Abstract
Exosomes are small membrane vesicles with size of 30-100 nm, which were found in bodily fluids including amniotic fluid and saliva. The biological materials in exosomes, such as proteins and RNA, can be used as novel potential biomarkers for diagnostic assays. The purpose of this study was to assess whether exosomal microRNAs (miRNAs) could be used as biomarkers to prenatally diagnose congenital hydronephrosis and to evaluate fetal kidney function. Transmission electron microscopy (TEM), flow cytometry (FACS), and western-blot were applied to identify exosomes in the amniotic fluid from fetuses with congenital hydronephrosis and healthy controls. Exosomal miRNA was extracted according to the manufacturer’s protocol and used for microarray. The differentially expressed miRNAs were selected for further study. The miRNA targets were analyzed to assess their possible function in the pathophysiology of obstructive nephropathy, and the miRNA array results were confirmed by qPCR. Amniotic fluid exosomes were identified based on CD24 and CD9 expression. The has-miR-942, has-miR-4289, has-miRPlus-A1073, and has-miR-195-3p were up-regulated in amniotic fluid exosomes from fetuses with congenital hydronephrosis comparing with those in healthy controls, and 35 had reduced expression levels. These results were confirmed by using qPCR. After integrating the miRNAs targets predicted via three databases and subjecting those target genes to KEGG pathway analysis, we found that the target genes of hsa-miR-300 and hsa-miR-299-5p were determined to be part of the Wnt signaling pathway. In addition, DVL2, PP2R5A, SRFP2, and SIAH1 predicted as target genes of has-miR-300 and has-miR-299-5p are informative for further exploration of congenital hydronephrosis pathologies. The reduced expression of hsa-miR-300 and hsa-miR-299-5p in the amniotic fluid of congenital hydronephrosis could be a biomarker for kidney fibrosis associated with congenital obstructive nephropathy.
Keywords: Congenital obstructive nephropathy, miRNA, exosome, prenatal diagnosis
Introduction
Congenital hydronephrosis could cause acute or chronic damage to the fetal kidney. As one of the most common abnormalities of urinary system, congenital hydronephrosis is associated with pathological changes in the kidney and leads to postnatal infant mortality. Approximately 15-20% of infants with congenital hydronephrosis require surgery [1], particularly when both of the kidneys were obstructed, resulting in rapid deterioration of kidney function. Ulti-mately, dialysis or a renal transplant may be required for the infant to survive. In other cases, hydronephrosis may disappear over time if there is no sign of urinary obstruction. It is a great challenging for both fetal medicine physicians and pediatric surgeons to prevent the loss of kidney function from obstructive urinary diseases. However, evaluation of kidney function and early diagnosis of prenatal urinary diseases have been stumbled due to lack of biomarkers.
Exosomes have no cytoplasmic organoids, such as the lysosome, mitochondrion, endoplasmic reticulum, nucleolus, and Golgi apparatus. Exosomes are also more stable and have a more homogeneous modality than other larger vesicles, such as apoptotic bodies [2]. The production of exosomes is initiated when cell membrane proteins transfer to early endosomes by inward budding. In early endosomes, molecules are either recycled to the plasma membrane or enwrapped into internal vesicles, commonly called multi-vesicular bodies (MVBs). Exosomes could be secreted by variety types of cells, such as dendritic cells, macrophages, lymphocytes, salivary gland epithelial cells, and tumor cells [3]. They have also been found in saliva, plasma, urine, amniotic fluid, malignant ascites, and bronchial veolar lavage fluid [4]. Many important biological materials, such as messenger RNA (mRNA), microRNAs (miRNAs), proteins, lipids, and DNA were found in Exosomes. Exosomes released from different cell types contain different biological information and therefore have different functions [4]. Interestingly, recent studies have demonstrated that exosomes can transfer their contents, such as mRNA and miRNAs to other cells, indicating that intracellular messages can be transmitted through exosome uptake. Exosomes could also transmit information via binding to receptors on the cell surface and activating cell signaling pathways [5,6]. As a highly glycosylated glycosyl phosphatidylinositol (GPI)-anchor protein, CD24 was believed to be involved in cell-cell adhesion and signaling, which has been found in various cancers including nasopharyngeal carcinoma, glioma cancer, breast cancer, and renal cell carcinoma [7]. CD9 is a member of tetra-span in superfamily with a highly conserved tertiary structure consisting of four trans-membrane domains, which is associated with many tissue-specific trans-membrane proteins [8]. Keller et al. showed that CD24 and CD9 were detected in exosomes secreted form amniotic fluid, which inspired researchers to developing methods utilizing this biomarker for prenatal diagnosis [9]. Recently, exosomes have been found to contain biological messages, such as proteins and RNA, which could influence gene expression and signal transduction in cells [10]. Exosomal proteins and RNA can also be used as biomarkers to diagnose disease and predict disease outcome [11]. The discovery of exosomes in human amniotic fluid could provide a novel avenue to study fetal kidney diseases.
The miRNAs are small single stranded RNAs (21-25 nucleotides). They regulate gene expression in both human and plant cells. The miRNAs generally exert their regulatory effects at the post-transcriptional level by binding to the 3’-untranslated region of the target mRNA. The degree of complementarity between the miRNA and the target mRNA determines the negatively regulation mechanism for gene expression. A perfect match between the miRNA and the target mRNA results in the cleaved mRNA by the RISC complex, while an imperfect match leads to translational repression of the mRNA. The suppressed mRNAs can be released to induce translation if required [12]. The discovery and characterization of miRNAs in the last decade has revolutionized our understanding of the regulatory functions of RNA in tissue and organ development and in the pathophysiology of cellular aging. There is also evidence that miRNAs have important functions in a variety of kidney diseases, such as renal cancer, polycystic kidney disease, EMT, IgA renal disease, lupus nephritis, and kidney allograft rejection [13-18]. However, there is rare contribution devoted to the relationship between miRNAs and obstructive nephropathy. In this study, we intend to make a first step elaborate about the relationship between miRNAs and congenital obstructive nephropathy before the children were born.
Material and methods
Chemicals and antibodies
FITC Mouse Anti-Human CD24 (555427, HAS, 1:50, BD), PE Mouse Anti-Human CD9 (555372, HAS, 1:50, BD), CD24 antibody (sc-11406, SANTA CRUZ, 1:200), CD9 antibody (sc-9148, SANTA, 1:200).
Human samples
This study was approved by the ethics commission of the first affiliated hospital of SUN YAT-SEN UNIVERSITY. Routine amniocentesis was performed to acquire 10 mL of amniotic fluid from eight pregnant women who had indications for a prenatal diagnosis of congenital hydronephrosis during the second trimester of pregnancy from September 2010 to September 2012. None of the pregnant woman had complications resulting from the amniocentesis. All of the acquired amniotic fluid was stored at -80°C until preforming isolation and identification. For microarray analysis, four fetuses with hydronephrosis in one or both kidneys were enrolled into the experimental group and fetuses with no congenital abnormalities were enrolled into control group. None of the fetuses had dilated ureter(s).
Isolation of exosomes
Amniotic fluid samples (10 mL) were vortexed for 2 min immediately after thawing at 4°C. To isolate the micro-vesicles, the amniotic fluid was centrifuged for 20 min at 300×g to remove cells and 20 min at 10,000×g to remove cellular debris. The vesicles were pelleted by centrifugation for 2.5 h at 100,000×g in a Beckmann ultracentrifuge. The vesicle pellet was re-suspended in 2 mL of PBS sample buffer for analysis or processed by sucrose density centrifugation. Isolated exosomes were loaded onto a sucrose step gradient with the layers: 2 M, 1.3 M, 1.16 M, 0.8 M, 0.5 M and 0.25 M sucrose as described in reference [19]. The gradients were centrifuged for 2.5 h at 100,000×g in a Beckman ultracentrifuge. 200 μL fractions were collected from the top of the gradient.
Transmission electron microscopy (TEM)
Briefly, about 20 μL of the exosome preparation was layered onto formvar-coated 200 mesh copper grids, and allowed to dry under an incandescent lamp for 2 min. Then the copper grids were negatively stained with phosphato-tungstic acid. Imaging was obtained under a transmission electron microscope operated between 60 kV.
Flow cytometry (FACS)
Exosomes were incubated overnight with beads at 4°C with gentle shaking. Blocking was performed by adding 300 μL of 200 mM glycine and incubating for 30 min. The exosome-bead complexes were washed three times in staining buffer. Then the PE Mouse Anti-Human CD9 and FITC Mouse Anti-Human CD24 were added separately to the exosome-bead complexes and incubated for 40 min with gentle shaking. The exosome-bead complexes were used for FACS analysis after three times of washing with staining buffer.
Western blot analysis
The exosome pellets were dissolved in protein lysis buffer, pipetted thoroughly, and then vortexed. To further lyse the exosomes, the samples were sonicated in a water bath three times for 5 min and vortexed between sonication steps. Finally, the samples were centrifuged at 14 000×g for 3 min at room temperature. The supernatant was transferred to a new eppendorf tube. The proteins were separated and transferred to a membrane by gel electrophoresis and electroblotting. The membrane was blocked and washed before immunestaining for detection of proteins enriched in the exosomes. Immunoreactive bands were detected by chemiluminescence using a digital imaging system and analysis software.
Extraction of exosomal RNA
TRIzol was used to extract the exosomal RNA according to the manufacturer’s protocol. The quality of the total RNA was examined using an ultraviolet spectrophotometer.
miRNA microarray analysis
The 6th generation of Exiqon’s miRNA microarray contains more than 1891 capture probes, including all of human, mouse, and rat miRNAs annotated in miRBase 16.0, as well as all of the viral miRNAs for these species. In addition, this array contains capture probes for 66 new miRPlus™ human miRNAs. These are proprietary miRNAs not found in miRBase. Exiqon’s miRNA arrays feature Tm-normalized LNA™-enhanced capture probes, designed for excellent specificity and sensitivity, even for AT-rich miRNAs. The arrays are also highly reproducible and have a 99% correlation between arrays and a dynamic range greater than five orders of magnitude.
KEGG pathway analysis of miRNA target genes
We have integrated miRNA targets in our proprietary database from three miRNA target databases, including Miranda, Mirbase and Targetscan. By using this database, once a miRNA that was confirmed to be up- or down-regulated, genes with target sites for at least two co-expressed miRNAs (simultaneously up- or down-regulated) could be identified as a potential cooperative target gene set and their possible functions in the pathophysiology of obstructive nephropathy would be also suggested. To facilitate the functional annotation and analysis of large lists of genes in the regulatory network, we input the target genes for KEGG term enrichment analysis.
qPCR analysis to confirm miRNA expression profiles
Routine amniocentesis was performed to acquire 10 mL of amniotic fluid from another 16 pregnant women who had indications for prenatal diagnosis. Eight fetuses with hydronephrosis in one or both kidneys were enrolled into experimental group. None of the fetus had a dilated ureter or ureters. The other 8 fetuses with no congenital abnormalities were enrolled into the control group. None of the pregnant women experienced complications related to the amniocentesis. The amniotic fluid was stored at -80°C until further use. Differential centrifugation was applied to obtain purified exosomes from the amniotic fluid. TRIzol was used to extract RNA from exosomes according to the manufacturer’s protocol. The qPCR was then performed to verify the reliability of the miRNA microarray results. The cDNA was synthesized with High Capacity RNA-to-cDNA Kit (Life technologies), and then amplified with SYBR qPCR Mix (TOYOBO, Japan). Real-time PCR was performed in Applied Biosystems 7300 Sequence Detection system. The 10 μl mixture containing 0.70 μl RT product, 1× TaqMan Universal PCR master mix, and 1 μl of primers and probe mix of the TaqMan MicroRNA Assay protocol (PE Applied Biosystems) was incubated in a 96-well optical plate at 95°C for 10 min, followed by 37 cycles of 94°C for 15 s and 62°C for 7 min. Ct values for each sample were determined and normalized to GAPDH.
Results
Morphology of exosome under TEM
To ensure the existence of exosomes in amniotic fluid, we applied the TEM to investigate the morphology of exosomes. The exosomes with size of 40 nm-100 nm had a typical cup-shaped morphology, and were surrounded by a two-layer lipid membrane. The exosomes were not disorganized so that it could reflect the bona fide molecular profiles of the fetus. Both the areas surrounding and inside the exosomes had a negative staining effect (Figure 1).
Figure 1.

Representative TEM image of exosomes isolated from amniotic fluid. The exosomes (indicated with arrows) with size of 40 nm-100 nm had atypical cup-shaped morphology, and were surrounded by a two-layer lipid membrane. Both surrounding and inside areas of the exosomes have negative staining effect.
Exosomal expression of CD24 and CD9 in amniotic fluid
The presence of exosomes in the amniotic fluid was confirmed using the exosome specific markers CD24 (Figure 2) and CD9 (Figure 3) by FACS. CD24 positive beads were concentrated in Q4, and 92.23%±3.8% of events was CD24+. Similarly, CD9 positive beads were concentrated in Q1, and 68.3%±5.9% of events was CD9+. In addition, both CD24 and CD9 were detected by western blot in exosomes from amniotic fluid (Figure 4).
Figure 2.

Representative FACS analysis plots for CD24 expression on exosome-bead conjugates.
Figure 3.
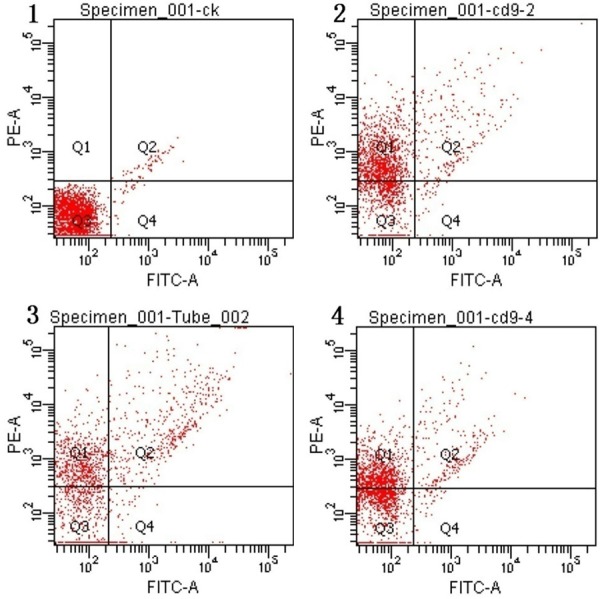
Representative FACS analysis plots for CD9 expression on exosome-bead conjugates.
Figure 4.
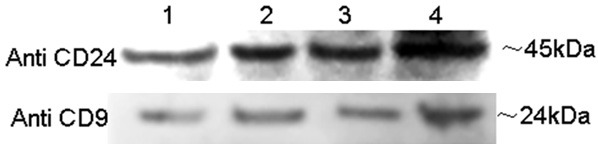
Representative western blot images of CD24 and CD9 expression in purified exosomes.
RNA quality analysis
To ensure that RNA content between experimental and control group is not an influence factor for expression level of specific miRNAs, the RNA quality of each sample was quantified. The concentrations of exosomal RNA isolated from the control samples were 28.67 ng/μL, 30.02 ng/μL, 37.57 ng/μL, and 21.08 ng/μL. The concentrations of exosomal RNA harvested from the experimental group samples were 49.41 ng/μL, 24.57 ng/μL, 24.24 ng/μL, and 26.77 ng/μL. The average concentration of purified exosomal RNA was not significantly different among groups. On average, 30.28 ng/μL of exosomal RNA was collected from the control and hydronephrosis groups.
Hierarchical clustering of differentially expressed miRNAs separated from experimental and control group
Since no differences were detected in RNA content between the exosomes in experimental and control group, miRNA profiling was conducted by using microarray chip. The differentially expressed miRNAs were defined as those with log transformed absolute fold change beyond 1.5. In total, we identified 39 miRNAs that were differentially expressed between hydronephrosis and control group, which was hierarchically clustered and visualized in a heat map (Figure 5). Each row represents an individual miRNA and each column represents an individual sample. The miRNA clustering tree is shown on the left, and the sample clustering tree appears at the top. Cluster analysis arranged the samples and miRNAs into groups based on their expression levels, in which the relationships between miRNAs and samples were apparent and intuitive. It is obvious that the experimental group was separated from control group.
Figure 5.
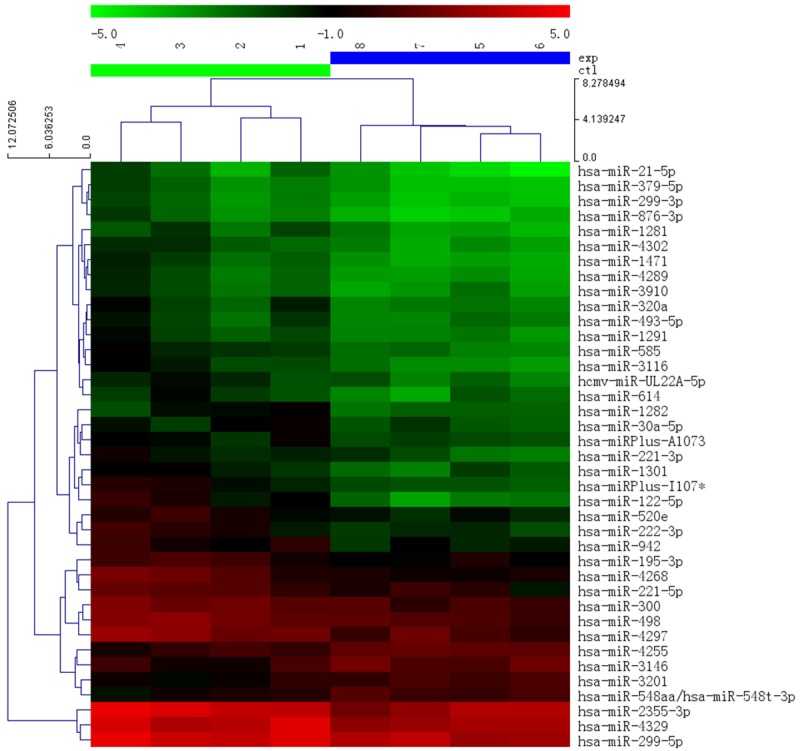
Cluster analysis for 39 selected miRNAs from amniotic fluid exosomes. Left cluster represents control and right cluster represents samples with hydronephrosis. The top bar is legend indicating colors corresponding to expression levels.
KEGG pathway analysis of miRNA target genes
In the 39 differentially expressed miRNAs, four miRNAs were elevated in the congenital hydronephrosis samples comparing with the controls, and 35 miRNAs were reduced in the congenital hydronephrosis samples. Based on previous research and the results of our microarray analysis, we selected 12 of the differentially expressed miRNAs for further analysis: hsa-miR-3201, hsa-miR-4255, hsa-miR-3146, hsa-miR-548aa/hsa-miR-548t-3p, hsa-miRPlus-A1073, hsa-miR-299-5p, hsa-miR-2355-3p, hsa-miR-195-3p, hsa-miR-300, hsa-miR-4289, hsa-miR-942, and hsa-miR-4329. Among them, has-miR-942, has-miR-4289, has-miRPlus-A1073, and has-miR-195-3p were up-regulated in the samples with hydronephrosis and the others were down-regulated. Then, potential target genes were searched by using Mirbase, Miranda, and Targetscan. The 12 miRNAs had 27266 possible target genes in Miranda, 2812 in Mirbase, and 3729 in Targetscan. For these possible target genes, 145 were identified by all three databases. The 145 potential targets were analyzed using the KEGG pathways. Since the Wnt signaling pathway has been reported to play a critical role in the process of kidney fibrosis, we focused on the genes involved in this pathway. Finally, four genes implicated in this pathway, DVL2, PPP2R5A, SRFP2, and SIAH1 were identified. By TargetScan prediction, two miRNAs were found to be up-regulated in hydronephrosis samples as potential regulator of these genes, has-miR-300 and has-miR-299-5p.
hsa-miR-300 and hsa-miR-299-5p were reduced in congenital hydronephrosis
Consistent with the results from the microarray analysis, qPCR showed that the expressions of hsa-miR-300 and hsa-miR-299-5p were reduced in the congenital hydronephrosis samples comparing with the normal samples (Figure 6; Table 1).
Figure 6.
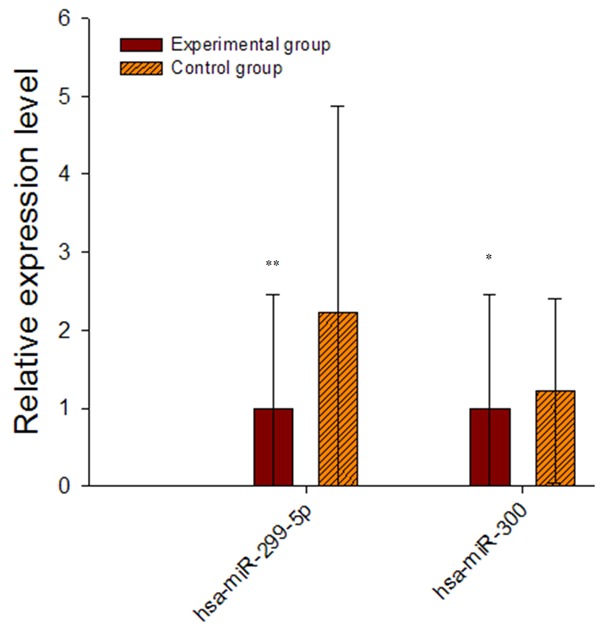
qPCR for expression levels of hsa-miR-300 and hsa-miR-299-5p in samples from fetuses with congenital hydronephrosis and healthy controls. The results confirmed that expression of both miRNAs was reduced in the hydronephrosis samples comparing with the controls.
Table 1.
The expressions of hsa-miR-300 and hsa-miR-299-5p in experiment and control groups
| miRNA | exp | ctl |
|---|---|---|
| hsa-miR-299-5p | 0.756±1.375 | 1.000±1.152 |
| hsa-miR-300 | 0.128±0.226 | 1.000±1.045 |
Discussion
Congenital urinary diseases are abnormalities that lead to pathophysiological changes in the fetal kidneys and cause acute or chronic damage. Congenital urinary diseases are also associated with kidney pathology and infant mortality [20]. Currently, there are no biomarkers for prenatal evaluation of kidney function and diagnosis of urinary diseases before the child is born. Recently, biological messages, such as proteins and RNA, have been found in exosomes, which could influence gene expression and signal transduction in cells. Exosomal proteins and RNA can also be applied as biomarkers for disease diagnose and outcome prediction [21,22]. The discovery of exosomes in human amniotic fluid could provide a novel avenue to study fetal kidney diseases [21]. We demonstrated the presence of exosomes in the amniotic fluid using TEM, FACS, and western-blot. Amniotic fluid exosomes expressed both CD24 and CD9. These exosomes contain almost equal quality of RNA in hydronephrosis samples and control ones, and the differentially expressed miRNAs identified from them are informative with regards to positivity of disease. Therefore, these exosomes may provide a novel basis for further research.
Using CD24 as a marker, Keller et al. [23] have demonstrated that the amniotic fluid exosomes was generated from fetus’ kidneys. The exosomes used in this study showed positive for both CD24 and CD9, which is consistent with the published findings at http://www.exocarta.org/. Therefore, the materials in the exosomes, such as mRNA, miRNAs, proteins, lipids, and DNA can be used as biomarkers of prenatal kidney function for fetal kidney disease diagnose and prenatal intervention guidance.
Analysis of human urinary exosomal mRNA could provide novel insights about genetic kidney diseases, such as Autosomal Dominant Polycystic Kidney Disease (ADPKD) [24]. After purifying exosomes from the amniotic fluid, the RNA could be extracted at concentration of 30.29 ng/μL, suggesting RNA from amniotic fluid exosomes could be applied as biomarkers to diagnose fetal kidney diseases and evaluate fetal kidney function. Alterations of miRNAs in urinary exosomes are associated with kidney function, indicating various chronic kidney diseases [25,26].
Obstructive uropathies account for the majority of cases and occur in approximately 1:2000 pregnancies [27]. Obstruction at the level of the urethra must be bilateral and it might involve some or all of the urinary tract. The majority of genitourinary abnormalities are diagnosed around 18-20 weeks. However, with the increasing use of first-trimester screening, more severe renal anomalies are being noted between 11 and 14 weeks by ultrasound. An accurate antenatal diagnosis allows caregivers and parents to plan for appropriate prenatal and postnatal care, consider prenatal intervention, and for parents to make psychological adjustments. Prenatal radiology can usually provide information only pertaining to changes in kidney modality, and not pathophysiological changes to the fetal kidneys or deterioration of fetal kidney function. Therefore, an objective biomarker that can be used to evaluate and monitor fetal kidney function and to guide therapy would be a valuable clinical tool, particularly for guiding early intervention for obstruction, which could promote the recovery of kidney function [28]. Obstructions in the fetal urinary tract can lead to antenatal obstructive uropathies. Ureterpelvic junction obstruction is the most common lesion of the fetal urinary tract. The causes of an ureterpelvic junction obstruction include abnormal recanalization of the ureter during development, a typical mucosal valves, ureteric duplication, and ureteral strictures. For consistency, in our experiment, all of the fetuses in the congenital hydronephrosis group were diagnosed with an ureterpelvic junction obstruction. The fetuses in the control group had no congenital or chromosomal abnormalities.
The prognosis of obstructive uropathies depends on the development of useful biomarkers to evaluate and monitor fetal kidney function, and defining the optimal time for surgical intervention to stop the deterioration of fetal kidney function. After birth, ultrasounds and radionuclide renal scans can help accurately grade the severity of hydronephrosis on ureteropelvic junction obstruction (UPJO). However, it is still difficult to establish a prognosis and decide when surgery is warranted. Thus, there is an urgent need to establish biomarkers from bodily fluids, such as blood, urine, and amniotic fluid to diagnose obstructive uropathies and the deterioration of kidney function. We acquired fetal body cells from routine amniocentesis, which was used to diagnosis congenital diseases. However, this method is not specific to urinary diseases because the cells harvested are from every part of the body. We hypothesized that the miRNAs from exosomes in the amniotic might be specific to urinary diseases, because they come from the fetal kidney [21].
UPJO has led the effort to define the pathophysiology of obstructive nephropathy. Following urinary tract obstruction and tubular dilatation, a cascade of events resulted in up-regulation of the intrarenal renin-angiotensin system, tubular apoptosis, and macrophage infiltration of the interstitium. This is followed by accumulation of interstitial fibroblasts through proliferation of resident fibroblasts and the epithelial-mesenchymal transformation (EMT) of renal tubular cells [29]. EMT describes a phenotypic change in epithelial cells that lose their defined cell-cell-basement membrane contacts and their structural polarity to become spindle-shaped and morphologically similar to mesenchymal/myofibroblast cells. The progression of obstructive nephropathy has been described as damage to structure and function of the tubular epithelial cells of the kidney. The cells lose their conglutination and morphology and undergo EMT, where they differentiate to fibroblasts [30]. The combination of these processes leads to kidney fibrosis. Studies using the mouse as a unilateral ureteral obstruction (UUO) model have shown the proliferation of myofibroblasts during EMT [31]. Alison J. Kriegel et al. [32]. Established a model of EMT that is important for understanding the development of renal interstitial fibrosis by treating human renal epithelial cells with TGF-β. EMT can promote the repair of tissue, but it is also associated with chronic inflammation, cancer transformation and development, and organization fibrosis. EMT of tubular epithelial cells underlies the formation of renal fibroblast [33].
We identified four putative target genes (DVL2, PPP2R5A, SRFP2, and SIAH1), which were seemly regulated by two differentially regulated miRNAs (hsa-miR-299-5p and hsa-miR-300) in this study. The identified genes are components of the Wnt signaling pathway, which has been associated with the process of kidney fibrosis [34,35]. There are three Wnt signaling pathways: the classical Wnt/β-catenin pathway (canonical Wnt/nonatenin), the planar cell polarity pathway, and the Wnt/Ca2+ pathway [36,37]. When the urinary tract is obstructed, increasing of compressive stress on the renal tubular epithelial cells activates the JNK gene, which is part of the planar cell polarity pathway, to inhibit renal cell growth and to induce apoptosis [38,39], suggesting that kidney dysplasia and tubular epithelial cell apoptosis during congenital obstructive nephropathy could be mediated by the planar cell polarity pathway [34,35]. It has also been suggested that the classical Wnt/β-catenin pathway (canonical Wnt signals) is involved in EMT associated with congenital obstructive uropathy. In this case, β-catenin and SFRP are up-regulated and wnt-4 is down-regulated, inducing kidney fibrosis. The miRNAs have also been reported to take part in the classical Wnt/β-catenin pathway [40-42]. In our experiment, two miRNAs hsa-miR-299-5p and hsa-miR-300 were down-regulated. Their putative target genes were PPP2R5A, SIAH1, DVL2, and SFRP2, all of which take part in the classical Wnt/β-catenin pathway. DVL2 is also part of the planar cell polarity pathway, and SFRP2 has been shown to contribute to kidney fibrosis during obstructive uropathy. Therefore, we hypothesize that hsa-miR-299-5p and hsa-miR-300 could contribute to renal fibrosis in obstructive uropathy. hsa-miR-300 may also play a role in kidney dysplasia and apoptosis of tubular epithelial cells. Further research is required to explore the molecular mechanism underlying these processes. As the investigation of mechanisms underlying those predicted target genes remain scarce, our studies also provide valuable indications for further researches exploring the role of those genes in fundamental biology or diagnosis of nephrosis.
Conclusion
We have proven that exosomes exist in the amniotic fluid. Furthermore, the exosomes express CD9 and CD24, which are biological markers of exosomes derived from fetal kidneys. The miRNAs were also identified to be differentially expressed between congenital hydronephrosis samples and control samples. Reduced expression of hsa-miR-300 and hsa-miR-299-5p in the amniotic fluid of fetuses with congenital hydronephrosis may serve as biomarkers of kidney fibrosis during congenital obstructive nephropathy.
Acknowledgements
This work was supported by Guangdong province science and technology plan projects No. 2012B031800297. The authors express their appreciation to Dr. Lin Hao-tian at the Zhongshan Ophthalmic Center, SUN YAT-SEN UNIVERSETY for reviewing our results.
Disclosure of conflict of interest
None.
References
- 1.Madsen MG. Urinary biomarkers in hydronephrosis. Dan Med J. 2013;60:B4582. [PubMed] [Google Scholar]
- 2.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 3.Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One. 2010;5:e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 5.Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr. 2007;1:156–158. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 7.Kristiansen G, Sammar M, Altevogt P. Tumour biological aspects of CD24, amucin-like adhesion molecule. J Mol Histol. 2004;35:255–262. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 8.Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci. 2001;114:4143–4151. doi: 10.1242/jcs.114.23.4143. [DOI] [PubMed] [Google Scholar]
- 9.Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 10.Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 11.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 12.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 13.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, Baffa R. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 14.Torres VE, Harris PC. Mechanisms of disease: autosomal dominant and recessive polycystic kidney diseases. Nat Clin Pract Nephrol. 2006;2:40–55. doi: 10.1038/ncpneph0070. [DOI] [PubMed] [Google Scholar]
- 15.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int. 2009;29:749–754. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 16.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Microarray analysis of micro-ribonucleic acid expression in primary immunoglobulin A nephropathy. Saudi Med J. 2008;29:1388–1393. [PubMed] [Google Scholar]
- 17.Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009;106:5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolfschoten IG, Roggli E, Nesca V, Regazzi R. Role and therapeutic potential of microRNAs in diabetes. Diabetes Obes Metab. 2009;11:118–129. doi: 10.1111/j.1463-1326.2009.01118.x. [DOI] [PubMed] [Google Scholar]
- 19.Cantin R, Diou J, Bélanger D. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Nwoko R, Plecas D, Garovic VD. Acute kidney injury in the pregnant patient. Clin Nephrol. 2012;78:478–486. doi: 10.5414/cn107323. [DOI] [PubMed] [Google Scholar]
- 21.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 24.Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, Tammachote R, Huang BQ, Leontovich AA, Beito TG, Madden BJ, Charlesworth MC, Torres VE, LaRusso NF, Harris PC, Ward CJ. Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol. 2009;20:278–288. doi: 10.1681/ASN.2008060564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akkina S, Becker BN. MicroRNAs in kidney function and disease. Transl Res. 2011;157:236–240. doi: 10.1016/j.trsl.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenzen JM, Haller H, Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol. 2011;7:286–294. doi: 10.1038/nrneph.2011.26. [DOI] [PubMed] [Google Scholar]
- 27.Scherz HC, Kaplan GW. Etiology, diagnosis, and management of urethral strictures in children. Urol Clin North Am. 1990;17:389–394. [PubMed] [Google Scholar]
- 28.Smith JM, Stablein DM, Munoz R, Hebert D, McDonald RA. Contributions of the Transplant Registry: the 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS) Pediatr Transplant. 2007;11:366–373. doi: 10.1111/j.1399-3046.2007.00704.x. [DOI] [PubMed] [Google Scholar]
- 29.Chevalier RL. Obstructive nephropathy: towards biomarker discovery and gene therapy. Nat Clin Pract Nephrol. 2006;2:157–168. doi: 10.1038/ncpneph0098. [DOI] [PubMed] [Google Scholar]
- 30.Carew RM, Wang B, Kantharidis P. The role of EMT in renal fibrosis. Cell Tissue Res. 2012;347:103–116. doi: 10.1007/s00441-011-1227-1. [DOI] [PubMed] [Google Scholar]
- 31.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, Liang M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: a novel role of miR-382. Nucleic Acids Res. 2010;38:8338–8347. doi: 10.1093/nar/gkq718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oba S, Kumano S, Suzuki E, Nishimatsu H, Takahashi M, Takamori H, Kasuya M, Ogawa Y, Sato K, Kimura K, Homma Y, Hirata Y, Fujita T. miR-200b precursor can ameliorate renal tubulointerstitial fibrosis. PLoS One. 2010;5:e13614. doi: 10.1371/journal.pone.0013614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hermens JS, Thelen P, Ringert RH, Seseke F. Alterations of selected genes of the Wnt signal chain in rat kidneys with spontaneous congenital obstructive uropathy. J Pediatr Urol. 2007;3:86–95. doi: 10.1016/j.jpurol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Liu Y. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am J Pathol. 2001;159:1465–1475. doi: 10.1016/S0002-9440(10)62533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huelsken J, Behrens J. The Wnt signalling pathway. J Cell Sci. 2002;115:3977–8. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- 37.Miller JR. The Wnts. Genome Biol. 2002;3:REVIEWS3001. doi: 10.1186/gb-2001-3-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 39.Nagafuchi A, Shirayoshi Y, Okazaki K, Yasuda K, Takeichi M. Transformation of cell adhesion properties by exogenously introduced E-cadherin cDNA. Nature. 1987;329:341–343. doi: 10.1038/329341a0. [DOI] [PubMed] [Google Scholar]
- 40.Strillacci A, Valerii MC, Sansone P, Caggiano C, Sgromo A, Vittori L, Fiorentino M, Poggioli G, Rizzello F, Campieri M, Spisni E. Loss of miR-101 expression promotes Wnt/beta-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229:379–389. doi: 10.1002/path.4097. [DOI] [PubMed] [Google Scholar]
- 41.Cha YH, Kim NH, Park C, Lee I, Kim HS, Yook JI. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle. 2012;11:1273–1281. doi: 10.4161/cc.19618. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, Wu F. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/betacatenin pathway. Liver Int. 2012;32:752–760. doi: 10.1111/j.1478-3231.2011.02750.x. [DOI] [PubMed] [Google Scholar]


