Abstract
Long non-coding RNA (lncRNA) X inactivate-specific transcript (XIST) acts as an important regulator in tumor progression. However, its expression and the underlying mechanism in glioma remain unclear. The aim of this study was to explore the potential function of XIST in glioma progression. In the present study, our data showed that the expression of XIST was significantly up-regulated in glioma tissues and enhanced the proliferation of glioma cells. The expression of miR-137 was significantly decreased in glioma tissues. Further correlation analysis demonstrated that there was a negative correlation between XIST expression and miR-137 expression. Bioinformatics prediction and luciferase reporter assays demonstrated that miR-137 could directly bind to XIST and negatively regulated the expression of miR-137. Additionally, our data further showed that XIST could up-regulate the expression of miR-137 targeted gene Rac1 through acting as an endogenous sponge of miR-137. In addition, we found that Rac1 inhibition or miR-137 overexpression could suppress glioma cells proliferation induced by XIST overexpression. Thus, a novel XIST-miR-137-Rac1 pathway regulatory axis in glioma pathogenesis was revealed in the present study. Overall, our study indicated that XIST could be a potential therapeutic target in the treatment of glioma.
Keywords: Glioma, long non-coding RNA, XIST, miR-137, Rac1
Introduction
Glioma accounts for the great majority of primary tumors in the central nervous systems of adults [1]. Although the diagnosis and treatment strategies for glioma have been improved prominently, the prognosis of glioma is still poor because of significant malignant proliferation and invasion, and the majority of patients suffered from glioma were diagnosed at the advanced stages [2]. Although some advances in comprehensive treatment as well as early diagnosis have been made, the five year survival rates remain poor [3,4]. Therefore, it is necessary to elucidate new molecular mechanisms associated with glioma development as well as on investigating the effective therapeutic targets for the treatment of glioma.
Long non-coding RNAs (lncRNAs), a group of non-coding genes with more than 200 nucleotides in length, have been reported to play important roles in types of malignant tumors, including glioma [5-8]. Mounting evidence indicated that lncRNAs could act as oncogenes or tumor suppressors by regulating the expression of tumor-related genes and the function of tumor-related pathways in tumorigenesis [9,10]. X-inactive specific transcript (XIST), a kind of lncRNA derived from XIST gene, was up-regulated in several types of tumors, including gastric cancer, non-small cell lung cancer, breast cancer and nasopharyngeal carcinoma [11-14]. Recent study showed that silencing XIST could inhibit hepatocellular carcinoma cell growth, metastasis as well as induce cell apoptosis, and repressed tumor growth and facilitated high survival in nude mice [15]. Those data indicated that XIST exerted an essential role on the progression ad development of tumor. However, there is no related study elaborating the relevance between XIST expression and glioma progression, the expression and underlying mechanisms of XIST were largely poor understood in glioma.
MicroRNAs (miRNAs) are another small non-coding RNA molecules, 18-25 nucleotides in length, which mainly post-transcriptionally regulate gene expression by translational repression or mRNA degradation through binding to the 3’-untranslated region (3’-UTR) of target mRNAs [16]. MiRNAs involved in various critical biological processes, including cell differentiation, proliferation and apoptosis [17]. Accumulating evidence showed that dysregulated expression of miRNA played important roles in carcinogenesis. Among these miRNAs, miR-137 was found to be down-regulated in human glioma and suppress glioma cells growth by targeting Rac1, indicating miR-137 was a potential tumor-suppressor factor in glioma [18]. Recent study showed that lncRNAs served as competing endogenous RNA, also known as sponge, which interacted with miRNAs and modulated the expression of miRNA target genes [19].
In the present study, we aimed to discover the underlying molecular mechanism of XIST on glioma progression. The expression of XIST and miR-137 were detected in glioma tissues. Then bioinformatics analysis revealed a potential interaction between XIST and miR-137, indicating XIST might function as a competing endogenous RNA for miR-137 in human glioma. In addition, the role of XIST in the regulation of Rac1 expression by directly binding to miR-137 was further determined in human glioma cells.
Materials and methods
Clinical sample collection
A total of 30 glioma tissues and 18 normal brain tissues were obtained from patients undergoing surgery at the Department of Neurosurgery, Zhumadian Central Hospital. Tissues were immediately frozen in liquid nitrogen until use. None of the patients were treated with radiotherapy or chemotherapy before surgery. The use of tissues for this study was approved by the ethics committee of Zhumadian Central Hospital. Before using these clinical materials for research purposes, all the patients have the written informed consent.
Cell culture and transfection
The human glioma cell lines, U251 and LN229, were purchased from the Chinese Academy of Science Cell Bank (Shanghai, China). Cell lines were cultured in DMEM medium (Invitrogen, USA) supplemented with 10% fetal bovine serum (Invitrogen, USA) at a humidified atmosphere incubator of 37°C and 5% CO2. Plasmid for lncRNA-XIST was constructed by introducing the cDNA sequence of XIST or Rac1 into the pcDNA3.1 expression vector (Invitrogen, USA). The miR-137 mimics, miR-137 inhibitor and small interference RNAs (siRNAs) for the knockdown of XIST or Rac1 expression were purchased from GenePharma (Shanghai, China). Transfection was performed by using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Quantitative real-time PCR assay
Total RNA was extracted from tissues and cell lines by TRIzol Reagent (Invitrogen, USA) according to the manufacturer’s protocol. Both mRNA and miRNA were then reverse transcribed into cDNA by using PrimeScript RT Reagent Kit (Takara, Japan). The real-time PCR were performed on ABI 7500 Fast Real-Time PCR system (Applied Biosystems, USA) by using SYBR Green PCR Kit (Takara, Japan) with specific primers according to the manufacturer’s instructions. U6 and GAPDH were used as an internal control respectively for normalization. Experiments were done independently in three times. The relative expression of genes was calculated by using the 2-ΔΔCt method.
Western blot analysis
Total proteins of cell lines were lysed with RIPA buffer with 1% PMSF, and the concentrations were detected by using a BCA Protein Assay Kit (Beyotime, Shanghai, China). Equal amounts of protein samples were separated by 10% SDS-PAGE and then transferred onto PVDF membranes. Membranes were probed with primary antibodies against Rac1 and GAPDH (Abcam, MA, USA) at 4°C overnight, followed by incubation with HRP-conjugated secondary antibodies. GAPDH was used as a loading control. Protein bands were analyzed using an enhanced chemiluminescence (ECL) kit (Pierce Biotechnology, Rockford, USA) with imaging system (Bio-Rad, CA, USA).
Cell proliferation assay
Cell proliferation was measured by using Cell Counting Kit-8 (CCK-8) assay kit (Dojindo Laboratory, Japan) according to the manufacturer’s protocol. After plating cells in 96-well microtiter plates at 1.0 × 103/well, CCK-8 reagents were added to each well at different time points. 1 h after adding CCK-8, cellular viability was determined by measuring the absorbance at the wavelength of 450 nm.
Colony formation assay
For colony formation experiments, transfected cells were plated in six-well plates at 1.0 × 103/well and incubated in DMEM medium containing 10% FBS at 37°C for approximately 2 weeks. Then washed twice with PBS and fixed with 10% formalin, Colony formation was determined by counting the number of visible colonies after staining with 0.1% crystal violet (Sigma, USA).
Cell cycle analysis
The transfected cells were harvested and then fixed with 500 μl of 70% cold ethanol for 2 h. The cells were added with 100 μl of RNase and incubated at 37°C for 30 min. Then, 400 μl of PI was added, and the cells were incubated at 4°C for 30 min away from light. The samples were immediately subjected to flow cytometer (FACScan, BD Biosciences, USA). The results were analyzed using CELL Quest 3.0 software.
Luciferase reporter assays
XIST fragment containing the predicted miR-137 binding site, the wide-type or mutant putative sequences of the binding site were cloned into a pmirGLO Dual-luciferase Vector (Promega, USA) to form the reporter vector pmiRGLO-XIST-wild-type (XIST-WT) or pmiRGLO-XIST-mutant (XIST-MUT). XIST-WT or XIST-MUT was co-transfected with miR-137 mimics or negative control into glioma cells by using Lipofectamie 2000. 48 hours after transfection, the relative luciferase activities were measured by a dual-luciferase reporter assay system (Promega, USA) according to the manufacturer’s instruction.
Similarly the 3’UTR of Rac1 containing the miR-137 binding site, the wide-type or mutant putative sequences of the binding site were cloned into a pmirGLO Dual-luciferase Vector to form the reporter vector pmiRGLO-Rac1-wild-type (Rac1-WT) or pmiRGLO-Rac1-mutant (Rac1-MUT). The resulting constructs were co-transfected with miR-137 mimics, negative control, and XIST plasmids into glioma cells by using Lipofectamine 2000. 48 hours after transfection, the relative luciferase activities were measured.
Statistical analysis
All data are presented as the mean ± standard deviation (SD) from three independent experiments. All statistical analyses were performed by using SPSS 18.0 software and GraphPad Prism 5.0. Differences between groups were analyzed by using Student’s t test or one-way ANOVA analysis. A chi-square test was applied to determine the association of XIST levels with clinicopathological features. A value of P<0.05 was considered statistically significant.
Results
Expression of lncRNA XIST and miR-137 in human glioma tissues
The expression levels of LncRNA XIST and miR-137 were detected by qRT-PCR analysis in glioma tissues and normal brain tissues. Our data revealed that XIST expression was significantly up-regulated in glioma tissues compared with normal brain tissues (Figure 1A). MiR-137 expression was significantly down-regulated in glioma tissues compared with normal brain tissues (Figure 1B). To further elucidate the significance of XIST in glioma, we calculated the correlation of XIST expression with clinicopathological features of glioma (Table 1), we found that high XIST expression was positively associated with tumor grade. However, no association of XIST expression with age, gender, tumor size, and tumor nodule number was detected. Thus, those data indicated that XIST played important implications in the progression of glioma.
Figure 1.
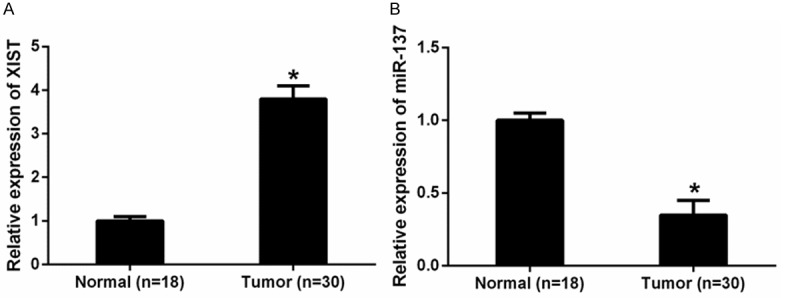
Expression of lncRNA XIST and miR-137 in human glioma tissues. A. The expression levels of XIST in glioma tissues were significantly lower than those in normal brain tissues. B. The expression levels of miR-137 were significantly higher in glioma tissues compared with normal brain tissues. *P<0.05.
Table 1.
Correlation between lncRNA XIST expression and clinicopathological features of glioma patients
| Parameters | Total | XIST expression | P value | |
|---|---|---|---|---|
|
|
||||
| Low expression | High expression | |||
| Age (years) | 0.182 | |||
| <50 | 8 | 2 | 6 | |
| >50 | 22 | 7 | 15 | |
| Gender | 0.306 | |||
| Male | 16 | 5 | 11 | |
| Female | 14 | 4 | 10 | |
| Clinical grade | 0.014* | |||
| Low grade I-II | 9 | 6 | 3 | |
| High grade III-IV | 21 | 3 | 18 | |
| Tumor size (cm) | 0.751 | |||
| <4.5 | 17 | 6 | 11 | |
| >4.5 | 13 | 3 | 10 | |
| Tumor nodule number | 0.230 | |||
| Multiple | 5 | 1 | 4 | |
| Single | 25 | 8 | 17 | |
XIST, X inactivate-specific transcript; lncRNA, long non-coding RNA;
P<0.05.
LncRNA XIST promoted glioma cell proliferation in vitro
To explore the biological function of XIST in glioma cells, XIST plasmid and siRNA (si-XIST) were separately transfected into U251 and LN229 cell lines. Successful transfection was confirmed by qRT-PCR analysis (Figure 2A). CCK-8 and colony formation assays were performed to determine the effect of XIST on glioma cell proliferation, we found that overexpression of XIST significantly promoted glioma cell proliferation in comparison to negative control. On the other hand, XIST inhibition exhibited a significant decrease in cell proliferation (Figure 2B-D). Next, flow cytometric analysis was performed to further determine whether the function of XIST on glioma cell proliferation was by altering cell-cycle progression. Cell-cycle analysis showed that XIST overexpression significantly decrease in the cellular population in G0/G1 phase, while XIST inhibition arrested glioma cells in G1/G0 phase (Figure 2E). These results demonstrated that XIST markedly promoted cell growth by regulated cell cycle progression in glioma cells.
Figure 2.
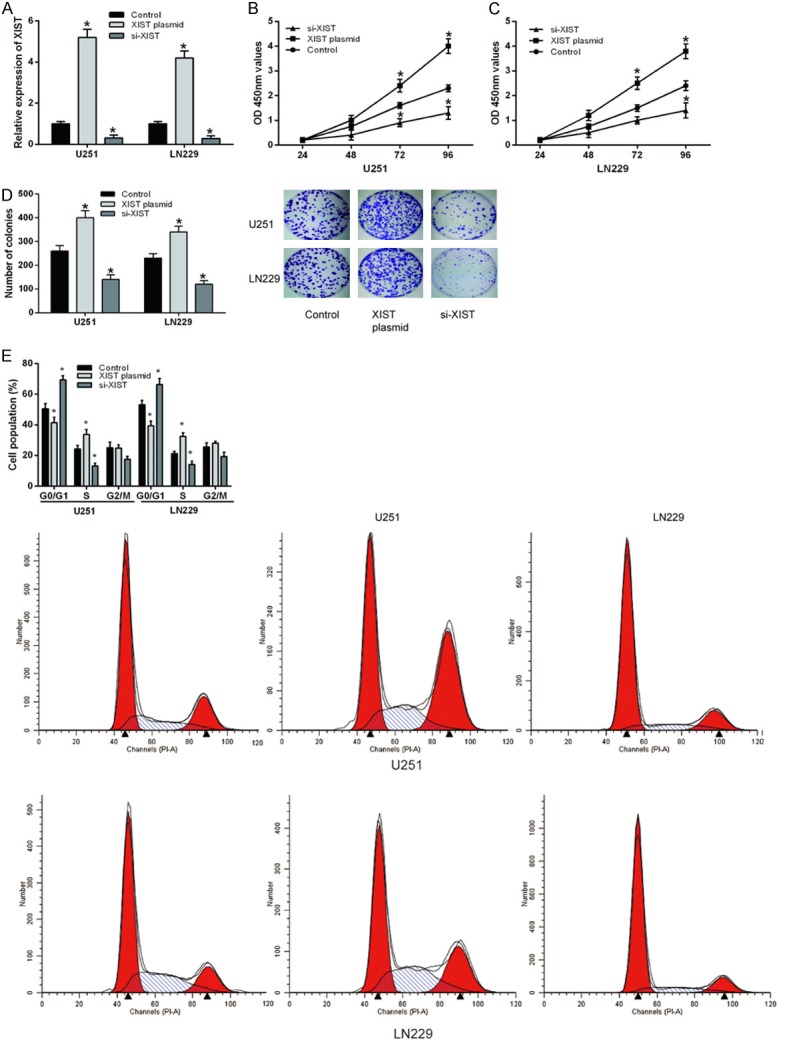
LncRNA XIST promoted glioma cell proliferation in vitro. A. The expression levels of XIST in glioma cell lines transfected with XIST plasmid or siRNA were determined by qRT-PCR. B and C. CCK-8 assay was used to determine glioma cell prolife ration after ransfected with XIST plasmid or siRNA. D. Colony formation assay was performed to measure glioma cell proliferation after ransfected with XIST plasmid or siRNA. E. Flow cytometer was used to explore gliomacell cycle after ransfected with XIST plasmid or siRNA. *P<0.05.
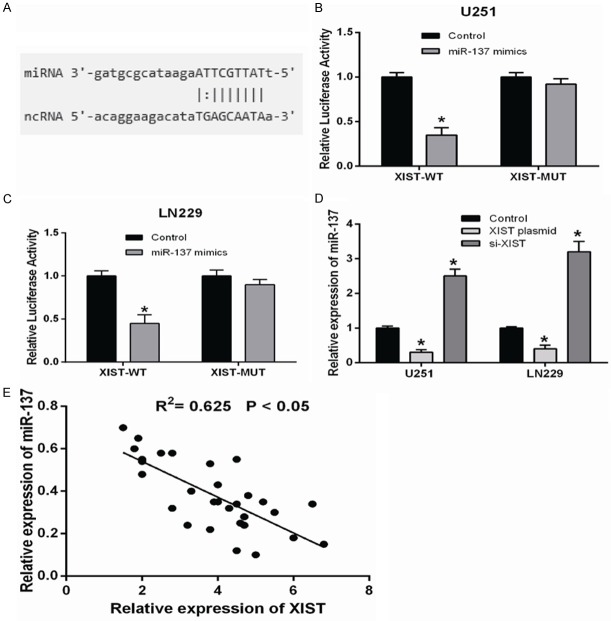
LncRNA XIST directly interacted with miR-137 in glioma cells
Recent studies showed that lncRNAs could act as a competing endogenous RNA or a molecular sponge in regulating the biological functions of miRNAs. As expected, bioinformatics tools (starBase2.0) for the predicted lncRNA-miRNA interactions revealed potential binding domains within XIST and miR-137 (Figure 3A). Then luciferase reporter assay was performed to confirm the bioinformatical prediction. Results revealed that miR-137 significantly decreased the luciferase activity of XIST-WT compared to negative control, while miR-137 did not affect the luciferase activity of XIST-MUT (Figure 3B and 3C). In addition, our data showed that XIST overexpression significantly inhibited miR-137 expression and XIST inhibition markedly increased miR-137 expression in U251 and LN229 cells (Figure 3D). Further correlation analysis demonstrated that there is a negative correlation between XIST expression and miR-137 in glioma tissues (R2=0.625, Figure 3E). Taken together, these data indicated that miR-137 could directly bind to XIST in glioma cells.
Figure 3.
LncRNA XIST directly interacted with miR-137 in glioma cells. A. Bioinformatics tool (starBase2.0) revealed the predicted binding sites between XIST and miR-137. B and C. Luciferase reporter assay showed that miR-137 overexpression significantly decreased the luciferase activity of XIST-WT in glioma cells compared to controls, while miR-137 did not affect the luciferase activity of XIST-MUT. D. The expression of miR-137 in glioma cells transfected with XIST plasmid or siRNA were examined by qRT-PCR. E. Correlation analysis revealed the negative relationship between miR-137 and XIST in glioma tissues. *P<0.05.
LncRNA XIST regulated Rac1 expression by inhibiting miR-137
Recent study reported that miR-137 inhibited human glioma cells growth by targeting Rac1 [18], but the potential regulation role of XIST on the expression of Rac1 in human glioma cell lines is still unclear. Our data showed that Rac1 mRNA expression was significantly increased in XIST plasmid transfected U251 and LN229 cells, while it was decreased in XIST siRNA transfected cells (Figure 4A), in line with the tendency shown in the corresponding changes in Rac1 protein expression (Figure 4B). Furthermore, luciferase reporter assay showed that overexpression of XIST could restore the luciferase activity of miR-137-mediated suppression of Rac1-WT compared with negative control (Figure 4C and 4D). In order to validate the correlation between Rac1 and XIST, Rac1 mRNA expression was also detected in glioma tissues as well as in normal brain tissues. qRT-PCR showed that Rac1 expression was significantly up-regulated in glioma tissues compared to normal brain tissues (Figure 4E), and further correlation analysis revealed that there was a positive relationship between Rac1 and XIST (R2=0.582, Figure 4F). In conclusion, these findings indicated that XIST regulated Rac1 expression through inhibiting miR-137 in human glioma cells.
Figure 4.
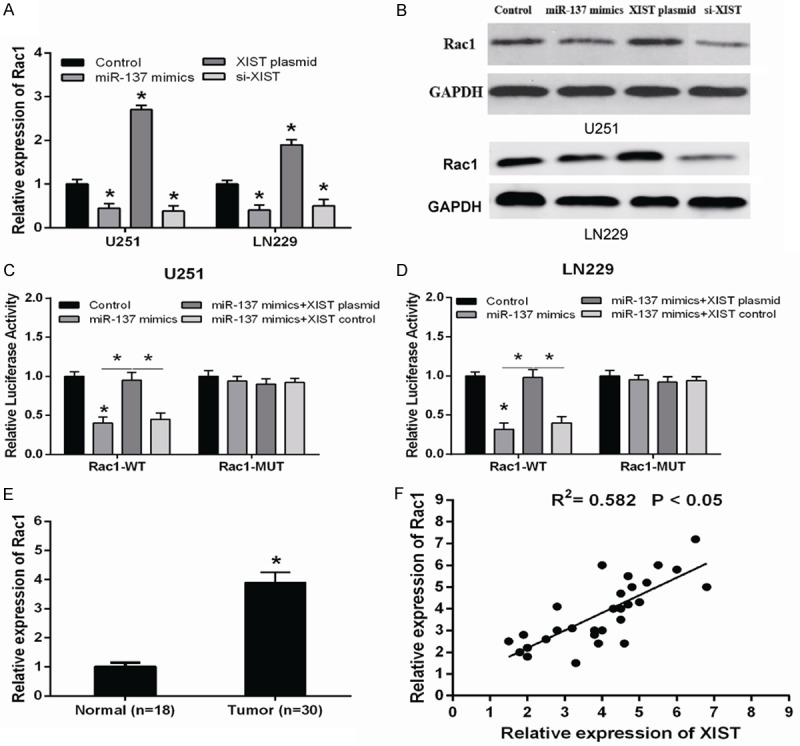
LncRNA XIST up-regulated Rac1 by inhibiting miR-137 expression in glioma cells. The mRNA (A) and protein (B) levels of Rac1 in glioma cells transfected with miR-137 mimics, XIST plasmid or XIST siRNA were detected by qRT-PCR and western blot. (C and D) The relative luciferase activity of Rac1 3’UTR in glioma cells transfected with miR-137 mimics or miR-137 mimics + XIST plasmid. (E) Rac1 mRNA expression was significantly up-regulated in glioma tissues compared to normal brain tissues. (F) The correlation between XIST mRNA and Rac1 expression in 30 glioma tissues. *P<0.05.
Rac1 knockdown or miR-137 overexpression suppressed the proliferation of glioma cells induced by XIST
To further investigate whether Rac1 knockdown or miR-137 overexpression involved in the effect of XIST on glioma cell proliferation, U251 and LN229 cells were co-transfected with XIST plasmid and Rac1 siRNA or miR-137 mimics. After transfection, the expression of Rac1 or miR-137 was significantly down-regulated or up-regulated compared with the negative controls in both U251 and LN229 cell lines (Figure 5A and 5B). As shown in Figure 5C-E, CCK-8 and colony formation assay showed that Rac1 knockdown and miR-137 overexpression significantly suppressed the cell proliferation of XIST overexpression glioma cells. Our results elucidated a novel XIST-miR-137-Rac1 pathway regulatory axis in the progression of glioma.
Figure 5.
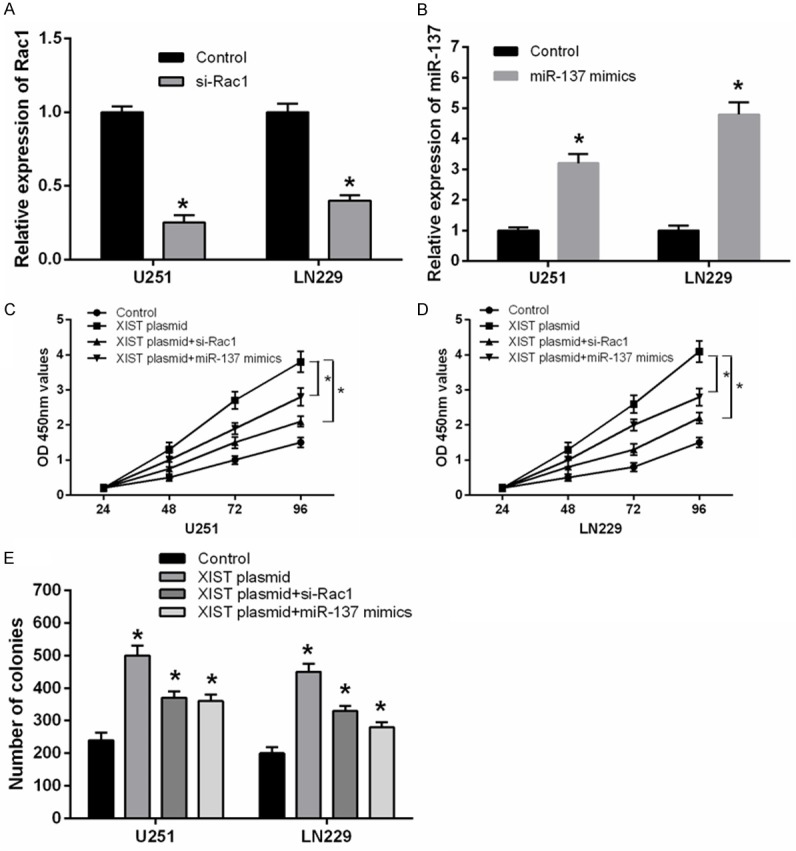
Rac1 knockdown and miR-137 overexpression suppressed the proliferation of glioma cells transfected with XIST plasmid. (A) Rac1 mRNA expression was significantly decreased in glioma cell lines transfected with Rac1 siRNA by using qRT-PCR. (B) The expression of miR-137 was significantly increased in glioma cell lines transfected with miR-137 mimics. (C-E) Rac1 knockdown or miR-137 overexpression significantly suppressed the proliferation of XIST overexpression glioma cells by using CCK-8 (C and D) and colony formation assay (E). *P<0.05.
Discussion
In the present study, we identified that XIST was significantly up-regulated in glioma tissues compared with normal brain tissues. XIST markedly promoted cell proliferation in human glioma cell lines. Then bioinformatics analysis presented the complementary binding sites between XIST and miR-137. The expression of miR-137 was significantly down-regulated in glioma tissues and further correlation analysis revealed the negative correlation between XIST and miR-137. In addition, luciferase reporter assay was performed to demonstrate that miR-137 might be a direct target of XIST in glioma cells. XIST also showed positive regulation on Rac1, which has been defined as a direct and functional target of miR-137. Furthermore, Rac1 knockdown significantly suppressed the cell proliferation of XIST overexpression glioma cells. Taken together, XIST exerted oncogenic functions and involved in the up-regulation of Rac1 in human glioma cells through inhibiting the expression of miR-137.
Recent studies showed that lncRNAs play important roles in the regulation of various molecular and cellular functions [20]. Increasing evidence has suggested that lncRNAs are closely associated with various types of human malignant tumors, including glioma [21]. Zhang et al indicated that lncRNA CCND2-AS2 was significantly up-regulated in malignant glioma, and promoted glioma cell proliferation through Wnt/β-catenin signaling [22]. LncRNA CRNDE was significantly increased in glioma tissues, and up-regulated CRNDE expression was associated with poor overall survival of glioma patients [23]. Specially, lncRNA XIST has been supposed to act as an oncogene and up-regulated in many cancers [11,24]. For example, Song et al reported that XIST expression in nasopharyngeal carcinoma (NPC) tissues was remarkably higher than that of in adjacent normal nasopharyngeal tissues, in addition, XIST significantly increased NPC cell viability and growth in vitro [14]. However, the mechanisms of XIST in glioma have not been thoroughly elucidated. In this study, we found XIST was significantly up-regulated in glioma tissues and promoted cell proliferation in human glioma cell lines, suggesting XIST acted as an oncogenic role in glioma progression.
Although XIST has been suggested to act as an oncogene, the underlying mechanism participated in tumorigenesis remains unclear. Mounting evidence revealed that lncRNA might function as a competing endogenous RNA or a molecular sponge in regulating the expression and biological functions of miRNA. For example, lncRNA HOTAIR was demonstrated to be an oncogene and negatively regulated the expression of miRNA-1 in hepatocellular carcinoma [25]. LncRNA EWSAT1 promoted human nasopharyngeal carcinoma cell growth in vitro through up-regulating cyclin D1 partially via sponging miR-326/330-5p clusters [26]. In the present study, we revealed a potential binding domain within XIST and miR-137 by bioinformatics prediction, and luciferase reporter assay demonstrated miR-137 was a direct target of XIST in glioma cells. XIST negatively regulated the expression of miR-137, and correlation analysis demonstrated the negative correlation of XIST and miR-137 expression in glioma tissues. Our results indicated that XIST functioned as endogenous miRNA sponges to bind to miR-137 and regulated its function.
MiRNAs are small non-coding RNA molecules, which mainly post-transcriptionally regulate gene expression through binding to the 3’-UTR of target mRNAs. Accumulating evidence showed that miRNAs play critical roles in tumor progression and development. Aberrant expression of miR-137 was found in several types of human cancers including glioma. For example, Qin et al demonstrated miR-137 was significantly downregulated in multiple myeloma and overexpression of miR-137 could reduce drug resistance and overcome chromosomal instability of the multiple myeloma cells [27]. Dong et al showed that miR-137 was decreased in ovarian cancer tissues, furthermore, the inhibition of miR-137 with antisense oligonucleotides promoted EMT and ovarian cancer cell invasion [28]. Our study revealed that the expression level of miR-137 was significantly down-regulated in glioma tissues. Moreover, miR-137 overexpression markedly suppressed the proliferation of XIST overexpression glioma cells.
LncRNAs can also affect the expression and biological functions of miRNA targets. Studies reported miR-137 could regulate numerous of target genes. Li et al demonstrated that miR-137 promoted apoptosis in ovarian cancer cells through the regulation of XIAP [29]. Du et al showed that miR-137 played tumor suppressor roles in gastric cancer by targeting KLF12 and MYO1C [30]. But among all of the predicted target genes for miR-137, we found that Rac1 acted as a crucial effector of miR-137 in glioma. Ras-related C3 botulinum toxin substrate1 (Rac1) is a member of the Rho family, which belong to the Ras superfamily of GTP enzymes. Rac1 played a key role in regulating the recombination of cytoskeletal proteins as well as proliferation, differentiation and apoptosis of tumor cells [31]. Aberrant expression of Rac1 has been associated to several types of cancers [32,33], and it has been reported miR-137 inhibited the growth of glioma cells by directly targeting Rac1 [18]. In order to investigate the role of XIST as a miR-137 sponge, the potential regulation of XIST on Rac1 expression was also analyzed in human glioma cell lines. In our study, Rac1 expression was significantly up-regulated in glioma tissues compared to normal brain tissues, and further correlation analysis revealed the positive relationship between Rac1 and XIST. Furthermore, XIST up-regulated Rac1 expression, and restored the inhibition of miR-137 on Rac1 expression and luciferase activity in glioma cells. These findings indicated that XIST regulated Rac1 expression through inhibiting miR-137 in human glioma cells.
In conclusion, a novel XIST-miR-137-Rac1 regulatory axis in glioma pathogenesis was revealed in the present study, providing a further understanding of the role of XIST in glioma development. XIST was an oncogenic lncRNA that promoted the tumorigenesis of glioma by functioning as a competing endogenous RNA, which regulated the target gene of miR-137 through directly sponging miR-137. Our study indicated that XIST could be a potential therapeutic target in the treatment of glioma.
Acknowledgements
The project was supported by innovation plan funding project of Science and Technology of Zhumadian (No. 20604).
Disclosure of conflict of interest
None.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan UA, Bhavsar A, Asif H, Karabatsou K, Leggate JR, Sofat A, Kamaly-Asl ID. Treatment by specialist surgical neurooncologists improves survival times for patients with malignant glioma. J Neurosurg. 2015;122:297–302. doi: 10.3171/2014.10.JNS132057. [DOI] [PubMed] [Google Scholar]
- 3.Ma CC, Xiong Z, Zhu GN, Wang C, Zong G, Wang HL, Bian EB, Zhao B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35:90. doi: 10.1186/s13046-016-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, Wang N, Long Z, Ren X. Long noncoding RNA BANCR promotes endometrial cancer cell proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK signaling pathway. Cell Physiol Biochem. 2016;40:644–656. doi: 10.1159/000452577. [DOI] [PubMed] [Google Scholar]
- 6.Liu B, Pan CF, He ZC, Wang J, Wang PL, Ma T, Xia Y, Chen YJ. Long noncoding RNA-LET suppresses tumor growth and EMT in lung adenocarcinoma. Biomed Res Int. 2016;2016:4693471. doi: 10.1155/2016/4693471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B, Liu C, Wu Q, Zhang J, Min Q, Sheng T, Wang X, Zou Y. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482:828–834. doi: 10.1016/j.bbrc.2016.11.120. [DOI] [PubMed] [Google Scholar]
- 8.Guo H, Hu G, Yang Q, Zhang P, Kuang W, Zhu X, Wu L. Knockdown of long non-coding RNA CCAT2 suppressed proliferation and migration of glioma cells. Oncotarget. 2016;7:81806–81814. doi: 10.18632/oncotarget.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y, Li L, Zheng Z, Chen S, Chen E, Hu Y. Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation. J Cell Mol Med. 2017;21:955–967. doi: 10.1111/jcmm.13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang C, Zan J, Yue B, Liu C, He C, Yan D. Long noncoding RNA ZFAS1 promotes the progression of colonic cancer by modulating ZEB1 expression. J Gastroenterol Hepatol. 2016 doi: 10.1111/jgh.13646. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 13.Huang YS, Chang CC, Lee SS, Jou YS, Shih HM. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget. 2016;7:43256–43266. doi: 10.18632/oncotarget.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592:8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang L, Yang Y, Ma X, Han B, Wang Z, Zhao Q, Wu L, Qu Z. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi E, Hornicek FJ, Duan Z. MicroRNA involvement in osteosarcoma. Sarcoma. 2012;2012:359739. doi: 10.1155/2012/359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 18.Sun G, Cao Y, Shi L, Sun L, Wang Y, Chen C, Wan Z, Fu L, You Y. Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother Radiopharm. 2013;28:327–334. doi: 10.1089/cbr.2012.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 20.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Zhang XQ, Leung GK. Long non-coding RNAs in glioma: functional roles and clinical perspectives. Neurochem Int. 2014;77:78–85. doi: 10.1016/j.neuint.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Wei DL, Wan L, Yan SF, Sun YH. Highly expressed lncRNA CCND2-AS1 promotes glioma cell proliferation through Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2017;482:1219–1225. doi: 10.1016/j.bbrc.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 23.Jing SY, Lu YY, Yang JK, Deng WY, Zhou Q, Jiao BH. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur Rev Med Pharmacol Sci. 2016;20:3992–3996. [PubMed] [Google Scholar]
- 24.Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, Liu Y. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 25.Su DN, Wu SP, Chen HT, He JH. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol Lett. 2016;12:4061–4067. doi: 10.3892/ol.2016.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song P, Yin SC. Long non-coding RNA EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro by targeting miR-326/-330-5p. Aging (Albany NY) 2016;8:2948–2960. doi: 10.18632/aging.101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin Y, Zhang S, Deng S, An G, Qin X, Li F, Xu Y, Hao M, Yang Y, Zhou W, Chang H, Qiu L. Epigenetic silencing of miR-137 induces drug resistance and chromosomal instability by targeting AURKA in multiple myeloma. Leukemia. 2017 doi: 10.1038/leu.2016.325. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Dong P, Xiong Y, Watari H, Hanley SJ, Konno Y, Ihira K, Yamada T, Kudo M, Yue J, Sakuragi N. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J Exp Clin Cancer Res. 2016;35:132. doi: 10.1186/s13046-016-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Chen W, Zeng W, Wan C, Duan S, Jiang S. microRNA-137 promotes apoptosis in ovarian cancer cells via the regulation of XIAP. Br J Cancer. 2017;116:66–76. doi: 10.1038/bjc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Y, Chen Y, Wang F, Gu L. miR-137 plays tumor suppressor roles in gastric cancer cell lines by targeting KLF12 and MYO1C. Tumour Biol. 2016;37:13557–13569. doi: 10.1007/s13277-016-5199-3. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Liao Q, Han Y, Chen J, Liu Z, Ling H, Zhang J, Yang W, Oyang L, Xia L. Rac1 overexpression is correlated with epithelial mesenchymal transition and predicts poor prognosis in non-small cell lung cancer. J Cancer. 2016;7:2100–2109. doi: 10.7150/jca.16198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Algayadh IG, Dronamraju V, Sylvester PW. Role of Rac1/WAVE2 signaling in mediating the inhibitory effects of gamma-tocotrienol on mammary cancer cell migration and invasion. Biol Pharm Bull. 2016;39:1974–1982. doi: 10.1248/bpb.b16-00461. [DOI] [PubMed] [Google Scholar]
- 33.Manara MC, Terracciano M, Mancarella C, Sciandra M, Guerzoni C, Pasello M, Grilli A, Zini N, Picci P, Colombo MP. CD99 triggering induces methuosis of Ewing sarcoma cells through IGF-1R/RAS/Rac1 signaling. Oncotarget. 2016;7:79925–79942. doi: 10.18632/oncotarget.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]