Latella et al. provide evidence of DNA damage response-mediated functional antagonism between senescence and MYOD-activated gene expression and indicate a previously unrecognized requirement of cell cycle progression for the activation of the myogenic program.
Keywords: MYOD, gene expression, senescence, DNA damage, cell cycle
Abstract
The molecular determinants of muscle progenitor impairment to regenerate aged muscles are currently unclear. We show that, in a mouse model of replicative senescence, decline in muscle satellite cell-mediated regeneration coincides with activation of DNA damage response (DDR) and impaired ability to differentiate into myotubes. Inhibition of DDR restored satellite cell differentiation ability. Moreover, in replicative human senescent fibroblasts, DDR precluded MYOD-mediated activation of the myogenic program. A DDR-resistant MYOD mutant could overcome this barrier by resuming cell cycle progression. Likewise, DDR inhibition could also restore MYOD's ability to activate the myogenic program in human senescent fibroblasts. Of note, we found that cell cycle progression is necessary for the DDR-resistant MYOD mutant to reverse senescence-mediated inhibition of the myogenic program. These data provide the first evidence of DDR-mediated functional antagonism between senescence and MYOD-activated gene expression and indicate a previously unrecognized requirement of cell cycle progression for the activation of the myogenic program.
Age-associated decline in muscle mass, contractile and metabolic performance, and regenerative ability coincides with the functional exhaustion of muscle stem (satellite) cells (MuSCs) (Brack and Rando 2007). Given the tremendous impact that global decline in muscle function has on organismal aging and life span, a great effort is currently directed toward elucidating pathways and networks that contribute to the progressive impairment of MuSCs in aged muscles and can be targeted by pharmacological interventions. Studies in the last decade have revealed that both intrinsic and extrinsic mechanisms contribute to the impairment of MuSC regenerative potential during aging (Conboy et al. 2005; Carlson et al. 2008; Sacco et al. 2010; Chakkalakal et al. 2012; Bentzinger and Rudnicki 2014; Bernet et al. 2014; Cosgrove et al. 2014; Price et al. 2014; Sinha et al. 2014; Sousa-Victor et al. 2014; Tierney et al. 2014). While some of these studies have established a correlation between aging, replicative senescence, and functional exhaustion of muscle regeneration, the molecular link between age-related MuSC impairment and cellular senescence remains elusive (Madaro and Latella 2015; Sacco and Puri 2015).
In particular, the potential antagonism between replicative senescence and the reduced regeneration ability of MuSCs has been suggested by previous studies (Sacco et al. 2010; Sousa-Victor et al. 2014). However, the relative contributions of senescence-mediated inhibition of the cell cycle, telomere shortening-induced DNA damage response (DDR) (Arnoult and Karlseder 2015), and/or potential mechanism of senescence-mediated inhibition of MYOD activity have not been investigated.
Results
Replicative senescence impairs activation of skeletal myogenesis
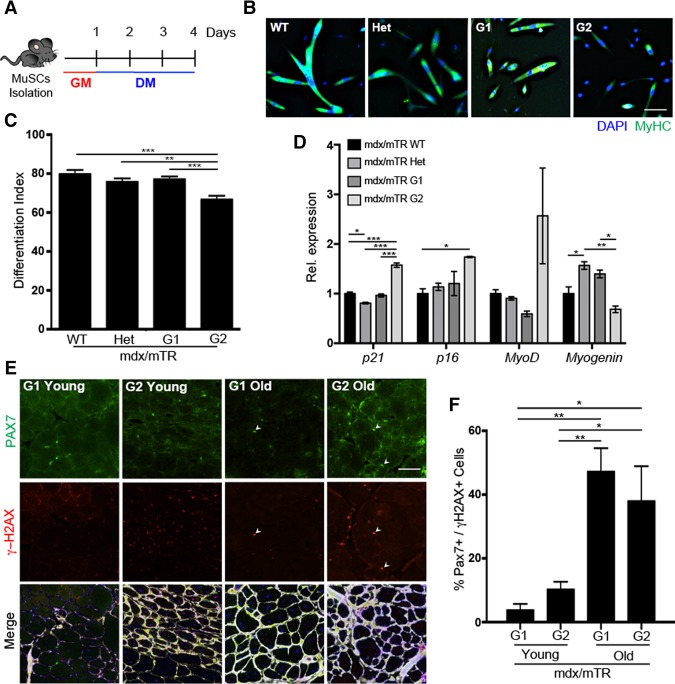
A Duchenne muscular dystrophy (DMD) mouse mutant lacking the RNA component of telomerase (mdx/mTR) provides a model of functional exhaustion of muscle regeneration due to the replicative senescence of telomerase-deficient MuSCs (Sacco et al. 2010). Progressive telomerase shortening is typically achieved by intercrossing mdx/mTRG1 to produce second-generation mdx/mTRG2 mice, in which the decline of the regeneration potential of MuSCs coincides with their replicative senescence. We used these mice to investigate whether the acquisition of a senescence-related phenotype in MuSCs alters their ability to differentiate into skeletal myotubes. MuSCs were isolated by fluorescent-activated cell sorting (FACS) (Gromova et al. 2015) and cultured in growth medium (GM) first and then in differentiation medium (DM) for three more days (Fig. 1A). Mdx/mTRG2-derived MuSCs displayed a compromised ability to differentiate, as indicated by the defective formation of myosin heavy chain (MyHC)-expressing myotubes (Fig. 1B,C) and reduced levels of Myogenin expression despite higher MyoD expression levels (Fig. 1D). Mdx/mTRG2-derived MuSCs also expressed higher levels of the cyclin-dependent kinase (CDK) inhibitors p21 (Cdkn1a) and p16 (Ink4a)—typical markers of cell cycle arrest and replicative senescence (Fig. 1D; Salama et al. 2014). Interestingly, in situ staining of muscles from mdx/mTRG2 mice revealed a progressive increase in the number of cells expressing PAX7 (a MuSC identification marker) (Seale et al. 2000) that were also positive for the DNA damage marker H2AX phosphorylated in Ser139 (γH2AX), as compared with mdx/mTRG1 mice, at either 2 mo of age (young) or 18–24 mo of age (old) (Fig. 1E,F). A persistent DDR is typically triggered in cells undergoing replicative senescence by telomere erosion (Arnoult and Karlseder 2015). γH2AX-positive nuclei have been observed in MuSCs of aged muscles (Sinha et al. 2014; Zhang et al. 2016) and, in our model, coincided with an impaired differentiation ability of senescent MuSCs from mdx/mTRG2 mice (Fig. 1B–D).
Figure 1.
MuSCs isolated from mdx/mTR mice display impaired differentiation, up-regulation of senescence markers, and DNA damage accumulation through disease progression. (A) Schematic representation of a MuSC differentiation experiment. (B) Immunofluorescence for MyHC (green) and DAPI (4′,6′-diamino-2-phenylindole) (blue) in MuSCs isolated from 9.5- to 11-mo-old mice and induced to differentiate for 3 d. (C) Differentiation index. (D) Quantitative PCR (qPCR) on isolated MuSCs from mdx/TR wild-type (WT), heterozygous (Het), G1, and G2 mice normalized for GAPDH. (E) Immunofluorescence for PAX7 (green), γH2AX (red), Laminin (white), and DAPI (blue) in gastrocnemius muscles isolated from 2-mo-old (young) and 18- to 24-mo-old (old) mdx/mTR mice. (F) Quantification of PAX7- and γH2AX-coexpressing cells. Bar, 25 µm.
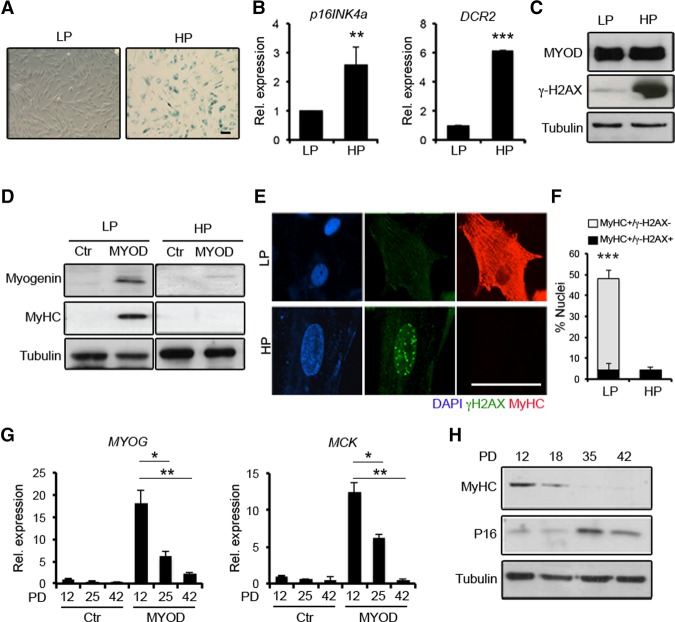
The increased MyoD expression and decreased levels of its immediate downstream target gene, Myogenin, in mdx/mTRG2-derived MuSCs suggests that replicative senescence could interfere with MYOD-mediated activation of the myogenic program. To further investigate the relationship between replicative senescence and activation of the myogenic program, we exploited the established model of MYOD-dependent activation of skeletal myogenesis in human primary fibroblasts, also referred to as myogenic conversion (Tapscott et al. 1988; Weintraub et al. 1989). This experimental setting is particularly suited to measure the efficiency of MYOD-dependent myogenic conversion in human primary fibroblasts as compared with their progeny undergoing replicative senescence in culture by serial passaging (Hayflick 1984). A progressive loss of proliferative ability was observed in human BJ-CRL2522 fibroblasts (BJs) along with the number of passages, with the large majority of the cells adopting typical morphological (i.e., flat phenotype) and biochemical (i.e., spontaneous positivity for β-galactosidase [β-Gal] activity) features of cellular senescence at population doubling (PD) 42 (Fig. 2A). At this stage, BJs expressed higher transcript levels of two senescence-associated genes: P16/INK4a and DCR2 (Fig. 2B; Collado et al. 2005). We therefore compared the ability of MYOD to activate the myogenic program in low passage (LP) proliferating BJs cultured at PD12 versus high passage (HP) BJs in which replicative senescence was induced by serial passages (PD42). Similar levels of MYOD protein were detected in LP and HP BJs following ectopic expression of MYOD by adenoviral infection (Fig. 2C). However, while MYOD was able to induce Myogenin and MyHC expression as well as promote the formation of MyHC-expressing myotubes in LP BJs, once cultured in DM, it failed to do so in HP BJs (Fig. 2D,E). Of note, elevated global γH2AX levels were detected in control (data not shown) as well as MYOD-expressing HP BJs (Fig. 2C), indicating a correlation between persistent DDR and impaired myogenic potential in senescent cells. Indeed, HP BJ resistance to conversion into MyHC-positive myotubes upon the ectopic expression of MYOD coincided with the presence of γH2AX-positive nuclear foci (Fig. 2E,F). Similarly, human primary IMR90 fibroblasts showed resistance to MYOD-mediated conversion (Supplemental Fig. S1) upon undergoing replicative senescence (PD55). Since the process of replicative senescence occurs gradually, being preceded by prereplicative senescence, we sought to monitor the ability of MYOD to convert fibroblasts once delivered at specific time points along with the transition toward senescence during serial passaging of BJs. We observed a progressive reduction of MYOD's ability to activate muscle gene expression throughout PD18–PD25 that culminated with a drastic inhibition at PD42 (Fig. 2G,H). These data indicate that the progressive acquisition of replicative senescence in fibroblasts coincides with activation of DDR and a proportional resistance to MYOD-mediated activation of the myogenic program.
Figure 2.
The resistance of human fibroblasts to MYOD-mediated conversion coincides with the activation of DDR by replicative senescence. (A) Primary human BJs at LP and HP PD tested for β-Gal staining. (B) Quantitative RT–PCR (qRT–PCR) for P16/Ink4a and DCR2 in LP and HP BJs. (C) Immunoblot for MYOD, γH2AX, and Tubulin in LP and HP BJs expressing MYOD. (D) Immunoblot for Myogenin, MyHC, and Tubulin in LP and HP BJs infected with mock (Ctr) and MYOD adenoviruses exposed to DM for 72 h. (E) Immunofluorescence for MyHC (red), γH2AX (green), and DAPI (blue) in LP and HP BJs expressing MYOD and exposed to DM for 72 h. (F) Quantification of the percentage of MyHC-positive/γH2AX-negative (white bars) and MyHC-positive/γH2AX-positive nuclei (black bars). (G) qRT–PCR for Myogenin (MYOG) and MCK in BJs at PD12, PD25, and PD42 Ctr- and MYOD-infected and shifted to DM for 48 h. (H) Immunoblot for MyHC, P16, and Tubulin in MYOD-expressing BJs at PD12, PD18, PD35, and PD42 shifted to DM for 48 h. Bar, 50 µm.
The correlation between resistance to MYOD-mediated conversion and persistent activation of DDR in senescent human fibroblasts is reminiscent of a previously described differentiation checkpoint by which muscle gene expression is transiently inhibited in muscle cells exposed to genotoxic stress via Tyr30 phosphorylation in MYOD by DNA damage-activated nuclear ABL kinase (Puri et al. 2002; Simonatto et al. 2011, 2013). Indeed, the active (autophosphorylated) form of ABL was detected in nuclei of senescent fibroblasts, which also displayed γH2AX nuclear foci; conversely, unphosphorylated ABL was detected almost exclusively in the cytoplasm of LP BJs, which do not exhibit DDR (Supplemental Fig. S2A,B). Similar to senescent fibroblasts, myoblasts from aged individuals showed reduced proliferation potential (Supplemental Fig. S3A,B) and nuclear expression of P16 (Supplemental Fig. S3C,D)—two hallmarks of cellular senescence not observed in myoblasts from young controls. Moreover, we observed a concomitant activation of DDR, as shown by the presence of nuclear foci positive for the DNA damage marker 53BP1 (Supplemental Fig. S3E) and nuclear accumulation of active ABL (Supplemental Fig. S3F) that correlated with an impaired ability to differentiate into myotubes (Supplemental Fig. S3E–G). In contrast, skeletal myoblasts from young individuals did not show 53BP1-positive foci or phosphorylated ABL and could differentiate into MyHC-positive myotubes (Supplemental Fig. S3E–G).
The functional antagonism between cellular senescence and differentiation is mediated by DDR via ABL-dependent phosphorylation of MYOD
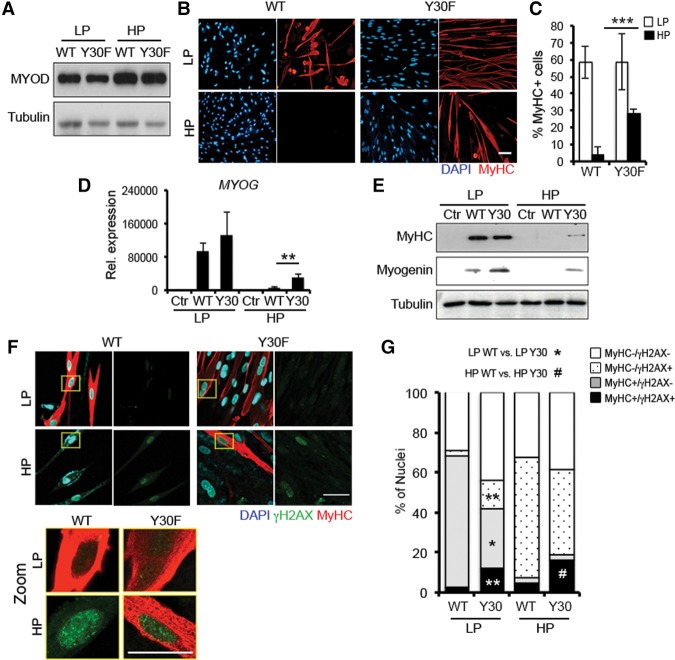
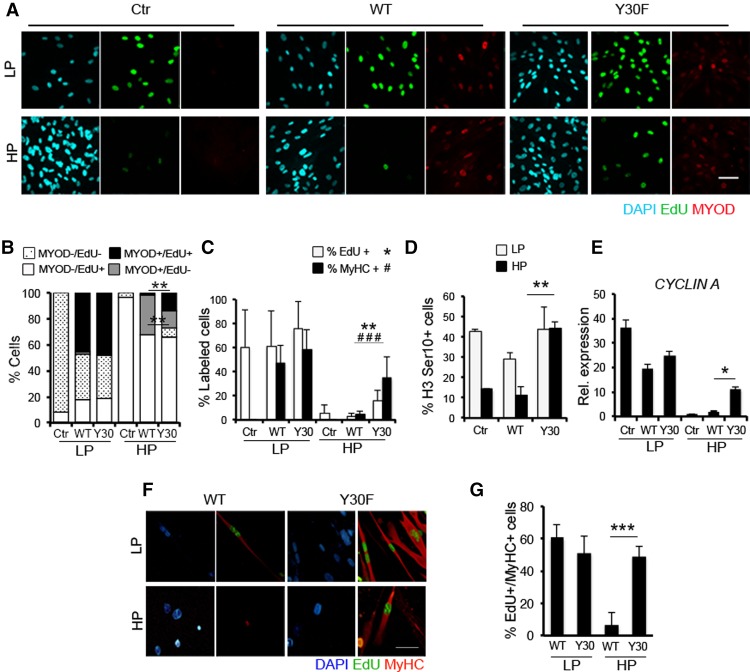
To determine the contribution of ABL signaling to the resistance of senescent fibroblasts to MYOD-mediated conversion, we compared the ability of wild-type MYOD versus the ABL phosphorylation-resistant MYOD/Y30F mutant (Puri et al. 2002) to convert LP or HP BJs. Wild-type MYOD or MYOD/Y30F was expressed at comparable levels in BJs (Fig. 3A). While both wild-type MYOD and MYOD/Y30F converted LP BJs into MyHC-positive myotubes with comparable efficiency, only the MYOD/Y30F mutant could convert HP BJs, which were otherwise resistant to wild-type MYOD (Fig. 3B,C). Indeed, the MYOD/Y30F mutant, but not wild-type MYOD, was able to induce the expression of MYOGENIN and MyHC in HP BJs (Fig. 3D,E). Of note, nuclei of MyHC-positive myotubes derived from MYOD/Y30F-converted HP BJs retained γH2AX-positive foci (Fig. 3F,G), which indicates persistent unrepaired DNA damage and DDR that can potentially lead to genomic instability. Indeed, these myotubes exhibited an increase in morphological nuclear abnormalities, including micronucleation, macronucleation, nuclear fragmentation, deformation, or polylobulation (Supplemental Fig. S4A,B), which have been associated previously with genomic instability (Yang et al. 2015).
Figure 3.
The DNA damage-resistant MYOD/Y30F mutant overcomes the resistance of senescent fibroblasts to myogenic conversion. (A) Immunoblot for MYOD in LP and HP BJs expressing wild-type MYOD (WT) or the MYOD/Y30F mutant (Y30F). (B) Immunofluorescence for MyHC (red) and DAPI (blue) in wild-type-expressing and Y30F-expressing LP and HP BJs shifted to DM for 72 h. (C) Quantification of the percentage of MyHC-positive nuclei in LP and HP BJs expressing wild-type MYOD and MYOD/Y30F. (D) qRT–PCR for Myogenin (MYOG) in Ctr, wild-type-expressing, and Y30F-expressing LP and HP BJs shifted to DM for 72 h. (E) Immunoblot for MyHC, Myogenin, and Tubulin in the same conditions as in D. (F, top) LP and HP BJs expressing wild-type MYOD or MYOD/Y30F shifted to DM for 72 h immunostained with MyHC (red), γH2AX (green), and nuclei (blue). (Bottom) Areas marked by the yellow boxes are shown at higher magnification (zoom). (G) Quantification of the percentage of γH2AX- and MyHC-positive cells. Bar 50 µm.
The ability of MYOD/Y30F to recover myogenic conversion in senescent cells coincides with the activation of cell cycle-associated genes
To further determine the differences in the transcription profiles that underlie the different phenotypes observed upon ectopic expression of wild-type MYOD versus the MYOD/Y30F mutant in HP BJs, we performed genome-wide transcriptome analysis by RNA sequencing (RNA-seq). We performed the analysis in LP and HP BJs expressing either wild-type MYOD or the MYOD/Y30F mutant after 36 h of culture in DM post-infection in order to capture early differences in gene expression associated with reversal of senescence-mediated repression of myogenesis by the MYOD/Y30F mutant (see Fig. 3A–E). A comparison of the transcriptional changes induced by wild-type MYOD and the MYOD/Y30F mutant in LP BJs revealed a substantial overlap of up-regulated (3011 transcripts) and down-regulated (2306 transcripts) genes (Supplemental Fig. S5A,B). Of note, more than approximately half of differentially expressed genes were shared with wild-type MYOD-expressing LP IMR90 fibroblasts (Supplemental Fig. S5A,B), indicating common transcriptional perturbations induced by MYOD in two distinct types of LP human primary fibroblasts. Common up-regulated genes were enriched in skeletal muscle-specific transcripts (Supplemental Fig. S5A), while common down-regulated genes were enriched in transcripts implicated in cell migration, cell death, and cell communications (Supplemental Fig. S5B). Within the differentially expressed genes, functional analysis by upstream prediction showed an enrichment of downstream targets of muscle-specific transcriptional activators (including myogenic basic helix–loop–helix [bHLH] and MEF2 factors) (Puri and Sartorelli 2000), cell cycle regulators (e.g., the retinoblastoma gene product RB1) (Sellers and Kaelin 1996), and muscle-specific microRNA (the myomiR miR-1) (Supplemental Fig. S5C; Horak et al. 2016). Gene ontology also revealed the induction of common genes associated with skeletal myogenesis and myofiber-specific activities (Supplemental Fig. S5D).
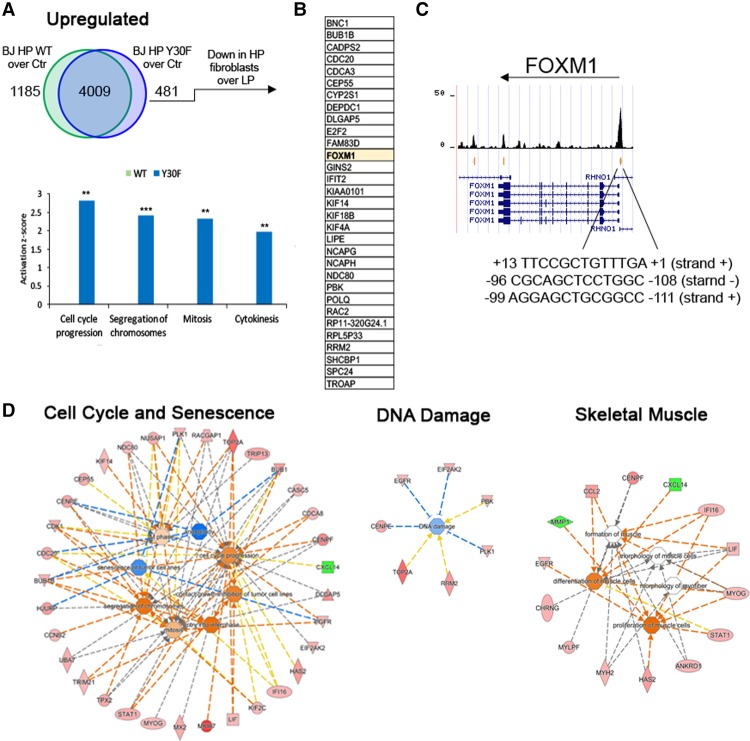
When expressed in HP BJs, both wild-type MYOD and MYOD/Y30F showed unique clusters of up-regulated genes compared with mock-infected HP BJs (Fig. 4A). Because of the unique ability of MYOD/Y30F to resume the myogenic program in senescent fibroblasts, we focused on the genes that were up-regulated in MYOD/Y30F and not in wild-type MYOD. Most of these genes were enriched in the categories related to cell proliferation and cytokinesis (Fig. 4A). We then further searched for those genes that are repressed by cellular senescence (i.e., down-regulated in HP BJs as compared with LP BJs) and whose expression was recovered by MYOD/Y30F in HP BJs. We identified a cluster of genes with this pattern of expression, and among them we found FOXM1—a gene that counters senescence by promoting cell cycle progression (Anders et al. 2011; Khongkow et al. 2014). Interestingly, ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data from MYOD-converted LP IMR90 fibroblasts (A Dall'Agnese and PL Puri, unpubl.) suggest that MYOD may directly activate FOXM1 transcription by binding Ebox sequences on the FOXM1 promoter (Fig. 4C). Gene ontology of all differentially expressed genes in HP BJs overexpressing MYOD/Y30F as compared with wild-type MYOD further identified activation of genes implicated in cell proliferation, skeletal muscle differentiation, and DNA repair (Fig. 4D). Among the genes involved in cell cycle activation, we noticed several putative FOXM1 targets (such as CENPE, TOP2A, and PLK1) and observed an enrichment of genes implicated in G1/S and G2/M transitions (Fig. 4C; Supplemental Fig. S6). Again, ChIP-seq data from MYOD-converted LP IMR90 fibroblasts show MYOD binding at Ebox sequences near the transcription start site (Supplemental Fig. S7) of the large majority of genes differentially expressed in HP BJs expressing MYOD/Y30F.
Figure 4.
MYOD/Y30F-specific modulation of gene expression in HP fibroblasts. (A, top) Venn diagram representing the genes common or uniquely up-regulated in HP BJs ectopically expressing wild-type MYOD (WT) or MYOD/Y30F (Y30F) compared with HP control mock BJs (Ctr). (Bottom) Functional comparison of the genes up-regulated only in HP BJs expressing wild-type MYOD (green) or MYOD/Y30F (blue) based on Ingenuity Pathway Analysis (IPA [Qiagen, http://www.qiagen.com/ingenuity]; shown at the left). The Y-axes represent the z-score. (B) List of genes up-regulated only in HP BJs ectopically expressing MYOD/Y30F compared with HP control mock BJs (Ctr) that are in common with genes down-regulated in HP control mock BJs compared with LP control mock BJs. (C) University of California at Santa Cruz Genome Browser screen shot of MYOD ChIP-seq on the FOXM1 gene. (D) Gene ontology of differentially expressed genes in HP BJs overexpressing MYOD/Y30F as compared with wild-type MYOD.
These results indicate that MYOD/Y30F's ability to rescue myogenic conversion in senescent fibroblasts is associated with a reversal of classical signatures of senescence, leading to resumption of the cell cycle. This evidence also suggests a general model by which transitioning through the cell cycle is a necessary event for the activation of the differentiation program in muscle progenitors. This model is consistent with MYOD expression typically restricted to activated proliferating MuSCs (Day et al. 2009) and the previously reported association of MYOD with transcriptional activation of S-phase genes (Zhang et al. 2010). Of note, the ability of MYOD to promote cell cycle progression is in apparent conflict with the previously reported cell cycle arrest properties of MYOD (Crescenzi et al. 1990; Sorrentino et al. 1990). However, it should be noted that MYOD's ability to impose cell cycle arrest has been so far observed in proliferating conditions. To clarify this issue, we evaluated the impact of MYOD on cell cycle in proliferating versus quiescent mouse and human fibroblasts. Mouse 10T1/2 fibroblasts and human BJs were either cultured in serum-containing proliferation medium or induced to quiescence by serum starvation for 48 h. Wild-type MYOD was expressed in these conditions followed by 4 h of EdU (5-ethynyl-2′-deoxyuridine) labeling. Supplemental Figure S8, A and B, shows a fivefold to fourfold reduction of EdU incorporation in quiescent fibroblasts as compared with their proliferating counterpart. In these conditions, while we observed a decrease in EdU-positive cells proliferating fibroblasts upon MYOD expression, the number of EdU-positive cells increased upon MYOD expression in quiescent fibroblasts. We sought to resolve this apparently contradictory relationship between MYOD-mediated fibroblast conversion and cell cycle activity by directly testing whether the inhibition of S phase by the DNA polymerase inhibitor aphidicolin (APH) could counter MYOD-induced myogenic conversion of fibroblasts. Indeed, treatment with APH could completely prevent EdU incorporation in either 10T1/2 fibroblasts or MYOD-expressing 10T1/2 fibroblasts (Supplemental Fig. S8C), and, despite comparable MYOD protein levels (Supplemental Fig. 8E), this invariably coincided with an inhibition of the activation of the myogenic program, as revealed by lack of expression of the downstream MYOD target Myogenin in 10T1/2 fibroblasts (Supplemental Fig. S8D,F). Likewise, APH treatment prevented MYOD-mediated conversion also in human fibroblasts (Supplemental Fig. S8G–I).
MYOD/Y30F-mediated cell cycle resumption precedes the ability of senescent cells to differentiate
We further investigated the relationship between MYOD–mediated activation of the cell cycle and the myogenic program by monitoring the EdU incorporation in LP and HP BJs expressing either wild-type MYOD or the MYOD/Y30F mutant. While the large majority of LP BJs incorporated EdU (Fig. 5A,B) and eventually differentiated into MyHC-positive myotubes (Fig. 5C) upon expression of either wild-type MYOD or the MYOD/Y30F mutant, wild-type MYOD-expressing HP BJs did not incorporate EdU and could not differentiate into MyHC-positive myotubes (Fig. 5C). In contrast, a significant proportion of MYOD/Y30F-expressing HP BJs incorporated EdU (Fig. 5A,B), and this coincided with the ability of the MYOD/Y30F mutant to override the inhibition of the myogenic program in these cells (Fig. 5C). The unique ability of MYOD/Y30F to reactivate the cell cycle in replicative senescent fibroblasts was further demonstrated by the increased frequency of nuclei positive for the mitotic marker phospho-H3 Ser10 (H3S10) and the up-regulation of the G1–S-phase marker Cyclin A in HP BJs expressing MYOD/Y30F but not wild-type MYOD (Fig. 5D,E). To demonstrate that the transition through the S-phase precedes the activation of the differentiation program in myogenic progenitors, we challenged LP and HP BJs expressing either wild-type MYOD or the MYOD/Y30F mutant with a long exposure to EdU (24 h) while inducing them to differentiate in DM. We reasoned that the presence of EdU-positive nuclei in MyHC-positive myotubes reflects the transition through the S phase in MYOD-expressing fibroblasts prior to their differentiation into skeletal myotubes. Indeed, a large proportion (∼60%) of LP BJs, which are competent to convert into skeletal muscle upon MYOD expression, displayed EdU-positive nuclei within MyHC-positive myotubes (Fig. 5F,G). In contrast, HP BJs, which are resistant to wild-type MYOD-mediated myogenic conversion, did not incorporate EdU (Fig. 5F,G). MYOD/Y30F enforced this resistance by inducing S phase in these cells, as indicated by the formation of myotubes with EdU-positive nuclei (Fig. 5F,G).
Figure 5.
The S-phase transition is coupled with the ability of the MYOD/Y30F mutant to bypass the differentiation block imposed by cellular senescence. (A) Immunofluorescence for MYOD (red), EdU (green), and DAPI (blue) in empty (Ctr) LP and HP BJs and LP and HP BJs expressing wild-type MYOD (WT) or MYOD/Y30F (Y30F) exposed to a 16-h EdU pulse 24 h after infection in GM. (B) Quantification of the percentage of MYOD- and EdU-positive cells. (C) Quantification of the percentage of EdU-positive (white bars) and MyHC-positive (black bars) nuclei in control (Ctr) LP and HP BJs and BJs expressing wild type or Y30F exposed to a 16-h EdU pulse in GM and then shifted to DM for 72 h. (D) Quantification of the percentage of H3S10 in LP and HP control, wild-type-expressing, or Y30F-expressing BJs in GM. (E) qRT–PCR analysis for Cyclin A in LP and HP control, wild-type-expressing, or Y30F-expressing BJs shifted to DM for 24 h. (F) Immunofluorescence for MyHC (red), EdU (green), and DAPI (blue) in LP and HP wild-type-expressing or Y30F-expressing BJs shifted to DM plus EdU for 72 h. (G) Quantification of the percentage of EdU-positive nuclei within MyHC-positive myotubes. Bar, 50 µm.
Cell cycle progression is required for MYOD/Y30F-mediated reactivation of the myogenic program in senescent fibroblasts
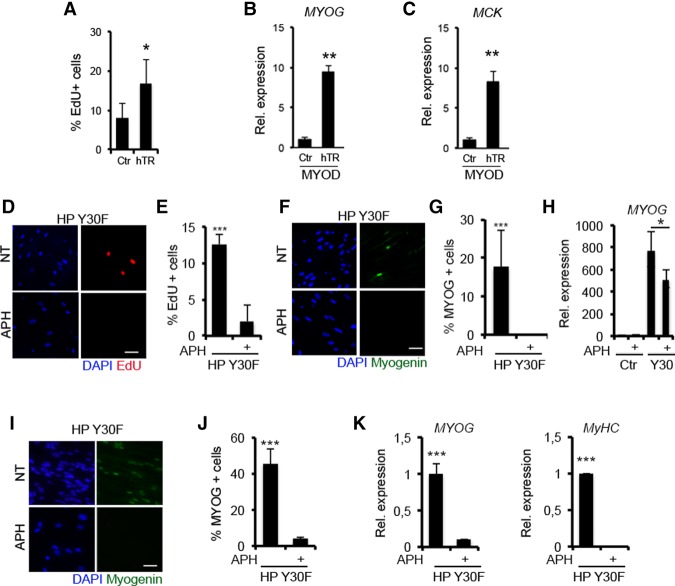
To conclusively demonstrate that re-entry into the cell cycle is a key event required for the activation of the myogenic program in replicative senescent fibroblasts, we first tested whether preventing the replicative senescence—by ectopically expressing human telomerase (hTERT)—was sufficient to enable wild-type MYOD-mediated activation of the myogenic program in HP BJs. Once expressed by retroviral infection in presenescent fibroblasts at PD35, hTERT could prevent the senescence-associated cell cycle arrest in ∼20% of the cells (Fig. 6A). In these cells, wild-type MYOD leads to Myogenin and MCK expression, which could not be detected in pBabe control wild-type MYOD HP BJs (Fig. 6B,C). In a reciprocal strategy, we showed that MYOD/Y30F's ability to activate the myogenic program in senescent fibroblasts required the cell cycle activity. Indeed, the S-phase inhibitor APH effectively prevented both S-phase progression (Fig. 6D,E) and activation of the myogenic program (Fig. 6F–H) in MYOD/Y30F-expressing HP BJs. Likewise, APH could block MYOD/Y30F-mediated recovery of the myogenic program in HP IMR90 fibroblasts (Fig. 6I–K).
Figure 6.
Cell cycle transitioning is needed to execute the myogenic program. (A–C) Presenescent fibroblasts at PD35 were infected with pBabe (Ctr) and hTERT (hTR) retroviruses and then kept in culture for an additional five PD. (A) Quantification of the percentage of EdU-positive cells. Myogenin (MYOG) (B) and MCK (C) RNA expression levels in empty (Ctr) and hTERT (hTR) BJs expressing MYOD shifted to DM for 48 h. Representative images of EdU (red) incorporation (D) and Myogenin expression (F) by immunofluorescence in HP BJs expressing the MYOD/Y30F mutant treated with APH in DM for 24 h. Quantification of the percentage of EdU-positive (E) and Myogenin-positive (G) cells. (H) Myogenin RNA expression levels in HP BJs (Ctr) and HP BJs expressing the MYOD/Y30F mutant treated with APH in DM for 24 h. (I) Representative images of Myogenin expression by immunofluorescence in HP IMR90 fibroblasts expressing the MYOD/Y30F mutant treated with APH in DM for 24 h. (J) Quantification of the percentage of Myogenin-positive cells. (K) Myogenin and MyHC RNA expression levels in HP IMR90 fibroblasts (Ctr) and HP IMR90 fibroblasts expressing the MYOD/Y30F mutant treated with APH in DM for 24 h. Bar, 50 µm.
DDR inhibition restores the myogenic program in wild-type MYOD-expressing replicative senescent fibroblasts and mdx/mTRG2 MuSCs
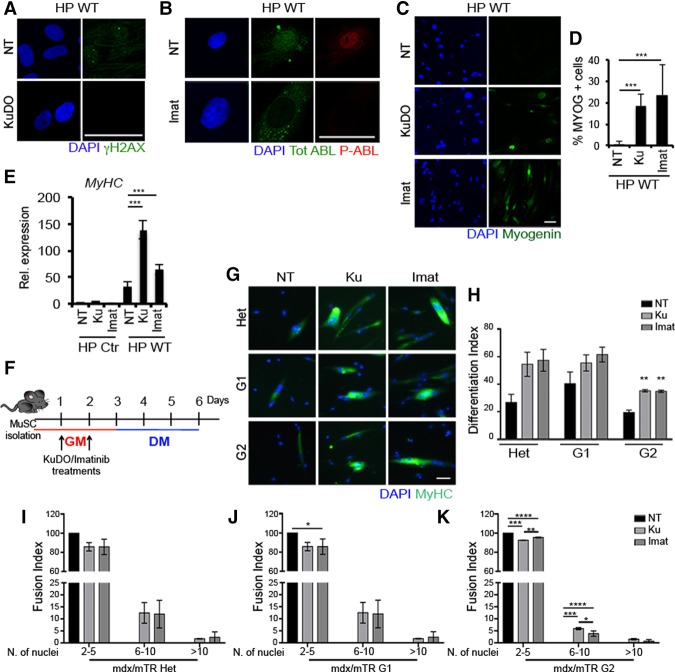
Finally, we tested whether inhibiting the senescence-activated DDR could enable wild-type MYOD to restore the myogenic program in senescent cells. We first tested whether soluble compounds that target the kinase activity of two key components of the DDR signaling to MYOD allowed wild-type MYOD to reverse the inhibition imposed by senescence in fibroblasts. To this purpose, HP BJs were exposed to either KuDO, which inhibits the DDR upstream activator ATM, or imatinib, which inhibits the terminal DDR effector ABL. Exposure to KuDO effectively inhibited the formation of γH2AX foci in HP BJs (Fig. 7A), while exposure to imatinib inhibited the activation of nuclear ABL (Fig. 7B). Moreover, treatment with either KuDO or imatinib could restore wild-type MYOD's ability to activate Myogenin (Fig. 7C,D) and MyHC (Fig. 7E) in HP BJs.
Figure 7.
DDR inhibition restores the differentiation ability in senescent fibroblasts expressing wild-type MYOD (WT) and in satellite cells from mdx/mTRG2 mice. (A) Representative images for γH2AX (green) and DAPI (blue) immunofluorescence of wild-type MYOD-expressing HP BJs treated with 10 µM KuDO for 24 h in GM. (B) Representative images for ABL (green), phospho-ABL (red), and DAPI (blue) immunofluorescence in wild-type MYOD-expressing HP BJs treated with 5 µM imatinib for 24 h in GM. (C) Representative images for Myogenin (green) and DAPI (blue) immunofluorescence of wild-type MYOD-expressing HP BJs treated with KuDO or imatinib for 24 h in GM and shifted to DM for 24 h. (D) Quantification of the percentage of Myogenin-positive cells. (E) MyHC RNA expression levels in HP BJs (Ctr) and HP BJs expressing wild-type MYOD treated with KuDO or imatinib in GM then shifted to DM for 48 h. (F) Schematic representation of DDR inhibition treatments in MuSCs. (G) Immunofluorescence for MyHC (green) and DAPI (blue) in MuSCs isolated from 8- to 20-mo-old mice treated for 2 d with KuDO or imatinib in GM and induced to differentiate for 4 d. (H) Differentiation index. Fusion index for MuSCs isolated from mdx/mTRHet (I), mdx/mTRG1 (J), and mdx/mTRG2 (K). Bar, 50 µm.
We next exposed mdx/mTR-derived MuSCs to the same DDR inhibitors. MuSCs were exposed to either KuDO or imatinib while cultured in GM and then switched to DM for four additional days without these compounds (Fig. 7F). Both compounds could reverse the differentiation block imposed by replicative senescence in mdx/mTRG2-derived MuSCs by increasing the formation of MyHC-positive myotubes as compared with control-treated cells (Fig. 7G,H). A similar trend could be observed in mdx/mTRHet- and mdx/mTRG1-derived MuSCs but was not statistically significant (Fig. 7G,H). Either KuDO- or imatinib-treated mdx/mTRG2-derived MuSCs showed a higher ability to fuse into MyHC-expressing multinucleated myotubes (Fig. 7I–K) and displayed a reduced number of γH2AX-positive nuclei (Supplemental Fig. S9).
Of note, the restoration of the myogenic program in KuDO- or imatinib-treated mdx/mTRG2-derived MuSCs appeared related to their ability to restore MYOD-mediated activation of myogenin, as suggested by the increased expression of myogenin but not Myod (Supplemental Fig. S10). This effect is consistent with the notion that senescence inhibits MYOD-mediated activation of muscle gene expression through the DDR.
Discussion
The functional antagonism between replicative senescence and the activation of the myogenic program described here provides a potential molecular mechanism responsible for the impairment of the regenerative potential observed in muscles of geriatric mice (Sousa-Victor et al. 2014). However, an earlier impairment of regenerative potential of aged muscles has been observed even before geriatric age (Chakkalakal et al. 2012; Bentzinger and Rudnicki 2014; Bernet et al. 2014; Cosgrove et al. 2014; Price et al. 2014; Sinha et al. 2014). It is possible that an initial exhaustion of MuSCs is due to deregulated symmetric cell division and is followed by the replicative senescence that impairs the myogenic potential of the residual pool of MuSCs in geriatric muscles. While correction of deregulated intracellular networks (such as the p38 or STAT3 signaling) has been shown to reverse age-related decline of muscle regeneration at pregeriatric stages (Bentzinger and Rudnicki 2014; Bernet et al. 2014; Cosgrove et al. 2014; Price et al. 2014; Tierney et al. 2014), replicative senescence appears to pose an additional barrier —such as the irreversible cell cycle arrest—to the activation of the myogenic program. In this regard, it is interesting that enforcing the reactivation of the cell cycle by MYOD/Y30F is sufficient to activate the myogenic program in senescent cells; however, our data show that the resulting myotubes continue to exhibit a persistent DDR and nuclear abnormalities that indicate genomic instability. This finding raises concerns about the potential detrimental effects of anti-aging treatments aimed at reversing cellular senescence, as they might lead to the formation of genetically unstable myofibers with impaired functions. An alternative approach has been proposed to prevent MuSC senescence by NAD repletion through an improvement of mitochondrial function, which is effective in countering the senescence-associated cell cycle arrest and activation of the DDR (Zhang et al. 2016). As the DDR-resistant MYOD/Y30F mutant overrides senescence-associated inhibition of the myogenic program, we propose that DNA damage signaling to MYOD is a critical mediator of the functional antagonism between replicative senescence and the activation of the myogenic program. While this signaling mediates a transient inhibition of the differentiation program in cells exposed to genotoxic stress, until the DNA lesions have been repaired (the so-called differentiation checkpoint [Puri et al. 2002]), in replicative senescent cells, a persistent activation of the DDR appears to continuously interfere with the activation of the myogenic program. We show here that DDR inhibition in senescent muscle progenitors is required for formation of terminally differentiated skeletal muscles that do not exhibit evidence of unrepaired DNA damage.
Our data also demonstrate that the senescence-associated cell cycle arrest provides an insurmountable barrier to the activation of the myogenic program by wild-type MYOD, which is otherwise sufficient to promote cell cycle entry and differentiation in conditions of reversible quiescence, such as in quiescent MuSCs. This finding reveals an intimate relationship between DNA replication, mitosis, and activation of the myogenic program by MYOD that has not been reported by previous studies. The relationship between cell cycle progression and activation of the myogenic program was anticipated by the ability of MYOD to directly activate S-phase genes (Zhang et al. 2010). Additional evidence in support of the importance of S phase for the activation of the myogenic program has also been suggested by Kosak and colleagues (Neems et al. 2016).
Because of the importance of cell cycle progression for the transcription of histones, related variants, and chaperons (Gurard-Levin et al. 2014) and given that decreased histone biosynthesis has been observed in aged MuSCs (Ivanov et al. 2013; Liu et al. 2013; Rai et al. 2014), we are tempted to speculate that histone depletion might contribute to the inhibition of muscle gene expression in senescent cells, presumably by depleting the nucleosomes from histone variants that could favor MYOD access to repressive chromatin. In this regard, the observed unique ability of MYOD/Y30F to restore the expression of histone variants and chaperons through the cell cycle activation further suggests that cell cycle-associated recovery of an optimal repertoire of histones enables the formation of nucleosomes amenable to MYOD access and the consequent activation of muscle gene expression.
Materials and methods
Animals
All protocols were approved by the Sanford Burnham Prebys Medical Discovery Institute Animal Care and Use Committee. mdx/mTR mice were kindly donated by H.M. Blau (Sacco et al. 2010). Male mice of 2–24 mo of age were used for this study.
Satellite cell isolation
MuSCs were isolated as described previously with minor revisions (Sacco et al. 2008; Gromova et al. 2015). Tibialis anterior, gastrocnemius, and quadriceps muscles of mice were minced and subjected to enzymatic dissociation (700 U/mL collagenase) for 1 h followed by another 30 min of incubation in 100 U/mL collagenase and 2 U/mL dispase. The cells were passed through a 20-gauge needle 10 times, subsequently filtered through a 70-μm nylon filter, and incubated with the following biotinylated rat antibodies: CD45 (clone 30-F11), CD11b (catalog no. 553309), CD31 (catalog no. 5011513), and Sca1 (clone E13-161.7) (BD Bioscience). Streptavidin beads (Miltenyi Biotech) were then added to the cells together with the antibodies integrin-α7-phycoerythrin (Ablab, catalog no. R2F2) and CD34-Alexa647 (eBioscience, clone RAM34), after which magnetic depletion of biotin-positive cells was performed. The (CD45−CD11b−CD31−Sca1−) CD34+integrin-α7+ population was then fractionated by flow cytometry (BD FACS Aria) followed by a purity check.
Cell culture and treatments
Primary human BJs were purchased from American Type Culture Collection and cultured in GM (DMEM supplemented with 10% FBS, nonessential amino acids). We passaged cells, measuring the PD until the proliferation rate slowed down and eventually ceased once cells reached replicative senescence. Muscle differentiation was induced by incubating cells in DMEM supplemented with 2% horse serum (DM). We obtained human IMR90 fibroblasts from the Coriell Institute for Medical Research. IMR90 fibroblasts were grown in GM (EMEM supplemented with 10% FBS). When cells were 95%–100% confluent, MYOD was induced with 200 ng/mL doxycycline (Sigma, D3072) in GM for 24 h, and then cells were differentiated in EMEM supplemented with 2% horse serum (Gibco, 16050), 1% ITS (Sigma, I2146), and 200 ng/mL doxycycline for 3 d for the DM time point. Medium with doxycycline was refreshed every 2 d. Muscle differentiation in IMR90 fibroblasts was induced in incubating cells in EMEM supplemented with 2% HS and 1% ITS (Sigma).
To block ATM, we used the ATM kinase inhibitor KU-55933 (Selleckchem) at concentrations of 10 µM in fibroblasts and 5 µM in MuSCs isolated from mdx/mTR mice. Pharmacological inhibition of the ABL kinase was achieved with imatinib at concentrations of 5 µM in fibroblasts and 2.5 µM in MuSCs isolated from mdx/mTR mice. Fibroblasts were treated with KuDO or imatinib 24 h before DM addition, whereas MuSCs were treated for 2 d before DM incubation.
To block the cells cycle, LP BJs, 10T1/2 fibroblasts expressing wild-type MYOD, and HP BJs expressing MYOD/Y30F were treated with 3.3 µL APH (Sigma) administered upon DM incubation.
Viral infections
Wild-type MYOD or MYOD/Y30F ectopical expression was achieved by infecting BJs at different passages with adeno-wild-type MYOD and adeno-MYOD/Y30F (or mock [control]) at a multiplicity of infection (MOI) of 800 for 1 h and then placing them in GM for 24 h; where indicated, differentiation was induced by incubating cells in DM.
Retroviral infection with retro-pBabe and retro-pLhTERT was performed in presenescent BJs (PD35). Cells were allowed to proliferate for additional five PDs and tested for their proliferative capacity or infected with MYOD and tested for their ability to express muscle-specific differentiation markers after exposure to DM for 3 d.
β-Gal staining
BJs were fixed in 4% formaldehyde and incubated at 37°C without CO2 with the following solution: 1 mg/mL X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2 in 40 mM citric acid/sodium phosphate (pH 6.0). Staining was evident within 24 h.
Immunoblot
Proteins were extracted with RIPA buffer separated on polyacrylamide gel and transferred to nitrocellulose filters. The following primary antibodies were used to detect protein levels: MF 20 (Developmental Studies Hybridoma Bank [DSHB] hybridoma product by D.A. Fischman) to detect MyHC, Myogenin (F5D; DSHB hybridoma product by D.A. Fischman), MYOD antibody (5.8 Dako; Santa Cruz Biotechnology, M-318), Ser139 H2AX (Millipore, 05-636), P16 (Santa Cruz Biotechnology, C-20), Tubulin (Neo Markers, Ab4), and GAPDH (Santa Cruz Biotechnology, 6C5). Primary antibodies were visualized with the ECL (Amersham) chemioluminescent kit following the manufacturer's instructions.
Immunofluorescence
For immunofluorescence, BJs and IMR90 fibroblasts were plated on glass coverslips and, when indicated, shifted to DM. Single or double immunofluorescence was performed with the following primary antibodies: MF20 (DSHB), Myogenin (DSHB), Ser139 H2AX (Millipore, 05-636), P16 (Santa Cruz Biotechnology, C-20), MYOD (Dako, 5.8), 53BP1 (Novus, NB100-304), H3S10 (Cell Signaling Technology, 9701S), ABL (Calbiochem, Ab3), and phospho-Y412-ABL (Abcam, ab4717). We used secondary antibodies from Alexa (Life Technologies). DNA synthesis was evaluated by EdU incorporation following the manufacturer's instructions (Invitrogen). Nuclei were visualized by DAPI (4′,6′-diamino-2-phenylindole). The images were analyzed with a LSM5 Pascal Zeiss confocal microscope.
RNA extraction and RT–PCR
Total RNA was extracted with Trizol (Invitrogen), according to the manufacturer's instructions. RNA (0.5–1 µg) was retrotranscribed using the TaqMan reverse transcription kit (Applied Biosystems). MuSC total RNA was extracted with miRNeasy microkit (Qiagen). Twenty nanograms of RNA was retrotranscribed using the SuperScript VILO transcription kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) was performed to analyze relative gene expression levels using SYBR Green master mix (Applied Biosystems). Relative gene expression was normalized for GAPDH or Rplpo expression value and calculated using the 2−δΔCT method. Primers sequences are listed in Supplemental Table S1.
RNA-seq procedure and data analysis
LP and HP BJs were infected with empty, wild-type MYOD, or MYOD/Y30F adenoviruses and shifted to DM for 36 h. IMR90 LP fibroblasts (PD28) expressing wild-type MYOD or not were induced to differentiate for 3 d prior to RNA extraction. Cells were collected, and RNA was isolated with Trizol. One microgram of BJ RNA was sent to the Institute of Applied Genomics (Udine, Italy) for deep sequencing, while IMR90 RNA was sequenced in HiSeq 2000 in Dr. Bing Ren's laboratory. PolyA RNA-seq was performed following Illumina's kit. Reads were mapped using TopHat2.1.1 (Kim et al. 2013) with options “–p 8 –g 1 –segment-length 12 –library-type fr-unstranded” to the Homo sapiens hg19 genomes. HTSeq-0.6.1p1 (Anders et al. 2015) with “–stranded=no option” was used to assigned mapped reads to the H. sapiens GRCh37.75 genes. Differential expression analysis was performed using DESeq2. Genes were considered differentially expressed if P < 0.05 and fold change was <0.54 or >1.83. Ingenuity Pathway Analysis (IPA [Qiagen, http://www.qiagen.com/ingenuity]) was used for gene ontology.
Statistical analysis and repeatability of experiments
Statistical significance and P-values were determined using one-way ANOVA (Figs. 1, 7H), two-way ANOVA (Fig 7I,L,M), and the two-tailed Student's t-test (P-value ≤ 0.05 [*]; P-value ≤ 0.01 [**]; P-value ≤ 0.001 [***]; P-value ≤ 0.0001 [****]) (Figs. 2–7). Two to five independent experiments were performed on qRT–PCR, immunostaining, and immunoblotting in Figures 1–3 and 5; two independent experiments were performed for hTERT reconstitution in Figure 6; and two independent experiments were performed on qRT–PCR and immunostaining in Figures 6 and 7.
Data are available under Gene Expression Omnibus accession number GSE98136.
Supplementary Material
Acknowledgments
We thank L. Boyd; B. Charbono; Amy Cortez; and the Cell Imaging Facility, Animal Facility, and Flow Cytometry Core Facility at Sanford Burnham Prebys Medical Discovery Institute for technical support. We thank Dr. Alessandro Rosa for sharing Helper and epB-Puro-TT plasmids; Dr. David Huhta for the administration of computer cluster; Dr. Bing Ren, Dr. Yarui Diao, Dr. Ah Young, and Dr. Samantha Kuan for library preparation and sequencing of MYOD ChIP; Dr S. Gatto for assistance in bioinformatic analysis; and Dr. M. Crescenzi and Dr. P. Adams for critical reading of the manuscript and suggestions. This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants R01 AR056712 and R01AR052779 to P.L.P.; R01 AR064873 to A.S., L.L., and P.L.P.; and P30 AR061303 to P.L.P. and A.S. This work was also supported by the Epigen Project Progetto Bandiera Epigenomica IFT to L.L. and P.L.P., Ellison Medical Foundation grant AG-NS-0843-11, the Glenn Award for Aging Research to A.S., and American Federation for Aging Research grant G16294 to A.D.A.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.293266.116.
References
- Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, Zhai H, Vidal M, Gygi SP, Braun P, et al. 2011. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 20: 620–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N, Karlseder J. 2015. Complex interactions between the DNA-damage response and mammalian telomeres. Nat Struct Mol Biol 22: 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Rudnicki MA. 2014. Rejuvenating aged muscle stem cells. Nat Med 20: 234–235. [DOI] [PubMed] [Google Scholar]
- Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB. 2014. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med 20: 265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Rando TA. 2007. Intrinsic changes and extrinsic influences of myogenic stem cell function during aging. Stem Cell Rev 3: 226–237. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Hsu M, Conboy IM. 2008. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature 454: 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, Brack AS. 2012. The aged niche disrupts muscle stem cell quiescence. Nature 490: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. 2005. Tumour biology: senescence in premalignant tumours. Nature 436: 642. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. 2005. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433: 760–764. [DOI] [PubMed] [Google Scholar]
- Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, Llewellyn ME, Delp SL, Blau HM. 2014. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med 20: 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzi M, Fleming TP, Lassar AB, Weintraub H, Aaronson SA. 1990. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc Natl Acad Sci 87: 8442–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day K, Paterson B, Yablonka-Reuveni Z. 2009. A distinct profile of myogenic regulatory factor detection within Pax7+ cells at S phase supports a unique role of Myf5 during posthatch chicken myogenesis. Dev Dyn 238: 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova A, Tierney MT, Sacco A. 2015. FACS-based satellite cell isolation from mouse hind limb muscles. Bio Protoc 5: e1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard-Levin ZA, Quivy JP, Almouzni G. 2014. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem 83: 487–517. [DOI] [PubMed] [Google Scholar]
- Hayflick L. 1984. Intracellular determinants of cell aging. Mech Ageing Dev 28: 177–185. [DOI] [PubMed] [Google Scholar]
- Horak M, Novak J, Bienertova-Vasku J. 2016. Muscle-specific microRNAs in skeletal muscle development. Dev Biol 410: 1–13. [DOI] [PubMed] [Google Scholar]
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. 2013. Lysosome-mediated processing of chromatin in senescence. J Cell Biol 202: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khongkow P, Karunarathna U, Khongkow M, Gong C, Gomes AR, Yague E, Monteiro LJ, Kongsema M, Zona S, Man EP, et al. 2014. FOXM1 targets NBS1 to regulate DNA damage-induced senescence and epirubicin resistance. Oncogene 33: 4144–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. 2013. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 4: 189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaro L, Latella L. 2015. Forever young: rejuvenating muscle satellite cells. Front Aging Neurosci 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neems DS, Garza-Gongora AG, Smith ED, Kosak ST. 2016. Topologically associated domains enriched for lineage-specific genes reveal expression-dependent nuclear topologies during myogenesis. Proc Natl Acad Sci 113: E1691–E1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price FD, von Maltzahn J, Bentzinger CF, Dumont NA, Yin H, Chang NC, Wilson DH, Frenette J, Rudnicki MA. 2014. Inhibition of JAK–STAT signaling stimulates adult satellite cell function. Nat Med 20: 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V. 2000. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol 185: 155–173. [DOI] [PubMed] [Google Scholar]
- Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY. 2002. A myogenic differentiation checkpoint activated by genotoxic stress. Nat Genet 32: 585–593. [DOI] [PubMed] [Google Scholar]
- Rai TS, Cole JJ, Nelson DM, Dikovskaya D, Faller WJ, Vizioli MG, Hewitt RN, Anannya O, McBryan T, Manoharan I, et al. 2014. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev 28: 2712–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Puri PL. 2015. Regulation of muscle satellite cell function in tissue homeostasis and aging. Cell Stem Cell 16: 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. 2008. Self-renewal and expansion of single transplanted muscle stem cells. Nature 456: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, et al. 2010. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell 143: 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama R, Sadaie M, Hoare M, Narita M. 2014. Cellular senescence and its effector programs. Genes Dev 28: 99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell 102: 777–786. [DOI] [PubMed] [Google Scholar]
- Sellers WR, Kaelin WG. 1996. RB [corrected] as a modulator of transcription. Biochim Biophys Acta 1288: M1–M5. [DOI] [PubMed] [Google Scholar]
- Simonatto M, Giordani L, Marullo F, Minetti GC, Puri PL, Latella L. 2011. Coordination of cell cycle, DNA repair and muscle gene expression in myoblasts exposed to genotoxic stress. Cell Cycle 10: 2355–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonatto M, Marullo F, Chiacchiera F, Musaro A, Wang JY, Latella L, Puri PL. 2013. DNA damage-activated ABL-MyoD signaling contributes to DNA repair in skeletal myoblasts. Cell Death Differ 20: 1664–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Jang YC, Oh J, Khong D, Wu EY, Manohar R, Miller C, Regalado SG, Loffredo FS, Pancoast JR, et al. 2014. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 344: 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V, Pepperkok R, Davis RL, Ansorge W, Philipson L. 1990. Cell proliferation inhibited by MyoD1 independently of myogenic differentiation. Nature 345: 813–815. [DOI] [PubMed] [Google Scholar]
- Sousa-Victor P, Gutarra S, Garcia-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V, Jardi M, Ballestar E, Gonzalez S, Serrano AL, et al. 2014. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506: 316–321. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. 1988. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242: 405–411. [DOI] [PubMed] [Google Scholar]
- Tierney MT, Aydogdu T, Sala D, Malecova B, Gatto S, Puri PL, Latella L, Sacco A. 2014. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat Med 20: 1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. 1989. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci 86: 5434–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZJ, Broz DK, Noderer WL, Ferreira JP, Overton KW, Spencer SL, Meyer T, Tapscott SJ, Attardi LD, Wang CL. 2015. p53 suppresses muscle differentiation at the myogenin step in response to genotoxic stress. Cell Death Differ 22: 560–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Sha J, Harter ML. 2010. Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J Cell Biol 188: 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D'Amico D, Ropelle ER, Lutolf MP, Aebersold R, et al. 2016. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352: 1436–1443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.