Abstract
MicroRNA-421 (miRNA-421) dysregulation has been found in various human tumors, however, the biological function and molecular mechanism of miR-421 in glioma remain unclear. In this study, we investigated the potential biological roles of miR-421 in glioma cell lines. First, we demonstrated that compared with that of low grade gliomas (LGG), miR-421 expression is much lower in high grade gliomas (HGG) within the CGGA (Chinese Glioma Genome Atlas) database. MiR-421 expression in 5 normal brain tissues and 20 glioma tissues were in agreement with the result in the CGGA. Second, exogenous expression of miR-421 inhibited glucose metabolism, invasion, angiogenesis and enhanced the radiosensitivity in glioma cell lines. Third, through an online database, myocyte enhancer factor 2D (MEF2D) was identified as a target of miR-421. Interestingly, down-regulation of MEF2D led to an inhibitory effect on glioma glucose metabolism, invasion, angiogenesis and enhancing effect on radiosensitivity, which was similar to the effects of the up-regulation of miR-421. Simultaneously, overexpression of MEF2D partially restored the effect of miR-421 on the glioma cell lines. Finally, in a xenograft model, overexpression of miR-421 suppressed tumorigenicity. These data collectively suggested miR-421 may suppress tumor-associated activity in gliomas by targeting MEF2D.
Keywords: miR-421, MEF2D, glucose metabolism, angiogenesis, radiosensitivity
Introduction
Glioma is the most common and aggressive primary brain tumor in the central nervous system and is based on a genetic abnormality [1,2]. Although much progress has been made in glioma treatment in recent years, patients with gliomas have a median survival time that rarely exceeds 18 months in both children and adults [3]. The major reason why patients survival has not improved is that the underlying molecular mechanisms of gliomagenesis have not yet been completely elucidated. Thus, it is critical to identify novel treatment options for patients with gliomas.
MicroRNAs (miRNAs) are a group of small noncoding RNAs, approximately 18-24 nucleotides in length, which are widely expressed in various tissues and involved in numerous biological processes including cell apoptosis, differentiation, and metabolism [4-6]. Accumulating evidence has indicated that miRNAs participate in the development and progression of human cancers through binding to complementary sequences at the 3’-untranslated regions (3’-UTRs) of their target mRNAs, which act as tumor promoters or suppressors [7-9]. In recent years, extensive investigation has shown that the dysregulation of miRNAs is involved in glioma initiation and progression, which provided a novel approach to therapy for glioma [10].
Aberrant expression of miR-421 has been reported in many types of cancer. For example, miR-421 is significantly up-regulated in human gastric cancer tissues and promotes the proliferation of gastric cancer cells by down-regulating caspase-3 expression [11]. Hao J et al. reported that miR-421 likely functions as a tumor promoter in pancreatic cancer by targeting DPC4/Smad4 [12]. Meanwhile, other studies have shown that miR-421 is down-regulated and acts as a tumor suppressor; miR-421 inhibits the proliferation and metabolism of prostate cancer cells by targeting Cullin 4B (CUL4B) [13] and suppresses breast cancer metastasis by targeting metastasis associated 1 (MTA1) [14]. Despite a study reporting that miR-421 could enhance the sensitivity to ionizing radiation in cells, the exact mechanism is still unclear [15].
Therefore, in the present study, we aimed to investigated the molecular mechanism that involves miR-421 in the regulation of the glucose metabolism, invasion, angiogenesis and radiosensitivity of glioma cells.
Materials and methods
Human tissue samples
Human glioma samples and normal tissues were obtained from the Department of Neurosurgery at Nanjing First Hospital, Nanjing Medical University. Ethical approval was obtained from the ethics committee of Nanjing First Hospital and written informed consent was obtained from all patients. Tissue samples were collected during surgery and frozen in liquid nitrogen and stored until total RNAs or proteins were extracted.
Cell culture
The human U87 and U251 glioma cell lines were purchased from the Chinese Academy of Sciences Cell Bank. Both cell lines were maintained in Dulbecco’s Modified Eagle’s Medium (Gibco, USA) with high glucose supplemented with 10% fetal bovine serum (ScienCell, USA). Human umbilical vein endothelial cells (HUVECs, purchased from GeneChem, China) were cultured in endothelial cell basal medium (Medium 200, Gibco, USA) supplemented with Low Serum Growth Supplement (Gibco, USA).
Plasmid constructs, Oligonucleotides and cell transfection
Oligonucleotides were purchased from GenePharma (Shanghai, China). The sequences were as follows: MEF2D-small interfering RNA-1 (si-MEF2D-1), GCG AGA TCG CAC TCA TCA TCT; and MEF2D-small interfering RNA-2 (si-MEF2D-2), GGT CTC CCA GTC TAC TCA TTC. The hsa-miR-421 mimic and hsa-miR-421 inhibitor were also purchased from GenePharma. The shRNAs were synthesized and inserted into the pHBLV-U6 lentivirus core vector (Hanbio, Shanghai, China).
Real-time quantitative PCR
RNA was isolated from harvested cells and tissue with TRIzol (Invitrogen, USA) following the manufacturer’s protocols. The MEF2D (qRT-PCR) reactions were performed using Fermentas reverse transcription reagents and SYBR Green PCR Master Mix (Applied Biosystems, USA) according to the manufacturer’s protocols. MiR-421 qRT-PCR reactions were performed using TaqMan miRNA assays (Applied Biosystems, USA). U6 was used for normalization. In addition, analysis was performed using the 2-ΔCt or 2-ΔΔCt method. Each experiment was performed in triplicate.
Western blot analysis
Total proteins were isolated from the indicated cells using RIPA lysis buffer (KeyGEN, Nanjing, Jiangsu, China) and quantified using a BCA Protein Assay Kit (Beyotime, Nanjing, Jiangsu, China). The obtained proteins were then separated by SDS-PAGE and transferred onto a 0.45-μm cellulose acetate membrane (Immobilon, USA). After blocking with 5% nonfat milk (Mengniu, Beijing, China), the membrane was incubated with primary antibodies recognizing MEF2D (1:1000, Abnova, China) or GAPDH (1:1000, YI FEI XUE BIOTECHNOLOGY, Nanjing, Jiangsu, China), followed by an incubation with a horseradish peroxidase (HRP)-conjugated secondary antibody (YI FEI XUE BIOTECHNOLOGY, Nanjing, Jiangsu, China). Finally, the bands were visualized under an Image Quant LAS 4000 mini (GE, USA).
Glucose uptake assay
The glucose uptake was determined using a 2-Deoxyglucose Glucose Uptake Assay Kit (Fluorometric, Abcam, USA) according to the protocol provided by the manufacturer. U87 and U251 cells were seeded into 96-well plates (1000 cells/well) overnight. After 24 hours, the cells were incubated in the darkness with 2-Deoxyglucose (10 mM) for 20 min at 37°C under a CO2 humidified atmosphere and subjected to the measurement of the 2-Doxyglucose uptake using a fluorescence microplate reader at Ex/Em = 535/587 nm.
Transwell assay
To measure cell invasion ability, 8 mm pore 24-well Matrigel invasion chambers (Coring, NY, USA) was used according to the protocol provided by manufacturer. 20000 cells were seeded into each well. The upper chambers were added with 200 µl DMEM contained with 0.1% FBS, while the lower chamber was supplemented with 400 µl DMEM contained 10% FBS. After incubated at 37°C in a 5% CO2 for 48 hours, non-invading cells were removed from the upper chamber. Then, the migrating cells were stained with 0.1% crystal violet (YI FEI XUE BIOTECHNOLOGY, Nanjing, Jiangsu, China). The migrated cells were quantified by photographing 3 independent visual fields under the microscope.
Angiogenesis assay
Matrigel (BD, USA) was dissolved at 4°C overnight, and each well of the prechilled 96-well plates was coated with 20 µl Matrigel. The plates were then incubated at 37°C for 2 hours to form a layer of Matrigel. The cells were cultured to 90-100% concentration, the old medium was discarded and replaced with serum-reduced medium (1% FBS) for 24 h. The medium was collected and stored at -80°C. HUVECs cells were switched to basic medium containing 0.2% FBS. In 24 h, the starved HUVECs were trypsinized, collected, counted, and resuspended in endothelial cell growth medium (Gibco, USA) supplemented with Gibco LSGS (Low Serum Growth Supplement, Gibco, USA). Then, the cells were mixed with equal volumes of the conditioned medium and seeded in the Matrigel-pretreated 96-well plate at 3×104 cells/well. After 12 h, tube formation was examined under light microscope. The length of the tubes was measured using the Soft Imaging System (Soft Imaging System GmbH, Germany).
Clone formation assay
Glioma cell lines in exponential growth phase were inoculated on 3.5 cm dishes and exposed to a range of irradiation doses (0, 2, 4, and 6 Gy) after adhesion. After incubation for 12 days at 37°C and 5% CO2, the colonies were washed with PBS, fixed with 10% formaldehyde and stained with 2 ml 1.0% crystal violet (YI FEI XUE BIOTECHNOLOGY, Nanjing, Jiangsu, China) overnight. The survival fractions (SF) were calculated as the number of colonies/(number of inoculated cells × plating efficiency) and the radiation survival curve was drawn.
Immunohistochemistry
Paraffin-embedded glioma tissues were incubated with 1:200-diluted primary antibodies against MEF2D overnight at 4°C, followed by incubated with a biotinylated secondary antibody (1:200 dilution, GeneTech, USA) at room temperature for 1 h. Subsequently, the sections were incubated with ABC-peroxidase for 1 h, washed with PBS and stained with diaminobenzidine for 5 min, counterstained with hematoxylin (GeneChem, Shanghai, China). Ten randomly selected visual fields per section were examined by light microscope to evaluate the MEF2D expression.
Xenograft tumor assay
Ten immunodeficient female nude mice (Beijing Laboratory Animal Center, Beijing, China) were used to test the effects of MEF2D on gliomas in vivo. These nude mice were randomly divided into two groups (5 mice per group). Approximately 2×106 logarithmically growing U87 cells stably expressing control shRNA or shRNA-MEF2D were subcutaneously injected into the nude mice. After 3 weeks, the subcutaneous tumors were stripped and weighed. Tumor volume was calculated according to the formula: V (mm3) = 0.5 * a* b2 (a represents the longest axis and b the shortest axis).
Statistical analysis
All the experiments except the animal experiments were conducted at least three times. All the values in this study are shown as the means ± SD. The difference between the groups was considered significant and very significant when P < 0.05 (* or #) and P < 0.01 (** or ##), respectively.
Results
miR-421 is down-regulated in glioma tissues and cell lines
First, we examined the expression patterns of miR-421 in the CGGA (Chinese Glioma Genome Atlas) database and found that miR-421 expression was significantly lower in HGG than in LGG (Figure 1A). Next, we measured the miR-421 expression in 5 normal brain tissues and 20 glioma tissues and observed a similar trend in that the expression of miR-421 was lower in the glioma tissues (Figure 1B). We also detected significantly discrepant expression levels of miR-421 in a normal astrocyte cell line and glioma cell lines (Figure 1C). Overall, these data suggest that miR-421 may play a critical role in glioma progression.
Figure 1.
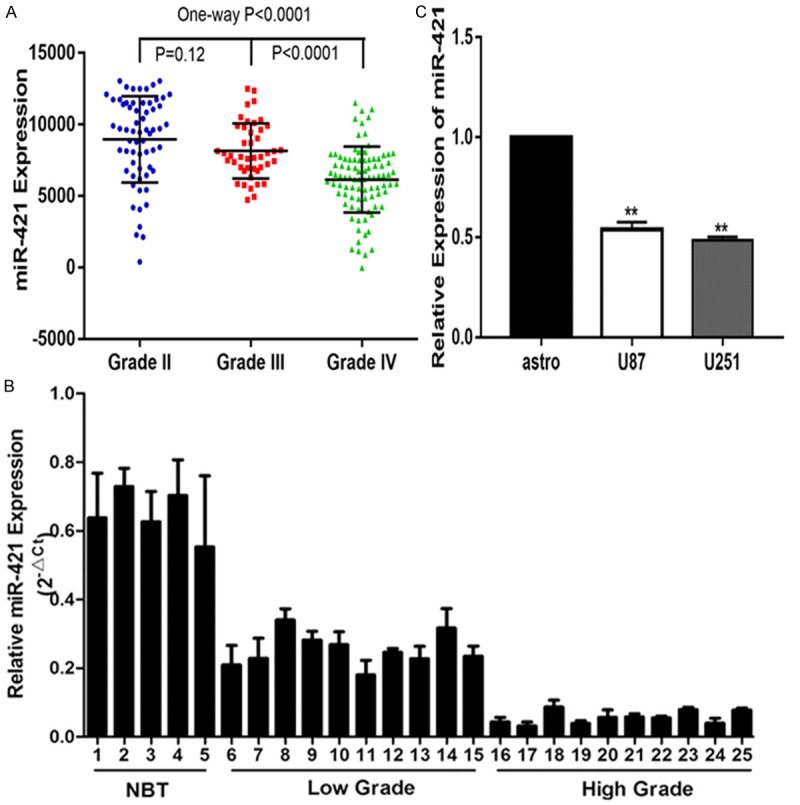
MiR-421 is down-regulated in glioma. A. Expression of miR-421 in CGGA public database. B. Relative expression of miR-421 in glioma tissues and normal brain tissues. C. Relative expressions of miR-421 in astrocyte and glioma cell lines U87, U251. *P < 0.05, **P < 0.01.
miR-421 inhibits glioma cell glucose metabolism in vitro
To explore the possible biological function of miR-421 in gliomas, miR-421 was overexpressed in glioma cell lines by transfecting a miR-421 mimic (Figure 2A). To support cell proliferation, cellular metabolism is reprogrammed to balance biosynthetic processes with the energy supply to tumor cells, which is a hallmark of tumors [16]. Thus, we first explored the effect of miR-421 on the glucose metabolism of glioma cells. To this aim, we treated two cell lines, U87 and U251, with different concentrations of glucose (0, 5, or 20 mM) and examined the levels of glucose uptake in both cell lines. We found that compared with that of the negative control, up-regulation of miR-421 by transfection with the miR-421 mimic significantly decreased the glucose uptake of the glioma cell lines (Figure 2B). This result suggested that miR-421 is involved in the regulation of glucose metabolism in gliomas.
Figure 2.
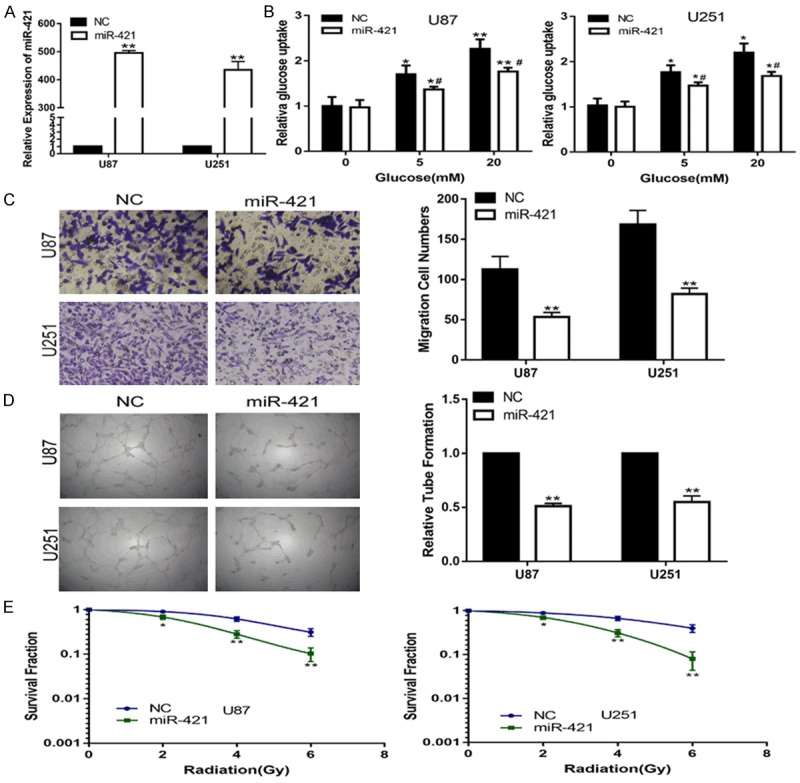
MiR-421 inhibits the glucose uptake, invasion, angiogenesis and enhances the radiosensitivity of glioma cells in vitro. A. The expression of miR-421 in U87 and U251 cells transfected with NC or miR-421 mimic. B. Relative glucose uptake in U87 and U251 cells transfected with NC or miR-421 mimic and treated with the indicated concentrations of glucose (0, 5, or 20 mM). *P < 0.05, **P < 0.01 compared to cells treated with 0 mM glucose, #p < 0.05 compared to control miR-421 transfected cells treated with the same glucose concentration. C. The transwell assay in U87 and U251 cells transfected with NC or miR-421 mimic. D. The tube formation assay in U87 and U251 cells transfected with NC or miR-421 mimic. E. The colony formation assay in U87 and U251 cells transfected with NC or miR-421 mimic and treated with ultraviolet ray. *P < 0.05, **P < 0.01, #P < 0.05.
miR-421 suppresses glioma cell invasion, angiogenesis and enhances the radiosensitivity in vitro
Given that miR-421 is associated with glioma progress, we assessed the effect of miR-421 on cell invasion, angiogenesis and radiosensitivity. The results of the Transwell assay showed that the invasion ability of the U87 and U251 cell lines was significantly suppressed compared with that of the control cell lines (Figure 2C). An angiogenesis assay showed that the up-regulation of miR-421 depressed the angiogenesis capacity of HUVECs (Figure 2D). In addition, a clone formation assay clearly demonstrated that the miR-421 mimic transfection significantly enhanced the radiosensitivity of the U87 and U251 cells (Figure 2E). Collectively, these results suggested that miR-421 functions as tumor suppressor in glioma.
MEF2D is a direct target of miR-421 in glioma cell lines
To elucidate the molecular mechanisms underlying the miR-421-mediated regulation of glioma cell lines, we searched the starBase v2.0 and found that MEF2D may be a target of miR-421 (Figure 3A). We validated the expression levels of MEF2D in glioma tissues by real-time RT-PCR. By categorizing all glioma samples into NBT (n = 5), LGG (n = 10) and HGG (n = 10), we found that there is an inverse relationship between MEF2D expression level and the grade of the glioma (Figure 3B). Immunohistochemical further verified the different expression of MEF2D in low grade glioma and high grade glioma (Figure 3C). Western blot analysis showed that MEF2D expression was increased/decreased in glioma cell lines with down-regulation/up-regulation of miR-421 (Figure 3D). To further confirm that MEF2D is a direct target of miR-421, luciferase activity assays were performed. The results showed that overexpression of miR-421 significantly suppresses the luciferase activity of cells transfected with wild-type MEF2D-3’UTR but had no significant effect on mutant-type MEF2D-3’UTR (Figure 3E). Based on these results, MEF2D was confirmed as a target of miR-421 in glioma cell lines.
Figure 3.
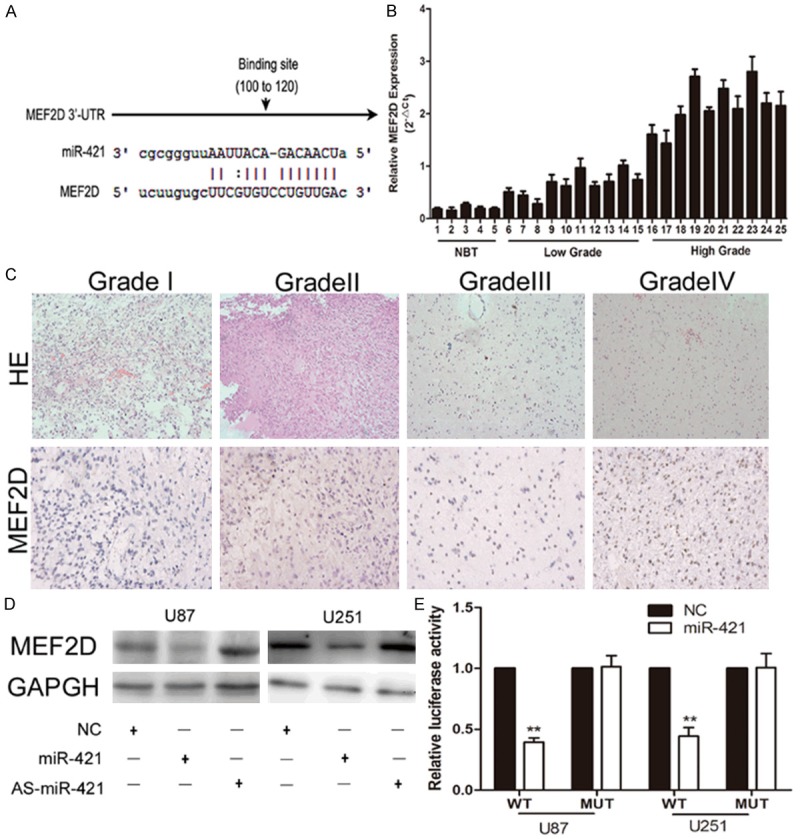
MEF2D was negatively regulated by miR-421 in glioma cells. A. Schematic of the MEF2D 3’UTR including the putative binding sites for miR-421, as predicted by starBase v2.0. B. The expression levels of MEF2D in glioma tissues and normal brain tissues. C. Immunohistochemica confirmation of increased MEF2D levels in HGG tissues compared with LGG tissues. D. Western blot assay showed the expression of MEF2D in U87 and U251 cells transfected with NC, mir-421 mimic or AS-miR-421. E. MiR-421 down-regulated luciferase activity of wild-type MEF2D 3’UTR expression vector, but did not reduce expression of a mutant MEF2D 3’UTR. **P < 0.01.
miR-421 regulates glucose metabolism, invasion, angiogenesis and radiosensitivity of glioma cell lines by targeting MEF2D
First, we transfected siRNA targeting MEF2D (si-MEF2D) into glioma cell lines to knockdown endogenous MEF2D. QT-PCR and a western blot assay were performed to confirm the expression of MEF2D (Figure 4A, 4B). Collectively, the down-regulation of MEF2D inhibited the glucose metabolism, invasion, angiogenesis and enhanced the radiosensitivity of the glioma cell lines (Figure 4C-F). To further confirm that miR-421 exerted its function by targeting MEF2D in the glioma cell lines, we transfected miR-421 together with a MEF2D expression plasmid into glioma cell lines, followed by a series of functional assays. PCR and Western blot analyses revealed that the glioma cell lines transfected with a MEF2D overexpression plasmid restored MEF2D expression (Figure 5A, 5B). Subsequently, we also validated that the restoration of MEF2D expression was capable of partially counteracting the effects of miR-421 on the glucose metabolism, invasion, angiogenesis and radiosensitivity of glioma cell lines (Figure 5C-F). Overall, these results suggested that MEF2D is a critical target of miR-421 in glioma cell lines.
Figure 4.
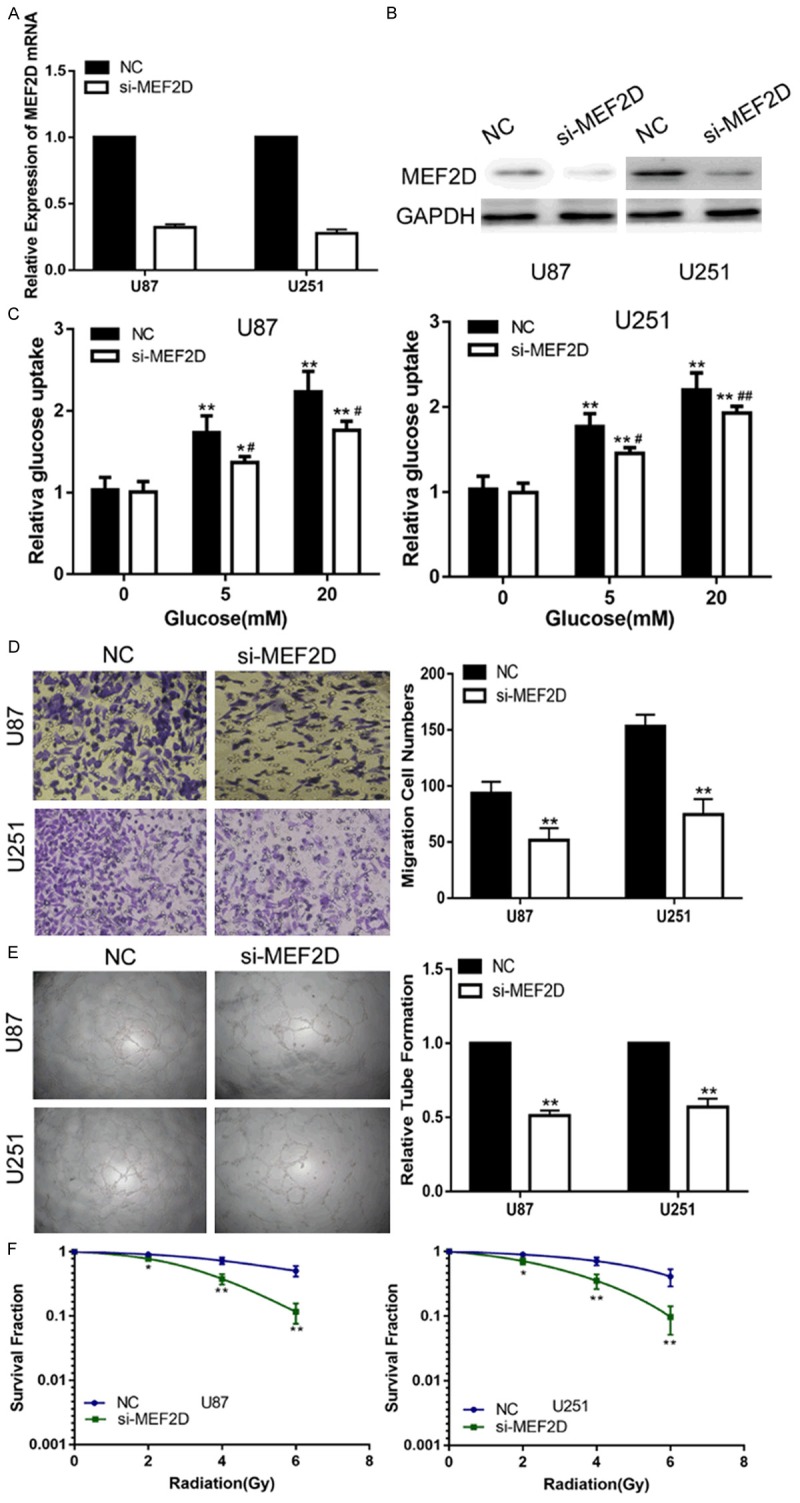
Down-regulation of MEF2D suppress the glucose uptake, invasion, angiogenesis and increase the radiosensitivity of glioma cells in vitro. A. The expression of MEF2D in U87 and U251 cells transfected with NC or si-MEF2D. B. Western blot assay showed the expression of MEF2D in U87 and U251 cells transfected with NC or si-MEF2D. C. Relative glucose uptake in U87 and U251 cells transfected with NC or si-MEF2D and treated with the indicated concentrations of glucose (0, 5, or 20 mM). *P < 0.05, **P < 0.01 compared to cells treated with 0 mM glucose, #P < 0.05, ##P < 0.01 compared to control miR-421 transfected cells treated with the same glucose concentration. D. The transwell assay in U87 and U251 cells transfected with NC or si-MEF2D. E. The tube formation assay in U87 and U251 cells transfected with NC or si-MEF2D. F. The colony formation assay in U87 and U251 cells transfected with NC or si-MEF2D and treated with ultraviolet ray. *P < 0.05, **P < 0.01, #P < 0.05, ##P < 0.01.
Figure 5.
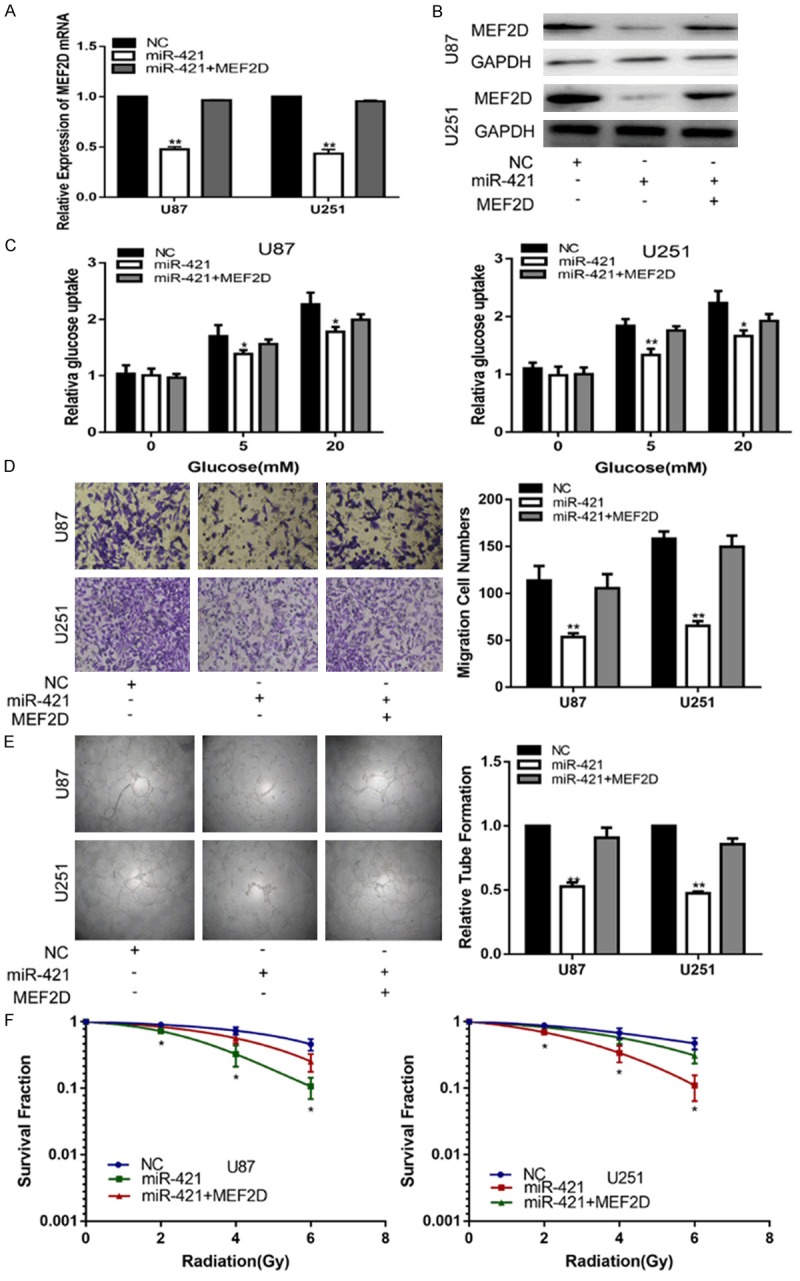
MEF2D is involved in the miR-421-induced inhibiting effects on glioma. A, B. The expression of MEF2D in U87 and U251 cells transfected with NC, miR-421 or miR-421+MEF2D. C. Relative glucose uptake in U87 and U251 cells transfected with NC, miR-421 or miR-421+MEF2D and treated with the indicated concentrations of glucose (0, 5, or 20 mM). *P < 0.05, **P < 0.01 compared to cells treated with miR-421+MEF2D. D. The transwell assay in U87 and U251 cells transfected with NC, miR-421 or miR-421+MEF2D. E. The tube formation assay in U87 and U251 cells transfected with NC, miR-421 or miR-421+MEF2D. F. The colony formation assay in U87 and U251 cells transfected with NC, miR-421 or miR-421+MEF2D and treated with ultraviolet ray. *P < 0.05, **P < 0.01.
MiR-421 inhibits glioma growth in vivo
To investigate the role of miR-421 in the tumor growth of gliomas in vivo, we extended our investigation by subcutaneous implantation of stably overexpressing miR-421 U87 cells in nude mice. The mice were sacrificed after 3 weeks. Then, the tumors were stripped and weighed. The average tumor weight in the control was significantly higher than that in the miR-421 mimic treated group. Meanwhile, the average tumor volume in the control group was significantly larger than that in the miR-421 mimic-treated group (Figure 6). Finally, these results indicated that increased miR-421 could inhibit glioma progression in vivo.
Figure 6.
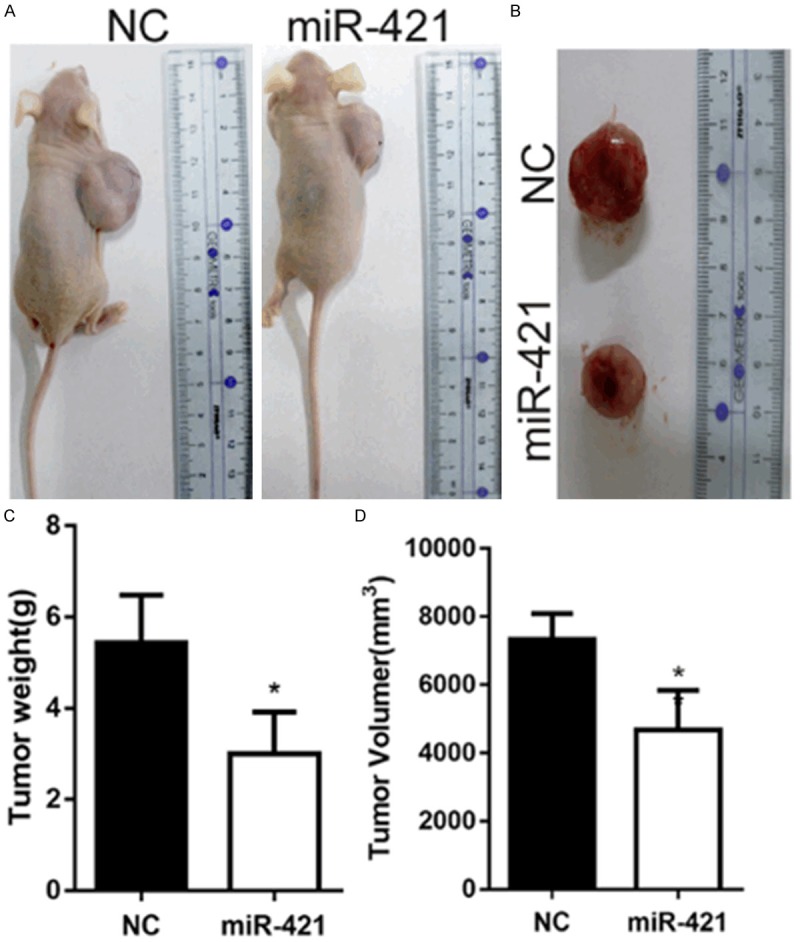
MiR-421 inhibits glioma growth in vivo. A, B. Tumor formation was performed in nude mice and the excision tumor of U87 xenografts. C. The weight of excision tumor. D. The volume of excision tumor by the formula: V (mm3) = 0.5 * a* b2 (a represents the longest axis and b the shortest axis). *P < 0.05.
Discussion
MiRNAs participate in various physiological and pathological process at the post-transcriptional level via binding to the 3’-untranslated regions (3’-UTR) of target mRNAs. Recent evidence has indicated that miRNAs are crucial determinants of glioma development. For example, Qian J et al. showed that miR-1224 acts as a tumor suppressor by directly targeting CREB1 in glioma [17]. Xu X et al. reported that miRNA-98 inhibits cell migration and invasion in malignant glioma by targeting pre-B cell leukemia homeobox 3 [18]. Hu S et al. found that miR-502c inhibits glioma cell migration and invasion by suppressing TGF-β2 [19].
Recently, a study reported that miR-421 could change the S-phase level of the cell cycle checkpoint and enhance the sensitivity of cells to ionizing radiation. However, the exact association between miR-421 and glioma has never been studied. In the present study, we found that compared to that in LGG, the expression of miR-421 in HGG is down-regulated. To further explore the role of miR-421 in glioma, we up-regulated the expression of miR-421 in U87 and U251. In vitro assays showed that the up-regulation of miR-421 dramatically suppresses glucose metabolism, invasion, angiogenesis and increase the radiosensitivity of glioma cells. These results are consistent with our xenograft tumor assay, which indicated that miR-421 can suppress glioma growth.
A number of reports have shown that miR-421 is involved in the development of human cancers, and many genes have been identified as targets of miR-421. For example, caspase-3 was identified to be responsible for the effect of miR-421 in gastric cancer [11]. In this study, through an online database, we identified MEF2D as a target of miR-421 in glioma cell lines.
The MEF2 family plays important roles in human physiological processes and has four members (MEF2A, MEF2B, MEF2C, and MEF2D) in humans [20,21]. MEF2D is the most famous member of the family because of its effects on the development of human cancers [22,23]. The first report of MEF2D in human cancer was in a hematological malignancy [24,25]. Subsequently, MEF2D has been identified in colorectal cancer and hepatocellular carcinoma. In our study, we found that compared with that in LGG, the expression of MEF2D in HGG was much higher. Luciferase activity analysis revealed the correlation between miR-421 and MEF2D. Meanwhile, the down-regulation MEF2D dramatically repressed glucose metabolism, invasion, angiogenesis and enhanced the radiosensitivity of the glioma cell lines, which is in agreement with the effect of miR-421 on glioma. Furthermore, introduction of ectopic expression of MEF2D significantly abolished miR-421-mediated suppression on glucose metabolism, invasion, angiogenesis and radiosensitivity of glioma cell lines. Thus, we conclude that miR-421 inhibits glucose metabolism, invasion, angiogenesis and enhances the radiosensitivity of the glioma cell lines by targeting MEF2D.
In summary, we have demonstrated that miR-421 is down-regulated and plays the role of a tumor suppressor in glioma via MEF2D. Therefore, our present study clarifies the importance of the miR-421/MEF2D molecular network in glioma development and may provide a new therapeutic approach for the treatment of glioma.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81502168, No. 81302180, No. 81602212), Natural Science Foundation of Jiangsu Province (BK20161119), Program Funding for Health Youth Talent Training Project of Nanjing (QRX11027), Scientific and Technologic Development Program of Nanjing City (No. 20160525), the Key Project supported by Medical Science and Technology Development Foundation, Nanjing Department of Health (Grant No. YKK16139 and YKK12139), Science and Technology Development Foundation of Nanjing Medical University (2015NJMU049). We thank General Clinical Research Center, Nanjing First Hospital, for technical support.
Disclosure of conflict of interest
None.
References
- 1.Li S, Zeng A, Hu Q, Yan W, Liu Y, You Y. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro Oncol. 2017;19:55–65. doi: 10.1093/neuonc/now129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrew RD, Fagan M, Ballyk BA, Rosen AS. Seizure susceptibility and the osmotic state. Brain Res. 1989;498:175–180. doi: 10.1016/0006-8993(89)90417-4. [DOI] [PubMed] [Google Scholar]
- 3.Galeano F, Tomaselli S, Locatelli F, Gallo A. A-to-I RNA editing: the “ADAR” side of human cancer. Semin Cell Dev Biol. 2012;23:244–250. doi: 10.1016/j.semcdb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Yuan L, Yuan P, Yuan H, Wang Z, Run Z, Chen G, Zhao P, Xu B. miR-542-3p inhibits colorectal cancer cell proliferation, migration and invasion by targeting OTUB1. Am J Cancer Res. 2017;7:159–172. [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Li Q, Li R, Ren P, Dong S. MicroRNA-363-3p inhibits papillary thyroid carcinoma progression by targeting PIK3CA. Am J Cancer Res. 2017;7:148–158. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Jiang R, Zhang C, Liu G, Gu R, Wu H. MicroRNA-101 inhibits proliferation, migration and invasion in osteosarcoma cells by targeting ROCK1. Am J Cancer Res. 2017;7:88–97. [PMC free article] [PubMed] [Google Scholar]
- 7.Zu Y, Yang Y, Zhu J, Bo X, Hou S, Zhang B, Qiu J, Zheng J. MiR-146a suppresses hepatocellular carcinoma by downregulating TRAF6. Am J Cancer Res. 2016;6:2502–2513. [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Zhang X, Li N, Liu Q, Chen D. miR-30b inhibits cancer cell growth, migration, and invasion by targeting homeobox A1 in esophageal cancer. Biochem Biophys Res Commun. 2017;485:506–512. doi: 10.1016/j.bbrc.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Tian W, Wang G, Liu Y, Huang Z, Zhang C, Ning K, Yu C, Shen Y, Wang M, Li Y, Wang Y, Zhang B, Zhao Y. The miR-599 promotes non-small cell lung cancer cell invasion via SATB2. Biochem Biophys Res Commun. 2017;485:35–40. doi: 10.1016/j.bbrc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Tao T, Wang Y, Luo H, Yao L, Wang L, Wang J, Yan W, Zhang J, Wang H, Shi Y, Yin Y, Jiang T, Kang C, Liu N, You Y. Involvement of FOSmediated miR-181b/miR-21 signalling in the progression of malignant gliomas. Eur J Cancer. 2013;49:3055–3063. doi: 10.1016/j.ejca.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Wu JH, Yao YL, Gu T, Wang ZY, Pu XY, Sun WW, Zhang X, Jiang YB, Wang JJ. MiR-421 regulates apoptosis of BGC-823 gastric cancer cells by targeting caspase-3. Asian Pac J Cancer Prev. 2014;15:5463–5468. doi: 10.7314/apjcp.2014.15.13.5463. [DOI] [PubMed] [Google Scholar]
- 12.Hao J, Zhang S, Zhou Y, Liu C, Hu X, Shao C. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem Biophys Res Commun. 2011;406:552–557. doi: 10.1016/j.bbrc.2011.02.086. [DOI] [PubMed] [Google Scholar]
- 13.Meng D, Yang S, Wan X, Zhang Y, Huang W, Zhao P, Li T, Wang L, Huang Y, Li T, Li Y. A transcriptional target of androgen receptor, miR-421 regulates proliferation and metabolism of prostate cancer cells. Int J Biochem Cell Biol. 2016;73:30–40. doi: 10.1016/j.biocel.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 14.Pan Y, Jiao G, Wang C, Yang J, Yang W. MicroRNA-421 inhibits breast cancer metastasis by targeting metastasis associated 1. Biomed Pharmacother. 2016;83:1398–1406. doi: 10.1016/j.biopha.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 15.Paolini A, Curti V, Pasi F, Mazzini G, Nano R, Capelli E. Gallic acid exerts a protective or an anti-proliferative effect on glioma T98G cells via dose-dependent epigenetic regulation mediated by miRNAs. Int J Oncol. 2015;46:1491–1497. doi: 10.3892/ijo.2015.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Qian J, Li R, Wang YY, Shi Y, Luan WK, Tao T, Zhang JX, Xu YC, You YP. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem. 2015;403:33–41. doi: 10.1007/s11010-015-2334-1. [DOI] [PubMed] [Google Scholar]
- 18.Xu X, Bao Z, Liu Y, Ji J, Liu N. MicroRNA-98 attenuates cell migration and invasion in glioma by directly targeting Pre-B cell leukemia homeobox 3. Cell Mol Neurobiol. 2017 doi: 10.1007/s10571-017-0466-4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Chen H, Zhang Y, Wang C, Liu K, Wang H, Luo J. MicroRNA-520c inhibits glioma cell migration and invasion by the suppression of transforming growth factor-beta receptor type 2. Oncol Rep. 2017;37:1691–1697. doi: 10.3892/or.2017.5421. [DOI] [PubMed] [Google Scholar]
- 20.Su L, Luo Y, Yang Z, Yang J, Yao C, Cheng F, Shan J, Chen J, Li F, Liu L, Liu C, Xu Y, Jiang L, Guo D, Prieto J, Avila MA, Shen J, Qian C. MEF2D transduces microenvironment stimuli to ZEB1 to promote epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res. 2016;76:5054–5067. doi: 10.1158/0008-5472.CAN-16-0246. [DOI] [PubMed] [Google Scholar]
- 21.Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, McCastlain K, Reshmi SC, Payne-Turner D, Iacobucci I, Shao Y, Chen IM, Valentine M, Pei D, Mungall KL, Mungall AJ, Ma Y, Moore R, Marra M, Stonerock E, Gastier-Foster JM, Devidas M, Dai Y, Wood B, Borowitz M, Larsen EE, Maloney K, Mattano LA Jr, Angiolillo A, Salzer WL, Burke MJ, Gianni F, Spinelli O, Radich JP, Minden MD, Moorman AV, Patel B, Fielding AK, Rowe JM, Luger SM, Bhatia R, Aldoss I, Forman SJ, Kohlschmidt J, Mrozek K, Marcucci G, Bloomfield CD, Stock W, Kornblau S, Kantarjian HM, Konopleva M, Paietta E, Willman CL, Loh ML, Hunger SP, Mullighan CG. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. doi: 10.1038/ncomms13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R, Zhang Y, Li H. miR-1244/Myocyte enhancer factor 2D regulatory loop contributes to the growth of lung carcinoma. DNA Cell Biol. 2015;34:692–700. doi: 10.1089/dna.2015.2915. [DOI] [PubMed] [Google Scholar]
- 23.Kong J, Liu X, Li X, Wu J, Wu N, Chen J, Fang F. Pokemon promotes the invasiveness of hepatocellular carcinoma by enhancing MEF2D transcription. Hepatol Int. 2016;10:493–500. doi: 10.1007/s12072-015-9697-y. [DOI] [PubMed] [Google Scholar]
- 24.Prima V, Gore L, Caires A, Boomer T, Yoshinari M, Imaizumi M, Varella-Garcia M, Hunger SP. Cloning and functional characterization of MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins created by a variant t(1;19)(q23; p13.3) in acute lymphoblastic leukemia. Leukemia. 2005;19:806–813. doi: 10.1038/sj.leu.2403684. [DOI] [PubMed] [Google Scholar]
- 25.Prima V, Hunger SP. Cooperative transformation by MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins generated by the variant t(1;19) in acute lymphoblastic leukemia. Leukemia. 2007;21:2470–2475. doi: 10.1038/sj.leu.2404962. [DOI] [PubMed] [Google Scholar]


