Abstract
Recent developments in genome-wide genetic analysis in B-acute lymphoblastic leukemia (B-ALL) have provided insight into disease pathogenesis and prognosis. B-ALL cases usually carry a primary genetic event, often a chromosome translocation, and a constellation of secondary genetic alterations that are acquired and selected dynamically in a nonlinear fashion. As far as we are aware of, for the first time, we studied centrosome aberration in patients with B-ALL to understand the progression of the disease. A cytogenetic study was carried out by GTG-banded karyotyping and fluorescence in situ hybridization. DNA index study was carried out with flow cytometry. Indirect immunostaining of centrosomes was performed on mononuclear cells using primary and corresponding secondary antibodies for centrosome-specific protein γ-tubulin. Three primary and corresponding secondary antibodies to three different centrosome-specific proteins, namely α-tubulin, γ-tubulin and pericentrin, were used for indirect immunostaining. The study was carried out on 50 patients with B-ALL. Centrosomal abnormalities were detected in 36 (72%) patients and the remainder (28%) had normal centrosome structure and numbers. Out of these 36 patients with abnormal centrosome, structural abnormalities were detected in 12 (33.3%) and numerical abnormalities in six (16.6%). Both structural and numerical aberrations were detected in 18 (50%) patients. When correlated with the cytogenetic and DNA index findings, 26/27 (96.2%) patients had centrosome defects concomitant with both abnormal karyotype and aneuploidy. Out of 50 patients with B-ALL , 17 (34%) had normal karyotype detected by both karyotype and DNA index, among these, seven (41.17%) patients had centrosome aberration. The morphological and structural abnormalities of the centrosome present in B-ALL cells have a role in disease development and can be considered as prognostic markers.
Keywords: B-ALL, chromosomal abnormalities, centrosome aberrations, hyperdiploidy, polyploidy, prognosis
Recurrent cytogenetic abnormalities such as hyperdiploidy and t(12;21), t(4;10), t(6;10), t(1;19), t(9;22), t(4;11), etc. are common in B-acute lymphoblastic leukemia (B-ALL) and are seen in around 60% of cases (1-3). Hyperdiploidy is the commonest cytogenetic abnormality seen in around 25-30% of childhood B-ALL and is associated with good prognosis. Mechanism of hyperdiploidy and aneuploidy in ALL remains conjectural with four possible mechanisms proposed: i) Initial near haploidy followed by doubling of the chromosomes; ii) Prior tetraploidization with subsequent loss of chromosomes; iii) Sequential gains of chromosomes in consecutive cell divisions; or iv) Simultaneous gain of all additional chromosomes in a single abnormal mitosis (4-6). However, what underlies the initial chromosomal instability is still uncertain.
The centrosome is a primary microtubule-organizing center of the cell, and regulates most microtubule-related functions, including transport of macromolecular complexes, positioning of cell organelles, cell motility, cell shape, polarity, and segregation of chromosomes during cell division (7). Each centrosome has a core of two centrioles, cylindrical structures consisting of nine microtubule triplets, surrounded by proteinaceous scaffold of the pericentriolar material .The centrosomes duplicate once per cell cycle and function as spindle poles to mediate the strictly balanced bipolar separation of chromosomes during mitosis (8,9). Here we studied centrosome proteins α-tubulin, γ-tubulin and pericentrin in patients with B-ALL in order to understand the centrosome aberration role in disease progression.
Centrosomal abnormalities lead to chromosomal instability and have been implicated in aneuploidy seen in various malignancies. Centrosomal amplification is the most common abnormality and correlates with the degree of aneuploidy and malignant behavior of tumor (10,11). Abnormalities of structure, function and localization within cells are also noted along with numerical abnormalities (12,13). Centrosomal amplification can arise from various aberrant processes such as cell to cell fusion, centrosomal overduplication during interphase, centrosomal fragmentation, de novo synthesis of centrosome or cytokinesis failure (14). Various tumor-suppressor genes and oncogenes are involved in the regulation of centrosomal duplication and their abnormalities result in centrosomal amplification (15,16). The importance of these centrosomal abnormalities in cancer cell survival, tumor progression and eventually as a potential tumor-selective therapeutic target is currently being extensively studied.
Centrosomal aberrations have been described as a possible cause of numerical chromosomal abnormalities in many solid human tumor types (17-19). However, in spite of an extensive literature search, we were unable to find any study of patients with B-ALL. This study was, therefore, undertaken to investigate whether (i) centrosomal aberrations occur in ALL and to correlate them with underlying cytogenetic abnormalities, and (ii) centrosomal pathology is a primary or secondary event on the road towards aneuploidy.
Materials and Methods
Patients and controls. Fifty consecutive patients with B-ALL aged between 2 and 72 years at presentation were included in this study. All the patients were diagnosed at our Institute between 2013 to 2014. Age- and sex-matched normal healthy persons were used as controls. The study was approved by Institutional Ethics Committee (NIIH/IEC/22-2013). Diagnosis of B-lineage ALL was established according to the European Group for the Immunological Characterization of Leukemia criteria (20). Standard GTG-banding technique was used for cytogenetic study. Chromosomal preparations were obtained from bone marrow sample by direct and short-term culture methods and were subjected to GTG-banding. Chromosomal analysis was carried out under microscopy using the 2009 International System for Cytogenetic Nomenclature (21). Fluorescence in situ hybridization (FISH) was carried out with centromeric probes for chromosomes X, 4, 6, 10, 14, 17, and 18. Locus-specific FISH probes were used for t(9;22)/breakpoint cluster region–abelson (BCR–ABL1), t(12;21)/ETS variant 6 gene and runt-related transcription factor 1 (ETV6-RUNX1) fusion genes and 11q23/mixed lineage leukemia (MLL), 14q32.3/immunoglobulin heavy locus gene (IGH) break-apart probes were used for FISH analysis (Vysis, Abbott Molecular, Des Plaines, IL, USA). Mononuclear cells were separated from 2 ml of heparinized blood by density gradient centrifugation using Ficoll-hypaque solution (Sigma-Aldrich, St.Louis, MO, USA).
DNA index study of the controls and patients was performed with the help of a BD FACScaliber flow cytometer. (Becton Dickinson, Franklin Lakes, NJ, USA). The calibration of the flow cytometer was carried by BD DNA QC Particle Kit (BD). CELL Quest Pro Software (BD) was used for calibration and the DNA index was obtained using BD Cycle TEST™ PLUS DNA Reagent Kit. (BD). A DNA index greater than 1.16 was considered as hyperdiploid and that less than 0.98 as hypodiploid.
Centrosomal staining. Indirect immunostaining of centrosomes was performed as described elsewhere (22,23).The mononuclear cells were mounted on a Teflon-coated slide and fixed with methanol. These cells were treated with 0.2% Tween 20 to increase the permeability of the cell membrane. To prevent non-specific binding of the antibodies, the slides were treated with 1% bovine serum albumin (Sigma-Aldrich) for 1 h. The primary and corresponding secondary antibodies for centrosome-specific protein γ-tubulin (Sigma-Aldrich, Saint Louis, MO, USA) was used for all 50 patients with B-ALL. Three primary and corresponding secondary antibodies to three different centrosome-specific proteins, namely α-tubulin, γ-tubulin and pericentrin (Sigma-Aldrich) were used on 11 patients for indirect immunostaining. This was to determine the nature of centrosomal aberration with each of the three proteins of the same patient. For each slide, one type of primary and its corresponding secondary antibody was used. Each antibody was stained by fluorescein isothiocyanate-labeled secondary anti rabbit IgG for 30 minutes at 20˚C. Then the slides were washed with phosphate buffered saline followed by counterstaining with 4’,6-diamidino-2-phenylindole. The slides were viewed under a fluorescence microscope (Nikon 90i; Towa Optics, New Delhi, India)and a minimum of 100 cells were scored for numerical and structural anomalies of centrosomes on each slide. The centrosome was considered to be abnormal when at least 10 cells exhibited the same centrosomal abnormality. Normal cells had two centrosomes of equal size (variation in the size between two centrosomes <20%). Hence centrosomal abnormalities were broadly classified as abnormality in number, morphology and both. If both the centrosomes of the same cell showed different results, this was also noted. Centrosomal pathology was correlated with DNA index, aneuploidy and karyotype. Non parametric test (χ2 test) was applied to test the significance at p<0.05.
Results
The study was carried out in 50 patients at the time of diagnosis of B-ALL. Twenty six patients were 2-15 years old and 24 were 16-72 years old. There were 32 and 18 males and females, respectively, in the whole group. Patients presented with hemoglobin, of 4.3-7.8 g/dl, total leukocyte count of 6.3-52.4×103/ml with lymphoblast varying between 26-85%.
The karyotyping was performed successfully in 44 (88%) B-ALL cases, including 27 (54%) cases with abnormal cytogenetics and 17(34%) with normal results. Hyperdiploidy was the commonest cytogenetic abnormality in both pediatric and adult age groups. Two patients (adult) had BCR–ABL1 fusion, one patient had IGH rearrangement and three pediatric patients had TEL–AML1 fusion. The DNA index was >1.16 in all chromosomally abnormal cases (27 cases) (Table I).
Table I. Centrosomal abnormalities detected in groups of patients (n=50) with B-cell acute lymphoblastic leukemia with different karyotypic abnormalities.
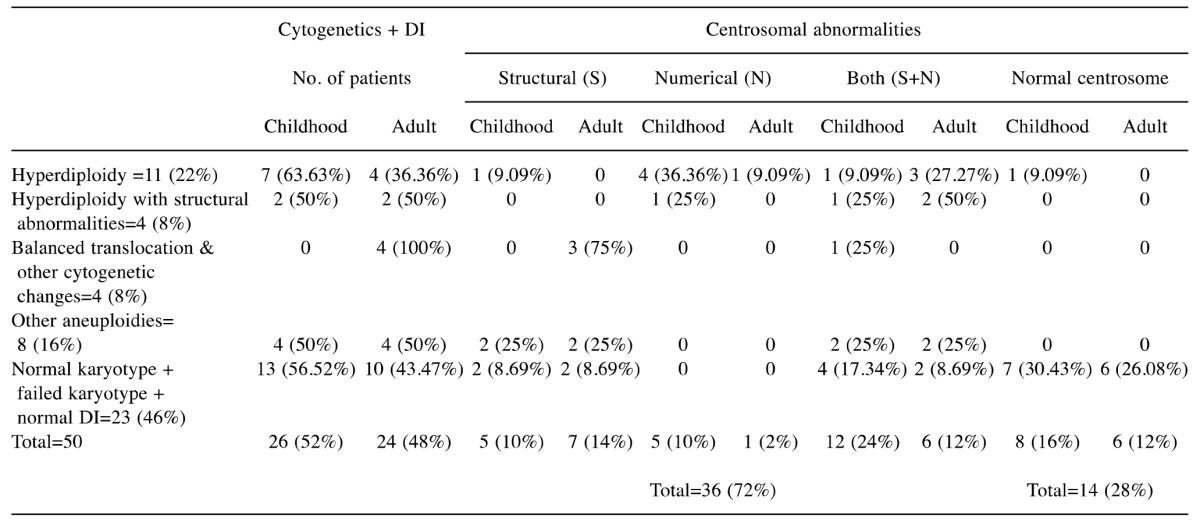
DI: DNA index.
The DNA index in mononuclear cells of B-ALL cases was between 1.03 and 2.2 (Morista-Horn similarity index, 0.99± 0.03) and that of age-matched controls was between 0.98 and 1.05.
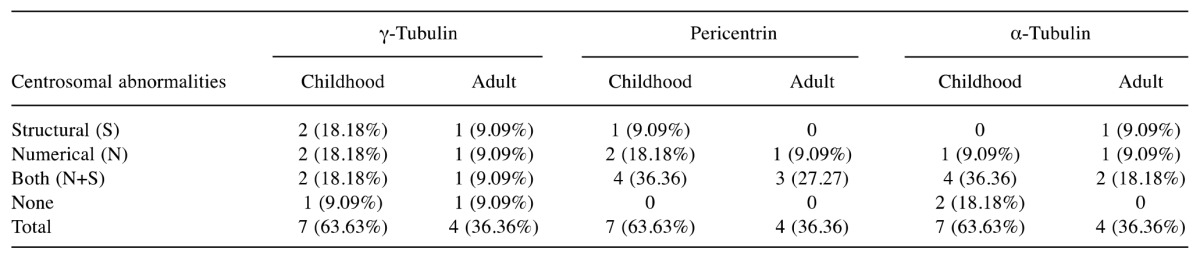
Centrosomal abnormalities in B-ALL. The centrosomal evaluation was carried out in 50 patients with B-ALL. Thirty-six (72%) patients had centrosomal abnormalities and 14 (28%) patients had normal centrosome number and structure with γ-tubulin (Table I). Out of these 36 patients, six (16.66%) had the extra centrosomes, 12 (33.33%) patients had structural abnormalities, and 18 (50%) patients had both structural and numerical abnormalities (Table II). Eleven samples stained with three antibodies showed 100% centrosomal abnormalities with pericentrin and 81.81% (9/11) abnormalities each with γ-tubulin and α-tubulin (Table III).
Table II. Centrosomal abnormalitie in 50 adult and pediatric patients with B cell acute lymphoblastic leukemia.
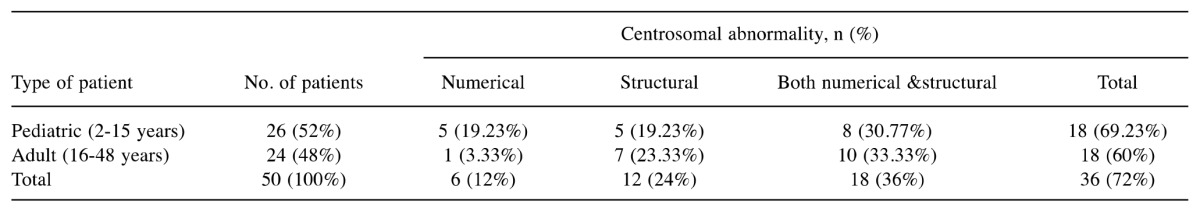
Table III. Centrosomal abnormalities in 11 patients with acute lymphoblastic leukemia stained for three different markers of centrosome.
Discussion
Chromosome abnormalities such as hyperdiploidy and other aneuploidies along with certain translocations are common genetic abnormalities seen in B-ALL. How this pathology arises has been an area of constant quest, producing several hypotheses (4-6). It is also believed that one theory may not be able to explain all the causes (7). The centrosome plays a crucial role in producing the mitotic apparatus, which eventually ensures the equal distribution of chromosomes between two dividing cells (8,9). Abnormalities of centrosomes or their function can lead to the uneven distribution of chromosomes in ALL. Centrosomal abnormalities via formation of multipolar spindles, or centrosome clustering resulting in chromosomal misaggregation, is proposed as a major mechanism for chromosomal instability and aneuploidy seen in cancer (14,24,25). Centrosomal amplification is frequently detected in solid tumors as well as hematological malignancies such as acute myeloid leukemia, Hodgkin’s and non-Hodgkin’s lymphoma multiple myeloma and mantle cell lymphoma (22,26-30). It is also now considered as a candidate hallmark of cancer cells. We were interested in studying the presence of different centrosomal aberrations using antibodies to three different centrosomal proteins and correlation of the extent of these abnormalities with the ploidy status of B-ALL.
The results presented here show, for the first time, presence of both structural and numerical abnormalities in a significant proportion of patients with B-ALL. A large number of our patients (72%) exhibited centrosomal pathology (Table I).
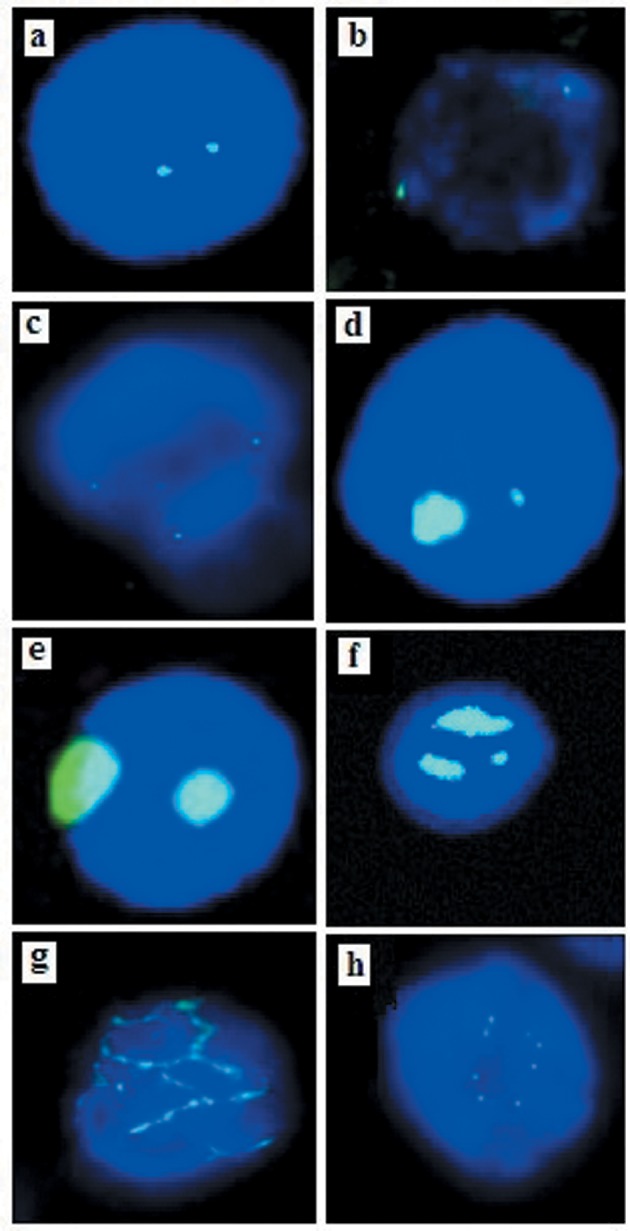
Various structural defects have been described in centrosomal abnormalities and they include changes in centrosomal shape (string-like elongated linear arrays, ring-like, amorphous, atypical filaments, corkscrew) or size (usually large patchy aggregates, but may range from tiny flecks). Defects in number i.e. more than two per cell), and position (diffuse patchy cytoplasmic staining, scattered, clustered) or composition (higher protein levels, inappropriate phosphorylation, absence of centrioles) are also described (18). In our study, we detected normal, perpendicular position tripolar cell division of centromere (Figure 1a-h) but also enlarged amorphous round and elongated centrosomes with uniform and crescentic margins Figure 1d). These structural abnormalities were detected in 12 (24%) patients. We also found extra centrosomes in samples from 6 (12%) patients, which are shown in Figure 1e and f. Lingle and Salisbury reported that cells with extra centrosomes in human breast cancer rarely exhibited abnormal mitosis (31). Pioneering studies by Nigg (11) and Brinkley demonstrated that centrosomal clustering enables cells to divide successfully despite the presence of extra centrosomes (32). The initial description of centrosomal clustering noted that cells with extra centrosomes pass through a transient multipolar spindle intermediate before centrosomal clustering and bipolar anaphase occurs (23). Recent data from several laboratories demonstrate that while passing through the transient multipolar state, merotelic kinetochore-microtubule attachment errors, defined by the persistent attachment of microtubules from both spindle poles to a single kinetochore, accumulate, and consequently increase the frequency of lagging chromosomes during bipolar anaphase after centrosomal clustering (33,34). Importantly, this finding implies that cells with amplified centrosomes do not necessarily need to divide in a multipolar fashion to allow low-level chromosomal missegregation that can fuel tumor progression. Such an interpretation also supports the emerging bimodal relationship between aneuploidy and tumorigenesis (35). In our case, out of six patients with extra centrosomes, five had childhood B-ALL with hyperdiploid karyotype, which comes under the good prognostic group. This tells us that these extra centrosomes in pediatric hyperdiploid B-ALL cases later develop clusters following normal cell division, which leads to disease with a good prognosis. Both structural and numerical abnormalities were observed in 18 (36%) of our patients, and in 11 (61%) this correlated with the poor prognostic group by karyotyping. It has been reported that this group of centrosomal aneuploidy correlates with high-risk karyotyping in patients with acute myeloid leukemia (22).
Figure 1. Centrosomal abnormalities detected with γ tubulin and α tubulin stain in patients with B-cell acute lymphoblastic leukemia. a: Normal centrosome. b: Centrosome in perpendicular position. c: Tripolar cell division. d, e: Structural abnormality (round) of centrosome. f: Numerical abnormality. g: Elongated centrosome abnormality. h: Numerical abnormality of centrosome.
When three different centrosome proteins were stained, three different kinds of abnormalities appeared with respective centrosome proteins in the same patient (Table III). These results suggests that the different abnormalities of the centrosome have a role in chromosomal instability and development of B-ALL. Studies on centrosomal abnormalities using more than one antibodies to centrosomal proteins by other authors showed different frequency of centrosomal abnormalities with different antibodies (22,36). However, a study on patients with chronic myeloid leukemia using pericentrin and γ-tubulin antibodies showed similar centrosomal abnormalities (30).
The frequency of centrosomal abnormalities was greater in patients with hyperploidy compared to patients with balanced chromosomal abnormalities and other aneuploidies. Centrosomal abnormality was also observed in eight (47%) patients who had neither abnormal DNA ploidy nor karyotypic abnormality. This shows that centrosomal abnormality precedes chromosomal abnormality in B-ALL. Other researchers also observed the same results with many other types of neoplasia (37). This shows that centrosomal abnormality may have predictive value in ALL and it may be used as a diagnostic and prognostic marker in this disease.
Centrosomal abnormality seems to be more frequent (96%, 26/27) of patients when both cytogenetic and DNA index abnormalities are present (p<0.01). This may be due to the association of centrosomal abnormalities with karyotypic abnormalities in neoplasia as many authors have already described in their reports (37). In our study, five patients showed centrosomal abnormalities (p<0.05) with abnormal DNA ploidy (>1.2) but had normal cytogenetics. We correlated the centrosomal abnormality patterns with the karyotypic results of the 50 with B-ALL (Table I). The frequency of aneuploidy detected by either karyotype or DNA index was 46% (23/50) and centrosomal aneuploidy in these patients was 95.65% (22/23). This finding shows that centrosomal abnormality is strongly associated with chromosomal instability as described by others (38). Only one patient of this group had normal centrosomes. In this single patient, this may have been due to the aberration of other centrosomal proteins excluding γ-tubulin. Centrosomal abnormalities (43.47%) were also detected in patients with B-ALL cytogenetically normal and with normal DNA index in our study. Patients who harbor diploid neoplastic cells should be closely followed-up to for development of any centrosomal aneuploidy, as we know from many other reports that centrosomal abnormalities precede karyotypic abnormality. In our study, karyotypic abnormalities other than aneuploidy were detected in four (8%) patients and centrosomal abnormality was detected in all of them. This may reflect the possibility for disease progression and development of aneuploidy in these tumor cells.
Immunostaining of centrosomal protein provides only a limited and static vision of centrosomal abnormality in a fixed cell. This cannot asses the functional attributes of centrosomes in a holistic way. Studies using molecular biology tools involving several centrosomal proteins have unraveled the genetic pathology in many of such proteins in different cases (13,22). However, in this small study we clearly showed the significant morphological and structural abnormalities of centrosomes which accompany affected cells in patients with B-ALL. The role of centrosomal pathology in the natural history and outcome treatment in patients with B-ALL remains to be revealed.
Conflicts of Interest
The Authors declare no conflict of interest in regard to this study.
Acknowledgements
The study was carried out with a core grant of the Institute funded by Indian Council of Medical Research, New Delhi, India.
The Authors thank Hematologists from the Department of Hematology, KEM Hospital, Mumbai for their help in diagnosis of the patients.
References
- 1.Meijerink JP, den Boer ML, Pieters R. New genetic abnormalities and treatment response in acute lymphoblastic leukemia. Semin Hematol. 2009;46:16–23. doi: 10.1053/j.seminhematol.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122:3407–3415. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dastugue N, Suciu S, Plat G, Speleman F, Cavé H, Girard S, Bakkus M, Pagès MP, Yakouben K, Nelken B, Uyttebroeck A, Gervais C, Lutz P, Teixeira MR, Heimann P, Ferster A, Rohrlich P, Collonge MA, Munzer M, Luquet I, Boutard P, Sirvent N, Karrasch M, Bertrand Y, Benoit Y. Hyperdiploidy with 58-66 chromosomes in childhood B-acute lymphoblastic leukemia is highly curable: 58951 CLG-EORTC results. Blood. 2013;121:2415–2423. doi: 10.1182/blood-2012-06-437681. [DOI] [PubMed] [Google Scholar]
- 4.Onodera N, McCabe NR, Nachman JB, Johnson FL, Le Beau MM, Rowley JD, Rubin CM. Hyperdiploidy arising from near-haploidy in childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 1992;4:331–336. doi: 10.1002/gcc.2870040410. [DOI] [PubMed] [Google Scholar]
- 5.Onodera N, McCabe NR, Rubin CM. Formation of a hyperdiploid karyotype in childhood acute lymphoblastic leukemia. Blood. 1992;80:203–208. [PubMed] [Google Scholar]
- 6.Paulsson K, Mörse H, Fioretos T, Behrendtz M, Strömbeck B, Johansson B. Evidence for a single-step mechanism in the origin of hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2005;44:113–122. doi: 10.1002/gcc.20222. [DOI] [PubMed] [Google Scholar]
- 7.Schatten H. The mammalian centrosome and its functional significance. Histochem Cell Biol. 2008;129:667–686. doi: 10.1007/s00418-008-0427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krämer A, Lukas J, Bartek J. Checking out the centrosome. Cell Cycle. 2004;3:1390–1393. doi: 10.4161/cc.3.11.1252. [DOI] [PubMed] [Google Scholar]
- 9.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Koutsami MK, Tsantoulis PK, Kouloukoussa M, Apostolopoulou K, Pateras IS, Spartinou Z, Drougou A, Evangelou K, Kittas C, Bartkova J, Bartek J, Gorgoulis VG. Centrosome abnormalities are frequently observed in non-small-cell lung cancer and are associated with aneuploidy and cyclin E overexpression. J Pathol. 2006;209:512–521. doi: 10.1002/path.2005. [DOI] [PubMed] [Google Scholar]
- 11.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 12.D’Assoro AB. Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 13.Nigg E.A. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 14.Kramer A, Muleir B, Bartek J. Centrosome clustering and chromosome (in) stability: A matter of life and death. Molecular Oncology. 2011;5:324–335. doi: 10.1016/j.molonc.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukasawa K. P53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim Biophys Acta. 2008;1786:15–23. doi: 10.1016/j.bbcan.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duensing A, Duensing S. Centrosomes, polyploidy and cancer. Adv Exp Med Biol. 2010;676:93–103. doi: 10.1007/978-1-4419-6199-0_6. [DOI] [PubMed] [Google Scholar]
- 17.Nigg EA. Centrosome aberrations: Cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 18.Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci. 2011;7:1122–1144. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emdad L.Sarkar D, Su ZZ, Fisher PB. Emerging roles of centrosomal amplification and genomic instability in cancer. Front Mol Biosci. 2005;10:728–742. doi: 10.2741/1567. [DOI] [PubMed] [Google Scholar]
- 20.Bene MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A, Van't Veer MB. Proposals for the immunological classification of acute leukemias, European Group for the Immunological Characterization of Leukemias (EGIL) Leukemia. 1995;9:1783–1786. [PubMed] [Google Scholar]
- 21.Shaffer LG, Slovak ML, Campbell LJ. An international system for human cytogenetic nomenclature (ISCN 2009) Hum Genet. 2009;126:603–604. [Google Scholar]
- 22.Neben K, Giesecke C, Schweizer S, Ho AD, Krämer A. Centrosome aberrations in acute myeloid leukemia are correlated with cytogenetic risk profile. Blood. 2003;10:289–291. doi: 10.1182/blood-2002-04-1188. [DOI] [PubMed] [Google Scholar]
- 23.Ring D, Hubble R, Kirschner M. Mitosis in a cell with multiple centrioles. J Cell Biol. 1982;94:549–556. doi: 10.1083/jcb.94.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogden A, Rida PCG, Aneja R. Let’s huddle to prevent a muddle: Centrosome clustering as an attractive anticancer strategy. Cell death and differentiation. 2012;19:1255–1267. doi: 10.1038/cdd.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leber B, Maier B, Fuchus F, Chi J, Reiffel P, Andrehub S. Protein requires for centrosome clustering in cancer cells. Sci Transl Med. 2010;2:33–38. doi: 10.1126/scitranslmed.3000915. [DOI] [PubMed] [Google Scholar]
- 26.Neben K, Ott G, Schweizer S, Kalla J, Tews B, Katzenberger T, Hahn M, Rosenwald A, Ho AD, Müller-Hermelink HK, Lichter P, Krämer A. Expression of centrosome-associated gene products is linked to tetraploidization in mantle cell lymphoma. Int J Cancer. 2007;120:1669–1677. doi: 10.1002/ijc.22404. [DOI] [PubMed] [Google Scholar]
- 27.Hensel M, Zoz M, Giesecke C, Benner A, Neben K, Jauch A, Stilgenbauer S, Ho AD, Krämer A. High rate of centrosome aberrations and correlation with proliferative activity in patients with untreated B-cell chronic lymphocytic leukemia. Int J Cancer. 2007;121:978–983. doi: 10.1002/ijc.22752. [DOI] [PubMed] [Google Scholar]
- 28.Krämer A, Neben K, Ho AD. Centrosome aberrations in hematological malignancies. Cell Biol Int. 2005;29:375–383. doi: 10.1016/j.cellbi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Krämer A, Schweizer S, Neben K, Giesecke C, Kalla J, Katzenberger T, Benner A, Müller-Hermelink HK, Ho AD, Ott G. Centrosome aberrations as a possible mechanism for chromosomal instability in non-Hodgkin's lymphoma. Leukemia. 2003;17:2207–2213. doi: 10.1038/sj.leu.2403142. [DOI] [PubMed] [Google Scholar]
- 30.Giehl M, Fabarius A, Frank O, Hochhaus A, Hafner M, Hehlmann R, Seifarth W. Centrosome aberrations in chronic myeloid leukemia correlate with stage of disease and chromosomal instability. Leukemia. 2005;19:1192–1197. doi: 10.1038/sj.leu.2403779. [DOI] [PubMed] [Google Scholar]
- 31.Lingle V, Salisbury JL. Altered centrosome structure is associated with abnormal mitosis in human breast tissues. Am J Pathol. 1999;155:1941–1951. doi: 10.1016/S0002-9440(10)65513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 33.Ganem NJ, Godinho SA, Pellman DA. Mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silkworth WT, NardiI K, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome missegregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weaver BA, Silk AD.Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Neben K, Tews B, Wrobel G, Hahn M, Kokocinski F, Giesecke C, Krause U, Ho AD, Krämer A, Lichter P. Gene expression patterns in acute myeloid leukemia correlate with centrosome aberrations and numerical chromosome changes. Oncogene. 2004;23:2379–2384. doi: 10.1038/sj.onc.1207401. [DOI] [PubMed] [Google Scholar]
- 37.Cosenza MR, Krämer A. Centrosome amplification, chromosomal instability and cancer: Mechanistic, clinical and therapeutic issues . Chromosome Res. 2016;24:105–126. doi: 10.1007/s10577-015-9505-5. [DOI] [PubMed] [Google Scholar]
- 38.Nolte F, Giehl M, Haass W, Nowak V, Schumann C, Nowak D, Mossner M, Popp HD, Schulze TJ, Klein S, Seifarth W, Hofmann WK, Fabarius A. Centrosome aberrations in bone marrow cells from patients with myelodysplastic syndromes correlate with chromosomal instability. Ann Hematol. 2013;92:1325–1333. doi: 10.1007/s00277-013-1772-7. [DOI] [PubMed] [Google Scholar]


