Abstract
MDM2 is a primary cellular inhibitor of p53. It inhibits p53 function by multiple mechanisms, each of which, however, is mediated by their direct interaction. It has been proposed that small-molecule inhibitors designed to block the MDM2–p53 interaction may be effective in the treatment of human cancer retaining wild-type p53 by reactivating the p53 tumor suppressor function. Through nearly two decades of intense efforts, a number of structurally distinct, highly potent, nonpeptide, small-molecule inhibitors of the MDM2–p53 interaction (MDM2 inhibitors) have been successfully designed and developed, and at least seven such compounds have now been advanced into human clinical trials as new anticancer drugs. This review offers a perspective on the design and development of MDM2 small-molecule inhibitors and discusses early clinical data for some of the MDM2 small-molecule inhibitors and future challenges for the successful clinical development of MDM2 inhibitors for cancer treatment.
Small-molecule inhibitors of the MDM2–p53 interaction—which effectively reactivate wild-type p53—may be effective in treating cancer. Significant progress in the development of such inhibitors has recently been made.
MDM2: A PRIMARY CELLULAR INHIBITOR OF p53
By controlling expression of a large number of genes, the transcription factor p53 plays a vital role in the regulation of many cellular processes, including cell-cycle progression, apoptosis, senescence, DNA repair, and metabolism, and functions as a powerful tumor suppressor (Vousden and Lu 2002; Toledo and Wahl 2006; Stiewe 2007; Brown et al. 2009; Wade et al. 2013). Mice lacking the p53 protein develop normally but are prone to the development of a variety of tumors (Kemp et al. 1993). Perhaps unsurprisingly, TP53, the gene encoding p53, is mutated or deleted in ∼50% of human cancers, rendering p53 nonfunctional as a tumor suppressor (Feki and Irminger-Finger 2004).
Because of the critical role of p53 in regulation of many cellular processes, the level and the activity of p53 are tightly controlled. The murine double minute 2 (MDM2) oncogene is a primary cellular regulator and inhibitor of p53. The role of MDM2 as a primary negative endogenous regulator of p53 is unambiguously established by the fact that MDM2-null is embryonically lethal in mice, and they can only be rescued by concurrent deletion of the TP53 gene (Jones et al. 1995; Montes de Oca et al. 1995).
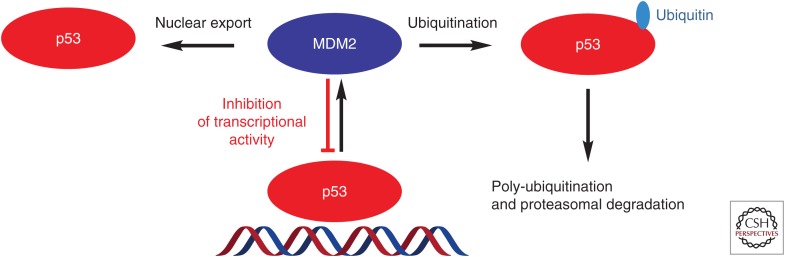
MDM2 and p53 regulate each other mutually through the autoregulatory feedback loop shown in Figure 1 (Wu et al. 1993; Freedman et al. 1999). In cells containing wild-type p53, on activation by a variety of stimuli, p53 transcribes the MDM2 gene, leading to an increase of MDM2 mRNA and protein. The MDM2 protein, in turn, binds to the p53 protein directly through their amino termini and inhibits p53 function through three major mechanisms: (1) On binding, MDM2 directly ubiquitinates p53 through its E3 ligase activity, promoting proteasomal degradation of p53; (2) the interaction of MDM2 with p53 blocks the binding of p53 to its targeted DNA, rendering p53 ineffective as a transcription factor; and (3) MDM2 promotes export of p53 out of the cell nucleus, making p53 inaccessible to its targeted DNA and further reducing its transcriptional ability (Wu et al. 1993; Freedman et al. 1999; Juven-Gershon and Oren 1999). Through these three inhibitory mechanisms, MDM2 functions as an effective p53 antagonist in cells with wild-type p53.
Figure 1.
Autoregulatory loop of p53 and MDM2. Activation of p53 transcribes MDM2 mRNA and increases MDM2 protein, which in turn inhibits p53 activity by three mechanisms.
Consistent with its role as an efficient inhibitor of the p53 tumor suppressor function, MDM2, when overexpressed, is oncogenic (Ganguli et al. 2000; see Oliner et al. 2016). In human tumors, overexpression of the MDM2 protein can be caused by gene amplification. The MDM2 gene is amplified in an average of 7% of human cancers based on an analysis of 28 different types of cancers involving ∼4000 human tumor samples (Momand et al. 1998), but a higher frequency of MDM2 gene amplification occurs in certain types of tumors, including well-differentiated liposarcomas (>80%), soft tissue tumors (20%), osteosarcomas (16%), and esophageal carcinomas (13%) (Weaver et al. 2008, 2009). In further support of its powerful inhibitory role of the p53 tumor suppressor function, MDM2 gene amplification and TP53 gene mutation are largely mutually exclusive in human cancers (Oliner et al. 1992; Shvarts et al. 1996; Wasylishen and Lozano 2016). In addition to MDM2 gene amplification, MDM2 overexpression can be the result of a variety of other mechanisms, such as single-nucleotide polymorphism, enhanced transcription, or increased translation (Capoulade et al. 1998; Momand et al. 2000; Bond et al. 2004, 2005). Pathologically, MDM2 overexpression has been correlated with poor clinical prognosis and poor response to current cancer therapies (Capoulade et al. 1998; Momand et al. 2000; Bond et al. 2004, 2005).
BLOCKING THE MDM2–p53 INTERACTION AND REACTIVATING p53 AS A NEW CANCER THERAPEUTIC STRATEGY
Because MDM2 functions as a primary inhibitor of the p53 tumor suppressor function, agents that target MDM2 can reactivate wild-type p53. MDM2 inhibits p53 through several mechanisms that are dependent on its direct interaction with p53. Hence, peptides or nonpeptide small molecules designed to block the MDM2–p53 protein–protein interaction can lead to an increase of p53 protein and transcriptional activation of p53. By harnessing the powerful tumor suppressor function of p53, such compounds may have a therapeutic potential for the treatment of human cancer retaining wild-type p53.
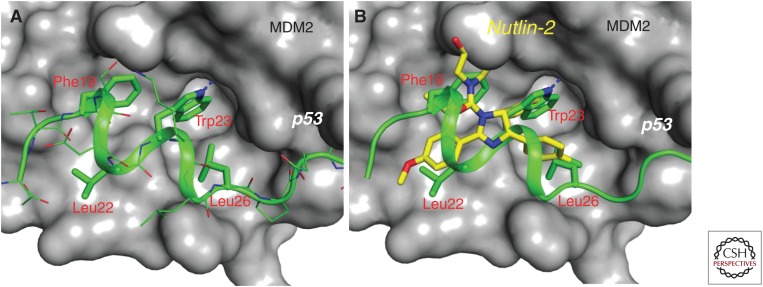
Biochemical studies have mapped the MDM2–p53 protein–protein interaction to the first ∼120 amino-terminal amino acid residues of MDM2 and the first 30 amino-terminal residues of p53 (Capoulade et al. 1998; Momand et al. 2000). The determination of a high-resolution cocrystal structure of MDM2 complexed with residues 15–29 of a p53 peptide (Fig. 2A) (Kussie et al. 1996) in 1996 provided the atomic details of their interaction and suggested the feasibility of the design of nonpeptide, drug-like, small-molecule inhibitors capable of blocking the MDM2–p53 interaction. Specifically, the cocrystal structure shows that the p53 peptide adopts an α-helical conformation and interacts with MDM2 primarily through three hydrophobic residues, Phe19, Trp23, and Leu26, which cluster together and bind into a well-defined hydrophobic pocket in MDM2. Although natural p53 peptides have only micromolar binding affinities to MDM2, peptides designed using unnatural amino acids can achieve low nanomolar binding affinities (García-Echeverría et al. 2000), further supporting the feasibility of designing high-affinity, nonpeptide, small-molecule inhibitors to block the MDM2–p53 interaction.
Figure 2.
Cocrystal structures. (A) Cocrystal structure of MDM2 (surface rendering) in complex with p53 (stick model). p53 protein uses primarily three key residues (Phe19, Trp23, and Leu26) to interact with a well-defined, surface hydrophobic pocket in MDM2. (B) Superposition of the cocrystal structures of nutlin-2/MDM2 complex and p53/MDM2 complex (PDBIDs: 1YCR and 4HG7). Nutlin-2 is shown by yellow sticks and the three key p53-binding residues are shown by green sticks with the MDM2 protein shown in the surface rendering.
NUTLINS: THE FIRST POTENT AND SPECIFIC SMALL-MOLECULE INHIBITORS OF THE MDM2–p53 INTERACTION
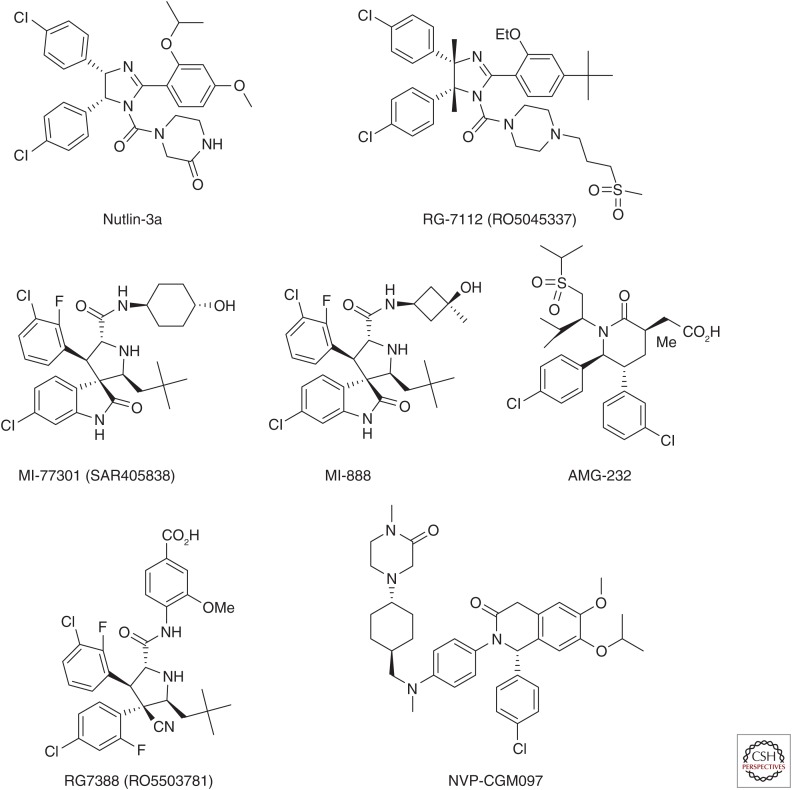
Despite intense research efforts in academic laboratories and pharmaceutical companies, design of highly potent, specific, nonpeptide small-molecule inhibitors of the MDM2–p53 interaction with a well-defined mechanism of action has proven to be much more difficult than originally anticipated. The breakthrough came in 2004 with the discovery of the nutlins by Vassilev et al. (2004) from Hoffmann-La Roche. Among the initial nutlins reported, nutlin-3a (Fig. 3) binds to MDM2 with an IC50 (half-maximal inhibitory concentration) value of 90 nm and shows a cellular mechanism of action consistent with targeting the MDM2–p53 interaction. Nutlin-3a effectively activates wild-type p53 in cancer cells, potently inhibits cell growth in cancer cell lines retaining wild-type p53 in a dose-dependent manner, and shows >10-fold selectivity over cancer cell lines harboring p53 mutation or with p53 deletion. Activation of p53 by nutlin-3a leads to transcription of p53-regulated genes, including MDM2 and the cell-cycle regulator p21. Interestingly, although nutlin-3a activates p53 in nontumorigenic NIH-3T3 cells and inhibits cell proliferation, it fails to kill the cells. Impressively, nutlin-3a is orally bioavailable. At high doses (100 and 200 mg/kg) and a twice daily dosing schedule, oral administration of nutlin-3a effectively inhibits tumor growth in the xenograft model of the human osteosarcoma SJSA-1 cell line containing an amplified MDM2 gene, while showing no signs of toxicity to mice. The cocrystal structure of nutlin-2, a close analog of nutlin-3a, in a complex with MDM2 clearly shows that the nutlins mimic the three key p53-binding residues (Phe19, Trp23, and Leu26) (Fig. 2B) (Vassilev et al. 2004). The preclinical data obtained using the nutlins show that highly potent and selective nonpeptide small-molecule inhibitors of the MDM2–p53 interaction may have a therapeutic potential for the treatment of human cancers retaining wild-type p53. The discovery of the nutlins has also inspired other research groups to design new MDM2 inhibitors with higher potency and selectivity and better pharmacokinetics.
Figure 3.
Chemical structures of representative MDM2 inhibitors.
RG7112 (RO5045337): THE FIRST MDM2 INHIBITOR ADVANCED INTO HUMAN CLINICAL TRIALS
Further optimization of nutlin-3a by scientists from Hoffmann-La Roche to improve its binding affinity to MDM2, cellular potency, pharmacokinetics, and chemical stability ultimately resulted in the discovery of RG7112 (RO5045337) (Fig. 3), the first MDM2 inhibitor to be advanced into human clinical trials (Vu et al. 2013; Siu et al. 2014). RG7112 has a binding affinity to MDM2 (IC50 = 18 nm), which is better than that of nutlin-3. It effectively inhibits cell growth in cancer cell lines with wild-type p53 (IC50 = 0.18–2.2 µm) and is several times more potent than nutlin-3. RG7112 shows good selectivity over cancer cell lines with a p53 mutation (IC50 = 5.7–20.3 µm). It effectively activates wild-type p53 in vitro and in vivo and shows good oral pharmacokinetic properties in mice. In two xenograft models of SJSA1 and MHM osteosarcoma cell lines with MDM2 gene amplification and overexpression of MDM2 protein, RG7122 dose-dependently inhibits tumor growth and is capable of achieving partial tumor regression with oral administration, without signs of toxicity in mice.
OTHER CLASSES OF HIGHLY POTENT AND SELECTIVE NONPEPTIDE, SMALL-MOLECULE INHIBITORS OF THE MDM2–p53 INTERACTION
Inspired by the discovery of the nutlins, several classes of potent and selective nonpeptide small-molecule inhibitors of the MDM2–p53 interaction have been designed and developed using different strategies. Some of them have achieved much higher affinities to MDM2 and better antitumor activity than nutlin-3 or RG7112 in animal models of human cancer.
Using a computational structure-based design strategy, our laboratory has designed spiro-oxindoles as a new class of nonpeptide small-molecule inhibitors (Ding et al. 2005). Extensive optimization has resulted in the discovery of MI-77301 (Fig. 3) (Wang et al. 2014) and MI-888 (Zhao et al. 2013a,b), which bind to MDM2 with Ki values of 0.88 and 0.44 nm, respectively, and are 50 times more potent than nutlin-3 in the same binding assay. MI-77301 and MI-888 show >10,000-fold selectivity over MDMX, a protein closely homologous with MDM2. In human cancer cell lines, both MI-77301 and MI-888 effectively activate wild-type p53 at concentrations of 30–100 nm. Consistent with their higher binding affinities to MDM2 and higher potencies in activation of p53, these compounds are more than 10 times more potent than nutlin-3 in inhibition of cell growth in cancer cell lines retaining wild-type p53, and they show >100-fold cellular selectivity over cancer cell lines harboring mutated p53 or with p53 deletion. In the SJSA-1 xenograft model, both compounds, orally administered daily, are capable of achieving complete and long-lasting tumor regression without signs of toxicity. A single, oral dose of MI-77301 is capable of achieving complete tumor regression of the SJSA-1 tumors in mice (Wang et al. 2014). The cocrystal structure of MI-77301 in a complex with human MDM2 protein (Wang et al. 2014) shows that consistent with its design, MI-77301 mimics all three of the key p53-binding residues and further is involved in additional hydrophobic and hydrogen-bonding interactions. A substituted phenyl group in MI-77301, for example, has a π-π stacking interaction with the His96 residue of MDM2. MI-77301 also induces refolding of the unstructured residues 10–25 of the MDM2 amino-terminal region, making these residues a part of the binding pocket and further enhancing the MDM2–MI-77301 binding affinity by 25 times.
AMG-232 (Fig. 3), which contains a piperidin-2-one scaffold, was discovered by Amgen scientists through structure-based design and extensive optimization (Rew et al. 2012; Sun et al. 2014). AMG-232 has a Kd value of 0.045 nm with MDM2 and is probably the most potent MDM2 inhibitor reported to date (Sun et al. 2014). It inhibits cell proliferation with IC50 values of 9.1 nm and 10 nm in the SJSA-1 and HCT-116 cell lines, respectively, and shows >1000-fold selectivity over the HCT-116 p53 knockout (p53−/−) cell line. AMG-232 effectively inhibits tumor growth in the SJSA-1 osteosarcoma model and achieves complete tumor regression in 10 of 12 animals orally administered 60 mg/kg daily. In the HCT-116 xenograft model, AMG-232 dose-dependently inhibits tumor growth and can achieve 100% tumor growth inhibition without tumor regression. Although no cocrystal structure of AMG-232 complexed with MDM2 is available, several high-resolution cocrystal structures of MDM2 complexed with analogs of AMG-232 have been published (Michelsen et al. 2012; Rew et al. 2012; Sun et al. 2014) and provide structural insights into their high-affinity binding to MDM2. These cocrystal structures show that this class of MDM2 inhibitors also nicely mimics all of the three key p53-binding residues for interactions with MDM2. Additionally, the sulfonyl isopropyl group of AMG-232 resides in the small pocket surrounding G58, an interaction not observed in the cocrystal structures of other classes of MDM2 inhibitors, and the carboxylic acid of AMG-232 forms a salt bridge with the His96 residue of MDM2. As with MI-77301, this class of MDM2 inhibitors also induces reorganization of the extreme amino terminus of the MDM2 protein and promotes additional hydrophobic contacts between the inhibitors and the Val14 and Thr16 residues in MDM2 (Michelsen et al. 2012).
Based on MI-219, an earlier analog of MI-77301, scientists from Hoffmann-La Roche have designed a new class of MDM2 inhibitors (Ding et al. 2013) and have advanced RG7388 (RO5503781) (Fig. 3) into clinical development. RG7388 binds to MDM2 with IC50 = 6 nm, potently inhibits cell growth in cancer cell lines containing wild-type p53 (average IC50 = 30 nm), and shows >100-fold selectivity over cancer cell lines containing mutated p53. It has good microsomal stability and good pharmacokinetic properties in animals. RO5503781 achieves complete tumor regression in the SJSA-1 osteosarcoma xenograft model in mice with oral administration. It is currently in three phase I clinical trials as a single agent and in combination with other chemotherapies for treatment of patients with solid tumors, acute myelogenous leukemia, or advanced malignancies.
Novartis has recently disclosed a new class of MDM2 inhibitors containing a dihydroisoquinolinone scaffold and advanced NVP-CGM097 (Fig. 3) into clinical development (Holzer et al. 2015). Merck has discovered MK-8242 (SCH 900242) as a potent MDM2 inhibitor and advanced it into phase I clinical trials. However, the chemical structure of MK-8242 and its characterizations have not been disclosed. Scientists from Daiichi Sankyo have identified DS-3032b as a highly potent MDM2 inhibitor and advanced it into clinical development, but no detailed information on DS-3032b has been disclosed.
RESULTS FROM CLINICAL TRIALS OF RG7112
RG7112 was the first MDM2 inhibitor advanced into human clinical trials. Because >80% of liposarcomas have an amplified MDM2 gene and overexpression of MDM2 protein, RG7112 was evaluated in chemotherapy-naïve liposarcoma patients and the phase I results have been reported (Ray-Coquard et al. 2012). A good human PK profile was achieved for RG7112 with oral administration. Clear activation of p53, an increase in p21 protein, and apoptosis induction in tumors were detected. Evidence of antitumor activity was observed in liposarcoma patients; 14 out of 20 patients were found to have stable disease and one had a confirmed partial response after treatment with RG7112. All patients treated with RG7112 had at least one adverse event, and 12 serious adverse events were observed in eight patients, including neutropenia (six patients) and thrombocytopenia (three patients). The plasma exposure of RG7112 in patients correlated with the observed hematological toxicity. Thus, the clinical data for RG7112 indicate that late hematological toxicity, particularly thrombocytopenia, should be considered in future clinical trials of MDM2 inhibitors (Ray-Coquard et al. 2012).
CLINICAL TRIALS OF OTHER MDM2 INHIBITORS
In addition to RG7112, at least six other MDM2 inhibitors have been advanced into clinical development, but results for only two of these six MDM2 inhibitors have been disclosed.
The initial phase I clinical data for RG7388 were reported recently (Siu et al. 2014). The maximum tolerated dose for RG7388 is 500 mg in a daily dosing (QD), 5-d schedule, 500 mg in a twice-daily (BID), 3-d schedule, and 1600 mg in a BID, weekly schedule. Activation of p53, as measured by MIC-1 concentration in plasma, was observed with either daily dosing for 5 d or with BID dosing for 3 d, but it is stronger with the former schedule. No clear p53 activation was observed with weekly BID dosing. Thrombocytopenia, neutropenia, febrile neutropenia, and diarrhea were the dose-limiting toxicities. Fluorothymidine positron emission tomography (FLT-PET) showed that RG7388 causes decreased proliferation in tumors. The recommended phase II dose is 500 mg with daily, 5-d scheduling.
MI-77301 (SAR405838) was first evaluated by Sanofi as a single agent to assess its safety, tolerability, pharmacokinetics, and biological activity in patients with advanced cancer. A second phase I trial was initiated in 2013 to evaluate SAR405838 in combination with pimasertib, an allosteric inhibitor of mitogen-activated protein kinase 1 and 2 (MEK1/2) in patients with solid tumors. The preliminary, single-agent phase I data of SAR405838 were recently presented in the 2015 51st Annual Meeting of American Society of Clinical Oncology (Watters 2015). The maximum tolerated daily dose for SAR405838 was 300 mg and activation of p53 by SAR405838 was shown at doses that have an acceptable safety profile. MDM2 inhibitor-induced p53 mutation was investigated for the first time. Analysis of plasma and tumor samples from 18 dedifferentiated liposarcoma (DDLPS) patients revealed that mutations of TP53, the gene encoding p53, appear in the circulating cell-free DNA (ccfDNA) of patients during treatment with SAR405838. All p53 mutations were found to be located in the DNA-binding domain of p53. Additionally, mutation variant allele frequency also increased during treatment, and multiple p53 mutations arose within individual patients. All patients undergoing five or more cycles of treatment showed evidence of p53 mutations in ccfDNA. Hence, the observation of p53 mutations in the circulating ccfDNA of patients during treatment with SAR405838 suggested that combinations of MDM2 inhibitors with agents that are effective against tumor cells with mutated p53 are needed for future clinical trials of MDM2 inhibitors.
CHALLENGES AND FUTURE DIRECTION IN THE DEVELOPMENT OF MDM2 INHIBITORS
Data from initial clinical trials of RG7112, RG7388, and SAR405838 have shown that MDM2 inhibitors can effectively activate p53 in patients with wild-type p53 tumors at dose schedules with acceptable safety profiles. Additionally, evidence of antitumor activity has been observed. These initial clinical data provide further support for the basic hypothesis that blocking the MDM2–p53 interaction will lead to activation of wild-type p53 and may represent a novel cancer therapeutic strategy.
Two significant challenges have been identified based on the initial clinical data. First, the dose-limiting toxicity for MDM2 inhibitors appears to be late hematological toxicity, including thrombocytopenia and neutropenia, because of activation of p53 in the bone marrow. Consequently, it is important to identify an optimal dose schedule for each MDM2 inhibitor so that robust p53 activation is achieved with minimal or at least manageable hematological toxicity. Second, detection of emergence of p53 mutations in liposarcoma patients treated with SAR405838 points out the importance of combination of MDM2 inhibitors with agents that effectively target tumor cells with mutated p53 for the successful clinical development of MDM2 inhibitors. To this end, our recent study (Hoffman-Luca et al. 2015) showed that when leukemia cell lines containing wild-type p53 develop resistance to either the MDM2 inhibitor SAR405838 or the Bcl-2 inhibitor ABT-263, the cells that became highly resistant to one agent remained very sensitive to the other agent both in vitro and in vivo, and the combination of the MDM2 inhibitor and the Bcl-2 inhibitor is capable of achieving longer tumor regression than either single agent in xenograft models of leukemia. Additionally, the antitumor activity of some traditional chemotherapies or newer molecularly targeted anticancer drugs is independent of the p53 status of tumors. Therefore, combination of an MDM2 inhibitor with agents that can effectively kill tumor cells harboring a mutated p53 should be explored in future clinical trials.
CONCLUSION
Intense research efforts from academic laboratories and pharmaceutical companies have led to the design and synthesis of several classes of highly potent and specific nonpeptide, small-molecule inhibitors that bind to MDM2 and block the MDM2–p53 interaction. At least seven such compounds have been advanced into clinical development for cancer treatment. These optimized MDM2 inhibitors have shown excellent pharmacokinetics on oral administration in animals, and several of them achieve complete tumor regression in animal models of human cancer, but two major issues related to future clinical development of MDM2 inhibitors have been identified.
Based on initial phase I clinical data reported for three of these compounds (RG7112, RG7388, and SAR405838), MDM2 inhibitors can activate wild-type p53 in patients and have acceptable safety profiles. Evidence of their clinical antitumor activity as single agents has also been reported. An issue reflecting on the use of MDM2 inhibitors, however, is their dose-limiting toxicity due to activation of p53 in the bone marrow, which must be mitigated with dose schedule optimization. The second issue is the emergence of p53 mutations in tumors initially containing wild-type p53 on treatment with MDM2 inhibitors. Because MDM2 inhibitors, based on preclinical data, are only effective against tumors containing wild-type p53, tumor cells harboring mutated p53 are expected to be resistant to MDM2 inhibitors. To overcome this drug-induced resistance issue, combination of MDM2 inhibitors with agents that are effective in targeting tumor cells harboring mutated p53 should be explored in both preclinical models and in clinical trials.
ACKNOWLEDGMENTS
We thank the present and past members of the Wang laboratory and our collaborators who have contributed to the design, synthesis, evaluation, and development of our MDM2 inhibitors. Funding from the National Cancer Institute (NCI), the National Institutes of Health (NIH), the Prostate Cancer Foundation, the Susan G. Komen Foundation, the Lymphoma and Leukemia Society, Ascenta Therapeutics, Inc., Ascentage Pharma Group, and Sanofi is greatly appreciated.
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. 2004. A single nucleotide polymorphism in the MDM2 promotor attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119: 591–602. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Levine AJ. 2005. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets 5: 3–8. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. 2009. Awakening guardian angels: Drugging the p53 pathway. Nat Rev Cancer 9: 862–873. [DOI] [PubMed] [Google Scholar]
- Capoulade C, Bressac-de Paillerets B, Lefrere I, Ronsin M, Feunteun J, Tursz T, Wiels J. 1998. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt’s lymphoma cells. Oncogene 16: 1603–1610. [DOI] [PubMed] [Google Scholar]
- Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding YS, Gao W, Stuckey J, Krajewski K, Roller PP, Tomita Y, et al. 2005. Structure-based design of potent non-peptide MDM2 inhibitors. J Am Chem Soc 127: 10130–10131. [DOI] [PubMed] [Google Scholar]
- Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, Chu XJ, Bartkovitz D, Podlaski F, Janson C, et al. 2013. Discovery of RG7388, a potent and selective p53–MDM2 inhibitor in clinical development. J Med Chem 56: 5979–5983. [DOI] [PubMed] [Google Scholar]
- Feki A, Irminger-Finger I. 2004. Mutational spectrum of p53 mutations in primary breast and ovarian tumors. Crit Rev Oncol Hematol 52: 103–116. [DOI] [PubMed] [Google Scholar]
- Freedman DA, Wu L, Levine AJ. 1999. Functions of the MDM2 oncoprotein. Cell Mol Life Sci 55: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli G, Abecassis J, Wasylyk B. 2000. MDM2 induces hyperplasia and premalignant lesions when expressed in the basal layer of the epidermis. EMBO J 19: 5135–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Echeverría C, Chène P, Blommers MJ, Furet P. 2000. Discovery of potent antagonists of the interaction between human double minute 2 and tumor suppressor p53. J Med Chem 43: 3205–3208. [DOI] [PubMed] [Google Scholar]
- Hoffman-Luca CG, Ziazadeh D, McEachern D, Zhao Y, Sun W, Debussche L, Wang S. 2015. Elucidation of acquired resistance to Bcl-2 and MDM2 inhibitors in acute leukemia in vitro and in vivo. Clin Cancer Res 21: 2558–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer P, Masuya K, Furet P, Kallen J, Valat-Stachyra T, Ferretti S, Berghausen J, Bouisset-Leonard M, Buschmann N, Pissot-Soldermann C, et al. 2015. Discovery of a dihydroisoquinolinone derivative (NVP-CGM097): A highly potent and selective MDM2 inhibitor undergoing phase 1 clinical trials in p53wt tumors. J Med Chem 58: 6348–6358. [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Oren M. 1999. Mdm2: The ups and downs. Mol Med 5: 71–83. [PMC free article] [PubMed] [Google Scholar]
- Kemp CJ, Donehower LA, Bradley A, Balmain A. 1993. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell 74: 813–822. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274: 948–953. [DOI] [PubMed] [Google Scholar]
- Michelsen K, Jordan JB, Lewis J, Long AM, Yang E, Rew Y, Zhou J, Yakowec P, Schnier PD, Huang X, et al. 2012. Ordering of the N-terminus of human MDM2 by small molecule inhibitors. J Am Chem Soc 134: 17059–17067. [DOI] [PubMed] [Google Scholar]
- Momand J, Jung D, Wilczynski S, Niland J. 1998. The MDM2 gene amplication database. Nucleic Acids Res 26: 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Wu HH, Dasgupta G. 2000. MDM2—Master regulator of the p53 tumor suppressor protein. Gene 242: 15–29. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358: 80–83. [DOI] [PubMed] [Google Scholar]
- *.Oliner JD, Saiki AY, Caenepeel S. 2016. The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a026336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J, Heil F, Rueger R, Graves B, Ding M, et al. 2012. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol 13: 1133–1140. [DOI] [PubMed] [Google Scholar]
- Rew Y, Sun DQ, De Turiso FGL, Bartberger MD, Beck HP, Canon J, Chen A, Chow D, Deignan J, Fox BM, et al. 2012. Structure-based design of novel inhibitors of the MDM2–p53 interaction. J Med Chem 55: 4936–4954. [DOI] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, et al. 1996. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J 15: 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Siu LL, Italiano A, Miller WH, Blay J-Y, Gietema JA, Bang Y-J, Mileshkin LR, Hirte HW, Reckner M, Higgins B, et al. 2014. Phase 1 dose escalation, food effect, and biomarker study of RG7388, a more potent second-generation MDM2 antagonist, in patients (pts) with solid tumors. J Clin Oncol 32(abstr 2535): 5s. [Google Scholar]
- Stiewe T. 2007. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer 7: 165–168. [DOI] [PubMed] [Google Scholar]
- Sun D, Li Z, Rew Y, Gribble M, Bartberger MD, Beck HP, Canon J, Chen A, Chen X, Chow D, et al. 2014. Discovery of AMG 232, a potent, selective, and orally bioavailable MDM2–p53 inhibitor in clinical development. J Med Chem 57: 1454–1472. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. 2002. Live or let die: The cell’s response to p53. Nat Rev Cancer 2: 594–604. [DOI] [PubMed] [Google Scholar]
- Vu B, Wovkulich P, Pizzolato G, Lovey A, Ding Q, Jiang N, Liu JJ, Zhao C, Glenn K, Wen Y, et al. 2013. Discovery of RG7112: A small-molecule MDM2 inhibitor in clinical development. ACS Med Chem Lett 4: 466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Li YC, Wahl GM. 2013. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun W, Zhao Y, McEachern D, Meaux I, Barrière C, Stuckey J, Meagher J, Bai B, Liu L, et al. 2014. SAR405838: An optimized inhibitor of MDM2–p53 interaction that induces complete and durable tumor regression. Cancer Res 74: 5855–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wasylishen AR, Lozano G. 2016. Attenuating the p53 pathway in human cancers: Many means to the same end. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters JW. 2015. TP53 mutations emerge in circulating cell-free DNA obtained from patients undergoing treatment with the HDM2 antagonist SAR405838. In 2015 American Society of Clinical Oncology Annual Meeting, Chicago. [Google Scholar]
- Weaver J, Downs-Kelly E, Goldblum JR, Turner S, Kulkarni S, Tubbs RR, Rubin BP, Skacel M. 2008. Fluorescence in situ hybridization for MDM2 gene amplification as a diagnostic tool in lipomatous neoplasms. Mod Pathol 21: 943–949. [DOI] [PubMed] [Google Scholar]
- Weaver J, Goldblum JR, Turner S, Tubbs RR, Wang WL, Lazar AJ, Rubin BP. 2009. Detection of MDM2 gene amplification or protein expression distinguishes sclerosing mesenteritis and retroperitoneal fibrosis from inflammatory well-differentiated liposarcoma. Mod Pathol 22: 66–70. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. 1993. The p53–mdm-2 autoregulatory feedback loop. Genes Dev 7: 1126–1132. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Liu L, Sun W, Lu J, McEachern D, Li X, Yu S, Bernard D, Ochsenbein P, Ferey V, et al. 2013a. Diastereomeric spirooxindoles as highly potent and efficacious MDM2 inhibitors. J Am Chem Soc 135: 7223–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu S, Sun W, Liu L, Lu J, McEachern D, Shargary S, Bernard D, Li X, Zhao T, et al. 2013b. A potent small-molecule inhibitor of the MDM2–p53 interaction (MI-888) achieved complete and durable tumor regression in mice. J Med Chem 56: 5553–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]