Shapira et al. show that EBF2 physically interacts with the chromatin remodeler BRG1 and the BAF chromatin remodeling complex in brown adipocytes. They identified the histone reader protein DPF3 as a brown fat-selective component of the BAF complex that was required for brown fat gene programming and mitochondrial function.
Keywords: EBF2, DPF3, BAF chromatin remodeling complex, brown adipogenesis, UCP1
Abstract
The transcription factor early B-cell factor 2 (EBF2) is an essential mediator of brown adipocyte commitment and terminal differentiation. However, the mechanisms by which EBF2 regulates chromatin to activate brown fat-specific genes in adipocytes were unknown. ChIP-seq (chromatin immunoprecipitation [ChIP] followed by deep sequencing) analyses in brown adipose tissue showed that EBF2 binds and regulates the activity of lineage-specific enhancers. Mechanistically, EBF2 physically interacts with the chromatin remodeler BRG1 and the BAF chromatin remodeling complex in brown adipocytes. We identified the histone reader protein DPF3 as a brown fat-selective component of the BAF complex that was required for brown fat gene programming and mitochondrial function. Loss of DPF3 in brown adipocytes reduced chromatin accessibility at EBF2-bound enhancers and led to a decrease in basal and catecholamine-stimulated expression of brown fat-selective genes. Notably, Dpf3 is a direct transcriptional target of EBF2 in brown adipocytes, thereby establishing a regulatory module through which EBF2 activates and also recruits DPF3-anchored BAF complexes to chromatin. Together, these results reveal a novel mechanism by which EBF2 cooperates with a tissue-specific chromatin remodeling complex to activate brown fat identity genes.
Adipose tissue plays a central role in regulating energy balance in mammals. Brown adipose tissue (BAT) is specialized to expend energy through the action of mitochondrial uncoupling protein UCP1 (Harms and Seale 2013). UCP1 dissipates the proton gradient generated by the electron transport chain, stimulating the breakdown of available substrates to release heat (Fedorenko et al. 2012). Additionally, beige UCP1+ adipocytes develop within white adipose depots in response to cold or β-adrenergic stimulation (Cannon and Nedergaard 2004; Seale et al. 2011). Increasing brown and beige fat mass or activity through genetic or chemical manipulation in mice suppresses obesity and associated metabolic disorders (Rosen and Spiegelman 2014). Additionally, mild cold exposure in humans also activates thermogenic fat cells, and this is associated with increased energy expenditure and reduced body fat mass (Yoneshiro et al. 2013). As such, there is great interest in developing approaches to increase the amount and/or function of brown fat to combat obesity and related diseases.
The helix–loop–helix transcription factor early B-cell factor 2 (EBF2) is required for the differentiation and function of brown and beige adipocytes (Rajakumari et al. 2013; Wang et al. 2014; Stine et al. 2016). The presumptive BAT in Ebf2 knockout animals has a white fat-like morphology and molecular profile. Ebf2 knockout animals also display severely impaired beige adipocyte recruitment in response to β3-adrenergic agonists. Notably, although EBF2 is expressed in white adipocytes, its activity and thus the brown/beige fat program are repressed by the transcription factor ZFP423 (Shao et al. 2016). In brown adipocytes, EBF2 is required for efficient binding of the adipogenic transcription factor PPARγ to Ucp1 and other brown fat-selective genes (Rajakumari et al. 2013). However, the mechanism by which EBF2 regulates chromatin accessibility at lineage-specific enhancers was unknown.
ATP-dependent chromatin remodeling complexes modulate histone–DNA contacts in a noncovalent manner and can therefore play both activating and repressive roles in gene transcription (Clapier and Cairns 2009; Ho and Crabtree 2010). The mammalian BAF (SWI/SNF) chromatin remodeling complex contains one of two related catalytic ATPases: either BRM or BRG1. These enzymes use energy from ATP hydrolysis to alter nucleosome positioning, eject histones, or expose occluded cis-regulatory elements (Fan et al. 2003; Kadam and Emerson 2003). Many lineage-instructive transcription factors, including EBF1, MYOD, and GATA1, recruit the BAF complex to tissue-specific promoters and enhancers (de la Serna et al. 2005; Gao et al. 2009; Hu et al. 2011). BAF complexes are polymorphic and also incorporate up to 14 noncatalytic regulatory subunits that contribute to anchoring the complex at specific target sites (Kadoch et al. 2013). Recently, BRG1 and two BAF regulatory subunits, ARID1A and BAF60b, have been shown to regulate Ucp1 expression in catecholamine-stimulated brown adipocytes (Abe et al. 2015).
In this study, we integrated genome-wide chromatin immunoprecipitation (ChIP) and RNA-seq (RNA sequencing) studies to discover that EBF2 globally orchestrates a brown fat transcriptional program by directly binding to lineage-specific enhancer regions. Moreover, EBF2 physically interacts with BRG1 and the BAF chromatin remodeling complex in brown fat cells to regulate brown fat cell identity and function. Interestingly, the BAF regulatory subunit double PHD fingers 3 (DPF3) is specifically expressed in brown fat relative to white fat cells. DPF3 is an evolutionarily conserved histone reader protein that is characterized by a tandem PHD finger capable of binding methylated and acetylated lysine residues on histones 3 and 4 (Zeng et al. 2010; Marmorstein and Zhou 2014). We found that DPF3 is specifically required for both basal and β-adrenergic-stimulated brown fat gene expression and function. Importantly, Dpf3 itself is a direct transcriptional target of EBF2 during brown adipogenesis. This establishes a feed-forward mechanism through which EBF2 drives Dpf3 expression and also directs DPF3-anchored BAF complexes to chromatin. Altogether, our studies reveal a novel model by which EBF2 cooperates with tissue-specific chromatin remodeling complexes to activate the thermogenic program in adipocytes.
Results
EBF2 regulates chromatin accessibility at brown fat-specific genes
EBF2 activates the expression of brown fat-selective genes in adipocytes, but whether EBF2 regulates these genes via direct binding was unknown. To address this question, we analyzed the genome-wide binding profile of EBF2 in BAT using ChIP followed by deep sequencing (ChIP-seq). We found that EBF2 binds to ∼28,000 sites in the genome (Supplemental Excel File 1), many of which are in regions near brown fat-specific genes that show characteristic enhancer marks in BAT but not white adipose tissue (WAT) (Fig. 1A; Harms et al. 2015). A de novo motif search within EBF2-binding sites identified EBF as the top enriched motif, which validates the specificity of our ChIP-seq results (Supplemental Fig. S1A). Genomic regions enrichment of annotations tool (GREAT) analysis of the top 5000 genes with nearby EBF2-binding sites revealed fatty acid metabolism, regulation of glucose metabolism, and brown fat cell differentiation as significantly enriched biological processes (Supplemental Fig. S1B.)
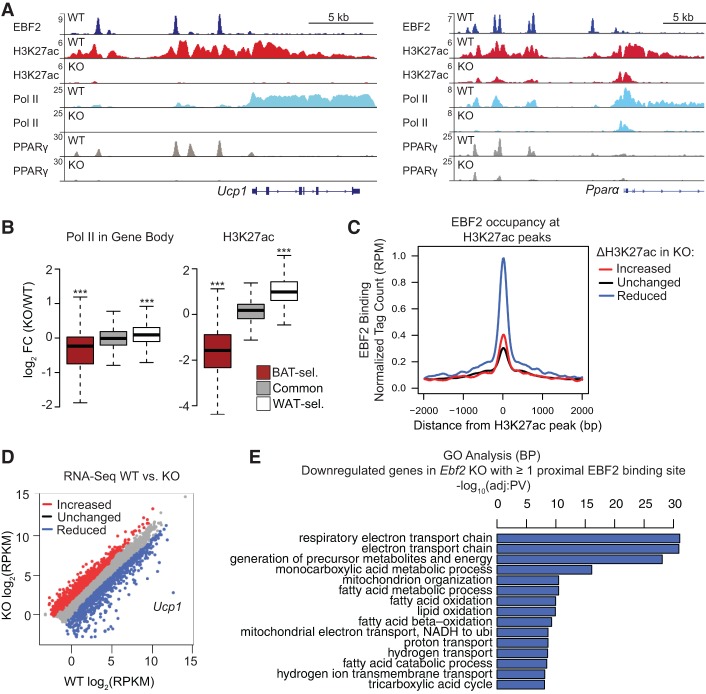
Figure 1.
EBF2 binds and regulates the chromatin state at brown fat-specific genes. (A) ChIP-seq profiles in reads per million total reads (RPM) for EBF2 (dark blue), H3K27ac (red), RNA polymerase II (Pol II; light blue), and PPARγ (gray) in Ebf2 wild-type and knockout BAT at Ucp1 and Pparα. (B) Box plot showing changes in Pol II levels within the gene body and H3K27ac levels within 100 kb of BAT-selective (BAT-sel.), common, and WAT-selective (WAT-sel.) genes in Ebf2 knockout/wild-type BAT. Wilcoxon rank sum test, (***) P < 10−16. (C) Correlation analysis between EBF2 occupancy and differentially regulated H3K27ac peaks in Ebf2 knockout relative to wild-type BAT. (D) Scatter plot analysis of differentially expressed genes in Ebf2 wild-type versus knockout BAT. Fold change >1.5; false discovery rate <0.01. (E) Gene ontology analysis of down-regulated genes in Ebf2 knockout relative to wild-type BAT with at least one proximal EBF2-binding site within a 50-kb window around the transcription start site.
To determine whether EBF2 is required for the activity of lineage-specific enhancers, we examined the levels of PPARγ, RNA polymerase II (Pol II), and H3K27ac at brown fat-specific genes in wild-type and Ebf2 knockout BAT. PPARγ binding was drastically reduced at enhancers of Ucp1 and Pparα in Ebf2 knockout relative to wild-type (control) BAT (Fig. 1A). The loss of Ebf2 also led to a striking reduction in the levels of the activating histone mark H3K27ac and RNA Pol II at many brown fat-selective genes, including Ucp1, Pparα, Cidea, Pgc1α, and Cox7a1 (Fig. 1A; Supplemental Fig. S1C–E). However, Ebf2 deletion did not affect the levels of these regulatory marks at common adipogenic genes such as Fabp4, which are not bound by EBF2 (Supplemental Fig. S1F). We performed genome-wide analysis anchoring on previously defined adipose depot-selective genes and regulatory regions (Supplemental Excel File 2; Harms et al. 2015). On a genome-wide scale, loss of Ebf2 reduced RNA Pol II gene body occupancy and H3K27ac levels at brown fat-selective regulatory regions and increased Pol II and H3K27ac levels at white fat genes and regulatory regions (Fig. 1B). We also performed an unbiased analysis of the relationship between EBF2 binding and global H3K27ac levels in wild-type and knockout tissue. In general, loss of EBF2 binding was highly correlated with loss of H3K27ac, suggesting that EBF2 predominantly acts as a transcriptional activator in BAT (Fig. 1C).
Global analyses of gene expression by RNA-seq identified close to 1000 differentially expressed genes between wild-type and Ebf2 knockout BAT (Fig. 1D), with loss of EBF2 leading to an increase in muscle-related processes and a decrease in respiratory processes (Supplemental Fig. S1G). A refined gene ontology analysis of putative EBF2 target genes that are down-regulated in knockout BAT and have one or more EBF2-binding sites within a 50-kb window of the transcription start site revealed that Ebf2 deletion reduced the expression of genes associated with the respiratory electron transport chain and oxidative metabolism, which are critical processes for brown fat thermogenesis (Fig. 1E). Many of the down-regulated genes with proximal EBF2-binding sites have well-established roles in BAT and mitochondrial function (Supplemental Excel File 3). Furthermore, integration of gene expression and ChIP-seq data revealed that genes with a greater number of proximal EBF2-binding sites generally tended to be down-regulated in knockout tissue (Supplemental Fig. S1H). Taken together, these analyses reveal that EBF2 is an activator of the brown fat gene program through direct binding to lineage-specific cis-regulatory elements.
EBF2 interacts with BRG1 and the BAF chromatin remodeling complex
We next sought to determine the mechanism through which EBF2 regulates chromatin structure at brown fat-specific genes. The BAF chromatin remodeling complex is known to play a critical role in many developmental and differentiation processes. In brown adipocytes, shRNA-mediated depletion of BRG1, a catalytic component of the BAF complex, dramatically reduced the expression of Ucp1 and other brown fat-specific genes, with no effect on general adipocyte genes (Supplemental Fig. S2A), as also reported by Abe et al. (2015). This suggested that EBF2 may cooperate with BRG1 and the BAF complex to remodel the chromatin structure of brown fat gene enhancers. Coimmunoprecipitation studies in 293T cells revealed a robust physical interaction between EBF2 and BRG1 (Fig. 2A). This interaction between EBF2 and BRG1 was also detected in mature brown adipocytes with both factors expressed at endogenous levels (Fig. 2B). Together, these findings show that EBF2 interacts with BRG1 and the BAF chromatin remodeling complex in brown adipocytes.
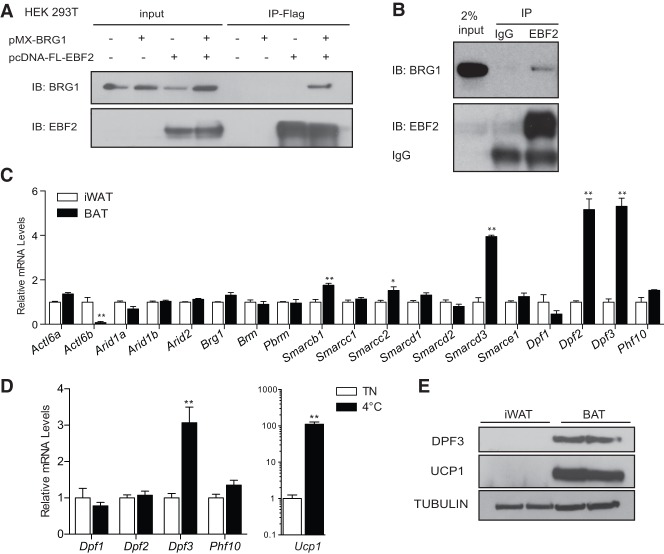
Figure 2.
EBF2 interacts with the BAF complex, which incorporates the subunit DPF3 in brown adipocytes. (A) Coimmunoprecipitation experiment in 293T cells transfected with pcDNA3.1-Flag-EBF2 ± pMX-BRG1 followed by Flag immunoprecipitation. (B) Endogenous coimmunoprecipitation in differentiated mature brown adipocytes. Sheep IgG was used as a negative control. (C) RT-qPCR analysis of BAF subunit expression in BAT and WAT from 6-wk-old male mice. Mean ± SE. n = 3. Two-sample Student's t-test, (*) P < 0.05; (**) P < 0.01. (D) RT-qPCR analysis of Dpf and Ucp1 expression in inguinal white adipose following 2 wk of cold exposure at 4°C. Mean ± SE. n = 5. Two-sample Student's t-test, (**) P < 0.01. (E) Western blot analysis of DPF3, UCP1, and Tubulin (loading control) in inguinal WAT (iWAT) and BAT.
DPF3 is a brown fat-enriched BAF regulatory subunit
The BAF complex and BRG1 are targeted to specific genes through interactions with tissue-specific transcription factors and via the function of its noncatalytic regulatory subunits. Many regulatory subunits are encoded by gene families, which allows for combinatorial assembly of these subunits to create diverse chromatin patterns in different tissues or at distinct developmental stages. (Lessard et al. 2007; Ho et al. 2009). To assess a potential role for subunit specificity in brown adipogenesis, we profiled the expression of 19 genes that encode the BAF subunits in BAT and inguinal WAT (iWAT). Interestingly, several members of the DPF/BAF45 family were differentially expressed, with Dpf1 (Baf45b) showing higher expression in iWAT, and Dpf2 (Baf45d) and Dpf3 (Baf45c) showing a fourfold to sixfold enrichment in BAT (Fig. 2C). However, beige adipocyte induction in WAT through chronic cold exposure (2 wk at 4°C) was associated only with preferential activation of Dpf3 expression (Fig. 2D), an effect also observed in inguinal fat following treatment with the β3-adrenergic agonist CL-316,243 (Supplemental Fig. S2B). Western blotting analysis showed that DPF3 protein expression was highly BAT-specific (Fig. 2E). These results identify DPF3 as a BAF regulatory subunit that is specifically expressed in thermogenic BAT and beige adipose tissue.
DPF3 is required for activation of the brown fat program
There are two known splice variants of Dpf3, Dpf3a and Dpf3b, which have different C termini (Lange et al. 2008; Zeng et al. 2010; Ishizaka et al. 2012). We interrogated the function of DPF3 in brown adipocytes using a lentiviral shRNA construct that targets both isoforms and reduces Dpf3 expression by ∼80% relative to control cells expressing a scrambled shRNA (shScr.) (Fig. 3A). shDpf3-expressing and control brown preadipocytes underwent efficient adipocyte differentiation as assessed by Oil-Red-O lipid staining (Fig. 3B). DPF3 knockdown increased the expression levels of pan-adipogenic genes (Pparγ, Fabp4, and AdipoQ) (Fig. 3C). Strikingly, DPF3 depletion also led to a significant reduction in basal and isoproterenol (iso)-induced expression of many brown fat-specific genes, including Ucp1, Pparα, Pgc1α, and Dio2 (Fig. 3D). In particular, Ucp1 expression was dramatically reduced at the mRNA level and protein level in DPF3-depleted brown adipocytes (Fig. 3E). DPF3 depletion did not significantly affect Ebf2 mRNA (Fig. 3D) or protein (Fig. 3E) levels. We confirmed these results with a second independent shRNA sequence, which caused a comparable and specific loss of brown fat-specific gene expression in brown adipocytes (Supplemental Fig. S3A).
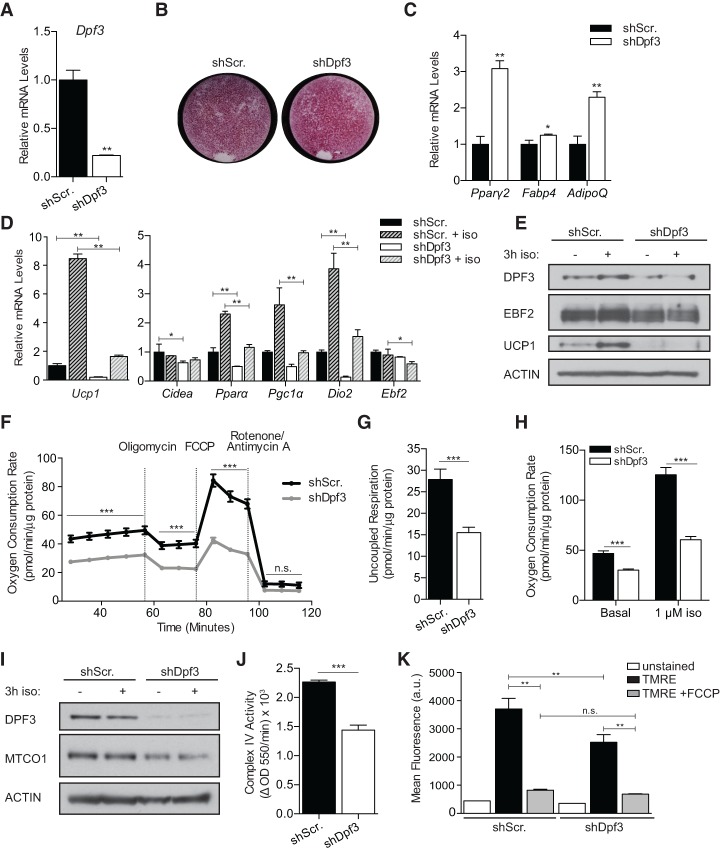
Figure 3.
DPF3 is required for activation of the brown fat program and mitochondrial function. (A) RT-qPCR analysis of day 7 mature brown adipocytes following shRNA-mediated Dpf3 depletion. (B) Oil-Red-O staining of shScr (control) and shDpf3 brown adipocytes. (C) RT-qPCR analysis of pan-adipogenic gene expression in control and DPF3-depleted mature brown adipocytes. (D) RT-qPCR analysis of brown fat-specific gene expression in control and DPF3-depleted mature brown adipocytes basally or following 3 h of stimulation with 1 µM iso. Mean ± SD. n = 3. Two-way ANOVA with Holm-Šídák multiple tests correction comparing shScr versus shDpf3 under basal or iso-stimlated conditions, (*) P < 0.05; (**) P < 0.01. (E) Western blot analysis of DPF3, EBF2, UCP1, and Actin (loading control) in control and DPF3-depleted brown adipocytes. (F) Oxygen consumption rate (OCR) in control and DPF3-depleted brown adipocytes; OCR was normalized to protein concentration. Mean ± SE. n = 23. Two-sample Student's t-test with Holm-Šídák multiple tests correction, (***) P < 0.001. (G) Uncoupled respiration in control and DPF3-depleted cells; OCR was normalized to protein concentration. Mean ± SE. n = 23. Two-sample Student's t-test, (***) P < 0.001 (H) OCR after acute iso stimulation in control and DPF3-depleted cells; OCR was normalized to protein concentration. Mean ± SE. n = 23. Two-sample Student's t-test, (***) P < 0.001. (I) Western blot analysis of DPF3, MTCO1, and Actin (loading control) in control and DPF3-depleted brown adipocytes. (J) Quantification of complex IV activity in control and DPF3-depleted brown adipocytes. Mean ± SE. n = 6. Two-sample Student's t-test, (***) P < 0.001. (K) Mean TMRE fluorescence assessed by flow cytometry in control and DPF3-depleted brown adipocytes. n = 3; 200,000 events recorded per sample. One-way ANOVA with Holm-Šídák correction for multiple comparisons, (**) P < 0.01.
We next evaluated whether DPF3 regulates the respiratory activity of mature brown adipocytes. DPF3 depletion in brown adipocytes resulted in markedly diminished basal, maximal, and uncoupled respiration (Fig. 3F,G). In addition, iso stimulation led to a 2.7-fold increase in the oxygen consumption rate (OCR) of control cells, an effect that was significantly blunted by DPF3 knockdown (Fig. 3H). The reduced OCR of DPF3-depleted cells under basal conditions suggested that DPF3 regulates overall mitochondrial number and/or function. Control and DPF3 knockdown cells had similar levels of mitochondrial DNA (Supplemental Fig. S3B). However, Western blot analysis of oxidative phosphorylation (OXPHOS) subunit levels revealed decreased MTCO1 (COX1) expression upon DPF3 depletion (Fig. 3I; Supplemental Fig. S3C). MTCO1 is the mitochondrial-encoded catalytic subunit of cytochrome c oxidase (complex IV) that catalyzes the reduction of molecular oxygen to water. We measured the oxidation rate of reduced cytochrome c by ELISA and observed a 40% reduction in complex IV activity in DPF3-depleted cells (Fig. 3J). Furthermore, staining with the cationic dye TMRE, which accumulates in the depolarized matrix of active mitochondria, showed that DPF3-depleted cells have reduced mitochondrial membrane potential (Fig. 3K). We thus conclude that DPF3 regulates mitochondrial activity and substrate flux through the respiratory chain in brown fat cells at least in part via regulating the expression levels of MTCO1.
In inguinal white adipocytes, knockdown of DPF3 blocked the rosiglitazone-mediated induction of several brown fat genes, including Ucp1 (Supplemental Fig. S3D), suggesting that DPF3 is also required for the adipocyte beigeing process. However, ectopic expression of DPF3A or DPF3B alone was not sufficient to induce a brown fat program in inguinal adipocytes (Supplemental Fig. S3E).
DPF3 regulates chromatin structure at brown fat-specific enhancers
We hypothesized that DPF3 is required for the modification of chromatin structure at brown fat-specific enhancer regions. To test this, we used formaldehyde-assisted identification of regulatory elements (FAIRE), which identifies nucleosome-depleted regions of DNA (Simon et al. 2012), followed by qPCR to analyze chromatin structure at brown fat-specific enhancer regions. Using this technique, we found that DPF3 knockdown reduced chromatin accessibility at Ucp1 and Pparα enhancers both in the basal state and following acute iso stimulation (Fig. 4A). H3K27ac levels were also reduced at Ucp1 and Pparα enhancers following DPF3 knockdown (Supplemental Fig. S4A). These data show that DPF3 is required for establishing a permissive chromatin state at brown fat-specific enhancers.
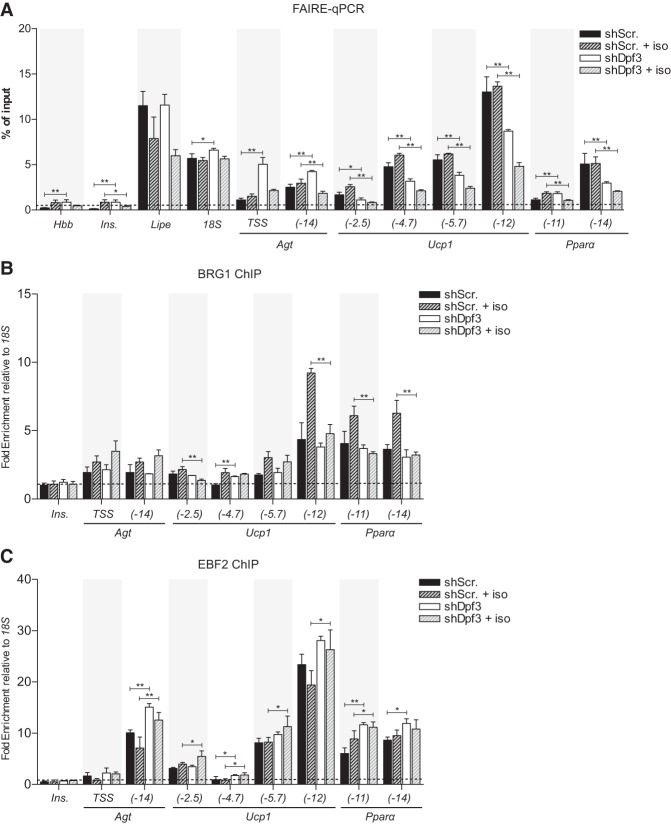
Figure 4.
DPF3 regulates the chromatin state at brown fat-specific genes. (A) FAIRE-qPCR analysis of control and DPF3-depleted mature brown adipocytes basally or following 3 h of stimulation with 1 µM iso. The insulin and β-globin promoters serve as negative controls; enrichment was analyzed as percentage of input. (B,C) ChIP-qPCR analysis of BRG1 (B) and EBF2 (C) binding in brown adipocytes ± iso; chromatin enrichment was analyzed as percentage of input recovery and normalized to 18S percentage of input to produce a fold enrichment. The insulin promoter served as a negative control. All data show mean ± SD. n = 3. Two-way ANOVA with Holm-Šídák multiple tests correction comparing shScr versus shDpf3 under basal or iso-stimulated conditions, (*) P < 0.05; (**) P < 0.01.
We next sought to determine whether DPF3 regulates localization of the BAF complex in brown adipocytes. ATP-dependent chromatin remodelers have dynamic and transient interactions with chromatin; therefore, binding events may be difficult to detect by ChIP assays. Previous work indicates that iso stimulates BRG1/BAF recruitment to brown fat-specific enhancers in mature brown adipocytes (Abe et al. 2015) and thus provides a defined time frame to observe BRG1-binding dynamics. Consistent with this, we observed increased BRG1 recruitment to Ucp1 and Ppara after 3 h of iso treatment (Fig. 4B). Notably, this iso-stimulated recruitment of BRG1 was blocked in DPF3 knockdown cells (Fig. 4B), indicating that DPF3 is required for BRG1 and BAF to support increased transcription of brown fat genes following β-adrenergic stimulation. Interestingly, EBF2 occupancy at cis-regulatory elements was not altered by DPF3 deficiency (Fig. 4C) or BRG1 deficiency (Supplemental Fig. S4B), suggesting that EBF2 binding precedes chromatin remodeling at these sites. Together, these results demonstrate that DPF3 is required downstream from EBF2 binding for chromatin remodeling at lineage-specific enhancers.
EBF2 transcriptionally activates Dpf3 expression
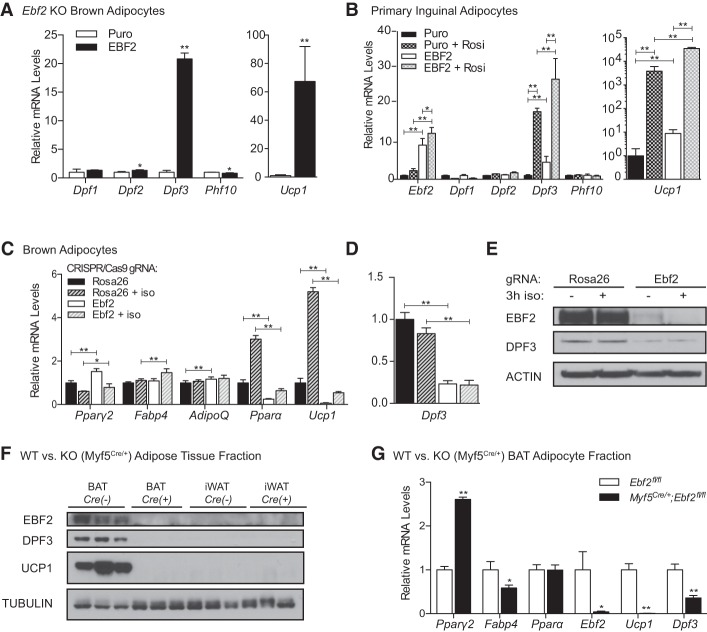
Given that EBF2 is recruited to chromatin in the absence of DPF3, we asked whether EBF2 could play a role in upstream activation of Dpf3 expression. Retroviral expression of EBF2 in Ebf2−/− brown adipocytes increased Dpf3 mRNA levels by 20-fold without affecting transcription of other DPF family members (Fig. 5A). EBF2 overexpression also increased Dpf3 expression in primary inguinal white adipocytes both alone and synergistically with rosiglitazone (Fig. 5B). Acute Ebf2 deletion in brown adipocytes blocked induction of the thermogenic program under basal and iso-stimulated conditions (Fig. 5C) and reduced the mRNA (Fig. 5D) and protein (Fig. 5E) levels of DPF3.
Figure 5.
EBF2 transcriptionally regulates Dpf3 expression. (A) RT-qPCR analysis of Ebf2−/− brown adipocytes following retroviral-mediated control (Puro) or EBF2 overexpression. (B) RT-qPCR analysis in primary inguinal adipocytes following retroviral-mediated control (Puro) or EBF2 overexpression with and without rosiglitazone included throughout differentiation. Mean ± SD. n = 3. To compare all groups in a pairwise fashion, data were analyzed using a two-way ANOVA with Holm-Šídák multiple tests correction, (*) P < 0.05; (**) P < 0.01. (C) RT-qPCR analysis following CRISPR-mediated gene knockout in control (Rosa26) and Ebf2 knockout mature brown adipocytes basally or following 3 h of stimulation with 1 µM iso. Two-way ANOVA with Holm-Šídák multiple tests correction comparing shScr versus shDpf3 under basal or iso-stimulated conditions, (*) P < 0.05; (**) P < 0.01. (D) RT-qPCR analysis of Dpf3 expression in control and Ebf2 knockout mature brown adipocytes basally or following 3 h of stimulation with 1 µM iso. Mean ± SD. n = 3. Two-way ANOVA with Holm-Šídák multiple tests correction comparing shScr versus shDpf3 under basal or iso-stimulated conditions, (**) P < 0.01. (E) Western blot analysis for EBF2, DPF3, and Actin (loading control) in control and Ebf2 knockout brown adipocytes. (F) Western blot analysis for EBF2, DPF3, UCP1, and Tubulin (loading control) in wild-type (Ebf2fl/fl) or knockout (Myf5Cre/+;Ebf2fl/fl) BAT and iWAT. (G) RT-qPCR analysis of the mature adipocyte fraction from wild-type (Ebf2fl/fl) or knockout (Myf5Cre/+;Ebf2fl/fl) animals. n = 3 animals pooled per genotype. Error bars show SD of technical replicates. Two-sample Student's t-test, (*) P < 0.05, (**) P < 0.01.
We also evaluated whether EBF2 regulates Dpf3 expression in BAT in vivo. To do this, we used the Myf5Credriver to delete Ebf2 in brown (but not beige) adipocytes and analyzed the adipose depots of 12-wk-old male mice. The presumptive interscapular BAT in these Ebf2 knockout (Myf5Cre) animals had a white fat-like morphology (Supplemental Fig. S5A,B). Gene expression analysis of wild-type and knockout BAT revealed a large reduction in the expression levels of brown fat-selective genes and increased levels of white fat-selective genes in knockout tissue (Supplemental Fig. S5C). Importantly, there was a complete loss of DPF3 protein expression in the BAT of knockout animals as assessed by Western blot (Fig. 5F) and a significant reduction in Dpf3 mRNA levels in the BAT adipocyte fraction (Fig. 5G). Together, these results demonstrate that EBF2 regulates Dpf3 expression both in vivo and in vitro.
EBF2 directly regulates Dpf3 through an intronic enhancer
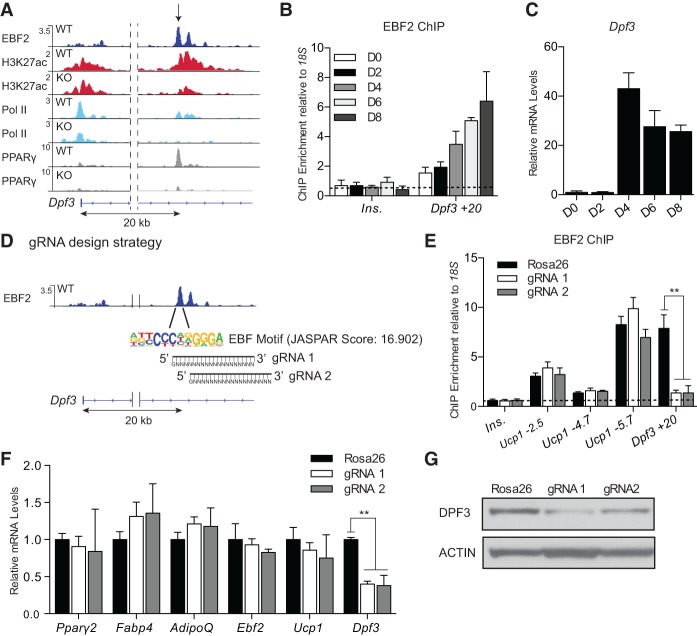
Examination of ChIP-seq data sets identified an EBF2-binding site in the first intron of Dpf3 located 20-kb downstream from the transcription start site. This locus is additionally marked by high levels of H3K27ac, RNA Pol II, and PPARγ in wild-type (but not Ebf2 knockout) BAT (Fig. 6A). ChIP-qPCR analyses revealed increased binding of EBF2 to this +20-kb site over the course of brown adipocyte differentiation (Fig. 6B), correlating with the transcriptional activation of Dpf3 expression at day 4 of differentiation (Fig. 6C). To investigate the role of EBF2 at this putative enhancer, we designed two guide RNAs (gRNAs) that target Cas9 to the central EBF motifs in the +20-kb site (Fig. 6D). Brown preadipocytes were infected (as pools) with control or gRNA-expressing retrovirus. Sequencing analysis of genomic DNA amplified from these cells showed that Cas9 introduced insertion/deletion (indel) and nucleotide substitutions in 100% of the clones transduced with the EBF motif gRNAs, while no mutations were identified in DNA isolated from control cells (Supplemental Fig. S6A). ChIP-qPCR in mature brown adipocytes confirmed that both gRNA–Cas9 constructs abolished EBF2 binding at the +20-kb site without affecting EBF2 recruitment to other genomic targets (Fig. 6E). General adipocyte differentiation was unchanged by expression of either gRNA. Furthermore, although Ucp1 expression was unaffected by the gRNAs, Dpf3 expression was significantly blunted, demonstrating that a single EBF-binding site in the +20-kb site is responsible for ∼60% of Dpf3 mRNA expression in brown adipocytes (Fig. 6F). Western blot analysis revealed that DPF3 protein was also reduced (but not absent) following CRISPR-mediated editing of the +20-kb locus (Fig. 6G). By separating Dpf3 expression from the brown adipogenic program, we thus were able to identify a specific element of Dpf3 regulation through direct EBF2 binding at a functional intronic enhancer. Together, these results suggest a regulatory mechanism through which EBF2 activates Dpf3 expression, resulting in the assembly of a tissue-specific chromatin remodeling complex, which in turn remodels brown fat-specific regulatory regions for efficient gene activation.
Figure 6.
EBF2 directly regulates Dpf3 expression via an intronic enhancer. (A) ChIP-seq profiles at Dpf3 in RPM for EBF2 (dark blue), H3K27ac (red), RNA Pol II (light blue), and PPARγ (gray) in Ebf2 wild-type and knockout BAT. (B) ChIP-qPCR analysis of EBF2 binding at the Dpf3 +20-kb site over the course of brown adipocyte differentiation. The insulin promoter served as a negative control. Mean ± SD. n = 3. (C) RT-qPCR analysis of Dpf3 expression over the course of brown adipocyte differentiation. Mean ± SD. n = 3. (D) CRISPR–Cas9-mediated genomic editing strategy at the EBF motif in the Dpf3 +20-kb enhancer. (E) ChIP-qPCR for EBF2 in control (Rosa26) and EBF gRNA-expressing pooled brown adipocytes. The insulin promoter served as a negative control. Mean ± SD. n = 3. Two-sample Student's t-test, (**) P < 0.01. (F) Gene expression analysis in control and EBF gRNA-expressing brown adipocytes. Mean ± SD. n = 3. Two-sample Student's t-test, (**) P < 0.01. (G) Western blot analysis of DPF3 and Actin (loading control) expression in control and EBF gRNA-expressing brown adipocytes.
Critical role for the histone-binding activity of DPF3 in brown adipocytes
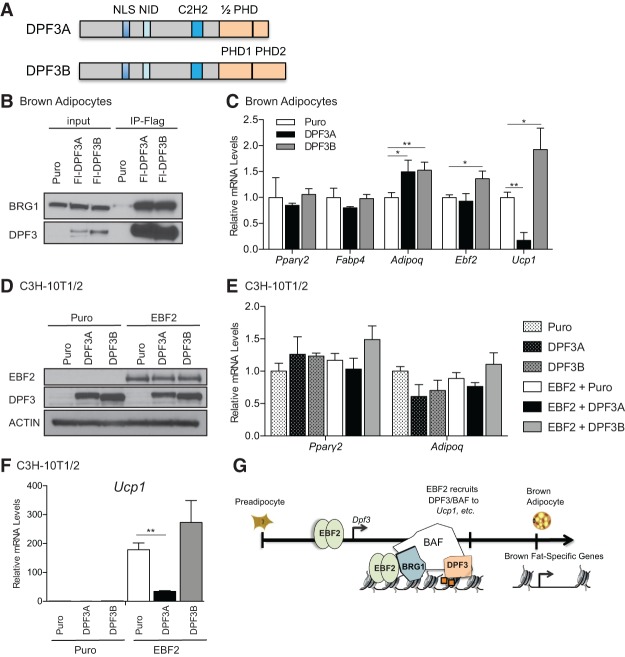
The full-length isoform of DPF3 (DPF3B) contains a predicted LXXLL nuclear receptor interaction domain, a C2H2 Krüppel-type zinc finger, and a unique tandem PHD finger domain (Lange et al. 2008). This tandem PHD finger binds methylated and acetylated lysines, allowing for combinatorial readout of histone modifications (Zeng et al. 2010). A naturally occurring C-terminal splice variant of DPF3 (DPF3A) is identical to DPF3B from amino acids 1–261 but contains a truncated PHD finger (Fig. 7A); this variant is unable to bind histones and instead can exert its function through interaction with sequence-specific transcription factors (Lange et al. 2008; Cui et al. 2016). We used retrovirus to express Flag-tagged versions of the truncated (DPF3A) and full-length (DPF3B) isoforms of DPF3 into brown adipocytes, with the reasoning that ectopic expression of these different DPF3 isoforms would disrupt the natural stoichiometry of the BAF complex. We confirmed that both isoforms were incorporated into the BAF complex through Flag immunoprecipitation followed by blotting for endogenous BRG1 (Fig. 7B). Expression of each isoform did not affect adipogenesis as assessed by pan-adipogenic gene expression (Fig. 7C). However, overexpression of DPF3A powerfully blocked induction of Ucp1 in mature brown adipocytes; conversely, overexpression of DPF3B modestly increased Ucp1 expression (approximately twofold) (Fig. 7C).
Figure 7.
Critical role for the histone-binding activity of DPF3 in brown adipocytes. (A) Schematic of the DPF3A and DPF3 domain structures. (NLS) Nuclear localization sequence; (NID) nuclear receptor interaction domain. (B) Flag immunoprecipitation of ectopically expressed control vector (Puro), DPF3A, or DPF3B followed by blotting for endogenous BRG1 in mature brown adipocytes. (C) Gene expression analysis in control (Puro), DPFA-expressing, and DPF3B-expressing mature brown adipocytes. Mean ± SD. n = 3. Two-sample Student's t-test, (*) P < 0.05; (**) P < 0.01. (D) Western blot analysis of control or EBF2-expressing C3H-10T1/2 cells infected with control (Puro), DPF3A, or DPF3B. (E,F) Gene expression analysis of common adipogenic genes (E) or Ucp1 (F) in C3H-10T1/2 cells. Mean ± SD. n = 3. Two-sample Student's t-test, (**) P < 0.01. (G) Model for EBF2-mediated regulation at brown fat genes.
We further investigated isoform-specific functions of DPF3 in C3H-10T1/2 mesenchymal cells, which are competent to undergo adipogenesis but have basally low levels of endogenous EBF2 and other brown fat-selective genes. To determine whether DPF3A could blunt EBF2-mediated conversion of 10T1/2 cells to brown adipocytes, we expressed either control or EBF2 retrovirus in combination with vector control, DPF3A, or DPF3B (Fig. 7D). All of the cultures differentiated into lipid-containing adipocytes that expressed equivalent levels of Pparγ2 and Adipoq (Fig. 7E). EBF2-expressing cells potently activated Ucp1 expression. Importantly, this EBF2-mediated activation of Ucp1 was significantly blunted by coexpression of DPF3A. Conversely, coexpression of DPF3B with EBF2 modestly potentiated Ucp1 activation (Fig. 7F; Supplemental Fig. S7A). Together, these data suggest that the histone-binding activity of DPF3B is critical for the activating function of the BAF complex in mediating EBF2 action at Ucp1.
Discussion
In this study, we identified a novel mechanism by which the brown fat lineage factor EBF2 regulates the chromatin state at brown fat genes via interaction with the chromatin remodeler BRG1 and a tissue-specific BAF complex. Several previous studies have demonstrated the importance of BRG1 for white and brown adipocyte differentiation (Salma et al. 2004; Madsen et al. 2014; Abe et al. 2015). The enzymatic remodeling function of BRG1 is targeted to chromatin through several mechanisms, including association with transcription factors, the C-terminal bromodomain within BRG1 itself, and the inclusion of regulatory subunits, which impart a combinatorial code to direct tissue-specific chromatin remodeling (Clapier and Cairns 2009). For example, the subunit BAF60c plays a critical role in regulating cardiac and skeletal muscle development (Lickert et al. 2004), whereas the paralogous factors BAF60a and BAF60b are repressed during myogenesis (Goljanek-Whysall et al. 2014). Similar mechanisms regarding BAF subunit inclusion or exclusion have been described in embryonic stem cell pluripotency as well as neuronal progenitor proliferation and differentiation (Ho and Crabtree 2010).
Investigation of BAF complex function in brown adipocytes led us to identify a novel role for the BAF regulatory subunit DPF3 (BAF45c) in brown adipogenesis. Dpf3 is preferentially expressed in BAT and beige adipose tissue (Fig. 2C), and DPF3 was specifically required for activating a thermogenic gene and functional program in adipocytes (Fig. 3). Furthermore, DPF3 regulates the expression of MTCO1, complex IV activity, and mitochondrial respiration in brown fat cells. Interestingly, BAT mitochondria from Ucp1 knockout animals also show reduced MTCO1 levels (Odegaard et al. 2016). Additional studies will be needed to establish the relationship between UCP1 and MTCO1 regulation as well as how DPF3 influences mitochondrial OXPHOS composition.
BRG1 binding was not detected at brown fat enhancers in unstimulated mature brown adipocytes. However, BRG1 was required for the differentiation-linked induction of brown fat-selective genes such as Ucp1 (Supplemental Fig. S2A). We speculate that BRG1/BAF is transiently recruited at an early time point in adipogenesis to remodel gene enhancers but that the complex is not retained at these sites. Interestingly, EBF2 binding to Ucp1 and other brown fat genes did not depend on DPF3 or BRG1 (Fig. 4C; Supplemental Fig. S4B), suggesting that EBF2 binding precedes BAF-mediated chromatin remodeling. This result highlights the potential role for EBF2 as a pioneer factor. Indeed, structural and functional studies show that the related family member EBF1 is capable of binding nucleosome-enriched DNA and establishing chromatin accessibility through its C-terminal domain (Buenrostro et al. 2013; Boller et al. 2016). The crystal structure of the EBF1 DNA-binding domain, which shares 92% amino acid sequence identity with EBF2 (Dubois and Vincent 2001), reveals a large 18-base-pair (bp) DNA footprint. EBF proteins are thus capable of making expansive chromatin contacts, which may underlie a potential pioneering function in cellular differentiation (Treiber et al. 2010). Future studies will be needed to determine whether EBF2 binds to nucleosomal DNA.
Dpf3 itself is a transcriptional target of EBF2 during brown adipogenesis (Fig. 5). Mutation of a single EBF2-binding site in intron 1 of Dpf3 reduced Dpf3 mRNA and protein levels by ∼60% (Fig. 6F,G). However, this level of Dpf3 depletion was not sufficient to reduce Ucp1 levels. Deletion of Ebf2 itself in BAT using the Myf5Cre driver led to a complete loss of Dpf3 expression, suggesting the presence of additional unidentified EBF2-regulated enhancers. Therefore, it would be interesting to perform chromatin conformation capture or similar assays to identify additional enhancers and associated factors that regulate Dpf3 expression.
EBF2 interacts with the brown fat-enriched long noncoding RNA (lncRNA) Blnc1. Similar to our findings with DPF3, EBF2 transcriptionally regulates the expression of Blnc1 and also forms a ribonucleoprotein complex with this lncRNA to regulate brown fat differentiation (Zhao et al. 2014). These findings suggest that EBF2 activates many components of the brown fat transcriptional machinery and further functions as a sequence-specific factor to direct these transcriptional regulatory components to the DNA. EBF2 could therefore cooperate with DPF3B to recognize specific histone modifications and anchor the BAF remodeling complex at brown fat genes.
Dpf3 is expressed in cardiac and skeletal muscles of mice in addition to its selective expression in brown versus white adipose. Additionally, Dpf3 was shown to be required for skeletal muscle development in zebrafish (Lange et al. 2008). BAT and skeletal muscle descend from a related precursor population in the somitic mesoderm (Atit et al. 2006; Seale et al. 2008) and share many functional properties: abundant mitochondria, high levels of sympathetic innervation, and high thermogenic capacity (Forner et al. 2009). Thus, DPF3 represents an epigenetic factor that has evolved to regulate key functions in oxidative tissues.
Elegant work from the Sperling laboratory (Lange et al. 2008; Cui et al. 2016) has highlighted different functions for the short isoform Dpf3a, which recruits BAF through interactions with tissue-specific transcription factors, and the long isoform Dpf3b, which can directly bind histones through its tandem PHD finger. Both Dpf3 splice isoforms are expressed in BAT; however, augmenting DPF3A levels in brown adipocytes repressed Ucp1 expression (Fig. 7C) and blocked EBF2-mediated conversion of 10T1/2 mesenchymal cells to brown adipocytes (Fig. 7F). We hypothesize that the histone reader function of DPF3B targets the BAF complex to facilitate chromatin remodeling and downstream recruitment of histone writers either directly or via BRG1, which is known to cooperate with histone acetyltransferases such as CBP and p300 (Naidu et al. 2009). Thus, we envision a regulatory module in which DPF3-anchored BAF complexes make chromatin more accessible to transcription factors and acetyltransferases for gene activation.
DPF3 is the most highly enriched DPF family member in brown/beige versus white fat cells. However, the domain organization of the DPF/BAF45 family is fairly conserved between paralogs (Lessard et al. 2007). Thus, Dpf2, which is also expressed at higher levels in BAT relative to WAT (Fig. 2C), may play a role redundant to that of DPF3 during BAT development in vivo. Establishing tissue-specific knockout mouse models will now be needed to examine the in vivo requirements for DPF3 activity in brown and beige fat.
In conclusion, we found that EBF2 is a global regulator of the brown fat gene program through direct binding to lineage-specific cis-regulatory elements. EBF2 functions at chromatin in part through interacting with the BAF chromatin remodeling complex that contains the histone reader DPF3 (Fig. 7G). Taken together, these results provide a more detailed understanding of how EBF2 functions to activate the brown fat gene program and thus have implications for therapeutically increasing thermogenesis to reduce metabolic disease.
Materials and methods
Animals
Myf5Cre/+ C57BL/6 mice were obtained from the Jackson laboratory (stock 007893). Ebf2fl/fl conditional null mice were generated by our laboratory using standard gene targeting techniques to insert loxP sites flanking exon 3 of the Ebf2 gene (Cyagen) in the C57BL/6 strain. Ebf2 whole-body knockout animals were obtained from Randall Reed (Johns Hopkins University) and have been described previously (Wang et al. 2004). All animal work was approved by the University of Pennsylvania's Institutional Animal Care and Use Committee.
Cell culture
Brown preadipocytes were immortalized through retroviral expression of SV-40 large T antigen (Harms et al. 2014). Cells were grown at subconfluence in 10% FBS/DMEM-F12 (Life Technologies). To induce differentiation, confluent cells were treated with 10% FBS/DMEM-F12 with 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 1 mM dexamethasone, 20 nM insulin, and 1 nM T3. After 48 h, cells were switched to medium containing 10% FBS, 20 nM insulin, and 1 nM T3 and harvested 7 d after induction. To stimulate thermogenesis, cells were treated with 1 µM iso for 3 h prior to harvest. C3H-10T1/2 mesenchymal cells were grown at subconfluence in 10% FBS/DMEM (Life Technologies) and differentiated as above with the inclusion of 1 µM rosiglitazone through differentiation. Primary white inguinal adipocytes were prepared from 6-wk-old male mice as described previously (Seale et al. 2011) using collagenase and dispase to isolate the stromal vascular fraction of the inguinal fat pad. Primary inguinal cells were grown and differentiated as above with the inclusion of 1 µM rosiglitazone through differentiation. Oil-Red-O, virus production, and transfection assays were performed as described previously (Ishibashi et al. 2012). The following constructs were used in cell culture: pMX-BRG1 (Hans Schöler; Addgene, plasmid no. 25855) (Singhal et al. 2010), pcDNA3.1-Fl-EBF2 (Rajakumari et al. 2013), MSCV-Fl-DPF3A, and MSCV-Fl-DPF3B, which were cloned from mouse cDNA into the MSCV backbone, and pLKO.1-shDpf3 (GE Healthcare, TRCN0000086344 or TRCN 0000086347). For CRISPR–Cas9 studies, gRNA constructs were designed using the Optimized CRISPR Design Web site (Zhang laboratory, http://crispr.mit.edu) and cloned into lentiCRISPR v2 (Feng Zhang; Addgene, plasmid no. 52961) (Sanjana et al. 2014).
Gene expression and protein analysis
Total RNA was extracted using TRIzol (Invitrogen) followed by purification using PureLink RNA columns (Invitrogen) and quantified using a NanoDrop. Five-hundred nanograms to 1 µg of RNA was reverse-transcribed using the high-capacity cDNA synthesis kit (Applied Biosystems) followed by real-time PCR with SYBR Green master mix (Applied Biosystems) on a 7900 HT machine (Applied Biosystems). Tata-binding protein (Tbp) was used as an internal normalization control. Mitochondrial DNA content was quantified as described previously (Harms et al. 2014). Data were analyzed using a two-sample two-sided Student's t-test and Microsoft Excel unless otherwise stated. Primer sequences are listed in Supplemental Table 1. Protein extracts were prepared by harvesting cells or tissue in NP-40 lysis buffer (20 mM Tris, 10% glycerol, 2 mM EDTA, 0.1% NP-40, 10 mM NaF, 150 mM NaCl). Protein concentration was measured using a detergent-compatible (DC) assay (Bio-Rad). Samples were separated on 4%–12% Bis-Tris NuPAGE gels (Invitrogen) and transferred to PVDF membranes for Western blot analysis. For coimmunoprecipitation analysis, cells were lysed as above, and 1 mg of cell lysate was used for immunoprecipitation with anti-Flag-conjugated beads (Sigma), sheep IgG (Santa Cruz Biotechnology, 2717), or EBF2 antibody. Immunoprecipitation samples were washed four times with NP-40 lysis buffer with 200 mM NaCl and eluted in 2× NuPage LDS sample buffer (Life Technologies) for Western blot analysis. The primary antibodies used were anti-BRG1 (1:1000; Abcam, 110641), anti-EBF2 (1:1000; R&D Systems, AF7006), anti-UCP1 (1:1000; R&D Systems, MAB6158), total rodent OXPHOS antibody cocktail (1:5000; Abcam, 110143), anti-MTCO1 (1:2000; Abcam, 14605), anti-Actin (1:10,000; Millipore, MAB 1501), and anti-Tubulin (1:5000; Sigma, DM1A). The anti-DPF3 (1:500) antibody is a polyclonal rabbit antibody generated using the peptide EALLRGEGVEKKVDAREE (Thermo Fisher Scientific).
Complex IV activity assay
Brown adipocytes were differentiated for 6 d, and cytochrome c oxidase activity was analyzed using the complex IV rodent enzyme activity microplate assay kit (Abcam, 109911). Briefly, 25 µg of cell lysate was immunocaptured on 96-well plates, and the OD550 was measured at 1-min intervals for 2 h. Complex IV activity rate was determined by calculating the gradient of the initial linear slope.
Mitochondrial membrane potential assay
FCCP-pretreated control brown adipocytes were incubated with 200 µM FCCP in culture medium for 10 min. Cells were then treated with FCCP + 400 nM TMRE (Abcam, 113852) or TMRE only and incubated for 30 min at 37°C. Cells were washed with PBS, trypsinized, pelleted, and washed once with FACS buffer (2% calf serum, 1 mM EDTA, 0.1% sodium azide in PBS). Cells were resuspended in FACS buffer and filtered through a 100-µm filter, and emission at 585 nm was measured on a BD FACS Canto.
ChIP and FAIRE analysis
For ChIP analysis, cells were fixed with 1% formaldehyde for 15 min at room temperature followed by quenching with 2.5 M glycine for 5 min at room temperature. Samples were pelleted and resuspended in ChIP lysis buffer (0.6% SDS, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, 20 mM Tris at pH 8) with protease inhibitors and 1 mM PMSF. Samples were sonicated using the Active Motif EpiShear platform for 8 min of active time (30 sec on, 20 sec off) at amplitude 25%. Chromatin was diluted with ChIP dilution buffer (1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, 20 mM Tris at pH 8) to a final concentration of 0.1% SDS. Antibodies were added for overnight incubation at 4°C with rotation. Antibodies used were 1 µg of anti-BRG1, 2 µg of anti-EBF2, and 2 µg of anti-H3K27ac (Abcam, ab4729). Protein A Sepharose beads were then added for 4 h at 4°C. Samples were washed twice with wash buffer 1 (0.1% SDS, 0.1% NaDOC, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, 20 mM Tris at pH 8), once with wash buffer 2 (0.1% SDS, 0.1% NaDOC, 1% Triton X-100, 0.5 M NaCl, 1 mM EDTA, 20 mM Tris at pH 8), once with wash buffer 3 (0.25 M LiCl, 0.5% NaDOC, 0.5% NP-40, 1 mM EDTA, 20 mM Tris at pH 8), and twice with wash buffer 4 (1 mM EDTA, 20 mM Tris at pH 8) followed by elution with 100 mM NaHCO3/1% SDS buffer. Samples were reverse cross-linked overnight at 65°C and treated with RNase A and proteinase K followed by column purification (Clontech NucleoSpin). ChIP enrichment was calculated as the percentage of input and normalized to 18S levels. For FAIRE analysis, cells were fixed and sonicated as for ChIP. Samples and inputs were then processed as described previously (Simon et al. 2012). Briefly, input DNA was treated with RNase A/proteinase K, reverse cross-linked overnight at 65°C, and then purified twice by phenol-chloroform extraction. FAIRE samples were purified twice by phenol-chloroform extraction, treated with RNase A/proteinase K, and reverse cross-linked overnight at 65°C. FAIRE DNA was then column-purified. FAIRE-qPCR enrichment was calculated as the percentage of input. Analysis of ChIP and FAIRE-qPCR results was performed using a two-way ANOVA with Holm-Šídák correction for multiple comparisons. Primer sequences are listed in Supplemental Table 1.
Oxygen consumption assays
Brown adipocytes were induced to differentiate and split on day 2 into 96-well Seahorse assay plates with 50,000 cells per well. At day 7 of differentiation, cells were washed twice with prewarmed assay medium (10 mM glucose, 2 mM L-glutamine, 1 mM sodium pyruvate in Seahorse XF base medium at pH 7.4) and then incubated in assay medium at for 10 min 37°C in a low CO2 incubator. OCR was measured using the Seahorse Wave software following injection with 1 µM iso or using the MitoStress kit with 2 µM oligomycin, 2 µM FCCP, and 0.5 µM rotenone/antimycin A. Protein concentration was measured using a standard BCA assay to normalize OCR. Data were analyzed using two-sample two-sided Student's t-test with Holm-Šídák correction for multiple comparisons.
ChIP-seq and RNA-seq processing and analysis
BAT from 8-wk-old Ebf2 wild-type and knockout mice was used for ChIP-seq and RNA-seq library preparation as described previously (Wang et al. 2014; Harms et al. 2015). Sequencing was performed on an Illumina HiSeq 2000 or 2500. ChIP-seq reads for EBF2, H3K27ac, RNA Pol II, and PPARγ were aligned to the mouse genome (mm9) using Bowtie. All redundant reads were discarded except for one per genomic position. Peak calling was performed using HOMER (Heinz et al. 2010). BigWig files were generated in RPM (reads per million total reads)-normalized scale for genome browser visualization. Depot-specific genes and H3K27ac peaks were defined previously (Harms et al. 2015) as follows: Depot-selective genes for Pol II gene body analysis were defined by differential gene analysis of BAT versus epididymal WAT (eWAT) microarray data (GSE8044). We compared Pol II changes within these groups of genes after Ebf2 knockout. Depot-selective regions for H3K27ac analysis were defined by BAT versus eWAT differential peak calling of H3K27ac (GSE63964): First, default H3K27ac peak calling was performed against a matching H3 ChIP-seq each for BAT and eWAT, and two peak sets were merged into a master peak set. Next, differential peak calling was performed using one depot as a foreground and the other as a background and vice versa. Each set of differential peaks was defined as a depot-specific peak. After subtracting depot-specific peaks, master peaks were defined as common peaks. We then compared H3K27ac changes at these groups of peaks after Ebf2 knockout. To identify Ebf2 knockout-regulated H3K27ac regions, histone peak calling for wild-type and knockout tissue was performed using HOMER with “-nfr” option to center on a putative nucleosome-free region (NFR) as a potential transcriptional binding site. After preparing a master peak set by pooling and merging overlapping ones, RPM-normalized H3K27ac signal was measured within a 2-kb window, and knockout-regulated H3K27ac peaks (repressed/unchanged/increased) were defined by twofold change criteria. EBF2 ChIP-seq signal from wild-type tissue was measured within the peaks in each group. The de novo motif search was done within a 300-bp window around the putative NFRs within each group of H3K27ac peaks using HOMER. RNA-seq reads were aligned to mm9 using the STAR pipeline (Dobin et al. 2013). Differentially expressed genes between wild-type and Ebf2 knockout tissue were identified via an exact test using the edgeR package (Robinson et al. 2010). Gene ontology analysis of differentially expressed genes was performed using Enrichr (Chen et al. 2013).
All ChIP-seq and RNA-seq data have been deposited in Gene Expression Omnibus (GEO) with accession number GSE97116.
Supplementary Material
Acknowledgments
We thank the University of Pennsylvania Diabetes Research Center for use of the Functional Genomics Core (P30-DK19525); the Institute of Diabetes, Obesity, and Metabolism for resources; and Dr. Silke Sperling, Dr. Huan Cui, Dr. Doug Epstein, and members of the Seale laboratory for advice and helpful discussion. We thank Rachel Stine and Chihiro Okada for technical assistance, and Anthony Angueira for assistance with statistical analyses. S.N.S. performed experiments and analyzed data. H.-W.L. and K.-J.W. performed computational analysis of ChIP-seq and RNA-seq data sets. A.P.S. and J.I. contributed new reagents and analytic tools. S.R. and M.J.H. generated sequencing libraries. S.N.S. and P.S. designed experiments and wrote the manuscript. This work was supported by National Institutes of Health grants 1F31DK108507-01 and 5T32GM008216-29 to S.N.S., 5R01DK106027-02 to K.J.W., and 5R01DK103008-02 to P.S.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.294405.116.
References
- Abe Y, Rozqie R, Matsumura Y, Kawamura T, Nakaki R, Tsurutani Y, Tanimura-Inagaki K, Shiono A, Magoori K, Nakamura K, et al. 2015. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat Commun 6: 7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. 2006. β-Catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol 296: 164–176. [DOI] [PubMed] [Google Scholar]
- Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, Schubeler D, Grosschedl R. 2016. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity 44: 527–541. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. 2004. Brown adipose tissue: function and physiological significance. Physiol Rev 84: 277–359. [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Cui H, Schlesinger J, Schoenhals S, Tonjes M, Dunkel I, Meierhofer D, Cano E, Schulz K, Berger MF, Haack T, et al. 2016. Phosphorylation of the chromatin remodeling factor DPF3a induces cardiac hypertrophy through releasing HEY repressors from DNA. Nucleic Acids Res 44: 2538–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol 25: 3997–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois L, Vincent A. 2001. The COE–Collier/Olf1/EBF–transcription factors: structural conservation and diversity of developmental functions. Mech Dev 108: 3–12. [DOI] [PubMed] [Google Scholar]
- Fan H-Y, He X, Kingston RE, Narlikar GJ. 2003. Distinct strategies to make nucleosomal DNA accessible. Mol Cell 11: 1311–1322. [DOI] [PubMed] [Google Scholar]
- Fedorenko A, Lishko PV, Kirichok Y. 2012. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 151: 400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. 2009. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab 10: 324–335. [DOI] [PubMed] [Google Scholar]
- Gao H, Lukin K, Ramírez J, Fields S, Lopez D, Hagman J. 2009. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci 106: 11258–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goljanek-Whysall K, Mok GF, Fahad Alrefaei A, Kennerley N, Wheeler GN, Munsterberg A. 2014. myomiR-dependent switching of BAF60 variant incorporation into Brg1 chromatin remodeling complexes during embryo myogenesis. Development 141: 3378–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M, Seale P. 2013. Brown and beige fat: development, function and therapeutic potential. Nat Med 19: 1252–1263. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Ishibashi J, Wang W, Lim HW, Goyama S, Sato T, Kurokawa M, Won KJ, Seale P. 2014. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab 19: 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Lim HW, Ho Y, Shapira SN, Ishibashi J, Rajakumari S, Steger DJ, Lazar MA, Won KJ, Seale P. 2015. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev 29: 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463: 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci 106: 5181–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K. 2011. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 21: 1650–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi J, Firtina Z, Rajakumari S, Wood KH, Conroe HM, Steger DJ, Seale P. 2012. An Evi1-C/EBPβ complex controls peroxisome proliferator-activated receptor γ2 gene expression to initiate white fat cell differentiation. Mol Cell Biol 32: 2289–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaka A, Mizutani T, Kobayashi K, Tando T, Sakurai K, Fujiwara T, Iba H. 2012. Double plant homeodomain (PHD) finger proteins DPF3a and -3b are required as transcriptional co-activators in SWI/SNF complex-dependent activation of NF-κB RelA/p50 heterodimer. J Biol Chem 287: 11924–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 11: 377–389. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, et al. 2008. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev 22: 2370–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. 2007. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, von Both I, Walls JR, McAuliffe F, Lee Adamson S, Mark Henkelman R, Wrana JL, Rossant J, Bruneau BG. 2004. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432: 107–112. [DOI] [PubMed] [Google Scholar]
- Madsen MS, Siersbaek R, Boergesen M, Nielsen R, Mandrup S. 2014. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol 34: 939–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Zhou MM. 2014. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6: a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. 2009. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene 28: 2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Lee MW, Sogawa Y, Bertholet AM, Locksley RM, Weinberg DE, Kirichok Y, Deo RC, Chawla A. 2016. Perinatal licensing of thermogenesis by IL-33 and ST2. Cell 166: 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. 2013. EBF2 determines and maintains brown adipocyte identity. Cell Metab 17: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. 2014. What we talk about when we talk about fat. Cell 156: 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salma N, Xiao H, Mueller E, Imbalzano AN. 2004. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor γ nuclear hormone receptor. Mol Cell Biol 24: 4651–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454: 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. 2011. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 121: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Ishibashi J, Kusminski CM, Wang QA, Hepler C, Vishvanath L, MacPherson KA, Spurgin SB, Sun K, Holland WL, et al. 2016. Zfp423 maintains white adipocyte identity through suppression of the beige cell thermogenic gene program. Cell Metab 23: 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Giresi PG, Davis IJ, Lieb JD. 2012. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc 7: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Schöler HR. 2010. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141: 943–955. [DOI] [PubMed] [Google Scholar]
- Stine RR, Shapira SN, Lim HW, Ishibashi J, Harms M, Won KJ, Seale P. 2016. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol Metab 5: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber N, Treiber T, Zocher G, Grosschedl R. 2010. Structure of an Ebf1:DNA complex reveals unusual DNA recognition and structural homology with Rel proteins. Genes Dev 24: 2270–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Lewcock JW, Feinstein P, Mombaerts P, Reed RR. 2004. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development 131: 1377–1388. [DOI] [PubMed] [Google Scholar]
- Wang W, Kissig M, Rajakumari S, Huang L, Lim HW, Won KJ, Seale P. 2014. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci 111: 14466–14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T, Aita S, Matsushita M, Kayahara T, Kameya T, Kawai Y, Iwanaga T, Saito M. 2013. Recruited brown adipose tissue as an antiobesity agent in humans. J Clin Invest 123: 3404–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. 2010. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466: 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Li S, Wang GX, Yu Q, Lin JD. 2014. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell 55: 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.