Abstract
Though it is widely known that hepatitis B virus X protein (HBx) is involved in the progression of hepatocellular carcinoma (HCC), the underlying mechanisms are not entirely clear. In recent years, metastasis associated with lung adenocarcinoma transcript 1 (MALAT1), which is an oncogenic long non-coding RNA (lncRNA), has been proved to be associated with many kinds of tumors, including liver cancer. In this study, we demonstrated that MALAT1 was involved in the HBx-mediated hepatocarcinogenesis. Firstly, we found that expression of MALAT1 was strongly up-regulated in HCC tissues and was directly proportional to the expression of HBx. Moreover, in HBx transfected LO2 and HepG2 cells, MALAT1 was also up-regulated compared with non-transfected cells. Then, we observed up-regulated MALAT1 in HepG2 cells could promote cell invasion and migration, whereas knockdown of MALAT1 in HBx-expressing hepatic cells (HepG2-HBx) resulted in a markedly inhibition of cell invasion and migration both in vitro and in vivo. To further obtain a deeper understanding of the effect of MALAT1, we took latent transforming growth factor β-binding protein 3 (LTBP3) into account by using several assays such as RNA interference, luciferase, transwell and wound healing. Results showed that MALAT1 could promote tumor growth and metastasis by activating LTBP3, which could also be up-regulated by HBx. Meanwhile, the similar results were detected in nude mice. These findings could demonstrate an important mechanism of hepatocarcinogenesis through the signaling of HBx-MALAT1/LTBP3 axis, and may give a potential target for treatment of HCC.
Keywords: HBx, MALAT1, LTBP3, hepatocellular carcinoma, cell metastasis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies as well as the second leading cause of worldwide cancer related death [1-3]. Though treatment strategies like chemotherapy have significantly improved the survival rate of the patients, many problems such as the high incidence of cancer metastasis still exist. One of the major risk factor of HCC is chronic hepatitis B virus (HBV). Among various cancer related proteins, hepatitis B virus X protein (HBx) is considered as the key protein in the process of HBV-induced hepatocarcinogenesis [4,5]. It has been detected with high frequency in patients who are suffering from liver cancer, chronic hepatitis and cirrhosis. HBx plays a multifunctional role in the development of HBV-related HCC, such as affecting cell growth, regulating cell apoptosis, and inhibiting nucleotide excision repair of damaged cellular DNA [6]. However, more detailed and deeper insights of HBx in HCC are still needed.
LncRNAs, which refer a class of non-protein-coding RNA transcripts longer than 200 nucleotides and do not contain an open reading frame [7], are involved in various physiological process, including cell proliferation, apoptosis, metastasis and invasion [8]. Several studies showed that lncRNAs are acted as oncogenes or tumor suppressors and closely related to HBx. With the development of high-throughput sequencing and bioinformatics, more and more lncRNAs have been discovered and are found to be abnormally expressed in HCC, such as PCNA-AS1 (antisense to PCNA) [9], HEIH (high expression in HCC) [10], MVIH (microvascular invasion in HCC) [11], HULC (highly up-regulated in liver cancer) [12] and metastasis associated with lung adenocarcinoma transcript 1 (MALAT1) [13]. MALAT1 is an oncogenic lncRNA which can stimulate cancer development, regulate cancer cell radio-sensitivity and chemo-sensitivity by diversity mechanisms, like acting as miRNA sponges, stimulating autophagy and enhancing epithelial-to-mesenchymal transition [14,15]. Studies show that MALAT1 is up-regulated in HCC, however either the deeper mechanisms or whether HBx is involved in the progress is still not clear [16].
Latent transforming growth factor β-binding proteins (LTBPs) are large secreted glycoproteins that regulate the bioavailability of transforming growth factor β (TGFβ) [17]. Among the four subtypes, LTBP1, LTBP3 and LTBP4 bind covalently to TGF-β and are needed for the correct folding and the secretion of TGF-β [18]. Studies showed that LTBPs were involved in lots of diseases such as esophageal cancer [19], glaucoma and pseudoexfoliation syndrome [20], malignant mesothelioma [21] and so on. Recently, several studies demonstrated that MALAT1 could positively regulate LTBP3 transcription in mesenchymal stem cells by recruiting Sp1 to the LTBP3 promoter [22]. However, up to now, no study focuses on whether MALAT1 regulate HBV-induced hepatocarcinogenesis through activation of LTBP3.
In the present study, we aimed to study the role of MALAT1 in the development of hepatocellular carcinoma mediated by HBx. Our results showed that MALAT1 could promote cell metastasis of HCC mediated by HBx via up-regulating LTBP3 expression. These findings may provide new insights to the roles of MALAT1 in the development of HCC mediated by HBx.
Methods and materials
Cell culture and tissues
The LO2, LO2-HBx, and HepG2 cell lines were obtained from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS, Hyclone) and 1% PS (100 IU/ml penicillin, 100 μg/ml streptomycin). All cells were incubated in a humidified atmosphere at 37°C with 5% CO2.
The 20 paired HBV-related HCC and the adjacent liver tissue samples used in this study were obtained from patients who underwent radical resection at Xiangya Hospital (Central South University, Changsha, China). All tissues were confirmed histologically. This study was approved by the Ethics Committee of Xiangya Hospital.
Construction of plasmid, siRNA and cell transfection
LO2 and HepG2 cells were infected with the Lv-HBx and Lv-control viruses (GeneChem, Shanghai, China). For constructing MALAT1 and LTBP3, the full length of MALAT1 and LTBP3 were amplified and cloned into pcDNA-3.1 vectors (Invitrogen, USA), respectively [23]. Sh-MALAT1, si-HBx and si-LTBP3 were used for further studies, which were synthesized by GenePharma (Shanghai, China). The sequences of shRNA and siRNA were listed in Table 1. Lipofectamine 2000 (Invitrogen, USA) was used to transfect oligonucleotides and constructs into cell lines according to the manufacturer’s instructions. The infection efficiency was confirmed by qRT-PCR or western blot 48 h after transfection.
Table 1.
The sequences for siRNA and qRT-PCR prime
| Name | Sequence |
|---|---|
| Sh-MALAT1 | 5’-CACCGCTGTGGAGTTCTTAAATATCTTCAAGAGAGATATTTAAGAACTCCACAGCTTTTTTG-3’ |
| 5’-GATCCAAAAAAGCTGTGGAGTTCTTAAATATCTCTCTTGAAGATATTTAAGAACTCCACAGC-3’ | |
| Si-LTBP3 | 5’-GCAAGCAGGGCUUCUACUATT-3’ |
| 5’-UAGUAGAAGCCCUGCUUGCTT-3’ | |
| HBx Forward | 5’-CGTCCTTTGTTTACGTCCCG-3’ |
| HBx Reverse | 5’-GCAGATGAGAAGGCACAGAC-3’ |
| MALAT1 Forward | 5’-AAAGCAAGGTCTCCCCACAAG-3’ |
| MALAT1 Reverse | 5’-GGTCTGTGCTAGATCAAAAGGCA-3’ |
| LTPB3 Forward | 5’-CGGAACGGAGTGTGTGAGAA-3’ |
| LTPB3 Reverse | 5’-CTCGTCCACGTCCATCTCT-3’ |
| GAPDH Forward | 5’-GCTGTAGCCAAATCGTTGT-3’ |
| GAPDH Reverse | 5’-CCAGGTGGTCTCCTCTGA-3’ |
Dual luciferase reporter assay
The LTBP3 promoter region (-1500/+200) was amplified and sub-cloned into the pGL3-basic luciferase vector (Promega, USA). LO2 and HepG2 cells cultured in 24-well plates were co-transfected with luciferase reporter plasmids and MALAT1 mimics/sh-MALAT1. After 48 h of transfection, luciferase activity was measured by using the Bright-Glo™ Luciferase Assay System (Promega, Cat. #E2610). All reactions were repeated in triplicate through at least three independent experiments.
RNA extraction and qRT-PCR
Detailed procedures used for total RNA extraction and were described elsewhere [24]. Primers used in qRT-PCR were listed in Table 1.
Western blotting (WB)
Western blotting was used to test the expression of MALAT1, HBx, LTPB3; and EMT related proteins E-cadherin, N-cadherin, vimentin. β-actin was used as a control. Detailed procedures of WB were described elsewhere [23].
In vitro cell invasion and migration assays
Transwell assay was used to determine cell invasion as described elsewhere [23]. Briefly, cell suspension containing 5×105 cells/ml was prepared in serum-free media, followed with 300 μl cell suspension. Then, 500 μl of DMEM containing 10% FBS was added and cells were incubated for 24 h, stained with 0.1% crystal violet, and then counted and photographed. Cell migration ability was tested using the scratch wound assay and then seeded into 6-well plates for 24 h. Cell layers were scratched using a 200 μl pipette tip to form wound gaps, and then maintained in DMEM with 10% FBS. The cells were photographed at 0 and 48 h to record the wound width. All experiments were independently repeated in triplicate.
Tumor growth analysis in vivo
Male BALB/C nude mice (4-6 weeks) were purchased from the Animal Center of Silaike Jingda (Hunan, China). All animals were housed in micro-isolator cages with free access to food and water according to the Guide for the Care and Use of Laboratory Animals. The whole study was approved by the Institutional Animal Care Committee of Third Xiangya Hospital. HepG2-HBx cells (1×107/ml, 0.1 ml) transfected with shMALAT1 (or siLTBP3) and sh/si-ctrl vector were subcutaneously injected into the left back and right back of 4-5 per group mice, respectively. The sizes of tumors were recorded every 5 days and all mice were sacrificed 1 month after injection. The tissues were collected, photographed and the weights of tumors were measured.
Statistical analysis
The measurement data was expressed by mean ± SD. Independent continuous variables was compared using the t-test and categorical data was compared using the chi square test or Fisher exact test. When P-value was less than 0.05 it was considered to be statistically significant. All calculations were made using SPSS 18.0.
Results
HBx could up-regulate the expression of MALAT1 in hepatic tissues and cells
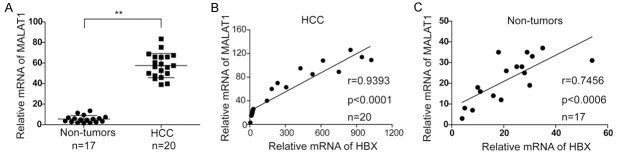
To examine whether MALAT1 was regulated by HBx, we detected the expression levels of HBx and MALAT1 in HCC (n=20) and non-tumorous (n=17) tissues by qRT-PCR. Results showed that the expression levels of MALAT1 was significantly increased in HCC tissues compared with non-tumorous tissues (Figure 1A). Meanwhile, we found that the mRNA levels of MALAT1 was positively correlated with HBx, indicating that HBx infection would up-regulate the expression of MALAT1 (Figure 1B and 1C). Thus, we constructed cell lines which could stably express HBx and found that MALAT1 was significantly up-regulated in cells stably expressing HBx (Figure 2A and 2B).
Figure 1.
HBx could up-regulate the expression of MALAT1 in hepatic tissues and cells. A. Expressions of HBx and MALAT1 in HCC and non-tumorous tissues; B. Relevance between HBx and MALAT1 in HCC tissues; C. Relevance between HBx and MALAT1 in non-tumorous tissues.
Figure 2.
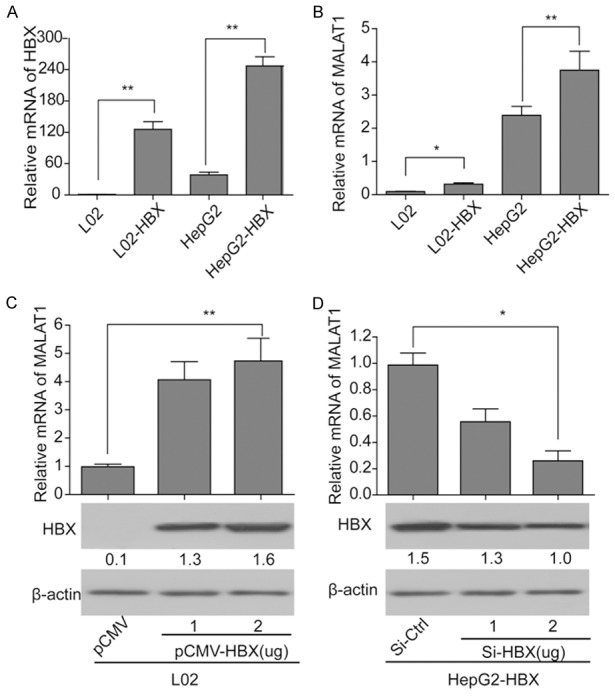
HBx could up-regulate the expression of MALAT1 in hepatic cells. (A, B) Expressions of HBx (A) and MALAT1 (B) in the four cell lines, LO2, LO2-HBx, HepG2 and HepG2-HBx; (C) Influence of over expression of HBx on MALAT1 in LO2 cells; (D) Influence of silencing of HBx on MALAT1 in HepG2-HBx cells. *P<0.05 and **P<0.01.
To further investigate the influence of HBx on MALAT1 in LO2 cells, pcDNA3.1-HBx was transfected into LO2 cells under concentrations of 1 μg/ml and 2 μg/ml respectively. Results of qRT-PCR and western blotting showed that expression of MALAT1 was significantly increased with the over expression of HBx, and the effect was dose-dependent (Figure 2C). Then si-HBx of different concentrations (1 μg/ml and 2 μg/ml) and si-ctrl was transfected into HepG2-HBx cells respectively. Results showed that when treated with si-HBx, expression of MALAT1 in HepG2-HBx cells was significantly decreased and the effect was also dose-dependent (Figure 2D). These results demonstrated that HBx in hepatocellular carcinoma cells could up-regulate the expression of MALAT1, which meant the expression of MALAT1 would significantly increase with the increasing expression of HBx. And the effect was in a dose dependent manner.
Influence of MALAT1 on cell invasion and migration in vitro
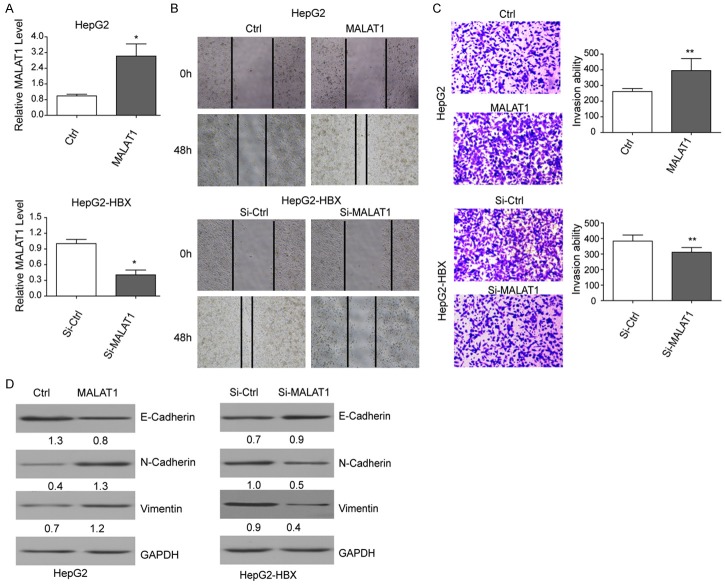
To further investigate the influence of MALAT1 on cell invasion and migration, MALAT1 over expression model was established through HepG2 cells transfected with pcDNA3.1-MALAT1. Also si-MALAT1 and si-control were transfected into HepG2-HBx to establish the down expression model of MALAT1. Results showed that expression of MALAT1 treated by pcDNA3.1-MALAT1 in HepG2 cells was significantly higher than that in the control cells (Figure 3A). And when treated with si-MALAT1, expression of MALAT1 in HepG2-HBx cells decreased significantly (Figure 3A). These results indicated the successful establishment of MALAT1 over/down-expression cells.
Figure 3.
MALAT1 could promote cell invasion and migration in vitro. A. Expression of MALAT1 in HepG2 cells treated with pcDNA3.1-MALAT1 and in HepG2-HBx cells treated with si-MALAT1 and si-ctrl, respectively; B. Cell migration for HepG2 HepG2-HBx cells when were transfected with pcDNA3.1-MALAT1 and Si-MALAT1 alone, pcDNA3.1 and si-ctrl as control, respectively; C. Cell invasion for HepG2 and HepG2-HBx cells when were transfected with pcDNA3.1-MALAT1 and si-MALAT1 alone, pcDNA3.1 and si-ctrl as control, respectively. D. Western blotting analysis of EMT maker proteins E-cadherin, N-cadherin, vimentin in different cell lines.
The above models were used in the in vitro assay of cell invasion and migration. Results showed that HBx could significantly enhance the cell migration of HepG2 cells compared with the HepG2-HBx cells. Additionally, cell migration was apparently promoted when MALAT1 was over expressed and was significantly inhibited when MALAT1 decreased (Figure 3B). Meanwhile, we found that the invasion ability of HepG2 cells was enhanced when HBx and MALAT1 were over expressed, but the invasion ability of HepG2-HBx cells was decreased when HBx was knockdown (Figure 3C). Western blotting results showed that EMT related protein levels of N-cadherin and vimentin significantly increased but level of E-cadherin significantly decreased when MALAT1 was over-expressed in the HepG2 cells. Meanwhile, expression of N-cadherin and vimentin also decreased with the decrease of MALAT1, while expression of E-cadherin increased when expression of MALAT1 was reduced in HepG2-HBx cells (Figure 3D). These results suggested that HBx and MALAT1 could enhance the cell invasion and migration of HCC cells in vitro.
MALAT1 could promote tumor growth in vivo
To further observe the effects of MALAT1 in tumor growth, we took in vivo xenograft model experiment. As shown in Figure 4A-C, silencing MALAT1 (si-MALAT1) significantly decreased the tumor volumes and weights compared with the control (si-NC). Meanwhile qRT-PCR confirmed that the expression levels of MALAT1 were down-regulated in the si-MALAT1 tumor tissues (Figure 4D). This finding suggested that MALAT1 could promote tumor growth in vivo.
Figure 4.
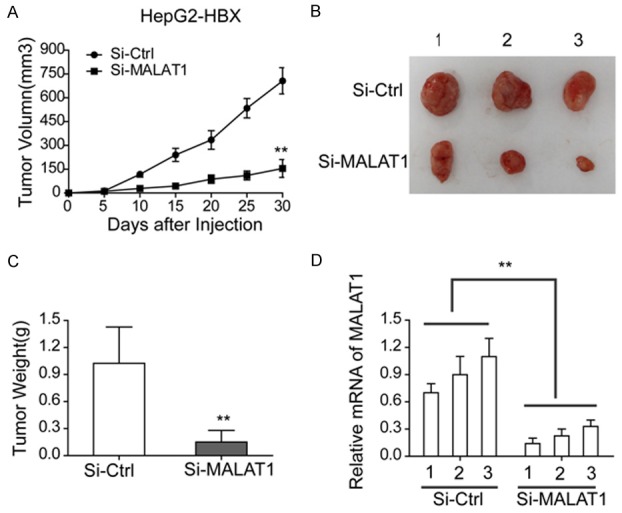
MALAT1 could promote tumor growth in vivo. A, B. Tumor volumes and size of mice treated with si-MALAT1 HepG2-HBx cells and the control cells, respectively; C. Tumor weights of mice treated with si-MALAT1 HepG2-HBx cells and the control cells, respectively; D. Expression of MALAT1 in mice treated with si-MALAT1 HepG2-HBx cells and the control cells, respectively.
MALAT1 could up-regulate the expression of LTBP3
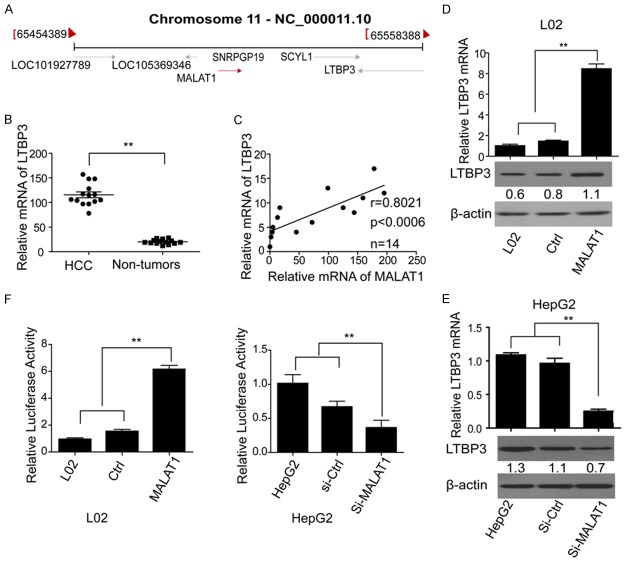
To study the possible mechanism of the above influence of MALAT1 in HCC, firstly, we analyzed the information of MALAT1 from NCBI gene database (www.ncbi.nlm.nih.gov/gene). Results showed that MALAT1 located at 11q13.1., which was 32.0 kb away from LTBP3 and possessed opposite transcriptional direction with it (Figure 5A). Then, we confirmed that the expression of LTBP3 was significantly higher in HCC tissues than in non-tumor tissues (Figure 5B) and was positively correlated with the expression of MALAT1 (Figure 5C). Similar results were confirmed in LO2 and HepG2 cells. As showed in Figure 5D and 5E, expression of LTBP3 significantly increased when MALAT1 was over expressed in LO2 cells (Figure 5D). However, when HepG2 cells were transfected with si-MALAT1, expression of LTBP3 significantly decreased compared with the control groups (Figure 5E). Then, dual luciferase assay showed that the over-expression of MALAT1 could increase the promoter activity of LTBP3 in LO2 cells, whereas knockdown of MALAT1 led to an obvious decrease of LTBP3 promoter activity in HepG2 cells (Figure 5F). Thus, we concluded that MALAT1 was able to up-regulate LTBP3 in hepatoma cells.
Figure 5.
MALAT1 could up-regulate the expression of LTBP3. A. Location of MALAT1 and LTBP3 analyzed by NCBI gene database; B. Expression of LTBP3 in HCC and non-tumorous tissues; C. Relevance between MALAT1 and LTBP3; D. Expression of LTBP3 in LO2 cells treated and untreated with MALAT1, respectively; E. Expression of LTBP3 in HepG2 cells treated with si-MALAT1 and the controls; F. Luciferase activity of HepG2 cells treated with si-MALAT1 and the controls; and LO2 cells treated with pcDNA3.1-MALAT1 and pcDNA3.1.
MALAT1 could promote tumor growth and metastasis by up-regulating LTBP3
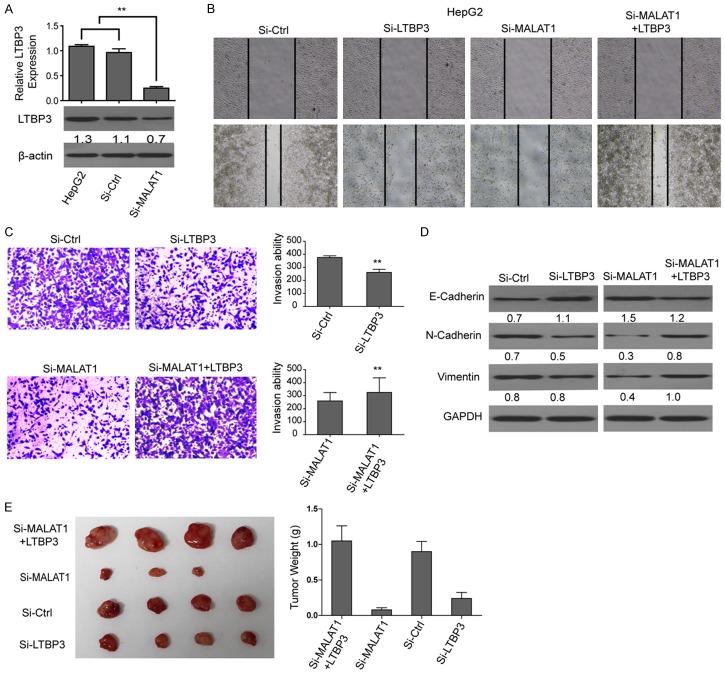
To investigate the mechanism of the promotion effect of MALAT1 on tumor growth and metastasis, we knocked down the LTBP3 in HepG2 cells (si-ctrl as the control, Figure 6A). Then, we observed that silencing LTBP3 significantly inhibited cell migration and invasion (Figure 6B and 6C). What’s more, when transfected with both si-MALAT1 and pcDNA3.1-LTBP3 in HepG2 cell line, cell migration and invasion ability were also enhanced significantly compared with cells only transfected with si-MALAT1, which demonstrated that LTBP3 was required for the MALAT1-enhanced cell invasion and migration of HepG2 (Figure 6B and 6C). Besides, western blotting results of EMT maker proteins E-cadherin, N-cadherin, vimentin showed that when silencing LTBP3, the expression of N-cadherin and vimentin decreased and expression of E-cadherin increased (Figure 6D). These results could further demonstrate that the promotion effect of MALAT1 on tumor growth and metastasis was through the regulation of LTBP3.
Figure 6.
MALAT1 could promote tumor growth and metastasis by up-regulating LTBP3. A. Expression of LTBP3 in different cell lines. B, C. Cell invasion and migration assay of HepG2 cells treated with si-LTBP3 (HepG2 cells treated with si-Ctrl as the control); and HepG2 cells treated with si-MALAT1 and si-MALAT1+pcDNA3.1-LTBP3. D. Western blotting analysis of EMT maker proteins E-cadherin, N-cadherin, vimentin in the above cells. E. Tumor growth assay for different group of mice treated by the above cells.
Furthermore, tumorigenicity assay showed that when LTBP3 was silenced in HepG2 cells, the tumor volumes and weights were significantly decreased compared with the control. Interestingly, the tumorigenicity inhibitory function of si-MALAT1 was partially attenuated by over-expressed LTBP3 (Figure 6E and 6F).
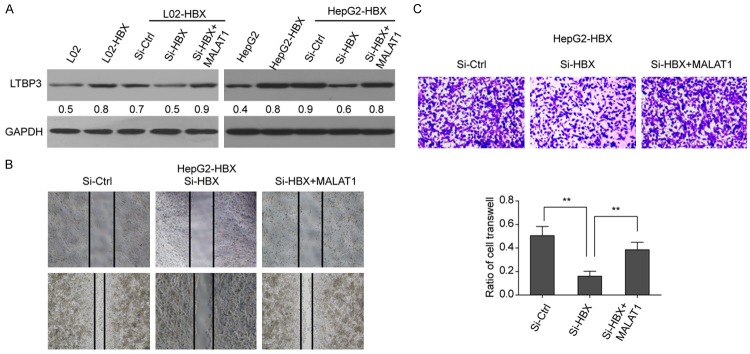
HBx enhanced cell metastasis through up-regulating LTBP3
Relationship between HBx and LTBP3 in the process of cell metastasis was also studied. si-HBx, si-LTBP3, si-HBx+LTBP3 and si-ctrl were used to transfect both LO2-HBx and HepG2-HBx cell lines respectively. Western blotting results showed that LTBP3 was significantly increased in LO2-HBx (or HepG2-HBx) cells compared with LO2 (or HepG2) cells. Meanwhile, silencing HBx in LO2-HBx and HepG2-HBx cells resulted in the decrease of LTBP3. Interestingly, the over-expression of MALAT1 was able to rescue the LTBP3 expression levels in both LO2-HBx and HepG2-HBx cells transfected with si-HBx (Figure 7A). The result suggested that HBx could up-regulate LTBP3 via MALAT1. Moreover, we found that silencing HBx could block the cell migration and invasion in HepG2-HBx cells, whereas it was partially rescued by the over-expression of MALAT1 (Figure 7B and 7C). Thus, we concluded that elevated MALAT1 by HBx could promote the metastasis of hepatoma cells through up-regulation of LTBP3.
Figure 7.
HBx enhanced cell metastasis through up-regulating LTBP3. A. Expression of LTBP3 in different cell lines, LO2-HBx and HepG2-HBx cell lines treated with si-HBx, si-HBx+MALAT1, si-ctrl respectively and untreated LO2, LO2-HBx, HepG2 and HepG2-HBx cells. B, C. Cell invasion and migration assay for si-Ctrl, si-HBx and si-HBx+MALAT1 cells.
Discussion
HBV is a major risk factor of HCC, and HBx plays a key role in the process of HBV-induced hepatocarcinogenesis, but how it affects the tumor development in HCC needs more illumination. Previous studies suggested that lots of lncRNAs were involved in HBx related HCC, including MALAT1 [4,5,25]. It has been proved that MALAT1 could promote tumor growth and cell metastasis in lots of cancers, such as malignant melanoma [22], osteosarcoma [26] and pancreatic cancer [27]. Recently, MALAT1 level was observed to be associated with liver damage and could predict development of HCC [13]. However, whether MALAT1 was involved in the hepatocarcinogenesis mediated by HBx was poorly understood. Thus in this study, for the first time we focused on investigating the role of MALAT1 in HBx-associated HCC and the possible mechanisms in tumorigenesis and metastasis of HCC. Firstly, we detected the expression of MALAT1 in hepatic tissues and HCC. Results showed that the expression of MALAT1 was significantly increased in HCC tissues and cells, which was in consistent with other studies [13]. Then we found that HBx could up-regulate the expression of MALAT1 in both hepatic tissues and HCC, which meant MALAT1 expression level was positively associated with the mRNA level of HBx in HCC cells. And this is the first time the relationship of HBx and MALAT1 in HCC was determined. Secondly, we investigated effects of MALAT1 on tumor growth and cell invasion and migration both in vitro and in vivo. Results showed that MALAT1 could promote HepG2-HBx cells migration and invasion both in vitro and in vivo. These results could further demonstrated the function of MALAT1 in tumor growth and metastasis in HBx-associated HCC.
Generally, lncRNAs regulated downstream genes expression through direct, indirect or epigenetic ways. As one of lncRNAs, previous studies also showed that MALAT1 could regulate several down-stream genes, including CD133 [28], miR-23c [29], and LTBP3 [22]. It has been proved that LTBPs are involved in cancer development, and MALAT1 could positively regulate LTBP3 transcription in mesenchymal stem cells [22]. Thus to further investigate possible mechanisms of the above influence of MALAT1 in HBx-associated hepatocarcinogenesis, we hypothesized whether MALAT1 affected HBx-associated HCC through regulation of LTBP3. Results showed that in both hepatic tissues and HCC, MALAT1 could up-regulate the expression of LTBP3. What’s more, the promotion effects on tumor growth and metastasis, cell invasion and migration were also found to be induced by LTBP3. At last, we preliminarily studied the relationship between HBx and LTBP3 and found expression of LTBP3 could be reduced by si-HBx and the effect could be moderated by the over-expression of MALAT1. Therefore, HBx is able to enhance cell metastasis through up-regulating LTBP3.
In conclusion, in this study we demonstrated that HBx could up-regulate long non-coding RNA MALAT1 in HCC, and MALAT1 could further influence the expression of LTBP3, resulting in promotion of development and metastasis of HCC. These results may provide new insights for the roles of lncRNAs in HBx-associated hepatocarcinogenesis.
Acknowledgements
This study was supported by the Central South University Xiangya Famous Doctor Foundation; Hunan provincial Natural Science Foundation (10JJ5034); Hunan Provincial Science and Technology Program (2010SK3093).
Disclosure of conflict of interest
None.
References
- 1.Han LL, Lv Y, Guo H, Ruan ZP, Nan KJ. Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J Gastroenterol. 2014;20:10249–10261. doi: 10.3748/wjg.v20.i30.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahbari NN, Mehrabi A, Mollberg NM, Müller SA, Koch M, Büchler MW, Weitz J. Hepatocellular carcinoma: current management and perspectives for the future. Ann Surg. 2011;253:453–469. doi: 10.1097/SLA.0b013e31820d944f. [DOI] [PubMed] [Google Scholar]
- 3.Osaki Y, Nishikawa H. Treatment for hepatocellular carcinoma in Japan over the last three decades: our experience and published work review. Hepatol Res. 2015;45:59–74. doi: 10.1111/hepr.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.JHu JJ, Song W, Zhang SD, Shen XH, Qiu XM, Wu HZ, Gong PH, Lu S, Zhao ZJ, He ML, Fan H. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep. 2016;6:23521. doi: 10.1038/srep23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 6.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang XW, Bu P, Liu L, Zhang XZ, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227–232. doi: 10.1016/j.bbrc.2015.04.121. [DOI] [PubMed] [Google Scholar]
- 8.Moyo B, Nicholson SA, Arbuthnot PB. The role of long non-coding RNAs in hepatitis B virusrelated hepatocellular carcinoma. Virus Res. 2016;212:103–113. doi: 10.1016/j.virusres.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 9.Yuan SX, Tao QF, Wang J, Yang F, Liu L, Wang LL, Zhang J, Yang Y, Liu H, Wang F, Sun SH, Zhou WP. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;394:87–94. doi: 10.1016/j.canlet.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 10.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Song Q, Yu S, Hu D, Zhuang X. Microvascular invasion in hepatocellular carcinoma overexpression promotes cell proliferation and inhibits cell apoptosis of hepatocellular carcinoma via inhibiting miR-199a expression. Onco Targets Ther. 2015;8:2303–2310. doi: 10.2147/OTT.S86807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu H, He Y, Lin L, Qi Z, Ma L, Li L, Su Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016;37:1683–1691. doi: 10.1007/s13277-015-3946-5. [DOI] [PubMed] [Google Scholar]
- 13.Konishi H, Ichikawa D, Yamamoto Y, Arita T, Shoda K, Hiramoto H, Hamada J, Itoh H, Fujita Y, Komatsu S, Shiozaki A, Ikoma H, Ochiai T, Otsuji E. Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci. 2016;107:149–154. doi: 10.1111/cas.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, Ji L, Kong R, Wang G, Jia YH, Bai XW, Sun B. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther. 2016;15:2232–43. doi: 10.1158/1535-7163.MCT-16-0008. [DOI] [PubMed] [Google Scholar]
- 15.Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, Zhang Y, Xing Y, Xi T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 16.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 17.Su CT, Huang JW, Chiang CK, Lawrence EC, Levine KL, Dabovic B, Jung C, Davis EC, Madan-Khetarpal S, Urban Z. Latent transforming growth factor binding protein 4 regulates transforming growth factor beta receptor stability. Hum Mol Genet. 2015;24:4024. doi: 10.1093/hmg/ddv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bultmann I, Conradi A, Kretschmer C, Sterner-Kock A. Latent transforming growth factor beta-binding protein 4 is downregulated in esophageal cancer via promoter methylation. PLoS One. 2013;8:e65614. doi: 10.1371/journal.pone.0065614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jelodari-Mamaghani S, Haji-Seyed-Javadi R, Suri F, Nilforushan N, Yazdani S, Kamyab K, Elahi E. Contribution of the latent transforming growth factor-β binding protein 2 gene to etiology of primary open angle glaucoma and pseudoexfoliation syndrome. Mol Vis. 2013;19:333–347. [PMC free article] [PubMed] [Google Scholar]
- 20.Mohanty K, Tanwar M, Dada R, Dada T. Screening of the LTBP2 gene in a north Indian population with primary congenital glaucoma. Mol Vis. 2013;19:78–84. [PMC free article] [PubMed] [Google Scholar]
- 21.Vehvilainen P, Koli K, Myllarniemi M, Lindholm P, Soini Y, Salmenkivi K, Kinnula VL, Keski-Oja J. Latent TGF-beta binding proteins (LTBPs) 1 and 3 differentially regulate transforming growth factor-beta activity in malignant mesothelioma. Hum Pathol. 2011;42:269–278. doi: 10.1016/j.humpath.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Chen P, Qu J, Shi L, Zhuang W, Fu J, Li J, Zhang X, Sun Y. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J Biol Chem. 2014;289:29365–29375. doi: 10.1074/jbc.M114.572693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luan W, Li L, Shi Y, Bu X, Xia Y, Wang J, Djangmah HS, Liu X, You Y, Xu B. Long noncoding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016;7:63901–63912. doi: 10.18632/oncotarget.11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou W, Zou B, Liu L, Cui K, Gao J, Yuan S, Cong N. MicroRNA-98 acts as a tumor suppressor in hepatocellular carcinoma via targeting SALL4. Oncotarget. 2016;7:74059–74073. doi: 10.18632/oncotarget.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, Ren J, Hou F, Li Q. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo W, He H, Xiao W, Liu Q, Deng Z, Lu Y, Wang Q, Zheng Q, Li Y. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget. 2016;7:54733–54743. doi: 10.18632/oncotarget.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 28.Latorre E, Carelli S, Raimondi I, D’Agostino V, Castiglioni I, Zucal C, Moro G, Luciani A, Ghilardi G, Monti E, Inga A, Di Giulio AM, Gorio A, Provenzani A. The ribonucleic complex HuRMALAT1 represses CD133 expression and suppresses epithelial-mesenchymal transition in breast cancer. Cancer Res. 2016;76:2626–2636. doi: 10.1158/0008-5472.CAN-15-2018. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Zeng L, Cao C, Lu C, Lian W, Han J, Zhang X, Zhang J, Tang T, Li M. Long noncoding rna malat1 regulates renal tubular epithelial pyroptosis by modulated mir-23c targeting of elavl1 in diabetic nephropathy. Exp Cell Res. 2016;350:327–335. doi: 10.1016/j.yexcr.2016.12.006. [DOI] [PubMed] [Google Scholar]