Abstract
Objective
Based on rapid advancement of genetic modification techniques, genomic editing is expected to become the most efficient tool for improvement of economic traits in livestock as well as poultry. In this study, we examined and verified the nickase of mutated CRISPR-associated protein 9 (Cas9) to modulate the specific target gene in chicken DF1 cells.
Methods
Chicken myostatin which inhibits muscle cell growth and differentiation during myogenesis was targeted to be deleted and mutated by the Cas9-D10A nickase. After co-transfection of the nickase expression vector with green fluorescent gene (GFP) gene and targeted multiplex guide RNAs (gRNAs), the GFP-positive cells were sorted out by fluorescence-activated cell sorting procedure.
Results
Through the genotyping analysis of the knockout cells, the mutant induction efficiency was 100% in the targeted site. Number of the deleted nucleotides ranged from 2 to 39 nucleotide deletion. There was no phenotypic difference between regular cells and knockout cells. However, myostatin protein was not apparently detected in the knockout cells by Western blotting. Additionally, six off-target sites were predicted and analyzed but any non-specific mutation in the off-target sites was not observed.
Conclusion
The knockout technical platform with the nickase and multiplex gRNAs can be efficiently and stablely applied to functional genomics study in poultry and finally adapted to generate the knockout poultry for agribio industry.
Keywords: Genome Editing, Cas9, Nickase, Myostatin, Knockout, Poultry
INTRODUCTION
Since transgenic mice were successfully created by introducing a foreign gene construct [1], genetic modification systems to generate transgenic, knockout and knockin animals have been the most promising approaches that help advance a comprehensive understanding of biology and lead to practical applications in agriculture and biopharmacy. Particularly, genetically modified livestock have a great potential for rapidly improving economic traits such as growth performance and disease resistance. In poultry, the recently developed transgenic technique utilizing piggyBac transposon and transposase is the most promising protocol to produce transgenic chickens with a stable transgene expression without tissue-specific repression as well as an efficient transgene insertion into genomic structure [2,3]. Due to the biosafety issue of virus-mediated transgene delivery, the non-viral production system of transgenic poultry is expected to be a reliable approach that could represent the best method for practical and industrial applications [3,4].
Knockout technical platforms have been developed and utilized for complete disruption of the specific gene or locus. Since knockout mice have been generated by conventional homologous recombination [5–8], the knockout system has revolutionized the research field of functional genomics by allowing the analysis of specific gene function(s) in animals. To date, the great biotechnological advancements such as zinc finger nuclease, transcription activator-like effector nuclease and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) make it possible to precisely alter the genetic information with higher efficiency and even large-scale outputs [9–11]. Genetically modified animals including avian species have become the most versatile experimental systems as models to study human diseases and develop pharmaceutical drugs [4]. Additionally, due to increasing interest from agricultural industry, practical strategies of precise genome editing in livestock have been sought for the last three decades.
Basically, the CRISPR/Cas9 expression plasmid systems strongly express the Cas9 enzyme but the expression vector as well as gRNA plasmids safely disappear after disruption of the targeted gene because of the transient expression. Thus, one of the great advantages of CRIPSR/Cas9-mediated genetic modification is that there are no transgenes integrated into the genome of the manipulated cells or animals. Nickase, which is a mutated Cas9 (Cas9-D10A) enzyme, was newly developed for precise genomic modification [12]. Cas9 enzyme which is an RNA-guided DNA endonuclease generates a double-strand DNA break and produces the mutation of nucleotide deletion or insertion during non-homologous end joining (NHEJ) repair process of the induced DNA break [10,11]. However, the mutant nickase creates a single-strand DNA break at the based on gRNA-defined target sequence [12]. The single-strand DNA break can apparently reduce the non-specific mutant induction without an off-target effect [12]. In this study, we firstly verified the mutation efficiency of nickase of mutated Cas9-D10A to disrupt the specific target gene in chicken.
MATERIALS AND METHODS
Chicken DF1 cell culture
The chicken DF1 cell line (American Type Culture Collection, Manassas, VA, USA) was maintained and sub-passaged in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and 1× antibiotic-antimycotic (Invitrogen, USA). DF1 cells were cultured in an incubator at 37°C in an atmosphere of 5% CO2 and 60% to 70% relative humidity.
Cas9-D10A nickase-mediated myostatin knockout and fluorescence-activated cell sorting
For knockout of the chicken myostatin (MSTN) gene, the nickase target loci were designed in exon 1 of chicken MSTN gene (Figure 1A). Two gRNAs (20 bp and 19 bp target sequences of left and right gRNA, respectively) were designed with +7 bp offset (Figure 1B). The gRNAs were controlled by U6 promoter and Cas9-D10A nickase was regulated by the cytomegalovirus promoter. For knockout of the MSTN gene, 7.5 μL Lipofectamine 3000 Reagent was diluted in 250 μL OPTI-MEM (Invitrogen, USA), and 2.5 μg each of the nickase (Cas9-D10A)-GFP co-expression plasmid (Sigma-Aldrich, St. Louis, MO, USA) and MSTN guide RNA (gRNA) was mixed with Lipofectamine P3000 Reagent in 250 μL OPTI-MEM at room temperature. After incubation for 5 min, the two mixtures were combined and incubated for an additional 20 min. The complex mixture was gently pipetted and dropped into a six-well plate containing chicken DF1 cells at 70% to 80% confluency. After incubation at 37°C in 5% CO2 for 4 h, cells were gently washed with phosphate-buffered saline (PBS) three times, and fresh culture medium was added. One day after lipofection, GFP-expressing cells were sorted using a FACSAria III cell sorter (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Following harvest using 0.05% trypsin-ethylenediaminetetraacetic acid (EDTA) (Invitrogen, USA), cells were resuspended in PBS containing 0.1% bovine serum albumin and strained through a 40 μm cell strainer for fluorescence-activated cell sorting (FACS) (BD Falcon; Becton, Dickinson and Company, Franklin Lakes, NJ, USA). After sorting, the cells were regrown in culture media for subsequent experiments.
Figure 1.
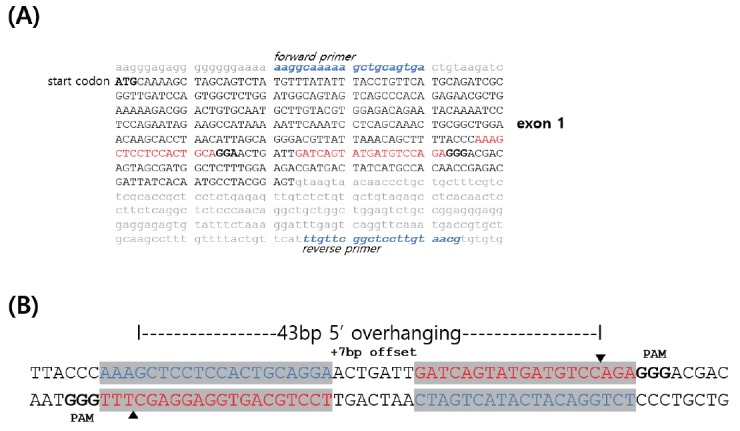
Chicken myostatin (MSTN) gene structure and targeted site design for nickase-mediated mutant induction. (A) Sequences of chicken MSTN exon1 and intron. The bold ATG indicates the start codon and the blue italic sequences present the forward and reverse primer for polymerase chain reaction amplification. The red sequences are the targeted sites of the CRISPR-associated protein 9 (Cas9)-D10A nickase. (B) The left and right target sequences (highlighted boxes) for the nickase. The target site has +7 bp offset and consequently, the targeted sites show 5′ overhanging 43 bp after the nickase-mediated DNA breaks. The red sequences are the targeted sites and the blue sequences are the complimentary sequences. The bold sequences indicate the protospacer adjacent motif (PAM). The arrow heads are the cleavage positions by the Cas-D10A nickase.
Off-target prediction and genotyping by T-vector cloning
To examine the non-specific mutation induced by the gRNAs of chicken MSTN gene, the off-targets of gRNAs were predicted by using the web-based program (Zhang Lab at Massachusetts Institute of Technology, http://crispr.mit.edu). Chicken genomic information (galGal4) of UCSC Genome Browser assembly was used for the off-target prediction.
Genomic polymerase chain reaction (PCR) was performed using an initial incubation at 94°C for 5 min, followed by 35 cycles of denaturation, annealing, and extension for each target gene or locus using the corresponding primer sets (Table 1). The reaction was terminated with a final incubation at 72°C for 7 min. To confirm the target locus mutation, PCR amplicons were cloned into the pGEM-T easy vector (Promega, Madison, WI, USA) and sequenced using an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
Table 1.
List of primer sequences for chicken myostatin (MSTN) gene and the predicted off-target sites for left and right guide RNA (gRNA) sequences
| Gene | Primer sequences | Locus | Annealing temp. (°C) | PCR product size (bp) |
|---|---|---|---|---|
| Chicken myostatin | Forward: 5′-aaggcaaaaagctgcagtga | 60 | 624 | |
| Reverse: 5′-cgttacaaggagccgaacaa | ||||
| Right gRNA off-target#1 | Forward: 5′-caggccagtctgaatgcctg | Chr3: −43439809 | 60 | 445 |
| Reverse: 5′-acacctgaagggaggatgca | ||||
| Right gRNA off-target#2 | Forward: 5′-ggtgctgtgaagccatgtca | Chr1:+172917848 | 60 | 422 |
| Reverse: 5′-gcccctggacaacactgttc | ||||
| Right gRNA off-target#3 | Forward: 5′-gtgaggagaggctgggagag | Chr2: −32334682 | 60 | 368 |
| Reverse: 5′-tccaggaaggttgaggtgtga | ||||
| Left gRNA off-target#1 | Forward: 5′-gaaggttcaccctctgctgc | Chr2:+95189546 | 60 | 415 |
| Reverse: 5′-cagaggtattgtgtgtggggg | ||||
| Left gRNA off-target#2 | Forward: 5′-aggccctgaaaaactggcat | Chr5: −50054848 | 60 | 405 |
| Reverse: 5′-actccctgagcagctaacca | ||||
| Left gRNA off-target#3 | Forward: 5′-gcagctgtgccagtgtatca | Chr2: −1000054090 | 60 | 497 |
| Reverse: 5′-ccggttcctctatgggctga |
Western blotting
Total protein was extracted with 1× radioimmunoprecipitation lysis buffer and separated on a 10% polyacrylamide gel followed by transfer to a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). The primary antibodies used were mouse anti-β-actin (Santa Cruz Biotechnology, Dallas, TX, USA) or anti-myostatin (Abcam, Cambridge, UK). Horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG (Bio-Rad, USA) were used as secondary antibodies. The blots were treated with enhanced chemiluminescence substrate solutions and exposed in a ChemiDoc XRS System (Bio-Rad, USA) to detect chemiluminescence.
RESULTS
Cas9-D10A nickase-mediated myostatin knockout in chicken DF1 cells
Based on the genome sequence information [NCBI (http://www.ncbi.nlm.nih.gov), Gallus gallus MSTN, NM_001001461 and UCSC Genomics Institute, chicken BLAT (http://genome.ucsc.edu)], the target guide RNA (gRNA) sites were designed following protospacer adjacent motif (PAM) sequences (Figure 1). The Cas9-D10A nickase target sites were designed in exon 1 of chicken MSTN gene (Figure 1A). Two gRNAs target 20 bp and 19 bp sequences of left and right gRNA, respectively were designed with a +7 bp offset and consequently, the targeted sites have 5′-overhanging of 43 bp after the Cas9-D10A nickase-mediated DNA breaks.
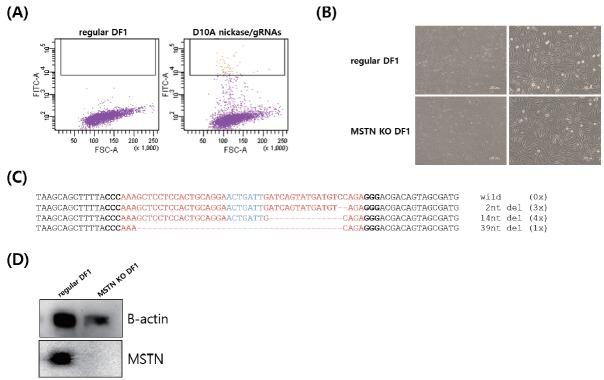
After co-transfection of the nickase-GFP and MSTN gRNA expression vectors into chicken DF1 cells, the highly GFP-positive cells were sorted out by FACS (Figure 2A). Subsequently, the stable MSTN knockout DF1 cells were established after sorting out and in vitro expansion culture. There was no significant phenotypic difference between regular and MSTN knockout DF1 cells (Figure 2B). In the MSTN knockout DF1 cells induced by the Cas9-D10A nickase, the various mutant genotypes of the mixed population were identified (Figure 2C). Surprisingly, all genotypes were identified as the deletion mutant and the number of the deleted nucleotides ranged from 2 to 39 nucleotides (Figure 2B). These mutant genotypes can induce a frameshift of the open reading frame or the deletion of 13 amino acids in myostatin protein (Figure 2B). To confirm the expression of myostatin protein, Western blotting was conducted with specific anti-chicken myostatin antibody. Comparing to regular DF1 cells, no positive signal was detected in the MSTN knockout DF1 cells indicating that the Cas9-D10A nickase-mediated knockout system efficiently and completely disrupted myostatin protein expression in chicken cells.
Figure 2.
Knockout of chicken myostatin (MSTN) gene and identification of mutant genotype in the targeted sites. (A) Fluorescence-activated cell sorting (FACS) of GFP-positive cells after co-transfection of the Cas9-D10A nickase expression vector with green fluorescent gene (GFP) gene and targeted multiplex guide RNAs (gRNAs). (B) Phenotypic comparison between regular and MSTN knockout DF1 cells. (C) Mutant genotypes of the targeted MSTN gene induced by the nickase. (D) Western blotting of chicken MSTN in regular and MSTN knockout DF1 cells.
Analysis of off-target effect in MSTN KO DF1 cells
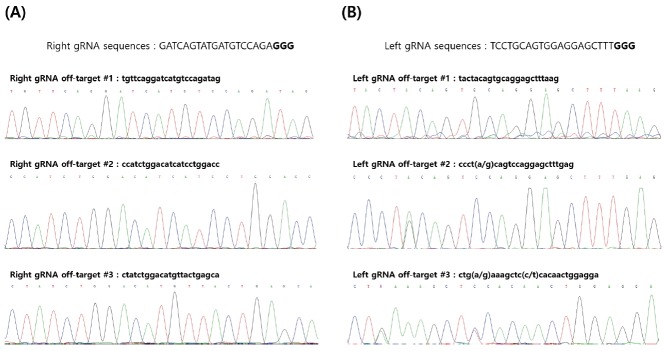
To investigate the non-specific off-target effect produced by the chicken myostatin knockout gRNAs, the predicted off-target sites were amplified and sequenced. Based on the chicken genomic sequences (galGal4), the off-target sites were analyzed and predicted on the web site of http://crispr.mit.edu (Zhang Lab at Massachusetts Institute of Technology). Finally, we selected six candidate sites (3 sites for each left and right of gRNA target) with higher score of the off-target effect (Table 1). These candidates of the off-target have 3–4 mismatch sequences compared to the original gRNA targets. Genotyping by amplification and sequencing of the predicted off-target sites showed that no mutant induction had occurred although a few of single nucleotide polymorphisms were detected in the targeted sites (Figure 3).
Figure 3.
Analysis of the predicted off-target sites. (A) Sequence analysis of the predicted off-target for right guide RNA (gRNA) sequences. (B) Sequence analysis of the predicted off-target for left gRNA sequences. Parentheses indicate single nucleotide polymorphism (SNP) in the targeted sites.
DISCUSSION
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system, which is the most efficient and reliable tool for precisely targeted genomic modification, can be applied and utilized for industrial applications as well as scientific research [10,11]. The most promising advantages of the CRISPR/Cas9 system are simplicity, large-scale output, and highly efficient genomic editing in various species [13–16]. Based on the somatic cell nuclear transfer (SCNT) cloning or direct injection technique into one-cell-stage embryos, CRISPR/Cas9-mediated knockout and knockin individuals have been successfully generated in various mammals [13,15,16]. Avian species have a distinguished history as a model system in the biological sciences [17]. However, in birds, the SCNT cloning technique and one-cell-stage microinjection method cannot be adapted for CRISPR/Cas9-mediated genome editing due to developmental differences and technical difficulties. Thus, combined with the genomic editing tools, the germ cell-mediated genetic modification system has been newly developed in poultry [2,3, 8,18].
Cas9 endonuclease was designed to direct a DNA double-strand break (DSB) with its guiding RNA at a desired genomic location. Subsequently, the cell activates an endogenous DNA repair pathway of NHEJ to ligate and fix the targeted DSB [10,11]. During this repairing process, the imprecise repair could lead to loss or insertion of nucleotides following by mutant induction of frameshift in the open reading frame of the specific target gene [10,11]. However, Fu et al [19] reported a high frequency of off-target mutagenesis induced by CRISPR/Cas9 in human cells. The off-target alterations generated by four out of six gRNAs targeted to other genomic loci in human cells were detected [19]. Off-target effect is the non-specific mutagenesis at an undesired genomic location and so these unwanted mutants could induce the frameshift in the structure of other functional genes [19–21]. Reduction of the off-target effect is one of major concern, especially for generation of genome-tailored livestock as well as therapeutic applications. The highly specific mutagenesis at the intended on-target site is a prerequisite and powerful option that will facilitate precision genome editing in livestock. In contrast, the newly developed Cas9-D10A nickase, a mutated Cas9 enzyme, generates only a single-strand DNA break rather than a DSB at the defined target sequences [12,21]. Single-strand breaks are preferentially repaired by homology directed repair (HDR) based on the intact opposite strand as a template [12,21]. Thus, HDR pathway shows high fidelity and rarely induces nucleotide deletion or addition during the repairing process [12,21]. In this study, we designed two adjacent (+7 bp offset, Figure 1B) but opposite single-strand breaks in chicken MSTN gene for Cas9-D10A nickase. As a result, the double-nicking system definitely did not induce any unwanted off-target effects in chicken cells (Figure 3).
Myostatin (also known as growth differentiation factor-8) is a secreted growth differentiation factor and a member of transforming growth factor beta protein family [22]. Myostatin protein negatively regulates skeletal muscle cell proliferation and differentiation during myogenesis [22]. Thus, mutations in MSTN gene are associated with the increased skeletal muscle mass in humans and other mammals [22–24]. Improving growth performances in livestock is one of the major research topics in the agricultural industry. Thus, a better understanding of skeletal muscle growth and differentiation mechanism could be directly applied to agribio industry. In this study, we examined and verified the Cas9-D10A nickase-mediated genomic editing in chicken cells to investigate the functional activity and regulatory mechanism. In the future, production of specific genome-tailored knockout and knockin cell lines will make important contributions via their use as in vitro models and in biofunctional studies in bioscience. Additionally, the potential of genome-edited chickens mediated by the CRISPR/Cas9 system reaches beyond basic research; locus-specific genome engineering will be widely used in agriculture and industry because of the lack of exogenous transgene integration.
ACKNOWLEDGMENTS
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01111401)” Rural Development Administration, Republic of Korea.
Footnotes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
REFERENCES
- 1.Palmiter RD, Norstedt G, Gelinas RE, Hammer RE, Brinster RL. Metallothionein-human GH fusion genes stimulate growth of mice. Science. 1983;222:809–14. doi: 10.1126/science.6356363. [DOI] [PubMed] [Google Scholar]
- 2.Park TS, Han JY. piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc Natl Acad Sci USA. 2012;109:9337–41. doi: 10.1073/pnas.1203823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macdonald J, Taylor L, Sherman A, et al. Efficient genetic modification and germ-line transmission of primordial germ cells using piggyBac and Tol2 transposons. Proc Natl Acad Sci USA. 2012;109:E1466–72. doi: 10.1073/pnas.1118715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park TS, Han JY. Genetic modification of chicken germ cells. Ann NY Acad Sci USA. 2012;1271:104–9. doi: 10.1111/j.1749-6632.2012.06744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson S, Clarke AR, Pow AM, Hooper ML, Melton DW. Germ line transmission and expression of a corrected HPRT gene produced by gene targeting in embryonic stem cells. Cell. 1989;56:313–21. doi: 10.1016/0092-8674(89)90905-7. [DOI] [PubMed] [Google Scholar]
- 6.Koller BH, Hagemann LJ, Doetschman T, et al. Germ-line transmission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homologous recombination in embryonic stem cells. Proc Natl Acad Sci USA. 1989;86:8927–31. doi: 10.1073/pnas.86.22.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zijlstra M, Li E, Sajjadi F, Subramani S, Jaenisch R. Germ-line transmission of a disrupted beta 2-microglobulin gene produced by homologous recombination in embryonic stem cells. Nature. 1989;342:435–8. doi: 10.1038/342435a0. [DOI] [PubMed] [Google Scholar]
- 8.Schwartzberg PL, Goff SP, Robertson EJ. Germ-line transmission of a c-abl mutation produced by targeted gene disruption in ES cells. Science. 1989;246:799–803. doi: 10.1126/science.2554496. [DOI] [PubMed] [Google Scholar]
- 9.Park TS, Lee HJ, Kim KH, Kim JS, Han JY. Targeted gene knockout in chickens mediated by TALENs. Proc Natl Acad Sci USA. 2014;111:12716–21. doi: 10.1073/pnas.1410555111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904–9. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–43. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 16.Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–5. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stern CD. The chick; a great model system becomes even greater. Dev Cell. 2005;8:9–17. doi: 10.1016/j.devcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 18.Schusser B, Collarini EJ, Yi H, et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc Natl Acad Sci USA. 2013;110:20170–5. doi: 10.1073/pnas.1317106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho SW, Kim S, Kim Y, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–41. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 23.Kambadur R, Sharma M, Smith TPL, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910–5. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- 24.Schuelke M, Wagner KR, Stolz LE, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350:2682–8. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]