Abstract
Long noncoding RNA activated by transforming growth factor-β (lncRNA-ATB) is a novel lncRNA, which is recently reported to have critical roles in carcinogenesis and progression of several cancers. However, the expression, clinical values, biological roles, and underlying molecular mechanisms of lncRNA-ATB in osteosarcoma are still known. In this study, we measured lncRNA-ATB expression in serum and osteosarcoma tissues of osteosarcoma patients, analyzed its diagnostic and prognostic values. Serum lncRNA-ATB is increased in osteosarcoma patients and could accurately discriminate osteosarcoma patients from healthy controls. LncRNA-ATB is also upregulated in osteosarcoma tissues and cell lines, and positively associated with Enneking stage, metastasis and recurrence. Increased lncRNA-ATB level indicates poor recurrence-free survival and overall survival. Functional experiments demonstrated that overexpression of lncRNA-ATB enhances osteosarcoma cells proliferation, migration, and invasion, and while depletion of lncRNA-ATB inhibits osteosarcoma cells proliferation, migration, and invasion. Mechanistically, we found that lncRNA-ATB inhibits miR-200s, and upregulates miR-200s target genes ZEB1 and ZEB2. Additionally, the roles of lncRNA-ATB on osteosarcoma cells proliferation, migration, and invasion in vitro, and osteosarcoma tumor growth in vivo are dependent on the regulation of miR-200s. Taken together, this study suggests that lncRNA-ATB may be a potential diagnostic and prognostic biomarker and a therapeutic target for osteosarcoma.
Keywords: Long noncoding RNA, ATB, osteosarcoma, proliferation, migration, invasion, miR-200s
Introduction
Osteosarcoma is one of the most common aggressive malignant bone tumor mainly affecting children and adolescents [1,2]. Although great improvements have been made in the treatment strategies, including surgical resection, neoadjuvant chemotherapy, and radiotherapy, the overall survival of osteosarcoma patients remains dismal, especially for those with advanced clinical stages at diagnosis [3,4]. Therefore, it is urgent to uncover the molecular mechanisms underlying osteosarcoma tumorigenesis and progression, identify new molecular biomarkers for early diagnosis, and develop novel therapeutic strategies for osteosarcoma [5-7].
With great progressions of genome and transcriptome sequencing technologies, many novel non-protein-coding transcripts were identified. About 80% of the genome is transcribed into transcripts, but only 2% of the genome codes for protein [8,9]. Among these numerous non-protein-coding transcripts, long noncoding RNA (lncRNA) is a novel class of non-protein-coding transcript greater than 200 nucleotides in length, with important regulatory roles in variety of pathophysiological processes [10-13]. Accumulating evidences revealed that many lncRNAs are dysregulated in multiple cancers, associated with patients’ prognosis, and can regulate tumor cells proliferation, cell cycle, apoptosis, drug resistance, migration, invasion, stem-like properties, et al [14-19]. The molecular mechanisms of lncRNAs in exerting their biological roles are complex and various, including binding to protein, RNA, or DNA, transcriptionally or post-transcriptionally regulating the expression of critical oncogenes or tumor suppressors [20-23]. Furthermore, several lncRNAs are detectable in peripheral blood and could be recognized as diagnostic biomarker for several cancers, such as PVT1 for cervical cancer, HIF1A-AS1 for non-small cell lung cancer [24,25]. However, the clinical values and biological roles of lncRNAs in osteosarcoma are still largely unknown.
LncRNA activated by transforming growth factor-β (lncRNA-ATB) has recently been reported to be upregulated, associated with poor prognosis, and promote tumor progression of hepatocellular carcinoma, colon cancer, and prostate carcinoma [14,26,27]. However, the roles and clinical significances of lncRNA-ATB in osteosarcoma are still unclear.
In this study, we measured serum lncRNA-ATB levels in osteosarcoma patients and healthy controls, analyzed its diagnostic values for osteosarcoma. Furthermore, lncRNA-ATB expression levels in osteosarcoma tissues and its association with clinicopathological features and prognosis were analyzed. Gain-of-function and loss-of-function experiments were performed to investigate the biological roles of lncRNA-ATB in osteosarcoma. The molecular mechanisms through which lncRNA-ATB exerts its biological roles were also explored.
Materials and methods
Serum and tissues samples
A total of 60 osteosarcoma patients and 60 age and sex matched healthy controls were recruited into this study. Peripheral blood was collected from these fasting participants before they received any treatment. Then the blood was centrifuged at 3000 rpm for 15 min at 4°C, and the supernatant was collected and stored at -80°C until use. Fresh osteosarcoma tissues and paired adjacent normal tissues were obtained from these 60 osteosarcoma patients at Zhumadian Central Hospital (Zhumadian, Henan, China). All the tissues were confirmed by pathological examination. This study was reviewed and approved by the Ethics Committee of Zhumadian Central Hospital. All participants signed written informed consent before inclusion into this study.
Cell culture
The human normal osteoblast cell line hFOB 1.19 and osteosarcoma cell lines MG63, U2OS, SAOS2, and HOS were obtained from American Type Culture Collection (ATCC) and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) in a humidified 5% CO2 incubator at 37°C.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from serum, tissues, and culturing cells using the TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using the M-MLV Reverse Transcriptase (Invitrogen) following the manufacturer’s instructions. QRT-PCR was carried out using SYBR® Premix Ex TaqTM II (Takara, Dalian, China) on ABI Prism 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. Β-actin was used as an endogenous control for lncRNA-ATB and mRNAs. The genes specific primers are as follows: For lncRNA-ATB, 5’-ACACAGAATAAAATAACAC-3’ (reverse transcription), 5’-TCTGGCTGAGGCTGGTTGAC-3’ (sense) and 5’-ATCTCTGGGTGCTGGTGAAGG-3’ (anti-sense); For ZEB1, 5’-ACTCTGATTCTACACCGC-3’ (sense) and 5’-TGTCACATTGATAGGGCTT-3’ (anti-sense); For ZEB2, 5’-TGAGGATGACGGTATTGC-3’ (sense) and 5’-ATCTCGTTGTTGTGCCAG-3’ (anti-sense); And for β-actin, 5’-GGGAAATCGTGCGTGACATTAAG-3’ (sense) and 5’-TGTGTTGGCGTACAGGTCTTTG-3’ (anti-sense). For miRNAs analysis, qRT-PCR was performed as above, using TaqMan microRNA assays (Applied Biosystems) following the manufacturer’s instructions. U6 was used as an endogenous control for miRNAs. The relative expression of RNAs was calculated using the comparative Ct method.
Vectors construction and transfection
LncRNA-ATB overexpression plasmid (pcDNA3.1-ATB), miR-200s targeting sites mutated lncRNA-ATB overexpression plasmid (pcDNA3.1-ATB-Mut), luciferase reporter containing ZEB1 3’UTR (pmirGLO-ZEB1), and luciferase reporter containing ZEB2 3’UTR (pmirGLO-ZEB2) were constructed as previously described [14]. Two independent shRNAs specifically targeting lncRNA-ATB were designed, synthesized, and inserted into the SuperSilencingTM shRNA expression vector pGPH1/Neo (GenePharma, Shanghai, China). The sh-ATB-1 target sequence was: 5’-CCTTATGGCCTAGATTACCTTTCCA-3’ and sh-ATB-2 target sequence was: 5’-CCTGTCTGTATTTGCGAATACCTTT-3’. Scrambled shRNA was used as negative control. MiR-200s mimics (the mixtures of the five members of the miR-200 family, including miR-200a mimics, miR-200b mimics, miR-200c mimics, miR-141 mimics, and miR-429 mimics), miR-200s inhibitors (the mixtures of the five members of the miR-200 family, including miR-200a inhibitors, miR-200b inhibitors, miR-200c inhibitors, miR-141 inhibitors, and miR-429 inhibitors), and their respective negative control were obtained from GenePharma. Transfection was carried out using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Stable cell lines construction
To construct lncRNA-ATB and lncRNA-ATB-Mut stably overexpressed osteosarcoma cells, pcDNA3.1-ATB, pcDNA3.1-ATB-Mut and pcDNA3.1 was transfected into MG63 cells, and selected with neomycin (800 µg/ml) for four weeks. To construct lncRNA-ATB stably depleted osteosarcoma cells, the lncRNA-ATB specific shRNAs sh-ATB-1 and sh-ATB-2, and control shRNAs were transfected into U2OS cells and selected with neomycin (800 µg/ml) for four weeks.
Cell proliferation assays
Cell proliferations were assessed by Cell Counting Kit-8 (CCK-8) assays and Ethynyl deoxyuridine (EdU) incorporation assays. For CCK-8 assays, equal number of indicated osteosarcoma cells was plated in 96-well plate. The absorbance at 450 nm was measured using Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) every 24 hours according to the manufacturer’s protocols. EdU incorporation assays were performed using an EdU Kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions.
Cell migration and invasion assays
For migration assays, indicated osteosarcoma cells suspended in FBS free medium with 1 μg/ml Mitomycin C were plated in the upper well of a 24-well poly-carbonate transwell insert (Millipore, Bedford, MA, USA). DMEM supplemented with 10% FBS was added to the lower well. After incubation for 48 hours, cells on the upper surface of inserts were scraped off, and cells on the lower surface were fixed, stained and counted. Cell invasion assays were performed using the Cell Invasion Assay Kit (ECM550, Millipore) following the manufacturer’s protocols.
Luciferase reporter assays
pmirGLO, pmirGLO-ZEB1 or pmirGLO-ZEB2 was cotransfected with miR-200s mimics into lncRNA-ATB stably overexpressed and control MG63 cells. PmirGLO, pmirGLO-ZEB1 or pmirGLO-ZEB2 was cotransfected with miR-200s inhibitors into lncRNA-ATB stably depleted and control U2OS cells. The co-transfection was performed using Lipofectamine 3000. The relative Firefly luciferase activity was normalized to Renilla luciferase activity 48 hours after transfection.
Western blotting
Proteins were retrieved from indicated osteosarcoma cells and equal amounts of proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to 0.22 μm nitrocellulose membrane (Millipore), and blocked with 5% bovine serum albumin. Then the membrane was incubated with ZEB1 (Abcam, Hong Kong, China), ZEB2 (Abcam), or β-actin (Abcam) specific primary antibodies. After being washed by TBST for three times, the membrane was incubated with IRdye 800-conjugated goat anti-rabbit IgG or IRdye 700-conjugated goat anti-mouse IgG, and detected using an Odyssey infrared scanner (Li-Cor Biosciences, Lincoln, NE, USA).
Animal experiments
2×106 lncRNA-ATB or lncRNA-ATB-Mut stably overexpressed or control MG63 cells were subcutaneously injected into athymic BALB/c nude mice. Subcutaneously tumors growth was measured weekly with a caliper for 28 days, and tumor volume was calculated as 1/2 a×b2 (a, long axes; b, short axes). The animal studies were approved by the Ethics Committee of Zhumadian Central Hospital.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism Software. For comparison, Mann-Whitney U test, ROC curve analysis, Wilcoxon signed-rank test, Student’s t test, Pearson chi-square test, Log-rank test, or Pearson correlation analysis were performed as indicated. P<0.05 was considered as statistically significant.
Results
Serum lncRNA-ATB is increased in osteosarcoma patients and may be a potential diagnostic biomarker for osteosarcoma
To investigate the clinical values of lncRNA-ATB in osteosarcoma, we measured lncRNA-ATB expression in the serum of 60 osteosarcoma patients and 60 age and sex matched healthy controls using qRT-PCR. The result showed that serum lncRNA-ATB expression is significantly increased in osteosarcoma patients compared with that in healthy controls (Figure 1A). To investigate the diagnostic power of serum lncRNA-ATB for osteosarcoma, Receiver Operating Characteristic (ROC) curve analysis was carried out. Compared with healthy controls, the area under the curve (AUC) was 0.9236 (95% CI: 0.8756-0.9716), with 83.33% sensitivity and 90% specificity for osteosarcoma (Figure 1B). This result demonstrated that serum lncRNA-ATB can accurately discriminate osteosarcoma patients from healthy controls and may be a potential novel non-invasive biomarker for osteosarcoma.
Figure 1.
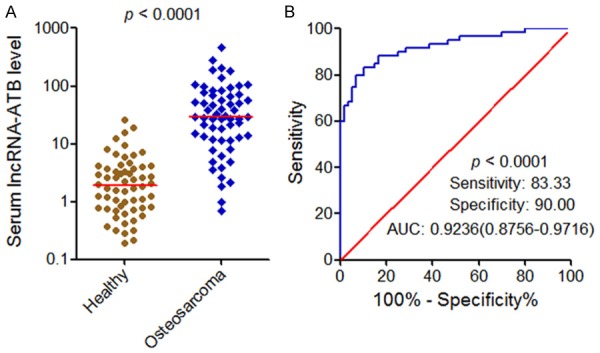
Serum lncRNA-ATB expression levels and its diagnostic values for osteosarcoma. A. Serum lncRNA-ATB expression levels in 60 osteosarcoma patients and 60 healthy controls. P<0.0001 by Mann-Whitney U test. B. ROC curve analysis for serum lncRNA-ATB in discriminating osteosarcoma patients from healthy controls. P<0.0001.
LncRNA-ATB is upregulated in osteosarcoma tissues and associated with poor prognosis of osteosarcoma patients
To further investigate the clinical values of lncRNA-ATB in osteosarcoma, we measured lncRNA-ATB expression in 60 pairs of osteosarcoma tissues and adjacent normal tissues using qRT-PCR. The result showed that lncRNA-ATB is significantly upregulated in osteosarcoma tissues compared with adjacent normal tissues (Figure 2A). LncRNA-ATB expression level was also measured in human normal osteoblast cell line hFOB 1.19 and osteosarcoma cell lines MG63, U2OS, SAOS2, and HOS. The result showed that lncRNA-ATB expression level is much higher in osteosarcoma cells than that in normal osteoblast cell (Figure 2B). Next, we analyzed the correlation between lncRNA-ATB expression level and clinicopathological features in these osteosarcoma patients. As shown in Table 1, increased expression of lncRNA-ATB is correlated with advanced Enneking stage, metastasis and recurrence. LncRNA-ATB expression level is significantly higher in osteosarcoma tissues with metastasis than that without metastasis (Figure 2C). Kaplan-Meier survival analysis was performed to evaluate the prognostic values of lncRNA-ATB for osteosarcoma patients. As shown in Figure 2D and 2E, osteosarcoma patients with higher lncRNA-ATB levels had a worse recurrence-free survival and overall survival. These data indicated that lncRNA-ATB is upregulated in osteosarcoma, correlated with the progression of osteosarcoma, and may be a potential prognostic biomarker for osteosarcoma.
Figure 2.
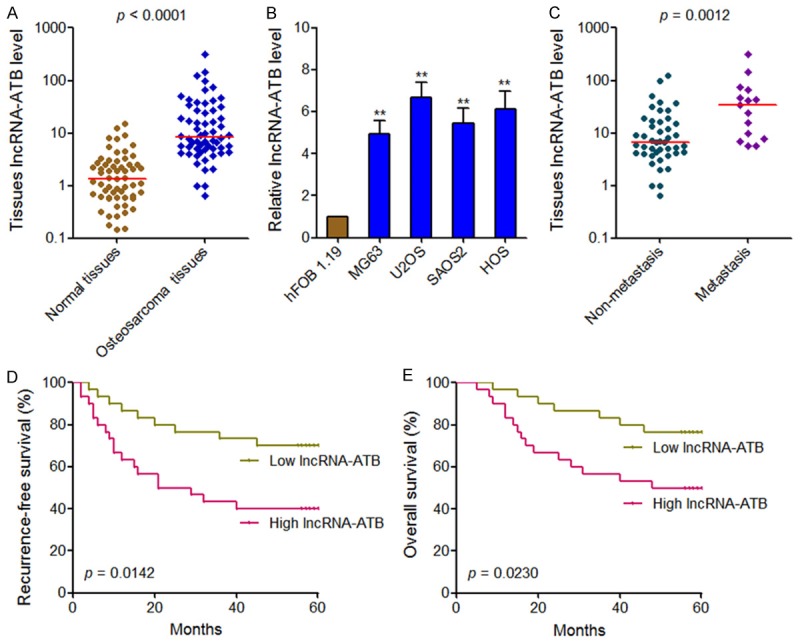
LncRNA-ATB expression levels in osteosarcoma and its association with osteosarcoma patients’ prognosis. (A) lncRNA-ATB expression levels in 60 pairs of osteosarcoma tissues and adjacent normal tissues. P<0.0001 by Wilcoxon signed-rank test. (B) lncRNA-ATB expression levels in human normal osteoblast cell line hFOB 1.19 and osteosarcoma cell lines MG63, U2OS, SAOS2, and HOS. Results are present as mean ± SD. **P<0.01 by Student’s t test. (C) lncRNA-ATB expression levels in osteosarcoma tissues with or without metastasis. P=0.0012 by Mann-Whitney U test. (D, E) Kaplan-Meier analyses of the association between lncRNA-ATB expression level and recurrence-free survival (P=0.0142, log-rank test) (D) or overall survival (P=0.0230, log-rank test) (E).
Table 1.
Correlation between lncRNA-ATB expression level and clinicopathological features in osteosarcoma patients (P-value was acquired by Pearson chi-square test)
| Parameters | lncRNA-ATB expression | P-value | |
|---|---|---|---|
|
| |||
| Low (n=30) | High (n=30) | ||
| Sex | 0.605 | ||
| Male | 13 | 15 | |
| Female | 17 | 15 | |
| Age | 0.795 | ||
| <18 | 17 | 16 | |
| ≥18 | 13 | 14 | |
| Tumor Site | 0.542 | ||
| Femur/Tibia | 22 | 24 | |
| Elsewhere | 8 | 6 | |
| Enneking stage | 0.017 | ||
| I | 16 | 6 | |
| II | 10 | 13 | |
| III | 4 | 11 | |
| Metastasis | 0.037 | ||
| Yes | 4 | 11 | |
| No | 26 | 19 | |
| Recurrence | 0.020 | ||
| Yes | 9 | 18 | |
| No | 21 | 12 | |
Overexpression of lncRNA-ATB enhances osteosarcoma cell proliferation, migration, and invasion
To investigate the biological roles of lncRNA-ATB in osteosarcoma, we stably overexpressed lncRNA-ATB in MG63 cells by transfecting lncRNA-ATB overexpression plasmid (Figure 3A). CCK-8 assays revealed that ectopic expression of lncRNA-ATB significantly enhanced MG63 cell proliferation (Figure 3B). EdU incorporation assays further verified the proliferation promoting roles of lncRNA-ATB (Figure 3C). Transwell migration assays revealed that ectopic expression of lncRNA-ATB drastically enhanced MG63 cellmigration (Figure 3D). The same results were found in invasion assays (Figure 3E). Collectively, these data indicated that overexpression of lncRNA-ATB enhances osteosarcoma cells proliferation, migration, and invasion.
Figure 3.
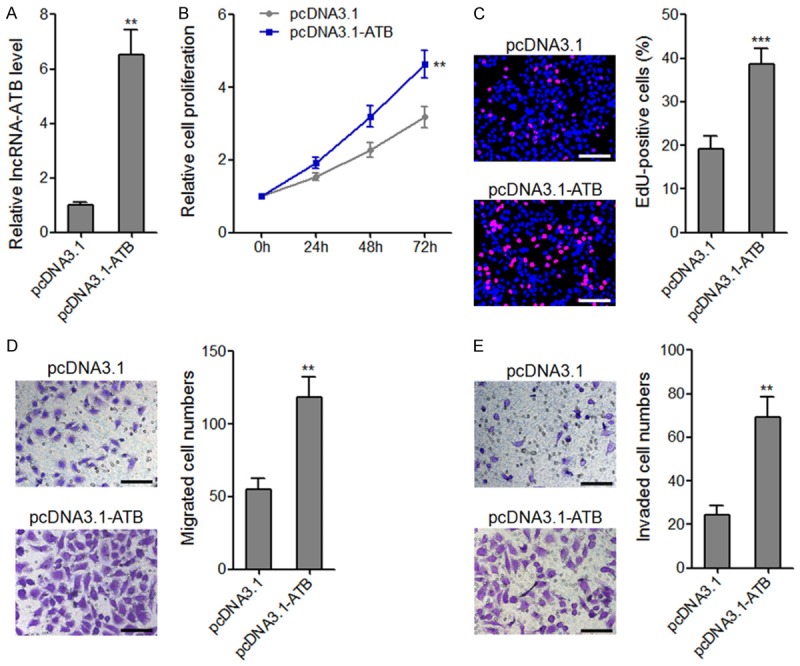
The effects of lncRNA-ATB overexpression on osteosarcoma cell proliferation, migration, and invasion. A. lncRNA-ATB expression levels in lncRNA-ATB stably overexpressed and control MG63 cells. B. Cell proliferation of lncRNA-ATB stably overexpressed and control MG63 cells were assessed by CCK-8 assays, and the relative numbers of cells to 0 h are shown. C. Cell proliferation of lncRNA-ATB stably overexpressed and control MG63 cells were assessed by EdU incorporation assays. Scale bars, 100 μm. D. The migrated cell numbers of lncRNA-ATB stably overexpressed and control MG63 cells per field were counted 48 h after seeding. Scale bars, 100 μm. E. The invaded cell numbers of lncRNA-ATB stably overexpressed and control MG63 cells per field were counted 48 h after seeding. Scale bars, 100 μm. For all panels, results are present as mean ± SD. **P<0.01, ***P<0.001 by Student’s t-test.
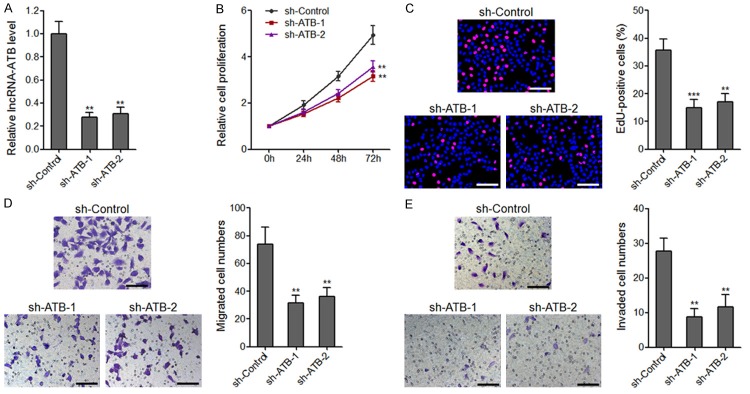
Silencing of lncRNA-ATB drastically inhibits osteosarcoma cell proliferation, migration, and invasion
To further characterize the biological roles of lncRNA-ATB in osteosarcoma, we stably silenced lncRNA-ATB expression in U2OS cells through transfecting two independent lncRNA-ATB specific shRNAs (Figure 4A). CCK-8 assays and EdU incorporation assays revealed that silencing of lncRNA-ATB drastically attenuated U2OS cell proliferation for both shRNAs (Figure 4B and 4C). Transwell migration assays and invasion assays revealed that silencing of lncRNA-ATB drastically attenuated U2OS cell migration and invasion for both shRNAs (Figure 4D and 4E). These data further confirmed the critical roles of lncRNA-ATB in osteosarcoma.
Figure 4.
The effects of lncRNA-ATB depletion on osteosarcoma cell proliferation, migration, and invasion. A. lncRNA-ATB expression levels in lncRNA-ATB stably depleted and control U2OS cells. B. Cell proliferation of lncRNA-ATB stably depleted and control U2OS cells were assessed by CCK-8 assays, and the relative numbers of cells to 0 h are shown. C. Cell proliferation of lncRNA-ATB stably depleted and control U2OS cells were assessed by EdU incorporation assays. Scale bars, 100 μm. D. The migrated cell numbers of lncRNA-ATB stably depleted and control U2OS cells per field were counted 48 h after seeding. Scale bars, 100 μm. E. The invaded cell numbers of lncRNA-ATB stably depleted and control U2OS cells per field were counted 48 h after seeding. Scale bars, 100 μm. For all panels, results are present as mean ± SD. **P<0.01, ***P<0.001 by Student’s t-test.
LncRNA-ATB upregulates ZEB1 and ZEB2 expression through inhibiting miR-200s
As lncRNA-ATB has been reported to bind and inhibit miR-200s expression, and further upregulate the miR-200s targets ZEB1 and ZEB2 expression in hepatocellular carcinoma and glioma [14,28], we next investigate whether lncRNA-ATB also regulate miR-200s, ZEB1, and ZEB2 expression in osteosarcoma. miR-200s expressions in lncRNA-ATB stably overexpressed and control MG63 cells, and lncRNA-ATB stably depleted and control U2OS cells were measured by qRT-PCR. The results showed that ectopic expression of lncRNA-ATB decreased miR-200s expression in MG63 cells (Figure 5A). Whereas, silencing of lncRNA-ATB increased miR-200s expression (Figure 5B). As the effects of miR-200s on ZEB1 and ZEB2 are dependent on the regulation of 3’UTR, we constructed luciferase reporter containing ZEB1 or ZEB2 3’UTR. Then the luciferase reporter plasmids were transfected into lncRNA-ATB stably overexpressed or control MG63 cells, and lncRNA-ATB stably depleted or control U2OS cells. The results showed that ectopic expression of lncRNA-ATB significantly upregulated the luciferase activities of the reporter containing ZEB1 or ZEB2 3’UTR, which were completely abolished by concurrent overexpression of miR-200s (Figure 5C). Reciprocally, silencing of lncRNA-ATB significantly inhibited the luciferase activities of the reporter containing ZEB1 or ZEB2 3’UTR, which were also completely rescued by concurrent depletion of miR-200s (Figure 5D). To further confirm the effects of lncRNA-ATB on ZEB1 and ZEB2, we measured ZEB1 and ZEB2 mRNA and protein levels in lncRNA-ATB stably overexpressed or control MG63 cells, and lncRNA-ATB stably depleted or control U2OS cells. The results showed that ectopic expression of lncRNA-ATB significantly increased ZEB1 and ZEB2 mRNA and protein levels, which were abolished by concurrent overexpression of miR-200s (Figure 5E and 5F). Whereas, silencing of lncRNA-ATB significantly decreased ZEB1 and ZEB2 mRNA and protein levels, which were abolished by concurrent depletion of miR-200s (Figure 5G and 5H). These data demonstrated that lncRNA-ATB upregulates ZEB1 and ZEB2 expression through inhibiting miR-200s.
Figure 5.
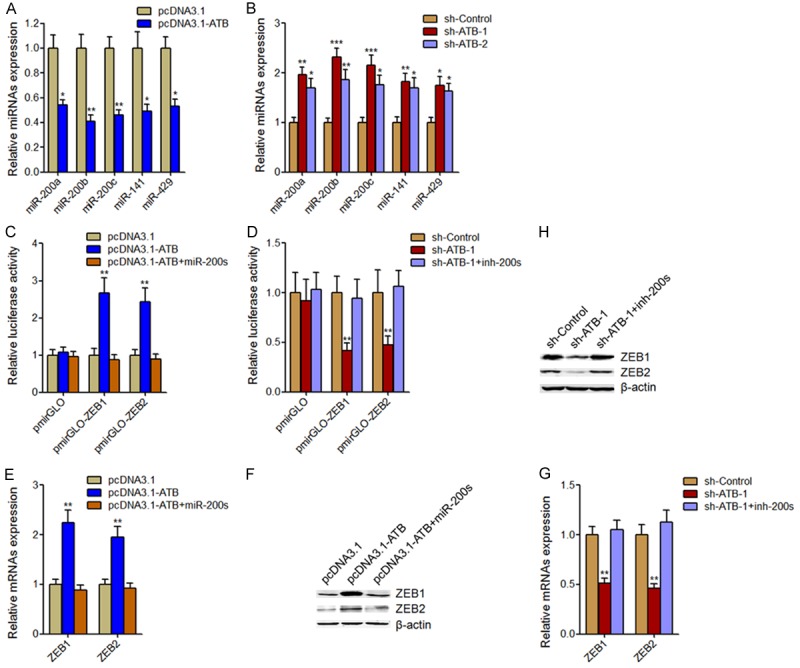
The effects of lncRNA-ATB on miR-200s, ZEB1, and ZEB2. A. MiR-200s expression levels in lncRNA-ATB stably overexpressed and control MG63 cells. B. MiR-200s expression levels in lncRNA-ATB stably depleted and control U2OS cells. C. Luciferase activity in lncRNA-ATB stably overexpressed and control MG63 cells co-transfected with miR-200s mimics and luciferase reporters containing ZEB1 3’UTR, ZEB2 3’UTR, or nothing. D. Luciferase activity in lncRNA-ATB stably depleted and control U2OS cells co-transfected with miR-200s inhibitors and luciferase reporters containing ZEB1 3’UTR, ZEB2 3’UTR, or nothing. E. ZEB1 and ZEB2 mRNA levels in lncRNA-ATB stably overexpressed and control MG63 cells transfected with miR-200s mimics. F. ZEB1 and ZEB2 protein levels in lncRNA-ATB stably overexpressed and control MG63 cells transfected with miR-200s mimics. G. ZEB1 and ZEB2 mRNA levels in lncRNA-ATB stably depleted and control U2OS cells transfected with miR-200s inhibitors. H. ZEB1 and ZEB2 protein levels in lncRNA-ATB stably depleted and control U2OS cells transfected with miR-200s inhibitors. Results are present as mean ± SD. *P<0.05, **P<0.01, ***P<0.001 by Student’s t-test.
The correlation between lncRNA-ATB and miR-200s, ZEB1, or ZEB2 expression levels in osteosarcoma
To investigate whether the regulation of miR-200s, ZEB1, and ZEB2 by lncRNA-ATB also exit in vivo, we measured miR-200b, ZEB1, and ZEB2 expression in the same 60 osteosarcoma tissues as shown in Figure 2A. The results showed that the expression level of lncRNA-ATB was significantly negatively correlated with miR-200b expression level in osteosarcoma (Figure 6A). Whereas, lncRNA-ATB expression level was significantly positively correlated with ZEB1 and ZEB2 expression level in osteosarcoma (Figure 6B and 6C). These data support the regulation of miR-200s, ZEB1, and ZEB2 by lncRNA-ATB in osteosarcoma.
Figure 6.
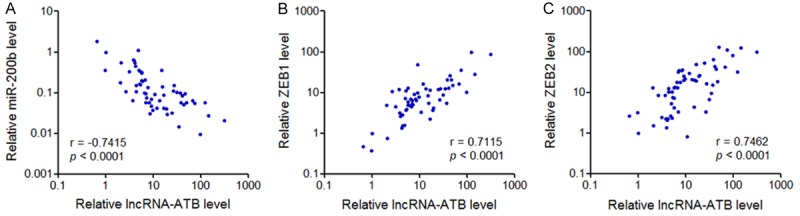
The correlation between lncRNA-ATB and miR-200s, ZEB1, or ZEB2 expression levels in osteosarcoma. A. The correlation between lncRNA-ATB and miR-200b expression levels in 60 osteosarcoma tissues was measured by qRT-PCR and subjected to Pearson correlation analysis. B. The correlation between lncRNA-ATB and ZEB1 expression levels in 60 osteosarcoma tissues was measured by qRT-PCR and subjected to Pearson correlation analysis. C. The correlation between lncRNA-ATB and ZEB2 expression levels in 60 osteosarcoma tissues was measured by qRT-PCR and subjected to Pearson correlation analysis.
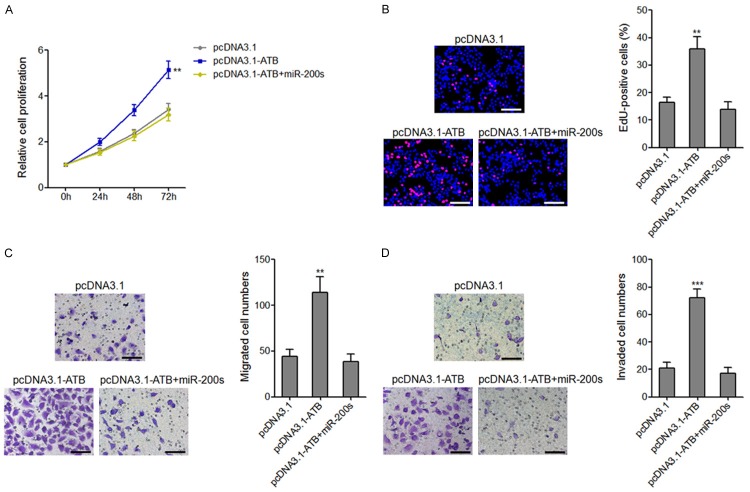
The effects of lncRNA-ATB on osteosarcoma cells proliferation, migration, and invasion are dependent on the inhibition of miR-200s
To investigate whether the effects of lncRNA-ATB on osteosarcoma cells proliferation, migration, and invasion are dependent on the regulation of miR-200s, we transiently overexpressed miR-200s in lncRNA-ATB stably overexpressed MG63 cells. CCK-8 assays and EdU incorporation assays demonstrated that overexpression of miR-200s completely abolished the pro-proliferativeroles of lncRNA-ATB on MG63 cells (Figure 7A and 7B). Transwell migration assays and invasion assays demonstrated that overexpression of miR-200s completely abolished the pro-migratory and pro-invasive roles of lncRNA-ATB on MG63 cells (Figure 7C and 7D). These data demonstrated that the biological roles of lncRNA-ATB on osteosarcoma cell proliferation, migration, and invasion are dependent on the inhibition of miR-200s.
Figure 7.
Overexpression of miR-200s reversed the effects of lncRNA-ATB on osteosarcoma cell proliferation, migration, and invasion. A. Cell proliferation of lncRNA-ATB stably overexpressed and control MG63 cells transfected with miR-200s mimics were assessed by CCK-8 assays, and the relative numbers of cells to 0 h are shown. B. Cell proliferation of lncRNA-ATB stably overexpressed and control MG63 cells transfected with miR-200s mimics were assessed by EdU incorporation assays. Scale bars, 100 μm. C. The migrated cell numbers of lncRNA-ATB stably overexpressed and control MG63 cells transfected with miR-200s mimics per field were counted 48 h after seeding. Scale bars, 100 μm. D. The invaded cell numbers of lncRNA-ATB stably overexpressed and control MG63 cells transfected with miR-200s mimics per field were counted 48 h after seeding. Scale bars, 100 μm. For all panels, results are present as mean ± SD. **P<0.01, ***P<0.001 by Student’s t-test.
Overexpression of lncRNA-ATB promotes osteosarcoma tumor growth in vivo in an miR-200s dependent manner
To further investigate the effects of lncRNA-ATB on osteosarcoma in vivo and the underlying molecular mechanisms, we stably overexpressed miR-200s targeting sites mutated lncRNA-ATB (lncRNA-ATB-Mut) in MG63 cells. Then lncRNA-ATB, lncRNA-ATB-Mut stably overexpressed or control MG63 cells were subcutaneously injected into nude mice. Subcutaneously tumors growth was measured weekly, and the tumors were excised and weighed at 28 days after injection. As shown in Figure 8A and 8B, overexpression of lncRNA-ATB, but not lncRNA-ATB-Mut significantly promoted tumor growth in vivo. LncRNA-ATB, ZEB1, and ZEB2 expressions in subcutaneous tumors were also measured. The results showed that overexpression of lncRNA-ATB upregulated ZEB1 and ZEB2 expression in vivo (Figure 8C). Although lncRNA-ATB-Mutstably overexpressed MG63 cells has similar overexpression efficiency with lncRNA-ATB stably overexpressed MG63 cells, enhanced expression of lncRNA-ATB-Mut had no effects on ZEB1 and ZEB2 expression (Figure 8C). These results suggested that lncRNA-ATB upregulates ZEB1 and ZEB2 expression and promotes osteosarcoma growth in vivo in an miR-200s dependent manner.
Figure 8.
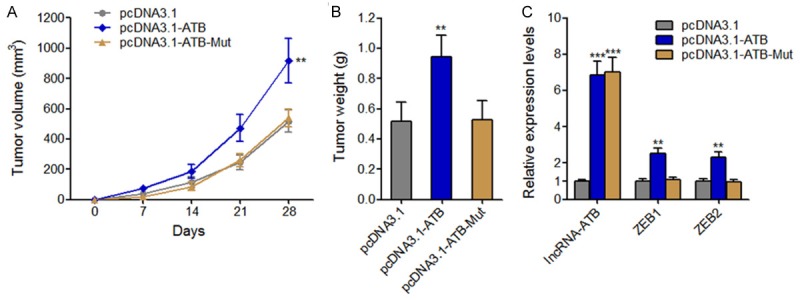
The effects of lncRNA-ATB on osteosarcoma tumor growth in vivo are dependent on miR-200s. (A) After subcutaneously injecting lncRNA-ATB, lncRNA-ATB-Mut stably overexpressed or control MG63 cells into nude mice, tumors growth curves were measured weekly. (B) Tumors weight was measured 28 days after injection. (C) LncRNA-ATB, ZEB1, and ZEB2 expression levels in tumors derived from (B). **P<0.01, ***P<0.001 by Mann-Whitney U test.
Discussion
With the great progression of neoadjuvant chemotherapy for osteosarcoma, the 5-year survival rate of osteosarcoma patients has been improved to 60%-70% for localized diseases [29,30]. But, the 5-year survival rate of osteosarcoma patients with metastases at diagnosis remains very poor [31,32]. Hence, the early diagnosis of osteosarcoma is necessary for efficiently improving the outcome of osteosarcoma patients. Searching for sensitive and specific non-invasive serum biomarkers for early diagnosis of osteosarcoma would great benefit the routine screening and early diagnosis of osteosarcoma. In this study, we identified lncRNA-ATB as a new serum diagnostic biomarker for osteosarcoma. Although several miRNAs, such as miR-221, miR-191, and miR-421 have been reported to be potential serum biomarkers for osteosarcoma [33-35], to our knowledge this is the first report that lncRNA could also be non-invasive serum biomarker for osteosarcoma. The combination of these reported non-coding RNAs would improve the diagnostic efficiency for osteosarcoma, and this needs further investigation.
The main causes of osteosarcoma patient’s death are recurrence and metastases [36]. Accurately predicting the patients in high risk for recurrences would give proper monitoring and treatment for osteosarcoma patients. In this study, in addition to measuring serum lncRNA-ATB levels in osteosarcoma patients, we also measured the expression of lncRNA-ATB in osteosarcoma tissues and analyzed its association with clinicopathological features and prognosis. Our results revealed that increased expression of lncRNA-ATB is associated with advanced Enneking stage, metastasis, recurrence, and poor prognosis, and suggested that lncRNA-ATB would be a novel prognostic biomarker for osteosarcoma. These data also implied that lncRNA-ATB may have critical roles in the progression of osteosarcoma.
To investigate the biological role of lncRNA-ATB in osteosarcoma, gain-of-function and loss-of-function experiments were performed. All these results revealed that lncRNA-ATB promotes osteosarcoma cell proliferation, migration, and invasion in vitro, and osteosarcoma tumor growth in vivo. In addition to lncRNA-ATB, other lncRNAs have been reported to function as oncogene or tumor suppressor in osteosarcoma, such as MALAT1, HOTTIP, HULC, LINC00161, and HOTAIR [37-41]. Combination with these reports, our study further demonstrates the critical roles of lncRNAs in osteosarcoma. Furthermore, we found lncRNA-ATB inhibits miR-200s and upregulates miR-200s target genes ZEB1 and ZEB2. Functional experiments showed that the roles of lncRNA-ATB on osteosarcoma cell proliferation, migration, and invasion in vitro, and osteosarcoma tumor growth in vivo are dependent on miR-200s.
Collectively, our study demonstrated that lncRNA-ATB is upregulated in osteosarcoma patient’s serum and tissues, promotes osteosarcoma cell proliferation, migration, and invasion in vitro, and osteosarcoma tumor growth in vivo through inhibiting miR-200s pathway. Our results implied that lncRNA-ATB may be a novel diagnostic and prognostic biomarker, and a potential therapeutic target for osteosarcoma.
Disclosure of conflict of interest
None.
References
- 1.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–718. [PubMed] [Google Scholar]
- 2.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 3.Mir O, Ropert S, Goldwasser F. Neoadjuvant chemotherapy with high-dose methotrexate in osteosarcoma. Lancet Oncol. 2008;9:1198. doi: 10.1016/S1470-2045(08)70309-7. [DOI] [PubMed] [Google Scholar]
- 4.Jaffe N, Puri A, Gelderblom H. Osteosarcoma: evolution of treatment paradigms. Sarcoma. 2013;2013:203531. doi: 10.1155/2013/203531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC, Cowley GS, Bhatt AS, Rheinbay E, Pedamallu CS, Helman E, Taylor-Weiner A, McKenna A, DeLuca DS, Lawrence MS, Ambrogio L, Sougnez C, Sivachenko A, Walensky LD, Wagle N, Mora J, de Torres C, Lavarino C, Dos Santos Aguiar S, Yunes JA, Brandalise SR, Mercado-Celis GE, Melendez-Zajgla J, Cardenas-Cardos R, Velasco-Hidalgo L, Roberts CW, Garraway LA, Rodriguez-Galindo C, Gabriel SB, Lander ES, Golub TR, Orkin SH, Getz G, Janeway KA. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111:E5564–5573. doi: 10.1073/pnas.1419260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang J, Shen L, Yang Q, Zhang C. Overexpression of metadherin mediates metastasis of osteosarcoma by regulating epithelial-mesenchymal transition. Cell Prolif. 2014;47:427–434. doi: 10.1111/cpr.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar RM, Arlt MJ, Kuzmanov A, Born W, Fuchs B. Sunitinib malate (SU-11248) reduces tumour burden and lung metastasis in an intratibial human xenograft osteosarcoma mouse model. Am J Cancer Res. 2015;5:2156–2168. [PMC free article] [PubMed] [Google Scholar]
- 8.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan CX, Zhang D, Yuan H, Hua K, Lu Y, Katsaros D, Huang Q, Montone K, Fan Y, Coukos G, Boyd J, Sood AK, Rebbeck T, Mills GB, Dang CV, Zhang L. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 12.Chen F, Mo J, Zhang L. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumour Biol. 2016;37:13403–13412. doi: 10.1007/s13277-016-5256-y. [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res. 2016;8:4095–4105. [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Ostensson M, Akyurek LM, Abrahamsson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 17.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung MC, Lin C, Yang L. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellinger J, Alam J, Rothenburg J, Deng M, Schmidt D, Syring I, Miersch H, Perner S, Muller SC. The long non-coding RNA lnc-ZNF180-2 is a prognostic biomarker in patients with clear cell renal cell carcinoma. Am J Cancer Res. 2015;5:2799–2807. [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Wang J, Chen Y, Li S, Jin M, Wang H, Chen Z, Yu W. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6:1099–1107. [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, Wang J, Liang H, Gan B. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18:431–442. doi: 10.1038/ncb3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, Leucci E, Lapouge G, Beck B, van den Oord J, Nakagawa S, Hirose T, Sablina AA, Lambrechts D, Aerts S, Blanpain C, Marine JC. P53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 22.Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739–3754. doi: 10.1111/febs.13839. [DOI] [PubMed] [Google Scholar]
- 23.Sun J, Wang X, Fu C, Wang X, Zou J, Hua H, Bi Z. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol Biol Rep. 2016;43:427–436. doi: 10.1007/s11033-016-3975-1. [DOI] [PubMed] [Google Scholar]
- 24.Yang JP, Yang XJ, Xiao L, Wang Y. Long noncoding RNA PVT1 as a novel serum biomarker for detection of cervical cancer. Eur Rev Med Pharmacol Sci. 2016;20:3980–3986. [PubMed] [Google Scholar]
- 25.Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 26.Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, Peng Z, Yan D. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31:595–603. doi: 10.1111/jgh.13206. [DOI] [PubMed] [Google Scholar]
- 27.Xu S, Yi XM, Tang CP, Ge JP, Zhang ZY, Zhou WQ. Long non-coding RNA ATB promotes growth and epithelial-mesenchymal transition and predicts poor prognosis in human prostate carcinoma. Oncol Rep. 2016;36:10–22. doi: 10.3892/or.2016.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma CC, Xiong Z, Zhu GN, Wang C, Zong G, Wang HL, Bian EB, Zhao B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35:90. doi: 10.1186/s13046-016-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heare T, Hensley MA, Dell’Orfano S. Bone tumors: osteosarcoma and ewing’s sarcoma. Curr Opin Pediatr. 2009;21:365–372. doi: 10.1097/MOP.0b013e32832b1111. [DOI] [PubMed] [Google Scholar]
- 30.Porter KA, Calne RY, Zukoski CF. Treatment of osteosarcoma. Lancet. 1964;2:1001. [PubMed] [Google Scholar]
- 31.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment-where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Benjamin RS. Osteosarcoma: better treatment through better trial design. Lancet Oncol. 2015;16:12–13. doi: 10.1016/S1470-2045(14)71186-6. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z, Zhang Y, Zhang X, Zhang M, Liu H, Zhang S, Qi B, Sun X. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed Pharmacother. 2015;75:153–158. doi: 10.1016/j.biopha.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Wang T, Ji F, Dai Z, Xie Y, Yuan D. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer Biomark. 2015;15:543–550. doi: 10.3233/CBM-150493. [DOI] [PubMed] [Google Scholar]
- 35.Zhou S, Wang B, Hu J, Zhou Y, Jiang M, Wu M, Qin L, Yang X. MiR-421 is a diagnostic and prognostic marker in patients with osteosarcoma. Tumour Biol. 2016;37:9001–7. doi: 10.1007/s13277-015-4578-5. [DOI] [PubMed] [Google Scholar]
- 36.Mirabello L, Koster R, Moriarity BS, Spector LG, Meltzer PS, Gary J, Machiela MJ, Pankratz N, Panagiotou OA, Largaespada D, Wang Z, Gastier-Foster JM, Gorlick R, Khanna C, de Toledo SR, Petrilli AS, Patino-Garcia A, Sierrasesumaga L, Lecanda F, Andrulis IL, Wunder JS, Gokgoz N, Serra M, Hattinger C, Picci P, Scotlandi K, Flanagan AM, Tirabosco R, Amary MF, Halai D, Ballinger ML, Thomas DM, Davis S, Barkauskas DA, Marina N, Helman L, Otto GM, Becklin KL, Wolf NK, Weg MT, Tucker M, Wacholder S, Fraumeni JF Jr, Caporaso NE, Boland JF, Hicks BD, Vogt A, Burdett L, Yeager M, Hoover RN, Chanock SJ, Savage SA. A genome-wide scan identifies variants in NFIB associated with metastasis in patients with osteosarcoma. Cancer Discov. 2015;5:920–931. doi: 10.1158/2159-8290.CD-15-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Zhao L, Wang Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/beta-catenin pathway. Am J Transl Res. 2016;8:2385–2393. [PMC free article] [PubMed] [Google Scholar]
- 39.Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8:2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Zhang L, Zheng X, Zhong W, Tian X, Yin B, Tian K, Zhang W. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. 2016;382:137–146. doi: 10.1016/j.canlet.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 41.Zheng H, Min J. Role of long noncoding RNA HOTAIR in the growth and apoptosis of osteosarcoma cell MG-63. Biomed Res Int. 2016;2016:5757641. doi: 10.1155/2016/5757641. [DOI] [PMC free article] [PubMed] [Google Scholar]