Abstract
Background: the association between infectious agents and tumour aetiology is relevant in about 20% of cases. Patients and Methods. We tested high-grade glioma tissues from 45 patients for the presence of viral nucleic acids of six herpes viruses, human adenoviruses (A-G), and two neurotropic human viruses (enteroviruses, tick-borne encephalitis virus). Real-time polymerase chain reaction was used with immunolabelling. Results: Three species of herpes viruses were detected: HSV-2, Epstein-Barr virus (EBV), HHV-6, and one human enterovirus. Plasma of these patients was not infected with viruses. In sera of patients, low HSV-1 and HSV-2 immunoreactivity were found in five cases, although these were not detected in their tumour tissue. Conclusion: Certain common viruses (HSV-1, HSV-2, EBV, human cytomegalovirus) are chronically present in the sera of patients with glioblastoma, but not necessarily in their tissues. Possibly both are associated with glioma progression, as we only found viruses in glioblastoma multiforme, but not in lower stages of glioma. Low titres of viruses in the blood indicate chronic viral virulence.
Keywords: Glioblastoma, oncogenic viruses, neurotropic viruses, serum biomarkers, HSV, VZV, EBV, HCMV, HHV-6, AdV, hEV, TBEV
Gliomas are the most common malignant brain tumours, and they comprise of glioblastoma (GBM; WHO grade IV), anaplastic astrocytoma, mixed anaplastic oligoastrocytoma, and anaplastic oligodendroglioma (all WHO grade III) (1,2). The highest incident rate is for GBM, which is associated with particularly poor prognosis (3), with only 27% of patients reaching 2-year survival (2). Aetiologically, a confirmed risk factor for glioma is exposure to ionizing radiation (4-7). The association of infectious agents with neoplasms, such as mycoplasma, viruses and bacteria, has only been considered recently (8). The most common and prevalent cancer-causing DNA viruses are human papillomaviruses 16 and 18, Epstein-Barr virus (EBV), Kaposi sarcoma-associated herpes virus (KSHV), hepatitis B virus, hepatitis C virus, human adult T-cell leukaemia virus type 1, and Merkel cell polyomavirus (9). In primary malignancies of the central nervous system (CNS), polyomavirus and herpes virus have been detected with varied frequencies in paediatric patients and adults with histologically diverse CNS malignancies. However, establishing a link between chronic viral infection and primary CNS malignancy has been an area of considerable controversy (10,11). The most commonly reported associations for tumours in the brain have been with polyomaviruses, such as John Cunningham virus (JCV), BK virus (BKV), and simian virus 40 (SV40) (12-14), although two studies in a large patient cohort of 225 paediatric and adult brain tumour tissues revealed relatively low JCV, BKV and SV40 frequencies (15). The second most common associations are with herpes viruses, which have comprised herpes simplex virus 1 and 2 (HSV-1, 2/HHV-1, 2), varicella zoster virus (VZV/HHV-3), EBV(/HHV-4), human cytomegalovirus (HCMV/HHV-5), and human herpes viruses 6, 7, and 8 (HHV-6, 7, 8) (10). Since Cobbs and co-workers first described the potential association between HCMV and malignant glioma in 2001 (16), which is believed to modulate the malignant GBM phenotype (17), no consensus has yet been reached (18-22). This is partly due to the inadequate testing of HCMV by standard diagnostic assays (23) and partly to large variations in the HCMV seropositivity of related ethnic groups (21,24). A meta-analysis of 16 reports by Salomon et al. (25) confirmed the concept that there is no clear-cut evidence for a role of HCMV in GBM tumorigenesis. Similarly, specific roles of other herpes viruses in glioma initiation have not been demonstrated to date (11,26-30).
Adenoviruses were also found to be associated with CNS malignancies (31). In the present study, human enteroviruses (hEV) and tick-borne encephalitis virus (TBEV) were included, as they are both neurotropic, although not oncogenic, in order to define their possible association with brain tumours. TBEV can cause the relatively common flaviviral infection in Europe, including Slovenia (32), and Palus et al. (33) reported on the link between TBEV infection and astrocyte injury. Taken together, the presence of this plethora of viruses in glioma, their carcinogenic activities, and their roles in glioma progression all remain unclear.
Here, we aimed to test 45 tissue samples from patients with high-grade glioma (HGG) for the presence of the oncomodulatory viral nucleic acids of six herpes viruses, human adenoviruses (species A-G) and neurotrophic hEV, and TBEV. In addition, the serum samples from 30 out of these 45 patients were tested for the presence of antibodies against HSV-1, HSV-2, TBEV, and hEV, based on their ability to cause persistent infection, which was studied in GBM for the first time here. To the best of our knowledge, this is the first clinical study that has evaluated viral nucleic acids in glioma tissue specimens and in the blood of the same patients in the Slovenian region.
Patients and Methods
Patients. Patients from the Department of Neurosurgery, UCC Maribor, Slovenia, who were undergoing neurosurgery for malignant glioma from May 2013 to December 2015 were included in the present study. Informed consent was obtained from the patients, and the study was approved by the Medical Ethics Committee, UCC Maribor, Slovenia (Document No. 123/13). Pre-operatively, 8 mg dexamethasone twice daily was administered. The patient personal and clinical data were obtained through interviews and their medical records. The data collected included patient age, sex, date and type of operation, tumour location, and preoperative C-reactive protein (CRP) level. The histological slides of all cases were reviewed by two pathologists and classified according to WHO (34).
Tissue sample preparation. Forty-five tumour tissue samples were obtained during craniotomies for tumour reduction (26/45) or during stereotactic biopsies (19/45). Histological diagnoses were initially made during surgery, on frozen tissue sections by staining smear preparations with methylene blue. Serial biopsy specimens were obtained stepwise along the trajectory through the entire lesions. Fresh tumour fragments were immediately transferred to physiological solution and kept at 4˚C until they were processed, within 24 h for viral analysis. The remaining tissue fragments were fixed in formaldehyde for histological analysis.
Serum analysis. Patient peripheral venous blood samples were taken in K2 EDTA tubes before surgery. CRP was measured in the serum by turbidimetry (Hitachi 912 analyser; Boehringer Mannheim, Germany). The preoperative CRP values were defined as low (≤5 mg/l) or high (>5 mg/l). In 30 cases, the blood samples were tested for the presence of antibodies against specific viruses.
Extraction and detection of viral DNA/RNA in tissue. Nucleic acids were extracted from tumour tissues using RTP® DNA/RNA virus mini kits (Stratec Biomedical AG, Germany), according to the manufacturer’s instructions. Briefly, disruption of the tissues and cells was performed with non-chaotropic lysis buffer and proteinase K. This followed by DNA/RNA adsorption to filter membranes and removal of the cell components and polymerase chain reaction (PCR) inhibitors using washing buffers. The viral nucleic acids were eluted in 60 μl elution buffer and stored at −20˚C.
Viruses were detected with commercial and in-house real-time PCR kits. A portion of the DNA from six herpes viruses was amplified using LightMix® kits (TIB Molbiol GmbH, Berlin, Germany), following the manufacturer instructions (Figure 1). Detection of HSV-1/-2 was based on the amplification of a 215 bp fragment of the polymerase gene, containing a variable region, which is used for the differentiation between HSV-1 and 2 based on melting analysis. Using the same PCR kit, a 196 bp of orf29 gene of VZV was amplified but the fluorescence was read in different channel (705 nm). For EBV, HCMV and HHV-6 a portion of ebna gene (166 bp), viral glycoprotein B gene (UL55; 226 bp) and 101K gene (U11; 272 bp) are amplified, respectively. For all LightMix kits, the detection of viruses is based on the amplification with hybridization probes, which allows melting analysis to confirm the obtained results. Temperature profiles for reactions are different for each of the detected viruses, as described in (http://www.tib-molbiol.de/index.html).
Figure 1. Amplification plot and standard curve from commercial LightMix PCR kits used for detection of herpes viruses.
PCR kits (RealStar® Adenovirus 1.0; Altona Diagnostics, Hamburg, Germany) were used for detection of human adenovirus, including the seven virus species from A to G. The manufacturer did not reveal the part of the genome amplified by the kit and stated that the amplification and detection of adenoviruses is based on the use of the dual labelled hydrolysis probes.
Human enterovirus and TBEV RNAs were amplified using in-house-validated PCR assays. The protocol for the detection of TBEV has been described previously (35) and it is based on the amplification of 3’ non-coding region of the TBEV genome with primers F-TBE1: GGG CGG TTC TTG TTC TCC and R-TBE1: ACA CAT CAC CTC CTT GTC AGA CT and probe TBE-P-WT: 6FAM-TGA GCC ACC ATC ACC CAG ACA CA-DB. Enteroviral RNA was detected using primers and dual labelled hydrolysis probe for amplification of 200 base pair in the 5’ non-coding region. Primers and probe sequences were: hEV-S: GGC TGC GTT GGC GGC CT, hEV-As: CAA AGT AGT CGG TTC CGC, hEV-P: 6FAM-GGC CCC TGA ATG CGG CTA ATC-BBQ. All primers and probes listed have sequence orientation of 5’ to 3’. Both PCRs were performed with the QIAGEN One-Step RT-PCR Kit (Qiagen, Hilden, Germany), which included enzymes Sensiscript and Omniscript reverse transcriptases and HotStarTaq DNA polymerase. Cycling was performed under following conditions: 50˚C for 30 min, 95˚C for 15 min, followed by 45 cycles of 95˚C for 10 s and 60˚C for 40 s. Fluorescence was read in the green channel (530 nm). PCR reactions were run on PCR cyclers (Rotor Gene Q; Qiagen, Hilden, Germany; and LightCycler 2.0; Roche Diagnostics, Indianapolis, IN, USA).
To avoid false-positive results because of contamination, negative controls were included in PCR runs. When specific viral targets were amplified, the assays were performed in independent duplicate.
Quantification and sequencing of viral DNA. Detected herpes viral DNA was quantified using LightMix® kits, allowing for quantification of the amplified DNA, expressed as target molecules or copies per 5 μl DNA, based on the amplification curve. Where applicable, nucleotide sequences were retrieved from amplicons by Sanger sequencing by a genetic analyser (model 3500; Applied Biosystems, Foster City, California, USA), using BigDye® Terminator v3.1 Cycle Sequencing kits. The amplicons were obtained using previously published protocols (36,37).
Detection of circulating viral antibodies. Patient sera samples were tested for the presence of antibodies against HSV-1, HSV-2, TBEV, and hEV. The antibodies were detected using commercial enzyme immunoassays (ELISA) (Enzygnost anti-FSME-IgG; Siemens Healthcare GmbH, Erlangen, Germany) for TBEV, and complement fixation tests (CFT) (Virion/Serion, Würzburg, Germany) for HSV and hEV.
IgG and IgM antibodies against TBEV were analysed in 100 μl serum in 1:20 dilution in sample buffer in plates coated with the TBEV antigen. The assays were performed automatically in a BEP III system (Siemens Healthcare GmbH, Erlangen, Germany). For detection of IgM, sera were pre-treated with an RF absorbent reagent to avoid false-positive results. The data are given as IgG concentrations (IU/ml), as provided by the BEP III system. Specific antibodies against HSV and hEV were detected by CFT. Serum dilutions (1:10 up to 1:160) were first subjected to 30 min incubation at 56˚C to inactivate endogenous complement, which may disturb the test calibration. The test is performed in microtiter plates where mixture of antigen, complement and indicator system is added. In the absence of immune complexes formation, a haemolysis reddish clear solution indicated negative result, whereas a cluster of red cells due the presence of immune complexes indicated a positive result.
Statistical analysis. Descriptive statistical methods (e.g. median, frequency tables) and independent Student’s t-tests were used for defining any correlations between the viral DNA/RNA and the patient data.
Results
Patient demographics. Among 45 HGG specimens from 21 males and 24 females, 12 were classified as WHO grade III and 33 as WHO grade IV (Table I). The patient ages ranged from 22 to 86 years (median=60.0 years). With respect to the locations of the tumours, most (n=38) cases affected the cerebral lobe at frontal, temporal, parietal, or occipital sites, with four located in the corpus callosum, two in the thalamus /or basal ganglia, and one in the cerebellum.
Table I. Demographic and clinical data of glioma patients and the levels of the nine tested viruses in their tissues and sera.
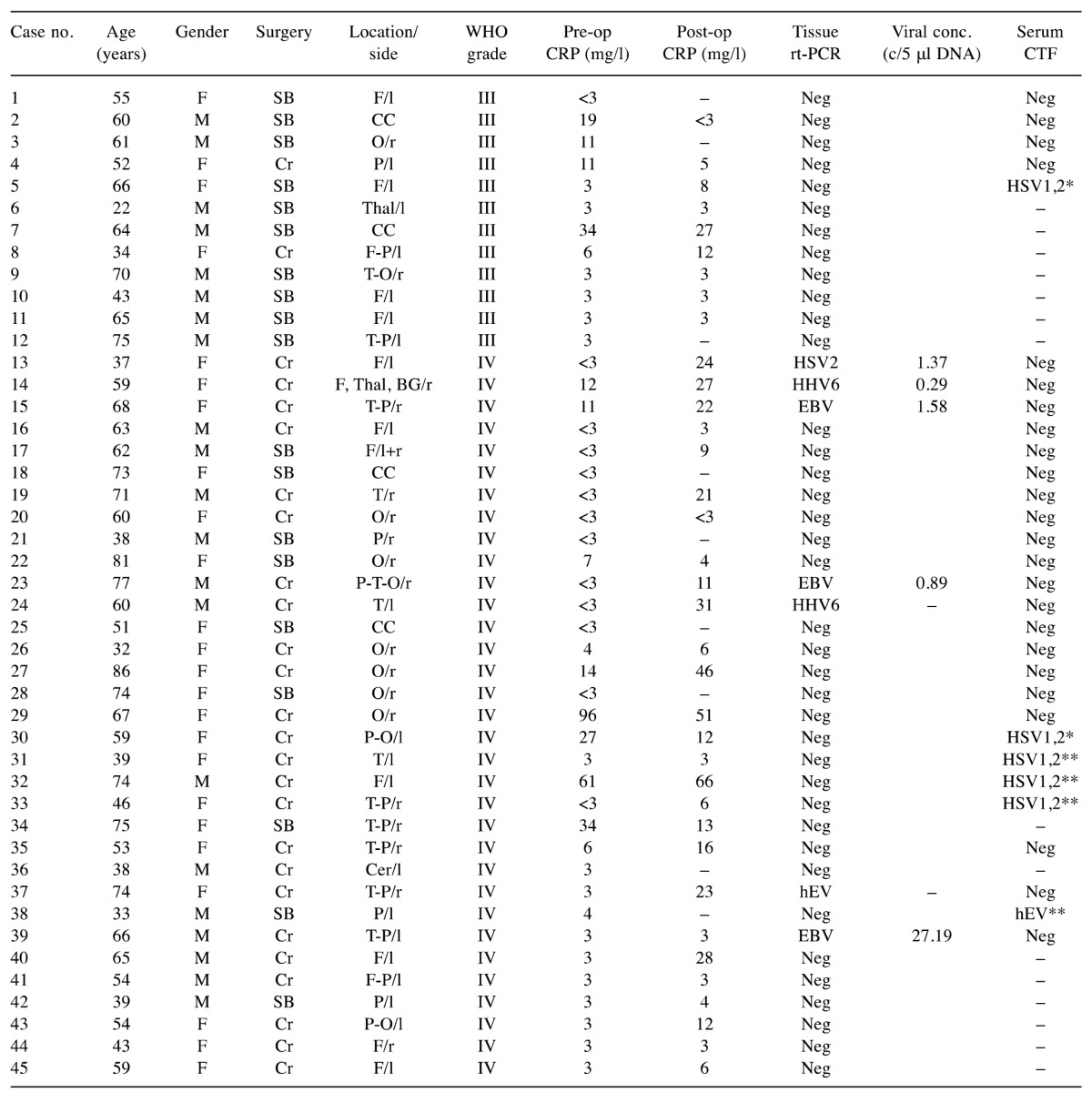
Conc. (c/5 μl DNA): number of herpes viral copies per 5 μl DNA; CTF: complement fixation test; F: female; M: male; Cr: craniotomy; SB: stereotactic biopsy; F: frontal lobe; T: temporal lobe; P: parietal lobe; O: occipital lobe; Thal: thalamus; BG: basal ganglia; CC: corpus callosum; Cer: cerebellum; l: left; r: right; neg: negative; –: no data; HSV-1: -2: herpes simplex virus-1: -2; HHV-6: human herpes virus-6; EBV: Epstein- Barr virus; hEV: human enteroviruses; CRP: C-reactive protein; *1:20 dilution, **1:40 dilution
Detection of viruses in tissue samples. To reveal the presence of viruses in the HGG tissues, the viral nucleic acids (DNA/RNA) were extracted directly from fresh resected samples, amplified and subjected to analysis using commercial real-time PCR kits or in-house assays. Positivity was confirmed by independent duplicate PCR runs, and the reactions were considered valid as the internal control was amplified, and hence no inhibitors were present in these DNA/RNA extracts. In addition, no amplification was noted for the negative controls, which excluded contamination. We selected nine distinct viruses with oncogenic or neurotropic potential (Table II). Overall, using real-time PCR, viral DNA/RNA was detected in only seven out of 45 tissue samples tested (Table I). Three species of herpes viruses were detected: HSV-2, EBV, and HHV-6, and one hEV. These viruses were detected only in the WHO IV samples. The presence of HSV-1, HCMV, and VZV was not confirmed in any of 45 samples tested. DNA extracts were also screened for the presence of six adenovirus subtypes, A, B, C, D, E, and G, and TBEV, but none of these viruses were detected in these samples from our cohort of patients.
Table II. Detection of viruses in glioma tissue samples, grouped according to the clinical characteristics of the patients (n=45).
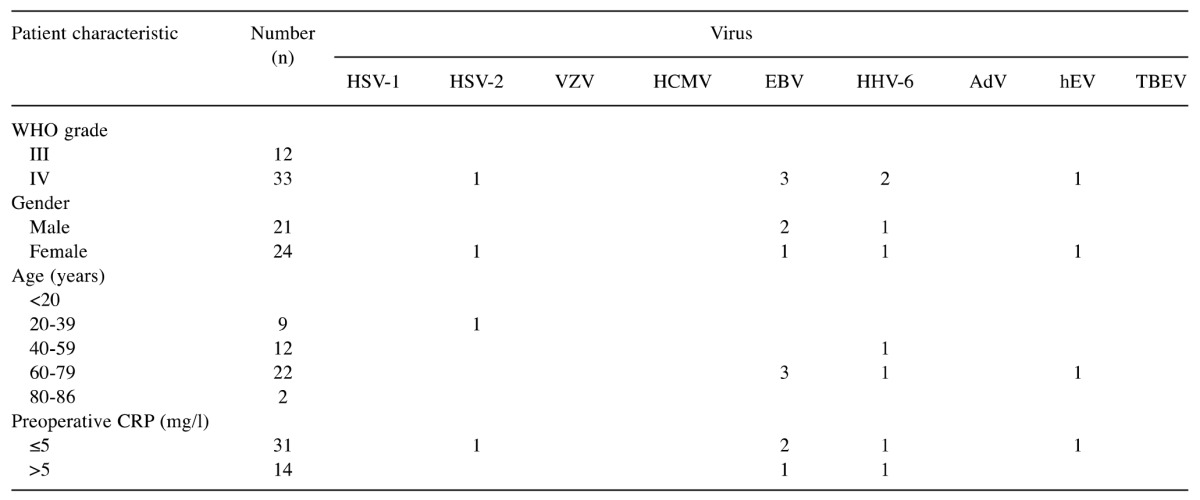
HSV-1, -2: Herpes simplex virus-1, -2; VZV: varicella zoster virus; HCMV: human cytomegalovirus; EBV: Epstein-Barr virus; HHV-6: human herpes virus-6; AdV: adenoviruses; hEV: human enteroviruses; TBEV: tick-borne encephalitis virus; CRP: C-reactive protein
Quantification of the amplified herpes viral DNA revealed low viral copy numbers per 5 μl DNA (Table I). Consequently, the nucleotide sequencing was successful only for three EBV-positive samples, while HSV-2, HHV-6, and hEV could not be sequenced. The 235-bp-long fragment of the EBV BamM region was identical across three samples sequenced, and provided a 100% match to the EBV strains under Accession numbers KT823509, APO15016, and others. The sequences were submitted to Gene Bank, and can be assessed as Accession numbers (in progress).
Analysis of peripheral blood. The preoperative CRP values in peripheral blood ranged from <3 mg/l to 96 mg/l, and they were low (≤5 mg/l) in 31 patients and high (>5 mg/l) in 14 patients. The high levels were detected in five out of 12 patients with WHO III grade tumour and nine out of 33 patients with WHO IV grade tumour (Figure 2). These CRP data show no significant differences between WHO grades III and IV.
Figure 2. Analyses of the peripheral blood in patients with glioma WHO grades III (a) and IV (b). The pre- and postoperative C-reactive protein(CRP) values were measured in serum by turbidimetry and were categorized as low (≤5 mg/l) or high (>5 mg/l). In 30 cases, the sera samples from patients were tested for the presence of specific antibodies against four viruses. Specific antibodies against herpes simplex virus -1, -2 (HSV-1, -2 and human enteroviruses (hEV) were detected by complement fixation test (CFT).
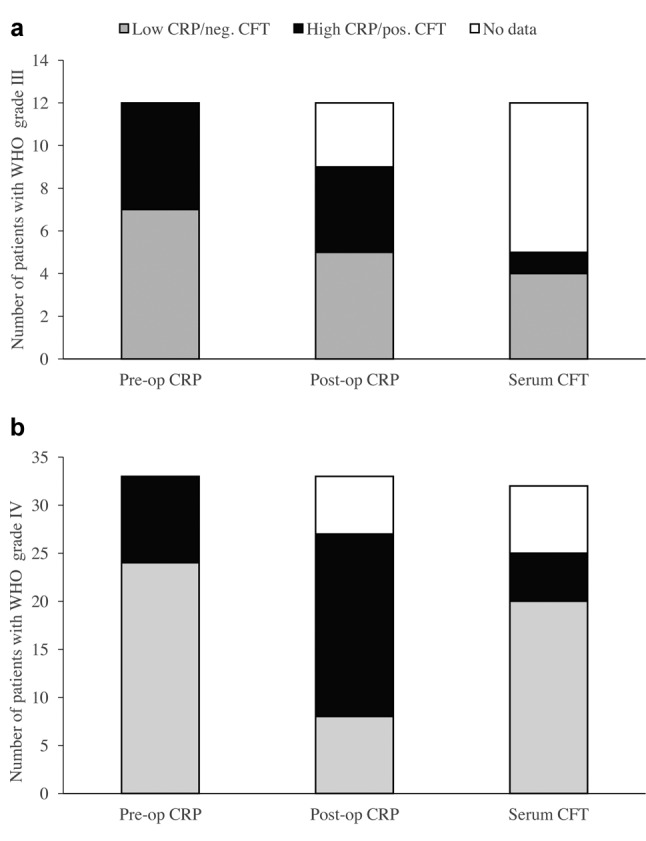
The presence of antibodies against viruses in peripheral blood of patients with HGG was determined by enzyme immunoassays and complement fixation tests in 30 samples (Figure 2). Five samples showed relatively low antibody titres (1:40) against HSV, and one sample had a low antibody titre (1:40) against hEV, as shown in Table I. Antibodies against TBEV were not detected in any of these samples tested.
Due to the relatively low frequency of positivity, statistical analyses of the viral infections in this group of patients were not performed, although we speculate on some of the trends, as discussed below.
Discussion
In their recent review, White et al. indicated at least seven viruses that have been causally linked to human oncogenesis, stating that viruses can be involved in both tumour initiation and tumour progression (9). In general, DNA viruses affect cells by binding of the viral proteins to tumour-suppressor genes, and they also produce dsRNA-binding proteins that can act as suppressors of RNA silencing, thereby favouring tumour development. Pro-oncogenic modulation is a typical characteristic of herpes viruses, for example with HSV-1, EBV, HCMV, and KSHV, and it is tumour-type specific (38).
Viruses can also change the host miRNA expression patterns, and make the infected cells more prone to oncogenic transformation. Previously, we identified 11 viral miRNAs that were differentially expressed in the plasma of patients with GBM, compared to those of healthy volunteers (39). These viral miRNAs induced signalling were mostly related to impaired immune responses in the progression of GBM. Six of these belonged to EBV miRNAs, and the rest were of HSV, KSHV, and HCMV origin. Therefore, in the present study, we analysed 45 patients with HGG for the presence of these and additional viral DNA of six herpes viruses (HSV-1, HSV-2, VZV, EBV, HCMV, HHV-6), human adenoviruses (A-G), hEV, and TBEV, which were measured in tumour tissues and plasma. In our cohort of patients with HGG, we only detected one HSV-2, three EBV, and two HHV-6 among the oncogenic viruses, and one hEV among the two neurotropic viruses.
As latent viral neurotropism can be seen with a variety of viruses and with widespread infection, i.e. there is seropositivity among the population, it has been difficult to establish any associations between viral infections and CNS malignancy based on epidemiology alone (9). The HSV-1, HSV-2, HCMV, and HHV-6 human herpes viruses are currently attracting much attention as possible factors in initiating human brain tumours (11). Wrensch et al. used serological IgG antibody binding using ELISA assays to demonstrate that about 80% (45/55) of their patients with GBM were seropositive for HSV-1 and HSV-2 (26). This is in line with the present study, where more GBM sera samples were positive for HSV-1 and HSV-2, but no viral RNA was detected in the tissues. In contrast, HSV-2 nucleic acids were detected in one patient with GBM where the serological study of the blood was negative. Taken together, it is not clear if there is a correlation between tissue and blood levels of HSV-1 and HSV-2 as a causative relation to possible GBM initiation.
Human herpes virus-6 is widespread in the population, and it is linked to persistent and most often asymptomatic infection in humans. The two variants, HHV-6A and HHV-6B, are recognized as two distinct viruses (40). HHV-6 PCR positivity has been reported in 45% of patients with GBM (14/31) (41), whereas Crawford et al. detected HHV-6 in 47% of patients from a large cohort of adult brain tumours using nested PCR (42), confirming their previous data in a series of paediatric primary brain tumours (43). Such relatively high frequencies of HHV-6A and HHV-6B in patients with HGG was not reflected in the present study, with the detection of only two positive cases out of 45 patients. This discrepancy cannot be explained by a low sensitivity of our detection method for this virus, as the real-time PCR used in the present study and the nested PCR used in the previous reports (42,43) have comparable sensitivities. However, nested PCR is a method that is prone to higher rates of contamination when compared to real-time PCR (44). Recently, the potential roles of both HHV-6A and HHV-6B in neuroinflammatory pathologies were reported, and the mechanisms that might explain virus-induced neuropathogenesis were reviewed by Reynaud et al., who indicated the immunomodulatory role of viral infections potentiating their oncogenesis in GBM (45).
Viruses evade host immune responses and a variety of these strategies have been discussed elsewhere (9). In a similar manner, tumours overcome immune surveillance during their malignant progression, as has also been demonstrated in the case of EBV infection, which is present in about 90% of the world population. EBV has been associated with selected human tumours, including Burkitt’s lymphoma and Hodgkin’s lymphoma, and less frequently for some cases of gastric carcinoma and nasopharyngeal carcinoma, depending on the geographic and ethnic distribution of the populations (10,29). In glioma, EBV was detected in about 30% of samples (9/35) obtained from patients with pilocytic astrocytoma (45). Recently, Fonseca et al. studied 75 fresh frozen glioma tissues of different histological subtypes using conventional PCR, and they reported 14.7% prevalence of EBV DNA in WHO III (2/11) and WHO IV (1/11) gliomas (29). Similarly, in the present study, we found EBV DNA in three out of 45 tissues, but not in the corresponding sera, possibly due to the very low amounts of EBV. In general, low levels of viral RNA in serum would indicate chronic infection, and in line with this, we found numerous viral miRNAs in the sera of 16 patients with GBM (39). Presumably in tissues, miRNAs allow for viral persistence and appear to inhibit cell-mediated immunity, apoptosis, and the cell cycle, favouring the latency of infection.
HCMV is relatively widespread in immunocompromised patients (11). The clinical relevance of HCMV infection in GBM pathogenesis has been investigated since 2002 (16,25), but there has been no evidence to date for a role of HCMV in gliomagenesis. Our observations agree with this, as no HCMV DNA sequences were found in these glioma tissues.
Adenovirus variants have been detected by PCR in different tumours, including GBM and some normal brain samples (31). In the present study, none of the six adenovirus subtypes was detected in the HGG tissues by PCR. The role of adenovirus infection in primary CNS tumours thus remains unclear.
The most interesting study of the role of VZV in brain malignancy comes from epidemiological studies, which have revealed possible inverse associations between VZV infection and glioma risk (26,27). Moreover, VZV DNA was detected in two out of 35 pilocytic astrocytoma by Neves et al. (46). None of the samples in the present study were positive for VZV DNA.
Human enterovirus has not been investigated to date as far as we are aware. We detected hEV nucleic acids in one GBM sample, but not in this plasma of this patient. In contrast, hEV was found in another GBM blood sample with low preoperative and postoperative CRP levels. However, the presence of hEV in HGG deserves further investigation, as persistent hEV infections are most likely the cause of meningoencephalitis (47). Another neurotropic virus without known oncogenic potential is TBEV, which causes encephalitis (48), but was not detected in any sample and is therefore possibly not related to gliomagenesis.
In conclusion, to our knowledge this is the first study on viral nucleic acids in glioma specimens linked also to their presence in the blood of the same patients, in a population of the Slovenian region. As nine distinct types of viruses were tested simultaneously this allowed for more direct comparisons. This, and our previous study on miRNA, showed presence of RNA/DNA, miRNAs, and protein of certain common viruses, namely HSV, EBV and HCMV, in the plasma of patients with GBM. Significantly higher levels of HSV-1 and HSV-2 in sera versus other tested viruses were found, but HSV did not infect the tumour tissue of the same patients. Furthermore, as we detected HSV-2 and hEV in the tissue, but not in the plasma of these patients, it appears that there is no correlation between the plasma and tissue levels. Possibly both are associated with glioma progression, as we only found viruses in GBM, not in the lower stages of glioma. Finally, low titres of viruses in the blood do not indicate an acute infection, but rather chronic viral virulence.
Acknowledgements
The Authors acknowledge Assistant Professor Dr. Rajko Kavalar, Head, Department of Pathology, University Clinical Centre Maribor, Slovenia, and his group for performing the histopathological diagnoses. They are also grateful to the Head of the Institute of Microbiology and Immunology at the Faculty of Medicine of the University of Ljubljana, Prof. Dr. Miroslav Petrovec for serological analyses, and Tomaž Šmigoc, MD, for collecting the patient data. This study was supported by the Research Agency of the Republic of Slovenia, Grant Number P3 - 0327 (T.S.).
References
- 1.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19:764–772. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Brada M, van den Bent MJ, Tonn JC, Pentheroudakis G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(suppl 3):iii93–iii101. doi: 10.1093/annonc/mdu050. [DOI] [PubMed] [Google Scholar]
- 3.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 4.Bondy ML, Scheurer ME, Malmer B, Barnholtz-Sloan JS, Davis FG, Il’yasova D, Kruchko C, McCarthy BJ, Rajaraman P, Schwartzbaum JA, Sadetzki S, Schlehofer B, Tihan T, Wiemels JL, Wrensch M, Buffler PA; Brain Tumour Epidemiology Consortium. Brain Tumour Epidemiology: Consensus from the Brain Tumour Epidemiology Consortium (BTEC) Cancer. 2008;113:1953–1968. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reuss D, von Deimling A. Hereditary tumour syndromes and gliomas. Recent Resu Cancer Res. 2009;171:83–102. doi: 10.1007/978-3-540-31206-2_5. [DOI] [PubMed] [Google Scholar]
- 6.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lönn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, LaChance DH, McCoy L, O’Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alibek K, Kakpenova A, Baiken Y. Role of infectious agents in the carcinogenesis of brain and head and neck cancers. Infect Agent Cancer. 2013;8:7. doi: 10.1186/1750-9378-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White MK, Pagano JS, Khalili K. Viruses and human cancers: a long road of discovery of molecular paradigms. Clin Microbiol Rev. 2014;27:463–481. doi: 10.1128/CMR.00124-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saddawi-Konefka R, Crawford JR. Chronic viral infection and primary central nervous system malignancy. J Neuroimmune Pharmacol. 2010;5:387–403. doi: 10.1007/s11481-010-9204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kofman A, Marcinkiewicz L, Dupart E, Lyshchev A, Martynov B, Ryndin A, Kotelevskaya E, Brown J, Schiff D, Abounader R. The roles of viruses in brain tumour initiation and oncomodulation. J Neuro-Oncol. 2011;105:451–466. doi: 10.1007/s11060-011-0658-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Valle L, Gordon J, Assimakopoulou M, Enam S, Geddes JF, Varakis JN, Katsetos CD, Croul S, Khalili K. Detection of JC virus DNA sequences and expression of the viral regulatory protein T-antigen in tumours of the central nervous system. Cancer Res. 2001;61:4287–4293. [PubMed] [Google Scholar]
- 13.Piña-Oviedo S, De León-Bojorge B, Cuesta-Mejías T, White MK, Ortiz-Hidalgo C, Khalili K, Del Valle L. Glioblastoma multiforme with small cell neuronal-like component: association with human neurotropic JC virus. Acta Neuropathol. 2006;111:388–396. doi: 10.1007/s00401-006-0050-3. [DOI] [PubMed] [Google Scholar]
- 14.Mazzoni E, Gerosa M, Lupidi F, Corallini A, Taronna AP, D’Agostino A, Bovenzi M, Ruggeri G, Casali F, Rotondo JC, Rezza G, Barbanti-Brodano G, Tognon M, Martini F. Significant prevalence of antibodies reacting with simian virus 40 mimotopes in sera from patients affected by glioblastoma multiforme. Neuro-Oncol. 2014;16:513–519. doi: 10.1093/neuonc/not217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rollison DE, Utaipat U, Ryschkewitsch C, Hou J, Goldthwaite P, Daniel R, Helzlsouer KJ, Burger PC, Shah KV, Major EO. Investigation of human brain tumours for the presence of polyomavirus genome sequences by two independent laboratories. Int J Cancer. 2005;113:769–774. doi: 10.1002/ijc.20641. [DOI] [PubMed] [Google Scholar]
- 16.Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, Nabors LB, Cobbs CG, Britt WJ. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. [PubMed] [Google Scholar]
- 17.Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, McGregor Dallas SR, Smit M, Soroceanu L, Cobbs CS; HCMV and Gliomas Symposium. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro-Oncol. 2012;14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poltermann S, Schlehofer B, Steindorf K, Schnitzler P, Geletneky K, Schlehofer JR. Lack of association of herpesviruses with brain tumours. J Neurovirol. 2006;12:90–99. doi: 10.1080/13550280600654573. [DOI] [PubMed] [Google Scholar]
- 19.Cimino PJ, Zhao G, Wang D, Sehn JK, Lewis JS Jr., Duncavage EJ. Detection of viral pathogens in high-grade gliomas from unmapped next-generation sequencing data. Exp Mol Pathol. 2014;96:310–315. doi: 10.1016/j.yexmp.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Yamashita Y, Ito Y, Isomura H, Takemura N, Okamoto A, Motomura K, Tsujiuchi T, Natsume A, Wakabayashi T, Toyokuni S, Tsurumi T. Lack of presence of the human cytomegalovirus in human glioblastoma. Mod Pathol. 2014;27:922–929. doi: 10.1038/modpathol.2013.219. [DOI] [PubMed] [Google Scholar]
- 21.Hashida Y, Taniguchi A, Yawata T, Hosokawa S, Murakami M, Hiroi M, Ueba T, Daibata M. Prevalence of human cytomegalovirus, polyomaviruses, and oncogenic viruses in glioblastoma among Japanese subjects. Infect Agent Cancer. 2015;10:3. doi: 10.1186/1750-9378-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priel E, Wohl A, Teperberg M, Nass D, Cohen ZR. Human cytomegalovirus viral load in tumour and peripheral blood samples of patients with malignant gliomas. J Clin Neurosci. 2015;22:326–330. doi: 10.1016/j.jocn.2014.06.099. [DOI] [PubMed] [Google Scholar]
- 23.Baumgarten P, Michaelis M, Rothweiler F, Starzetz T, Rabenau HF, Berger A, Jennewein L, Braczynski AK, Franz K, Seifert V, Steinbach JP, Allwinn R, Mittelbronn M, Cinatl J Jr. Human cytomegalovirus infection in tumour cells of the nervous system is not detectable with standardized pathologico-virological diagnostics. Neuro-Oncol. 2014;16:1469–1477. doi: 10.1093/neuonc/nou167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehrer S, Green S, Ramanathan L, Rosenzweig K, Labombardi V. No consistent relationship of glioblastoma incidence and cytomegalovirus seropositivity in Whites, Blacks, and Hispanics. Anticancer Res. 2012;32:1113–1115. [PubMed] [Google Scholar]
- 25.Solomon IH, Ramkissoon SH, Milner DA Jr, Folkerth RD. Cytomegalovirus and glioblastoma: a review of evidence for their association and indications for testing and treatment. J Neuropathol Exp Neurol. 2014;73:994–998. doi: 10.1097/NEN.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 26.Wrensch M, Weinberg A, Wiencke J, Miike R, Barger G, Kelsey K. Prevalence of antibodies to four herpesviruses among adults with glioma and controls. Am J Epidemiol. 2001;154:161–165. doi: 10.1093/aje/154.2.161. [DOI] [PubMed] [Google Scholar]
- 27.Pundole X, Amirian ES, Scheurer ME. Role of varicella zoster virus in glioma risk: current knowledge and future directions. OA Epidemiology. 2014;2:6. [Google Scholar]
- 28.Chi J, Gu B, Zhang C, Peng G, Zhou F, Chen Y, Zhang G, Guo Y, Guo D, Qin J, Wang J, Li L, Wang F, Liu G, Xie F, Feng D, Zhou H, Huang X, Lu S, Liu Y, Hu W, Yao K. Human herpesvirus 6 latent infection in patients with glioma. J Infect Dis. 2012;206:1394–1398. doi: 10.1093/infdis/jis513. [DOI] [PubMed] [Google Scholar]
- 29.Fonseca RF, Rosas SL, Oliveira JA, Teixeira A, Alves G, Carvalho Mda G. Frequency of Epstein-Barr virus DNA sequences in human gliomas. Sao Paulo Med J. 2015;133:51–54. doi: 10.1590/1516-3180.2013.1912814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosulin K, Haberler C, Hainfellner JA, Amann G, Lang S, Lion T. Investigation of adenovirus occurrence in pediatric tumour entities. J Virol. 2007;81:7629–7635. doi: 10.1128/JVI.00355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogovic P, Strle F. Tick-borne encephalitis: a review of epidemiology, clinical characteristics, and management. World J Clin Cases. 2015;3:430–441. doi: 10.12998/wjcc.v3.i5.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palus M, Bílý T, Elsterová J, Langhansová H, Salát J, Vancová M, Růžek D. Infection and injury of human astrocytes by tick-borne encephalitis virus. J Gen Virol. 2014;95:2411–2426. doi: 10.1099/vir.0.068411-0. [DOI] [PubMed] [Google Scholar]
- 34.Kleihues P, Burger PC, Scheithauer BW. The new WHO classification of brain tumours. Brain Pathol. 1993;3:255–268. doi: 10.1111/j.1750-3639.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 35.Schwaiger M, Cassinotti P. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J Clin Virol. 2003;27:136–145. doi: 10.1016/s1386-6532(02)00168-3. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Chan RK, Tan SH, Ng PP. Detection and genotyping of human herpes simplex viruses in cutaneous lesions of erythema multiforme by nested PCR. J Med Virol. 2003;71:423–428. doi: 10.1002/jmv.10502. [DOI] [PubMed] [Google Scholar]
- 37.Achour A, Malet I, Le Gal F, Dehée A, Gautheret-Dejean A, Bonnafous P, Agut H. Variability of gB and gH genes of human herpesvirus-6 among clinical specimens. J Med Virol. 2008;80:1211–1221. doi: 10.1002/jmv.21205. [DOI] [PubMed] [Google Scholar]
- 38.Pfeffer S, Voinnet O. Viruses, microRNAs and cancer. Oncogene. 2006;25:6211–6219. doi: 10.1038/sj.onc.1209915. [DOI] [PubMed] [Google Scholar]
- 39.Herman A, Gruden K, Blejec A, Podpečan V, Motaln H, Rožman P, Hren M, Zupančič K, Veber M, Verbovšek U, Lah Turnšek T, Porčnik A, Koršič M, Knežević M, Jeras M. Analysis of glioblastoma patients’ plasma revealed the presence of microRNAs with a prognostic impact on survival and those of viral origin. PLoS ONE. 2015;10:e0125791. doi: 10.1371/journal.pone.0125791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuomo L, Trivedi P, Cardillo MR, Gagliardi FM, Vecchione A, Caruso R, Calogero A, Frati L, Faggioni A, Ragona G. Human herpesvirus 6 infection in neoplastic and normal brain tissue. J Med Virol. 2001;63:45–51. [PubMed] [Google Scholar]
- 42.Crawford JR, Santi MR, Cornelison R, Sallinen SL, Haapasalo H, MacDonald TJ. Detection of human herpesvirus-6 in adult central nervous system tumours: predominance of early and late viral antigens in glial tumours. J Neurooncol. 2009;95:49–60. doi: 10.1007/s11060-009-9908-2. [DOI] [PubMed] [Google Scholar]
- 43.Crawford JR, Santi MR, Thorarinsdottir HK, Cornelison R, Rushing EJ, Zhang H, Yao K, Jacobson S, MacDonald TJ. Detection of human herpesvirus-6 variants in pediatric brain tumours: association of viral antigen in low grade gliomas. J Clin Virol. 2009;46:37–42. doi: 10.1016/j.jcv.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weidmann M, Meyer-König U, Hufert FT. Rapid detection of herpes simplex virus and varicella-zoster virus infections by real-time PCR. J Clin Microbiol. 2003;41:1565–1568. doi: 10.1128/JCM.41.4.1565-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynaud JM, Horvat B, Hufert FT. Human herpesvirus 6 and neuroinflammation. ISRN Virology. 2013. Available from: https://www.medtext.com/hdcn.htm.
- 46.Neves AM, Thompson G, Carvalheira J, Trindade JC, Rueff J, Caetano JM, Casey JW, Hermouet S. Detection and quantitative analysis of human herpesvirus in pilocytic astrocytoma. Brain Res. 2008;1221:108–114. doi: 10.1016/j.brainres.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Rhoades RE, Tabor-Godwin JM, Tsueng G, Feuer R. Enterovirus infections of the central nervous system. Virology. 2011;411:288–305. doi: 10.1016/j.virol.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindquist L, Vapalahti O. Tick-borne encephalitis. Lancet. 2008;371:1861–1871. doi: 10.1016/S0140-6736(08)60800-4. [DOI] [PubMed] [Google Scholar]