HIV-1 patients attenuated the responsiveness to LPS stimulation. Furthermore, we revealed that the miR-126-5p-CYLD-JNK pathway is engaged in and plays an important role in the regulation of monocyte-mediated inflammatory responses during chronic HIV-1 infection.
RESULTS
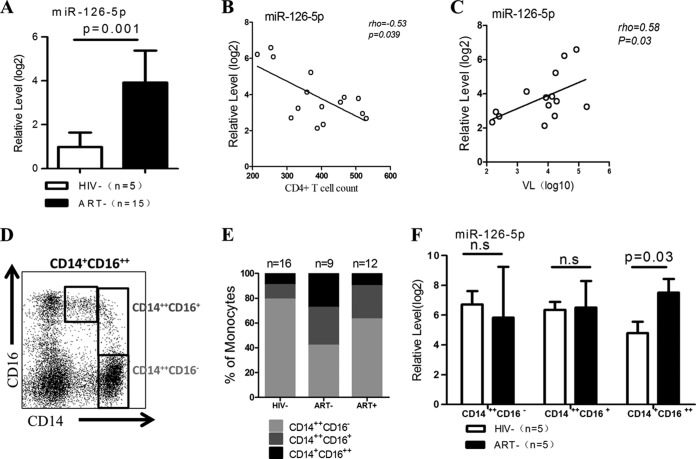
Upregulation of miR-126-5p in monocytes correlates with HIV-1 disease progression.
We profiled miRNA expression using Ion Torrent PGM and observed that the majority of the known miRNAs were dysregulated in purified CD14+ monocytes from chronic HIV-1 patients compared with those from healthy donors (data not shown). Eight miRNAs showed a 50-fold change (adjusted P value of <0.05); miR-126-5p was one of the most upregulated miRNAs. Because previous studies have demonstrated that miR-126-3p is engaged in the TLR4 signaling pathway and miR-126-5p usually plays a synergistic role with miR-126-3p (33–35), we postulated that miR-126-5p plays a role similar to that of miR-126-3p in the LPS/TLR4 pathway. Given that the LPS/TLR4 pathway plays an important role in the pathogenesis of HIV-1 infection, we considered the priority was to focus on deciphering the role of miR-126-5p in all eight miRNAs. Therefore, we further validated the significant differences in miR-126-5p expression between chronic HIV-1 patients and healthy donors using a quantitative PCR (qPCR) assay (Fig. 1A). We also examined miR-126-5p expression in CD14+ monocytes in relation to the clinical parameters of HIV-1 disease progression. In a cross-sectional study, miR-126-5p expression in purified CD14+ monocytes was inversely correlated with the absolute CD4+ T cell counts and positively correlated with the viral loads (the viral load of one patient was not detected) (Fig. 1B and C). As the purity of CD14+ monocytes in our study exceeded 97% based on CD14 staining, the predominant miR-126-5p quantified was likely derived from monocytes, although we were unable to exclude the possibility of contamination by predominant endothelial cells (ECs), which have been reported as the main source of the miR-126-3p/5p cluster (35–38).
FIG 1.
miR-126-5p is significantly upregulated in primary monocytes during chronic HIV-1 infection and correlated with disease progression. (A) miR-126-5p expression is significantly increased during chronic HIV-1 infection. The expression of miR-126-5p was quantified by qPCR and normalized to the expression of U6 in each sample. (B) The inverse correlation of miR-126-5p expression in monocytes with CD4+ T cell counts. The expression levels of miR-126-5p were used to perform the correlation analysis with CD4+ T cell counts, and a significant negative correlation was observed (rho = −0.53, P = 0.039). (C) The correlation of miR-126-5p expression in monocytes with plasma viral loads. The expression levels of miR-126-5p were used to perform the association with plasma viral loads, and a significant association was observed (rho = 0.58, P = 0.03). (D) An example of a dot plot analysis of three monocyte subsets: CD14++ CD16− (classical) monocytes, CD14+ CD16++ (nonclassical) monocytes, and CD14++ CD16+ (intermediate) monocytes. (E) HIV-1 infection resulted in increased levels of nonclassical and intermediate monocyte subsets, and ART mainly reduces the level of nonclassical monocytes. The percentages of the three monocyte subsets were obtained from HIV− (HIV-1 seronegative; n = 16), ART− (HIV-1 infected, naïve to ART; n = 9), and ART+ (HIV-1 infected, ART treated; n = 12) subjects and averaged into groups; the constituent ratios then were calculated using the averaged data and plotted. (F) miR-126-5p is primarily upregulated in nonclassical monocytes. qPCR was performed on three monocyte subsets (CD14++ CD16−, CD14+ CD16++, and CD14++ CD16+) derived from the ART− (n = 5) and HIV− (n = 5) groups, and the data were averaged by groups. n.s, not significant.
As noted above, human monocyte subsets can be classified by flow cytometry based on the expression of CD14 and CD16. The majority of monocytes express CD14 but not CD16 (CD14++ CD16−), and those expressing CD16 can be further subdivided into two subpopulations as CD14++ CD16+ cells and CD14+ CD16++ cells (Fig. 1D). CD16+ monocytes are well known to play a pivotal role in TNF-α overproduction in response to LPS stimulation, and higher frequencies of CD14++ CD16+ and CD14+ CD16++ cells were identified in chronic HIV-1 patients than in healthy donors (Fig. 1E). We quantified the expression of miR-126-5p in three monocyte subsets to exclude the possibility that the elevated miR-126-5p level was due to the perturbation of monocyte subsets and identified that miR-126-5p was significantly upregulated in CD14+ CD16++ monocytes but not CD14++ CD16− monocytes and CD14++ CD16+ monocytes in HIV-1 patients (Fig. 1F). Previous studies have demonstrated that miR-126-5p is an intronic product of the vascular endothelial epidermal growth factor-like domain 7 (EGFL7) in ECs (38, 39). The expression of EGFL7 and its coexpression with miR-126-5p in primary monocytes requires further investigation.
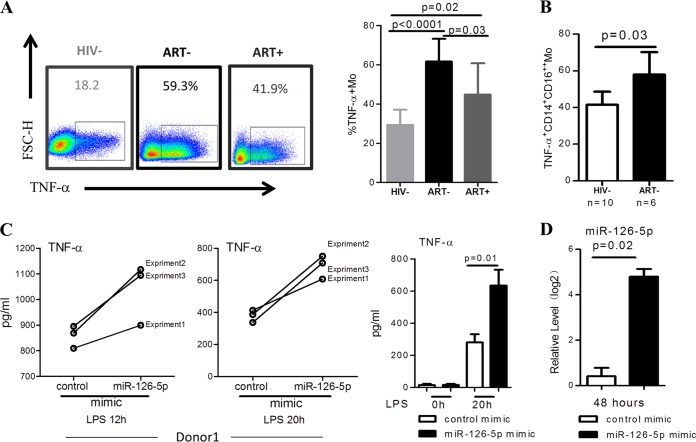
miR-126-5p upregulates TNF-α production in LPS-stimulated monocytes.
Hyperactivation of CD14+ monocytes upon LPS/TLR4 stimulation during chronic HIV-1 infection has been reported previously. We compared the production of TNF-α in monocytes purified from healthy donors (HIV−), HIV-1+ naive to ART (ART−), and ART-treated HIV-1 patients (ART+) in the absence or presence of LPS stimulation. As shown in Fig. 2A, the percentage of TNF-α+ monocytes that responded to LPS stimulation was profoundly increased in ART-naive patients compared with healthy donors (P < 0.001) and was less significantly increased in patients who were effectively treated with ART (P = 0.02). We further confirmed that CD14+ CD16++ monocytes from ART-naive HIV-1 patients produced significantly more TNF-α in response to LPS than those from HIV-1 seronegative healthy controls (Fig. 2B). To determine whether the overexpression of miR-126-5p influenced the responses of monocytes to LPS stimulation, we electrotransfected primary monocytes purified from healthy donors with miR-126-5p mimic (Fig. 2C, left) and then stimulated the monocytes with LPS. Interestingly, miR-126-5p-overexpressing monocytes produced significantly higher levels of TNF-α than the control mimic-transfected monocyte cells (Fig. 2C, right). The efficiency of miR-126-5p overexpression was determined by qPCR as shown in Fig. 2D. These data demonstrated that higher expression of miR-126-5p in monocytes during HIV-1 infection resulted in the upregulation of TNF-α in response to LPS stimulation.
FIG 2.
miR-126-5p upregulates TNF-α production in LPS-stimulated primary monocytes. (A) HIV-1 infection resulted in increased TNF-α production of monocytes in response to LPS stimulation. Examples of TNF-α production of monocytes with and without LPS stimulation are shown on the left. PBMCs from the HIV− (n = 9), ART− (n = 8), and ART+ (n = 8) groups were stimulated with LPS (100 ng/ml) for 20 h. TNF-α production in monocytes was quantified by gating CD14−-positive cells in a flow cytometry-based intracellular staining assay and statistically calculated by groups. TNF-α production is significantly higher in the ART− (P < 0.0001) and ART+ (P = 0.02) than in the HIV− group. ART treatment significantly reduces TNF-α production (P = 0.03 for the ART− group versus the ART+ group). (B) TNF-α production is significantly upregulated in nonclassical monocytes. TNF-α production was quantified as described above; nonclassical monocytes were gated as CD14+ CD16++, and TNF-α production in this subset was compared between the HIV-1-seronegative and HIV-1-infected groups. (C) Overexpression of miR-126-5p in primary monocytes derived from the HIV− subjects profoundly enhances TNF-α production. Here, 1 × 105 transfected monocytes were stimulated with LPS (100 ng/ml) for 20 h, and the supernatants were collected for TNF-α analysis. The level of TNF-α was quantified by ELISA. The results shown on the left were from three independent experiments in one donor, and the production of TNF-α in response to LPS stimulation in three donors is shown on the right. (D) Effective transfection of miR-126-5p mimic in primary monocytes was observed. Primary monocytes derived from the HIV− group were electrotransfected with control or miR-126-5p mimic, and the transfection efficacy was determined by qPCR.
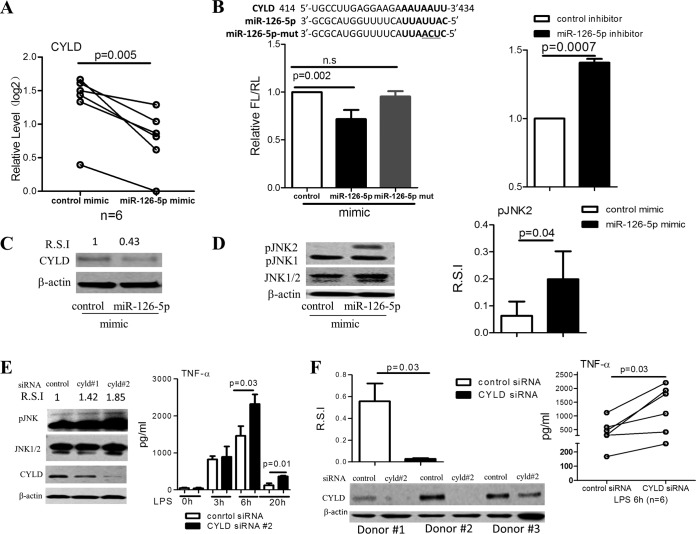
CYLD is a candidate target of miR-126-5p.
We next sought to identify the target genes of miR-126-5p. In silico analyses provided several gene candidates, including CYLD (a protein that belongs to the A20 family), NLR family CARD domain-containing 3 (NLRC3), suppressor of cytokine signaling 1 (SOCS1), TNFAIP3 interacting protein 1 (TNIP1), and protein tyrosine phosphatase, nonreceptor type 6 (SHP1), which are also involved in regulating the LPS/TLR4/nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB)/mitogen-activated kinase (MAPK) signaling pathways. We found that overexpression of miR-126-5p significantly decreased the expression of CYLD (Fig. 3A) and SOCS1 but not NLRC3, TNIP1, or SHP1 in monocytes purified from healthy donors (data not shown). We next concentrated on the regulatory role of miR-126-5p on CYLD and SOCS1. We constructed reporter plasmids by replacing the 3′-untranslated region (3′UTR) of firefly luciferase with the 3′UTR of CYLD or SOCS1. A three-base-pair mutation (mutant mimic) was introduced into miR-126-5p mimic as a further control (shown at the top of Fig. 3B). After transfection of 293T cells with the miR-126-5p mimic, miR-126-5p mutant mimic, control inhibitor, or miR-126-5p inhibitor, we observed that the miR-126-5p mimic markedly decreased the expression of luciferase in 293T cells cotransfected with pMIR-REPORT vector containing the 3′UTR of CYLD (Fig. 3B) or SOCS1 (data not shown), whereas miR-126-5p inhibitor profoundly increased luciferase expression in 293T cells cotransfected with pMIR-REPORT vector containing the CYLD 3′UTR (Fig. 3B). In contrast, there was no significant difference between the expression of luciferase in 293T cells transfected with miR-126-5p mutant mimic and control mimic. Furthermore, immunoblot analyses showed that forced expression of miR-126-5p repressed the expression of CYLD in primary monocytes (Fig. 3C).
FIG 3.
CYLD is targeted by miR-126-5p and is involved in the regulation of the inflammatory response to LPS stimulation in monocytes. (A) Overexpression of miR-126-5p resulted in a significant decrease in CYLD expression. Overexpression of miR-126-5p for 72 h was performed in primary monocytes (n = 6), and the expression of CYLD, the predicted target gene of miR-126-5p, was quantified by qPCR. As shown, overexpression of miR-126-5p resulted in a significant decrease in CYLD expression. (B) miR-126-5p regulates CYLD 3′UTR activity. The CYLD 3′UTR was placed into a reporter plasmid carrying a luciferase-encoding gene, and 293T cells were cotransfected with reporter plasmids and miR-126-5p mimic or miR-126-5p inhibitor for 48 h. (Left) As shown, overexpression of miR-126-5p resulted in a reduction in luciferase activity, whereas overexpression of the miR-126-5p mutant (containing three unmatched base pairs) failed to do so. (Right) In contrast, inhibition of miR-126-5p increased the luciferase activity. (C) Overexpression of miR-126-5p resulted in a decrease in CYLD protein expression in monocytes. CYLD protein expression was determined by Western blotting in primary monocytes that were electrotransfected with miR-126-5p mimic or control for 72 h. Overexpression of miR-126-5p resulted in a decrease in CYLD protein expression in monocytes. R.S.I., relative signal intensity. (D) Overexpression of miR-126-5p causes an increase in JNK2 phosphorylation in primary monocytes. Western blot analyses were performed to assess JNK phosphorylation in primary monocytes that were electrotransfected with miR-126-5p mimic or control mimic for 72 h and are shown as an example (left) and as R.S.I. (right). Experiments were performed in three individuals. (E) Transient knockdown of CYLD in THP-1 cells causes enhanced TNF-α production. (Left) CYLD expression and JNK phosphorylation in THP-1 cells with transient knockdown and their parental cells were analyzed by Western blotting 72 h after transient transfection with CYLD siRNA. (Right) TNF-α production in supernatants collected at 3, 6, and 20 h after LPS (100 ng/ml) stimulation was quantified by ELISA, and 2 × 105 THP-1 cells with transient knockdown of CYLD produced significantly more TNF-α than their controls. (F) Transient knockdown of CYLD in primary CD14+ monocytes (n = 6) resulted in increased TNF-α production. (Right) TNF-α production in the supernatants collected at 6 h after LPS (100 ng/ml) stimulation in 1 × 105 transfected monocytes was quantified by ELISA. (Left, upper) CYLD expression in the transient knockdown primary monocytes and their parental cells was analyzed by Western blotting 72 h after transient transfection with CYLD siRNA. (Left, lower) The representative pattern is shown.
Because CYLD, a target of miR-126-5p, has been previously shown to be involved in the negative regulation of JNK signaling (40), we hypothesized that the CYLD-mediated attenuation of pJNK could be released by miR-126-5p. To test this hypothesis, we isolated primary monocytes from healthy donors and overexpressed miR-126-5p in these cells. Importantly, the overexpression of miR-126-5p significantly increased pJNK2 levels in primary monocytes from healthy donors (Fig. 3D). Overall, these data demonstrated that miR-126-5p could activate the MAPK signaling pathway by upregulating pJNK through the inhibition of CYLD expression, thereby enhancing TLR4 responsiveness.
CYLD negatively regulates TNF-α secretion in monocytes in response to LPS stimulation.
CYLD acts as a negative regulator for Toll-like receptor 2 signaling via negative cross talk with TNF receptor-associated factor 6 (TRAF6) and TRAF7 in 293T cell lines and other cell types (41–44). Whether CYLD regulates TLR4 signaling in monocytes remains unclear. Transient knockdown of CYLD by siRNA resulted in a significant increase in TNF-α secretion in THP-1 cells (Fig. 3E). We also established an in vitro model via stable knockdown of CYLD expression in THP-1-shCYLD cell lines. Significantly higher levels of TNF-α secretion in response to LPS stimulation were observed in THP-1-shCYLD cells as early as 3 h after LPS treatment and through 20 h (data not shown). The production of other inflammatory cytokines, especially interleukin-1β (IL-1β), was also significantly increased in THP-1-shCYLD cells (data not shown). In addition, the function of CYLD in regulating TLR4 responsiveness was further confirmed in primary monocytes (Fig. 4F).
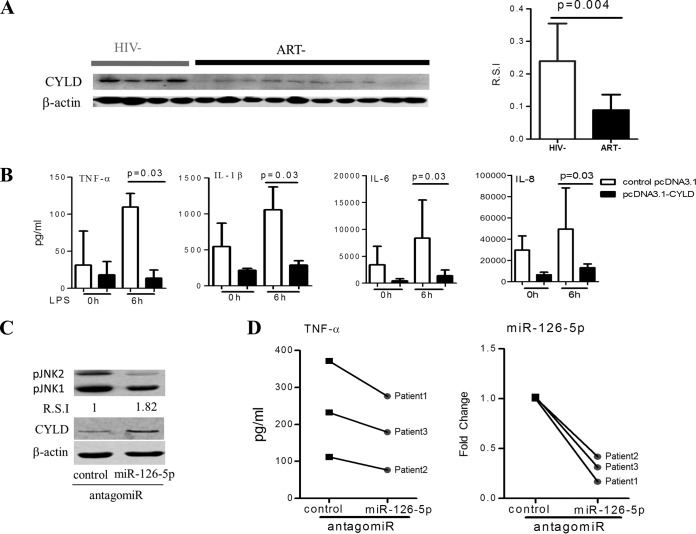
FIG 4.
miR-126-5p/CYLD/JNK axis regulates the responses of monocytes to LPS stimulation during chronic HIV-1 infection. (A) HIV-1 infection resulted in a reduction in CYLD protein. Western blot analyses of CYLD protein in monocytes from ART− individuals (n = 10) were compared with that from HIV− individuals (n = 8), shown as an example (left) and as R.S.I. (right). (B) Overexpression of CYLD in primary monocytes (n = 5) derived from the ART− group suppresses their inflammatory responses to LPS stimulation. The supernatants were collected at 6 h after LPS (100 ng/ml) stimulation of 5 × 104 primary monocytes transfected with pcDNA3.1-CYLD (codon optimization) plasmids or pcDNA3.1 plasmids for 72 h. The levels of TNF-α, IL-6, IL-1β, and IL-8 were quantified using BioLegend LEGENDplex CBA. (C) Inhibition of miR-126-5p increases CYLD protein expression and decreases JNK phosphorylation in monocytes from the ART− group. Expression of CYLD protein and JNK phosphorylation were analyzed by Western blotting in primary monocytes that were electrotransfected with control antagomir or miR-126-5p antagomir for 72 h. Transfection of the miR-126-5p antagomir resulted in an increase in CYLD protein levels and a decrease in phosphorylated JNK levels. (D) Downregulation of miR-126-5p in monocytes from the ART− group resulted in decreased TNF-α production. Monocytes from the ART− group (n = 3) were electrotransfected with either the control antagomir or the miR-126-5p antagomir; supernatants were collected at 12 h after LPS (100 ng/ml) stimulation of 5 × 104 transfected monocytes. The downregulation of miR-126-5p was detected by qPCR and is shown on the right.
The CYLD/miR-126-5p/JNK axis is involved in monocyte hyperactivation during HIV-1 infection.
As miR-126-5p is dramatically increased in monocytes during HIV-1 infection and CYLD is the candidate primary target gene for miR-126-5p, we rationalized that CYLD should be downregulated during HIV-1 infection. To confirm this hypothesis, we determined the expression of CYLD in HIV-1 patients who were naive to ART by Western blotting and observed that CYLD was significantly downregulated in monocytes from HIV-1 patients who were naive to ART compared to monocytes from healthy donors (Fig. 4A). To confirm the regulatory role of CYLD in monocytes from HIV-1 patients, we overexpressed CYLD by electrotransfecting monocytes from HIV-1 patients with CYLD-encoding plasmids (pCDNA3.1-CYLD) according to transfection methods reported previously (45). As shown in Fig. 4, overexpression of CYLD in primary monocytes from HIV-1 patients who were naive to ART resulted in a significant reduction of not only TNF-α but also IL-1β, IL-6, and IL-8 production in response to LPS stimulation (Fig. 4B). To corroborate the regulatory role of miR-126-5p in monocytes during HIV-1 infection, we transfected monocytes from ART-naive patients with the miR-126-5p antagomir and observed that pJNK1/2 was decreased along with increased expression of CYLD (Fig. 4C), and the production of TNF-α was blunted upon LPS stimulation (Fig. 4D).
Altogether, our data demonstrated that CYLD is a negative regulator of the LPS/TLR4 signaling pathway, and suppression of CYLD by miR-126-5p in chronic HIV-1 infection could lead to an upregulation of LPS/TLR4 pathway responses via JNK phosphorylation.
DISCUSSION
HIV-1 infection results in damage to the host immune system with a progressive loss of CD4+ T cells and ultimately the development of AIDS, which has been associated with persistent immune activation (1, 46). Microbial and LPS translocation from the gastrointestinal tract to the blood has been considered to be the causative factor for the persistent immune activation (1, 2, 4, 47, 48). Monocytes, as the primary target cells for LPS, have been observed to be hypersensitive to LPS stimulation and to produce markedly more inflammatory cytokines during HIV-1 infection than monocytes derived from HIV-1-seronegative subjects (7–9, 11, 14, 15); however, the underlying mechanism remains elusive. In the current study, we identified miR-126-5p as an important regulator of monocyte-mediated inflammatory responses to LPS stimulation. The upregulation of miR-126-5p expression during HIV-1 infection is likely to result in the downregulation of CYLD and thereby lead to removal of the blockade of the MAPK pathway in primary monocytes. Overall, our data demonstrated that the miRNA can heavily engage in the regulation of innate immune responses and thereby influence disease progression.
Preexposure of monocytes to the TLR7/8 pathway triggered by HIV-1 ssRNA-derived ligands has been considered the causative factor for the increased LPS/TLR4 responsiveness (15). Other studies have focused on the skewed differentiation of monocyte subgroups during HIV-1 infection and observed that proinflammatory CD16+ monocytes and TNF-α-producing M-DC8+ CD14+ CD16++ monocytes were more expanded in chronic HIV-1 infection than in healthy subjects (7, 9, 17, 49). Interestingly, the expansion levels of these cells appear to vary among different reports (7, 8, 49). Active viral replication is likely to push the differentiation of the CD14++ CD16− subset toward CD14++ CD16+ and CD14+ CD16++ subsets, and prolonged ART is required to inhibit this tendency (8). Although the hyperreactivity of CD14+ CD16++ subsets to LPS stimulation has been widely observed, the underlying mechanism is poorly understood. miRNAs have been shown to exercise their function to regulate HIV-1 replication and are associated with HIV-1 latency by directly targeting the genome of HIV-1 or indirectly targeting its host dependency factors (HDFs) in monocytes, dendritic cells, and CD4+ T cells (24–27, 50). HIV-1, in turn, can hijack Dicer, which is responsible for miRNA processing, to escape from miRNA-silencing pathway-mediated host defense (51–53). Let-7 family miRNAs and miR-9 have been confirmed to modulate IL-10 and IL-2 production, respectively, in CD4+ T cells (54, 55). These data clearly indicate that miRNAs can engage in the regulation of immune responses in HIV-1 infection.
To further reveal the mechanism responsible for monocyte-mediated hyperinflammatory responses, we hypothesized that miRNAs are involved in this mechanism by removing the inhibitory molecules and thereby increasing the sensitivity of monocytes to LPS stimulation. Indeed, a significant increase in several miRNAs in monocytes was observed in HIV-1 infection, among which miR-126-5p was the most upregulated member, especially in proinflammatory CD14+ CD16++ monocytes. The miR-126-3p/5p cluster is expressed predominately in ECs (35–38) and can exert their functions in non-ECs (34, 56, 57). miR-126-5p has been shown to correlate with metastasis-free survival in breast cancer patients (33) and to promote endothelial proliferation and prevent atherosclerosis by suppressing delta-like 1 (Dlk1) (58). Vascular endothelial growth factor-induced phosphorylation of ERK (pERK) can be blocked by the inhibition of miR-126-3p (37, 56), and forced expression of miR-126-3p leads to a substantial increase in pERK in leukemogenesis (56). These results suggest that miR-126-3p is involved in modulating the MAPK pathway. miR-126-3p and its complement, miR-126-5p, consistently jointly exert the same function (33, 34). miR-126-3p has been shown to be engaged in promoting the innate immune response (35, 59–61), but the role of miR-126-5p in regulating the innate immune response, especially TLR4 responsiveness, which is always accompanied by activation of the MAPK pathway, remains unknown. To determine whether miR-126-5p is engaged in regulating the LPS/TLR4 pathway, we overexpressed miR-126-5p in monocytes derived from healthy donors and observed a significant increase in TNF-α production when stimulated with LPS. In contrast, inhibition of miR-126-5p expression in monocytes derived from HIV-1 patients reduced the inflammatory response to LPS stimulation. These data suggested the presence of a novel posttranscriptional regulatory mechanism that modulates the LPS/TLR4 pathway in monocytes.
miRNAs generally exert their functions by negatively regulating their targets posttranscriptionally (62). Dlk1, C-X-C motif chemokine ligand 12 (CXCL12 or SDF-1α), matrix metallopeptidase 13 (MMP-13), and parathyroid hormone-related protein (PTHrP) have been identified as targets of miR-126-5p in previous reports (33, 58, 63, 64). However, none of these molecules were significantly engaged in the regulation of the responses of monocytes to LPS stimulation. In fact, we identified CYLD as the primary target of miRNA-126-5p; overexpression of miR-126-5p in monocytes resulted in CYLD downregulation and thereby allowed activation of the MAPK pathway. CYLD is a deubiquitination enzyme and regulates cell proliferation, apoptosis, cell movement, and differentiation through the adjustment of signal transduction and the cytoskeleton (65, 66). Recent studies have shown that CYLD directly regulates multiple key signaling cascade pathways, such as the NF-κB and the MAPK pathways, and acts as a negative regulator for Toll-like receptor 2 signaling via negative cross talk with TRAF6 and TRAF7 (40, 41). The inhibition of phosphodiesterase 4B (PDE4B) markedly enhances CYLD expression in response to bacteria and resulted in suppressed inflammation (44). Moreover, a very recent study by the same group showed that CYLD acts as a negative regulator of bacterium nontypeable Haemophilus influenzae (NTHi)-induced inflammation by suppressing K63-linked ubiquitination of myeloid differentiation factor 88 (MyD88) (42). Other studies have indicated that CYLD enhances severe listeriosis by impairing IL-6/signal transducer and activator of transcription 3 (STAT3)-dependent fibrin production (43) and by controlling HIV-1 transcription in an NF-κB-dependent manner (67). CYLD was recently deemed to be involved in hematopoietic stem cell quiescence and function via inhibition of the p38-MAPK pathway (68). Taking into consideration the observation that circulating monocytes in HIV-1-infected subjects exhibit a differential gene signature (69, 70), CYLD may serve as an important regulator in monocytes, as it is significantly downregulated in monocytes from HIV-1-infected subjects who are naive to ART in comparison to monocytes from healthy donors.
Similar to miR-126-5p, the suppression of CYLD expression by CYLD siRNA led to a hyperresponsiveness of monocytes to LPS stimulation. In contrast, in our study, forced CYLD expression in monocytes from HIV-1 patients attenuated the production of inflammatory cytokines with engagement of LPS/TLR4 stimulation. These results reveal a new role of CYLD in the TLR4 signaling pathway in human innate immune cells. In addition, the reduction of CYLD in monocytes from healthy donors also caused hyperresponsiveness to Pam3Csk4 (TLR2 ligand) stimulation (data not shown).
Overall, our data validate a hypothesis that CYLD is downregulated by miR-126-5p during chronic HIV-1 infection, and its reduction fuels monocyte activation/inflammation by enhancing the initial pJNK2 in primary monocytes. These data suggest that the miR-126-5p/CYLD/JNK axis is an important component of the inflammatory responses: increased expression of miR-126-5p and decreased expression of CYLD in monocytes during chronic HIV-1 infection resulted in the hypersensitivity of these cells to LPS stimulation and fuels the persistent immune activation, thereby facilitating disease progression during HIV-1 infection. The inhibition of miR-126-5p and promotion of CYLD expression in monocytes may represent a practical immune intervention strategy to contain HIV-1-facilitated disease progression. Taking into consideration the versatility of miR-126-5p in different cells types and its distinct role in different types of disease (33–38, 56, 57), the potential risks of the application of miR-126-5p in clinical therapy to inhibit inflammation in chronic HIV-1 infection should be carefully evaluated.
MATERIALS AND METHODS
Human subjects.
The basic characteristics of the studied subjects have been previously published (13, 71). All of the HIV-1 patients were infected by blood transmissions during 1995 or 1996 and were enrolled in the study and have been followed up six times per year since 2009. For this study, 15 out of 37 ART-naive (ART−) patients and 12 out of 19 ART-treated (ART+) patients were randomly selected based on the classification described in previous research (13, 14, 71). HIV-1-seronegative subjects were recruited from the Shanghai Public Health Clinical Center as healthy controls.
Ethics statement.
This study was conducted in accordance with the guidelines of the Ethics Committee of the Shanghai Public Health Clinical Center to ensure protection of the donors, and written informed consent was obtained from all human subjects.
Phenotypic analyses.
In the cell phenotyping assay, peripheral blood mononuclear cells (PBMCs) from human subjects were stained with antibodies (Abs) against CD14-allophycocyanin (APC), purchased from BioLegend (San Diego, CA), and CD16-fluorescein isothiocyanate (FITC), purchased from BD Biosciences (San Jose, CA). FlowJo software (FlowJo LLC, Ashland, OR) was used to perform flow cytometry analysis.
Cells, cell culture, and reagents.
PBMCs were isolated from human subjects using Ficoll-Hypaque (General Electric Company, Chicago, IL) density gradient centrifugation. Primary monocytes were isolated from PBMCs using the EasySep human monocyte enrichment kit without CD16 deletion (STEMCELL Technologies, Vancouver, Canada), and purity of >97% was achieved as determined by flow cytometry based on CD14 staining. The primary monocytes were further genotyped into three monocyte subgroups, CD14++ CD16−, CD14++ CD16+, and CD14+ CD16++, by flow cytometry based on surface staining with Abs against CD14-APC and CD16-FITC.
We obtained THP-1 cells (a monocytic cell line) from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Primary or THP-1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS (fetal bovine serum) (HyClone Laboratories, Logan, UT), penicillin (100 IU/ml), and streptomycin (100 μg/ml) (Life Technologies/BRL, Carlsbad, CA) at 37°C with 5% CO2. HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone Laboratories, Logan, UT) supplemented with 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml) at 37°C with 5% CO2. LPS derived from Escherichia coli O55:B5, a ligand of TLR4, was purchased from Sigma-Aldrich (St. Louis, MO) and used to stimulate monocytes. Synthetic triacylated lipoprotein (Pam3CSK4), a ligand of TLR2, was purchased from Invivogen (San Diego, CA).
RNA isolation and RT-PCR.
Total RNA was isolated from purified primary monocytes or cell lines using a Direct-zol RNA miniprep kit (Zymo Research, Orange, CA), and then 1 μg of total RNA was reverse transcribed using a reverse transcription system product with oligo(dT)15 primer (Promega, Fitchburg, WI) or a specific miRNA RT primer from the Bulge-Loop miRNA quantitative reverse transcription-PCR (qRT-PCR) primer set (RiboBio Co. Ltd., Guangzhou, China). The mRNA primers used for PCR amplification are listed in Table 1. miRNAs and mRNAs were quantified using a Mastercycler ep realplex (Eppendorf, Hamburg, Germany), and the PCR procedures were the following: 95°C for 5 min, followed by 40 cycles at 95°C for 15 s and 60°C for 30 s. A melting curve was added at the final step to ensure the specificity of the amplified products. miRNA and mRNA quantification were normalized against housekeeper controls, U6 snRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively. The relative expression of miRNAs and mRNAs was determined by the 2−ΔΔCT method according to the manufacturer's instructions (Eppendorf, Hamburg, Germany). PCR was performed in 96-well plates from Axygen (Life Technologies, Carlsbad, CA) with a total reaction volume of 20 μl per well.
TABLE 1.
Primers for qPCR
| Primer | Sequence |
|
|---|---|---|
| Forward | Reverse | |
| CYLD | 5′-TCTATGGGGTAATCCGTTGG-3′ | 5′-CAGCCTGCACACTCATCTTC-3′ |
| NLRC3 | 5′-AAACCACATTGGGGACTCCG-3′ | 5′-TTCTCCTGATCCGTCCACCA-3′ |
| SOCS1 | 5′-TGAACTCGCACCTCCTACCTCT-3′ | 5′-CAACCCCTGGTTTGTGCAA-3′ |
| TNIP1 | 5′-TGGCTGAGCTCACAGGAAAG-3′ | 5′-ACACAGGGTCTGCAGAATGG-3′ |
| GAPDH | 5′-ACGGATTTGGTCGTATTGGG-3′ | 5′-ATCTCGCTCCTGGAAGATGG-3′ |
Microarray data.
Total RNA was isolated from monocytes from five heathy controls or five HIV-1-seropositive patients in the absence of combined ART treatment. The total RNA from five heathy controls or five HIV-1 patients was then mixed as one sample and evaluated in parallel using Ion Torrent PGM (Life Technologies, Carlsbad, CA) for small RNA sequencing.
Intracellular cytokine staining.
To determine the intracellular expression of TNF-α, PBMCs from heathy controls or HIV-1-seropositive patients at a density of 5 × 105 cells/ml were seeded in 96-well plates with 200 μl of fresh culture medium per well and were then stimulated with LPS at a concentration of 100 ng/ml. After 1 h, brefeldin A (1 μg/ml) and monensin (1 μM), purchased from Sigma-Aldrich (St. Louis, MO), were added to the cell cultures. After 20 h of incubation, the cells were washed with 2% newborn calf serum (NCS) (HyClone Laboratories, Logan, UT) in PBS for one-step cleaning and then stained with Abs against surface markers of CD14 and CD16 for 15 min. Cells washed three times were harvested, fixed, and permeabilized according to the manufacturers' instructions for fixation/permeabilization solution (BD Biosciences, San Jose, CA) and then stained with anti-TNF-α–phycoerythrin (PE)–Cy7 (BD Biosciences, San Jose, CA) for 30 min. All procedures were performed at room temperature (25°C). FlowJo software (FlowJo LLC, Ashland, OR) was used to perform the flow cytometry analysis.
Cytokine measurements.
To measure the cytokines, 2 × 105 cultured THP-1 cells or 1 × 105 monocytes from healthy donors were seeded in 96-well plates with 200 μl of fresh culture medium per well and then treated with fresh culture medium or LPS at 100 ng/ml. Monocytes from HIV-1 patients at a density of 5 × 105 cells/ml were seeded in 96-well plates with 100 μl of fresh culture medium per well and then treated with fresh culture medium or LPS at 100 ng/ml. The cell supernatant was collected upon stimulation and stored at −80°C until the levels of the inflammatory cytokines (IL-1β, IL-6, IL-8, IL-10, IL-12, MCP-1, and TNF-α) in the supernatant were measured simultaneously using a BioLegend LEGENDplex (BioLegend, San Diego, CA) or separately by enzyme-linked immunosorbent assay (ELISA; BD Biosciences, San Jose, CA).
Construction of the Dual-Luciferase reporter system.
The 3′-untranslated region (3′UTR) of CYLD containing the miR-126-5p binding sites was first amplified by PCR from total RNA extracted from THP-1 cells and then cloned into the pMIR-REPORT vector (Applied Biosystems, Carlsbad, CA). We used restriction enzyme cloning sites (underlined) for the restriction enzymes SpeI and HindIII. The primers for PCR amplification were the following: CYLD 3′UTR forward primer, 5′-GACTAGTCAAGGATATGAAATCATTTT; CYLD 3′UTR reverse primer, 5′-CCCAAGCTTAACACCATTAAGGGAATTTA; SOCS1 3′UTR forward primer, 5′-GACTAGTCCGGCAGCGCCCGCCGTGCA; SOCS1 3′UTR reverse primer, 5′-CCCAAGCTTAAAAAAAAAACTTTCATAAT.
Oligonucleotide and plasmid transfection.
MicrON miR-126-5p mimic, miR-126-5p mutant mimic containing three altered nucleotides in the seed sequence of miR-126-5p, and micrON miRNA mimic negative controls (control mimic, with minimal homology to all known miRNAs) were used for the miRNA gain-of-function studies in cell lines and primary cells; micrOFF miR-126-5p inhibitor and micrOFF miRNA inhibitor negative controls (control inhibitor) were used for the miRNA loss-of-function studies in cell lines. The micrOFF miR-126-5p antagomir, a chemically modified miR-126-5p inhibitor, and micrOFF miRNA antagomir control (control antagomir) were used for miRNA loss-of-function studies in primary monocytes. Scrambled small interfering RNA (control siRNA) and CYLD-specific siRNA were used for transient silencing of the expression of CYLD. All oligonucleotides were synthesized by RiboBio (RiboBio Co. Ltd. Guangzhou, China) and delivered to 1.5 × 106 to ∼3 × 106 THP-1 cells or primary monocytes at a final concentration of 1.5 × 106/200 pmol oligonucleotides using the Amaxa Cell Line Nucleofector kit or the Amaxa Human Monocyte Nucleofector kit (Lonza, Basel, Switzerland) according to the directions supplied and previous studies (13, 31, 45, 72, 73). The cells were cultured in a 24-well plate for 48 h or 72 h with 2 ml of fresh culture medium (RPMI 1640 medium supplemented with 10% fetal bovine serum) per well and harvested, and the transfection efficiency was confirmed by qPCR or Western blotting.
To confirm the presence of miR-126-5p targeting sequences in the 3′UTR of the candidate gene, HEK293T cells were obtained from the Cell Bank of the Chinese Academy of Sciences, and 1 × 104 cells/well were seeded into a 96-well plate 1 day before transfection. The cells were then cotransfected with the CYLD 3′UTR reporter, a control plasmid, pRL-CMV (Promega, Fitchburg, WI), and a final concentration of 100 nM miR-126-5p mimic, miR-126-5p-mutant mimic, control mimic, control inhibitor, or miR-126-5p inhibitor using TurboFect in vitro transfection reagent (Thermo Fisher Scientific, Waltham, MA). Luciferase activity was measured 48 h posttransfection using a Dual-Luciferase reporter assay system (Promega, Fitchburg, WI) according to the manufacturer's instructions.
Primary cells (1.5 × 106) from HIV-1 patients were electrotransfected for 72 h with 2.5 μg of endotoxin-free pCDNA3.1 or pCDNA3.1-CYLD (codon optimization), constructed by Genewiz (Suzhou, China). The experimental procedures were optimized according to previous studies (28, 45, 74, 75).
Lentiviral transduction.
Lentiviruses were produced by cotransfecting 293T cells in a 6-well plate with, per well, 1 μg of pVSV-G (envelop-encoding plasmid), 4 μg of psPAX2 (packing plasmid), and 4 μg of TRC2-pLKO-puro constructs containing short hairpin RNA directed against human CYLD (lentiviral vectors), all purchased from Sigma-Aldrich (St. Louis, MO), in a 2-ml volume using TurboFect transfection reagent (Thermo Fisher Scientific, Waltham, MA). DMEM was refreshed 6 h after transfection, and the lentivirus was harvested 48 h later, filtered through a 0.45-μm filter, and stored at −80°C until transduction. THP-1 cells were transduced with vesicular stomatitis virus glycoprotein-pseudotyped lentivirus to generate CYLD-reduced THP-1-shCYLD cells and THP-1-shCtrl cells. The efficiency of transfection was confirmed by Western blotting every 4 days until stable silencing of the expression of CYLD.
Immunoblot analysis.
After transfection with miRNA mimic or siRNA, 1 × 106 to ∼1.5 × 106 cells were washed, harvested in 100 μl of PBS, and lysed in 20 μl of 6× loading buffer (TransGen Biotech, Beijing, China). Equal amounts of transfected cell lysates were loaded onto a 10% SDS-PAGE gel, separated at a constant voltage, and transferred electrophoretically to polyvinylidene difluoride (PVDF) membranes at a constant current. PVDF membranes were blocked, incubated with primary Abs, and further incubated with horseradish peroxidase (HRP)-conjugated secondary Abs. The Abs used in the current study were the following: rabbit anti-CYLD polyclonal Ab, rabbit anti-SAPK/JNK monoclonal Ab (MAb), and rabbit anti-phosphor-SAPK/JNK MAb, which were purchased from Cell Signaling Technology (Danvers, MA), as well as mouse anti-β-actin MAb, HRP-conjugated goat anti-mouse, and HRP-conjugated donkey anti-rabbit secondary Abs, which were from Santa Cruz Biotechnology (Santa Cruz, CA).
Statistical analyses.
The data obtained from the different treatments in the same sample were evaluated using the two-tailed Student's t test, and all data were expressed as the means ± standard deviations. Correlations were tested with the Spearman rank correlation test. Differences between healthy controls and HIV-1-seropositive patients or cellular subsets were determined with a Mann-Whitney nonparametric test. All data were processed using GraphPad Prism, version 5.0 (GraphPad Software, San Diego, CA). A P value of <0.05 was defined as statistically significant.
ACKNOWLEDGMENTS
We thank all of the donors suffering from HIV-1 infection from Shanxi and Anhui for their blood donations. We thank Anli Zhang, Weihui Fu, Jingjing Yan, Yuan Dong, and Yongquan He for processing the samples and performing pretreatment. We thank Chenli Qiu and Jun Sun for their contribution to the flow cytometry assay.
We also thank the following people for their assistance in coordinating the patients for sample collection: Ximing Shi, Jinsheng Wang, Jun Wei, Shaoxian Jia, Shaoping Ning, and Xiujie Lu at the Yuncheng Municipal Center for Disease Control and Prevention; Wangqian Jia, Shaoling Dong, Wei Zhao, Qinghai Yang, Xiaoli Zhao, Lingbo Wang, Hui Wang, and Ailing Zhong at the Wenxi County Center for Disease Control and Prevention; Baoxing Gao, Xiuzhen Li, Xia Zhang, Weina Feng, Lingling Qi, Bin Wei, Junmiao Zhu, and Qifeng Xu at the Xia County Center for Disease Control and Prevention; and Jun Ji, Xia Zhang, Tingting Ma, Linna Li, Yanxia Li, and Qinghua Li at the Jiang County Center for Disease Control and Prevention, Shanxi, China.
This work was supported by Chinese Ministry of Health and Chinese Ministry of Science and Technology National Grand Program on Key Infectious Disease Control grant 2013ZX10001-002, Chinese Ministry of Science and Technology 973 National Key Research Project on Basic Sciences grant 2014CB542502, and National Natural Science Foundation of China grant 81561128008.
REFERENCES
- 1.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 12:1365–1371. [DOI] [PubMed] [Google Scholar]
- 2.Brenchley JM, Douek DC. 2008. HIV infection and the gastrointestinal immune system. Mucosal Immunol 1:23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Neuhaus J, Nixon D, Paton NI, Neaton JD, INSIGHT SMART Study Group. 2008. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med 5:e203. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redd AD, Dabitao D, Bream JH, Charvat B, Laeyendecker O, Kiwanuka N, Lutalo T, Kigozi G, Tobian AA, Gamiel J, Neal JD, Oliver AE, Margolick JB, Sewankambo N, Reynolds SJ, Wawer MJ, Serwadda D, Gray RH, Quinn TC. 2009. Microbial translocation, the innate cytokine response, and HIV-1 disease progression in Africa. Proc Natl Acad Sci U S A 106:6718–6723. doi: 10.1073/pnas.0901983106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zevin AS, McKinnon L, Burgener A, Klatt NR. 2016. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS 11:182–190. doi: 10.1097/COH.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ancuta P, Kamat A, Kunstman KJ, Kim EY, Autissier P, Wurcel A, Zaman T, Stone D, Mefford M, Morgello S, Singer EJ, Wolinsky SM, Gabuzda D. 2008. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS One 3:e2516. doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutertre CA, Amraoui S, DeRosa A, Jourdain JP, Vimeux L, Goguet M, Degrelle S, Feuillet V, Liovat AS, Muller-Trutwin M, Decroix N, Deveau C, Meyer L, Goujard C, Loulergue P, Launay O, Richard Y, Hosmalin A. 2012. Pivotal role of M-DC8(+) monocytes from viremic HIV-infected patients in TNFalpha overproduction in response to microbial products. Blood 120:2259–2268. doi: 10.1182/blood-2012-03-418681. [DOI] [PubMed] [Google Scholar]
- 8.Funderburg NT, Zidar DA, Shive C, Lioi A, Mudd J, Musselwhite LW, Simon DI, Costa MA, Rodriguez B, Sieg SF, Lederman MM. 2012. Shared monocyte subset phenotypes in HIV-1 infection and in uninfected subjects with acute coronary syndrome. Blood 120:4599–4608. doi: 10.1182/blood-2012-05-433946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manuzak JA, Dillon SM, Lee EJ, Dong ZM, Hecht DK, Wilson CC. 2013. Increased Escherichia coli-induced interleukin-23 production by CD16+ monocytes correlates with systemic immune activation in untreated HIV-1-infected individuals. J Virol 87:13252–13262. doi: 10.1128/JVI.01767-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons RP, Scully EP, Groden EE, Arnold KB, Chang JJ, Lane K, Lifson J, Rosenberg E, Lauffenburger DA, Altfeld M. 2013. HIV-1 infection induces strong production of IP-10 through TLR7/9-dependent pathways. AIDS 27:2505–2517. doi: 10.1097/01.aids.0000432455.06476.bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell JH, Hearps AC, Martin GE, Williams KC, Crowe SM. 2014. The importance of monocytes and macrophages in HIV pathogenesis, treatment, and cure. AIDS 28:2175–2187. doi: 10.1097/QAD.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgener A, McGowan I, Klatt NR. 2015. HIV and mucosal barrier interactions: consequences for transmission and pathogenesis. Curr Opin Immunol 36:22–30. doi: 10.1016/j.coi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Xu X, Qiu C, Zhu L, Huang J, Li L, Fu W, Zhang L, Wei J, Wang Y, Geng Y, Zhang X, Qiao W, Xu J. 2014. IFN-stimulated gene LY6E in monocytes regulates the CD14/TLR4 pathway but inadequately restrains the hyperactivation of monocytes during chronic HIV-1 infection. J Immunol 193:4125–4136. doi: 10.4049/jimmunol.1401249. [DOI] [PubMed] [Google Scholar]
- 14.Chang JJ, Lacas A, Lindsay RJ, Doyle EH, Axten KL, Pereyra F, Rosenberg ES, Walker BD, Allen TM, Altfeld M. 2012. Differential regulation of toll-like receptor pathways in acute and chronic HIV-1 infection. AIDS 26:533–541. doi: 10.1097/QAD.0b013e32834f3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mureith MW, Chang JJ, Lifson JD, Ndung'u T, Altfeld M. 2010. Exposure to HIV-1-encoded Toll-like receptor 8 ligands enhances monocyte response to microbial encoded Toll-like receptor 2/4 ligands. AIDS 24:1841–1848. doi: 10.1097/QAD.0b013e32833ad89a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez JC, Stevenson M, Latz E, Urcuqui-Inchima S. 2012. HIV type 1 infection up-regulates TLR2 and TLR4 expression and function in vivo and in vitro. AIDS Res Hum Retrovir 28:1313–1328. doi: 10.1089/aid.2011.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegler-Heitbrock L. 2007. The CD14+ CD16+ blood monocytes: their role in infection and inflammation. J Leukoc Biol 81:584–592. [DOI] [PubMed] [Google Scholar]
- 18.Rodger AJ, Fox Z, Lundgren JD, Kuller LH, Boesecke C, Gey D, Skoutelis A, Goetz MB, Phillips AN, INSIGHT Strategies for Management of Antiretroviral Therapy (SMART) Study Group. 2009. Activation and coagulation biomarkers are independent predictors of the development of opportunistic disease in patients with HIV infection. J Infect Dis 200:973–983. doi: 10.1086/605447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, Pedersen C, Ruxrungtham K, Lewin SR, Emery S, Neaton JD, Brenchley JM, Deeks SG, Sereti I, Douek DC, INSIGHT SMART Study Group. 2011. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis 203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan S, Wilson EM, Sheikh V, Rupert A, Mendoza D, Yang J, Lempicki R, Migueles SA, Sereti I. 2014. Evidence for innate immune system activation in HIV type 1-infected elite controllers. J Infect Dis 209:931–939. doi: 10.1093/infdis/jit581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson EM, Singh A, Hullsiek KH, Gibson D, Henry WK, Lichtenstein K, Onen NF, Kojic E, Patel P, Brooks JT, Sereti I, Baker JV, Study to Understand the Natural History of HIV/AIDS in the Era of Effective Therapy (SUN Study) Investigators. 2014. Monocyte-activation phenotypes are associated with biomarkers of inflammation and coagulation in chronic HIV infection. J Infect Dis 210:1396–1406. doi: 10.1093/infdis/jiu275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Provost P, Barat C, Plante I, Tremblay MJ. 2006. HIV-l and the microRNA-guided silencing pathway: an intricate and multifaceted encounter. Virus Res 121:107–115. doi: 10.1016/j.virusres.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen HS, Daher A, Soye KJ, Frankel LB, Alexander MR, Laine S, Bannwarth S, Ong CL, Chung SW, Campbell SM, Purcell DF, Gatignol A. 2007. Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production. J Virol 81:5121–5131. doi: 10.1128/JVI.01511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung TL, Rice AP. 2009. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog 5:e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, Ho WZ. 2009. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood 113:671–674. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen CJ, Jia YH, Tian RR, Ding M, Zhang C, Wang JH. 2012. Translation of Pur-alpha is targeted by cellular miRNAs to modulate the differentiation-dependent susceptibility of monocytes to HIV-1 infection. FASEB J 26:4755–4764. doi: 10.1096/fj.12-209023. [DOI] [PubMed] [Google Scholar]
- 29.Swaminathan G, Rossi F, Sierra LJ, Gupta A, Navas-Martin S, Martin-Garcia J. 2012. A role for microRNA-155 modulation in the anti-HIV-1 effects of Toll-like receptor 3 stimulation in macrophages. PLoS Pathog 8:e1002937. doi: 10.1371/journal.ppat.1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y, O'Neill LA. 2010. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol 11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 31.Taganov KD, Boldin MP, Chang KJ, Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. 2007. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, Li C, Chong M, Ibrahim T, Mercatali L, Amadori D, Lu X, Xie D, Li QJ, Wang XF. 2013. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol 15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. 2008. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A 105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oglesby IK, Bray IM, Chotirmall SH, Stallings RL, O'Neill SJ, McElvaney NG, Greene CM. 2010. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J Immunol 184:1702–1709. doi: 10.4049/jimmunol.0902669. [DOI] [PubMed] [Google Scholar]
- 36.van Solingen C, Seghers L, Bijkerk R, Duijs JM, Roeten MK, van Oeveren-Rietdijk AM, Baelde HJ, Monge M, Vos JB, de Boer HC, Quax PH, Rabelink TJ, van Zonneveld AJ. 2009. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J Cell Mol Med 13:1577–1585. doi: 10.1111/j.1582-4934.2008.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. 2008. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. 2008. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. 2008. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development 135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 40.Reiley W, Zhang M, Sun SC. 2004. Negative regulation of JNK signaling by the tumor suppressor CYLD. J Biol Chem 279:55161–55167. doi: 10.1074/jbc.M411049200. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida H, Jono H, Kai H, Li JD. 2005. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 and TRAF7. J Biol Chem 280:41111–41121. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- 42.Lee BC, Miyata M, Lim JH, Li JD. 2016. Deubiquitinase CYLD acts as a negative regulator for bacterium NTHi-induced inflammation by suppressing K63-linked ubiquitination of MyD88. Proc Natl Acad Sci U S A 113:E165–E171. doi: 10.1073/pnas.1518615113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishanth G, Deckert M, Wex K, Massoumi R, Schweitzer K, Naumann M, Schluter D. 2013. CYLD enhances severe listeriosis by impairing IL-6/STAT3-dependent fibrin production. PLoS Pathog 9:e1003455. doi: 10.1371/journal.ppat.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komatsu K, Lee JY, Miyata M, Hyang Lim J, Jono H, Koga T, Xu H, Yan C, Kai H, Li JD. 2013. Inhibition of PDE4B suppresses inflammation by increasing expression of the deubiquitinase CYLD. Nat Commun 4:1684. doi: 10.1038/ncomms2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. 2009. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A 106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallo RC. 1988. HIV–the cause of AIDS: an overview on its biology, mechanisms of disease induction, and our attempts to control it. J Acquired Immune Defic Syndr 1:521–535. [PubMed] [Google Scholar]
- 47.Marchetti G, Bellistri GM, Borghi E, Tincati C, Ferramosca S, La Francesca M, Morace G, Gori A, Monforte AD. 2008. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS 22:2035–2038. doi: 10.1097/QAD.0b013e3283112d29. [DOI] [PubMed] [Google Scholar]
- 48.Sandler NG, Douek DC. 2012. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol 10:655–666. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- 49.Han J, Wang B, Han N, Zhao Y, Song C, Feng X, Mao Y, Zhang F, Zhao H, Zeng H. 2009. CD14(high)CD16(+) rather than CD14(low)CD16(+) monocytes correlate with disease progression in chronic HIV-infected patients. J Acquired Immune Defic Syndr 52:553–559. doi: 10.1097/QAI.0b013e3181c1d4fe. [DOI] [PubMed] [Google Scholar]
- 50.Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. 2007. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem 282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- 51.Bennasser Y, Jeang KT. 2006. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology 3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennasser Y, Yeung ML, Jeang KT. 2006. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J Biol Chem 281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- 53.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 54.Seddiki N, Phetsouphanh C, Swaminathan S, Xu Y, Rao S, Li J, Sutcliffe EL, Denyer G, Finlayson R, Gelgor L, Cooper DA, Zaunders J, Kelleher AD. 2013. The microRNA-9/B-lymphocyte-induced maturation protein-1/IL-2 axis is differentially regulated in progressive HIV infection. Eur J Immunol 43:510–520. doi: 10.1002/eji.201242695. [DOI] [PubMed] [Google Scholar]
- 55.Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, Clancy JL, Murray DD, Mendez C, Gelgor L, Anderson B, Roth N, Cooper DA, Kelleher AD. 2012. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol 188:6238–6246. doi: 10.4049/jimmunol.1101196. [DOI] [PubMed] [Google Scholar]
- 56.Li Z, Chen P, Su R, Li Y, Hu C, Wang Y, Arnovitz S, He M, Gurbuxani S, Zuo Z, Elkahloun AG, Li S, Weng H, Huang H, Neilly MB, Wang S, Olson EN, Larson RA, Le Beau MM, Zhang J, Jiang X, Wei M, Jin J, Liu PP, Chen J. 2015. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood 126:2005–2015. doi: 10.1182/blood-2015-04-639062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, Chen J. 2011. In vitro functional study of miR-126 in leukemia. Methods Mol Biol 676:185–195. doi: 10.1007/978-1-60761-863-8_13. [DOI] [PubMed] [Google Scholar]
- 58.Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C. 2014. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med 20:368–376. doi: 10.1038/nm.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agudo J, Ruzo A, Tung N, Salmon H, Leboeuf M, Hashimoto D, Becker C, Garrett-Sinha LA, Baccarini A, Merad M, Brown BD. 2014. The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids. Nat Immunol 15:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferretti C, La Cava A. 2014. miR-126, a new modulator of innate immunity. Cell Mol Immunol 11:215–217. doi: 10.1038/cmi.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mattes J, Collison A, Plank M, Phipps S, Foster PS. 2009. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A 106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou W, Yin H, Wang T, Liu T, Li Z, Yan W, Song D, Chen H, Chen J, Xu W, Yang X, Wu Z, Xiao J. 2014. miR-126-5p regulates osteolysis formation and stromal cell proliferation in giant cell tumor through inhibition of PTHrP. Bone 66:267–276. doi: 10.1016/j.bone.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 64.Wu Z, Yin H, Liu T, Yan W, Li Z, Chen J, Chen H, Wang T, Jiang Z, Zhou W, Xiao J. 2014. miR-126-5p regulates osteoclast differentiation and bone resorption in giant cell tumor through inhibition of MMP-13. Biochem Biophys Res Commun 443:944–949. doi: 10.1016/j.bbrc.2013.12.075. [DOI] [PubMed] [Google Scholar]
- 65.Reiley WW, Zhang M, Jin W, Losiewicz M, Donohue KB, Norbury CC, Sun SC. 2006. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol 7:411–417. doi: 10.1038/ni1315. [DOI] [PubMed] [Google Scholar]
- 66.Gao J, Sun L, Huo L, Liu M, Li D, Zhou J. 2010. CYLD regulates angiogenesis by mediating vascular endothelial cell migration. Blood 115:4130–4137. doi: 10.1182/blood-2009-10-248526. [DOI] [PubMed] [Google Scholar]
- 67.Manganaro L, Pache L, Herrmann T, Marlett J, Hwang Y, Murry J, Miorin L, Ting AT, Konig R, Garcia-Sastre A, Bushman FD, Chanda SK, Young JAT, Fernandez-Sesma A, Simon V. 2014. Tumor suppressor cylindromatosis (CYLD) controls HIV transcription in an NF-B-dependent manner. J Virol 88:7528–7540. doi: 10.1128/JVI.00239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tesio M, Tang Y, Mudder K, Saini M, von Paleske L, Macintyre E, Pasparakis M, Waisman A, Trumpp A. 2015. Hematopoietic stem cell quiescence and function are controlled by the CYLD-TRAF2-p38MAPK pathway. J Exp Med 212:525–538. doi: 10.1084/jem.20141438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giri MS, Nebozyhn M, Raymond A, Gekonge B, Hancock A, Creer S, Nicols C, Yousef M, Foulkes AS, Mounzer K, Shull J, Silvestri G, Kostman J, Collman RG, Showe L, Montaner LJ. 2009. Circulating monocytes in HIV-1-infected viremic subjects exhibit an antiapoptosis gene signature and virus- and host-mediated apoptosis resistance. J Immunol 182:4459–4470. doi: 10.4049/jimmunol.0801450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. 2010. Interferon-alpha drives monocyte gene expression in chronic unsuppressed HIV-1 infection. AIDS 24:1415–1423. doi: 10.1097/QAD.0b013e32833ac623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian X, Zhang A, Qiu C, Wang W, Yang Y, Qiu C, Liu A, Zhu L, Yuan S, Hu H, Wang W, Wei Q, Zhang X, Xu J. 2015. The upregulation of LAG-3 on T cells defines a subpopulation with functional exhaustion and correlates with disease progression in HIV-infected subjects. J Immunol 194:3873–3882. doi: 10.4049/jimmunol.1402176. [DOI] [PubMed] [Google Scholar]
- 72.Rossato M, Curtale G, Tamassia N, Castellucci M, Mori L, Gasperini S, Mariotti B, De Luca M, Mirolo M, Cassatella MA, Locati M, Bazzoni F. 2012. IL-10-induced microRNA-187 negatively regulates TNF-alpha, IL-6, and IL-12p40 production in TLR4-stimulated monocytes. Proc Natl Acad Sci U S A 109:E3101–E3110. doi: 10.1073/pnas.1209100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. 2013. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci U S A 110:11499–11504. doi: 10.1073/pnas.1219852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martinez-Nunez RT, Louafi F, Friedmann PS, Sanchez-Elsner T. 2009. MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN). J Biol Chem 284:16334–16342. doi: 10.1074/jbc.M109.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Z, Kim SJ, Chamberlain ND, Pickens SR, Volin MV, Volkov S, Arami S, Christman JW, Prabhakar BS, Swedler W, Mehta A, Sweiss N, Shahrara S. 2013. The novel role of IL-7 ligation to IL-7 receptor in myeloid cells of rheumatoid arthritis and collagen-induced arthritis. J Immunol 190:5256–5266. doi: 10.4049/jimmunol.1201675. [DOI] [PMC free article] [PubMed] [Google Scholar]