Abstract
Long non-coding RNAs (lncRNAs) serve critical roles in the tumorigenesis and development of multiple human malignancies. Herein, we aimed to explore the biological and clinical significance of lncRNA CRNDE in human breast cancer (BC). The expression of CRNDE in BC tissues and cell lines was detected, and the association between CRNDE expression and clinicopathologic features of BC patients was also analyzed. Novel targets of CRNDE were identified through a bioinformatics search and confirmed using a dual-luciferase reporter system. Gain and loss-of-function studies were carried out to verify whether CRNDE exerts its biological functions through its downstream target. CCK-8, colony formation, wound-healing, and transwell assays were applied to detect the altered phenotypes of BC cell lines in vitro after transfection. Tumor xenografts were created to detect the function of CRNDE in vivo tumorigenesis. CRNDE expression is remarkably up-regulated in BC tissue specimens and cell lines in comparison to corresponding normal tissues and normal human breast epithelial cells. Up-regulated CRNDE expression was greatly associated with larger tumor size, advanced TNM stage and unfavorable prognosis of BC patients. We uncovered that miR-136 is a bona fide binding target of CRNDE, and that up-regulation of CRNDE promoted the mRNA and protein expressions of β-catenin, c-myc and cyclinD1. Overexpressed CRNDE facilitated in vitro cell proliferation, migration and invasion of BC cells. In vivo assay showed that the average tumor volume and weight were largest in the group of CRNDE overexpression. CRNDE might hyperactivate the Wnt/β-catenin signaling pathway through directly repressing miR-136 expression in BC; CRNDE could be considered as a prognostic biomarker and therapeutic target in BC diagnosis and treatment.
Keywords: Breast cancer, CRNDE, microRNA-136, Wnt/β-catenin signaling pathway, cell proliferation, tumorigenesis, prognosis
Introduction
Breast cancer (BC) is featured as one of the most wide spread malignant tumors and the main reason of cancer-related death among female population around the world [1]. In USA, one in eight women will develop BC in her lifetime and approximately 230,000 new cases and 40,000 BC-associated deaths occurred in 2013 [2]. In spite of great advances in diagnostic and therapeutic strategies during the past decades, morbidity and mortality of BC still remain high. Accordingly, it is of critical importance for us to elucidate novel mechanisms correlated to BC development and establish promising therapeutic targets for BC treatment.
Recent articles have suggested that more than 90% of the transcripts from the human genome could not code for proteins [3]. Long noncoding RNAs (lncRNAs), a kind of non-protein coding transcripts more than 200 nucleotides in length, are widely expressed in human cells and serve crucial roles in various biological events, such as cell-cycle regulation [4], genomic expression [5], and cell differentiation [6]. Recently, mounting evidence revealed that aberrant expression of lncRNAs might be closely associated with multiple types of tumors [7]. As a well-known member of lncRNAs, CRNDE (Colorectal neoplasia differentially expressed), located on the long arm of chromosome 16 (16q12.2) of the human genome, was originally identified as an lncRNA in human colorectal cancer [8]. In a wide range of human malignant diseases, such as glioma [9,10], ovarian cancer [11] and hepatocellular carcinoma [12], CRNDE expression is evidently increased and is closely correlated with unfavorable survival and aggressive clinical features. However, the expression profiles and biological functions of CRNDE in BC remain largely elusive.
The Wnt family, which contains 19 glycoproteins, is frequently implicated in cell proliferation [13], differentiation [14] and migration [15]. The Wnt signaling pathway, which could be divided into the canonical pathway and the noncanonical pathway, has been extremely well studied in the past decades. As an element of intracellular signal transduction, β-catenin is regarded as one of the leading components in the cadherin protein complex and is critical for the modulation of the Wnt/β-catenin signaling pathway [16]. In addition, Shao et al. reported that in renal cell carcinoma, CRNDE is significantly up-regulated and exerts its functions through regulating Wnt/β-catenin signaling pathway [17]. However, it is still unclear about the correlation between Wnt/β-catenin signaling pathway and CRNDE expression in BC.
Recently, accumulating articles revealed that one potential function of lncRNAs was to directly interact with microRNAs (miRNAs) as a sponge and regulate their expression and activity [18,19]. MiRNAs, a class of short (18-24 nt), single stranded and noncoding RNA molecules, are involved in regulating gene transcription and expression via directly binding with the target mRNAs [20]. For example, Wang et al. demonstrated that miR-326, regulated by HOTAIR, could modulate cell proliferation and migration via targeting phox2a in lung cancer [21].
The present article aimed to explore the underlying mechanisms of CRNDE function in BC. Moreover, we predicted that CRNDE could modulate the Wnt/β-catenin signaling pathway through directly inhibiting miR-136 to exert its oncogenic function in BC cells.
Materials and methods
Tissue samples
BC tissue samples and their matched non-cancerous tissue samples, dissected at least 2 cm away from the tumor border, were randomly collected from a total of 103 patients who were diagnosed as BC and had underwent complete or partial surgical resection in The Eighth People’s Hospital of Shanghai. All samples were histologically verified via two clinical pathologists with at least five years experiences. None of patients underwent chemotherapy or postmenopausal hormone therapy prior to surgery. The fresh tissue specimens were collected and frozen in liquid nitrogen tanks after they were removed from the body immediately and stored at -80°C. Prior to enrollment, written informed consent was obtained from each participant. Research Ethics Board of The Eighth People’s Hospital of Shanghai approved this study procedure.
Cell lines and cell culture
One normal breast epithelial cell line (HBL-100) and four human BC cell lines (MCF-7, MDA-MB-231, MDA-MB-468 and BT-549) were obtained from the Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 100 U/mL penicillin/streptomycin (Invitrogen, Shanghai, China) and 10% fetal bovine serum (FBS; Gibco) in a humidified culture chamber of 5% CO2 at 37°C.
Cell transfection
A full-length CRNDE sequence was synthesized and inserted into pcDNA3.1 vector (Invitrogen) to generated pcDNA3.1-CRNDE. Plasmid vectors (pcDNA3.1-CRNDE and pcDNA3.1-NC) were prepared using DNA Midiprep or Midiprep kits (Qiagen, Hilden, Germany). siRNA for CRNDE and scrambled control siRNA (si-NC) were acquired from Invitrogen. MiR-136 mimic was purchased from Dharmacon (Lafayette, USA). When reaching approximately 80% confluence, MDA-MB-231 cells were transfected with plasmid vectors, mimic or siRNA using Lipofectamine 2000 reagent (Invitrogen) as recommended by the manufacturer’s protocol. 48 h after transfection, cells were harvested for further analyses.
Luciferase assay
Potential lncRNA targets were predicted and analyzed by using starBase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php). The wild-type CRNDE (WT) and mutant CRNDE (MUT) containing the seeding site of miR-136 were established and incorporated into the Firefly luciferase expressing psiCHECK-2 vector (Promega, Madison, WI, USA). MDA-MB-231 cells were seeded into a 24-well plate 24 h before transfection. They were transfected with 0.4 µg of the pMiR-REPORT-CRNDE-WT or pMiR-REPORT-CRNDE-MUT, 20 µM miR-136 mimic or control oligo, together with 0.02 µg of the Renilla luciferase vector pRL-TK (Promega), using Lipofectamine 2000. After 48 h, the luciferase activities were detected consecutively through Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) following the manufacturer’s instructions.
RNA isolation and qRT-PCR
Total RNAs were extracted from tissue samples and cells using Trizol® Reagent (Invitrogen). The purity of total RNA was determined through measuring the absorbance at 260 and 280 nm. Only samples with an A260:A280 ratio between 1.8 and 2.1 was appropriate for further analysis. The first-strand cDNA was synthesized from 1 μg of total RNA using the Reverse Transcription System Kit (Takara, Dalian, China). Quantitation of lncRNAs and mRNAs was performed using SYBR® Green PCR Master Mix (Applied Bsiosystems, USA) on the ABI 7900 Sequence Detection System (Life Technologies, NY, USA). For miRNA investigation, total RNA was reverse transcribed using miRNA specific primers and stem-loop real-time qPCR was conducted using Taqman miRNA assays (Applied Biosystems). GAPDH mRNA and U6 were applied as an internal standard. Fold changes were calculated by relative quantification (2-ΔΔCt) method as described previously [22]. The sequences of the primers, synthesized by Shanghai GenePharma, Co., Ltd., were recorded in Table 1.
Table 1.
The sequences of primers used for qRT-PCR
| Gene name | Primer sequences |
|---|---|
| CRNDE | |
| Forward | 5’-ATATTCAGCCGTTGGTCTTTGA-3’ |
| Reverse | 5’-TCTGCGTGACAACTGAGGATTT-3’ |
| miR-136 | |
| Forward | 5’-CGCCACTCCATTTGTTTTGA-3’ |
| Reverse | 5’-GTGCAGGGTCCGAGGT-3’ |
| β-catenin | |
| Forward | 5’-AAAGCGGCTGTTAGTCACTGG-3’ |
| Reverse | 5’-GACTTGGGAGGTATCCACATCC-3’ |
| c-myc | |
| Forward | 5’-ATGGCCCATTACAAAGCCG-3’ |
| Reverse | 5’-TTTCTGGAGTAGCAGCTCCTAA-3’ |
| cyclinD1 | |
| Forward | 5’-GCTGCGAAGTGGAAACCATC-3’ |
| Reverse | 5’-CCTCCTTCTGCACACATTTGAA-3’ |
| U6 | |
| Forward | 5’-CGGGTTTGTTTTGCATTTCT-3’ |
| Reverse | 5’-AGTCCCAGCATGAACAGCTT-3’ |
| GAPDH | |
| Forward | 5’-TGCACCACCAACTGCTTAGC-3’ |
| Reverse | 5’-GGCATGGACTGTGGTCATGAG-3’ |
Protein isolation and western blot analysis
Standard western blotting was performed for protein expression analyses. The cultured MDA-MB-231 cells were lysed by RIPA buffer (Beyotime, China) containing protease inhibitors (Sigma, USA) on ice. Protein lysates were isolated by 10% SDS-PAGE, and then electrophoretically transferred onto a PVDF membrane (Millipore, Bedford, MA, USA). The membranes were blocked by 5% skim milk soluted in TBST buffers and incubated with the specific primary antibodies against β-catenin (BD, Transduction Laboratories, KY, USA), c-myc (Roche Applied Sciences, Indianapolis, IN, USA) and cyclinD1 (Santa Cruz Biotechnology, CA, USA) overnight at 4°C, and probed with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology). β-actin antibody (Calbiochem, San Diego, CA) was applied as an internal loading control. Chemiluminescent investigation was conducted using the ECL PlusWestern Blotting System (Thermo Fisher Scientific, Waltham, MA). The signal was quantified using the Image J software (http://rsb.info.nih.gov/ij).
CCK-8 assay
Cell Counting Kit-8 (CCK-8, Beyotime Institute of Biotechnology, Jiangsu, China) assay was performed to detect cell proliferation. MDA-MB-231 cells were seeded into 96-well cell culture plates. 48 h after transfection, CCK-8 was added into each well and incubated for another 4 h, and the absorbance was finally investigated at the wavelength of 450 nm using a microplate reader (Molecular Devices, Menlo Park, CA).
Colony formation assay
Approximately 500 cells were plated into 6-well plates and incubated in DMEM with 10% FBS at 37°C. 10 days later, the cells were fixed, stained with 0.1% crystal violet and observed.
Transwell assay
Matrigel (BD, Franklin Lakes, NJ, USA) was diluted with serum-free DMEM, and the membrane was coated with matrigel to form a matrix barrier. 48 h after transfection, a total of 5 × 104 MDA-MB-231 cells were harvested and suspended in the upper chamber containing 200 μL serum-free DMEM. In each lower chamber, 500 μL DMEM with 10% FBS was added. After 24 h incubation and removal of the cells on the surface of upper chamber, the migrated or invaded cells were fixed with 4% paraformaldehyde for 10 min, and stained with 0.1% crystal violet (Sigma) in 20% ethanol for 10 min, and counted in five randomly assigned fields under a microscope (Olympus, Tokyo, Japan).
Wound-healing assay
48 h after transfection, MDA-MB-231 cells (1 × 106/well) were placed in 6-well plates and cultured overnight. Upon reaching confluency, the cell layer was scratched manually with a 200 μL pipette tip and then immediately washed with growth medium twice and cultured again. Photo images of the plates were captured under a microscope at 0, 12 and 24 h.
Tumor xenografts
Four-week-old female athymic BALB/C nude mice (Shanghai SLAC Laboratory Animal Co. Ltd., Shanghai, China) were randomly allocated into six groups (n=6 per group). 1 × 107 transfected MDA-MB-231 cells were suspended in 100 μl PBS and then administrated into the posterior flank of BALB/C nude mice by subcutaneous injection. On the day 26, the mice were euthanized and their tumors were collected for analysis. Tumor volumes in mice were calculated with a slide caliper according to the following formula: tumor volume (mm3)=0.5 × length × width2. All experimental procedures involving animals were performed following approval by the Animal Care and Experiment Committee of The Eighth People’s Hospital of Shanghai.
Statistical analysis
Statistical analysis was carried out using SPSS 16.0 software (SPSS Ltd., UK) and graphs were generated with Graphpad Prism 6.0 (Graphpad Software Inc, USA). The relationship between CRNDE expression and clinicopathological features of BC patients were evaluated by the chi-square test. Comparison of experimental results was analyzed by Student’s t-test. Pearson correlation analysis was used to estimate the association between CRNDE and miR-136 expression. Overall survival curve was plotted by Kaplan-Meier survival analysis and the significance was compared by the log rank test. Statistical tests and P-values were two-sided. A P-value < 0.05 was regarded to suggest a statistically significant difference in this study.
Results
CRNDE is increased in BC and correlated to aggressive features and poor prognosis of BC patients
To evaluate CRNDE expression in BC tissues, qRT-PCR was conducted in 103 paired samples of BC patients. Our results demonstrated that CRNDE expression in BC tissues was evidently higher than in corresponding non-tumor tissues (Figure 1A, P < 0.001). In addition, CRNDE expression was dramatically increased in all four BC cell lines (MCF-7, MDA-MB-231, MDA-MB-468 and BT-549) in comparison toHBL-100 normal breast cell line (Figure 1B, all P < 0.001). The MDA-MB-231 cell line exhibited the highest CRNDE expression and was thus selected for the subsequent experiments.
Figure 1.
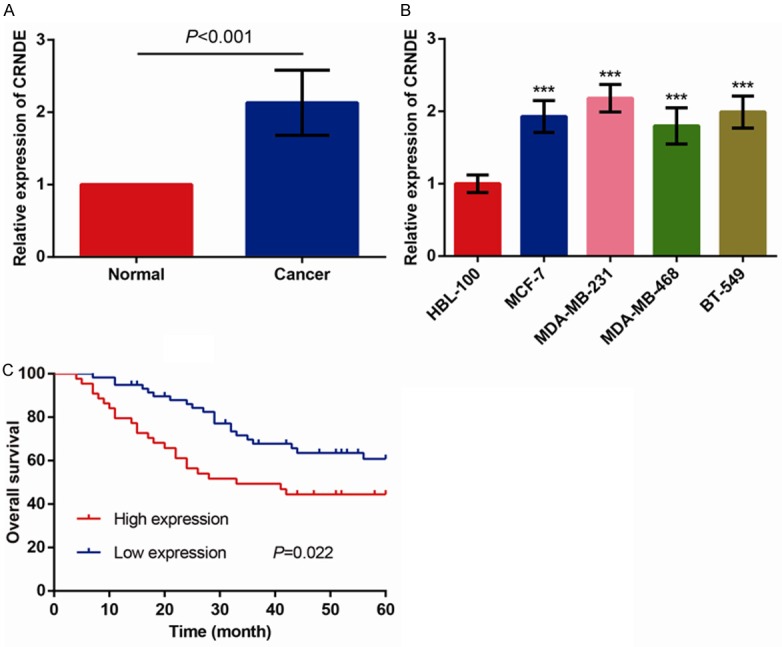
CRNDE expression is frequently elevated in BC. A. Relative expression of CRNDE was determined in 103 pairs of BC tissues and paired adjacent normal breast tissues by qRT-PCR and normalized to GAPDH expression. B. Relative expression of CRNDE in BC cell lines (MCF-7, MDA-MB-231, MDA-MB-468 and BT-549) and normal breast epithelial cell line (HBL-100). C. Correlation between CRNDE expression and overall survival of BC patients through Kaplan-Meier analysis and log-rank test. The data are presented as mean ± S.D. from three independent experiments. ***P < 0.001 as determined by Student’s t-test.
The correlations between CRNDE expression and clinicopathological variables of 103 BC patients were recorded in Table 2. 103 patients was allocated into high CRNDE expression (n=44) and low CRNDE expression (n=59) according to CRNDE expression levels in their BC tissues. The results revealed that up-regulation of CRNDE was obviously correlated to tumor size (P=0.016) and TNM stage (P=0.013), whereas no significantly correlated to age, menopause, histological grade, ER status, PR status and Her-2 status (all P > 0.05).
Table 2.
Association between CRNDE expression and clinicopathological characteristics of 103 BC patients
| Characteristics | Total number | CRNDE expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low (n=59) | High (n=44) | |||
| Age (years) | 0.320 | |||
| ≤ 55 | 67 | 36 | 31 | |
| > 55 | 36 | 23 | 13 | |
| Menopause | 0.786 | |||
| No | 39 | 23 | 16 | |
| Yes | 64 | 36 | 28 | |
| Tumor size (cm) | 0.016 | |||
| ≤ 2.5 | 63 | 42 | 21 | |
| > 2.5 | 40 | 17 | 23 | |
| Histological grade | 0.134 | |||
| I-II | 82 | 50 | 32 | |
| III | 21 | 9 | 12 | |
| TNM stage | 0.013 | |||
| I-II | 74 | 48 | 26 | |
| III-IV | 29 | 11 | 18 | |
| ER status | 0.372 | |||
| Positive | 58 | 31 | 27 | |
| Negative | 45 | 28 | 17 | |
| PR status | 0.1 | |||
| Positive | 42 | 20 | 22 | |
| Negative | 61 | 39 | 22 | |
| Her-2 status | 0.972 | |||
| Positive | 63 | 36 | 27 | |
| Negative | 40 | 23 | 17 | |
The prognostic value of CRNDE expression was determined for OS in 103BC patients through the Kaplan-Meier method and log-rank test. As shown in Figure 1C, BC patients with higher CRNDE expression have significantly shorter OS than those with lower CRNDE expression (P=0.022, log-rank test), thus indicating that there might be a close association between CRNDE expression and prognosis of BC patients.
CRNDE acts as a molecular sponge for miR-136
Through bioinformatics algorithms, we considered that miR-136 might function as a potential binding target of CRNDE, and the 5’ region of miR-136 contains a highly conserved binding site for CRNDE (Figure 2A). In order to verify the prediction, we firstly performed a dual luciferase assay and observed that miR-136 overexpression significantly decreased the luciferase activity of CRNDE-WT but not that of the vector and CRNDE-MUT in MDA-MB-231 cells (Figure 2B).
Figure 2.
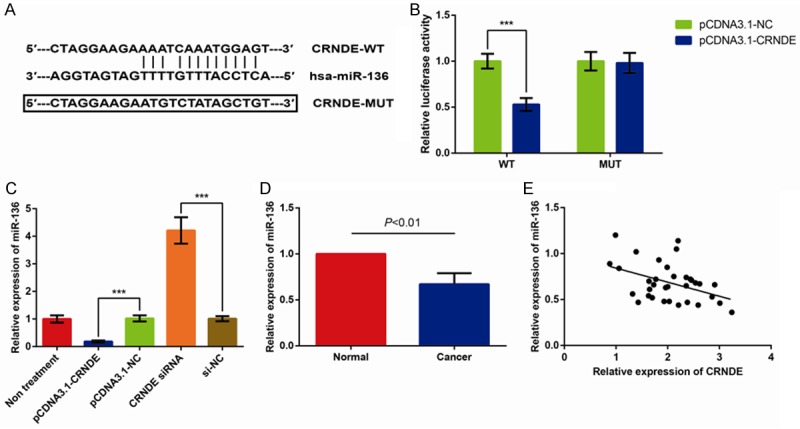
CRNDE acts as a molecular sponge for miR-136. A. Alignment of potential CRNDE base pairing with miRNA-136 as investigated by Starbase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php). B. The relative luciferase activities were measured in MDA-MB-231 cells. C. miR-136 expression in response to CRNDE overexpression and CRNDE inhibition was determined by qRT-PCR. D. Expression of miR-136 in 34 pairs of BC tissues and paired adjacent normal breast tissues. E. The correlation between CRNDE and miR-136 level was measured in 34BC tissues by Pearson correlation analysis. The data are presented as mean ± S.D. from three independent experiments. ***P < 0.001 as determined by Student’s t-test.
Next, we observed that MDA-MB-231cells transfected with pcDNA3.1-CRNDE exhibited significantly increased CRNDE expression (more than 80 folds), and cells transfected with CRNDE siRNA exhibited obviously reduced CRNDE expression (more than 3 folds) (Data not shown). To investigate the interaction of CRNDE and miR-136, the relative miR-136 expression in MDA-MB-231 cell transfected with pcDNA3.1-CRNDE, pcDNA3.1-NC, CRNDE siRNA, and si-NC were detected. As shown in Figure 2C, pcDNA3.1-CRNDE decreased miR-136 expression and CRNDE siRNA increased miR-136 expression compared to NC, respectively (all P < 0.001). Further experiments demonstrated that overexpression of miR-136 significantly also inhibited CRNDE expression, and the levels of CRNDE were greatly increased in cells transfected with miR-136 inhibitors (Data not shown).
Next, we detected miR-136 expression in BC tissues by qRT-PCR. As shown in Figure 2D, miR-136 expression was significantly reduced in BC tissues compared with that in normal tissues (P < 0.01). Furthermore, we assessed the correlation between CRNDE and miR-136 expression in 34BC tissues, and the results indicated that CRNDE and miR-136 expression exhibited a remarkably negative correlation by Pearson correlation analysis (r2=0.179, P=0.013) (Figure 2E). Although the interaction between CRNDE and miR-136 was confirmed, the biological behaviors of BC cells regulated by CRNDE and miR-136 are required to be further verified.
CRNDE activates Wnt/β-catenin signaling through inhibiting miR-136 in BC cell lines
Wnt/β-catenin pathway is a critical regulator of tumor initiation and progression. As expected, the mRNA and protein levels of several downstream targets of the Wnt/β-catenin signaling pathway, including β-catenin, c-myc and cyclinD1, were remarkably up-regulated in the CRNDE-overexpressed MDA-MB-231 cells and decreased in the CRNDE-silenced MDA-MB-231 cells; However, co-transfection with miR-136 mimic reversed the hyperactivation ofthe Wnt/β-catenin signaling pathway in CRNDE-overexpressed cells (Figure 3A, 3B, all P < 0.001). Collectively, these data clearly illustrated that CRNDE overexpression hyper activated the Wnt/β-catenin signaling pathway through regulating miR-136 expression in BC cells.
Figure 3.
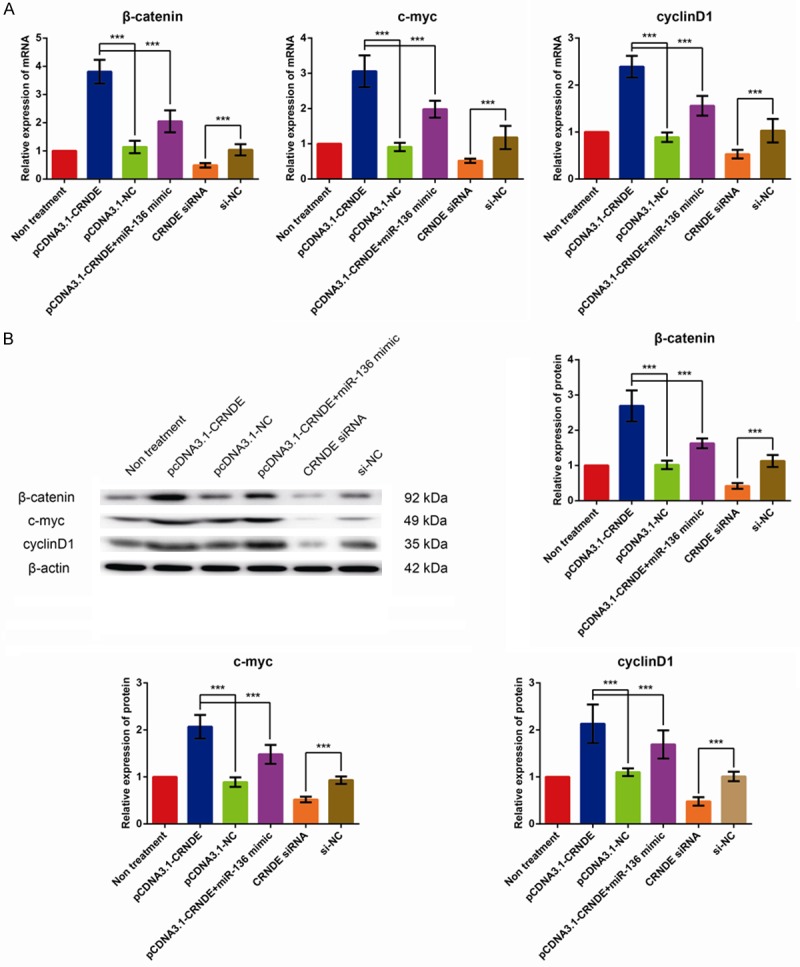
CRNDE activates Wnt/β-catenin signaling through inhibiting miR-136 in BC cell lines. A. The effect of CRNDE expression on β-catenin, c-myc and cyclinD1 mRNA expression in MDA-MB-231 cells. B. The effect of CRNDE expression on β-catenin, c-myc and cyclinD1 protein expression in MDA-MB-231 cells. The data are presented as mean ± S.D. from three independent experiments. ***P < 0.001 as determined by Student’s t-test.
CRNDE overexpression promotes BC cell proliferation, migration and invasion in vitro
We investigated the effects of CRNDE on BC cell proliferation through performing CCK-8 assay. As illustrated in Figure 4A, up-regulation of CRNDE led to a significantly increased proliferation of MDA-MB-231 cells, and co-transfection with miR-136 mimic reversed the promoted proliferation of CRNDE-overexpressed cells. In addition, the proliferation of MDA-MB-231 cells transfected with CRNDE siRNA was dramatically reduced. The results of colony formation assay were also consistent with CCK-8 assay as CRNDE-overexpressed MDA-MB-231 cells formed an increased number of colonies, and co-transfection with miR-136 mimic reversed the increased colonies (Figure 4B).
Figure 4.
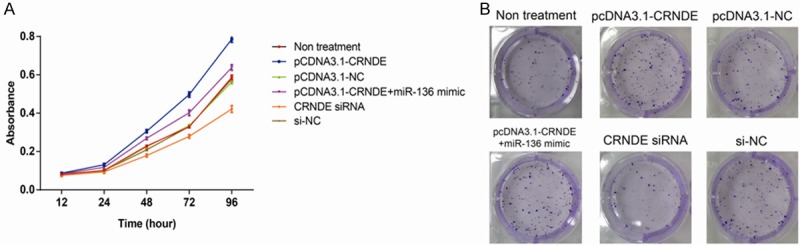
CRNDE promotes the proliferation of BC cells in vitro. A. Cell proliferation was assessed using CCK-8 assay in MDA-MB-231 cells. B. A colony formation assay was performed in MDA-MB-231 cells. The data are presented as mean ± S.D. from three independent experiments.
Furthermore, the effects of CRNDE on the migratory ability and invasiveness of BC cells were assessed by transwell assays and wound-healing assay. As illustrated in Figure 5A, the number of invaded and migrated MDA-MB-231 cells were remarkably increased after overexpression of CRNDE (all P < 0.001), and co-transfection with miR-136 mimic reversed the promoted invasion and migration of CRNDE-overexpressed cells. In addition, the invaded and migrated MDA-MB-231 cells were significantly reduced after transfection with CRNDE siRNA (all P < 0.001). Wound-healing assay also demonstrated that treatment with pcDNA3.1-CRNDE significantly enhanced the motility of MDA-MB-231 cells, and co-transfection with miR-136 mimic reversed the enhanced motility of CRNDE-overexpressed cells, as shown in Figure 5B.
Figure 5.
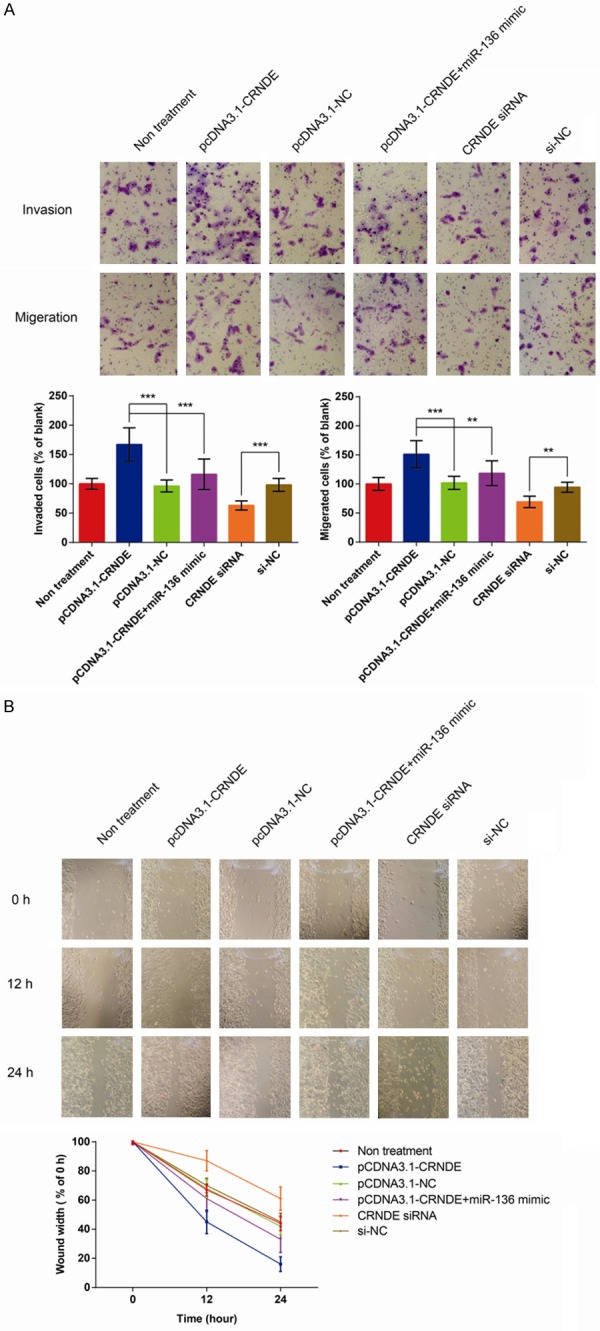
CRNDE promotes the migration, invasion and mobility of BC cells in vitro. A. Results of transwell assays showing the effect of CRNDE expression on the migration and invasion of MDA-MB-231 cells in vitro. B. Results of wound-healing assay showing the effect of CRNDE expression on the mobility of MDA-MB-231 cells in vitro. The data are presented as mean ± S.D. from three independent experiments. ***P < 0.001, **P < 0.01 as determined by Student’s t-test.
Increased CRNDE expression promotes tumor growth in vivo
In order to further verify our findings in vivo, we examined the effects of CRNDE on tumor growth in nude mice. As shown in Figure 6, the average tumor volume and weight were largest in the group of CRNDE overexpression (P < 0.001), whereas the nude mice injected with cells co-transfected with pcDNA3.1-CRNDE and miR-136 mimic exhibited smaller tumor volume and weight than that in the group of CRNDE overexpression (P < 0.001). Besides, in the CRNDE silence group, the average tumor volume and weight were significantly smaller than that in the other group (P < 0.001). Accordingly, we conclude that CRNDE could promote tumorigenesis through regulating miR-136 in vivo.
Figure 6.
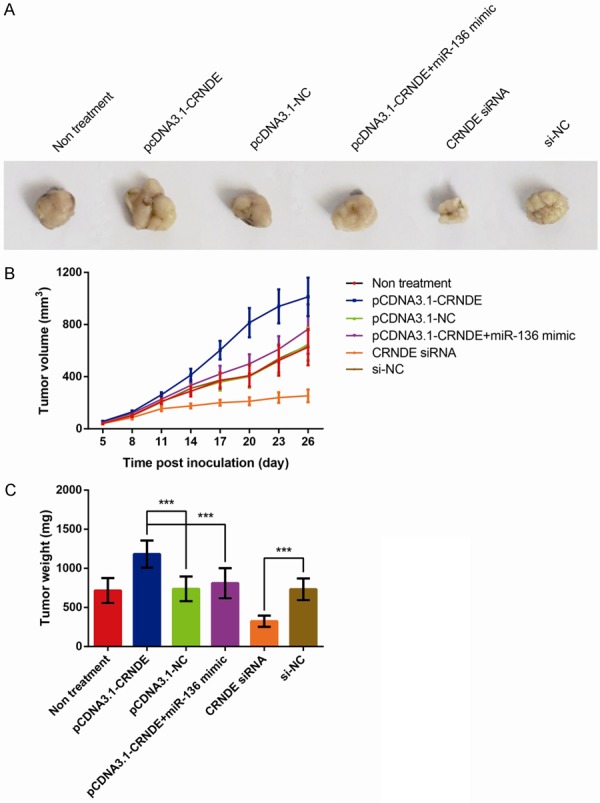
CRNDE promotes BC cells growth by inhibiting miR-136 in vivo. A. Representative pictures of the tumors. B. The growth curves of tumor sizes. C. The tumor weights. The data are presented as mean ± S.D. from three independent experiments. ***P < 0.001 as determined by Student’s t-test.
Discussion
BC is one of the most aggressive malignancies caused by multiple factors [23]. With the rapid development of molecular biotechnology, biological targeted therapy has gradually become a crucial assistant treatment for BC [24]. Multiple evidence illustrates that aberrant lncRNA expression is engaged in various cancers, and the targeted therapies applying lncRNA as a novel diagnostic and therapeutic tool has obtained more and more attention, such as ATB [25], PVT1 [26] and MALAT1 [27]. To our knowledge, the current research is the first time to offer evidence for a novel mechanistic correlation between CRNDE and the Wnt/β-catenin signaling pathway in human BC. We found that CRNDE expression was remarkably increased in BC tissues and was inversely associated with unfavorable prognosis of BC patients. Overexpression of CRNDE facilitated, while CRNDE silencing suppressed, BC cell viability both in vitro and in vivo. Moreover, we showed that overexpression of CRNDE hyperactivated the Wnt/β-catenin signaling pathway via directly targeting miR-136. Collectively, our results exhibited that CRNDE functions as one of the oncogenes in BC and might be regarded as a critical target for therapeutic tool for BC in clinical application.
The canonical Wnt/β-catenin signaling is triggered by the secreted Wnt ligands, which bind to the Frizzled (FZD) family receptors and LRP5/LRP6 co-receptor to stimulate the β-catenin signaling cascade, and thus facilitates cyclinD1 and c-myc expressions [28]. Dsyregulated activation of the Wnt/β-catenin signaling pathway is commonly uncovered in various human cancers and might promote tumor progression, including BC [29]. Understanding the underlying mechanisms is critical for the improvement of cancer therapy. Shao et al. found that CRNDE promotes cancer cell proliferation and growth through regulating Wnt/β-catenin signaling; this exhibited a representative molecular mechanism involved in renal cell carcinoma, which could be considered as a promising therapeutic method for these patients [17]. However, to date, the upstream factors of the Wnt/β-catenin signaling pathway and their function role in modulating carcinogenesis and metastasis in BC is still not fully elucidated.
Current analyses of the mammalian transcriptome have indicated that approximately 70% of the genome is transcribed into RNA that does not function as templates for protein, while only 2% of the genome play roles as coding transcripts [30]. Dysregulated expressions of various lncRNAs play critical roles in cancer origination and progression, and the regulatory roles of lncRNAs in cancer attract wide attentions globally. Previous articles have documented that CRNDE is overexpressed in multiple human cancers, including glioma in which it might be the most upregulated lncRNA [31]. Intriguingly, despite its little to no expression in adult colorectal mucosa and white blood cells, CRNDE is high expressed in testis, skin and parotid gland [32]. Herein, our findings exhibited that CRNDE overexpression could promote the proliferation, migration and invasion of BC cells through activating the Wnt/β-catenin signaling pathway. Chen et al. showed that in hepatocellular carcinoma, CRNDE accelerated the expression levels of NF-κB and p-AKT though acting as a sponge for miR-384 [12]. Other article found that in gallbladder cancer, CRNDE promotes PI3K-AKT pathway as a scaffold to recruit the DMBT1 and c-IAP1 [33]. Thus we further focused on the direct target of CRNDE in BC.
The underlying functional mechanisms of lncRNA are quite complicated actually. One important mechanism is that it can play a role as a ceRNA or a molecular sponge and direct interact with miRNA so as to restrain its inhibitory effect on downstream target mRNA [34,35]. An example for this kind of regulation is exemplified by HOST2 (human ovarian cancer-specific transcript 2), an lncRNA specifically expressed at high level in human ovarian cancer, which could bind with miRNA let-7b and thus forms a regulatory interaction [36]. Aberrant expression of miR-136 has been found in miRNA profile studies of different cancer types, including non-small cell lung cancer [37], glioma [38], chemoresistant ovarian cancer [39] and BC [40]. We predicted the miR-136 binding site of CRNDE by using the bioinformatics databases (Starbase v2.0). Meanwhile, our experimental findings demonstrated that up-regulation of CRNDE inhibited miR-136 expression, and overexpression of miR-136 could re-inactivate the Wnt/β-catenin signaling pathway hyperactivated by up-regulation of CRNDE; therefore suggested that CRNDE could directly inhibiting miR-136, thus activating the Wnt pathway in BC. We speculated that CRNDE might sequester miR-136 as a molecular “sponge” or as a ceRNA and thereby regulate miRNA function. So far we have preliminarily clarified the relationship between CRNDE and miR-136.
Up to date, the molecular mechanisms underlying the pathogenesis of BC still remain elusive. Our present study demonstrated that CRNDE could promote tumor growth in BC through acting as a molecular sponge of miR-136, thus hyperactivating the Wnt/β-catenin signaling pathway. These findings uncover a novel mechanism of the Wnt pathway hyperactivation in BC, and CRNDE might serve as a potential therapeutic tool for the treatment of BC in the near future.
Acknowledgements
The work of our research was supported by key research project of Shanghai Health Bureau (NO. SH2012483).
Disclosure of conflict of interest
None.
References
- 1.Fu DY, Tan HS, Wei JL, Zhu CR, Jiang JX, Zhu YX, Cai FL, Chong MH, Ren CL. Decreased expression of SOX17 is associated with tumor progression and poor prognosis in breast cancer. Tumour Biol. 2015;36:8025–8034. doi: 10.1007/s13277-015-3547-3. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 3.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu L, Xu H, Luo F, Liu X, Lu X, Yang Q, Xue J, Chen C, Shi L, Liu Q. Epigenetic silencing of miR-218 by the lncRNA CCAT1, acting via BMI1, promotes an altered cell cycle transition in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2016;304:30–41. doi: 10.1016/j.taap.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Ballantyne RL, Zhang X, Nunez S, Xue C, Zhao W, Reed E, Salaheen D, Foulkes AS, Li M, Reilly MP. Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits. Hum Mol Genet. 2016;25:3125–3141. doi: 10.1093/hmg/ddw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Song Z, Huang S, Wang R, Qin W, Guo J, Lin Z. lncRNA DANCR suppresses odontoblast-like differentiation of human dental pulp cells by inhibiting wnt/beta-catenin pathway. Cell Tissue Res. 2016;364:309–318. doi: 10.1007/s00441-015-2333-2. [DOI] [PubMed] [Google Scholar]
- 7.Mendell JT. Targeting a long noncoding RNA in breast cancer. N Engl J Med. 2016;374:2287–2289. doi: 10.1056/NEJMcibr1603785. [DOI] [PubMed] [Google Scholar]
- 8.Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, Lapointe LC, Molloy PL. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2:829–840. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu Y, Li Z, Li ZQ, Wang ZH, Liu YH. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6:25339–25355. doi: 10.18632/oncotarget.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing SY, Lu YY, Yang JK, Deng WY, Zhou Q, Jiao BH. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur Rev Med Pharmacol Sci. 2016;20:3992–3996. [PubMed] [Google Scholar]
- 11.Szafron LM, Balcerak A, Grzybowska EA, Pienkowska-Grela B, Podgorska A, Zub R, Olbryt M, Pamula-Pilat J, Lisowska KM, Grzybowska E, Rubel T, Dansonka-Mieszkowska A, Konopka B, Kulesza M, Lukasik M, Kupryjanczyk J. The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget. 2015;6:43897–43910. doi: 10.18632/oncotarget.6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Yu C, Zhan L, Pan Y, Chen L, Sun C. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6:2299–2309. [PMC free article] [PubMed] [Google Scholar]
- 13.Monin MB, Krause P, Stelling R, Bocuk D, Niebert S, Klemm F, Pukrop T, Koenig S. The anthelmintic niclosamide inhibits colorectal cancer cell lines via modulation of the canonical and noncanonical Wnt signaling pathway. J Surg Res. 2016;203:193–205. doi: 10.1016/j.jss.2016.03.051. [DOI] [PubMed] [Google Scholar]
- 14.Rong S, Zhao X, Jin X, Zhang Z, Chen L, Zhu Y, Yuan W. Vascular calcification in chronic kidney disease is induced by bone morphogenetic protein-2 via a mechanism involving the Wnt/beta-catenin pathway. Cell Physiol Biochem. 2014;34:2049–2060. doi: 10.1159/000366400. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, Yan W, Liu X, Jia Y, Cao B, Lv Y, Brock MV, Herman JG, Licchesi J, Yang Y, Guo M. DACT2 is frequently methylated in human gastric cancer and methylation of DACT2 activated Wnt signaling. Am J Cancer Res. 2014;4:710–724. [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Q, Krause M, Samoylenko A, Vainio S. Wnt signaling in renal cell carcinoma. Cancers (Basel) 2016;8 doi: 10.3390/cancers8060057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao K, Shi T, Yang Y, Wang X, Xu D, Zhou P. Highly expressed lncRNA CRNDE promotes cell proliferation through Wnt/beta-catenin signaling in renal cell carcinoma. Tumour Biol. 2016 doi: 10.1007/s13277-016-5440-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 20.Zununi Vahed S, Barzegari A, Rahbar Saadat Y, Mohammadi S, Samadi N. A microRNA isolation method from clinical samples. Bioimpacts. 2016;6:25–31. doi: 10.15171/bi.2016.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Chen X, Xu T, Xia R, Han L, Chen W, De W, Shu Y. MiR-326 regulates cell proliferation and migration in lung cancer by targeting phox2a and is regulated by HOTAIR. Am J Cancer Res. 2016;6:173–186. [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Wheler JJ, Atkins JT, Janku F, Moulder SL, Yelensky R, Stephens PJ, Kurzrock R. Multiple gene aberrations and breast cancer: lessons from super-responders. BMC Cancer. 2015;15:442. doi: 10.1186/s12885-015-1439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaidat OO, Fitzsimmons BF, Woodward BK, Wang Z, Killer-Oberpfalzer M, Wakhloo A, Gupta R, Kirshner H, Megerian JT, Lesko J, Pitzer P, Ramos J, Castonguay AC, Barnwell S, Smith WS, Gress DR. Effect of a balloonexpandable intracranial stent vs medical therapy on risk of stroke in patients with symptomatic intracranial stenosis: the VISSIT randomized clinical trial. JAMA. 2015;313:1240–1248. doi: 10.1001/jama.2015.1693. [DOI] [PubMed] [Google Scholar]
- 25.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colombo T, Farina L, Macino G, Paci P. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int. 2015;2015:304208. doi: 10.1155/2015/304208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jadaliha M, Zong X, Malakar P, Ray T, Singh DK, Freier SM, Jensen T, Prasanth SG, Karni R, Ray PS, Prasanth KV. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7:40418–40436. doi: 10.18632/oncotarget.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Hwang MS, Yu N, Stinson SY, Yue P, Newman RJ, Allan BB, Dornan D. miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One. 2013;8:e66502. doi: 10.1371/journal.pone.0066502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 31.N L, GP Z, SM L. Stenting versus medical therapy for symptomatic intracranial arterial stenosis. Chinese Journal of Cerebrovascular Diseases. 2012:483–485. [Google Scholar]
- 32.Ellis BC, Molloy PL, Graham LD. CRNDE: a long non-coding RNA involved in cancer, neurobiology, and development. Front Genet. 2012;3:270. doi: 10.3389/fgene.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen S, Liu H, Wang Y, Wang J, Ni X, Ai Z, Pan H, Shao Y. Long non-coding RNA CRNDE promotes gallbladder carcinoma carcinogenesis and as a scaffold of DMBT1 and C-IAP1 complexes to activating PI3K-AKT pathway. Oncotarget. 2016;7:72833–72844. doi: 10.18632/oncotarget.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 36.Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao T, Liu Y, Ou J, Wang D, Yao L, Hui N. LncRNAHOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24:841–852. doi: 10.1093/hmg/ddu502. [DOI] [PubMed] [Google Scholar]
- 37.Shen S, Yue H, Li Y, Qin J, Li K, Liu Y, Wang J. Upregulation of miR-136 in human nonsmall cell lung cancer cells promotes Erk1/2 activation by targeting PPP2R2A. Tumour Biol. 2014;35:631–640. doi: 10.1007/s13277-013-1087-2. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Wu J, Guan H, Cai J, Fang L, Li J, Li M. MiR-136 promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2. FEBS Lett. 2012;586:3608–3612. doi: 10.1016/j.febslet.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Liu S, Wang G, Wu X, Ding Y, Guo G, Jiang J, Cui S. Expression of miR-136 is associated with the primary cisplatin resistance of human epithelial ovarian cancer. Oncol Rep. 2015;33:591–598. doi: 10.3892/or.2014.3640. [DOI] [PubMed] [Google Scholar]
- 40.Yan M, Li X, Tong D, Han C, Zhao R, He Y, Jin X. miR-136 suppresses tumor invasion and metastasis by targeting RASAL2 in triple-negative breast cancer. Oncol Rep. 2016;36:65–71. doi: 10.3892/or.2016.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]


