Abstract
MicroRNAs are associated with different types of cancers. In this study, we found that miR-1468-5p could inhibit growth and cell cycle progression in glioma by targeting ribonucleotide reductase large subunit M1 (RRM1). First, we analyzed miR-1468-5p expression in different glioma grades and the prognostic significance of its expression in glioblastoma multiform patients from the Chinese Glioma Genome Atlas. Then, we expressed miR-1468-5p in U87 and U251 cells and assessed the effects on proliferation and cell cycle progression using cell counting kit-8, colony formation, EdU and flow cytometry assays. Western blotting and luciferase reporter assays identified RRM1 as a novel direct target of miR-1468-5p. Experiments to determine the role of RRM1 in glioma showed that RRM1 expression was significantly higher in glioma than in normal brain tissues, and silencing RRM1 with small-interfering RNAs decreased proliferation and suppressed cell cycle progression, which indicated that RRM1 had pro-tumor functions. miR-1468-5p overexpression suppressed RRM1 expression, reduced glioma cell proliferation and induced cell cycle arrest, which was partially rescued by forced RRM1 expression. In summary, our study revealed that the regulatory mechanism of miR-1468-5p in glioma cell cycle progression involved direct regulation of RRM1 expression, suggesting that RRM1 may be a potential therapeutic target for glioma.
Keywords: miR-1468-5p, RRM1, glioma, proliferation, cell cycle
Introduction
Gliomas are the most common nervous system malignancy, accounting for more than 70% of brain tumors [1,2]. The most common subtype of glioma is glioblastoma multiform (GBM), which has an age-adjusted incidence rate ranging from 0.59 to 3.69 per 100,000 persons [3]. Although great progress in standard therapies, including surgical resection, chemotherapy and radiotherapy have been made, the prognosis of GBM remains poor [4,5]. Thus, there is an urgent need to develop new molecular targets and treatment strategies for this disease.
MicroRNAs (miRs) are short single-stranded RNA molecules, composed of approximately 20 nucleotides, that act as key regulators of gene expression by binding to the 3’-untranslated region (UTR) of target mRNAs [6-8]. miRs can lead to translational repression or mRNA degradation, and thus, generally suppress the protein expression of their targets [9,10]. By mediating target-gene expression, miRNAs have been implicated in the regulation of important cellular functions such as proliferation, invasion, apoptosis, death, stress response, differentiation and development [11-15]. miRNAs downregulate multiple target genes, including oncogenes and tumor suppressors, making some miRNAs function as tumor suppressors and others as oncogenes [16-18].
Ribonucleotide reductase (RNR) is the rate-limiting enzyme for converting ribonucleoside diphosphates (NDPs) to deoxyribonucleoside diphosphates (dNDPs), which are the building blocks of DNA [19,20]. In mammalian cells, the active form of the RNR holoenzyme is a tetramer composed of two large RRM1 subunits and two small RRM2 or RRM2B subunits [19]. These two subtypes of RNR, RRM1-RRM2 and RRM1-RRM2B, provide dNTPs for DNA replication and repair, respectively [21,22]. RRM1 contains the catalytic and allosteric regulation sites while RRM2 and RRM2B contain the diiron-tyrosyl radical cofactor required for enzyme activity [23-25]. RRM1 plays a vital role in DNA synthesis and cell proliferation, therefore, its expression and activity are tightly controlled in cells. When RRM1 is inhibited, the cell cycle is also arrested [26,27]. RRM1 dysregulation has been closely associated with many types of cancers [28,29]; therefore, RRM1 inhibition may be a useful strategy for cancer therapy. However, the specific role of RRM1 in different cancers remains unclear. To date, there have been no reports on the role and clinical significance of RRM1 expression in glioma.
In this study, we identified a novel tumor-suppressive miRNA, miR-1468-5p, and investigated its functional roles and therapeutic effects in glioma. Moreover, we identified RRM1 as a direct miR-1468-5p target. As the function of RRM1 has not been adequately studied in glioma, we also investigated its role in this malignancy for the first time. Our results not only revealed that RRM1 overexpression in glioma could be the result of downregulation of miR-1468-5p, but also suggested an important role for the loss of miR-1468-5p in proliferation and cell cycle progression in glioma, highlighting its potential as a therapeutic target.
Materials and methods
Databases and human tissue samples
miRNA expression profiles of 198 glioma samples of different grades were downloaded from the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn). Among the 198 cases, 49 were diagnosed as astrocytoma (A), 14 as oligodendroglioma (O), 12 as anaplastic astrocytoma (AA), 13 as anaplastic oligodendroglioma (AO), 19 as anaplastic oligoastrocytoma (AOA), and 91 as GBM. The mRNA expression microarray data were downloaded from the CGGA, which included 5 normal brain tissues, 58 A, 22 O, 21 oligoastrocytomas (OA, WHO Grade II), 8 AA, 11 AO, 15 AOA, 85 GBM. Gene expression data (21 normal brain tissues, 99 WHO II, 84 WHO III, 188 GBM) were downloaded from The Repository for Molecular Brain Neoplasia Data (REMBRANDT, http://caintegrator-info.nci.nih.gov/rembrandt).
The human glioma tissue samples were obtained from patients undergoing standard surgical procedure at the Department of Neurosurgery of the First Affiliated Hospital of Nanjing Medical University, China. NBTs were collected as negative controls from patients undergoing decompressive craniectomy for traumatic brain injury. All study procedures were approved by the Institutional Review Board of the hospital. Informed consent was given by all participants. Tissue fragments were immediately frozen in liquid nitrogen after surgery. Total protein and RNA of paired tissues were extracted and stored at -80°C.
Cell culture
The human glioma cell lines U87, U251, T98, U118, LN229, H4 and A172 were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Normal human astrocytes (NHAs) were obtained from Lonza (Basel, Switzerland) and cultured in the provided astrocyte growth media supplemented with recombinant human EGF, insulin, ascorbic acid, GA-1000, L-glutamine and 5% fetal bovine serum.
Lentiviral packaging and the establishment of stable cell lines
A lentiviral packaging kit was purchased from GenePharma (Shanghai, China). Lentiviruses carrying has-miR-1468-5p or hsa-miR-negative control (miR-ctrl) were packaged following the manufacturer’s instructions. Stable cell lines were established by infecting U87 and U251 cells, followed by puromycin selection.
Oligonucleotides, plasmids and transfection
siRRM1 and control si-non-coding (siNC) oligonucleotides were purchased from GenePharma. pcDNA3(+)-RRM1 vector was obtained from Sangon Biotech (Shanghai, China). All oligonucleotides and plasmids were transfected into U87 and U251 cells using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Quantitative real-time PCR
Total RNA from glioma cells was isolated using TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (Roche, Basel, Switzerland). cDNA synthesis was performed using the BcaBest RNA PCR kit from TaKaRa (Dalian, China) according to the manufacturer’s instructions. Quantitative rea-time PCR was performed using the iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with Real-time SYBR Green PCR Master Mix. Endogenous U6 snRNA was used as an internal control. The PCR conditions for relative quantification were as follows: initial denaturation at 95°C for 5 min, then 40 cycles consisting of 95°C for 10 s, 60°C for 30 s and 72°C for 30 s. The relative expression of each gene was calculated and normalized using the 2-ΔΔCt method. Each sample was tested in triplicate.
Western blotting
Total proteins were extracted from human glioma cells in radioimmunoprecipitation assay buffer (Applygen, Beijing, China) and quantified using the bicinchoninic acid protein assay kit (KenGEN, Nanjing, China). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed on 20 mg of protein from each sample. The electrophoresed proteins were transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA), which were incubated with diluted primary antibodies against RRM1 (1:1000; Abcam, Cambridge, UK) phosphorylated (p)-ERK1/2, ERK1/2, p-Akt or Akt (1:1000; Cell Signaling Technology, Danvers, MA, USA); anti-β-actin antibody was obtained from Beyotime (Haimen, China) and used as a loading control. Secondary antibody incubation and the visualization of immunoreactive bands were performed using standard laboratory procedures.
Cell proliferation assays
U87 and U251 cells were seeded in 96-well plates at a density of 2000 cells per well. Cell proliferation was assayed using Cell Counting Kit 8 (CCK8; Beyotime) at 24, 48, 72 and 96 h after transfection. Briefly, 10 μL CCK8 solution was added into each well, and cells were incubated for 1.5 h in a humidified incubator. Optical density was measured at 450 nm. Five replicate wells were setup in each group and three independent experiments were performed.
Colony formation assays were performed to assess the proliferation ability of transfected U87 and U251 cells. Briefly, U87 and U251 cells in logarithmic phase were digested in 0.25% Trypsin (Sigma-Aldrich, Shanghai, China). Cells were added to pre-warmed medium in new six-well plates (200 cells/well) and incubated in a humidified atmosphere with 5% CO2 at 37°C for 2 weeks. Then, the medium was discarded, and the cells were washed twice with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 15 min, stained by Giemsa (Vetec, Shanghai, China) for 30 min, washed in running water and dried. The colony formation rate was used to calculate the post-transfection survival rate.
For the 5-ethynyl-2’-deoxyuridine (EdU) proliferation assay, we used a Cell-Light EdU imaging detection kit purchased from RiboBio (Guangzhou, China). Cells that had been transfected with lentivirus were incubated with 10 μM EdU for 24 h, fixed, permeabilized and stained with both the Apollo567 reaction cocktail for EdU and 4’,6-diamidino-2-phenylindole (DAPI) for cell nuclei, according to the manufacturer’s protocol. Finally, samples were imaged under a fluorescent microscope.
Cell cycle analysis
Cells were harvested, washed twice with PBS, fixed with 75% ethanol and stored at -20°C for 12 h. The cells were then resuspended in PBS containing 25 mg/mL propidium iodide, 0.1% Triton and 10 mg/mL RNase, incubated for 30 min in the dark and analyzed by flow cytometry.
MiRNA target prediction and luciferase assay
miRNA target prediction and analysis were performed with the algorithms from TargetScan (http://www.targetscan.org/), miRanda (http://www.microrna.org/), miRDB (http://www.mirdb.org/) and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html).
The pGL3-RRM1-3’UTR-wild-type or pGL3-RRM1-3’UTR-mutant luciferase plasmids were transfected into U87 and U251 cells using Lipofectamine 2000 (Invitrogen). At 36 h after transfection, Luciferase activity was measured using the Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
Immunohistochemistry
Immunohistochemical assays were conducted on human brain tissue microarrays and nude mouse xenograft tumor tissues to detect RRM1, Ki-67 and CD31 expression using previously described methods [30].
Fluorescence in situ hybridization (FISH)
The expression of miR-1468-5p in GBM samples and NBTs was detected by FISH. The mature human miR-1468-5p sequence is: 3’-GUCGCUUUGUCCGUUUGCCUC-5’. We used (LNA)-based probes directed against the full length mature miRNA sequence. The 5’-FAM-labelled miR-1468-5p probe sequence is: 5’-CAGCGAAACAGGCAAACGGAG-3’, and was purchased from BioSense (Guangzhou, China). The FISH procedure followed the BioSense instructions. Briefly, frozen sections were fixed with 4% paraformaldehyde for 30 min, then washed twice with PBS. Fixed slides were then treated with proteinase K at 37°C for 10 min, followed by dehydration in 70%, 85% and 100% ethanol for 5 min. The probe was then added to the slides, which were denatured at 78°C for 5 min. Hybridization was then carried out overnight at 42°C in a humid chamber. The next day, post-hybridization washes were performed with 50% formamide with 2× SSC at 43°C, followed by 2× SSC washes at room temperature to remove non-specific and repetitive RNA hybridization. Finally, slides were counterstained with DAPI (Sigma) for 10 min and examined with a Zeiss LSM 700 Meta confocal microscope (Oberkochen, Germany).
Nude mouse intracranial glioma model
Animal experiments were approved by the Animal Management Rule of the Chinese Ministry of Health (document 55, 2001) and were in conducted in accordance with the approved guidelines and experimental protocols of Nanjing Medical University. Sixteen nude mice 3-4 weeks of age were purchased from the Shanghai Experimental Animal Center of the Chinese Academy of Sciences and randomly divided into two groups. U87 cells were co-infected with lentiviruses expressing luciferase with miR-ctrl or miR-1468-5p. Then 2×105 infected U87 cells were stereotactically implanted using cranial guide screws to establish intracranial gliomas. Mice were imaged for firefly Luciferase activity using a bioluminescence imaging system on days 7, 14, 21 and 28. The Living Images software package (Caliper Life Sciences, Waltham, MA, USA) was used to determine the integrated flux of photons (photons per second) within each region of interest. The error bars indicate standard deviations. Mice were sacrificed after observation, and their brains were extracted, fixed in 10% formalin for 24 h and embedded in paraffin for immunohistochemistry.
Statistical analysis
All experiments were performed in triplicate, and data were analyzed with GraphPad Prism 5 (San Diego, CA, USA). Statistical evaluation of data was performed using Student’s t-test with P<0.05 considered statistically significant.
Results
miR-1468-5p is downregulated in glioma tissues and cell lines
To explore miR-1468-5p expression in glioma, we initially analyzed the CGGA database, which included 198 glioma tissues (Grade II: 63; Grade III: 44 and Grade IV: 91). We found that miR-1468-5p expression was significantly lower in high-grade glioma tissues compared with low-grade glioma tissues, indicating that miR-1468-5p expression correlated with malignancy (Figure 1A). Next, the prognostic value of miR-1468-5p expression in 82 GBM cases was investigated by Kaplan-Meier survival analysis. The results indicated that patients with high miR-1468-5p expression (n=41) had longer median overall survival (OS) times than patients with low miR-1468-5p expression (n=41) (Figure 1B).
Figure 1.
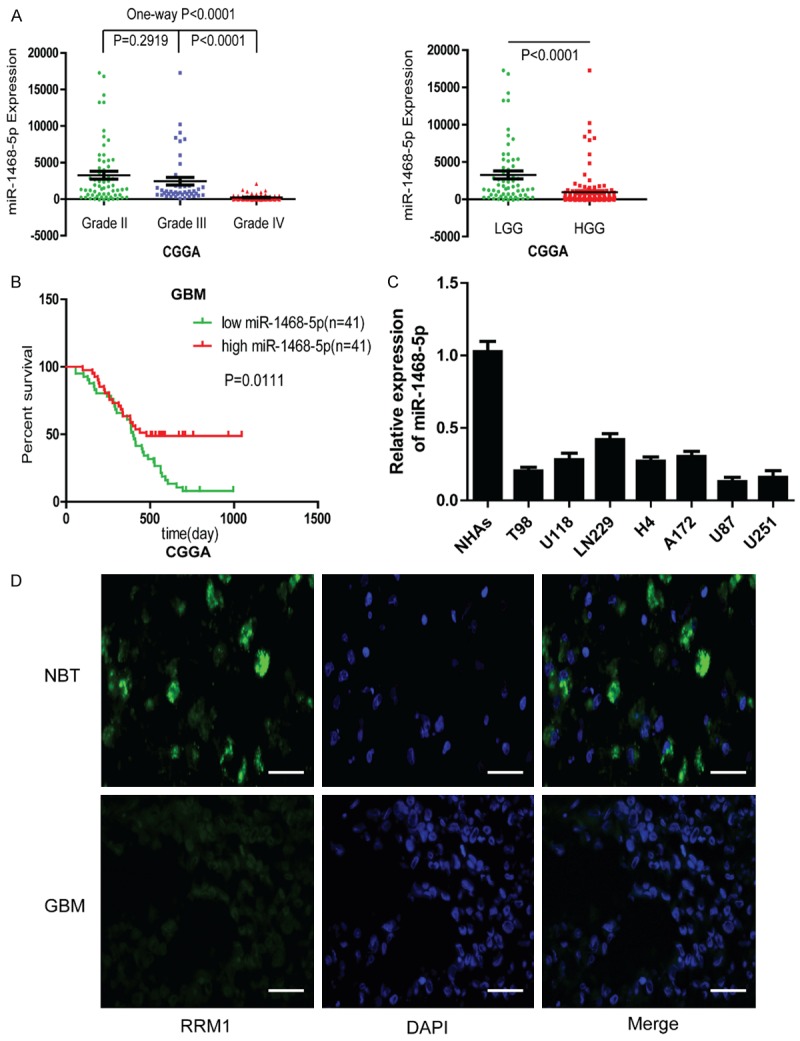
miR-1468-5p expression levels in human gliomas and cell lines. A: CGGA database showing miR-1468-5p expression in different grades of glioma. B: Kaplan-Meier survival analysis of overall survival duration in 82 GBM patients according to miR-1468-5p expression using CGGA. C: qRT-PCR analysis of miR-1468-5p in NHAs and different glioma cell lines. D: The different expressions of miR-1468-5p were determined by FISH in GBM and NBT, respectively. Scale bar: 50 μm.
Quantitative real-time PCR was used to analyze miR-1468-5p levels in NHAs and glioma cell lines. We found that miR-1468-5p was downregulated in different kinds of cell lines, especially in U87 and U251 cells compared with NHAs (Figure 1C). Therefore, we chose to further study U87 and U251 cells. Finally, we chose one representative GBM specimen and one NBT for FISH analysis. The results were consistent with the CGGA database (Figure 1D). Together, these results suggested that low miR-1468-5p expression was associated with glioma malignancy, and that miR-1468-5p may serve as a prognostic marker for glioma patients.
miR-1468-5p expression suppresses glioma cell proliferation and induces a G1/S arrest
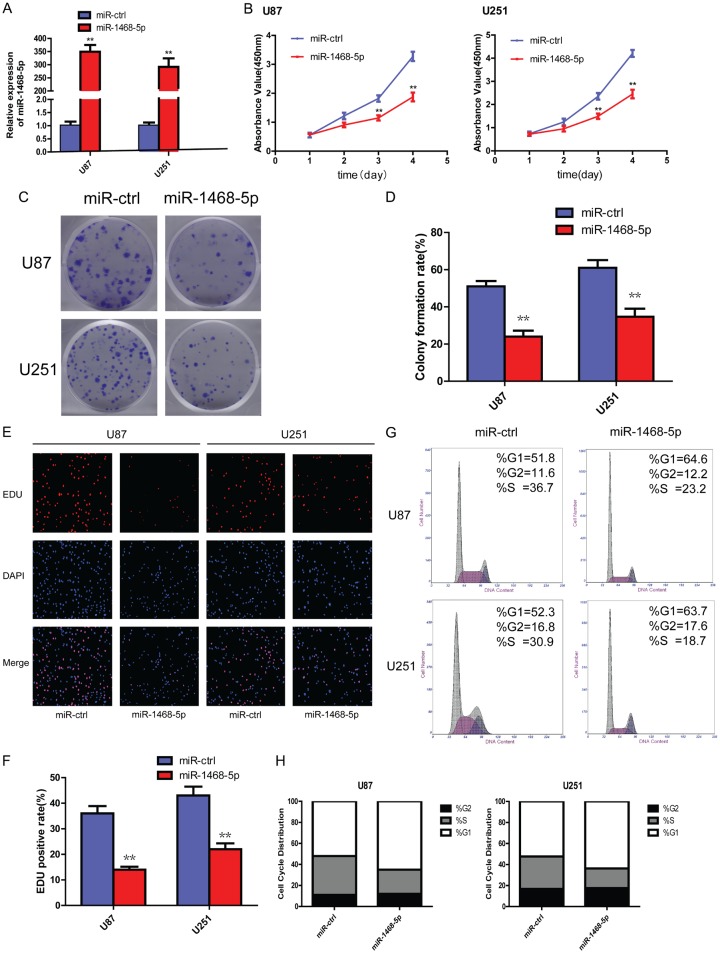
The reduced expression of miR-1468-5p in gliomas and glioma cell lines led us to investigate its biological roles. To study the function of miR-1468-5p in glioma cells, U87 and U251 cells were infected with lentiviruses containing miR-1468-5p or miR-ctrl. qRT-PCR showed that miR-1468-5p expression was significantly increased in the miR-1468-5p-transfected U87 and U251 cells compared with the negative control group (Figure 2A).
Figure 2.
Changes in glioma cell proliferation and cell cycle progression following miR-1468-5p overexpression. A: qRT-PCR showed miR-1468-5p expression in U87 and U251 cells after infection with lentiviruses containing miR-ctrl or miR-1468-5p; **P<0.01. B: CCK-8 assay revealed the proliferation ability of U87 and U251 cells transduced with miR-ctrl or miR-1468-5p following 4 days of culture; **P<0.01. C, D: Colony formation assays performed in U87 and U251 cells after miR-ctrl or miR-1468-5p transduction revealed long-term cell viability. The experiments were performed 3 times, and average scores are indicated with error bars on the histogram; **P<0.01. E, F: EdU assay was performed in U87 and U251 cells transduced with miR-ctrl or miR-1468-5p. Representative images are shown (original magnification, 200×); **P<0.01. G, H: Cell cycle analysis of U87 and U251 cells transduced with miR-ctrl or miR-1468-5p and analyzed by flow cytometry.
We then evaluated the effect of miR-1468-5p overexpression on cell proliferation using CCK-8, colony formation and EdU assays in U87 and U251 cells. The CCK-8 assay revealed that miR-1468-5p-overexpressing U87 and U251 cells exhibited significantly slower growth rates than the miR-ctrl group on days 3 and 4 after plating (Figure 2B). The colony formation assay consistently showed that the colony formation rate of the experimental group was reduced after 12-d culture compared with the miR-ctrl group (Figure 2C and 2D). Furthermore, EdU experiments showed that the EdU-positive rate of the miR-1468-5p group was significantly decreased in both U87 and U251 cells (Figure 2E and 2F). Moreover, important signaling pathway genes, such as p-AKT and p-ERK1/2 levels, were significantly reduced when miR-1468-5p was overexpressed in U87 and U251 cells (Figure 3B). AKT and ERK1/2 are known to be key roles in controlling cell proliferation [31]. In conclusion, the proliferation ability of two glioma cell lines, U87 and U251, was reduced when miR-1468-5p was upregulated.
Figure 3.
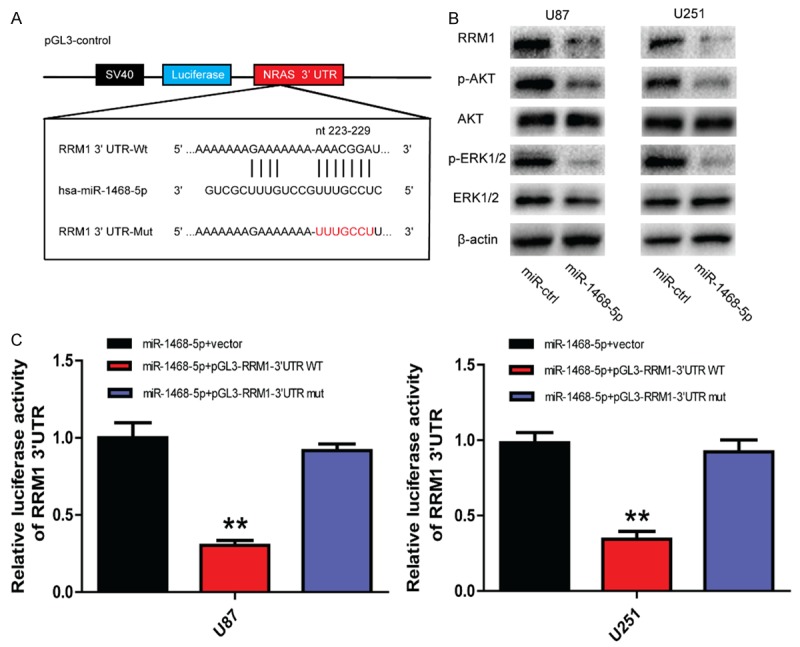
miR-1468-5p directly and negatively regulates RRM1 in glioma cells. A: The miR-1468-5p binding sites in the 3’UTR of RRM1 with the wild type (Wt) and mutated (Mut) sequences highlighted. B: Western blot analysis of RRM1, p-AKT, AKT, p-ERK1/2 and ERK1/2 in U87 and U251 cells after transduction with miR-ctrl or miR-1468-5p. C: U87 and U251 cells were co-transfected with miR-1468-5p and luciferase reporter constructs containing either pGL3-RRM1-3’-UTR-WT or pGL3-RRM1-3’-UTR-Mut; **P<0.01.
A reduction in cell proliferation is often accompanied by changes in cell cycle progression [32]. Therefore, we next performed a cell cycle analysis, which indicated that following miR-1468-5p transfection, there was a marked increase in the G1 fraction and a decrease in the S fraction (Figure 2G). Together, these results suggest that miR-1468-5p may serve as a tumor suppressor in glioma.
RRM1 is a direct target of miR-1468-5p in glioma
It is generally accepted that miRNAs do not cause direct changes in cellular functions, rather they exert their functions by regulating the expression of downstream target genes. Therefore, we searched for miR-1468-5p targets using four algorithms: TargetScan, miRanda, miRDB and miRWalk. According to these algorithms, RRM1, which might be a potential target of miR-1468-5p, attracted our attention. We identified a miR-1468-5p binding site in the 3’UTR of RRM1 using the microRNA algorithms (Figure 3A). Western blot analysis showed that expression of RRM1 protein was downregulated in miR-1468-5p-transfected cells (Figure 3B).
To identify whether RRM1 is a direct target of miR-1468-5p, plasmids containing the potential RRM1 miR-1468-5p binding sites (Wt) or a mutated RRM1 3’UTR were synthesized (Figure 3A). Overexpression of miR-1468-5p inhibited Wt RRM1 reporter activity but not the activity of the mutated reporter construct in both U87 and U251 cells, demonstrating that miR-1468-5p can specifically target the RRM1 3’UTR by binding to the seed sequence (Figure 3C). These data indicated that miR-1468-5p is a post-transcriptional regulator of RRM1 via direct binding to its 3’UTR.
RRM1 is overexpressed in glioma and indicates poor prognosis
RRM1 has been rarely studied in glioma. First, we analyzed RRM1 expression in NBTs and low and high grade glioma tissues using the CGGA and Rembrandt databases. We found that RRM1 levels were upregulated in both disease groups compared with the normal brain group. Moreover, the levels of RRM1 expression in high grade glioma tissues (WHO clinical stage III-IV) were higher than in low grade glioma tissues (WHO clinical stage I-II), indicating that RRM1 expression correlated with glioma malignancy (Figure 4A). The mean OS of the low RRM1 expression group was longer than high RRM1 expression group in GBM, using both the CGGA and Rembrandt databases (Figure 4B). We then chose one representative GBM specimen and one NBT for immunohistochemistry; these results were also consistent with the above database results (Figure 4C).
Figure 4.
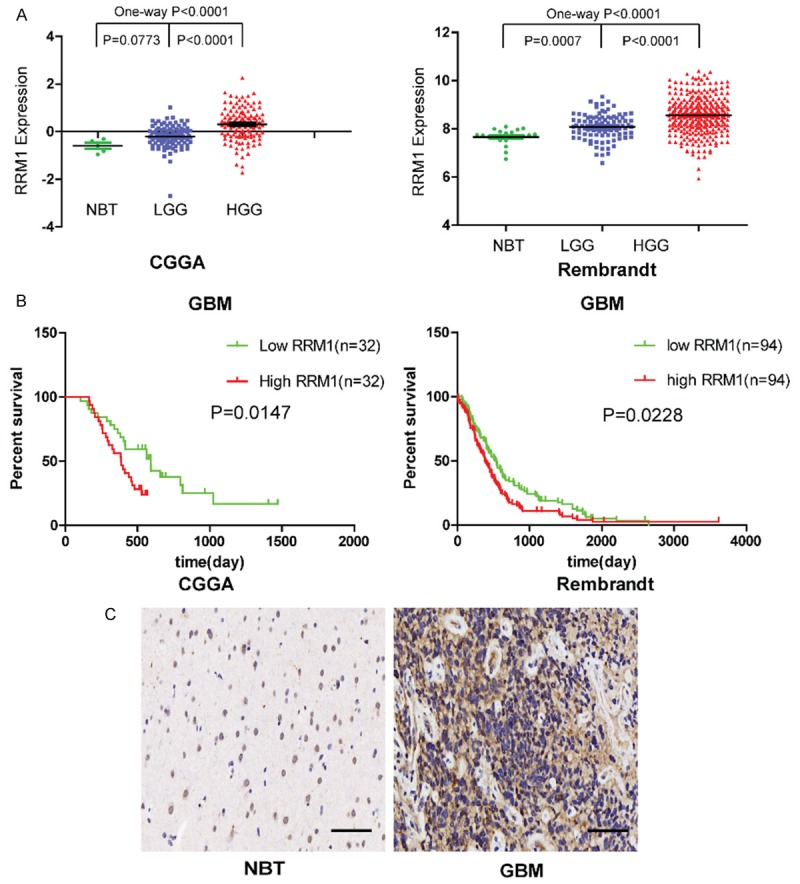
RRM1 is overexpressed in human gliomas and high RRM1 expression is correlated with poor prognosis in GBM patients. A: RRM1 expression in normal brain tissues, low grade glioma tissues and high grade glioma tissues using the CGGA and Rembrandt databases. B: The overall survival of high and low RRM1 expression GBM cases using the CGGA and Rembrandt databases. C: Immunohistochemistry analysis of RRM1 in two representative tissues (one NBT and one GBM specimen). Scale bar: 50 μm.
Silencing RRM1 inhibits glioma cell proliferation and induces a G1/S cell cycle arrest
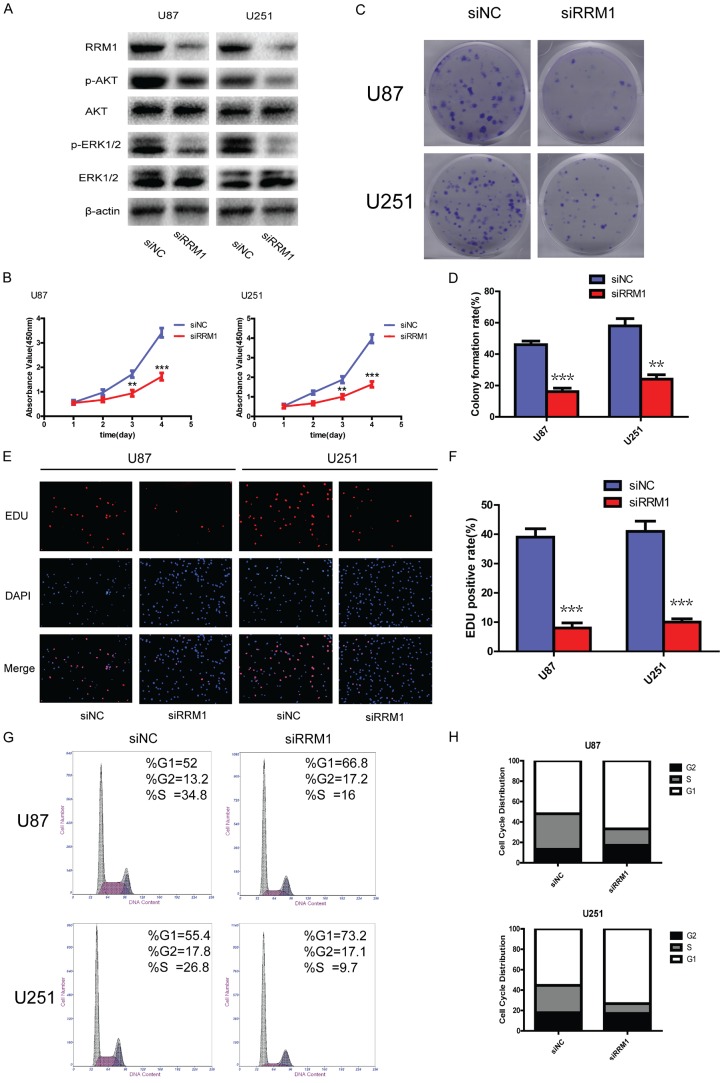
To further confirm the role of RRM1 in glioma cell proliferation and cell cycle progression, we tested whether silencing RRM1 by siRNA could alter proliferation and suppress cell cycle progression in glioma cells. To this end, we transfected siNC and siRRM1 into U87 and U251 cells; western blotting showed effective RRM1 protein down-regulation following manipulation. Following knockdown, there were reduced p-AKT and p-ERK1/2 levels compared with cells stably expressing siNC, while no significant reduction in AKT or ERK1/2 levels was detected (Figure 5A). We also found that silencing RRM1 strongly decreased the proliferation of U87 and U251 cells compared with controls (Figure 5B-F). Moreover, a substantial proportion of the cell cycle arrest in G1/S phase was observed in the siRRM1 group compared with the siNC group (Figure 5G and 5H), suggesting that silencing RRM1 could suppress cell cycle progression, restraining cells in the G1 phase. Together, these data confirm that RRM1 stimulates proliferation and promotes G1/S transition in glioma cells.
Figure 5.
RRM1 knockdown suppressed proliferation and cell cycle progression of glioma cells. A: Western blot showing the levels of RRM1, p-AKT, AKT, p-ERK1/2 and ERK1/2 in U87 and U251 cells after transfection with siRRM1. B: The proliferation ability was determined using CCK-8 assay following 4 days of culture; **P<0.01 and ***P<0.001. C, D: Long-term cell viability was evaluated using the colony formation assay; **P<0.01 and ***P<0.001. E, F: Proliferating cells were examined using the EdU assay; representative images are shown (original magnification: 200×); ***P<0.001. G, H: The cell cycle distributions of U87 and U251 cells transfected with siNC or siRRM1, as analyzed by flow cytometry.
RRM1 reintroduction attenuates the inhibitory effects of miR-1468-5p
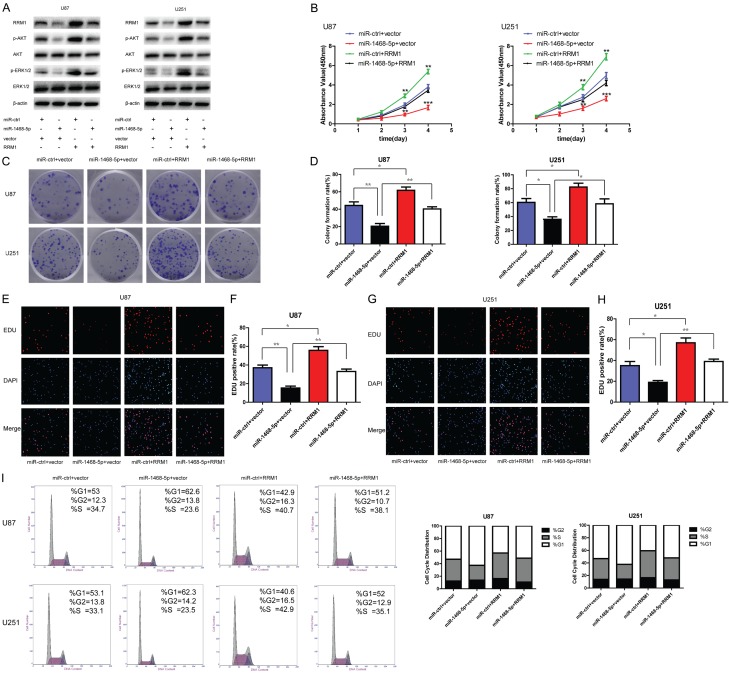
Having demonstrated that RRM1 is a miR-1468-5p target, we further investigated whether RRM1 was a functional miR-1468-5p target by co-transfecting miR-1468-5p lentivirus and RRM1 plasmid into glioma cells. First, we used western blot analysis to determine whether RRM1 was involved in changes induced by miR-1468-5p expression. Western blot analysis proved that compared with the two other groups, RRM1 was significantly downregulated in the miR-1468-5p-transfected group, but upregulated in the RRM1-transfected group (Figure 6A). Subsequently, we examined the effect of miR-1468-5p overexpression on proliferation and cell cycle progression with regards to its target RRM1. Both cell proliferation and cell cycle assay data indicated that the reintroduction of RRM1 antagonized the inhibitory effects of miR-1468-5p (Figure 6B-I). Meanwhile, p-AKT and p-ERK1/2 levels also recovered after exogenous introduction of RRM1 (Figure 6A). These results suggested that RRM1 is a functional miR-1468-5p target in glioma cells.
Figure 6.
RRM1 reintroduction reverses the inhibitory effects of miR-1468-5p. Rescue experiments were performed by introducing or RRM1 overexpression vectors or empty pcDNA3 in the presence or absence of miR-1468-5p in U87 and U251 cells. A: Western blot analysis of RRM1, p-AKT, AKT, p-ERK1/2 and ERK1/2 in the indicated cells. B: CCK8 assay with miR-ctrl- or miR-1468-5p-transduced U87 and U251 cells transfected with vector or RRM1; **P<0.01 and ***P<0.001. C, D: Co-transfected U87 and U251 cells were analyzed by the colony formation assay; *P<0.05 and **P<0.01. E-H: The EdU assay was performed 48 h after co-transfection; *P<0.05 and **P<0.01. I: Flow cytometry was conducted to determine the cell cycle distribution in each group.
miR-1468-5p upregulation impedes tumor growth in vivo
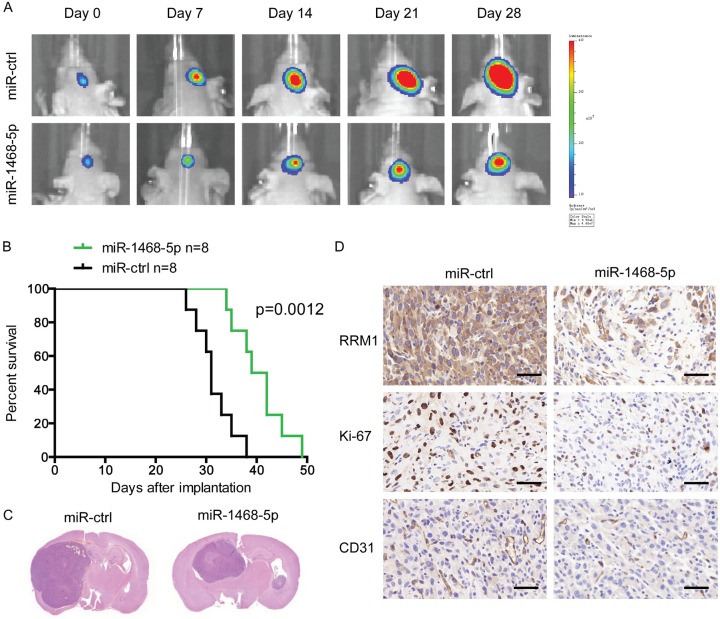
Considering the remarkable glioblastoma-inhibiting effects of miR-1468-5p in vitro, we extended our investigation to examine if miR-1468-5p could impede glioblastoma growth in vivo using nude mice. Before implantation, U87 glioblastoma cells were co-infected with lentiviruses expressing luciferase with miR-ctrl or miR-1468-5p. The intracranial tumor volumes of the miR-1468-5p group were significantly reduced compared with those of the miR-ctrl group. On days 7, 14, 21, and 28 after implantation, the growth of intracranial tumors was significantly inhibited in association with overexpression of miR-1468-5p (Figure 7A). Hematoxylin and eosin staining results were also consistent with the in vitro results (Figure 7C). Survival analysis also demonstrated significantly better outcomes for the animals injected with miR-1468-5p-overexpressing U87 cells (Figure 7B). Furthermore, when miR-1468-5p was upregulated in the U87 cells, it significantly decreased RRM1, Ki-67 and CD31 expression in the tumor tissues, which was consistent with the in vitro results (Figure 7D). Together, these findings demonstrate that miR-1468-5p inhibits the proliferation of glioma cells in vivo.
Figure 7.
miR-1468-5p upregulation inhibits tumor growth and is associated with good prognosis in a murine intracranial glioma xenograft model. A: U87 cells pretreated with a lentivirus with miR-1468-5p or miR-ctrl and a lentivirus containing luciferase were implanted in the brains of nude mice, and tumor formation was assessed by bioluminescence imaging. Changes in the bioluminescence signals were measured on post-implantation days 7, 14, 21 and 28. B: Overall survival was determined by Kaplan-Meier survival curves, and a log-rank test was used to assess the statistical significance of the differences. C: Tissue slices from representative tumors of the two groups were stained with hematoxylin-eosin-saffron. D: Images show representative immunohistochemical staining for RRM1, Ki67 and CD31 in the two groups. Scale bar: 50 μm.
Discussion
miRNA dysregulation is a common feature of human cancers, including glioma [33,34]. miRNAs can function as tumor suppressors or oncogenes, and the expression of more than one-third of the protein-coding genes in the human genome is thought to be controlled by miRNAs [35,36]. In recent years, the expression of tumor-suppressive miRNAs in glioma has been a topic of interest for antineoplastic research, and accumulating evidence has demonstrated the potential of these antineoplastic miRNAs as prognostic indicators and therapeutic targets [37]. Considering that miRNA research has advanced from the identification of an initial association with glioma to the commercial development of miRNA-based therapeutics in less than a decade, the anticipation of significant developments in this field that ultimately improve patient outcomes is reasonable [38]. Several recent reports have confirmed that numerous highly-expressed miRNAs, such as miR-10b [39], miR-21 [40-42], miR-210 [42] and miR-221/222 [27] are predictive of poor prognosis in glioma patients. However, an increasing number of tumor-suppressive miRNAs have also been discovered, including miR-637 [43], miR-663 [44], miR-218 [45], miR-128 [46] and miR-34a [47]. Here, we identified miR-1468-5p as a novel tumor-suppressive miRNA that was rarely reported in any cancers including glioma.
In this study, we found that miR-1468-5p was downregulated in human glioma compared with NBTs. On the basis of bioinformatic analyses, we further predicted RRM1 to be a miR-1468-5p target. We also demonstrated that miR-1468-5p overexpression in glioma cells led to reduced p-Akt and p-ERK1/2 levels via directly targeting the RRM1 3’UTR; and for the first time, we showed that RRM1 was upregulated in glioma specimens and played a pro-tumor role in glioma. As the large subunit of human RNR, RRM1 is involved in cell proliferation and tumor development by supplying dNTPs for DNA synthesis [14]. Abnormal RRM1 expression has been found in several types of cancer [48-52], but contrasting roles of RRM1 have been reported in different human cancers. For example, Gautam et al. suggested that RRM1 acted as a tumor suppressor in lung cancer, as decreased RRM1 expression was significantly associated with metastasis and poor survival [53]. In contrast, RRM1 was upregulated in papillary thyroid carcinoma, where RRM1 levels were positively correlated with aggressiveness [29]. Thus, the role of RRM1 is tumor-specific. Our study is the first to clarify the role of RRM1 in glioma. The results of cell viability and EdU incorporation assays showed that RRM1 contributed to DNA synthesis and proliferation in both U87 and U251 cells, which was consistent with previous studies in other cancers [54,55]. Moreover, we observed that siRNA-mediated RRM1 knockdown had a negative effect on proliferation and cell cycle progression in vitro, which was similar to the effects of restoring miR-1468-5p, whereas RRM1 overexpression blocked the inhibitory effects of miR-1468-5p on proliferation and cell cycle progression. Maintenance of a balanced dNTP pool is fundamental for cellular function because the consequences of imbalances in the substrates for DNA synthesis and repair include mutagenesis and cell death. RNR expression and activity is therefore tightly regulated both in cell cycle and the DNA damage checkpoints [22,56]. Targeted inhibition of RRM1 depletes dNTPs and could lead to aberrant replication forks, G1/S checkpoint activation and cell cycle arrest, which is consistent with the results of our study that RRM1 inhibition caused G1/S arrest of U87 and U251 cells [22]. Our findings also provide the first evidence that RRM1 is a predominant mediator of miR-1468-5p-induced tumor-suppressive function.
We also confirmed that miR-1468-5p overexpression in glioma cells decreased both p-AKT and p-ERK1/2 levels, which play vital functions in regulating tumorigenesis. Furthermore, RRM1 overexpression in stable miR-1468-5p-expressing cell lines restored p-AKT and p-ERK1/2 levels. As a consequence, restoring RRM1 expression can partially, or even totally, restore the miR-1468-5p-induced inhibition of proliferation and cell cycle progression. These results show that miR-1468-5p is a tumor suppressor that inhibits tumor growth and causes cell cycle arrest via targeting RRM1.
In summary, we found that miR-1468-5p was significantly downregulated in glioma, and reduced miR-1468-5p expression was associated with poor survival. Moreover, we demonstrated, for the first time, that the role of the miR-1468-5p/RRM1 axis was to regulate glioma proliferation and cell cycle progression. This newly identified miR-1468-5p/RRM1 link provides new insight into the mechanisms underlying glioma development, and suggests that targeting the miR-1468-5p/RRM1 axis may represent a promising therapeutic strategy for glioma treatment. Nevertheless, further studies are needed to determine the exact mechanism of decreased miR-1468-5p expression during the progression of glioma and to further explore other possible targets of miR-1468-5p in glioma. Additionally, a large cohort study, incorporating RRM1 expression and function should also be further investigated.
Acknowledgements
This work was supported by grants from the Research Special Fund For Public Welfare Industry of Health (201402008), the National Key Research and Development Plan (2016YFC0902500), National Natural Science Foundation of China (81672501, 81472362, 81302184), Jiangsu Province’s Natural Science Foundation (20131019, 20151585), the Program for Advanced Talents within Six Industries of Jiangsu Province (2015-WSN-036, 2016-WSW-013), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–342. doi: 10.1007/978-1-60327-492-0_14. [DOI] [PubMed] [Google Scholar]
- 3.Ostrom QT, Gittleman H, Stetson L, Virk SM, Barnholtz-Sloan JS. Epidemiology of gliomas. Cancer Treat Res. 2015;163:1–14. doi: 10.1007/978-3-319-12048-5_1. [DOI] [PubMed] [Google Scholar]
- 4.Marumoto T, Saya H. Molecular biology of glioma. Adv Exp Med Biol. 2012;746:2–11. doi: 10.1007/978-1-4614-3146-6_1. [DOI] [PubMed] [Google Scholar]
- 5.Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang X, Jiang C, Kang C, Li X, Chen L, Qiu X, Wang W, Li W, Yao Y, Li S, Li S, Wu A, Sai K, Bai H, Li G, Chen B, Yao K, Wei X, Liu X, Zhang Z, Dai Y, Lv S, Wang L, Lin Z, Dong J, Xu G, Ma X, Cai J, Zhang W, Wang H, Chen L, Zhang C, Yang P, Yan W, Liu Z, Hu H, Chen J, Liu Y, Yang Y, Wang Z, Wang Z, Wang Y, You G, Han L, Bao Z, Liu Y, Wang Y, Fan X, Liu S, Liu X, Wang Y, Wang Q. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375:263–273. doi: 10.1016/j.canlet.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 6.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and function. Thromb Haemost. 2012;107:605–610. doi: 10.1160/TH11-12-0836. [DOI] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 11.Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotaja N. MicroRNAs and spermatogenesis. Fertil Steril. 2014;101:1552–1562. doi: 10.1016/j.fertnstert.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Imbar T, Eisenberg I. Regulatory role of microRNAs in ovarian function. Fertil Steril. 2014;101:1524–1530. doi: 10.1016/j.fertnstert.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 14.Garg D, Cohen SM. miRNAs and aging: a genetic perspective. Ageing Res Rev. 2014;17:3–8. doi: 10.1016/j.arr.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Dogini DB, Pascoal VD, Avansini SH, Vieira AS, Pereira TC, Lopes-Cendes I. The new world of RNAs. Genet Mol Biol. 2014;37:285–293. doi: 10.1590/s1415-47572014000200014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med. 2006;79:131–140. [PMC free article] [PubMed] [Google Scholar]
- 17.Esquela-Kerscher A, Slack FJ. OncomirsmicroRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 18.Ruvkun G. Clarifications on miRNA and cancer. Science. 2006;311:36–37. doi: 10.1126/science.311.5757.36d. [DOI] [PubMed] [Google Scholar]
- 19.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 20.Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 21.Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19:4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 23.Cory JG, Sato A. Regulation of ribonucleotide reductase activity in mammalian cells. Mol Cell Biochem. 1983;53-54:257–266. doi: 10.1007/BF00225258. [DOI] [PubMed] [Google Scholar]
- 24.Cooperman BS, Kashlan OB. A comprehensive model for the allosteric regulation of Class Ia ribonucleotide reductases. Adv Enzyme Regul. 2003;43:167–182. doi: 10.1016/s0065-2571(02)00035-3. [DOI] [PubMed] [Google Scholar]
- 25.Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 2003;42:1696–1706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- 26.Kim SH, Park ER, Joo HY, Shen YN, Hong SH, Kim CH, Singh R, Lee KH, Shin HJ. RRM1 maintains centrosomal integrity via CHK1 and CDK1 signaling during replication stress. Cancer Lett. 2014;346:249–256. doi: 10.1016/j.canlet.2013.12.031. [DOI] [PubMed] [Google Scholar]
- 27.Parker NJ, Begley CG, Fox RM. Human R1 subunit of ribonucleotide reductase (RRM1): 5’ flanking region of the gene. Genomics. 1994;19:91–96. doi: 10.1006/geno.1994.1017. [DOI] [PubMed] [Google Scholar]
- 28.Aye Y, Li M, Long MJ, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34:2011–2021. doi: 10.1038/onc.2014.155. [DOI] [PubMed] [Google Scholar]
- 29.Fang Z, Song R, Gong C, Zhang X, Ren G, Li J, Chen Y, Qiu L, Mei L, Zhang R, Xiang X, Chen X, Shao J. Ribonucleotide reductase large subunit M1 plays a different role in the invasion and metastasis of papillary thyroid carcinoma and undifferentiated thyroid carcinoma. Tumour Biol. 2016;37:3515–3526. doi: 10.1007/s13277-015-4175-7. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Wu W, Wang HW, Wang S, Chen Y, Zhang X, Yang J, Zhao S, Ding HF, Lu D. Analysis of specialized DNA polymerases expression in human gliomas: association with prognostic significance. Neuro Oncol. 2010;12:679–686. doi: 10.1093/neuonc/nop074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Luca A, Maiello MR, D’Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets. 2012;16(Suppl 2):S17–27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 34.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 35.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 36.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 37.Yan W, Li R, Liu Y, Yang P, Wang Z, Zhang C, Bao Z, Zhang W, You Y, Jiang T. MicroRNA expression patterns in the malignant progression of gliomas and a 5-microRNA signature for prognosis. Oncotarget. 2014;5:12908–12915. doi: 10.18632/oncotarget.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tivnan A, McDonald KL. Current progress for the use of miRNAs in glioblastoma treatment. Mol Neurobiol. 2013;48:757–768. doi: 10.1007/s12035-013-8464-0. [DOI] [PubMed] [Google Scholar]
- 39.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM, Tannous BA, Krichevsky AM. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 41.Yang CH, Yue J, Pfeffer SR, Fan M, Paulus E, Hosni-Ahmed A, Sims M, Qayyum S, Davidoff AM, Handorf CR, Pfeffer LM. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3) J Biol Chem. 2014;289:25079–25087. doi: 10.1074/jbc.M114.593863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbano R, Palumbo O, Pasculli B, Galasso M, Volinia S, D’Angelo V, Icolaro N, Coco M, Dimitri L, Graziano P, Copetti M, Valori VM, Maiello E, Carella M, Fazio VM, Parrella P. A miRNA signature for defining aggressive phenotype and prognosis in gliomas. PLoS One. 2014;9:e108950. doi: 10.1371/journal.pone.0108950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Que T, Song Y, Liu Z, Zheng S, Long H, Li Z, Liu Y, Wang G, Liu Y, Zhou J, Zhang X, Fang W, Qi S. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34:4952–4963. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, Chen C, Zhang X, Liu Q, Xu JL, Zhang HR, Yao XH, Jiang T, He ZC, Ren Y, Cui W, Xu C, Liu L, Cui YH, Yu SZ, Ping YF, Bian XW. Primatespecific miR-663 functions as a tumor suppressor by targeting PIK3CD and predicts the prognosis of human glioblastoma. Clin Cancer Res. 2014;20:1803–1813. doi: 10.1158/1078-0432.CCR-13-2284. [DOI] [PubMed] [Google Scholar]
- 45.Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin W, Zhang Y. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stemlike cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013;73:6046–6055. doi: 10.1158/0008-5472.CAN-13-0358. [DOI] [PubMed] [Google Scholar]
- 46.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Specks J, Lecona E, Lopez-Contreras AJ, Fernandez-Capetillo O. A single conserved residue mediates binding of the ribonucleotide reductase catalytic subunit RRM1 to RRM2 and is essential for mouse development. Mol Cell Biol. 2015;35:2910–2917. doi: 10.1128/MCB.00475-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tantraworasin A, Saeteng S, Lertprasertsuke N, Patumanond J, Arreyakajohn N, Kasemsarn C. The prognostic value of ERCC1 and RRM1 gene expression in completely resected non-small cell lung cancer: tumor recurrence and overall survival. Cancer Manag Res. 2013;5:327–36. doi: 10.2147/CMAR.S52073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulker M, Duman BB, Sahin B, Gumurdulu D. ERCC1 and RRM1 as a predictive parameter for non-small cell lung, ovarian or pancreas cancer treated with cisplatin and/or gemcitabine. Contemp Oncol (Pozn) 2015;19:207–213. doi: 10.5114/wo.2015.52656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie H, Jiang W, Jiang J, Wang Y, Kim R, Liu X, Liu X. Predictive and prognostic roles of ribonucleotide reductase M1 in resectable pancreatic adenocarcinoma. Cancer. 2013;119:173–181. doi: 10.1002/cncr.27715. [DOI] [PubMed] [Google Scholar]
- 52.Yuan ZJ, Zhou WW, Liu W, Wu BP, Zhao J, Wu W, He Y, Yang S, Su J, Luo Y. Association of GSTP1 and RRM1 polymorphisms with the response and toxicity of gemcitabine-cisplatin combination chemotherapy in chinese patients with non-small cell lung cancer. Asian Pac J Cancer Prev. 2015;16:4347–4351. doi: 10.7314/apjcp.2015.16.10.4347. [DOI] [PubMed] [Google Scholar]
- 53.Gautam A, Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66:6497–6502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- 54.Qi H, Lou M, Chen Y, Liu X, Chen N, Shan J, Ling Z, Shen J, Zhu L, Yen Y, Zheng S, Shao J. Non-enzymatic action of RRM1 protein upregulates PTEN leading to inhibition of colorectal cancer metastasis. Tumour Biol. 2015;36:4833–4842. doi: 10.1007/s13277-015-3137-4. [DOI] [PubMed] [Google Scholar]
- 55.Reid G, Wallant NC, Patel R, Antonic A, Saxon-Aliifaalogo F, Cao H, Webster G, Watson JD. Potent subunit-specific effects on cell growth and drug sensitivity from optimised siRNA-mediated silencing of ribonucleotide reductase. J RNAi Gene Silencing. 2009;5:321–330. [PMC free article] [PubMed] [Google Scholar]
- 56.Cerqueira NM, Fernandes PA, Ramos MJ. Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents. Recent Pat Anticancer Drug Discov. 2007;2:11–29. doi: 10.2174/157489207779561408. [DOI] [PubMed] [Google Scholar]