Abstract
Objectives:
The rationale for directing targeted biopsy towards the centre of lesions has been questioned in light of prostate cancer grade heterogeneity. In this study, we assess the assumption that the maximum cancer Gleason grade (Gleason grade hotspot) lies within the maximum dimension (volume hotspot) of a prostate cancer lesion.
Methods:
3-D histopathological models were reconstructed using the outputs of the 5-mm transperineal mapping (TPM) biopsies used as the reference test in the pilot phase of Prostate Mri Imaging Study (PROMIS), a paired validating cohort study investigating the performance of multi-parametric magnetic resonance imaging (MRI) against transrectal ultrasound (TRUS) biopsies. The prostate was fully sampled with 5 mm intervals; each core was separately labelled, inked and orientated in space to register 3-D cancer lesions location. The data from the histopathology results were used to create a 3-D interpolated reconstruction of each lesion and identify the spatial coordinates of the largest dimension (volume hot spot) and highest Gleason grade (Gleason grade hotspot) and assess their concordance.
Results:
Ninety-four men, with median age 62 years (interquartile range, IQR= 58–68) and median PSA 6.5 ng ml−1 (4.6–8.8), had a median of 80 (I69–89) cores each with a median of 4.5 positive cores (0–12). In the primary analysis, the prevalence of homogeneous lesions was 148 (76% 95% confidence interval (CI) ±6.0%). In all, 184 (94±3.2%) lesions showed concordant hotspots and 11/47 (23±12.1%) of heterogeneous lesions showed discordant hotspots. The median 3-D distance between discordant hotspots was 12.8 mm (9.9–15.5). These figures remained stable on secondary analyses using alternative reconstructive assumptions. Limitations include a certain degree of error within reconstructed models.
Conclusions:
Guiding one biopsy needle to the maximum cancer diameter would lead to correct Gleason grade attribution in 94% of all lesions and 79% of heterogeneous ones if a true hit was obtained. Further correlation of histological lesions, their MRI appearance and the detectability of these hotspots on MRI will be undertaken once PROMIS results are released.
Introduction
Correct risk attribution is of key importance to guide appropriate management for men with prostate cancer. Our current diagnostic pathway based on transrectal ultrasound (TRUS) guided biopsy can result in inaccurate risk stratification in up to half of all men diagnosed.1 This can then result in missed diagnoses and under-treatment as well as the more commonly recognised issues of over-diagnosis and over-treatment.2, 3, 4, 5, 6
Recent research has focused on using multi-parametric magnetic resonance imaging (mpMRI) to improve the diagnostic accuracy by introducing tumour location at the time of biopsy.7, 8, 9 The provision of information on tumour location means that biopsies can be directed to the region of interest rather than be solely spread across the prostate as currently done with random TRUS biopsies. Some have argued that the ‘targeted approach' might result in employing fewer needle deployments than we have previously used.10, 11 If this is to happen—and it would be desirable if it could—then we would need to know where within the ‘target' to direct those needles.
It is customary when presented with a target to direct the needle to the centre of the target. However, some have raised concerns that this strategy may not be optimal.12, 13 Instead, it has been argued that information acquired from imaging can identify a particular area to target in order to obtain the most aggressive component of one lesion which may not be.
To further explore this question, we have analysed data obtained from the pilot phase of the MRC (Medical Research Council)/HTA (Health Technology Assessment) Prostate Mri Imaging Study (PROMIS), in which biopsy-naive men underwent mpMRI followed by a transperineal template prostate mapping (TPM) biopsy and TRUS biopsy. In this study, we attempted to assess the validity of the premise that the largest dimension of a tumour (volume hotspot) harbours the highest Gleason grade (Gleason grade hotspot).
Materials and methods
Design of the PROMIS trial
PROMIS is a multicentre paired validating cohort study (Trial registry identifiers: ISRCTN16082556, NCT01292291) funded by the UK NIHR (National Institute of Health Research)—HTA programme and designed to assess the diagnostic accuracy of an mpMRI-dependent pathway in detection of clinically significant prostate cancer.14
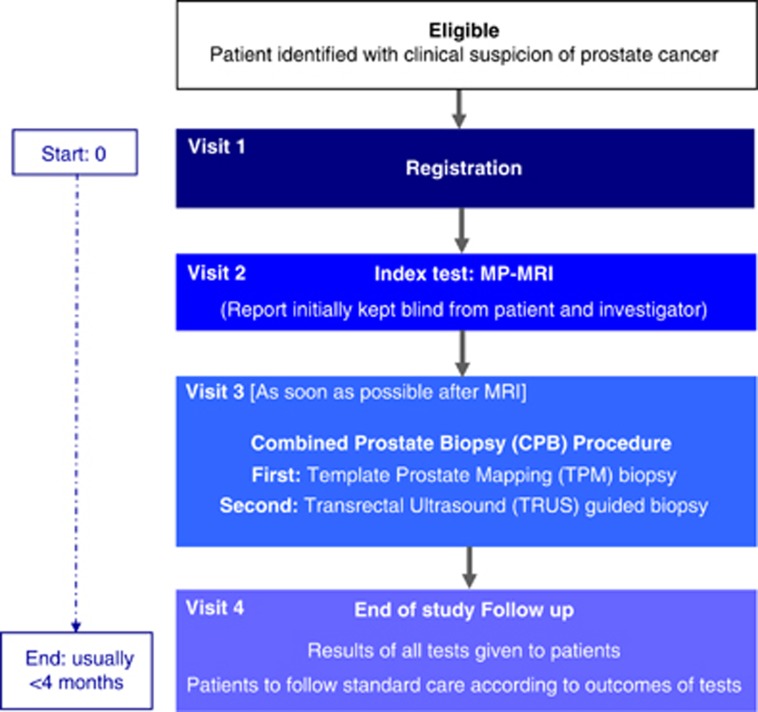
Biopsy-naive men who have been recommended a biopsy for suspicion of prostate cancer (elevated PSA, abnormal digital rectal examination, family history and/or ethnic risk group) are offered participation to the trial (Table 1). After an informed consent is obtained, patients undergo a standardised mpMRI (index test) protocol compliant to the European Society of Urogenital Radiology guidelines15 followed by a combined biopsy procedure involving TPM biopsy (reference test) and TRUS biopsy (standard test) under general anaesthetic (Figure 1). MpMRI results are blinded to clinicians performing the biopsies and to pathologists reporting the biopsy results.
Table 1. PROMIS inclusion and exclusion criteria.
| Patient inclusion criteria |
| Men at least 18 years or over at risk of prostate cancer who have been advised to have a prostate biopsy |
| Serum PSA ⩽15ng ml−1 within previous 3 months |
| Suspected stage ⩽T2 on rectal examination (organ confined) |
| Fit for general/spinal anaesthesia |
| Fit to undergo all protocol procedures including a transrectal ultrasound |
| Signed informed consent |
| Patient exclusion criteria |
| Treated using 5-alpha-reductase inhibitors at time of registration or during the prior 6 months |
| Previous history of prostate biopsy, prostate surgery or treatment for prostate cancer (interventions for BPH/bladder outflow obstruction are acceptable) |
| Evidence of a urinary tract infection or history of acute prostatitis within the last 3 months |
| Contraindication to MRI (e.g., claustrophobia, pacemaker, estimated GFR ⩽50) |
| Any other medical condition precluding procedures described in the protocol |
| Previous history of hip replacement surgery, metallic hip replacement or extensive pelvic orthopaedic metal work. |
Abbreviations: GFR, glomerular filtration rate; MRI, magnetic resonance imaging.
Figure 1.
Prostate Mri Imaging Study (PROMIS) trial schema. MP-MRI, multi-parametric magnetic resonance imaging.
For the purpose of this work, we have used the histological outputs from TPM biopsies conducted within the pilot phase of the trial. The pilot phase was unique in permitting such an analysis as each individual core was potted, processed and reported separately as well as oriented in space (cranio-caudal and x-y plane). The full trial incorporated 5 mm sampling but potted a number of cores together within prostate zones so would not permit such an analysis as this. Imaging results and its correlation to histology form the primary objective of the trial, we do not make any reference to imaging findings within this paper as our aim is to asses the validity of the premise that the highest Gleason score resides in the lesion's largest dimension, not if is consistent with a specific, identifiable imaging phenotype.
TPM biopsy
TPM biopsy has been chosen as the reference standard to validate mpMRI in PROMIS. It produces a histological map of the entire prostate in 3-dimensions with an estimated sensitivity and negative predictive value of around 95% (relative to prostatectomy) for clinically significant cancer.16, 17
In TPM, the prostate is sampled every 5 mm. This procedure has been described in detail elsewhere.18 Only within the pilot phase of PROMIS, each biopsy core was individually 3-D oriented in space in concordance to its location in the prostate by recording the brachytherapy template grid (‘x' and ‘y' planes) coordinates whilst the apical end of each core was stained with India ink to identify its cranio-caudal orientation (‘z' plane).
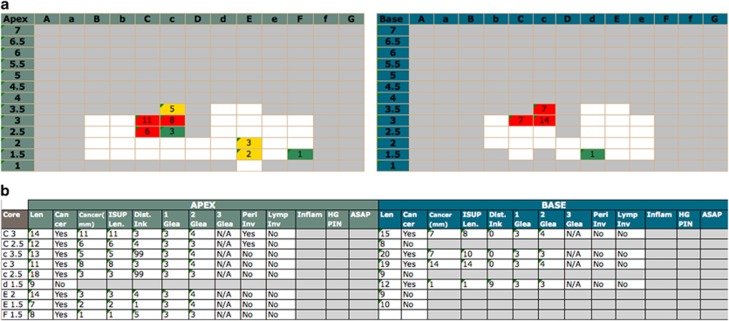
Each core was examined by an experienced uro-pathologist and reported in terms of core length, Gleason score, high-grade prostatic intraepithelial neoplasia and inflammation. The cancer core length (CCL) was reported with the distance from the apical aspect of the core. Results were plotted on a visual map with colour-coding to reflect the risk stratification derived from each core (Figures 2 and 3).19
Figure 2.
Transperineal mapping (TPM) histological findings per patient within the Prostate Mri Imaging Study (PROMIS) trial are resumed by a full report and by a visual report. In the full report (a), each core is labelled per coordinate and the following details are displayed: core length, cancer status, cumulative and separate cancer core length (CCL), cancer position, primary, secondary and tertiary Gleason grade, perineural and lymphovascular invasion as well as the presence of inflammation, high-grade PIN and ASAP. The TPM visual report (b) provides immediate zonal location within the gland. Maximum CCL and colour-coded risk attribution are displayed per coordinate with white boxes representing prostate biopsies with no cancer. ASAP, atypical small acinar proliferation; PIN, prostatic intraepithelial neoplasia.
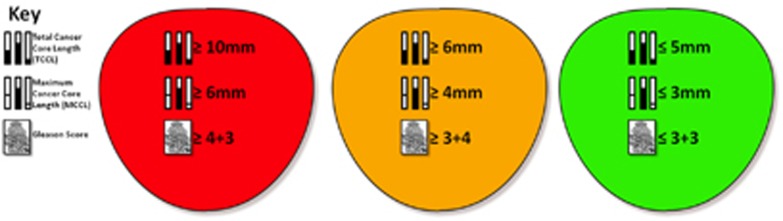
Figure 3.
UCL definitions of clinical significance on transperineal mapping (TPM) biopsy. Red indicates UCL definition 1 for significant disease (maximum cancer core length (MCCL)⩾6mm and/or Gleason score⩾4+3). Yellow indicates UCL definition 2 for significant disease (MCCL⩾4mm and/or Gleason score⩾3+4) and green defines insignificant disease (MCCL⩽3mm and Gleason score⩽3+3). UCL, University College London.
This reporting format presents unprecedented detail and clarity in recording the spatial position and relationship between positive cores, their Gleason grades and estimated lesion volumes enabling us to interpolate the results into a 3-D model.
Histological definitions
Volume hotspot
Volume hotspot is the coordinate in which if a biopsy needle is deployed it will sample the largest dimension of the lesion and return the longest CCL. The relationship between lesion volume and CCL is well demonstrated in Ahmed et al.'s19 previous work, and it is the basis of the lesion volume interpolation. It is determined across the sampling plane (cranio-caudal) of a template biopsy rather than on maximum dimension of the interpolated lesion, which may not be accessible from a TRUS or a template approach hence does not contribute to patient risk stratification.
Calculation of CCL
For the purpose of this study, decisions had to be taken to define how the CCL is calculated. There is no consensus with respect to which is the best method to define the CCL when discontinuous foci of cancer are present within the same core. Based on a recent survey around half pathologists consider that intervening benign tissue is not part of the cancer (separate count), whereas the remaining half count CCL from the initial part of the core with cancer to the end of the last cancer foci, regardless of the amount of benign tissue in between (cumulative count).20 In this study, we used separate counts within our primary analysis but also secondarily evaluated the impact of using the cumulative count.
Gleason hotspot
Gleason hotspot is the coordinate in which if a biopsy needle is deployed, it will capture the highest Gleason grade in the lesion independent of the overall lesion volume or Gleason score.
A homogeneous lesion is defined as a lesion compromised of only one Gleason pattern, and hence both Gleason and volume hotspots are inherently considered as concordant.
Aheterogeneous lesion is defined as a lesion composed of more than one Gleason pattern, and hence the volume and Gleason grade hotspots may not be at the same biopsy coordinates (non-Concordance).
On reporting biopsies, primary and secondary Gleason grade are reported on the basis of relative percentage rather than on fixed quantitative thresholds. Therefore, in the case of one lesion generated by the combination of various cores with the same total Gleason scores (ex: Gleason 7) but different amounts of each grade pattern per core (ex: Gleason 3+4, 40% grade 4), it is difficult to determine whether there is or there is not total Gleason score heterogeneity. For the purpose of our primary analysis, we considered the presence of Gleason 3+4 and Gleason 4+3 in different areas of the same lesion was a criterion for heterogeneity as there is some evidence that such a differentiation matters.21 For secondary analyses, we also assumed heterogeneity within one lesion was present only when different total Gleason scores were present on biopsy. In other words, Gleason 4+3 and Gleason 3+4 within the same lesion were considered as homogeneous.
3-Dimensional model/map
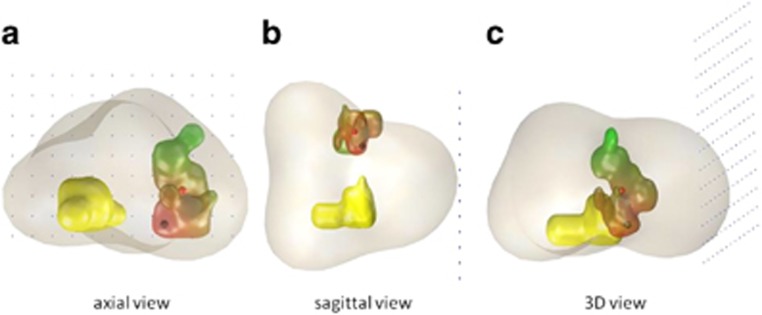
The 3-dimensional disease maps (Figures 4a and c) were reconstructed using the detailed pathological results from all specimens. This enabled the creation of a 3-D map which potentially has 13 × 13 × 40 sections of pathological results (in terms of Gleason scores), as 13 × 13 (5 mm) template grid holes are combined with two (apex and base) needle lengths of 20 mm. Individual lesions were delineated on the reconstructed 13 × 13 × 40 map by using the rule of 26 connectivity. This means that any block of positive samples is connected to 26 potential neighbour blocks to form a single lesion. This map was then further reconstructed into a finer spatial resolution (0.5 × 0.5 × 0.5 mm3) by linear interpolation followed by a Gaussian smoothing, whose single parameter of isotropic variation was tuned so that the original histology results when re-sampled at the same template grid sites will be preserved. This is the simplest reconstruction algorithm from relatively sparse data maintaining the clinical validity of reconstructed lesion maps. Simulations were performed on the reconstructed map to get CCLs and Gleason scores and determine their concordance. An animation of this modelling is available at https://sites.google.com/site/yipenghu/gallery/template-biopsy-animation.
Figure 4.
(a–c) Disease maps showing two different lesions within the same patient. One lesion (right side of the prostate; yellow) shows homogeneous grade; therefore, the hotspots are considered as concordant. The second lesion (left side of the prostate; scale of colours) shows grade heterogeneity with the Gleason grade hotspot located in the inferior right side of the lesion; therefore, the hotspots are considered as discordant.
Statistics
Descriptive statistics with continuous and categorical variables were analysed using median with interquartile range (IQR), and frequencies with percentages, respectively. Binomial 95% confidence intervals (CIs) were calculated. The data were analysed by statistical functions and procedures for descriptive statistics and significance testing, implemented in MATLAB 2014 (The MathWorks, Cambridge, UK) with Statistics Toolbox.
Results
Overall, 129 men were enrolled in the pilot phase of PROMIS. Thirty-five were excluded for various reasons: 19 chose to withdraw; 12 because it was deemed not possible to sample the entire prostate with a 5-mm density due to gland size; 3 due to co-morbidities that developed before biopsy; and finally 1 due to accidental un-blinding of his mpMRI. Therefore, 94 were included in the present study. Patients' characteristics are reported in Table 2. Median age was 62 years (IQR= 58–68) and median PSA was 6.5 ng ml−1 (4.6–8.8). A median of 80 cores (69–89) were taken per patient with a median of 4.5 positive cores (0–12). Median maximum cancer core length (MCCL) was 3 mm, both when using a cumulative (0–8) and a separate (0–7) CCL count. An example of a TPM report is given in Figure 2.
Table 2. Patients' characteristics.
| Variable | Value |
|---|---|
| No. of patients | 94 |
| Age, years, median (IQR) | 62 (58, 68) |
| PSA, ng ml−1, median (IQR) | 6.5 (4.6, 8.8) |
| Total no. of cores, median (IQR) | 80 (69, 89) |
| No. of positive cores, median (IQR) | 4.5 (0, 12) |
| Cancer core length (separate count), median (IQR) | 3 (0, 8) |
| Cancer core length (cumulative count), median (IQR) | 3 (0, 7) |
Abbreviation: IQR, interquartile range.
Primary analyses: for our primary analyses, which used the separate criteria for defining MCCL and Gleason grades, 195 independent lesions were detected (Table 3). The overall prevalence of homogeneous lesions was 148 (95%CI 76±6.0%). Most of these lesions had a Gleason score 3+3 (n=119; 61±6.9%), fewer had a Gleason score 3+4 (n=66; 34±6.6%), and a minority had a Gleason score 4+3 (n=10; 5±3.1%). Median lesion volume was 0.075 ml (0.025–0.225). Discordant hotspots were present in 11/47 (23±12.1%). The median 3-D distance between the hotspots when they were discordant was 12.8 mm (9.9–15.5).
Table 3. Results of analysis after interpolation.
| Variable | Primary analysis |
Secondary analysis |
||
|---|---|---|---|---|
| Definitions of CCL and heterogeneity | Separate count/Gleason grades | Separate count/Gleason score | Cumulative count/Gleason grades | Cumulative count/Gleason score |
| No. of independent lesions | 195 | 195 | 190 | 190 |
| Gleason score, no (±95% CI) | ||||
| 3+3 | 119 (61±6.9%) | 119 (61±6.9%) | 118 (62±6.9%) | 118 (62±6.9%) |
| 3+4 | 66 (34±6.6%) | 66 (34±6.6%) | 64 (34±6.7%) | 64 (34±6.7%) |
| 4+3 | 10 (5±3.1%) | 10 (5±3.1%) | 8 (4±2.9%) | 8 (4±2.9%) |
| Lesion volume, ml, median | 0.075 | 0.075 | 0.075 | 0.075 |
| IQR | (0.025–0.225) | (0.025–0.225) | (0.025–0.275) | (0.025–0.275) |
| Range | (0.025–6.200) | (0.025–6.200) | (0.025–8.275) | (0.025–8.275) |
| Homogeneous lesions, no. (±95% CI) | 148 (76±6.0%) | 152 (78±5.8%) | 144 (76±6.1%) | 148 (78±5.9%) |
| Heterogeneous lesions, no. (±95% CI) | 47 (24±6.0%) | 43 (22±5.8%) | 46 (24±6.1%) | 42 (22±5.9%) |
| Heterogeneous lesions with concordant hotspots, no. (±95% CI) | 36/47 (77±12.1%) | 33/43 (77±12.6%) | 34/46 (74±12.7%) | 33/42 (79±12.4%) |
| Heterogeneous lesions with no concordant hotspots, no. (±95% CI) | 11/47 (23±12.1%) | 10/43 (23±12.6%) | 12/46 (26±12.7%) | 9/42 (21±12.4%) |
| 3-D hotspots distance in heterogeneous non-concordant lesions, mm, median (IQR) | 12.8 (9.9–15.5) | 12.5 (9.9–15.8) | 11.5 (9.9–14.0) | 9.9 (9.8–15.3) |
| Total number of concordant lesions (±95% CI) | 184/195 (94±3.2%) | 185/195 (95±3.1%) | 178/190 (94±3.5%) | 181/190 (95±3.0%) |
Abbreviations: CCL, cancer core length; CI, confidence interval; IQR, interquartile range.
Overall, considering both homogeneous and heterogeneous lesions together, 184/195 (94±3.2%) of lesions harboured the Gleason grade hotspot in the volume hotspot.
Secondary analyses: Using separate criteria for defining the MCCL and the overall score to define heterogeneity, the results remained stable. Discordant hotspots were present in 10/43 (23±12.6%); the median 3-D distance between hotspots when they were discordant was 12.5 mm (9.9–15.8).
When the histology outputs were reconstructed to determine the 3-D models using the cumulative method to assign CCL, 190 independent lesions were found (Table 3). Most of these lesions had a Gleason score 3+3 (n=118; 62±6.9%), fewer had a Gleason score 3+4 (n=64; 34±6.7%), and a minority a Gleason score 4+3 (n=8; 4±2.9%). Median lesion volume was 0.075 ml (0.025–0.275). Between 144 (76±6.1%) and 148 (78±5.9%) lesions were considered as homogeneous, according to the definition of grade heterogeneity used. Of the remaining heterogeneous lesions, 33/42 (79±12.4%) and 34/46 (74±12.7%) had Gleason grade hotspots that were concordant to the volume hotspots. The median 3-D distance in the discordant lesions was 9.9 mm (9.8–15.3) and 11.5 mm (9.9–14), respectively.
The overall concordance rates of all secondary analyses when including all lesions were not different compared with the primary analysis.
Discussion
In summary, we have shown that the Gleason grade hotspot for a lesion is concordant with the volume hotspot in over 9 in 10 of all lesions. We also found that in biopsy-naive men, about one in five lesions are heterogeneous in grade. For these lesions, the Gleason grade and volume hotspot are discordant in about 2 in 10 lesions with approximately 10 mm distance between the two.
Before discussing the clinical implications of our findings, there are some limitations that need to be addressed. First, while this is a computer reconstruction based on precise 3-D pathology data, a certain degree of error is inevitable. It is possible that some very small lesions might have been missed, and the clustering of some lesions might have been incorrect. To minimise these errors, we used two methods for determining whether positive biopsies belonged to one specific lesion or not; this had minimal impact on our findings.
Second, these findings may be valid in this study population of biopsy-naive men with early suspicion of prostate cancer and PSA less than 15 ng ml−1, but it is likely, and it has been indeed previously shown that greater heterogeneity is present in men with more advanced disease.22, 23
Third, some may argue that radical prostatectomy specimen analysis should be used as a reference test within the trial. Although we have already clarified the reasons for choosing TPM biopsy as the reference test within the PROMIS trial elsewhere,14 we would argue that TPM biopsy represents a more valid tool as it can avoid the selection bias towards higher disease burden associated with the use of radical prostatectomy as a reference test. This is especially true in this subgroup of biopsy-naive men, in which a minority are expected to undergo radical prostatectomy.
Finally, our study was limited to the 94 men included in the pilot phase of the trial. Although this is an embedded study with no power calculation upfront, the PROMIS TMG approved this study as these very detailed 3-D histopathological maps could be built only for this subgroup of men, and awaiting trial completion would not add additional data to this study. Indeed, while patients recruited after the pilot phase had the same TPM procedure, there was a lack of some spatial information which are of key importance for the purpose of this study (core inking and precise 3-D orientation). This change in the histopathological analysis was due to resources and cost implications.
Clinical implications
Precise risk stratification remains a challenge. In a disease such as prostate cancer that exhibits such a degree of heterogeneity, and in which the course of the disease appears to be defined by the dominant Gleason pattern, the provision of tissue that enables its identification remains a key.24 The role of image-targeted biopsies has come to the fore as a way of possibly improving risk stratification. Targeted biopsies can be carried out in three possible ways: first, visually targeted, second using image-fusion software and third, in-bore (within the scanner). Recently, some have challenged the use of MRI to ultrasound registration, which currently directs the urologist's needle to the centre of a lesion by assuming that the most aggressive part of the tumour might not be in the centre of the lesion.12 One study showed that the most aggressive part of the prostate is not necessarily in the centre, and therefore the authors suggested targeted biopsies are best carried out in-bore. Our findings are somewhat contrasting to this report. We believe there might be some reasons for this discrepancy. First, the authors selected only men undergoing radical prostatectomy. We know that the larger the tumour the greater the heterogeneity.22, 23 This can be due to branching of a clone into different distinct phenotypes, but it can also be due to two adjacent, but clonally independent tumours, merging to form one.22, 25 Second, the authors based their results on retrospective radiological findings of diffusion coefficient heterogeneity, which does not always correspond to true histological heterogeneity.26, 27
Our study implies that the method of targeting and deploying a needle towards the centre of a lesion would lead to correct risk stratification in the vast majority. This reinforces the idea that MRI to ultrasound registration might be adequate to achieve correctly sampling, although the number of needles to be used to achieve a true hit in the centre of the tumour is yet to be determined taking into account that registration and operator errors can occur as well as dynamic swelling in between each needle deployment. Some early evidence from our group points to increasing accuracy of detection using up to five needle deployments per target.28
Conclusion
Our study demonstrates that guiding one biopsy needle to the maximum cancer diameter would lead to correct grade attribution in the majority of all lesions and approximately 80% of lesions heterogeneous for Gleason grade. Correlation of these histological lesions to their MRI appearance as well as the optimal biopsy needle deployment protocol requires further research.
Acknowledgments
PROMIS trial funding is provided by National Institute of Health Research-Health Technology Assessment: (Project number 09/22/67). The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the health technology assessment programme, NIHR, NHS or the Department of Health. Prostate Cancer UK supported funding for blood and urine collection and processing for the translational aspect of PROMIS (PROMIS-T). PROMIS is also supported and partially funded by UCLH/UCL Biomedical Research Centre and The Royal Marsden and Institute for Cancer Research Biomedical Research Centre and is coordinated by the Medical Research Council Clinical Trials Unit (MRC CTU) at UCL. It is sponsored by University College London (UCL).
Footnotes
ME and HUA would like to acknowledge funding from the Medical Research Council (UK), the Pelican Cancer Foundation charity, Prostate Cancer UK, St Peters Trust charity, Prostate Cancer Research Centre, the Wellcome Trust, National Institute of Health Research-Health Technology Assessment programme, and the US National Institute of Health-National Cancer Institute. ME receives funding in part from the UK National Institute of Health Research UCLH/UCL Comprehensive Biomedical Research Centre. ME and HUA receive funding from USHIFU, GSK, AngioDynamics and Advanced Medical Diagnostics for clinical trials. ME is a paid consultant to AngioDynamics, Steba Biotech and SonaCare Medical (previously called USHIFU). Both have previously received consultancy payments from Oncura/GE Healthcare and Steba Biotech. None of these sources had any input whatsoever into this article. YH is funded by the Health Technology Innovation Fund, a parallel funding partnership between the Wellcome Trust and the Department of Health. RGH has previously received payments from SonaCare Medical (formerly USHIFU) and Nuada Medical for consultancy work. The remaining authors declare no conflicts of interest.
References
- Shaw GL, Thomas BC, Dawson SN, Srivastava G, Vowler SL, Gnanapragasam VJ et al. Identification of pathologically insignificant prostate cancer is not accurate in unscreened men. Br J Cancer 2014; 110: 2405–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djulbegovic M, Beyth RJ, Neuberger MM, Stoffs TL, Vieweg J, Djulbegovic B et al. Screening for prostate cancer: systematic review and meta-analysis of randomised controlled trials. BMJ 2010; 341: c4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugosson J, Carlsson S, Aus G, Bergdahl S, Khatami A, Lodding P et al. Mortality results from the Goteborg randomised population-based prostate-cancer screening trial. Lancet Oncol 2010; 11: 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V et al. Prostate-cancer mortality at 11 years of follow-up. The New Engl J Med 2012; 366: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg L, Bill-Axelson A, Steineck G, Garmo H, Palmgren J, Johansson E et alResults from the Scandinavian Prostate Cancer Group. Trial Number 4: a randomized controlled trial of radical prostatectomy versus watchful waiting. J Natl Cancer Inst Monogr 2012; 2012: 230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt TJ. The Prostate Cancer Intervention Versus Observation Trial:VA/NCI/AHRQ Cooperative Studies Program #407 (PIVOT): design and baseline results of a randomized controlled trial comparing radical prostatectomy with watchful waiting for men with clinically localized prostate cancer. J Natl Cancer Inst Monogr 2012; 2012: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeks CM, Barentsz JO, Hambrock T, Yakar D, Somford DM, Heijmink SW et al. Prostate cancer: multiparametric MR imaging for detection, localization, and staging. Radiology 2011; 261: 46. [DOI] [PubMed] [Google Scholar]
- Sciarra A, Barentsz J, Bjartell A, Eastham J, Hricak H, Panebianco V et al. Advances in magnetic resonance imaging: how they are changing the management of prostate cancer. Eur Urol 2011; 59: 962–977. [DOI] [PubMed] [Google Scholar]
- de Rooij M, Hamoen EH, Futterer JJ, Barentsz JO, Rovers MM. Accuracy of multiparametric MRI for prostate cancer detection: a meta-analysis. AJR Am J Roentgenol 2014; 202: 343–351. [DOI] [PubMed] [Google Scholar]
- Kaufmann S, Kruck S, Kramer U, Gatidis S, Stenzl A, Roethke M et al. Direct comparison of targeted MRI-guided biopsy with systematic transrectal ultrasound-guided biopsy in patients with previous negative prostate biopsies. Urol Int 2014; 94: 319–325. [DOI] [PubMed] [Google Scholar]
- Valerio M, Donaldson I, Emberton M, Ehdaie B, Hadaschik BA, Marks LS et al. Detection of clinically significant prostate cancer using magnetic resonance imaging-ultrasound fusion targeted biopsy: a systematic review. Eur Urol 2014; 68: 8–19. [DOI] [PubMed] [Google Scholar]
- van de Ven WJ, Hulsbergen-van de Kaa CA, Hambrock T, Barentsz JO, Huisman HJ. Simulated required accuracy of image registration tools for targeting high-grade cancer components with prostate biopsies. Eur Radiol 2013; 23: 1401–1407. [DOI] [PubMed] [Google Scholar]
- Hambrock T, Hoeks C, Hulsbergen-van de Kaa C, Scheenen T, Fütterer J, Bouwense S et al. Prospective assessment of prostate cancer aggressiveness using 3- T diffusion-weighted magnetic resonance imaging–guided biopsies versus a systematic 10-core transrectal ultrasound prostate biopsy cohort. Eur Urol 2012; 61: 177–184. [DOI] [PubMed] [Google Scholar]
- El-Shater Bosaily A, Parker C, Brown LC, Gabe R, Hindley RG, Kaplan R et al. PROMIS - Prostate MR imaging study: a paired validating cohort study evaluating the role of multi-parametric MRI in men with clinical suspicion of prostate cancer. Contemp Clin Trials 2015; 42: 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barentsz JO, Richenberg J, Clements R, Choyke P, Verma S, Villeirs G et al. ESUR prostate MR guidelines 2012. Eur Radiol 2012; 22: 746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecornet E, Ahmed HU, Hu Y, Moore CM, Nevoux P, Barratt D et al. The accuracy of different biopsy strategies for the detection of clinically important prostate cancer: a computer simulation. J Urol 2012; 188: 974–980. [DOI] [PubMed] [Google Scholar]
- Crawford ED, Rove KO, Barqawi AB, Maroni PD, Werahera PN, Baer CA et al. Clinical-pathologic correlation between transperineal mapping biopsies of the prostate and three-dimensional reconstruction of prostatectomy specimens. Prostate 2013; 73: 778–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onik G, Barzell W. Transperineal 3D mapping biopsy of the prostate: an essential tool in selecting patients for focal prostate cancer therapy. Urol Oncol 2008; 26: 506–510. [DOI] [PubMed] [Google Scholar]
- Ahmed HU, Hu Y, Carter T, Arumainayagam N, Lecornet E, Freeman et al. Characterizing clinically significant prostate cancer using template prostate mapping biopsy. J Urol 2011; 186: 458–464. [DOI] [PubMed] [Google Scholar]
- Egevad L, Allsbrook WC Jr., Epstein JI. Current practice of diagnosis and reporting of prostate cancer on needle biopsy among genitourinary pathologists. Hum Pathol 2006; 37: 292–297. [DOI] [PubMed] [Google Scholar]
- Reese AC, Cowan JE, Brajtbord JS, Harris CR, Carroll PR, Cooperberg MR. The quantitative Gleason score improves prostate cancer risk assessment. Cancer 2012; 118: 6046–6054. [DOI] [PubMed] [Google Scholar]
- Qian J, Bostwick DG, Takahashi S, Borell TJ, Herath JF, Lieber MM et al. Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer Res 1995; 55: 5408–5414. [PubMed] [Google Scholar]
- Ruijter ET, van de Kaa CA, Schalken JA, Debruyne FM, Ruiter DJ. Histological grade heterogeneity in multifocal prostate cancer. Biological and clinical implications. J Pathol 1996; 180: 295–299. [DOI] [PubMed] [Google Scholar]
- Van der Kwast TH, Roobol MJ. Defining the threshold for significant versus insignificant prostate cancer. Nat Rev Urol 2013; 10: 473–482. [DOI] [PubMed] [Google Scholar]
- Algaba F, Montironi R. Impact of prostate cancer multifocality on its biology and treatment. J Endourol 2010; 24: 799–804. [DOI] [PubMed] [Google Scholar]
- Oto A, Yang C, Kayhan A, Tretiakova M, Antic T, Schmid-Tannwald C et al. Diffusion-weighted and dynamic contrast-enhanced MRI of prostate cancer: correlation of quantitative MR parameters with Gleason score and tumor angiogenesis. AJR Am J Roentgenol 2011; 197: 1382–1390. [DOI] [PubMed] [Google Scholar]
- Itou Y, Nakanishi K, Narumi Y, Nishizawa Y, Tsukuma H. Clinical utility of apparent diffusion coefficient (ADC) values in patients with prostate cancer: can ADC values contribute to assess the aggressiveness of prostate cancer? J Magn Reson Imaging 2011; 33: 167–172. [DOI] [PubMed] [Google Scholar]
- Robertson NL, Hu Y, Ahmed HU, Freeman A, Barratt D, Emberton M. Prostate cancer risk inflation as a consequence of image-targeted biopsy of the prostate: a computer simulation study. Eur Urol 2014; 65: 628–634. [DOI] [PMC free article] [PubMed] [Google Scholar]