Abstract
MicroRNAs (miRNAs) are noncoding single-stranded RNAs of ~22 nucleotides suppressing a wide range of gene expression by direct degradation or translational inhibition of their target mRNAs. Acute myocardial infarction (AMI), a common cardiovascular disease mainly induced by coronary artery occlusion, can lead to the development of heart failure. Several recent findings have indicated that miRNAs might play vital roles in AMI, and some miRNAs have even been proposed as potential candidates for intervening AMI. However, the pathophysiological functions of miRNAs implicated in MI are still largely unidentified. Here, we show that miR-1231 is overexpressed both in human hearts after MI insults and in rat hearts with experimental MI compared with their healthy counterparts. Next, by using predictive strategy and gene expression array, cacna2d2 is identified as the target of miR-1231. In human and rat ischemic hearts, cacna2d2 expression is indeed suppressed by miR-1231. In addition, the inhibition of miR-1231 in vivo ameliorates arrhythmias in rat MI hearts; conversely, the forced overexpression of miR-1231 promotes arrhythmias. Furthermore, cacna2d2 knockdown alone induced arrhythmias in ischemic hearts, despite knockdown of miR-1231. Thus, these results indicate that miR-1231 exacerbates arrhythmia by inhibiting cacna2d2 in ischemic heart, which shed light on the important arrhythmogenic function of miR-1231 in MI and suggest it may serve as a potential antiarrhythmic target.
Keywords: Myocardial infarction, miRNAs, miR-1231, cacna2d2, arrhythmia
Introduction
MicroRNAs (miRNAs) are small noncoding single-stranded RNAs of ~22 nucleotides constituting a novel class of regulators that participate in controlling a wide range of gene expression patterns [1]. Mechanically, through physically binding to the specific sites of 3-untranslated region (3’-UTR) in their mRNA targets, the mature miRNAs post-transcriptionally downregulate target expression via direct degradation or translational inhibition [2]. Among their well-recognized functional versatilities, several miRNAs have been shown involved in important processes contributing to the pathophysiological consequences of acute myocardial infarction (AMI) [3]. AMI, one common cardiovascular disease, has long been one of the leading causes of morbidity and mortality for humans in developed countries, therefore, it’s urgent to develop new therapeutic approaches [4]. MI is mainly caused by coronary artery occlusion, followed by the deficiency of blood flow supplies to the myocardium region of heart, which results in the death of cardiac myocytes, impaired contractility, loss of pump function and susceptibility to abnormal ventricular heart rhythm (arrhythmias), eventually the development of chronic heart failure [5].
Ample studies support critical roles of miRNAs in MI, such as: 1) either promote or inhibit the cell death of cardiomyocytes [6,7]; 2) regulate postischaemic angiogenesis [8]; 3) control cardiac regeneration [9]; 4) direct reprogramming of cardiofibroblasts into cardiomyocytes [10]. Based on these findings, several miRNAs have been proposed as potential candidates for developing therapeutic approaches to MI. However, questions still remain, such as which miRNAs are implicated in MI and their specific roles in MI are still largely uncharacterized [3]. More recently, miR-1231 in whole blood and serum sample was found increased and significantly correlated with the severity of non-ischemic systolic heart failure as illustrated by left ventricular ejection fraction [11], which points to a connection between miR-1231 and cardiovascular disease.
In this study, we aimed to examine whether miR-1231 might have pathophysiological functions in MI. Firstly, we noticed that miR-1231 was abnormally overexpressed in both human MI hearts and rat experimental MI hearts compared with their healthy counterparts; in addition, we also found miR-1231 was predominantly expressed in cardiac myocytes not in cardiac fibroblasts. Next, combine predictive strategy and gene expression array, we identified CACNA2D2, one calcium channel gene, as the target of miR-1231. Further results showed that cacna2d2 expression was indeed suppressed by miR-1231 in ischemic hearts of human and rat. Moreover, in vivo inhibition of miR-1231 ameliorated arrhythmias in rat MI hearts; conversely, forced overexpression of miR-1231 promoted arrhythmias. At last, cacna2d2 knockdown induced arrhythmias in ischemic hearts, regardless of the level of miR-1231. Thus, we propose here that miR-1231 exacerbates arrhythmia by inhibiting cacna2d2 in ischemic hearts.
Materials and methods
Clinical samples
Five heart tissue samples of border zones of anonymous patients diagnosed with myocardial infarction were obtained by biopsy after catheterization treatment in Sir Run Run Shaw Hospital. In addition, eight control heart tissue samples of left ventricular endocardium of healthy anonymous individuals were obtained from Myogen. All the human clinical heart samples in this study were investigated under the protocols approved by the Ethnic Committee for Human Sample Application of Sir Run Run Shaw Hospital.
Model of rat myocardial infarction and animal care
Experimental myocardial infarction was established through permanent occlusion of the left coronary artery (LCA) in 9-12 week old adult male Wistar rats. The surgical procedures were performed as previously reported [12]. Briefly, rats were randomly divided into myocardial infarction group and sham operation group who received the same procedures but without LCA occlusion. All the rats were anesthetized with 2.4% isoflurane and placed in a supine position and carefully catheterized with a stump needle. A nearly 1.5 cm cut was made along the left side of sternum, then the thorax was opened and heart was exposed from pleural cavity. The LCA was ligated via using a 6-0 prolene suture under the visualization of a microscope. Finally, the chest was closed. All the surgical procedures were conducted under sterile condition. 7 or 14 days after operation, hearts were harvested. Eventually, the ischemia zone, border zone and non-ischemic zone were identified by visual inspection under a dissecting microscope and separately dissected for experiments. All the animal protocols and procedures were approved by the Institutional Animal Care and Ethics Committee of the University of Sir Run Run Shaw Hospital, Zhejiang University of Medicine.
Cardiomyocytes and cardiofibroblasts in vitro culture
Cardiomyocytes and cardiofibroblasts were isolated and cultured normally as described in ref 5 and 13-14 [5,13,14]. For hypoxic incubation, briefly, cells were cultured with low oxygen levels in a hypoxic chamber continuously maintained at 10 mmHg (1 mmHg=133 Pa).
Northern blotting analysis and real-time qPCR
Total RNA were isolated from human and rat samples or from cultured myocytes or fibroblasts by using TRIzol reagent (Sigma-Aldrich). The amount of total RNA in each sample was preliminary quantified by Nanodrop (Thermo Scientific) and then equal loading was confirmed by staining with ethidium bromide in Northern gels. Measurement of miRNAs by Northern blot were displayed as described in ref 15 in detail [7]. The probes were generated using PCR and labeled with a random oligonucleotide priming kit (Roche Life Science). For miRNA level detection. RT-PCR was carried out using Taqman MicroRNA reverse Transcriptase kit (Thermo Scientific) according to the manufacturer’s instructions. The expressions of genes of interests were measured by real-time qPCR through using Taqman probes purchased from Applied Biosystems. The expression level of miR-1231 and β-Actin was assessed as described previously [15]. All value were normalized to β-Actin detected via human or rat specific probe.
Western blotting
The protein samples were extracted from human or rat tissues or in vitro cultured cells in accordance with the protocols as depicted in ref 17 [16]. The protein concentration was determined by Pierce BCA Protein Assay Kit (Pierce). Nearly 50 ng protein in each sample was fractionated by SDS-PAGE with 10% polyacrylamide gels and subsequently transferred to PVDF membrane (Millipore) at constant 350 mA current for 2 hours. Then, PVDF membrane was block in 5% BSA for 1 hour at room temperature. After that, PVDF membrane was incubated overnight at 4°C with primary antibodies forβ-Actin (Santa Cruz, SC-47778) and cacna2d2 (Novus Biologicals, NBP1-81501). The band signal was detected using the chemiluminescent substrate reagent (Thermo Scientific). The intensity of Western blotting bands was quantified using ImageJ software and all value were normalized to endogenous β-Actin.
Small interference RNA (siRNA)
We delivered siRNAs targeting cacna2d2 or control in vivo via using the Ambion® In Vivo siRNA System (Thermo Scientific) into rat ventricular myocytes before LCA occlusion according to the manufacturer’s protocol. The sequence of siRNA targeting cacna2d2 used in this study were: 5’-GACCAACGUUCUGAUCUGC(dTdT)-3’ (Genebank accession # NM_175592.2). The scrambled siRNA targeting luciferase was used as the negative control siRNA.
Oligonucleotides sequences
The sequences of anti-miR-1231 and anti-mutated of which cholesterol-linked oligonucleotides were complementary to the sequence of miR-1231 or containing a four-base mismatch mutation. Each residue nucleoside is modified with a 2’-OMe.S is a phosphorothioateinter nucleoside bond and oligonucleotides were linked with one cholesterol through a hydroxyproline linker. In detail, the sequence of anti-miR-1231 in human: AsAsGGGAAGACCAAGGCUUGCsUsGsUsCs-cholesterol; the sequence of anti-mutated-miR-1231 in human: AsCsUGGCCGACCAAGGCUUGCsUsGsUsCs-cholesterol. The sequence of anti-miR-1231 in rat: AsGsAAGGGCAGGGCCGGACAUsCsUsCsUs-cholesterol; the sequence of anti-mutated-miR-1231 in rat: AsGsAGGAGCGGGGCGGGACAUsCsUsCsUs-cholesterol.
Cardiac arrhythmia assessment
Rat arrhythmia events were measured using standard lead II ECG and then categorized according to the Curtis and Walker arrhythmia scoring system [17]. For example, in briefly, 0= no arrhythmia; 1=VT with<10 s duration; 2=VT with 11-30 s duration; 3=VT with 31-90 s duration; 4=VT with 91-180 s duration, or reversible VF with<10 s duration; 5=VT with>180 s duration, or reversible VF with>10 s duration; 6= irreversible VF. Both the incident rate of VT and VF were calculated as percentage of rats suffering arrhythmias in the total number of subjects studied as previously described [18].
Transfection and luciferase assay
A 1531-bp size genomic fragment with regions coding for miR-1231 was amplified by PCR and then ligated into pCMV6 vector. Genomic fragment of human or rat 3’-UTR containing miR-1231 binding sites were amplified by PCR and ligated into the firefly luciferase reporter vector (pMIR-REPORTTM; Ambion). The sequence of hsa-miR-1231: CGUCGACAGGCGGGUCUGUG; hsa-cacna2d2 3’UTR: AGAGGAACCCCCACCAGACAU; rno-miR-1231: AGAGAGGACGUCGAGACGGGUCU; rno-cacna2d2 3’UTR: UGUGUGUAUGGUCCAUGCCCAGU. HEK293 cells were cultured in 24-well plate with the number of 1×105/well. Before transfection treatment, cells were starved overnight in serum-free culture medium, cells were then transfected with lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Experiments were performed 24 h after transfection.
Cell culture
HEK293 (human embryonic kidney) cell line was purchased from American Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS and 100 g/ml penicillin/streptomycin in a standard cell culture condition (37°C, humidified, 5% CO2/95% air composition).
Data statistical analysis
Data in each group were expressed as mean ± s.e.m. Data statistical comparison was performed using unpaired Student’s t test between two groups, unless indicated otherwise. A value of P<0.05 indicated a statistically significant difference; NS indicated no significant difference.
Results
Upregulation of miR-1231 expression after MI
In an effort to identify whether miR-1231 is involved in the pathological processes of MI, we first compared its expression between MI heart and healthy heart samples in human individuals. The results showed that the expression of miR-1231 was upregulated nearly 3.5-fold in RNA samples from individuals with MI hearts compared with those from healthy control heart samples (Figure 1A). To examine whether this manifestation of elevated expression of miR-1231 in MI hearts could also be observed in animal models with disease similar to MI, we then compared the RNA samples among non-ischemic hearts and non-ischemic zone, border zone and ischemic zone from ischemic myocardium isolated from rats with MI hearts which were induced by experimental occlusion of left coronary artery for 7 days and 14 days. As depicted in Figure 1A, a similar tendency of increased expression, obviously 2.9-fold and 4.4-fold at 7 days and 14 days post-MI respectively, of miR-1231 in the RNA samples of ischemic zone but not in the non-ischemic hearts or non-ischemic zone was found as well (Figure 1A). To verify these real-time qPCR evidence, we randomly selected two pairs of human control non-ischemic hearts and MI hearts and rat control non-ischemic hearts and three different zones of MI hearts at 14 days post-MI, then the expression of miR-1231 was measured by Northern blot analysis. Consistent with the results displayed in Figure 1A, the expression of miR-1231 was indeed highly expressed in MI hearts of human and ischemic zone of rat hearts compared with their control non-ischemic hearts (Figure 1B). Combining these findings, we draw the conclusion that miR-1231 is overexpressed in human and rat hearts after MI, in addition, they also suggests that miR-1231 may play roles in the pathological processes of MI, whether it is protective or harmful.
Figure 1.
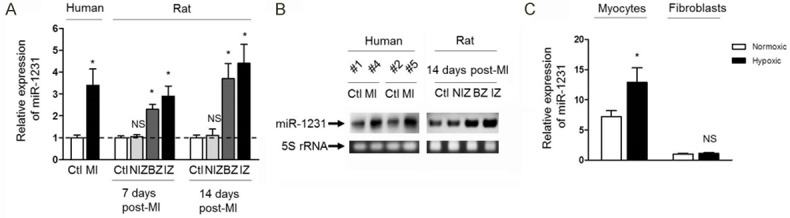
miR-1231 is overexpressed in human and rat hearts after MI. A. miR-1231 levels were aberrantly elevated in RNA samples of hearts from both human with MI and from rats subjected to experimental MI. Real-time PCR analysis was used to measure miR-1231 levels. In human samples, MI hearts (n=5) were compared with control non-ischemic hearts (Ctl, n=8); for rat samples, ctl samples as well as three different sections from MI hearts induced for 7 days or 14 days were compared (n=8. NIZ, nonischemic zone; BZ, border zone; IZ, ischemic zone). Data were presented as mean ± s.e.m. and normalized to Ctl. *, P<0.05 compared with Ctl; NS, not significant compared with Ctl. Unpaired Student’s t-test. B. Northern blot analysis of miR-1231 verified the real-time PCR expression data. Northern blot analysis of randomly selected two pairs of human control non-ischemic hearts and MI hearts as indicated (left panel) and rat ctl and three different regions from MI hearts at 14 days post-MI (right panel) showed a consistent increase in miR-1231 in human and rat heart samples in response to MI. 5S rRNA bands stained with ethidium bromide were used as a loading control. C. miR-1231 levels were determined by real-time PCR analysis of RNA samples from neonatal rat cardiac myocytes and fibroblasts cultured under normoxic or hypoxic condition for 48 h. Predominately high expression of miR-1231 in cardiac myocytes compared with fibroblasts was shown, and its expression was even more upregulated in response to hypoxia condition. Comparable amounts of RNA were used in each reaction. *, P<0.05 compared with normoxic cardiac myocytes. NS, not significant compared with normoxic fibroblasts.
To discover the physiological functions of miR-1231, we first characterized its expression pattern in heart tissues to understand where it may preferably exert functions, therefore we isolated cardiac myocytes and cardiac fibroblasts and culture them in vitro. The results indicated that miR-29 was expressed predominantly in cardiac myocytes, furthermore, its expression in myocytes was even more upregulated to nearly 2-fold under hypoxia culture condition compared with normal condition (Figure 1C). This line of evidence suggests that miR-1231 is induced in cardiac myocytes when subjected to hypoxia-mediated injure, a similar scenario reminding of blood flow deprivation (ischemia) taking place in MI patients [14].
miR-1231 suppresses cacna2d2 expression by targeting its mRNA
In order to define the possible functions of miR-1231 in the pathological processes of heart MI, we identified putative miR-1231 targets by computational predictions using TargetScan software [19]. Among the in silico predicted targets, the TargetScan (version 7.1) prediction program displayed an unexpected array of 14 mRNAs encoding ion channel proteins participating in the regulation of ion transportations, such as calcium, sodium and potassium, as possible targets for miR-1231, which aroused our much interests (Table 1). To confirm which one is indeed the target gene of miR-1231, we conducted mRNA gene expression array among 14 mRNAs in human and rat heart samples as mentioned before. The analysis of the expression of predicted 14 ion channel target genes showed that the mRNA level of cacna2d2 was significantly decreased in ischemic heart samples compared with the healthy control heart samples in both human and rats, however with other candidates unaffected (Figure 2A). In fact, the 3’-untranslated regions (3’-UTRs) of cacna2d2 contain seven sequential nucleotides that are totally complementary to the seven nucleotides starting from the 5’ end of miR-1231 in both human and mouse species Figure 2B), together pointing out a post-transcriptional control mechanism by which miR-1231 regulates cacna2d2 expression.
Table 1.
Putative target genes for miR-1231
| Ortholog of target gene | Representative transcript | Gene name | 3P-seq tags + 5 | Total sites | 8mer sites | 7mer-m8 sites | 7mer-A1 sites | 6mer sites | Representative miRNA | Cumulative weighted context++ score | Total context++ score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CACNA2D2 | ENST00000423994.2 | Calcium channel, voltage-dependent, alpha 2/delta subunit 2 | 19 | 3 | 0 | 1 | 2 | 1 | hsa-miR-1231 | -0.23 | -0.23 |
| SCN8A | ENST00000354534.6 | Sodium channel, voltage gated, type VIII, alpha subunit | 44 | 3 | 0 | 2 | 1 | 2 | hsa-miR-1231 | -0.21 | -0.21 |
| KCNJ9 | ENST00000368088.3 | Potassium inwardly-rectifying channel, subfamily J, member 9 | 5 | 3 | 0 | 3 | 0 | 1 | hsa-miR-1231 | -0.15 | -0.15 |
| CACNG8 | ENST00000270458.2 | Calcium channel, voltage-dependent, gamma subunit 8 | 5 | 3 | 1 | 1 | 1 | 2 | hsa-miR-1231 | -0.13 | -0.13 |
| KCNA1 | ENST00000382545.3 | Potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) | 5 | 2 | 1 | 1 | 0 | 0 | hsa-miR-1231 | -0.11 | -0.11 |
| KCNJ15 | ENST00000328656.4 | Potassium inwardly-rectifying channel, subfamily J, member 15 | 5 | 1 | 0 | 1 | 0 | 4 | hsa-miR-1231 | -0.31 | -0.31 |
| KCNE2 | ENST00000290310.3 | Potassium voltage-gated channel, Isk-related family, member 2 | 7 | 1 | 0 | 1 | 0 | 0 | hsa-miR-1231 | -0.3 | -0.3 |
| CACNG5 | ENST00000533854.1 | Calcium channel, voltage-dependent, gamma subunit 5 | 5 | 1 | 1 | 0 | 0 | 0 | hsa-miR-1231 | -0.3 | -0.3 |
| SCNN1G | ENST00000300061.2 | Sodium channel, non-voltage-gated 1, gamma subunit | 5 | 1 | 1 | 0 | 0 | 0 | hsa-miR-1231 | -0.29 | -0.29 |
| KCNC4 | ENST00000369787.3 | Potassium voltage-gated channel, Shaw-related subfamily, member 4 | 138 | 1 | 1 | 0 | 0 | 2 | hsa-miR-1231 | -0.17 | -0.17 |
| KCNK9 | ENST00000520439.1 | Potassium channel, subfamily K, member 9 | 5 | 1 | 0 | 1 | 0 | 1 | hsa-miR-1231 | -0.15 | -0.18 |
| SCN3A | ENST00000360093.3 | Sodium channel, voltage-gated, type III, alpha subunit | 5 | 1 | 0 | 0 | 1 | 0 | hsa-miR-1231 | -0.14 | -0.14 |
| CACNA2D1 | ENST00000356860.3 | Calcium channel, voltage-dependent, alpha 2/delta subunit 1 | 418 | 1 | 0 | 0 | 1 | 0 | hsa-miR-1231 | -0.14 | -0.14 |
| KCND1 | ENST00000376477.1 | Potassium voltage-gated channel, Shal-related subfamily, member 1 | 149 | 1 | 0 | 1 | 0 | 0 | hsa-miR-1231 | -0.12 | -0.12 |
A list of potential ion channel target genes for miR-1231 was predicted using TargetScan7.1 and miRanda software. The in silico putative targets included 14 mRNAs encoding ion channel regulators participating in the regulation of different ions’ transportation, such as calcium, sodium and potassium. The genes were listed according to the number of total binding sites.
Figure 2.
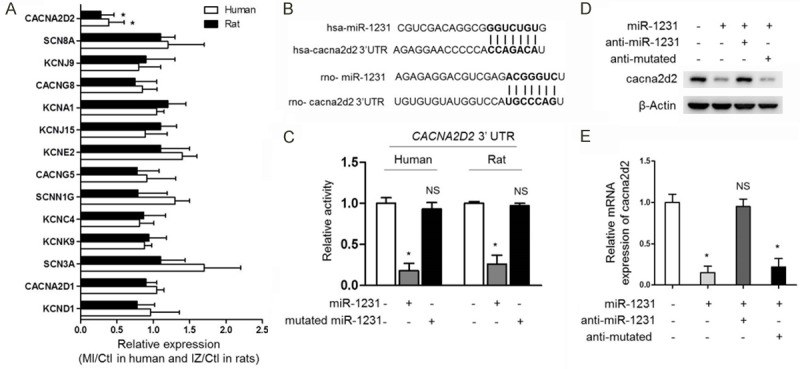
miR-1231 suppresses cacna2d2 expression by targeting its mRNA. (A) The mRNA level of cacna2d2 was significantly decreased in both human and rat ischemic heart samples. Real-time PCR analysis of the expression of predicted 14 ion channel target genes in ischemic hearts compared with the healthy control heart samples in both human and rats as depicted in Figure 1. In human samples, the randomly selected three pairs of human control non-ischemic hearts and MI hearts were compared, and in rat samples, three randomly chosen pairs of NIZ, BZ, IZ and Ctl samples from rats at 14 days post-MI were compared as well. Results of MI or IZ relative to Ctl were shown (*, P<0.05 compared with Ctl group, n=3). (B) Schematic illustration of sequence complimentarity between miR-1231 and the 3’-UTRs of cacna2d2 mRNAs in human (hsa) and rat (rno), provided with computational and bioinformatics-based approach using TargetScan [2]. Watson-Crick complementarity was presented in bold text and linked with its paired nucleotide. (C) 3’-UTR of human or rat cacna2d2 was target for human or rat miR-1231 in HEK293 cells, respectively. HEK293 Cells were transiently transfected with luciferase reporters linked to the 3’-UTR sequences of human or rat CACNA2D2 gene as indicated. The luciferase reporter activity was measured after 1 day co-expression. Human or rat miR-1231 repressed luciferase reporter gene activity, however, mutatedmiR-1231 was unable to decrease luciferase activity. Mean ± SD; n=8; *P<0.05; NS, not significant compared with Ctl). Unpaired Student’s t-test. (D, E) The mRNA and protein expression of cacna2d2 were downregulated by miR-1231 in cultured myocytes. The protein (D) and mRNA (E) levels of cacna2d2 were determined by Western blotting and qRT-PCR in cultured neonatal rat cardiac myocytes, which were transfected with miR-1231 alone or in combination with anti-miR-1231or anti-mutated. Left, β-Actin was used as a loading control and representative results of Western blotting bands were shown; Right, mRNA levels in four groups relative to Ctl were shown (*, P<0.05; NS, not significant compared with Ctl group, n=3).
Previous studies have demonstrated that miRNA-binding sites are artificially transferable and sufficient to confer the functions of miRNA-mediated gene silencing [18]. Next, we therefore inserted a fragment of the cacna2d2 3’-UTR sequence belonging to human or rat into the 3’-UTR downstream of a luciferase reporter plasmid and then examined luciferase activity after cotransfected them with miR-1231 in human embryonic kidney (HEK293) cells. The coexpression of miR-1231 with constructed plasmid containing human or rat cacna2d2 3’-UTR sequence into HEK293 cells reduced about 5-fold or 4-fold luciferase activity than that of the transfection of control plasmid alone, whereas the mutantcounterpart of miR-1231 did not downregulate luciferase activity (Figure 2C), which strongly proves that the mRNA of cacna2d2 is a direct target of miR-1231 in both human and rat.
To further verify that cacna2d2 is one target gene post-transcriptionally repressed by miR-1231, we determined the effects of miR-1231 on both the expression level of transcripts and protein product of CACNA2D2 gene in cultured neonatal rat cardiac myocytes by real-time qPCR and Western blotting analysis. The sequence of miR-1231 was transfected alone or combined with anti-miR-1231 or anti-mutated sequence of nucleotides to interfere its expression in myocytes. The results confirmed that both the mRNA and protein expression of cacna2d2 were indeed downregulated by miR-1231, moreover, anti-miR-1231 reversed the inhibiting effects of miR-1231 on cacna2d2, whereas anti-mutated did not show rescue effect (Figure 2D, 2E), these evidence further support the notion that cacna2d2 expression is repressed by miR-1231 via targeting its mRNA.
miR-1231 suppresses cacna2d2 expression in ischemic hearts
Based on the situation that CACNA2D2, as one target gene of miR-1231, was screened out firstly using human and rat heart samples as described as before, which extends our understanding that the repressing effects of miR-1231 on cacna2d2 expression could also exist in vivo in the context of heart tissues suffering MI. In an attempt to testify whether or not cacna2d2 is repressed by miR-1231 in ischemic hearts, in the first place, we compared the protein level of cacna2d2 between healthy control hearts and ischemic hearts in both human and rat samples we used in before experiments. Indeed, the protein levels of cacna2d2 were reduced in both human and rat ischemic hearts compared with their healthy control samples (Figure 3A), in accordance with the results of comparisons in CACNA2D2 gene transcripts we have seen in Figure 2A.
Figure 3.
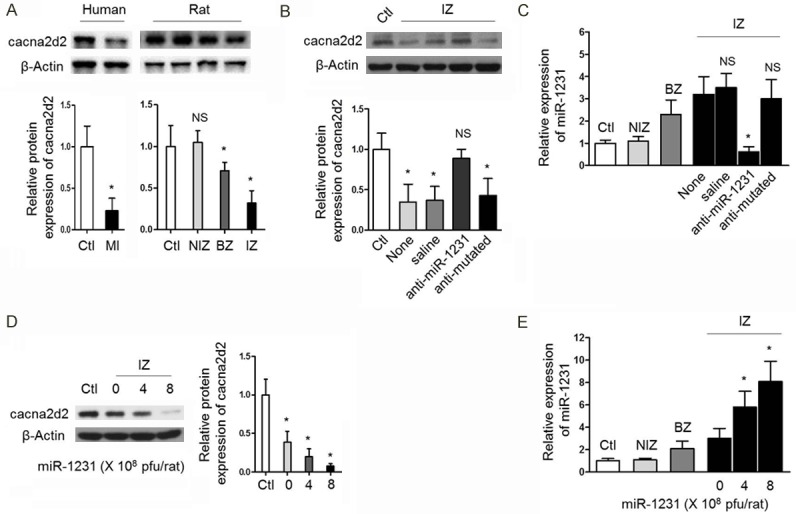
miR-1231 suppresses cacna2d2 expression in ischemic hearts. A. The protein levels of cacna2d2 were reduced in both human and rat ischemic hearts. As depicted in Figure 1A, three pairs of human IZ and Ctl samples or rat NIZ, BZ, IZ and Ctl samples were subjected to Western blotting assay to detect cacna2d2 protein expression. β-Actin was used as a loading control. Top, representative results of Western blotting bands; bottom, quantitation relative to Ctl as mean ± SD (n=3). (*, P<0.05; NS, not significant compared with Ctl group). Unpaired Student’s t-test. B. The protein level of cacna2d2 in IZ zone recovered upon in vivo miR-1231 knockdown. Relative protein levels of cacna2d2 in rat hearts after in vivo transfer of saline, anti-miR-1231 or anti-mutated were measured by Western blotting analysis. β-Actin was used as a loading control. Top, representative results of Western blotting bands; bottom, quantitation relative to Ctl as mean ± SD (n=3). (*P<0.05; NS, not significant compared with Ctl). Unpaired Student’s t-test. C. miR-1231 levels in IZ zone decreased in response to in vivo knockdown. Relative levels of miR-1231 in rat hearts after in vivo transfer of saline, anti-miR-1231 or anti-mutated were measured by real-time PCR analysis. All values were normalized to non-ischemic hearts (Ctl, n=4). (*P<0.05; NS, not significant compared with group receiving no injection (None). Unpaired Student’s t-test. D. The protein level of cacna2d2 in IZ zone decreased after in vivo enforced expression of miR-1231. Relative protein level of cacna2d2 in rat hearts after in vivo transfer of adenovirus control or different amount of adenovirus-miR-1231 as indicated were analyzed by Western blotting. β-Actin was used as a loading control. Left, representative results of Western blotting bands; right, quantitation relative to adenovirus Ctl as mean ± SD (n=3). (*P<0.05; NS, not significant compared with adenovirus control). Unpaired Student’s t-test. E. miR-1231 levels in IZ zone increased after in vivo enforced expression. Relative levels of miR-1231 in rat hearts after in vivo transfer of adenovirus control or different amount of adenovirus-miR-1231 were measured by real-time PCR analysis. All values were normalized to adenovirus control (Ctrl, n=4). *P<0.05 compared with adenovirus control. NS, not significant compared with adenovirus control. Unpaired Student’s t-test.
Since miR-1231 is overexpressed in ischemic hearts, which is accompanied by the downregulation of cacna2d2, and more importantly, we have proved that miR-1231 is able to suppress cacna2d2 expression through post-transcriptional mechanism of targeting its mRNA, therefore, we ask whether the downregulation of cacna2d2 in ischemic hearts is directly caused by miR-1231 overexpression. To address this issue, we examined the effects of interfering miR-1231 on cacna2d2. A designed specific anti-miR-1231 sequence, which contains single-stranded RNA oligonucleotides complementary to miR-1231 and is chemically modified for the improvement of stability and cell delivery efficiency [20], was delivered into the infarcted myocardium of rats by in vivo gene transfer to inhibit miR-1231, and one anti-mutated sequence with a four-base mismatch was also transferred as negative control. The results revealed that the protein level of cacna2d2 in IZ zone almost totally recovered upon in vivo miR-1231 knockdown, as controls, the transfer of saline or anti-mutated did not show antagonizing effects (Figure 3B, 3C), demonstrating that cacna2d2 is downregulated in ischemic hearts directly by miR-1231. To further strengthen this conclusion, we applied enforced overexpression strategy to investigate whether cacna2d2 could be more reduced in response to artificial overexpression of miR-1231 in ischemic hearts. Adenovirus-mediated miR-1231 gene was transferred with different dosages in vivo into the infarcted myocardium of rats to render it overexpressed locally. Opposite to the recovery expression of cacna2d2 by miR-1231 knockdown, the enforcement of miR-1231 expression reduced cacna2d2 expression to a lower level, even though miR-1231 had already elevated in ischemic hearts (Figure 3D, 3E). The collection of these results together strongly prove that cacna2d2 expression was suppressed in response to miR-1231 overexpression occurring in ischemic hearts.
miR-1231 promotes detrimental arrhythmias in ischemic hearts
cacna2d2 (voltage-dependent calcium channel subunit alpha2delta-2) is the protein product encoded by the voltage-dependent calcium channel α2δ auxiliary subunit gene (CACNA2D2), which is supposed to be responsible for regulating calcium transportation and signal responses in cardiomyocytes [21]. Some studies have reported that some voltage-gated calcium channel mutations or deficiencies are related to sudden arrhythmogenic death and heart failure, moreover, the abnormalities in calcium current density, protein and transcripts expression have also been connected with a trial fibrillation occurrence [22-25], which was well-documented by the study that KCNJ2 (which encodes the K+ channel subunit Kir2.1) and GJA1 (which encodes connexin 43) possess arrhythmogenic potential [18]. Given the above evidence that cacna2d2 expression was suppressed by miR-1231, in order to understand the pathophysiological functions of miR-1231 overexpressed in ischemic hearts, we tested the possibility of whether or not it could affect arrhythmias in MI hearts. As exhibited in Figure 4A, 4B, the knockdown of miR-1231 by anti-miR-1231 ameliorated arrhythmias in rat ischemic hearts, as presented by the decreased incidence of both ventricular tachycardia and ventricular fibrillation. Expectedly, the anti-mutated could not reduce the incidence of arrhythmias. On the other hand, by contrast, the enforced expression of miR-1231 in the infarcted myocardium promoted detrimental arrhythmias incidence compared with vehicle control (Figure 4C, 4D). Notably, nearly all the experimental rats were subjected with ischemic arrhythmias when injected with adenovirus-miR-1231. Overall, these results intensely point out a positive feedback loop existing in which miR-1231 is an arrhythmogenic factor induced by ischemic status, which in turn exacerbates arrhythmias that is detrimental to ischemic hearts.
Figure 4.
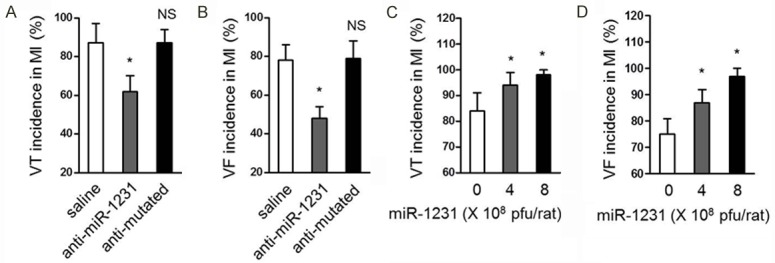
miR-1231 promotes cardiac arrhythmias in ischemic hearts. (A, B) Inhibition of miR-1231 ameliorates ischemic arrhythmias in MI hearts of rats. Incidence of ventricular tachycardia (VT) (A) and ventricular fibrillation (VF) (B) in MI declined following miR-1231 knockdown in vivo by using oligonucleotides complementary to the miR-1231 sequence (anti-miR-1231), and injection of saline or a mutant oligonucleotide with base mismatches (anti-mutated) were used as negative controls. Data are presented as percent incidence (mean ± s.e.m.) in rat hearts at 14 days post-MI. Three independent experiments were conducted and each group contained 8 rats. *P<0.05 compared with saline. NS, not significant compared with saline χ2-test. (C, D) Overexpression of miR-1231 promotes arrhythmias in ischemic hearts. MI hearts were induced for 11 days and then subsequent three days of enforced expression of miR-1231 in vivo by using the local delivery method at doses: 0 (adenovirus control), 4×108 or 8×108 pfu/rat was carried out, the incidence of VT (C) and VF (D) in MI hearts of rats was recorded. Data are presented as percent incidence (mean ± s.e.m.). Three independent experiments were conducted and each group contained 8 rats. *P<0.05 compared with adenovirus control. NS, not significant compared with adenovirus control χ2-test.
Cacna2d2 knockdown induces arrhythmias in ischemic hearts
We suppose that if the arrhythmogenic function of miR-1231 is caused by its repressing effect on downregulating cacna2d2 expression, then the direct knockdown of cacna2d2 to paralyze its function should also induce arrhythmias, despite of the status of miR-1231. Hence, RNA interference technique was used to further investigate whether cacna2d2 is the intermediate connecting miR-1231 and its arrhythmias promoting act. siRNA-mediated specific knockdown of cacna2d2 induces arrhythmias in ischemic hearts of rats, as proved by the increased incidence of VT and VF in the presence or absence of anti-miR-1231 injection, and scrambled siRNA exhibited no similar effects predictably (Figure 5A, 5B). The desired functions of anti-miR-1231 and sicacna2d2 were demonstrated in Figure 5C, 5D. In conclusion, these data indicate that miR-1231 exacerbates arrhythmia by inhibiting cacna2d2 in ischemic hearts.
Figure 5.
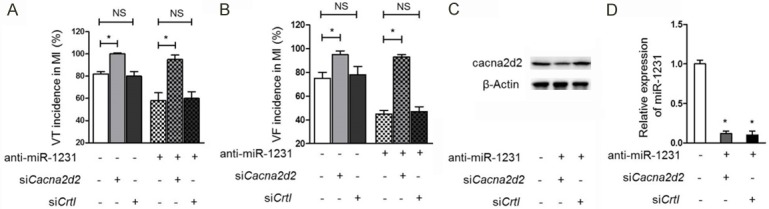
Silence of cacna2d2 induces arrhythmias in ischemic hearts. (A, B) Knockdown of cacna2d2 by siRNA induces arrhythmias in ischemic hearts of rats. Both the incidence of VT (A) and VF (B) in ischemic hearts increased following siRNA-mediated cacna2d2 silencing in the presence or absence of anti-miR-1231 treatment. Scrambled siRNA was used as a negative control (siCtrl). Data are presented as percent incidence (mean ± s.e.m.) in rat hearts at 14 days post-MI. Three independent experiments were conducted and each group contained 8 rats. *P<0.05; NS, not significant compared with control group without siRNA administration χ2-test. (C) The protein level of cacna2d2 in IZ zone was dramatically downregulated upon siRNA targeting. The protein level of cacna2d2 was measured by Western blotting analysis. β-Actin was used as a loading control and representative results of Western blotting bands were shown. (D) The level of miR-1231 was effectively reduced when anti-miR-1231 was utilized. Values were normalized to ischemic hearts receiving no treatment of anti-miR-1231. *, P<0.05. Unpaired Student’s t-test.
Discussion
The results of our study unravel a new pathophysiological function of miR-1231 in promoting arrhythmia in ischemic hearts suffering MI disease, the mechanism of which is mediated by repressing the expression of cacna2d2, one calcium channel protein essential for modulating calcium signaling responses in cardiomyocytes and is closely related to sudden arrhythmogenic death and heart failure problem. Actually, this detrimental effect of miR-1231 on MI progression in ischemic hearts is intensified by a positive feedback loop wherein miR-1231 is an arrhythmogenic factor induced by ischemic status, which in turn exacerbates arrhythmias by targeting cacna2d2 function. Therefore, interfering endogenous miR-1231 level might represent a new potential approach for inhibiting arrhythmias in ischemic hearts with MI.
In the first place, we show that miR-1231 is overexpressed in ischemic hearts in both humans and rats. It’s interesting to test whether miR-1231 is also ubiquitously elevated in other animal MI disease models. To date, plenty of researches have used microRNA microarray analysis to screen miRNAs with aberrant expression in response to MI [5,8]. Consequently, a large number of miRNAs have been identified as regulators in MI pathogenesis, such as cardiac fibrosis [5], or angiogenesis and functional recovery [8]. These previous reports undoubtedly clarify the huge numbers of miRNAs and their functional diversities involved in MI. Recently, some studies have showed that miR-1231 is associated with the susceptibility of hepatocellular carcinoma and pancreatic cancer by targeting IFNAR1 or LINC00673 [26], besides, it can also suppresses hepatitis B virus replication by repressing core mRNA [27]. Another reported function of miR-1231 is that it could serve as biomarker of non-ischemic systolic heart failure [11]. These non-cardiovascular or cardiovascular related functions of miR-1231 demonstrate its functional versatility, which seems largely depends on the targets it regulates. Our study uncovers the arrhythmogenic function of miR-1231 in ischemic hearts. It is important to note that in our study, we only investigate the functions of miR-1231 in ischemic hearts after MI, whether it is also arrhythmogenic in healthy hearts is unclear deduced from our present evidence. Likewise, considering the distinct physiological environments, no direct relationship could be connected between the arrhythmogenic function of miR-1231 in ischemic hearts and its indicative function in non-ischemic systolic heart failure. Further studies are needed to clarify these clinically relevant questions.
On the other hand, we discover the muscle-specific distribution of miR-1231 in heart tissue as indicated by its exclusive expression in cardiomyocytes, whereas in cardiac fibroblasts, it is nearly undetectable. This expression pattern may confine its functions in cardiomyocytes. Therefore, we speculate here that miR-1231 might not have direct effects on the functions associated with cardiofibroblasts, such as cardiac fibrosis, which requires the abnormal proliferation of cardiofibroblasts and increased deposition of extracellular matrix components [5]. However, it cannot exclude the possibility that indirect relationship may exist. Noteworthily, an earlier study describe the regulatory effect of miR-1 on regulating cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2, which encodes the connexin 43 and K+ channel subunit Kir2.1, respectively [18]. Combine this line of evidence with the findings in our study, it appears that ion channel genes are vulnerable targets for miRNAs during MI progression, and it is plausible there is a common paradigm existing in which miRNAs regulate arrhythmia in ischemic hearts after MI is through acting on ion channel genes. A more profound mechanism should be addressed in the future is how exactly heart rate is uncoordinated by ion channel proteins in cardiomyocytes to lead to arrhythmia when the complicated network of miRNAs is involved.
Overall, our results suggest that the suppression of miR-1231 contributes to arrhythmias amelioration and this strategy may be beneficial in the settings of MI diseases. A more comprehensive knowledge of miR-1231 is needed to understand the mechanisms of other functions and the safety of interfering it, even though no obvious side effects of in vivo knockdown of miR-1231 were observed throughout this study.
Acknowledgements
This study was financially supported by National Natural Science Foundation of China (81000064) and Zhejiang Provincial Natural Science Foundation (LY16H020005 and LY17H020007).
Disclosure of conflict of interest
None.
References
- 1.Zhao Y, Ransom JF, Li A, Vedantham V, Von DM, Muth AN, Tsuchihashi T, Mcmanus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 2.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR. Targeted deletion reveals essential and overlapping functions of the miR-17~92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiedler J, Thum T. MicroRNAs in myocardial infarction. Nat Rev Cardiol. 2014;12:135–142. doi: 10.1038/nrcardio.2014.207. [DOI] [PubMed] [Google Scholar]
- 4.Fox CS, Coady S, Sorlie PD, Sr DAR, Pencina MJ, Vasan RS, Meigs JB, Levy D, Savage PJ. Increasing cardiovascular disease burden due to diabetes mellitus: the Framingham Heart Study. Circulation. 2007;115:1544–1550. doi: 10.1161/CIRCULATIONAHA.106.658948. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij E, Sutherland LB, Thatcher JE, Dimaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernardo BC, Mcmullen JR. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci U S A. 2012;109:17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 9.Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, Popescu LM, Torre T, Siclari F, Moccetti T, Vassalli G. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res. 2014;103:530–41. doi: 10.1093/cvr/cvu167. [DOI] [PubMed] [Google Scholar]
- 10.Heinrich EM, Dimmeler S. MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circ Res. 2012;110:1014–22. doi: 10.1161/CIRCRESAHA.111.243394. [DOI] [PubMed] [Google Scholar]
- 11.Vogel B. Multivariate miRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur Heart J. 2013;34:2812–2822. doi: 10.1093/eurheartj/eht256. [DOI] [PubMed] [Google Scholar]
- 12.Yang B, Lin H, Xu C, Liu Y, Wang H, Han H, Wang Z. Choline produces cytoprotective effects against ischemic myocardial injuries: evidence for the role of cardiac m3 subtype muscarinic acetylcholine receptors. Cell Physiol Biochem. 2005;16:163–174. doi: 10.1159/000089842. [DOI] [PubMed] [Google Scholar]
- 13.Yue P, Zhang Y, Du Z, Xiao J, Pan Z, Wang N, Yu H, Ma W, Qin H, Wang W. Ischemia impairs the association between connexin 43 and M3 subtype of acetylcholine muscarinic receptor (M3-mAChR) in ventricular myocytes. Cell Physiol Biochem. 2006;17:129–136. doi: 10.1159/000092074. [DOI] [PubMed] [Google Scholar]
- 14.Kubasiak LA, Hernandez OM, Bishopric NH, Webster KA. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc Natl Acad Sci U S A. 2002;99:12825–12830. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meder B, Keller A, Vogel B, Haas J, Sedaghathamedani F, Kayvanpour E, Just S, Borries A, Rudloff J, Leidinger P. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res Cardiol. 2011;106:13–23. doi: 10.1007/s00395-010-0123-2. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Zhang Y, Cao L, Han H, Wang J, Yang B, Nattel S, Wang Z. HERG K+ channel, a regulator of tumor cell apoptosis and proliferation. Cancer Res. 2002;62:4843–4848. [PubMed] [Google Scholar]
- 17.Curtis MJ, Walker MJ. Quantification of arrhythmias using scoring systems: an examination of seven scores in an in vivo model of regional myocardial ischaemia. Cardiovasc Res. 1988;22:656–65. doi: 10.1093/cvr/22.9.656. [DOI] [PubMed] [Google Scholar]
- 18.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 19.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krutzfeldt J, Rajewsky N, Braich R. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 21.Gao B, Sekido Y, Maximov A, Saad M, Forgacs E, Latif F, Wei MH, Lerman M, Lee JH, Perezreyes E. Functional properties of a new voltage-dependent calcium channel α2δ auxiliary subunit gene (CACNA2D2) J Biol Chem. 2000;275:12237–12242. doi: 10.1074/jbc.275.16.12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soon JL, Ping L, Chua YL, Soong TW, Sin KY. Absence of calcium channel alpha1C-subunit mutation in human atrial fibrillation. Asian Cardiovas Thoracic Ann. 2010;18:349–353. doi: 10.1177/0218492310375749. [DOI] [PubMed] [Google Scholar]
- 23.Nattel S, Burstein B, Dobrev D. Atrial remodeling and atrial fibrillation: mechanisms and implications. Circ Arrhythm Electrophysiol. 2008;1:62–73. doi: 10.1161/CIRCEP.107.754564. [DOI] [PubMed] [Google Scholar]
- 24.Allessie M, Ansma J, Schotten V. Electrical, contractile and structural remodeling during atrial fibrillation. Cardiovasc Res. 2002;54:230–246. doi: 10.1016/s0008-6363(02)00258-4. [DOI] [PubMed] [Google Scholar]
- 25.Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kääb S, Hinterseer M. Reprogramming of the human atrial transcriptome in permanent atrial fibrillation expression of a ventricular-like genomic signature. Circ Res. 2005;96:1022–1029. doi: 10.1161/01.RES.0000165480.82737.33. [DOI] [PubMed] [Google Scholar]
- 26.Zhou C, Yu Q, Chen L, Wang J, Zheng S, Zhang J. A miR-1231 binding site polymorphism in the 3’ UTR of IFNAR1 is associated with hepatocellular carcinoma susceptibility. Gene. 2012;507:95–98. doi: 10.1016/j.gene.2012.06.073. [DOI] [PubMed] [Google Scholar]
- 27.Kohno T, Tsuge M, Murakami E, Hiraga N, Abe H, Miki D, Imamura M, Ochi H, Hayes CN, Chayama K. Human microRNA hsa-miR-1231 suppresses hepatitis B virus replication by targeting core mRNA. J Viral Hepat. 2014;21:e89–97. doi: 10.1111/jvh.12240. [DOI] [PubMed] [Google Scholar]


