Abstract
Aberrant DNA methylation is a critically important modification in cancer cells, which, through promoter and enhancer DNA methylation changes, use this mechanism to activate oncogenes and silence of tumor-suppressor genes. Targeting DNA methylation in cancer using DNA hypomethylating drugs reprograms tumor cells to a more normal-like state by affecting multiple pathways, and also sensitizes these cells to chemotherapy and immunotherapy. The first generation hypomethylating drugs azacitidine and decitabine are routinely used for the treatment of myeloid leukemias and a next-generation drug (guadecitabine) is currently in clinical trials. This review will summarize preclinical and clinical data on DNA hypomethylating drugs as a cancer therapy.
DNA hypomethylating drugs reprogram tumor cells to a normal-like state and sensitize the cells to chemotherapy and immunotherapy. Azacitidine and decitabine are used to treat myeloid leukemias; more drugs are under development.
DNA methylation occurs through covalent addition of a methyl group by DNA methyltransferases (DNMTs) to the C5 position of cytosine to form 5-methylcytosine (5mC). There are three members of DNMTs that have catalytic activity for DNA methylation—DNMT1, DNMT3A, and DNMT3B. DNMT1 is a maintenance methyltransferase that methylates preexisting hemimethylated DNA, whereas DNMT3A and 3B are de novo methyltransferases that establish methylation on unmethylated DNA (Baylin and Jones 2011). Other related members include DNMT3L, which lacks a catalytic domain and can modulate the activity of other DNMTs (Wienholz et al. 2010), and DNMT2, which has activity as a RNA methyltransferase (Goll et al. 2006). Demethylation of 5mC occurs through a reaction mediated by the three members of the ten-eleven trans location (TET) family, TET1, TET2, and TET3. TETs catalyze the conversion of 5mC to 5-hydroxymethylcytosine (5hmC), and 5hmC can be converted back to unmethylated cytosines either through active demethylation by the base excision repair pathway or passive demethylation by loss of 5hmC during cell division (Williams et al. 2012; Jin et al. 2015). DNA methylation can be recognized and bound by the methyl-binding proteins MBD1, MBD2, MBD4, and MeCP2. These MBDs form complexes with other epigenetic enzymes such as histone deacetylases (HDAC) and histone methyltransferases that catalyze the addition of histone modifications, which lead to compaction of the chromatin and silencing of gene expression (Parry and Clarke 2011).
The majority of DNA methylation occurs at cytosines that are followed by guanines (CpG sites), and there are certain regions in the genome where there is a high density of CpG sites that are termed CpG islands (CGIs). Fifty percent of gene promoters contain CGIs, and methylation of these CGIs has been associated with repression of gene expression (Taby and Issa 2010). In normal tissues, CpG sites throughout the genome are usually methylated, whereas promoter CGIs are usually unmethylated, with exceptions being the inactive X-chromosome, the silenced alleles of imprinted genes, and tissue-specific genes (Shen et al. 2007a; Smith and Meissner 2013).
CANCER AND DNA METHYLATION
Although DNA methylation in normal adult tissues is relatively stable, extensive DNA methylation changes occur in cancer that lead to global hypomethylation of the genome along with focal hypermethylation at promoter CGIs (Jones and Baylin 2007; Taby and Issa 2010; Jelinek et al. 2012). Five to ten percent of promoter CGIs become hypermethylated in most cancers, leading to silencing of many critical tumor-suppressor genes such as VHL, RB1, CDKN2A, MGMT, GATA4, and MLH1 (Taby and Issa 2010; Baylin and Jones 2011). In any given cancer, the number of genes showing promoter DNA methylation-associated gene silencing in cancer is substantially higher than those genes inactivated by genetic mutations, indicating that the majority of tumor-suppressor gene silencing in cancer is through epigenetic mechanisms (Plass et al. 2013). Global hypomethylation of the genome usually occurs in intergenic areas and does not have large effects on gene expression, but, in some cases, profound hypomethylation can lead to genomic instability through increased frequency of mutations, deletions, amplifications, inversions, and translocations (Chen et al. 1998).
One of the most well-known examples of the effect of DNA methylation on cancer occurs in patients categorized as having the CGI methylator phenotype (CIMP). CIMP was originally identified in colon cancer and indicates an abundance of cancer-specific hypermethylated promoter CGIs (Toyota et al. 1999). In colon cancer, CIMP is tightly correlated with BRAF mutations (Weisenberger et al. 2006; Shen et al. 2007b; Yagi et al. 2010; Hinoue et al. 2012; TCGA 2012), whereas in glioblastoma, CIMP tumors are associated with a high rate of IDH1 mutations and a better outcome (Noushmehr et al. 2010; Brennan et al. 2013). Further supporting the critical role of DNA methylation in cancer development, mutations in enzymes that regulate DNA methylation frequently occur in many hematological malignancies. DNMT3A has been found to be one of the most frequently mutated genes in hematological cancers (Yang et al. 2015). Loss-of-function mutations of TET2 or gain-of-function mutations of IDH1 or IDH2 frequently occur in hematological malignancies and are mutually exclusive (Abdel-Wahab et al. 2009; Delhommeau et al. 2009; Langemeijer et al. 2009; Gaidzik et al. 2012; Plass et al. 2013). IDH usually converts isocitrate to α-ketoglutarate, but mutant IDH will convert α-ketoglutarate to 2-hydroxyglutarate (2-HG) (Cohen et al. 2013), and 2-HG is an inhibitor of the TET proteins that mediates DNA demethylation (Xu et al. 2011). Patients with mutations in TET2 or IDH have specific hypermethylation signatures in both acute myelogenous leukemia (AML) (Figueroa et al. 2010; TCGA 2013; Yamazaki et al. 2015, 2016) and glioblastoma (Noushmehr et al. 2010; Turcan et al. 2012; Brennan et al. 2013).
DNA HYPOMETHYLATING DRUGS IN THE CLINIC
Given the reversibility of epigenetic modifications and the substantial DNA methylation changes that occur in cancer, it was hypothesized that DNA hypermethylation at promoter CGIs could be reversed to reexpress silenced genes and reprogram cancer cells to a more normal-like state.
This led to the pursuit of DNMT inhibitors for the treatment of cancer, and two DNMT inhibitors have had significant success in the clinic. They are the nucleoside analogs 5-aza-2′-deoxycytidine (decitabine) and 5-azacytidine (azacitidine) (Table 1). These inhibitors incorporate into newly synthesized DNA where they form a covalent bond with DNMTs, leading to the degradation of these DNMTs and hypomethylation of the genome through passive demethylation as the cells replicate and DNA methylation is not maintained (Issa and Kantarjian 2009). The main difference between these drugs is that decitabine incorporates into DNA, whereas azacitidine can incorporate into both DNA and RNA.
Table 1.
Hypomethylating drugs
| Chemical name | Generic name | Mechanism | Drug status |
|---|---|---|---|
| 5-Azacytidine | Azacitidine | Cytosine analog | FDA approved for treatment of MDS |
| 5-Aza-2′-deoxycytidine | Decitabine | Cytosine analog | FDA approved for treatment of MDS |
| SGI-110 | Guadecitabine | Cytosine analog | Phase III clinical trial in AML |
| 5-Fluro-2′-deoxycytidine | FdCyd | Cytosine analog | Phase II clinical trial in refractory solid tumors |
| Zebularine | – | Cytosine analog | Preclinical |
| CP-4200 | – | Cytosine analog | Preclinical |
| RG108 | – | Small molecule inhibitor | Preclinical |
| Nanaomycin A | – | Small molecule inhibitor | Preclinical |
MDS, Myelodysplastic syndrome; AML, acute myelogenous leukemia.
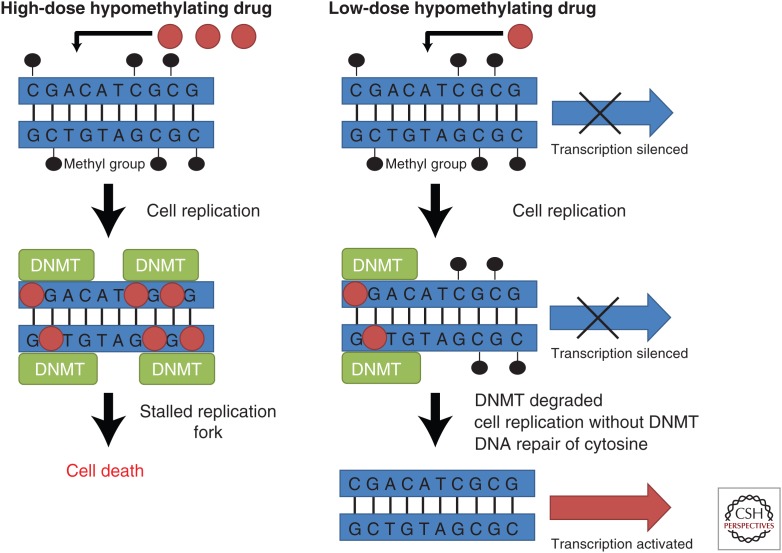
Decitabine and azacitidine were originally developed as cytotoxic anticancer agents in the 1960s, where they were used at high doses without clinical success (Taby and Issa 2010). However, further studies with these drugs led to the observation that they could cause differentiation of cells by inhibiting methylation of DNA (Jones and Taylor 1980). Importantly, it was discovered that hypomethylation after decitabine or azacitidine treatment did not occur at very high doses like the ones used previously in the clinic (Issa and Kantarjian 2009). These effects were attributed to the fact that high doses of decitabine and azacitidine would inhibit cell proliferation and DNA synthesis, and the incorporation of these drugs into DNA as well as their passive DNA demethylation effect were dependent on cell replication (Fig. 1) (Qin et al. 2007). Therefore, it was hypothesized that giving lower doses of the drug would lead to more effective DNA demethylation and improved clinical results (Issa and Kantarjian 2009). This hypothesis was supported by early phase clinical trials, which showed that repeated exposure with low doses of the inhibitors led to DNA demethylation and better responses than using the drugs at high doses (Wijermans et al. 2000; Issa et al. 2004).
Figure 1.
Comparison of the effects of high dose and low dose of hypomethylating drugs on cancer cells. When cells are treated with high doses of hypomethylating drugs, the drug gets incorporated in the DNA in a cell-replication-dependent manner, then binds and traps DNA methyltransferases (DNMTs). This causes the formation of bulky adducts, leading to a stalled replication fork and inhibition of DNA replication, which causes cell death. When cells are treated with low doses of hypomethylating drugs, the drugs still incorporate into DNA and bind the DNMTs, but the DNMTs end up being degraded. Without DNMTs to maintain DNA methylation, CpG sites lose their methylation after cell replication, and transcription of genes silenced by promoter methylation is restored.
These results eventually led to a phase III clinical trial of azacitidine in treating myelodysplastic syndrome (MDS) (Silverman et al. 2002). Patients that received azacitidine had an increased response rate (60% vs. 5%) and delayed progression to leukemia (21 mo vs. 13 mo) when compared with patients receiving supportive care (Silverman et al. 2002). Following these results, the FDA approved the use of azacitidine in 2004 for the treatment of patients with MDS (Taby and Issa 2010). A follow-up international phase III clinical trial confirmed the efficacy of azacitidine in treating MDS, as patients treated with azacitidine had higher median overall survival (24.5 mo) compared with patients receiving conventional care (15 mo) (Fenaux et al. 2009).
Meanwhile, a phase III clinical trial of decitabine in treating MDS resulted in higher response rate (17% vs. 0%) and longer median time to leukemia or death (12.1 mo vs. 7.8 mo) in patients treated with decitabine compared with best supportive care (Kantarjian et al. 2006). These results led to the 2006 FDA approval of decitabine in the treatment of patients with MDS (Taby and Issa 2010). Follow-up studies that optimized the dosing schedule showed a very high response rate (>70% objective response) of MDS patients treated with decitabine and increased survival compared with patients treated with chemotherapy (22 mo vs. 12 mo) (Kantarjian et al. 2007a,b). A more recent phase III trial performed in elderly patients with MDS that are ineligible for intensive chemotherapy showed that patients treated with decitabine had increased progression-free survival (6.6 mo vs. 3 mo) and increased overall response rate (34% vs. 2%) when compared with patients receiving the best supportive care (Lübbert et al. 2011).
DNMT inhibitors have also shown effectiveness in treating patients with other leukemias. A phase II study in patients with chronic myelogenous leukemia (CML) showed complete response in 34% and partial response in 20% of patients treated with decitabine (Issa et al. 2005). Activity was also seen in AML, as a multicenter phase III trial performed on AML patients treated with decitabine showed an increase in overall survival (7.7 mo vs. 5.0 mo) and CR rate (17.8% vs. 7.8%) when compared with treatment choice (Kantarjian et al. 2012). These results led to the approval of decitabine in AML in Europe.
The effect of decitabine and azacitidine has a slower onset than traditional cytotoxic therapies, which usually require only one cycle of treatment to achieve a complete remission. For example, one study reported that although median time to first response to azacitidine was two cycles, continued treatment improved response in 48% of patients (Silverman et al. 2011). Another study reported that patients that show no evidence of response to decitabine after 3 mo can end up achieving complete remission with continued treatment (Oki et al. 2008). The delayed onset of response is consistent with the mechanism of action of decitabine and azacitidine, as the hypomethylation that occurs in response to inhibition of DNMTs is dependent on cell replication.
Treating solid tumors with DNMT inhibitors remains a challenge because solid cancers tend to have lower drug penetrance in larger tumors and often proliferate at slower rate than hematological cancers, which is problematic for DNMT inhibitors that are known to be S-phase-dependent and unstable in solution (Issa and Kantarjian 2009). However, there is evidence that DNMT inhibitors can be effective in inhibiting the growth of solid tumors in vitro (Bender et al. 1998; Qin et al. 2009; Tsai et al. 2012). Recent clinical trials of decitabine in solid tumors (Stewart et al. 2009) and azacitidine in combination with an HDAC inhibitor on non-small-cell lung cancer patients showed some objective responses (Juergens et al. 2011). There are ongoing clinical trials to determine whether using hypomethylating drugs as a monotherapy or in combination with other agents can be an effective therapy for solid tumors.
Collectively, these data indicate that hypomethylating drugs are an important therapy that acts through epigenetic mechanisms to target cancer instead of inducing general toxicity as seen with other chemotherapies. Although the mechanism of action of decitabine and azacitidine are proposed to be very similar, it is yet to be determined whether these two drugs will lead to different clinical outcomes because they have never been directly compared. There are some indications that the two drugs can have differing effects, as one study showed that some MDS patients resistant to azacitidine could be treated with decitabine and achieve objective responses (Borthakur et al. 2008). Regardless, both drugs remain important agents in the clinical treatment of MDS and leukemia, and quite possibly for solid tumors in the future.
RESISTANCE TO DNA HYPOMETHYLATING DRUGS
Despite the initial success of DNMT inhibitors in the treatment of MDS, lack of initial response to therapy (primary resistance), or acquired resistance after robust initial responses (secondary resistance) remains a major obstacle (Issa and Kantarjian 2009). Only ∼50% of MDS or AML patients treated with hypomethylating drugs achieve a clinical response (Treppendahl et al. 2014). Therefore, efforts have been made to find predictive biomarkers to DNMT inhibitors, but there has been limited correlation observed with biomarkers such as DNA methylation before therapy or hypomethylation of tumor-suppressor genes after therapy (Oki et al. 2007; Fandy et al. 2009). Because all cancer patients have methylation defects, the key to correlating DNA methylation with response may be to find the right set of methylated gene promoters to use as biomarkers or to study methylated sequences outside the promoter region that may control gene expression, such as enhancers. Interestingly, a recent study found responders and nonresponders to decitabine could be distinguished in 40 chronic myelomonocytic leukemia (CMML) patients by using 167 differentially methylated regions that were primarily located in distal intergenic regions and enhancers (Meldi et al. 2015). It will be important to determine whether this observation can be replicated in a larger set of patients and in other leukemias. On the other hand, measuring hypomethylation of genes after therapy may not be the best approach to use as biomarkers because cells that are most sensitive to therapy and become hypomethylated may be the first to die off, leaving behind only the cells resistant to hypomethylation (Qin et al. 2007).
Mutations in proteins involved in regulation of DNA methylation have been tested as biomarkers for response to hypomethylating drugs, but the results have been mixed (Treppendahl et al. 2014). For example, two studies have found that MDS or AML patients with TET2 mutations were more likely to respond to hypomethylating drugs (Itzykson et al. 2011; Bejar et al. 2014), but two other studies have found limited correlation between TET2 mutations and response in MDS and CMML patients (Braun et al. 2011; Voso et al. 2011). Another recent study on patients with MDS and related disorders found a correlation between response to azacitidine or decitabine with TET2 or DNMT3A mutations (Traina et al. 2014). Because of to these conflicting reports, more studies are needed to establish whether TET2 or DNMT3A mutations can serve as a biomarker for hypomethylating drugs.
One cause of primary resistance to hypomethylating drugs was discovered when it was observed that cell lines that were resistant to decitabine in vitro had low deoxycytidine kinase (DCK), which activates decitabine through phosphorylation, and high cytosine deaminase (CDA), which inactivates decitabine through deamination (Qin et al. 2009). When resistance to decitabine was investigated in MDS patients, it was found that patients with primary resistance had a threefold higher CDA/DCK ratio (Qin et al. 2011). Another factor for the primary resistance may be the instability of DNMT inhibitors, because decitabine and azacitidine are rapidly cleared from the body and have a half-life less than 1 h (Derissen et al. 2013). In terms of secondary resistance, it was observed that a stable clone that initially responded to decitabine but gained secondary resistance had mutations in DCK (Qin et al. 2009). However, this mutation was not observed in any of the MDS patients that were treated with decitabine and developed secondary resistance (Qin et al. 2011).
EXPERIMENTAL STUDIES ON THE EFFECT OF DNA HYPOMETHYLATING DRUGS
There has been major progress made in experimental settings to better understand the effects of DNMT inhibitors. Studies using 27K or 450K methylation arrays (Tsai et al. 2012; Klco et al. 2013; Pandiyan et al. 2013), as well as with whole genome bisulfite sequencing (Lund et al. 2014) have shown that DNA demethylating agents cause global hypomethylation throughout the entire genome. However, these studies have also shown that only a small percentage of genes that undergo promoter hypomethylation actually become reactivated (Tsai et al. 2012; Klco et al. 2013; Pandiyan et al. 2013; Lund et al. 2014). This is most likely because other factors such as expression of the right transcription factor, changes in histone acetylation, changes in histone methylation, or chromatin remodeling are required on top of DNA demethylation for reactivation of gene expression, giving DNMT inhibitors a degree of specificity (Kondo et al. 2004; Paul et al. 2010; Si et al. 2010; Pandiyan et al. 2013). In fact, genes that gained chromatin accessibility after decitabine treatment were more likely to be genes that are expressed in normal tissue and down-regulated in cancer (Pandiyan et al. 2013).
In contrast to DNA methylation in the promoter, methylation in the gene body is usually associated with activation of gene expression (Maunakea et al. 2010; Jones 2012; Kulis et al. 2012; Varley et al. 2013). Because decitabine and azacitidine lead to global hypomethylation of the genome, these drugs also affect gene body methylation. This was shown by a recent study showing that decitabine can down-regulate expression of genes regulated by c-MYC as well as genes involved in metabolic processes through demethylation of gene bodies (Yang et al. 2014). This study indicated that, in addition to the therapeutic benefit gained from reactivation of silenced tumor-suppressor genes, hypomethylating drugs can also have a dual benefit by down-regulating oncogenes and metabolic genes (Yang et al. 2014).
Hypomethylating drugs can have very long-term effects on gene expression even after removal of the drug. The expression of a silenced, stably integrated GFP could still be detected 3 mo after initial decitabine treatment in a colon cancer model (Raynal et al. 2012). Similarly, the expression of a subset of alleles remained demethylated for more than 3 mo after treatment with decitabine in a breast cancer model (Kagey et al. 2010). These findings were also observed in a third study, in which it was found that low-dose DNMT inhibitors cause long-term gene-expression changes of hypermethylated genes involved in key antitumor pathways such as apoptosis, increased lineage commitment, and down-regulation of cell cycling (Tsai et al. 2012). Importantly, these long-term gene changes led to inhibition of cancer-initiating cells and tumor growth (Tsai et al. 2012).
In addition to the effect that hypomethylating drugs have on cancer cells, these drugs also affect the tumor microenvironment. For example, one study observed decreased angiogenesis after decitabine treatment because of inhibition of endothelial cell proliferation (Hellebrekers et al. 2006). Another study found that decitabine treatment increased expression of THBS1, which is an antiangiogenesis factor commonly methylated and silenced in various tumors (Li et al. 1999). Consistent with these findings, combining azacitidine with lenalidomide, an FDA-approved therapy for MDS that has been shown to mediate some of its effects through affecting the tumor microenvironment, has shown promise in a phase II clinical trial on higher risk MDS patients (Sekeres et al. 2012).
IMMUNE RESPONSE MEDIATED BY DNA HYPOMETHYLATING DRUGS
Cancer cells avoid detection by the host immune system through altered expression of tumor-associated antigens and secretion of cytokines, which leads to deficient antigen presenting cells and cytolytic T cells (Heninger et al. 2015). DNA methylation is responsible for silencing of many immune-related genes, and DNMT inhibitors have been shown to induce expression of these genes in various experimental settings and clinical trial studies (Karpf et al. 1999; James et al. 2013; Wrangle et al. 2013; Odunsi et al. 2014; Heninger et al. 2015). For example, in a study looking at 63 cancer cell lines derived from breast, colorectal, and ovarian cancers that were treated with low-dose azacitidine for 3 days, it was found that genes commonly up-regulated by azacitidine were enriched for immunomodulatory genes and pathways such as cancer testis antigens (CTAs), interferon signaling, antigen presentation, inflammation, and cytokine signaling (Li et al. 2014). These immunomodulatory pathway genes were grouped together and termed the azacitidine immune gene set (AIMs). Patients in clinical trials that were treated with azacitidine and the HDAC inhibitor entinostat for 8 weeks had a higher expression of these AIM genes (Li et al. 2014).
Two recent studies have clarified the mechanism of the DNMT inhibitor-mediated immune response by showing that DNMT inhibitors could demethylate and induce endogenous retroviral sequences (ERVs), which led to activation of the cellular antiviral response (Chiappinelli et al. 2015; Roulois et al. 2015). One study looked at colorectal cancer cells treated with low-dose azacitidine, and found that there was a group of genes that were still highly expressed 42 days after drug withdrawal (Roulois et al. 2015). These genes were enriched in the interferon response and MDA5/MAVS RNA recognition pathway, and the up-regulation of these genes was mediated by hypomethylation of endogenous retroviral elements, which led to the expression of viral dsRNAs (Roulois et al. 2015). This viral recognition pathway was essential for the ability of azacitidine to inhibit cancer-initiating cells (Roulois et al. 2015). A second study studying ovarian cancer cells also found that the RNA-sensing pathways MAVS and TLR3 were up-regulated after azacitidine treatment, and this was a result of increased demethylation and expression of ERVs (Chiappinelli et al. 2015). Importantly, melanoma patients treated with immune checkpoint therapy with a high viral defense gene-expression signature had better clinical response, and mice pretreated with azacitidine had an amplified response to anti-CTLA4 immune checkpoint therapy, indicating that patients that lack the viral defense gene-expression signature may benefit from treatment with hypomethylating drugs before treatment with immunotherapy (Chiappinelli et al. 2015).
Given these findings relating cancer immune response to DNMT inhibitors, there is an ongoing phase II clinical trial to study the efficacy of the PD-1 inhibitor nivolumab on lung cancer patients pretreated with DNMT inhibitors (NCT01928576). These findings are also intriguing because biomarkers to predict response to DNMT inhibitors have been lacking as mentioned previously. Incorporating the expression of immune response genes affected by DNMT inhibitors with the expression of tumor-suppressor genes reactivated by DNMT inhibitors may give us a better ability to predict patients and cancers that will have the optimal response to hypomethylating drugs.
COMBINATION THERAPY WITH DNA HYPOMETHYLATING DRUGS
HDAC inhibitors have had success in the clinic for treating cutaneous T-cell lymphomas (Duvic et al. 2007; Whittaker et al. 2010), and combining DNMT inhibitors with HDAC inhibitors led to synergistic effects on reactivation of silenced tumor-suppressor genes (Cameron et al. 1999; Kalac et al. 2011). These observations led to the testing of this combination in numerous clinical trials. Because DNMT inhibitors are dependent on cell replication for its hypomethylating action and HDAC inhibitors lead to cell-cycle arrest, the effect of the combination is maximized when cells are treated with DNMT inhibitors first then with HDAC inhibitors, and clinical trials have mainly been tested using this dosing schedule. However, clinical trials have shown limited evidence of this combination being effective in improving patient survival (Garcia-Manero et al. 2006; Gore et al. 2006; Blum et al. 2007; Lin et al. 2009; Prebet et al. 2014; Issa et al. 2015a). For example, a recent phase II clinical trial in patients with MDS or AML showed that patients receiving only azacitidine had a median overall survival of 18 mo compared with 13 mo for the patients receiving the combination of azacitidine and entinostat (Prebet et al. 2014). Another phase II clinical trial in patients with MDS or AML showed that patients receiving only decitabine had a CR rate of 31% with overall response rate of 51%, compared with patients receiving decitabine along with valproic acid who had a CR rate of 37% and an overall response rate of 58%, neither of which were significantly improved over decitabine alone (Issa et al. 2015a). The median survival of patients receiving decitabine compared with decitabine plus valproic acid also did not have a significant improvement (11.2 mo vs. 11.9 mo). The lack of clinical success of combining these two agents may be because of the large, nonspecific gene-expression changes that occur when treating cells with HDAC inhibitors (Peart et al. 2005; LaBonte et al. 2009; Chueh et al. 2015).
Other agents that DNMT inhibitors have been combined with in clinical trials include lenalidomide, carboplatin, cisplatin, gemtuzumab ozogamicin, erythropoietin, filgrastim, romiplostim, bortezomib, arsenic trioxide, and sorafinib (Blum et al. 2012; Sekeres et al. 2012; Greenberg et al. 2013; Ravandi et al. 2013; Glasspool et al. 2014; Navada et al. 2014; Daver et al. 2015). These trials have produced mixed results about whether the combinations are beneficial and require further investigation, although the trials investigating the sensitization of cancers to platinum compounds by pretreatment with hypomethylating drugs have been particularly promising (Matei et al. 2012). The combination of decitabine with platinum compounds such as carboplatin is made even more intriguing by the fact that this combination showed increased epigenetic activity in reactivation of silenced tumor-suppressor genes such as MLH1 and PDLIM4 (Qin et al. 2015). This epigenetic synergy was found to be mediated through inhibition of HP1α expression by the platinum compounds, which led to reduced binding by MeCP2 and MBD2 (Qin et al. 2015). Similarly, combining arsenic trioxide or cardiac glycosides with hypomethylating drugs may have potential because a recent study showed that these drugs can also reactivate epigenetically silenced tumor-suppressor genes (Raynal et al. 2015). There are other epigenetic therapies in clinical development such as EZH2 inhibitors (McCabe et al. 2012; Knutson et al. 2013), LSD1 inhibitors (Mohammad et al. 2015), and BET inhibitors (Filippakopoulos et al. 2010), which have shown promising results in early clinical trials. It will be intriguing to explore whether these agents could have additive or synergistic effects in the clinic when combined with DNMT inhibitors because they may lead to synergistic gene reactivation as seen with the combination of hypomethylating drugs with HDAC inhibitors.
DEVELOPMENT OF NOVEL DNA HYPOMETHYLATING DRUGS
After the success of decitabine and azacitidine in the clinic, efforts were made to develop more effective hypomethylating drugs (Table 1). Zebularine is a cytidine analog similar to decitabine and azacitidine that incorporates into DNA and forms a reversible complex with DNMTs, effectively preventing methylation (Zhou et al. 2002; Champion et al. 2010). RG108 is a small molecular inhibitor discovered through virtual screening that binds to the catalytic site of DNMT1 and shows reactivation of silenced tumor-suppressor genes such as P16, SFRP1, and TIMP3 (Brueckner et al. 2005). CP-4200 is an elaidic acid derivative of azacitidine that allows entry into the cell independent of nucleoside transporters (Brueckner et al. 2010). Nanaomycin A is a small molecular inhibitor that has been shown to be selective for DNMT3B (Kuck et al. 2010). FdCyd is a nucleoside analog that has shown the ability to inhibit DNMTs and is undergoing early-stage clinical trials but requires simultaneous administration of tetahyrdrouridine to reduce enzymatic deamination (NCT00978250) (Newman et al. 2015). Further studies are needed to determine whether any of these inhibitors are more effective than azacitidine or decitabine in their ability to demethylate and reduce growth of tumors.
The most advanced of these novel DNA hypomethylating drugs has been SGI-110 (guadecitabine). Guadecitabine was designed to overcome one of the main limitations of decitabine and azacitidine, which is its instability in the human body because of degradation by CDA. This was accomplished by developing a compound with the structure of decitabine linked to a deoxyguanosine, which made it into a more stable compound resistant to this degradation (Griffiths et al. 2013). In experimental settings, guadecitabine had improved bioavailability and increased half-life while still maintaining the ability to act as a hypomethylating drug (Griffiths et al. 2013). In clinical trials, guadecitabine has shown promising results in a phase I clinical trial involving 93 patients with either AML or MDS (Issa et al. 2015b). Guadecitabine was well tolerated and maximal demethylation occurred at 60 mg/m2 with no additional demethylation at higher doses, similar to previous clinical data from decitabine and azacitidine. Six out of 19 MDS patients and six out of 74 AML patients had a clinical response to SGI-110. Importantly, there was a strong correlation between response and demethylation, indicating that guadecitabine is reducing tumor growth by acting as a hypomethylating drug (Issa et al. 2015b). Guadecitabine is now in phase II–III clinical trials in MDS and AML (NCT02096055 and NCT01261312).
CONCLUDING REMARKS
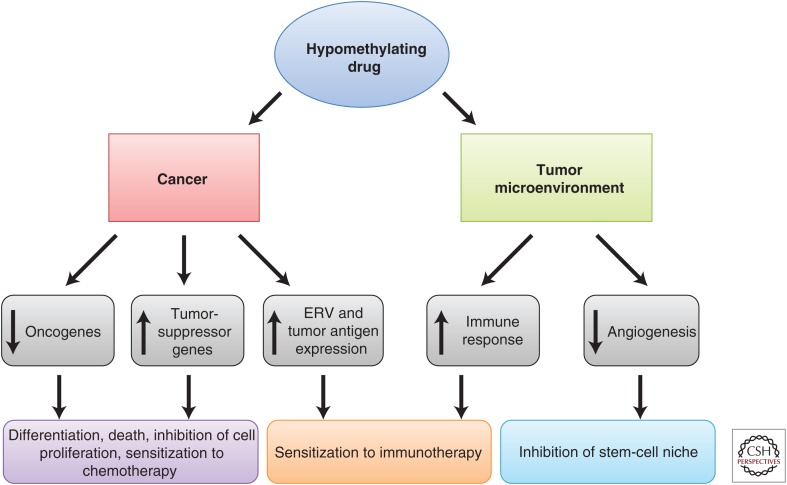
Modifications in DNA methylation is a critically important process in tumor initiation as well as progression. Targeting DNA methylation using DNMT inhibitors has changed the way we treat cancer, as it is an established therapy for MDS and AML. Hypomethylating drugs can be used to target cancer through effects on both the cancer itself and the surrounding tumor microenvironment (Fig. 2). Our increase in understanding of the mechanisms of the DNMT inhibitors as well as the degree and type of aberrant DNA methylation in different cancers should lead to even more clinical success in the future as we switch from using cytotoxic therapies to targeted therapies that can reverse the cancers back to a more normal-like state.
Figure 2.
Hypomethylating drugs have effects on both the cancer itself and the surrounding microenvironment. In cancer cells, hypomethylating drugs can lead to reactivation of tumor-suppressor genes through demethylation of gene promoters, reactivation of oncogenes through demethylation of gene bodies, and reactivation of endogenous retroviral sequence (ERV) and tumor antigen expression through demethylation of these elements. These events lead to cell differentiation, death, inhibition of cell proliferation, sensitization to chemotherapy, and sensitization to immunotherapy. In the tumor microenvironment, hypomethylating drugs lead to increases in immune response and decreases in angiogenesis, which can subsequently lead to sensitization to immunotherapy and inhibition of the stem-cell niche.
Footnotes
Editors: Scott A. Armstrong, Steven Henikoff, and Christopher R. Vakoc
Additional Perspectives on Chromatin Deregulation in Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. 2009. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114: 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome—Biological and translational implications. Nat Rev Cancer 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Lord A, Stevenson K, Bar-Natan M, Perez-Ladaga A, Zaneveld J, Wang H, Caughey B, Stojanov P, Getz G, et al. 2014. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood 124: 2705–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CM, Pao MM, Jones PA. 1998. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res 58: 95–101. [PubMed] [Google Scholar]
- Blum W, Klisovic RB, Hackanson B, Liu Z, Liu S, Devine H, Vukosavljevic T, Huynh L, Lozanski G, Kefauver C, et al. 2007. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol 25: 3884–3891. [DOI] [PubMed] [Google Scholar]
- Blum W, Schwind S, Tarighat SS, Geyer S, Eisfeld AK, Whitman S, Walker A, Klisovic R, Byrd JC, Santhanam R, et al. 2012. Clinical and pharmacodynamic activity of bortezomib and decitabine in acute myeloid leukemia. Blood 119: 6025–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthakur G, Ahdab SE, Ravandi F, Faderl S, Ferrajoli A, Newman B, Issa JP, Kantarjian H. 2008. Activity of decitabine in patients with myelodysplastic syndrome previously treated with azacitidine. Leuk Lymphoma 49: 690–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Itzykson R, Renneville A, de Renzis B, Dreyfus F, Laribi K, Bouabdallah K, Vey N, Toma A, Recher C, et al. 2011. Molecular predictors of response to decitabine in advanced chronic myelomonocytic leukemia: A phase 2 trial. Blood 118: 3824–3831. [DOI] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. 2013. The somatic genomic landscape of glioblastoma. Cell 155: 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. 2005. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res 65: 6305–6311. [DOI] [PubMed] [Google Scholar]
- Brueckner B, Rius M, Markelova MR, Fichtner I, Hals PA, Sandvold ML, Lyko F. 2010. Delivery of 5-azacytidine to human cancer cells by elaidic acid esterification increases therapeutic drug efficacy. Mol Cancer Ther 9: 1256–1264. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21: 103–107. [DOI] [PubMed] [Google Scholar]
- Champion C, Guianvarc’h D, Senamaud-Beaufort C, Jurkowska RZ, Jeltsch A, Ponger L, Arimondo PB, Guieysse-Peugeot AL. 2010. Mechanistic insights on the inhibition of c5 DNA methyltransferases by zebularine. PLoS ONE 5: e12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. 1998. DNA hypomethylation leads to elevated mutation rates. Nature 395: 89–93. [DOI] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh AC, Tse JW, Togel L, Mariadason JM. 2015. Mechanisms of histone deacetylase inhibitor-regulated gene expression in cancer cells. Antioxid Redox Signal 23: 66–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Holmen SL, Colman H. 2013. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep 13: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daver N, Kantarjian H, Ravandi F, Estey E, Wang X, Garcia-Manero G, Jabbour E, Konopleva M, O’Brien S, Verstovsek S, et al. 2015. A phase II study of decitabine and gemtuzumab ozogamicin in newly diagnosed and relapsed acute myeloid leukemia and high-risk myelodysplastic syndrome. Leukemia 30: 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- Derissen EJ, Beijnen JH, Schellens JH. 2013. Concise drug review: Azacitidine and decitabine. Oncologist 18: 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvic M, Talpur R, Ni X, Zhang C, Hazarika P, Kelly C, Chiao JH, Reilly JF, Ricker JL, Richon VM, et al. 2007. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL). Blood 109: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fandy TE, Herman JG, Kerns P, Jiemjit A, Sugar EA, Choi SH, Yang AS, Aucott T, Dauses T, Odchimar-Reissig R, et al. 2009. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood 114: 2764–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A, Schoch R, Gattermann N, Sanz G, List A, et al. 2009. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol 10: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidzik VI, Paschka P, Spath D, Habdank M, Kohne CH, Germing U, von Lilienfeld-Toal M, Held G, Horst HA, Haase D, et al. 2012. TET2 mutations in acute myeloid leukemia (AML): Results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol 30: 1350–1357. [DOI] [PubMed] [Google Scholar]
- Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, Yang H, Rosner G, Verstovsek S, Rytting M, Wierda WG, Ravandi F, Koller C, et al. 2006. Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood 108: 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasspool RM, Brown R, Gore ME, Rustin GJ, McNeish IA, Wilson RH, Pledge S, Paul J, Mackean M, Hall GD, et al. 2014. A randomised, phase II trial of the DNA-hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in combination with carboplatin vs carboplatin alone in patients with recurrent, partially platinum-sensitive ovarian cancer. Br J Cancer 110: 1923–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398. [DOI] [PubMed] [Google Scholar]
- Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, et al. 2006. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res 66: 6361–6369. [DOI] [PubMed] [Google Scholar]
- Greenberg PL, Garcia-Manero G, Moore M, Damon L, Roboz G, Hu K, Yang AS, Franklin J. 2013. A randomized controlled trial of romiplostim in patients with low- or intermediate-risk myelodysplastic syndrome receiving decitabine. Leuk Lymphoma 54: 321–328. [DOI] [PubMed] [Google Scholar]
- Griffiths EA, Choy G, Redkar S, Taverna P, Azab M, Karpf AR. 2013. SGI-110: DNA methyltransferase inhibitor oncolytic. Drugs Future 38: 535–543. [PMC free article] [PubMed] [Google Scholar]
- Hellebrekers DM, Jair KW, Vire E, Eguchi S, Hoebers NT, Fraga MF, Esteller M, Fuks F, Baylin SB, van Engeland M, et al. 2006. Angiostatic activity of DNA methyltransferase inhibitors. Mol Cancer Ther 5: 467–475. [DOI] [PubMed] [Google Scholar]
- Heninger E, Krueger TE, Lang JM. 2015. Augmenting antitumor immune responses with epigenetic modifying agents. Front Immunol 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, et al. 2012. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 22: 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Kantarjian HM. 2009. Targeting DNA methylation. Clin Cancer Res 15: 3938–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, Bayar E, Lyons J, Rosenfeld CS, Cortes J, et al. 2004. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 103: 1635–1640. [DOI] [PubMed] [Google Scholar]
- Issa JP, Gharibyan V, Cortes J, Jelinek J, Morris G, Verstovsek S, Talpaz M, Garcia-Manero G, Kantarjian HM. 2005. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol 23: 3948–3956. [DOI] [PubMed] [Google Scholar]
- Issa JP, Garcia-Manero G, Huang X, Cortes J, Ravandi F, Jabbour E, Borthakur G, Brandt M, Pierce S, Kantarjian HM. 2015a. Results of phase 2 randomized study of low-dose decitabine with or without valproic acid in patients with myelodysplastic syndrome and acute myelogenous leukemia. Cancer 121: 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Roboz G, Rizzieri D, Jabbour E, Stock W, O’Connell C, Yee K, Tibes R, Griffiths EA, Walsh K, et al. 2015b. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: A multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol 16: 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O, Quesnel B, Vey N, Gelsi-Boyer V, Raynaud S, et al. 2011. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia 25: 1147–1152. [DOI] [PubMed] [Google Scholar]
- James SR, Cedeno CD, Sharma A, Zhang W, Mohler JL, Odunsi K, Wilson EM, Karpf AR. 2013. DNA methylation and nucleosome occupancy regulate the cancer germline antigen gene MAGEA11. Epigenetics 8: 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek J, Liang S, Lu Y, He R, Ramagli LS, Shpall EJ, Estecio MR, Issa JP. 2012. Conserved DNA methylation patterns in healthy blood cells and extensive changes in leukemia measured by a new quantitative technique. Epigenetics 7: 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Qin T, Barton MC, Jelinek J, Issa JJ. 2015. Minimal role of base excision repair in TET-induced global DNA demethylation in HEK293T cells. Epigenetics 10: 1006–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. 1980. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20: 85–93. [DOI] [PubMed] [Google Scholar]
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. 2011. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 1: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey JD, Kapoor-Vazirani P, McCabe MT, Powell DR, Vertino PM. 2010. Long-term stability of demethylation after transient exposure to 5-aza-2′-deoxycytidine correlates with sustained RNA polymerase II occupancy. Mol Cancer Res 8: 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalac M, Scotto L, Marchi E, Amengual J, Seshan VE, Bhagat G, Ulahannan N, Leshchenko VV, Temkin AM, Parekh S, et al. 2011. HDAC inhibitors and decitabine are highly synergistic and associated with unique gene-expression and epigenetic profiles in models of DLBCL. Blood 118: 5506–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C, Ravandi F, et al. 2006. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer 106: 1794–1803. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O’Brien S, Cortes J, Faderl S, Bueso-Ramos C, Ravandi F, Estrov Z, et al. 2007a. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood 109: 52–57. [DOI] [PubMed] [Google Scholar]
- Kantarjian HM, O’Brien S, Huang X, Garcia-Manero G, Ravandi F, Cortes J, Shan J, Davisson J, Bueso-Ramos CE, Issa JP. 2007b. Survival advantage with decitabine versus intensive chemotherapy in patients with higher risk myelodysplastic syndrome: Comparison with historical experience. Cancer 109: 1133–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian HM, Thomas XG, Dmoszynska A, Wierzbowska A, Mazur G, Mayer J, Gau JP, Chou WC, Buckstein R, Cermak J, et al. 2012. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J Clin Oncol 30: 2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, Jones DA. 1999. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci 96: 14007–14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klco JM, Spencer DH, Lamprecht TL, Sarkaria SM, Wylie T, Magrini V, Hundal J, Walker J, Varghese N, Erdmann-Gilmore P, et al. 2013. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood 121: 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Porter Scott M, Chesworth R, Moyer MP, Copeland RA, et al. 2013. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci 110: 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Yan PS, Huang TH, Issa JP. 2004. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad Sci 101: 7398–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck D, Caulfield T, Lyko F, Medina-Franco JL. 2010. Nanaomycin A selectively inhibits DNMT3B and reactivates silenced tumor suppressor genes in human cancer cells. Mol Cancer Ther 9: 3015–3023. [DOI] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. 2012. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 44: 1236–1242. [DOI] [PubMed] [Google Scholar]
- LaBonte MJ, Wilson PM, Fazzone W, Groshen S, Lenz HJ, Ladner RD. 2009. DNA microarray profiling of genes differentially regulated by the histone deacetylase inhibitors vorinostat and LBH589 in colon cancer cell lines. BMC Med Genomics 2: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. 2009. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41: 838–842. [DOI] [PubMed] [Google Scholar]
- Li Q, Ahuja N, Burger PC, Issa JP. 1999. Methylation and silencing of the Thrombospondin-1 promoter in human cancer. Oncogene 18: 3284–3289. [DOI] [PubMed] [Google Scholar]
- Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, Topper MJ, Luo J, Connolly RM, Azad NS, et al. 2014. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 5: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Gilbert J, Rudek MA, Zwiebel JA, Gore S, Jiemjit A, Zhao M, Baker SD, Ambinder RF, Herman JG, et al. 2009. A phase I dose-finding study of 5-azacytidine in combination with sodium phenylbutyrate in patients with refractory solid tumors. Clin Cancer Res 15: 6241–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübbert M, Suciu S, Baila L, Rüter BH, Platzbecker U, Giagounidis A, Selleslag D, Labar B, Germing U, Salih HR, et al. 2011. Low-dose decitabine versus best supportive care in elderly patients with intermediate- or high-risk myelodysplasticcontrast syndrome (MDS) ineligible for intensive chemotherapy: Final results of the randomized phase III study of the European Organisation for Research and Treatment of Cancer Leukemia Group and the German MDS Study Group. J Clin Oncol 29: 1987–1996. [DOI] [PubMed] [Google Scholar]
- Lund K, Cole JJ, VanderKraats ND, McBryan T, Pchelintsev NA, Clark W, Copland M, Edwards JR, Adams PD. 2014. DNMT inhibitors reverse a specific signature of aberrant promoter DNA methylation and associated gene silencing in AML. Genome Biol 15: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. 2012. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res 72: 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III, Diaz E, et al. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–112. [DOI] [PubMed] [Google Scholar]
- Meldi K, Qin T, Buchi F, Droin N, Sotzen J, Micol JB, Selimoglu-Buet D, Masala E, Allione B, Gioia D, et al. 2015. Specific molecular signatures predict decitabine response in chronic myelomonocytic leukemia. J Clin Invest 125: 1857–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, Schneck JL, Carson JD, Liu Y, Butticello M, et al. 2015. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28: 57–69. [DOI] [PubMed] [Google Scholar]
- Navada SC, Steinmann J, Lubbert M, Silverman LR. 2014. Clinical development of demethylating agents in hematology. J Clin Invest 124: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EM, Morgan RJ, Kummar S, Beumer JH, Blanchard MS, Ruel C, El-Khoueiry AB, Carroll MI, Hou JM, Li C, et al. 2015. A phase I, pharmacokinetic, and pharmacodynamic evaluation of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine, administered with tetrahydrouridine. Cancer Chemother Pharmacol 75: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odunsi K, Matsuzaki J, James SR, Mhawech-Fauceglia P, Tsuji T, Miller A, Zhang W, Akers SN, Griffiths EA, Miliotto A, et al. 2014. Epigenetic potentiation of NY-ESO-1 vaccine therapy in human ovarian cancer. Cancer Immunol Res 2: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki Y, Aoki E, Issa JP. 2007. Decitabine—Bedside to bench. Crit Rev Oncol Hematol 61: 140–152. [DOI] [PubMed] [Google Scholar]
- Oki Y, Jelinek J, Shen L, Kantarjian HM, Issa JP. 2008. Induction of hypomethylation and molecular response after decitabine therapy in patients with chronic myelomonocytic leukemia. Blood 111: 2382–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiyan K, You JS, Yang X, Dai C, Zhou XJ, Baylin SB, Jones PA, Liang G. 2013. Functional DNA demethylation is accompanied by chromatin accessibility. Nucleic Acids Res 41: 3973–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry L, Clarke AR. 2011. The roles of the methyl-CpG binding proteins in cancer. Genes Cancer 2: 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul TA, Bies J, Small D, Wolff L. 2010. Signatures of polycomb repression and reduced H3K4 trimethylation are associated with p15INK4b DNA methylation in AML. Blood 115: 3098–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, Holloway AJ, Johnstone RW. 2005. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci 102: 3697–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. 2013. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 14: 765–780. [DOI] [PubMed] [Google Scholar]
- Prebet T, Sun Z, Figueroa ME, Ketterling R, Melnick A, Greenberg PL, Herman J, Juckett M, Smith MR, Malick L, et al. 2014. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: Results of the US Leukemia Intergroup trial E1905. J Clin Oncol 32: 1242–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T, Youssef EM, Jelinek J, Chen R, Yang AS, Garcia-Manero G, Issa JP. 2007. Effect of cytarabine and decitabine in combination in human leukemic cell lines. Clin Cancer Res 13: 4225–4232. [DOI] [PubMed] [Google Scholar]
- Qin T, Jelinek J, Si J, Shu J, Issa JP. 2009. Mechanisms of resistance to 5-aza-2′-deoxycytidine in human cancer cell lines. Blood 113: 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T, Castoro R, El Ahdab S, Jelinek J, Wang X, Si J, Shu J, He R, Zhang N, Chung W, et al. 2011. Mechanisms of resistance to decitabine in the myelodysplastic syndrome. PLoS ONE 6: e23372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T, Si J, Raynal NJ, Wang X, Gharibyan V, Ahmed S, Hu X, Jin C, Lu Y, Shu J, et al. 2015. Epigenetic synergy between decitabine and platinum derivatives. Clin Epigenet 7: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravandi F, Alattar ML, Grunwald MR, Rudek MA, Rajkhowa T, Richie MA, Pierce S, Daver N, Garcia-Manero G, Faderl S, et al. 2013. Phase 2 study of azacytidine plus sorafenib in patients with acute myeloid leukemia and FLT-3 internal tandem duplication mutation. Blood 121: 4655–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal NJ, Si J, Taby RF, Gharibyan V, Ahmed S, Jelinek J, Estecio MR, Issa JP. 2012. DNA methylation does not stably lock gene expression but instead serves as a molecular mark for gene silencing memory. Cancer Res 72: 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal NJ, Lee JT, Wang Y, Beaudry A, Madireddi P, Garriga J, Malouf GG, Dumont S, Dettman EJ, Gharibyan V, et al. 2015. Targeting calcium signaling induces epigenetic reactivation of tumor suppressor genes in cancer. Cancer Res 76: 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, et al. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162: 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekeres MA, Tiu RV, Komrokji R, Lancet J, Advani AS, Afable M, Englehaupt R, Juersivich J, Cuthbertson D, Paleveda J, et al. 2012. Phase 2 study of the lenalidomide and azacitidine combination in patients with higher-risk myelodysplastic syndromes. Blood 120: 4945–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. 2007a. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 3: 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, et al. 2007b. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci 104: 18654–18659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si J, Boumber YA, Shu J, Qin T, Ahmed S, He R, Jelinek J, Issa JP. 2010. Chromatin remodeling is required for gene reactivation after decitabine-mediated DNA hypomethylation. Cancer Res 70: 6968–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, Stone RM, Nelson D, Powell BL, DeCastro CM, et al. 2002. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: A study of the cancer and leukemia group B. J Clin Oncol 20: 2429–2440. [DOI] [PubMed] [Google Scholar]
- Silverman LR, Fenaux P, Mufti GJ, Santini V, Hellstrom-Lindberg E, Gattermann N, Sanz G, List AF, Gore SD, Seymour JF. 2011. Continued azacitidine therapy beyond time of first response improves quality of response in patients with higher-risk myelodysplastic syndromes. Cancer 117: 2697–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: Roles in mammalian development. Nat Rev Genet 14: 204–220. [DOI] [PubMed] [Google Scholar]
- Stewart DJ, Issa JP, Kurzrock R, Nunez MI, Jelinek J, Hong D, Oki Y, Guo Z, Gupta S, Wistuba II. 2009. Decitabine effect on tumor global DNA methylation and other parameters in a phase I trial in refractory solid tumors and lymphomas. Clin Cancer Res 15: 3881–3888. [DOI] [PubMed] [Google Scholar]
- Taby R, Issa JP. 2010. Cancer epigenetics. CA Cancer J Clin 60: 376–392. [DOI] [PubMed] [Google Scholar]
- TCGA. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. 1999. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci 96: 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traina F, Visconte V, Elson P, Tabarroki A, Jankowska AM, Hasrouni E, Sugimoto Y, Szpurka H, Makishima H, O’Keefe CL, et al. 2014. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia 28: 78–87. [DOI] [PubMed] [Google Scholar]
- Treppendahl MB, Kristensen LS, Gronbaek K. 2014. Predicting response to epigenetic therapy. J Clin Invest 124: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al. 2012. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21: 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, et al. 2013. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res 23: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voso MT, Fabiani E, Piciocchi A, Matteucci C, Brandimarte L, Finelli C, Pogliani E, Angelucci E, Fioritoni G, Musto P, et al. 2011. Role of BCL2L10 methylation and TET2 mutations in higher risk myelodysplastic syndromes treated with 5-azacytidine. Leukemia 25: 1910–1913. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. 2006. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38: 787–793. [DOI] [PubMed] [Google Scholar]
- Whittaker SJ, Demierre MF, Kim EJ, Rook AH, Lerner A, Duvic M, Scarisbrick J, Reddy S, Robak T, Becker JC, et al. 2010. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol 28: 4485–4491. [DOI] [PubMed] [Google Scholar]
- Wienholz BL, Kareta MS, Moarefi AH, Gordon CA, Ginno PA, Chedin F. 2010. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet 6: e1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijermans P, Lubbert M, Verhoef G, Bosly A, Ravoet C, Andre M, Ferrant A. 2000. Low-dose 5-aza-2′-deoxycytidine, a DNA hypomethylating agent, for the treatment of high-risk myelodysplastic syndrome: A multicenter phase II study in elderly patients. J Clin Oncol 18: 956–962. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. 2012. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep 13: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangle J, Wang W, Koch A, Easwaran H, Mohammad HP, Vendetti F, Vancriekinge W, Demeyer T, Du Z, Parsana P, et al. 2013. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget 4: 2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, et al. 2010. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res 16: 21–33. [DOI] [PubMed] [Google Scholar]
- Yamazaki J, Jelinek J, Lu Y, Cesaroni M, Madzo J, Neumann F, He R, Taby R, Vasanthakumar A, Macrae T, et al. 2015. TET2 mutations affect non-CpG island DNA methylation at enhancers and transcription factor binding sites in chronic myelomonocytic leukemia. Cancer Res 75: 2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki J, Taby R, Jelinek J, Raynal NJ, Cesaroni M, Pierce SA, Kornblau SM, Bueso-Ramos CE, Ravandi F, Kantarjian HM, et al. 2016. Hypomethylation of TET2 target genes identifies a curable subset of acute myeloid leukemia. J Natl Cancer Inst 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. 2014. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26: 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Rau R, Goodell MA. 2015. DNMT3A in haematological malignancies. Nat Rev Cancer 15: 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. 2002. Zebularine: A novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol 321: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]