ABSTRACT
MicroRNAs (miRNAs) are a class of small, single-stranded, noncoding, functional RNAs. Hepatitis B virus (HBV) is an enveloped DNA virus with virions and subviral forms of particles that lack a core. It was not known whether HBV encodes miRNAs. Here, we identified an HBV-encoded miRNA (called HBV-miR-3) by deep sequencing and Northern blotting. HBV-miR-3 is located at nucleotides (nt) 373 to 393 of the HBV genome and was generated from 3.5-kb, 2.4-kb, and 2.1-kb HBV in a classic miRNA biogenesis (Drosha-Dicer-dependent) manner. HBV-miR-3 was highly expressed in hepatoma cell lines with an integrated HBV genome and HBV+ hepatoma tumors. In patients with HBV infection, HBV-miR-3 was released into the circulation by exosomes and HBV virions, and HBV-miR-3 expression had a positive correlation with HBV titers in the sera of patients in the acute phase of HBV infection. More interestingly, we found that HBV-miR-3 represses HBsAg, HBeAg, and replication of HBV. HBV-miR-3 targets the unique site of the HBV 3.5-kb transcript to specifically reduce HBc protein expression, levels of pregenomic RNA (pgRNA), and HBV replication intermediate (HBV-RI) generation but does not affect the HBV DNA polymerase level, thus suppressing HBV virion production (replication). This may explain the low levels of HBV virion generation with abundant subviral particles lacking core during HBV replication, which may contribute to the development of persistent infection in patients. Taken together, our findings shed light on novel mechanisms by which HBV-encoded miRNA controls the process of self-replication by regulating HBV transcript during infection.
IMPORTANCE Hepatitis B is a liver infection caused by the hepatitis B virus (HBV) that can become a long-term, chronic infection and lead to cirrhosis or liver cancer. HBV is a small DNA virus that belongs to the hepadnavirus family, with virions and subviral forms of particles that lack a core. MicroRNA (miRNA), a small (∼22-nt) noncoding RNA, was recently found to be an important regulator of gene expression. We found that HBV encodes miRNA (HBV-miR-3). More importantly, we revealed that HBV-miR-3 targets its transcripts to attenuate HBV replication. This may contribute to explaining how HBV infection leads to mild damage in liver cells and the subsequent establishment/maintenance of persistent infection. Our findings highlight a mechanism by which HBV-encoded miRNA controls the process of self-replication by regulating the virus itself during infection and might provide new biomarkers for diagnosis and treatment of hepatitis B.
KEYWORDS: miRNA, hepatitis B virus, pgRNA, HBc
INTRODUCTION
MicroRNAs (miRNAs) represent a class of small RNAs ranging from 21 to 24 nucleotides (nt) that regulate complementary mRNAs by inducing translational repression or mRNA decay (1) or upregulating the expression of the target gene in a G-rich RNA sequence binding factor 1 (GRSF1)-dependent manner (53). miRNAs have been identified in all multicellular eukaryotes, and some are evolutionarily conserved among insects, nematodes, and humans. Human miRNAs play important roles in diverse cellular processes, including virus-host interactions (2–5). Like their hosts, many human and animal viruses have been identified as encoding miRNAs. The first report of virus-encoded miRNA was of Epstein-Barr virus-encoded miRNA (6). Subsequently, it was found that miRNAs were encoded by simian virus 40 (SV40) (7), herpes simplex virus (HSV) (8), human cytomegalovirus (HCMV) (9), Kaposi's sarcoma-associated herpesvirus (KSHV) (10), Marek's disease virus (MDV) (11), and retrovirus families (12–14). Although the functions of the majority of viral miRNAs have not been elucidated, emerging studies have demonstrated that some viral miRNAs regulate the viral life cycle itself, including the maintenance/establishment of persistent infection and productive infection (15). Therefore, viral miRNAs may profoundly affect the natural progression of infection of some viruses in vivo.
Hepatitis B virus (HBV) infection is a key risk factor for chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC) worldwide. Currently, HBV has infected an estimated 2 billion people, including 350 million people who are chronically infected with HBV worldwide (16). Compared with uninfected individuals, HBV-infected patients have an approximately 100-fold-increased risk for HCC (17). Additionally, because a cure for the virus is lacking, it is crucial to explore HBV pathogenesis. HBV is a small DNA virus that belongs to the hepadnavirus family. The genomic material of HBV is circular, partially double-stranded DNA that is only 3.2 kb in size and is transcribed into four transcripts, i.e., 3.5 kb, 2.4 kb, 2.1 kb, and 0.7 kb. There are four overlapping open reading frames (ORFs): S, P, C, and X, which encode hepatitis B viral surface antigen (HBsAg), DNA polymerase (HBV DNA Pol), core protein (HBcAg) and HBeAg, and HBV X (HBx) proteins, respectively. A subset of the 3.5-kb transcript (pregenomic RNA [pgRNA]) is packaged into core particles with DNA polymerase and serves as the template for synthesis of DNA by reverse transcription, which is then enveloped and released from the cell as progeny virions (18–21).
HBV utilizes viral factors for its successful replication and the subsequent establishment/maintenance of infection, especially for persistent infections. Many cellular miRNAs have been reported to modulate HBV replication by direct interaction with HBV transcripts to influence the HBV life cycle (22). Among them, our laboratory first demonstrated that human miR-199a-3p and miR-210 efficiently reduced HBsAg expression and HBV replication by directly targeting the HBsAg coding region and the pre-S1 region of the HBV genome, respectively (23). Meanwhile, HBV replication is also regulated by its own proteins. HBx is a modest transcriptional activator in regulating HBV's own replication (24). The HBc protein could positively regulate HBV transcription by binding to HBV covalently closed circular DNA (cccDNA) or by stimulating HBV enhancer activities (25, 26). However, experimental evidence for HBV-encoded miRNA is lacking.
In the present study, we obtained 5 novel small RNAs which matched the HBV genome by deep sequencing of HBV-positive (HBV+) HCC tissue. Here, we report that HBV encodes HBV-miR-3, as shown by deep sequencing in HBV+ hepatoma tissue, and we also confirmed its expression. HBV-miR-3 repressed HBV protein and HBV replication. We discovered that HBV-miR-3 targeted the HBV 3.5-kb transcript to reduce HBc and pgRNA levels, which resulted in attenuation of HBV replication.
RESULTS
Identification of HBV small noncoding RNAs by deep sequencing.
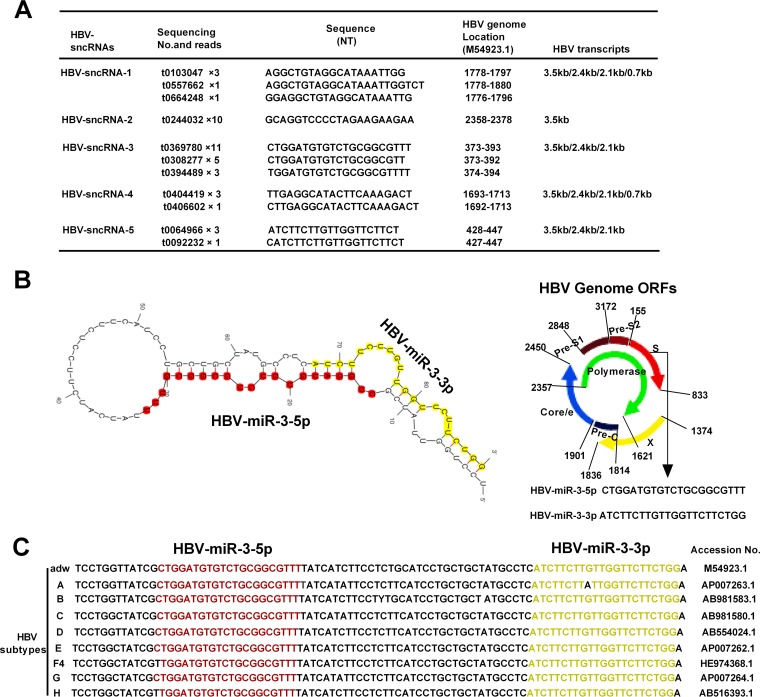
To identify small RNAs originating from HBV, we utilized deep sequencing to profile small RNAs isolated from HBV+ HCC tissues. Small RNAs isolated from HBV+ HCC tissues and HBV-negative (HBV−) HCC tissues were utilized as controls for sequencing. We obtained some small RNAs that were size selected for small (approximately 22-bp) transcripts. The sequences, nucleotides, and reads obtained from deep sequencing are shown in Fig. 1A. Five small RNAs were identified as homologues with HBV transcripts and were located in different regions of the HBV genome; they were designated HBV small noncoding RNAs (HBV-sncRNAs). By predicting the architecture and secondary structure of these RNAs, combined with 19 reads that were found to correspond to HBV-sncRNA-3, we found that the precursor sequence of HBV-sncRNA-3 and HBV-sncRNA-5 could form the classic stem-loop structure of miRNA, as predicted by the Mfold Web Server (27) (Fig. 1B). Mature HBV-sncRNA-3 and HBV-sncRNA-5 are located in the 5′-end stem and 3′-end stem of the predicted precursor, respectively. Therefore, we named HBV-sncRNA-3 HBV-miR-3-5p (referred to below as HBV-miR-3) and named HBV-sncRNA-5 HBV-miR-3-3p. HBV-miR-3 was in the HBsAg coding region of the HBV genome and overlapped the 2.1-kb, 2.4-kb, and 3.5-kb HBV transcripts (Fig. 1A). Alignment analysis revealed that HBV-miR-3 is highly conserved among different HBV subtypes (Fig. 1C). These results indicated that the HBV genome encodes miRNAs and that HBV-miR-3 is located in the highly conserved HBsAg coding regions of various HBV genotypes.
FIG 1.
Identification of HBV small noncoding RNAs by deep sequencing. (A) The primary sequence and reads of five HBV-sncRNAs matched the HBV genome at the expected location. (B) Secondary-structure prediction of HBV-miR-3 and schematic diagram of the HBV-miR-3 location on the HBV genome. (C) Conserved analysis of HBV-miR-3 among different HBV subtypes.
Experimental validation of HBV-miR-3.
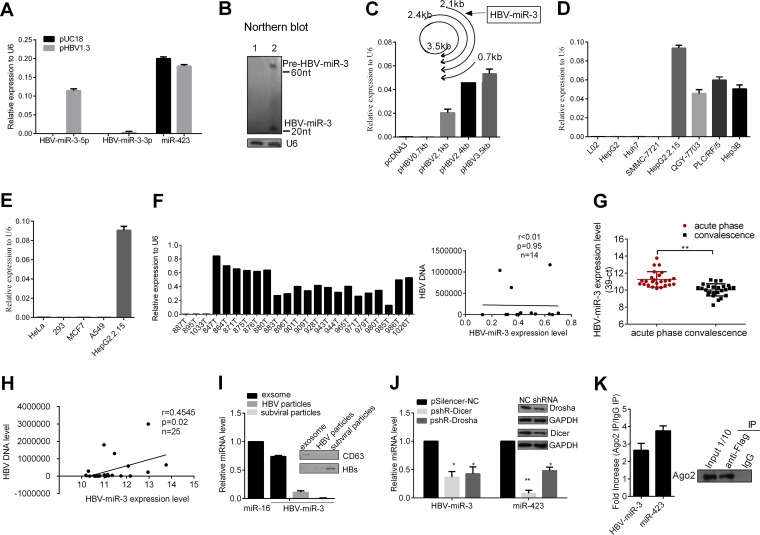
To validate HBV-miR-3 expression, we first examined its expression in HBV− Huh7 cells transiently transfected with HBV replication-competent plasmids (pHBV1.3) by stem-loop reverse transcription quantitative PCR (RT-qPCR). miR-423 was set as a negative control and was not affected by HBV infection (28). As shown in Fig. 2A, transfection of pHBV1.3 resulted in high expression of HBV-miR-3, while expression of HBV-miR-3-3p was low and the expression of miR-423 was not affected. In addition, Northern blotting also showed that small RNAs corresponding to both the pre-miRNA (60 nt) and the mature HBV-miR-3 (20 nt) were observed in Huh7 cells after transfection 72 h later but not in Huh7 cells transfected with the control vector, pUC18 (Fig. 2B). Furthermore, to examine which HBV transcripts might be processed into HBV-miR-3, we generated expression plasmids for the transcripts (pHBV3.5 kb, pHBV2.4 kb, pHBV2.1 kb, and pHBV0.7 kb), with pcDNA3 as a negative control. A stem-loop RT-qPCR assay showed that HBV-miR-3 was highly expressed in Huh7 cells transfected with pHBV 3.5 kb, pHBV 2.4 kb, and pHBV 2.1 kb, but not pHBV 0.7 kb (Fig. 2C), which indicated that HBV-miR-3 may be processed from all the transcripts overlapping the HBV-miR-3 coding region.
FIG 2.
Experimental validation of HBV-miR-3. (A and B) Huh7 cells were transfected with pHBV1.3 or an empty vector, pUC18, as a negative control. (A) Mature HBV-miR-3, HBV-miR-3-3p, and miR-423 were detected with stem-loop-RT-qPCR. (B) HBV-miR-3 was examined by Northern blotting. The probe was the biotin-conjugated antisense strand of HBV-miR-3. Lane 1, 45 μg of small RNA from Huh7 cells transfected with pUC18; lane 2, 45 μg of small RNA from Huh7 cells transfected with pHBV1.3. U6 was used as a control. (C) Stem-loop RT-qPCR was conducted on total RNAs isolated from Huh7 cells transfected with HBV transcripts: pHBV3.5 kb, pHBV2.4 kb, pHBV2.1 kb, or pHBV0.7 kb with primers for HBV-miR-3; the empty vector pcDNA3 was used as a control. (D) Cellular RNA was extracted and analyzed for HBV-miR-3 expression among hepatoma cell lines with a detectable integrated HBV genome (HepG2.2.15, 7703, PLC/RF/5, and Hep3B cells), HBV-negative hepatoma cells (7721, HepG2, and Huh7 cells), and L02 cells. miR-423 was set as a negative control, while U6 RNA expression was set as 1 unit as the internal control. (E) Cellular RNA was extracted, and HBV-miR-3 expression was detected among non-hepatoma cell lines. (F) (Left) RNA was extracted and analyzed for the expression of HBV-miR-3 in 20 specimens with HBV+ hepatoma tumors and 3 specimens with HBV− hepatoma tumors, (Right) Pearson's correlation analysis indicated a positive correlation between the expression of HBV-miR-3 and HBV DNA in HBV+ hepatoma tumors. (G) The HBV-miR-3 level was measured by RT-qPCR in the sera of 25 pairs of patients with acute-phase HBV infection that progressed to convalescence. The miRNA expression levels are presented as 39-Ct after normalization to miR-16. (H) Pearson's correlation analysis indicated positive correlation between the expression of HBV-miR-3 and HBV DNA in sera of patients with HBV infection in the acute phase. (I) RT-qPCR was performed to detect the expression of HBV-miR-3 in purified HBV particles and exosomes from the culture supernatants of HepG2.2.15 cells that were ultracentrifuged and immunoprecipitated to purify them. (J) Dicer and Drosha were knocked down with shRNAs in HepG2.2.15 cells, and HBV-miR-3 expression was detected by RT-qPCR. NC, normal control. (K) Western blot showing pulldown of AGO2 protein in Huh7 cells (right) and levels of HBV-miR-3 and miR-423 that immunoprecipitated with the AGO2 complex by RT-qPCR (left). The graphs represent averages of results from three experiments. The data represent means and SD. *, P < 0.05; **, P < 0.01.
Next, we applied stem-loop RT-qPCR to examine HBV-miR-3 expression in various hepatoma cell lines, 20 HBV+ hepatoma tumors, and 3 HBV− hepatoma tumors. HBV-miR-3 was highly expressed in hepatoma cell lines with an integrated HBV genome (HepG2.2.15, QGY-7703, PLC/RF/5, and Hep3B). In contrast, there was no expression of HBV miR-3 in HBV− hepatoma cells (HepG2, Huh7, and SMMC-7721) and L02 cells (Fig. 2D). Moreover, to determine whether HBV-miR-3 could be detected in other HBV− non-liver tissue cell lines, we examined HeLa (human cervical cancer cells), 293 (human embryo kidney cells), MCF7 (human breast cancer cells), and A549 (human lung cancer cells) cell lines and did not observe HBV-miR-3 expression (Fig. 2E).
In addition, as shown in Fig. 2F (left), HBV-miR-3 expression was detected in 20 HBV+ hepatoma tumors, while there was no expression of HBV-miR-3 in 3 HBV− hepatoma tumors. Next, we analyzed the correlation between HBV-miR-3 levels and HBV DNA levels in 14 HBV+ hepatoma tumors; the relationship was not significant (Fig. 2F, right), which may be due to the fact that HBV DNA in HCC tissue might contain integrated HBV DNA in genome and episomal HBV DNA, and HBV-miR-3 may not influence the integrated HBV DNA. Detailed information on hepatoma tumors is provided in Table 1. We also detected the expression levels of HBV-miR-3 in the sera of 25 patients with HBV infection in the acute phase or in convalescence by RT-qPCR. Detailed information on the patients is provided in Table 2. Using miR-16 as a normalization control (29), as shown in Fig. 2F, the HBV-miR-3 expression level was significantly higher in the sera of patients with HBV infection in the acute phase than in those in convalescence. Meanwhile, in the same patients, HBV-miR-3 expression was high in the serum in the acute phase of infection, while during convalescence after treatment, HBV-miR-3 expression was relatively low (Fig. 2G). We analyzed the correlation between HBV-miR-3 levels and HBV DNA levels, and it presented a positive correlation between HBV-miR-3 and HBV DNA levels in the sera of patients with HBV infection in the acute phase (Fig. 2H).
TABLE 1.
Details of hepatoma tumor specimens
| Specimen no. | Donor sex | Donor age (yr) | Collection date (yr/mo/day) | TNMa | HBV DNAb |
|---|---|---|---|---|---|
| 847T | Male | 20 | 2007/1/15 | T1N0M0 | + |
| 864T | Female | 68 | 2007/2/2 | T1N0M0 | 3.56 × 104 |
| 871T | Male | 45 | 2007/2/12 | T1N0M0 | 320.22 |
| 875T | Male | 30 | 2007/3/6 | T1N0M0 | 1.41 × 104 |
| 876T | Male | 46 | 2007/3/7 | T1N0M0 | + |
| 880T | Male | 55 | 2007/3/13 | T4N0M0 | 1.17 × 106 |
| 883T | Male | 36 | 2007/3/16 | T4N0M0 | HBsAg+ HBeAb+ HBcAb+ |
| 887T | Male | 38 | 2007/3/21 | T1N0M0 | − |
| 895T | Male | 35 | 2007/4/5 | T1N0M0 | − |
| 901T | Male | 35 | 2007/4/17 | T1N0M0 | HBsAg+ |
| 909T | Male | 44 | 2007/4/30 | T1N0M0 | HBsAg+ HBeAb+ HBcAb+ |
| 928T | Male | 42 | 2007/6/7 | T1N0M0 | 4.41 × 104 |
| 943T | Male | 34 | 2007/6/27 | T1N0M0 | 948.5 |
| 944T | Female | 65 | 2007/6/28 | T3N0M0 | 6.12 × 103 |
| 965T | Female | 51 | 2007/8/13 | T4N0M0 | 1.21 × 104 |
| 971T | Male | 40 | 2007/8/17 | T3N0M0 | 1.04 × 106 |
| 979T | Male | 36 | 2007/8/24 | T2N0M0 | 2.86 × 103 |
| 980T | Male | 50 | 2007/8/28 | T4N0M0 | 6.37 × 105 |
| 985T | Male | 53 | 2007/9/11 | T4N0M0 | 1.35 × 103 |
| 986T | Male | 41 | 2007/9/13 | T1N0M0 | 4.15 × 104 |
| 1026T | Male | 69 | 2007/11/28 | T1N0M0 | 1.63 × 104 |
| 1033T | Male | 34 | 2007/12/14 | T1N0M0 | − |
TNM, tumor, node, and metastasis, respectively.
+, positive; −, negative.
TABLE 2.
Details of patients with hepatitis B
| Specimen No. | Patient sex | Patient age (yr) | Disease phase | Detection date (yr.mo.day) | HBsAga | HBeAga | DNAb |
|---|---|---|---|---|---|---|---|
| 1 | Male | 58 | Acute | 13.12.11 | 474.5 | 23.12 | 9.8 × 104 |
| Convalescence | 14.3.11 | <0.05 | 0.06 | <20 | |||
| 2 | Male | 39 | Acute | 14.3.26 | 328.7 | 72.33 | 2.3 × 103 |
| Convalescence | 14.6.18 | <0.05 | 0.08 | <500 | |||
| 3 | Male | 37 | Acute | 13.10.22 | 260 | 0.18 | <500 |
| Convalescence | 13.11.4 | 21.95 | 0.11 | <500 | |||
| 4 | Female | 44 | Acute | 13.12.26 | 220.4 | 0.12 | 4.6 × 104 |
| Convalescence | 14.5.3 | <0.05 | 0.08 | ||||
| 5 | Male | 27 | Acute | 13.10.22 | 9367 | 708.5 | 5.8 × 105 |
| Convalescence | 13.11.25 | 2.23 | 0.11 | ||||
| 6 | Female | 74 | Acute | 14.7.8 | 223.7 | 0.13 | 2.3 × 105 |
| Convalescence | 14.9.30 | <0.05 | 0.09 | <500 | |||
| 7 | Male | 33 | Acute | 13.11.15 | 176.5 | 0.12 | <500 |
| Convalescence | 13.12.29 | 0.61 | 0.06 | ||||
| 8 | Male | 28 | Acute | 14.7.23 | 680.5 | 0.55 | 2.9 × 105 |
| Convalescence | 14.10.20 | <0.05 | 0.07 | ||||
| 9 | Male | 51 | Acute | 13.11.7 | 15730 | 155.5 | 6.5 × 105 |
| Convalescence | 14.4.9 | <0.05 | 0.07 | ||||
| 10 | Female | 42 | Acute | 13.12.23 | 15375 | 1145 | 3.0 × 106 |
| Convalescence | 14.2.25 | <0.05 | 0.11 | ||||
| 11 | Male | 32 | Acute | 14.6.29 | 3111 | 12.19 | 1.7 × 104 |
| Convalescence | 14.8.1 | 0.47 | 0.1 | <20 | |||
| 12 | Male | 42 | Acute | 13.11.7 | 93.67 | 0.16 | 4.5 × 103 |
| Convalescence | 13.11.25 | <0.050 | 0.13 | <500 | |||
| 13 | Female | 45 | Acute | 14.2.25 | 11786 | 9 | 4.6 × 102 |
| Convalescence | 14.5.19 | 32.76 | 0.08 | ||||
| 14 | Male | 27 | Acute | 14.3.28 | 1873 | 13.8 | 2.0 × 103 |
| Convalescence | 14.5.4 | 36.05 | 1.37 | 4.3 × 101 | |||
| 15 | Female | 59 | Acute | 14.1.16 | 294.9 | 61.67 | 1.7 × 104 |
| Convalescence | 14.2.8 | 0.06 | 0.1 | ||||
| 16 | Male | 26 | Acute | 14.2.6 | 350 | 87.56 | 1.3 × 106 |
| Convalescence | 14.2.26 | <0.050 | 0.06 | ||||
| 17 | Female | 46 | Acute | 13.11.2 | 463.7 | 0.33 | <500 |
| Convalescence | 13.12.17 | 0.13 | 0.06 | ||||
| 18 | Female | 71 | Acute | 13.11.30 | 461.4 | 283.6 | 1.8 × 106 |
| Convalescence | 14.1.2 | 33.8 | 5.95 | <500 | |||
| 19 | Male | 34 | Acute | 14.1.18 | 342.2 | 264.6 | 4.9 × 103 |
| Convalescence | 14.2.21 | 0.05 | <500 | ||||
| 20 | Female | 42 | Acute | 14.2.14 | 79.49 | 0.08 | 1.4 × 104 |
| Convalescence | 14.3.3 | <0.050 | 0.07 | <500 | |||
| 21 | Male | 40 | Acute | 14.3.2 | 917.6 | 3.68 | 4.8 × 103 |
| Convalescence | 14.3.17 | 0.08 | 0.06 | <500 | |||
| 22 | Female | 39 | Acute | 14.3.6 | 315.2 | 42.36 | 2.8 × 103 |
| Convalescence | 14.4.17 | <0.050 | 0.18 | <500 | |||
| 23 | Male | 37 | Acute | 14.3.18 | 985.2 | 1.08 | 2.5 × 103 |
| Convalescence | 14.4.15 | 4.67 | 0.06 | ||||
| 24 | Male | 52 | Acute | 14.3.18 | 649.6 | 0.07 | 7.0 × 104 |
| Convalescence | 14.3.26 | 0.43 | 0.05 | <500 | |||
| 25 | Male | 49 | Acute | 14.8.17 | 307.1 | 54.95 | 4.4 × 104 |
| Convalescence | 14.9.28 | 0.12 | 0.07 |
IU/ml.
copies/ml.
Furthermore, to determine how HBV-miR-3 was released from cells, we isolated exosomes and purified HBV-related particles from the culture supernatants of HepG2.2.15 cells by sucrose gradient ultracentrifugation; we then extracted RNA to detect the expression of HBV-miR-3. As shown in Fig. 2I, the expression of HBV-miR-3 was detected in both exosomes (CD63 is a known exosome marker [30]) and HBV core particles, but not in HBV subviral particles.
To determine whether HBV-miR-3 was generated via the canonical miRNA biogenesis pathway, two key components of miRNA precursor processing, Drosha and Dicer, were knocked down using short hairpin RNAs (shRNAs) in HepG2.2.15 cells. Deletion of Drosha and Dicer resulted in significantly decreased HBV-miR-3 expression, using miR-423 as a canonical control (Fig. 2J). In addition, to determine whether HBV-encoded HBV-miR-3 was associated with the RISC complex, we cotransfected pHBV1.3 and pcDNA3-Flag-AGO2 in Huh7 cells by RNA immunoprecipitation (RIP) using Flag-specific antibody or mouse IgG antibody as a negative control, and the eluted associated RNAs were used to detect HBV-miR-3 using RT-qPCR. As shown in Fig. 2K, RIP analysis showed that HBV-encoded HBV-miR-3 was bound to the AGO2 complex, using miR-423 as a control. These data indicated that HBV-miR-3 was found within the host RISC machinery, similar to the majority of host and viral miRNAs, and that the biogenesis of HBV-miR-3 is in the classic Dicer-Drosha-dependent mode and functions within the host RISC machinery.
HBV-miR-3 represses viral protein production and HBV replication.
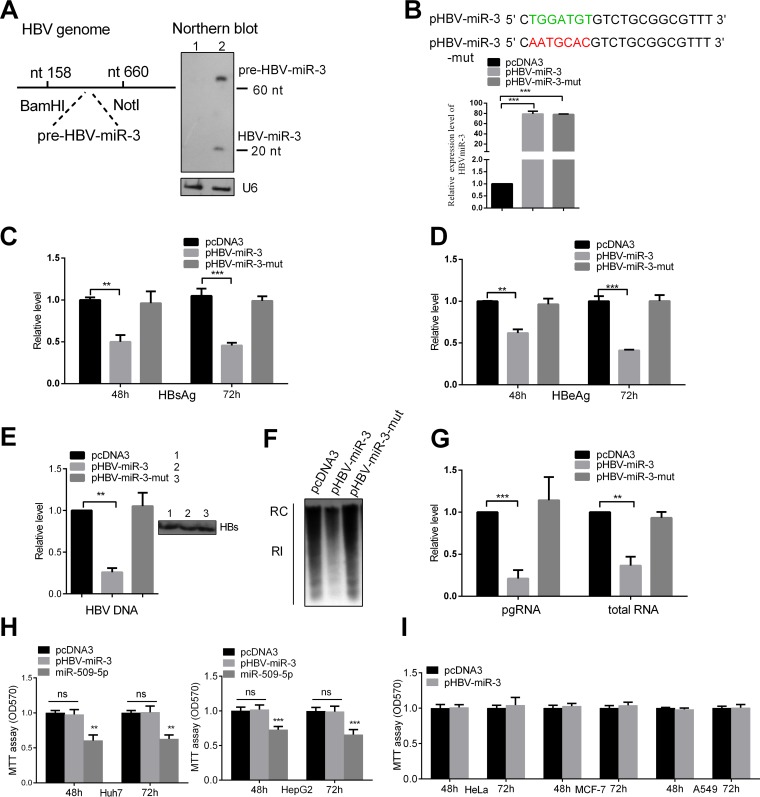
To investigate the role of HBV-miR-3 in HBV gene expression, we predicted the HBV-miR-3 precursor, and by cloning, the fragment was inserted into the BamHI-NotI sites of pCDNA3 (Fig. 3A, left); the resultant vector was named pHBV-miR-3. We validated the expression of HBV-miR-3 by Northern blotting (Fig. 3A, right). Then, we constructed an HBV-miR-3-mut plasmid (pHBV-miR3-mut) in which the HBV-miR-3 seed sequence was mutated (Fig. 3B). As shown in Fig. 3B, the expression efficiencies of these plasmids were confirmed by RT-qPCR using different stem-loop RT primers. The plasmids were then cotransfected with pHBV1.3 into Huh7 cells. The secreted viral protein levels (HBsAg and HBeAg) in the culture supernatants were reduced by ectopic expression of HBV-miR-3, but not HBV-miR-3-mut (Fig. 3C and D), suggesting that the repression of HBV protein expression was specifically due to HBV-miR-3. Furthermore, to detect secreted complete particles in the transfected cell culture supernatant, immunoprecipitation using HBsAb antibody was performed; then, we used DNase I treatment to remove free DNA from the complete viral particles in the culture supernatants. The extracted HBV DNA and qPCR were then used to evaluate the production of complete viral particles (31). As shown in Fig. 3E, ectopic expression of HBV-miR-3, but not HBV-miR-3-mut, significantly decreased HBV DNA in complete viral particles from the supernatants.
FIG 3.
HBV-miR-3 represses viral protein production and HBV replication. (A) (Left) Schematic diagram of a DNA fragment, including the predicted HBV-miR-3 precursor, and cloning of the fragment into pcDNA3. (Right) Huh7 cells were transfected with HBV-miR-3 or an empty vector, pcDNA3, as a negative control, and 10 μg RNA was used to detect the mature HBV-miR-3 by Northern blotting, with U6 as a control. (B) Mutant sequence of HBV-miR-3-mut compared to that of HBV-miR-3. HBV-miR-3 or HBV-miR-3-mut was transfected into Huh7 cells, with pcDNA3 as the negative control. The expression of the plasmids was tested using RT-qPCR. (C to E) Huh7 cells were cotransfected with pHBV1.3 and the indicated plasmids. HBsAg (C), HBeAg (D), and HBV DNA (E) levels in the culture supernatant were measured by ELISA and qPCR. Immunoprecipitation using HBsAb antibody was performed before extracting HBV DNA. (F) Southern blot analysis of HBV DNA. RC, relaxed circular DNA; RI, RI-DNA. (G) Total cellular RNA was extracted and analyzed as HBV transcripts by RT-qPCR. The effects of HBV-miR-3 on viabilities in different cell lines are shown. (H and I) Effects of HBV-miR-3, HBV-miR-3-mut, and miR-509-5p on Huh7 and HepG2 cells using an MTT assay, with miR-509-5p as a positive control (H), and in non-liver cell lines (HeLa, MCF7, and A549) using an MTT assay (I). The graphs represent the averages of results from three experiments. The data represent means and SD. **, P < 0.01; ***, P < 0.001; ns, not significant.
To further investigate the effect of HBV-miR-3 on HBV replication, Southern blotting and RT-qPCR were performed to examine the cellular replication intermediate DNA (RI-DNA) and HBV RNA. A Southern blot assay showed that HBV-miR-3 ectopic expression markedly reduced the levels of RI-DNA of the HBV genome (Fig. 3F). Moreover, the HBV mRNA, pgRNA, and total RNA levels were also significantly decreased in HBV-miR-3-transfected Huh7 cells compared to control cells (Fig. 3G). Meanwhile, HBV-miR-3 or HBV-miR-3-mut was transfected into Huh7 and HepG2 cells. Cell growth and vitality were detected by an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay, and the results showed that HBV-miR-3 and HBV-miR-3-mut had no obvious effects on cell growth or vitality in hepatoma cell lines; we used miR-509-5p as a positive-control plasmid, which was reported to inhibit hepatoma cell viability (32) (Fig. 3H). We also carried out the same experiment in HeLa, MCF7, and A549 cells, and the results indicated that HBV-miR-3 did not obviously influence cell viability in those cell lines (Fig. 3I). These data indicated that HBV-miR-3 repressed HBV pgRNA and RI-DNA levels and that this effect may not occur by influencing cytological changes.
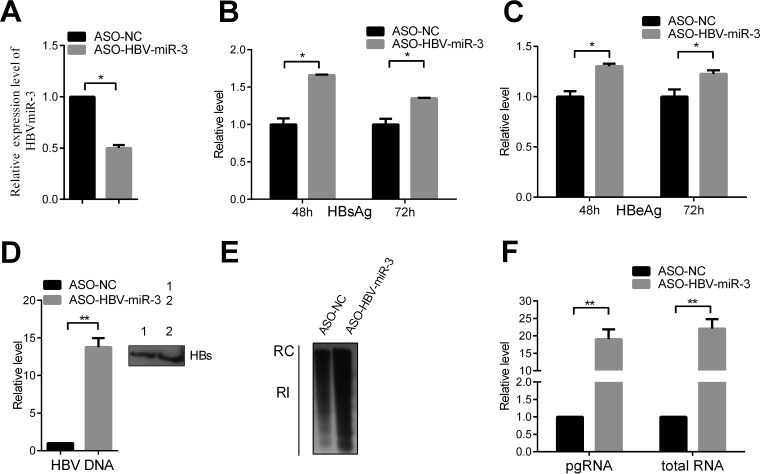
To further verify the effect of HBV-miR-3 on HBV replication, we examined the effect of blocking HBV-miR-3 in HepG2.2.15 cells using an HBV-miR-3-specific antisense oligonucleotide (ASO). Transfection of ASO-HBV-miR-3 could efficiently reduce HBV-miR-3 levels (Fig. 4A). Blocking HBV-miR-3 markedly increased the HBsAg and HBeAg protein levels (Fig. 4B and C) and HBV DNA in complete viral particles from supernatants (Fig. 4D). Moreover, transfection of ASO-HBV-miR-3 increased RI-DNA in HepG2.2.15 cells (Fig. 4E), and the HBV mRNAs, pgRNA, and total RNA were increased (Fig. 4F). Taken together, these results indicated that HBV-miR-3 represses HBsAg and HBeAg expression, HBV genome replication, and complete particle production.
FIG 4.
ASO-HBV-miR-3 increases viral protein production and HBV replication. (A) ASO-HBV-miR-3 was transfected into HepG2.2.15 cells. ASO-NC was used as a control. HBV-miR-3 expression was examined by RT-qPCR. (B to D) HBsAg (B), HBeAg (C), and HBV DNA (D) levels in the culture supernatant were measured by ELISA and qPCR in HepG2.2.15 cells transfected with ASO-HBV-miR-3. Immunoprecipitation using HBsAb antibody was performed before extracting HBV DNA. (E) Southern blot analysis of HBV DNA. (F) Total cellular RNA was extracted and analyzed for HBV transcripts by RT-qPCR. The graphs represent the average results of three experiments. The data represent means and SD. *, P < 0.05; **, P < 0.01.
HBV-miR-3 targets the ORFs of HBV core proteins in the 3.5-kb mRNA.
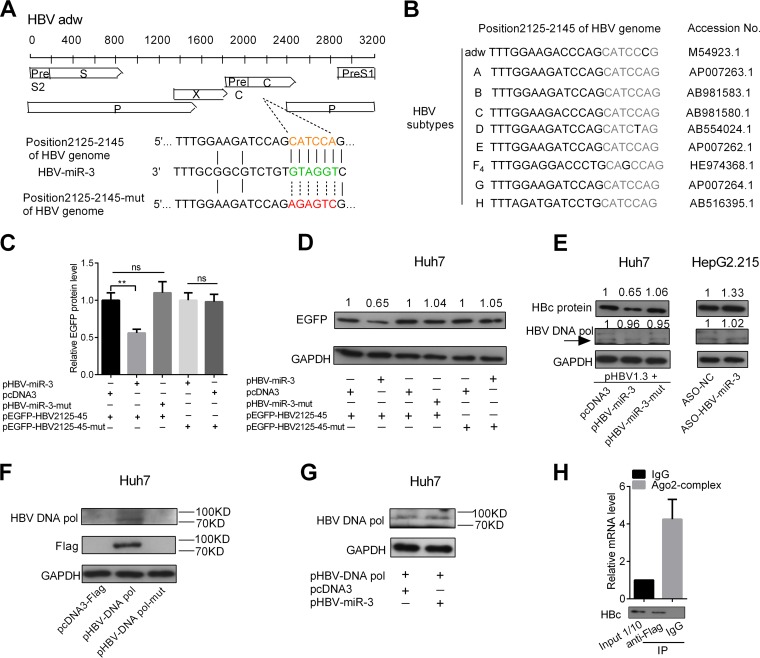
To explore the mechanisms by which HBV-miR-3 suppresses viral replication, we first utilized BLAST and the RNA-hybrid Web server (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) to predict the targeting site of HBV-miR-3 on the HBV transcripts. Sequence analysis revealed that there was a candidate HBV-miR-3-binding site in the HBV 3.5-kb mRNA that was located in the ORF of the HBV core gene (HBV nt 2125 to 2145). To determine the direct interaction between HBV-miR-3 and the predicted binding site, we constructed enhanced green fluorescent protein (EGFP) reporter plasmids carrying the wild type or a mutant in which the HBV-miR-3-binding site was mutated (Fig. 5A) and the ORF of the HBV core gene (HBV nt 2125 to 2145) was conserved in different subtypes of the HBV genome (Fig. 5B). EGFP reporter assays were performed, and Huh7 cells were cotransfected with either wild-type or mutant HBV2125-45 EGFP constructs and pHBV-miR-3 or pHBV-miR-3-mut. The intensity of EGFP fluorescence from the wild-type EGFP reporter was significantly reduced in the pHBV-miR-3-transfected group compared to the control group. However, reporter expression was not influenced by alteration of HBV-miR-3 levels when the miRNA seed sequence-binding site was mutated (Fig. 5C). In addition, the Western blot assay using EGFP antibodies was similar to that from the EGFP reporter assays (Fig. 5D). In addition to HBeAg, the 3.5-kb mRNA also encodes HBc and DNA Pol. We measured the effects of HBV-miR-3 on the endogenous expression levels of HBc and DNA Pol. As shown in Fig. 5E, HBc and HBeAg expression levels (Fig. 3D) were significantly repressed in Huh7 cells transfected with pHBV1.3 by HBV-miR-3, but not HBV-miR-3-mut. Conversely, HBc expression was increased by blocking HBV-miR-3 with ASO-HBV-miR-3 in HepG2.2.15 cells. However, DNA Pol expression was not affected by HBV-miR-3.
FIG 5.
HBV-miR-3 target region encoding the HBc protein of the HBV 3.5-kb mRNA. (A) Predicted binding site of HBV-miR-3 in a region encoding the HBc protein of the HBV 3.5-kb mRNA. The arrows indicate the mutant nucleotides. (B) Analysis of conserved HBV-miR-3-targeted site (nt 2125 to 2145) in subtype HBV genomes. (C and D) Huh7 cells were cotransfected with either wild-type or mutant HBV-EGFP nt 2125-to-2145 constructs and pHBV-miR-3 or pHBV-miR-3-mut for 48 h. pcDNA3 was used as a negative control. Cells were harvested to analyze endogenous protein expression by EGFP fluorescence or Western blot analysis. pDsRed2-N1 was used as normalization for fluorescence. GAPDH was used as an endogenous control for EGFP protein expression. (E) Huh7 cells were cotransfected with pHBV1.3 and pHBV-miR-3 or pHBV-miR-3-mut for 48 h. pcDNA3 was a negative control. HepG2.2.15 cells were transfected with ASO-HBV-miR-3, and ASO-NC (NC, normal control) was a negative control. HBV core protein and DNA Pol were detected by Western blotting. The arrow indicates the position of the protein expression. (F) Huh7 cells were transfected with pcDNA3, pHBV DNA Pol, and a pHBV DNA Pol mutant; 24 h posttransfection, cells were harvested for Western blotting to detect HBV DNA Pol using Flag and HBV DNA Pol antibodies. (G) Huh7 cells were transfected with pHBV DNA Pol and pcDNA3 or pHBV-miR-3; 24 h posttransfection, cells were harvested for Western blotting to detect HBV DNA Pol using HBV DNA Pol antibody. (H) HBV pgRNA (3.5-kb RNA) and HBc protein were detected in AGO2-related complexes by RT-qPCR and Western blotting. The graphs represent average results of three experiments. The data represent means and SD. **, P < 0.01.
In order to further validate the effect of HBV-miR-3 on DNA Pol, we constructed pcDNA3-Flag/HBV DNA Pol and pcDNA3-Flag/HBV DNA Pol-mutant with an ATG mutant that can generate HBV DNA Pol mRNA but cannot translate it into protein as a control. Huh7 cells were transfected with pcDNA3, pHBV DNA Pol, and pHBV DNA Pol-mutant, and we detected HBV DNA Pol using Flag and DNA Pol antibodies, as shown in Fig. 5F; the expression of HBV DNA Pol was detected using both Flag and DNA Pol antibodies in Huh7 cells transfected with pcDNA3-Flag/HBV DNA Pol but not HBV DNA Pol-mutant. Furthermore, we found that exogenous DNA Pol expression was not affected by HBV-miR-3 (Fig. 5G). This was because the target sites of HBV-miR-3 in the HBV genome were not in the ORF region of HBV DNA Pol. In addition, we used RNA immunoprecipitation to show that HBV pgRNA (3.5-kb RNA) and HBc protein were detected in AGO2-related complexes (Fig. 5H). These results demonstrated that HBV-miR-3 directly binds to the HBV 3.5-kb transcript to repress HBc expression.
The effects of HBV-miR-3 on HBV replication and viral protein production were blocked by pHBV1.3-mut.
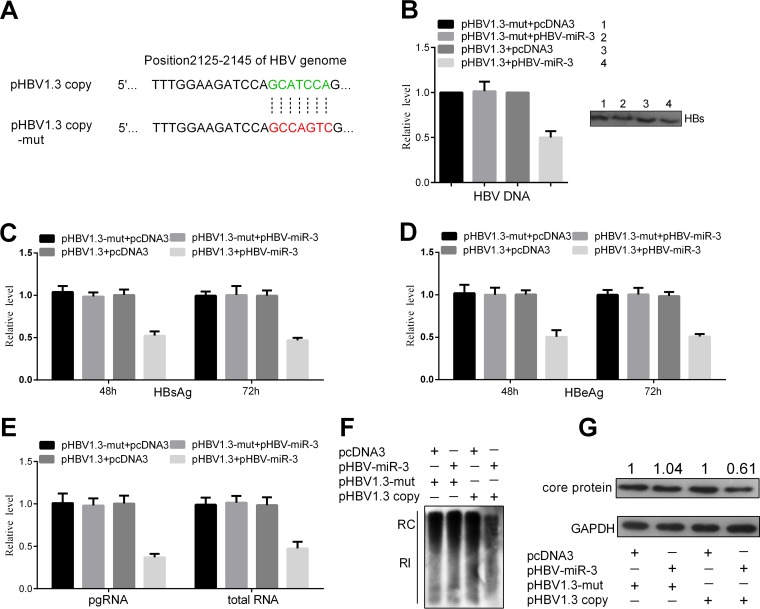
To further analyze the role of HBV-miR-3 in HBV gene expression, we constructed the plasmid pHBV1.3-mut, in which the target site (positions 2125 to 2145 of the HBV genome) of the HBV-miR-3 seed sequence was mutated (Fig. 6A). Huh7 cells were then cotransfected with pHBV1.3-mut and pHBV-miR-3 or pcDNA3. The secreted viral protein levels (HBsAg and HBeAg) and HBV DNA in the culture supernatants were not changed significantly by ectopic expression of HBV-miR-3 (Fig. 6B, C, and D). Immunoprecipitation using HBsAb antibody was performed before extracting HBV DNA. Meanwhile, RT-qPCR, Southern blotting, and Western blotting were performed to examine HBV RNA, cellular RI-DNA, and HBV core protein. The levels of pgRNA and total RNA (Fig. 6E), RI-DNA of the HBV genome (Fig. 6F), and HBV core protein (Fig. 6G) were not significantly changed in HBV-miR-3-transfected Huh7 cells compared to control cells. These results indicate that HBV-miR-3 indeed directly binds to the HBV 3.5-kb transcript (positions 2125 to 2145 of the HBV genome) to repress HBV replication.
FIG 6.
The effects of HBV-miR-3 on HBV replication and viral protein production were blocked by pHBV1.3-mut. (A) Mutant sequence of pHBV1.3-mut in positions 2125 to 2145 of the HBV genome compared to that in pHBV1.3. Huh7 cells were cotransfected with pHBV1.3-mut or pHBV1.3 and the indicated plasmids, (B to D) HBsAg (B), HBeAg (C), and HBV DNA (D) levels in culture supernatants were measured by ELISA and qPCR. (E) Total cellular RNA was extracted and analyzed as HBV transcripts by RT-qPCR. (F) Southern blot analysis of HBV DNA. (G) HBV core protein was detected by Western blotting. The graphs represent averages of results from three experiments. The data represent means and SD.
DISCUSSION
Interactions of virus-host cell molecules are pivotal processes that determine infection status during viral infection. Recent studies have revealed virus-encoded miRNAs that are key regulators of both cellular and viral transcripts and that may play roles in regulating the gene expression profiles of the cells or the virus itself (33). HBV is a major pathogen causing hepatitis in humans. HBV has a long incubation period and can easily develop into a persistent infection. HBV-encoded surface, core, DNA Pol, and X proteins have been intensively studied and play different roles in both HBV replication and host cells. Here, we presented a series of experiments highlighting three important points. First, we indicated that HBV encodes HBV-sncRNAs. Second, we demonstrated that HBV-encoded miRNA, i.e., HBV-miR-3, represses the generation of HBV protein and HBV replication. Finally, HBV-miR-3 targets the 3.5-kb transcript of HBV to inhibit HBc protein expression, which contributes to suppression of HBV replication and protein expression.
Chronic HBV infection is a major risk factor for hepatoma development; therefore, most patients with hepatoma are HBV positive. To identify whether HBV encodes miRNA, we first applied deep sequencing to screen the RNA from HBV-positive hepatoma tissue. Five small RNAs 20 to 21 nt in length were obtained. Although previous computational analysis predicted an HBV miRNA (34), we did not get the predicted miRNA sequence. Generally, the tools for identifying novel miRNAs were Northern blotting and stem-loop RT-qPCR (7, 35–38) and bioinformatics analysis for predicating the secondary structure of candidate miR precursors. Bioinformatics analysis predicted the precursor structure and found that one hairpin structure contained HBV sncRNA-3 and -5 at two stems, which were named HBV-miR-3-3p and HBV-miR-3, and they are conserved in various HBV strains. We then used Northern blotting to identify HBV-miR-3 in Huh7 cells transfected with HBV replication-competent plasmids (plasmid HBV1.3 copy). In addition, a stem-loop RT-qPCR assay showed that HBV-miR-3 was expressed at high levels in an additional 20 hepatoma tissues in HBV+ and HBV+ hepatoma cell lines; this high expression was not observed in HBV− hepatoma cell lines or other non-liver tissue cell lines. Furthermore, we showed that HBV-miR-3 levels in sera from patients in the acute phase of HBV infection were obviously higher than those in the convalescence phase. Next, we asked which transcripts HBV-miR-3 was derived from, because it was located in nt 373 to 393 of the HBV genome, which overlapped in the 3.5-kb, 2.4-kb, and 2.1-kb HBV transcripts. We expressed four transcripts (3.5 kb, 2.4 kb, 2.1 kb, and 0.7 kb) in Huh7 cells, and stem-loop RT-qPCR revealed that plasmids expressing the 3.5-kb, 2.4-kb, and 2.1-kb transcripts, but not the 0.7-kb transcript, were capable of HBV-miR-3 generation. In addition, RIP analysis indicated that HBV-miR-3 was in the RISC (AGO2) complex, which is the miRNA functional complex. Thus, we concluded that HBV encoded miRNA and that HBV-miR-3 is expressed in HBV+ tissues, cells, and sera from patients with HBV infection, and we confirmed that HBV-miR-3 is released into sera by exosomes and HBV core particles, not HBV subviral particles.
Subsequently, we addressed the role of HBV-miR-3. Ectopic expression of HBV-miR-3 did not affect cell viability but significantly suppressed HBsAg, HBeAg, and HBc protein expression in Huh7 cells that were transfected with pHBV1.3. Meanwhile, Huh7 cells were transfected with pHBV1.3-mut, in which the target site (positions 2125 to 2145 of the HBV genome) of the HBV-miR-3 seed sequence was mutated, the effect of HBV-miR-3 on HBV replication was blocked. Depletion of HBV-miR-3 by ASO increased HBsAg, HBeAg, and HBc protein expression in HepG2.2.15 cells. Furthermore, we demonstrated that HBV-miR-3 repressed HBV virion production, according to the immunoprecipitation using HBsAb antibody and the qPCR procedure that was used to quantitate HBV virions (31). In addition, a Southern blot assay showed that HBV-miR-3 reduced the replication intermediate of HBV. Similar phenomena have been reported in other viruses. For instance, HCMV-encoded miR-UL112-1 regulates a variety of genes by binding to the 3′ untranslated region (UTR) of viral transcripts and inhibiting translation (9). Human herpesvirus 6A (HHV-6A)-encoded miR-U86 targets the HHV-6A U86 immediate-early (IE) gene, thereby regulating lytic replication (35). These findings suggested that a virus-encoded miRNA modulates its own replication process and infection status. Therefore, HBV-encoded miRNA represses HBV protein expression and virion production, which may avoid damage to infected cells and could contribute to persistent infection.
miRNA plays its role by regulating the expression of the target gene (39). To reveal the mechanism by which HBV-miR-3 suppresses HBV replication, we used a bioinformatics assay to predict the targets of HBV-miR-3 on HBV transcripts and cellular mRNA. Surprisingly, HBV-miR-3 was predicted to target the 3.5-kb HBV transcript at nt 2125 to 2145 of the HBV genome, which were conserved in different subtypes of the HBV genome. The 3.5-kb transcript serves as a translational template for HBc (ORF, nt 1815 to 2452); for HBV DNA Pol (ORF, nt 2087 to 3215 and 1 to 1632); and for pgRNA, which is a template of HBV replication intermediates (HBV-RI) (21). An EGFP reporter assay identified the HBV-miR-3-binding site of the HBV 3.5-kb mRNA; however, Western blotting showed that HBV-miR-3 reduced the HBc protein expression level but did not decrease HBV DNA Pol expression, which may be due to the target site of HBV-miR-3, which is located in the HBc ORF. Simultaneously, HBV-miR-3 reduced the production of pgRNA and HBV-RI. During HBV replication, an abundance of subviral particles, including small spheres and filament particles lacking nucleocapsid, are produced and could reach amounts up to 103- to 106-fold higher than those of HBV virions in the blood of infected individuals (40). One hypothesis is that the autoassembly of the S protein is more efficient than its incorporation into virions (41), but the mechanism underlying this phenomenon is not clear. HBc and pgRNA are required for nucleocapsid assembly/maturation and virion formation (42–45). Thus, our finding that HBV-miR-3 reduced HBV core protein expression and pgRNA/HBV-RI production may provide a possible explanation for the phenomenon of relatively few complete virions.
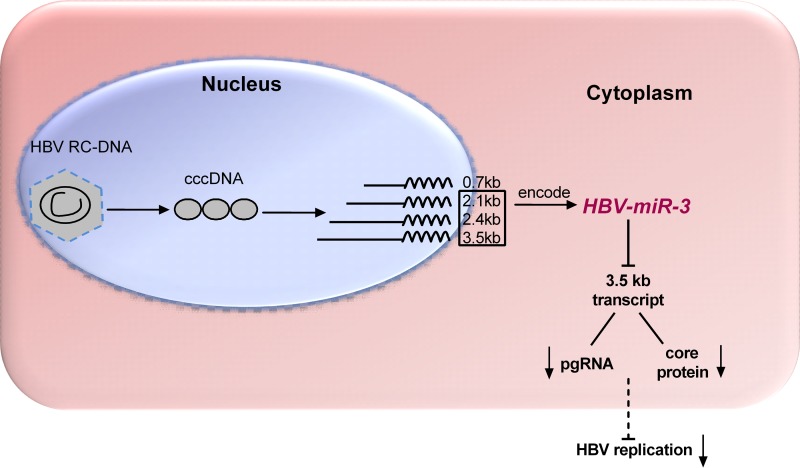
In summary, we determined that HBV encodes HBV-miR-3 is present in all HBV+ tissues and cells. HBV-miR-3 represses HBV replication by targeting the region of HBV 3.5-kb mRNA encoding HBV core protein (HBc) to reduce HBc protein and HBV pregenomic RNA (pgRNA), which in turn led to attenuated HBV replication (Fig. 7). Overall, these results indicate that HBV encodes miRNA which may provide new potential biomarkers for clinical HBV infection and shed light on a new mechanism of HBV replication regulation by which HBV-encoded miRNAs control self-replication by targeting viral transcripts.
FIG 7.
Proposed model describing HBV-encoded miRNA (HBV-miR-3) targeting HBV 3.5-kb mRNA to reduce HBc protein and pgRNA levels and to suppress HBV transcription and HBV replication.
MATERIALS AND METHODS
Human hepatoma tissue specimens and serum specimens from patients with HBV infection.
Twenty HBV+ hepatoma tumors and 3 HBV− hepatoma tumors were obtained from the cancer center of Sun Yat-Sen University. These diagnoses were verified by pathological analysis. The sera of 25 pairs of patients with HBV infection were obtained from the Second People's Hospital of Tianjin. Written informed consent was obtained from every patient, and ethics approval for the work was granted by the Ethics Committee of Tianjin Medical University. All the methods were carried out in accordance with the relevant guidelines, including any relevant details.
Cell cultures and transfections.
Human hepatoma cell lines Huh7 and HepG2.2.15 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The Huh7 and HepG2.2.15 cell lines were propagated and maintained in Dulbecco's modified Eagle's medium (DMEM) or minimal essential medium alpha (MEM-α) (Gibco, BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco), 20 mM HEPES, 100 units/ml penicillin, and 100 μg/ml streptomycin. MEM-α was additionally supplemented with 2 mM glutamine and 200 μg/ml G418. Cultures were incubated in a humidified atmosphere at 37°C with 5% CO2. All transfections were performed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's recommendations.
Deep sequencing and bioinformatics analysis.
Small RNA was extracted from 3 HBV+ (880T, 971T, and 980T) and 3 HBV− (887T, 895T, and 1033T) hepatoma tumors using the mirVana miRNA isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's protocol. The samples were qualified and sent to Beijing Genomics Institute Shenzhen Co. (BGI-Shenzhen) for sequencing using Solexa technology (58), and 18- to 30-nt sequences were identified. Sequencing reads that did not align with the human genome were analyzed to align with the HBV genome (GenBank accession number M54923.1), and thus, 5 HBV-sncRNAs were obtained. The secondary-structure analysis and prediction of the precursor and targets of the miRNA were performed online with RNAhybrid, TargetScan, and PicTar.
RT-qPCR.
For RNA detection, RNA was isolated from cells and HCC tissues using a mirVana miRNA isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. Serum RNA was extracted with a miRcute miRNA isolation kit (Tiangen Biotech, China) according to the manufacturer's instructions and was then subjected to cDNA synthesis using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI). qPCR was performed at 94°C for 10 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s in an iQ5 real-time PCR detection system (Bio-Rad). A SYBR Premix Ex Taq II kit (TaKaRa) was used, following the manufacturer's instructions. β-Actin mRNA or U6 RNA was used as the normal control. For miRNA detection, we used a special RT-qPCR (stem-loop RT-qPCR), as described by Chen et al. (46), and miR-16 was analyzed to serve as an internal control. The relative fold change in the transcripts was calculated using the 2−ΔCT method (47) and the 2−ΔΔCT method (46). The relative levels of HBV-miR-3 in serum were presented as a standardized threshold cycle value of 39 (39-Ct) after normalization to miR-16 (48). The specific primers are shown in Table 3.
TABLE 3.
Primers used in the construction of plasmids and RT-qPCR
| Use | Primer name | Sequence (5 ′–3 ′) |
|---|---|---|
| Construction of HBV-miR-3 | HBV-miR-3-S | CGGGGTACCGAGAGCACAACATCAGGATTC |
| HBV-miR-3-AS | GACGAATTCGGACTGAGGCCCCCTCCC | |
| RT-qPCR of HBV-miR-3 | HBV-miR-3/HBV-miR-3-mut RT | GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAACGCCG |
| HBV-miR-3 Forward | TGCGGCTGGATGTGTCTGCGG | |
| RT-qPCR of HBV-miR-3-3p | HBV-miR-3-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAGAAGAAC |
| HBV-miR-3 Forward | TGCGGATCTTCTTGTTGGTT | |
| Construction of HBV-miR-3mut | HBV-miR-3-mut-MS | TATCGCAATGCACGTCTGCGGCGTTTTATC |
| HBV-miR-3-mut-MA | GATAAAACGCCGCAGACGTGCATTGCGATA | |
| RT-qPCR of HBV-miR-3mut | HBV-miR-3-mut Forward | TGCGGCAATGCACGTCTGCG |
| RT-qPCR of miR-423 | miR-423 RT | GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAGTCTC |
| miR-423 Forward | TGCGGTGAGGGGCAGAGAG | |
| RT-qPCR of miR-16 | miR-16 RT | GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACCGCCAAT |
| miR-16 Forward | TGCGGTAGCAGCACGTAAATA | |
| RT-qPCR of U6 snRNA | U6 RT | GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGG |
| U6 Forward | TGCGGGTGCTCGCTTCGGCAGC | |
| miRNA Reverse primer | CCAGTGCAGGGTCCGAGGT | |
| Construction of HBV2139 EGFP reporters | EGFP-HBV2139-S | GATCCAGATCCAGCATCCAGGAAGCTTG |
| EGFP-HBV2139-AS | AATTCAAGCTTCCTGGATGCTGGATCTG | |
| EGFP-HBV2139-MS | GATCCAGATCCAGAGAGTCGGAAGCTTG | |
| EGFP-HBV2139-MA | AATTCAAGCTTCCGACTCTCTGGATCTG | |
| Construction of pHBV-1.3-mut | HBV1.3-mut-S | GGAAGATCCAGCCAGTCGAGACCTAGTAGTCAGT |
| HBV1.3-mut-AS | CTAGGTCTCGACTGGCTGGATCTTCCAAATTAAC | |
| RT-qPCR of β-actin | β-actin-S | CGTGACATTAAGGAGAAGCTG |
| β-actin-AS | CTAGAAGCATTTGCGGTGGAC | |
| Construction of HBV 3.5 kb | HBV3.5kb-S | GCAGGCAAGCTTGTTCATGTCCTACTGTTCAAG |
| HBV-mRNA-Reverse | ATAAGAATGCGGCCGCGCAAAAACGAGAGTAACTCC | |
| Construction of HBV 2.4 kb | HBV2.4kb-S | GCAGGCAAGCTTATAAGAGAGAAACAACACAT |
| HBV-mRNA-Reverse | ATAAGAATGCGGCCGCGCAAAAACGAGAGTAACTCC | |
| Construction of HBV 2.1 kb | HBV2.1kb-S | GCAGGCAAGCTTAGAAACACTCATCCTCAGGC |
| HBV-mRNA-Reverse | ATAAGAATGCGGCCGCGCAAAAACGAGAGTAACTCC | |
| Construction of HBV 0.7 kb | HBV0.7kb-S | GCAGGCAAGCTTCATGGC TGCTAGGCTGTGC |
| HBV-mRNA-Reverse | ATAAGAATGCGGCCGCGCAAAAACGAGAGTAACTCC | |
| RT-qPCR of pgRNA/HBV DNA copies | pgRNA-qRT-S | TCTTGCCTTACTTTTGGAAG |
| pgRNA-qRT-AS | AGTTCTTCTTCTAGGGGACC | |
| RT-qPCR of total RNA | Total RNA-qRT-S | CTCCCCGTCTGTGCCTTCTC |
| Total RNA-qRT-AS | TCGGTCGTTGACATTGCTGA | |
| Construction of shR-Dicer | shR-Dicer-S | GATCCCTCGAAATCTTACGCAAATACTCGAGTATTTGCGTAAGATTTCGAGTTTTTGA |
| shR-Dicer-AS | AGCTTCAAAAACTCGAAATCTTACGCAAATACTCGAGTATTTGCGTAAGATTTCGAGG | |
| Construction of shR-Drosha | shR-Drosha-S | GATCCCCAGATGAGACTGAAGACATCTCGAGATGTCTTCAGTCTCATCTGGTTTTTGA |
| shR-Drosha-AS | AGCTTCAAAAACCAGATGAGACTGAAGACATCTCGAGATGTCTTCAGTCTCATCTGGG | |
| Construction of DNA Pol | DNA pol-S | GCCGGTACCGCCATGCCCCTATCTTATCCACAC |
| DNA pol-AS | ATAAGAATGCGGCCGCTCACGGGGGTCTCCATGCGAC | |
| Construction of DNA Pol-mut | DNA pol-M-S | GCCGGTACCGCCATGCCCTATCTTATCCACAC |
| DNA pol-M-AS | ATAAGAATGCGGCCGCTCACGGGGGTCTCCATGCGAC |
ELISA for HBsAg and HBeAg detection.
Huh7 cells were cotransfected with pHBV1.3 (300 ng), HBV-miR-3 (600 ng), HBV-miR-3-mut (600 ng), or the control vector pcDNA3 (600 ng), and HepG2.2.15 cells were transfected with ASO-HBV-miR-3 (20 pmol) or scrambled oligomer (ASO-NC; 20 pmol) in 24-well plates. At 48 or 72 h after transfection, the supernatants of Huh7 cells were harvested or the supernatants of HepG2.2.15 cells were harvested and diluted 1:10 in phosphate-buffered saline (PBS). The harvested supernatants were used to examine HBsAg and HBeAg using ELISA kits (InTec Products, Xiamen, China), following the manufacturer's protocol. Absorbance was determined in a microtiter plate reader (Quant, Biotek, Germany) with dual-wavelength measurement (450 nm).
Plasmid construction.
The HBV replication-competent plasmid cloned in pUC18, pHBV1.3, containing 1.3 copies of the HBV genome that could result in HBV particles, was a generous gift from Z. H. Yuan's laboratory at Shanghai Medical School, Fudan University (49). pHBV1.3-mut, containing the HBV-miR-3-binding site of the HBV genome, was constructed by PCR with specific mutant primers.
To construct HBV transcripts, pHBV3.5 kb, -2.4 kb, -2.1 kb, and -0.7 kb were PCR amplified from pHBV1.3, pHBV-miR-3 vectors, and pHBV-miR-3 mutant vectors in which the seed sequence of HBV-miR-3 was mutated by PCR with the primers. The fragments were then cloned into the pcDNA3 vector.
The construction of the EGFP expression vector, pcDNA3/EGFP, was described previously (50). Nucleotides 2125 to 2145 of the HBV genome and mutations in the complementarity sites of HBV-miR-3 seed sequences were designed. The oligomers were synthesized and annealed into double-stranded fragments that were cloned into the BamHI and EcoRI sites downstream of the EGFP ORF in pcDNA3/EGFP.
All the specific primers are shown in Table 3. All the constructs were confirmed by DNA sequencing.
Fluorescent reporter assay.
Huh7 cells were seeded in 48-well plates the day before transfection and were then transfected with HBV-miR-3 (400 ng) or HBV-miR-3-mut (400 ng), together with the pEGFP-HBV2125-45 (200-ng) reporter vectors and the pEGFP-HBV2125-45-mut (200-ng) vectors. The red fluorescent protein (RFP) expression vector pDsRed2-N1 (Clontech; 20 ng) was used as an internal control. At 48 h after transfection, cells were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (10 mM Tris-HCl, pH 7.4, 1% Triton X-100, 0.1% SDS, 1% NP-40, 1 mM MgCl2), and the proteins were harvested. Subsequently, we detected the EGFP and RFP intensities using an F-4500 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). All experiments were performed more than three times.
Western blotting.
The transfected cells were lysed in RIPA buffer. Proteins were separated by SDS-10% PAGE, transferred to nitrocellulose membranes, and then detected using the appropriate antibodies. Rabbit anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase), -AGO2, -mouse IgG, -EGFP, -HBcAg, -HBsAg, -HBV DNA Pol, -Dicer, and -CD63 antibodies were from Saierbio (Tianjin, China). Anti-Drosha and anti-Flag antibodies were from Abcam (Cambridge, United Kingdom).
Northern blotting.
Total cellular RNA was extracted. Five micrograms of total RNA was resolved in a 1% agarose gel containing 2.2 M formaldehyde and was then transferred onto a positively charged nylon membrane (GE Healthcare, Waukesha, WI). To detect HBV transcripts, a biotin-labeled specific RNA probe corresponding to nt 373 to 393 of the HBV genome, which was synthetized directly, was used for the detection of biotin luminescence using the chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) according to the manufacturer's instructions. The U6 RNA was used as a loading control.
HBV DNA analysis.
Huh7 cells were cotransfected with pHBV1.3 (1 μg), HBV-miR-3 (2 μg), HBV-miR-3-mut (2 μg), or the control vector pcDNA3 (2 μg), and HepG2.2.15 cells were transfected with ASO-HBV-miR-3 (40 pmol) or scrambled oligomer (ASO-NC; 40 pmol) in 6-well plates. Seventy-two hours posttransfection, extracellular encapsulated HBV DNA was extracted according to the protocol described previously (51) with modifications. Briefly, 500 μl of culture supernatants was treated with DNase I and MgCl2 for 2 h at 37°C to remove free DNA. In the presence of EDTA, SDS, NaCl, and proteinase K and following incubation for 4 h at 55°C overnight, viral DNA was isolated by phenol-chloroform extraction and ethanol precipitation. The DNA pellet was rinsed with 75% ethanol and resuspended in 10 μl Tris-EDTA (TE). The viral DNA was then analyzed by qPCR.
Intracellular HBV DNA was isolated as described previously (52), and the cells were washed with cold PBS and lysed in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.5] and 0.5% NP-40) at 4°C for 30 min. Following centrifugation, the supernatants were digested with DNase I (Promega, Madison, WI) and RNase A (Promega, Madison, WI) at 37°C for 2 h. The following procedures were similar to the processes of extracellular DNA extraction. All of the extracted DNA was loaded and separated on a 1% agarose gel. After standard denaturation and neutralization procedures, the DNA was transferred onto a positively charged nylon membrane (GE Healthcare, Waukesha, WI) and probed with a biotin-labeled HBV RNA probe, similar to the method for Northern blotting.
RNA immunoprecipitation assay.
The RIP assay was carried out following the method described by Song et al. (53). Briefly, Huh7 cells were cotransfected with pHBV1.3 (10 μg) and pcDNA3-Flag-AGO2 (10 μg) in a 75-cm2 culture flask. After transfection for 72 h, the cells were washed with cold PBS before lysis in 1 ml lysis buffer (50 mM Tris-HCl [pH 7.5], 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, 140 mM NaCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], 100 U/ml RNasin, and 1× proteinase inhibitor cocktail [Roche]). The lysates were placed on ice for 10 min and then centrifuged at 16,000 × g at 4°C for 5 min. Fifty microliters of protein A/G slurry and 10 μg of anti-Flag antibody or nonspecific control mouse IgG (Tianjin Saierbio, Inc., Tianjin, China) were added to the supernatants. After rotation at 4°C overnight, the samples were washed twice with lysis buffer. The pellet was then washed twice with high-salt lysis buffer (the same as lysis buffer except that NaCl was at 700 mM). The RNAs in the pellets were released from the samples after proteinase K treatment for 30 min at 50°C, extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and then precipitated in alcohol overnight. The isolated RNA was used for RT-qPCR as described above.
Exosome and HBV virion particle preparation.
To prepare exosomes, the culture supernatants of HepG2.2.15 cells were harvested by differential centrifugation as previously described (54–56), with some modifications. Briefly, culture supernatants were harvested and centrifuged at 500 × g for 10 min to eliminate cells and at 16,500 × g for 20 min to remove cell debris. The supernatants were pelleted by ultracentrifugation at 100,000 × g for 70 min and then washed with PBS and filtered through 0.45-μm filters and centrifuged again at 100,000 × g for 100 min, and the pellets were resuspended in 1 ml PBS. Then, sucrose gradient sedimentation was performed as described by Song et al. with some modifications (53). Briefly, 4 ml of 40%, 43%, 45%, and 50% (1 ml per fraction) sucrose gradients in 50 mM NH4Cl, 50 mM Tris-acetate at pH 7.0, 12 mM MgCl2, 150 mM NaCl, 100 μg/ml cycloheximide (added before use), and 1 mM DTT (added before use) was prepared 1 day prior to culture supernatant harvest. The supernatant was loaded on top of the sucrose gradient and centrifuged at 25,000 rpm for 4 h at 4°C. The density of HBV core particles was 1.22 g/ml, that of HBV empty particles was 1.18 (57), and that of the exosomes was 1.13 to 1.19 (56). After centrifugation, the top fractions (exosomes and HBV subviral particles) and the bottom fractions (HBV particles) (1 ml per fraction) were collected, diluted 6-fold with double-distilled water (DDW), and centrifuged at 110,000 × g for 2 h at 4°C; the supernatant was poured off, the pellets were resuspended in 1 ml PBS, and then the HBV particle pellets were extracted with TRIzol or RIPA; the exosome and HBV subviral particle pellets were immunoprecipitated using HBs monoclonal antibody (Tianjin Saierbio, Inc., Tianjin, China) to obtain the HBV subviral particles; then, the exosomes and HBV subviral particles were extracted with TRIzol or RIPA, respectively.
Statistical analysis.
Statistical significance was determined using a two-tailed unpaired Student t test. The data represent means and standard deviations (SD). P values of ≤0.05 were considered statistically significant. The graphs represent the average results of three experiments.
Accession number(s).
The GenBank accession number for the HBV-miR-3 sequence is KY684291.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 91629302, 91029714, 31270818, and 81572790) and the Natural Science Foundation of Tianjin (grant 12JCZDJC25100).
We declare no competing financial interests.
REFERENCES
- 1.Krol J, Loedige I, Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 3.Axtell MJ, Westholm JO, Lai EC. 2011. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol 12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, Yee AJ, Ang LC, He C, Shan SW, Yang BB. 2011. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-beta8. Oncogene 30:806–821. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 5.Su Y, Li X, Ji W, Sun B, Xu C, Li Z, Qian G, Su C. 2014. Small molecule with big role: microRNAs in cancer metastatic microenvironments. Cancer Lett 344:147–156. doi: 10.1016/j.canlet.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 6.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 8.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol 80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grey F, Meyers H, White EA, Spector DH, Nelson J. 2007. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog 3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog 3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Sewer A, Zavolan M, Nair V. 2007. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J Virol 81:7164–7170. doi: 10.1128/JVI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. 2007. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol 8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whisnant AW, Kehl T, Bao Q, Materniak M, Kuzmak J, Lochelt M, Cullen BR. 2014. Identification of novel, highly expressed retroviral microRNAs in cells infected by bovine foamy virus. J Virol 88:4679–4686. doi: 10.1128/JVI.03587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwig A, Das AT, Berkhout B. 2014. Retroviral microRNAs. Curr Opin Virol 7:47–54. doi: 10.1016/j.coviro.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Tycowski KT, Guo YE, Lee N, Moss WN, Vallery TK, Xie M, Steitz JA. 2015. Viral noncoding RNAs: more surprises. Genes Dev 29:567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trepo C, Chan HL, Lok A. 2014. Hepatitis B virus infection. Lancet 384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 17.Chemin I, Zoulim F. 2009. Hepatitis B virus induced hepatocellular carcinoma. Cancer Lett 286:52–59. doi: 10.1016/j.canlet.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Tiollais P, Pourcel C, Dejean A. 1985. The hepatitis B virus. Nature 317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 19.Beck J, Nassal M. 2007. Hepatitis B virus replication. World J Gastroenterol 13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schadler S, Hildt E. 2009. HBV life cycle: entry and morphogenesis. Viruses 1:185–209. doi: 10.3390/v1020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479-480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitab B, Alj HS, Ezzikouri S, Benjelloun S. 2015. MicroRNAs as important players in host-hepatitis B virus interactions. J Clin Transl Hepatol 3:149–161. doi: 10.14218/JCTH.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. 2010. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res 88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Bouchard MJ, Schneider RJ. 2004. The enigmatic X gene of hepatitis B virus. J Virol 78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang A, Ren F, Lei X, Zhang J, Guo R, Lu Z, Guo Y. 2015. The hepatitis B virus (HBV) core protein enhances the transcription activation of CRE via the CRE/CREB/CBP pathway. Antiviral Res 120:7–15. doi: 10.1016/j.antiviral.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Kwon JA, Rho HM. 2002. Hepatitis B viral core protein activates the hepatitis B viral enhancer II/pregenomic promoter through the nuclear factor kappaB binding site. Biochem Cell Biol 80:445–455. doi: 10.1139/o02-133. [DOI] [PubMed] [Google Scholar]
- 27.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD. 2013. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem 288:18484–18493. doi: 10.1074/jbc.M113.458158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. 2011. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One 6:e28486. doi: 10.1371/journal.pone.0028486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thery C, Zitvogel L, Amigorena S. 2002. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2:569–579. [DOI] [PubMed] [Google Scholar]
- 31.Dandri M, Murray JM, Lutgehetmann M, Volz T, Lohse AW, Petersen J. 2008. Virion half-life in chronic hepatitis B infection is strongly correlated with levels of viremia. Hepatology 48:1079–1086. doi: 10.1002/hep.22469. [DOI] [PubMed] [Google Scholar]
- 32.Ren ZJ, Nong XY, Lv YR, Sun HH, An PP, Wang F, Li X, Liu M, Tang H. 2014. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death Dis 5:e1387. doi: 10.1038/cddis.2014.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin WB, Wu FL, Kong D, Guo AG. 2007. HBV-encoded microRNA candidate and its target. Comput Biol Chem 31:124–126. doi: 10.1016/j.compbiolchem.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Nukui M, Mori Y, Murphy EA. 2015. A human herpesvirus 6A-encoded microRNA: role in viral lytic replication. J Virol 89:2615–2627. doi: 10.1128/JVI.02007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kincaid RP, Burke JM, Cox JC, de Villiers EM, Sullivan CS. 2013. A human torque teno virus encodes a microRNA that inhibits interferon signaling. PLoS Pathog 9:e1003818. doi: 10.1371/journal.ppat.1003818. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A 102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganem D, Prince AM. 2004. Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med 350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 41.Garcia T, Li J, Sureau C, Ito K, Qin Y, Wands J, Tong S. 2009. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J Virol 83:11152–11165. doi: 10.1128/JVI.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ponsel D, Bruss V. 2003. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol 77:416–422. doi: 10.1128/JVI.77.1.416-422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koschel M, Oed D, Gerelsaikhan T, Thomssen R, Bruss V. 2000. Hepatitis B virus core gene mutations which block nucleocapsid envelopment. J Virol 74:1–7. doi: 10.1128/JVI.74.1.1-7.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruss V. 2004. Envelopment of the hepatitis B virus nucleocapsid. Virus Res 106:199–209. doi: 10.1016/j.virusres.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. 2015. Core protein: a pleiotropic keystone in the HBV lifecycle. Antiviral Res 121:82–93. doi: 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Fan M, Geng G, Liu B, Huang Z, Luo H, Zhou J, Guo X, Cai W, Zhang H. 2014. A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology 11:23. doi: 10.1186/1742-4690-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Chen HC. 2010. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One 5:e12745. doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin X, Yuan ZH, Wu L, Ding JP, Wen YM. 2001. A single amino acid in the reverse transcriptase domain of hepatitis B virus affects virus replication efficiency. J Virol 75:11827–11833. doi: 10.1128/JVI.75.23.11827-11833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiang R, Wang F, Shi LY, Liu M, Chen S, Wan HY, Li YX, Li X, Gao SY, Sun BC, Tang H. 2011. Plexin-B1 is a target of miR-214 in cervical cancer and promotes the growth and invasion of HeLa cells. Int J Biochem Cell Biol 43:632–641. doi: 10.1016/j.biocel.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Tian Y, Chen WL, Ou JH. 2011. Effects of interferon-alpha/beta on HBV replication determined by viral load. PLoS Pathog 7:e1002159. doi: 10.1371/journal.ppat.1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao R, He J, Liu X, Gao G, Liu D, Cui L, Yu G, Yu W, Chen Y, Guo D. 2015. Inhibition of hepatitis B virus gene expression and replication by hepatocyte nuclear factor 6. J Virol 89:4345–4355. doi: 10.1128/JVI.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song G, Wang R, Guo J, Liu X, Wang F, Qi Y, Wan H, Liu M, Li X, Tang H. 2015. miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively. Sci Rep 5:15793. doi: 10.1038/srep15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. 1996. B lymphocytes secrete antigen-presenting vesicles. J Exp Med 183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 56.Mincheva-Nilsson L, Baranov V, Nagaeva O, Dehlin E. 2016. Isolation and characterization of exosomes from cultures of tissue explants and cell lines. Curr Protoc Immunol 115:14.42.11–14.42.21. doi: 10.1002/cpim.17. [DOI] [PubMed] [Google Scholar]
- 57.Kimura T, Ohno N, Terada N, Rokuhara A, Matsumoto A, Yagi S, Tanaka E, Kiyosawa K, Ohno S, Maki N. 2005. Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J Biol Chem 280:21713–21719. doi: 10.1074/jbc.M501564200. [DOI] [PubMed] [Google Scholar]
- 58.Bentley DR. 2006. Whole-genome re-sequencing. 2006. Curr Opin Genet Dev 16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]