Abstract
Abstract: A virtual screening analysis of our library of phytochemical structures with dengue virus protein targets has been carried out using a molecular docking approach. A total of 2194 plant-derived secondary metabolites have been docked. This molecule set comprised of 290 alkaloids (68 indole alkaloids, 153 isoquinoline alkaloids, 5 quinoline alkaloids, 13 piperidine alkaloids, 14 steroidal alkaloids, and 37 miscellaneous alkaloids), 678 terpenoids (47 monoterpenoids, 169 sesquiterpenoids, 265 diterpenoids, 81 steroids, and 96 triterpenoids), 20 aurones, 81 chalcones, 349 flavonoids, 120 isoflavonoids, 74 lignans, 58 stilbenoids, 169 miscellaneous polyphenolic compounds, 100 coumarins, 28 xanthones, 67 quinones, and 160 miscellaneous phytochemicals. Dengue virus protein targets examined included dengue virus protease (NS2B-NS3pro), helicase (NS3 helicase), methyltransferase (MTase), RNA-dependent RNA polymerase (RdRp), and the dengue virus envelope protein. Polyphenolic compounds, flavonoids, chalcones, and other phenolics were the most numerous of the strongly docking ligands for dengue virus protein targets.
Keywords: Molecular docking, dengue fever virus, natural products drug discovery
1. INTRODUCTION
Dengue fever is an acute viral infection caused by all four serotypes (1, 2, 3, or 4) of the dengue virus (DENV), and is one of the seventeen “Neglected Tropical Diseases” (NTDs) defined by the World Health Organization [1]. The highest incidence of dengue fever is in Southeast Asia, India, and the Neotropics; where the principal vector, the Aedes aegypti mosquito, is found [2]. There are two well defined manifestations of dengue virus infection in humans, dengue fever and severe dengue (dengue hemorrhagic fever / dengue shock syndrome, DHF/DSS) [3]. Dengue viral infections account for around 100 million cases of dengue fever and more than 500,000 cases of DHF/DSS each year [4]. India accounts for around 34% of the total global dengue infections while tropical America contributes around 14% [1, 5]. Delhi, India has had several epidemic outbreaks of dengue fever (1970, 1982, 1988, 1996, 2006, 2010, and 2013) [6]. In the 2013 outbreak, 71% of suspected patients tested positive for dengue virus, mainly DENV-2, but co-infection by different serotypes was also detected [7]. Rio de Janeiro, Brazil, has suffered similar epidemic outbreaks of dengue fever (1986, 1990, 2001-02, 2007-08, 2011, and 2012) [8, 9]. Between 2010 and 2012, dengue virus infection was confirmed in 47.5% of the cases; DENV-2 the prevalent serotype in 2010, but DENV-1 re-emerged as the major serotype in 2011 [9]. In addition, between 2010 and 2012, 7.3% of the cases were severe (DHF/DSS), due mainly to secondary dengue infection. In 2007, 53% of DHF/DSS cases occurred in children under 15 years of age [10]. Furthermore, a correlation between increased dengue infection risk and socioeconomic status has been demonstrated; poorer inhabitants with poor access to health services showed a significantly higher seroprevelence compared to wealthy inhabitants [10, 11]. A serological survey carried out in 2007-2008 in three neighborhoods of Rio de Janeiro showed more than half the population was seropositive (60.3% in Higienópolis, a densely populated urban neighborhood, 56.1% in Tubiacanga, a suburban residential neighborhood, and 77.4% in Palmares, a recently settled slum) [8].
Currently, there are no treatment options for dengue fever; there is no known cure or vaccine. There are, however, several dengue virus proteins that have been identified as potential drug targets [12]. Natural products have been and continue to be excellent sources of medicinal agents, as themselves or as templates for synthetic agents [13]. The people of developing nations often have limited access to health care and inadequate resources for pharmaceuticals, and traditional herbal medicines offer the potential for treatments of diseases in these regions. In this work, we have screened our virtual library of phytochemicals against several protein targets of dengue virus [14], included dengue virus protease (NS2B-NS3pro), helicase (NS3 helicase), methyltransferase (MTase), RNA-dependent RNA polymerase (RdRp), and the dengue virus envelope protein (Ep), using a molecular docking approach.
1.1. Dengue Virus Protease
DENV protease (NS2B-NS3pro) is a trypsin-like serine protease that cleaves the dengue polyprotein into individual proteins necessary for viral replication. DENV protease has been identified as a primary target for the development of dengue antiviral drugs [15]. There are two potential targets for DENV protease inhibition: (1) the active site of the protease [16], and (2) block association of NS3pro with its protein cofactor NS2B [17].
1.2. Dengue Virus Helicase
DENV NS3 helicase is an essential enzyme for viral replication and has been identified as a potential drug target. The enzyme shows both ATP-hydrolysis activity as well as RNA duplex unwinding activity. These two enzymatic activities are likely coupled with ATPase activity providing the chemical energy for RNA unwinding. Thus, there are two potential targets for DENV NS3 helicase, the RNA binding site [18] and the ATP binding site [12].
1.3. Dengue Virus Methyltransferase
DENV MTase is located on the N-terminus of NS5 and catalyzes the N-7 and 2ʹ-O methylations of the viral RNA cap [19]. These modifications are necessary for formation of the mature RNA cap structure. The MTase uses S-adenosylmethionine (SAM) as the methylating cofactor. There are binding sites for GTP and the guanine moiety of the RNA cap (the GTP pocket) and for SAM. DENV MTase is a valid antiviral target, and the GTP binding site has been identified as a valid site for development of antivirals [20]. Although S-adenosylmethionine associates tightly with DENV MTase and displacement of SAM may be difficult, the SAM binding site may, however, be capable of binding other ligands [12, 21].
1.4. Dengue Virus RNA-Dependent RNA Polymerase
DENV RdRp is essential for viral replication and there are no similar enzymes found in the host. This enzyme, then, is considered to be a prime target for antiviral development [12]. Nucleoside-based prodrug antivirals have been investigated. These agents are phosphorylated in vivo, and then target the active site of the RdRp enzyme [20]. Non-nucleoside small molecules can inhibit viral RdRp by binding to allosteric sites of the protein [19].
1.5. Dengue Virus Envelope Protein
DENV E protein mediates the binding of the virus to the host cell surface receptors [19]. Thus, interference of this interaction would be a potential avenue for antiviral development. A small hydrophobic channel has been identified in the DENV E protein and this site has been identified as a target for small-molecule inhibitors [12].
The purpose of this work was to examine whether higher plants may serve as a source of antiviral agents for treatment of dengue fever. A molecular docking approach was used to examine a selection of phytochemicals from our virtual library (2194 different compound structures) with the protein targets of dengue virus.
2. COMPUTATIONAL METHODS
Protein-ligand docking studies were carried out based on the crystal structures (PDB 3U1J [22], 4M9K, 4M9M, and 4M9T [23]) and the NMR structures (PDB 2M9P and 2M9Q [24]) of dengue virus NS3 protease; crystal structures of dengue virus NS3 helicase (PDB 2BHR, 2BMF [25], 2JLQ, 2JLR, 2JLS, 2JLU, 2JLV, 2JLX [26], and 2WHX [27]); crystal structures of dengue virus NS5 RNA methyltransferase (PDB 3P8Z [28], 4CTJ, 4CTK, and 4R8S [21]); crystal structures of dengue virus NS5 RNA-dependent RNA polymerase (2J7U, 2J7W [29], 3VWS, and 4HHJ [30]); and crystal structures of dengue virus envelope protein (PDB 1OAN and 1OKE [31]). In order to examine selectivity for DEVN MTase, human RNA methyltransferase proteins (PDB 3BGV [32], 3EPP [33], and 4N49 [34]) were also used for docking. Similarly, in order to examine selectivity for DENV protease over mammalian serine proteases, we have carried out docking of the phytochemical ligands with crystal structures of rat trypsin (PDB 1BRB [35]), bovine trypsin (PDB 2ZDK [36] and 4MTB [37]), and porcine elastase (PDB 7EST [38]). Prior to docking, all solvent molecules and the co-crystallized ligands were removed from the structures. Molecular docking calculations for all compounds with each of the proteins were undertaken using Molegro Virtual Docker (version 6.0, Molegro ApS, Aarhus, Denmark) [39], with a sphere (15 Å radius) large enough to accommodate the cavity centered on the binding sites of each protein structure in order to allow each ligand to search. If a co-crystallized inhibitor or substrate was present in the structure, then that site was chosen as the binding site. If no co-crystallized ligand was present, then suitably sized (> 50 Å3) cavities were used as potential binding sites. Standard protonation states of the proteins based on neutral pH were used in the docking studies. Each protein was used as a rigid model structure; no relaxation of the protein was performed. Assignments of charges on each protein were based on standard templates as part of the Molegro Virtual Docker program; no other charges were necessary to be set. Overall, 2194 plant-derived secondary metabolites have been docked. This molecule set was comprised of 290 alkaloids (68 indole alkaloids, 153 isoquinoline alkaloids, 5 quinoline alkaloids, 13 piperidine alkaloids, 14 steroidal alkaloids, and 37 miscellaneous alkaloids), 678 terpenoids (47 monoterpenoids, 169 sesquiterpenoids, 265 diterpenoids, 81 steroids, and 96 triterpenoids), 20 aurones, 81 chalcones, 349 flavonoids, 120 isoflavonoids, 74 lignans, 58 stilbenoids, 169 miscellaneous polyphenolic compounds, 100 coumarins, 28 xanthones, 67 quinones, and 160 miscellaneous phytochemicals. While this list does not include all phytochemicals, it does represent a practical selection of phytochemical classes and structural types. Each ligand structure was built using Spartan ’14 for Windows (version 1.1.8, Wavefunction Inc., Irvine, California). For each ligand, a conformational search and geometry optimization
was carried out using the MMFF force field [40]. Flexible ligand models were used in the docking and subsequent optimization scheme. Variable orientations of each of the ligands were searched and ranked based on their re-rank score. For each docking simulation the maximum number of iterations for the docking algorithm was set to 1500, with a maximum population size of 50, and 100 runs per ligand. The RMSD threshold for multiple poses was set to 1.00 Å. The generated poses from each ligand were sorted by the calculated re-rank score.
3. RESULTS and DISCUSSION
In this analysis, we have rejected those phytochemical ligands that violate Lipinski’s rule of five [41]. That is, ligands with MW > 500 g/mol, hydrogen-bond-donating atoms > 5, hydrogen-bond-accepting atoms > 10, or ClogP > 5, even though they may have strong docking energies, were not considered.
3.1. Dengue Virus Protease
The phytochemical ligands in this investigation were docked to the DENV NS2B-NS3 protease as well as with three different mammalian trypsin-like proteases (bovine trypsin, rat trypsin, and porcine elastase). Those ligands with the best docking for the DENV NS2B-NS3 protease (Edock ≤ -130 kJ/mol, comparable to the inhibitor (4S,5S)-4-amino-6,6,6-trifluoro-5-hydroxyhexyl-1-guanidinium) that were also selective for the virus protease over mammalian trypsin (at least 15 kJ/mol stronger docking) are listed in Table 1. The binding site of DENV NS2B-NS3 protease involves the catalytic triad of Ser196, His112, and Asp136. Additional amino acids at the active site are Glu46, Ile97, and Glu47. The binding site of this serine protease, as evidenced by the NMR structure with bound dipeptide Lys-Arg (PDB 2M9Q) [24], is bounded by Glu46 and Glu47, which both form electrostatic ionic interactions with the -NH3+ moiety of Lys-Arg. Other amino acids at the binding site include Val113, Ile97, Gln88, and Ile91.
Table 1.
Docking energies (kJ/mol) of phytochemical ligands with DENV NS2B-NS3 protease and with mammalian trypsin.
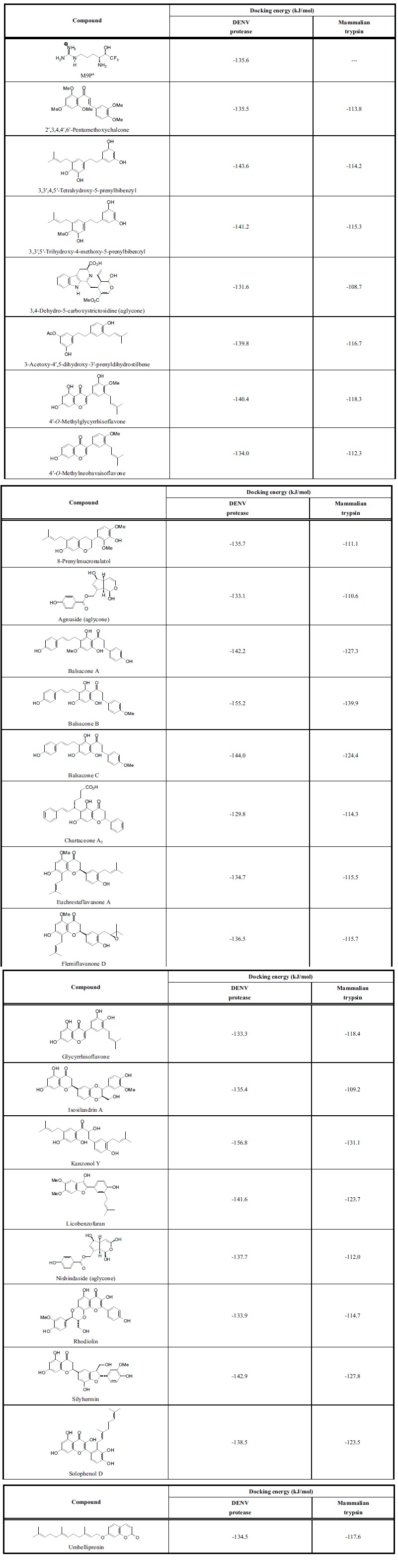
|
a M9P = the inhibitor (4S,5S)-4-amino-6,6,6-trifluoro-5-hydroxyhexyl-1-guanidinium [24].
As a class, the strongest docking ligands were the chalcones (average docking energy = -121.2 kJ/mol) and the stilbenoids (average docking energy = -119.5 kJ/mol). The worst docking compounds were the terpenoids, monoterpenoids (average Edock = -63.8 kJ/mol), sesquiterpenoids (average Edock = -89.6 kJ/mol), diterpenoids (average Edock = -89.7 kJ/mol), and triterpenoids (average Edock = -91.8 kJ/mol). The lowest-energy docking poses of the chalcone ligands balsacone A, balsacone B, and balsacone C, place these compounds into both the binding site of Lys-Arg as well as the catalytic site occupied by the co-crystallized inhibitor M9P (see Fig. 1).
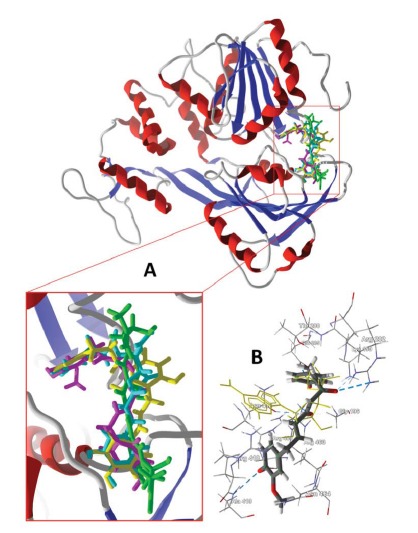
Fig. (1).
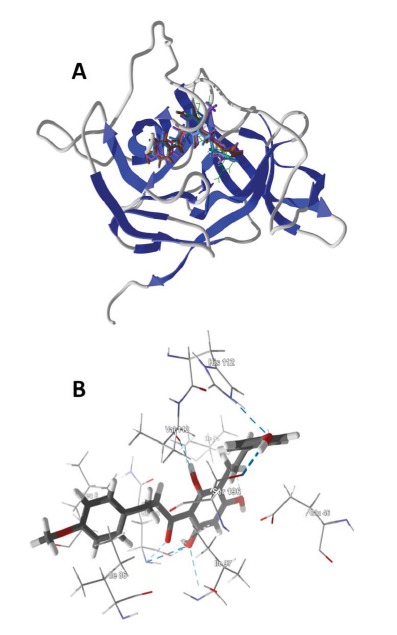
(A) Lowest-energy docking poses of balsacone A (magenta), balsacone B (brown), and balsacone C (aqua) with DENV NS2B-NS3 protease (PDB 2M9Q) [24]. The Lys-Arg co-crystallized ligand is shown as a green wire figure. (B) Balsacone B in the active site of DENV NS2B-NS3 protease (PDB 2M9Q) showing key intermolecular contacts, including hydrogen bonding (blue dashed lines) of the ligand with His112 and Ser196.
Previous docking studies with DENV NS2B-NS3 protease by de Sousa and co-workers had focused on flavonoids that had shown protease inhibition activity [42]. The most active compound in that study was agathisflavone, but in our docking analysis, this compound docks to DENV protease with higher energy (Edock = -126.1 kJ/mol) than it does with rat trypsin (Edock = -138. 6 kJ/mol). Additionally, this biflavone has one rule-of-five violation (MW = 522.46 g/mol). Other flavonoid ligands examined by de Sousa et al.,
myricetin, quercetin, and kaempferol, were relatively weak docking ligands (Edock = -105.5, -102.7, and -91.5 kJ/mol, respectively) compared to those shown in Table 1.
ul Qamar and co-workers had carried out a docking analysis with DENV NS2B-NS3 protease using a library of 940 phytochemicals [43]. These workers found Garcinia phytochemicals to be their best hits, including gossypol, mangostenone C, garcidepsidone A, and dimethyl-calabaxanthone. We have docked these compounds in this present study, but they do not dock as well as the ligands listed in Table 1 (Edock = -112.6, -116.8, -111.7 kJ/mol for gossypol, mangostenone C, and demethylcalabaxanthone, respectively). Garcidepsidone A had a docking energy of -127.4 kJ/mol, but this compound violates Lipinski’s rule of five (ClogP = 5.74).
A docking study with compounds from Murraya koenigii and DENV NS2B-NS3 protease revealed the bisindole alkaloid bismurrayafoline E to be a promising binding ligand [44]. This compound, however, violates Lipinski’s rule of 5 (MW = 724.98 g/mol), and was not considered in our study. In another docking study, panduratin A and 4-hydroxypanduratin A, two phenolic compounds from Boesenbergia rotunda with dengue-2 NS3 protease inhibitory activity [45], were found to dock well with the active site of DENV NS2B-NS3 protease [46]. In this present docking study, we find only modest docking energies for panduratin A and 4-hydroxypanduratin A (-97.1 and -90.9 kJ/mol, respectively) compared to those listed in Table 1, suggesting that much more potent dengue virus NS2B-NS3 protease inhibitors are available.
Glycyrrhiza extracts have shown antiviral activity against human immunodeficiency virus [47, 48], Newcastle disease virus [49], human respiratory syncytial virus [50] and herpes simplex virus 1 [51]. Prenylated stilbenoids (3,3ʹ,4,5ʹ-tetrahydroxy-5-prenylbibenzyl, 3,3ʹ,5ʹ-trihydroxy-4-methoxy-5-prenylbibenzyl, 3-acetoxy-4ʹ,5-dihydroxy-3ʹ-prenyldihydrostilbene, licobenzofuran) and isoflavonoids (glycyrrhisoflavone, 4ʹ-O-methylglycyrrhisoflavone) chalcones (kanzonol Y) from G. glabra have demonstrated outstanding docking properties to DENV protease (Table 1). Prenylated phenolics from G. glabra have inhibited HIV giant cell formation [47]. In addition, kanzonol Y docked well with DENV RNA-dependent RNA polymerase and DENV envelope protein (see below). Glabraisoflavanone, also from G. glabra, docked well with DENV methyltransferase (see below).
The balsacones A-C are antibacterial dihydrochalcones from Populus balsamifera [52]. We are unaware of any antiviral properties of these compounds, but they do exhibit excellent docking properties to DENV NS2B-NS3 protease (Table 1) and DENV helicase (see below, Table 2). The prenylated flavonoids glabranine and 7-O-methylglabranine, isolated from Tephrosia spp., had shown in-vitro antiviral activity against dengue virus serotype 2 [53]. Although these two flavonoids were not among the strongest docking, they did show docking selectivity with DENV protease (Edock = -113.4 and -118.8 kJ/mol, respectively). Two diprenylated flavanones, euchrestaflavanone A from Macaranga pleiostemona [54] and flemiflavanone D from Flemingia stricta [55] showed notably strong docking with DENV NS2B-NS3 protease (Table 1). These compounds had shown antibacterial activity, but to our knowledge have not been screened for anti-dengue activity.
Table 2.
Lowest-energy docking scores (re-rank scores) for phytochemical ligands with dengue virus helicase.
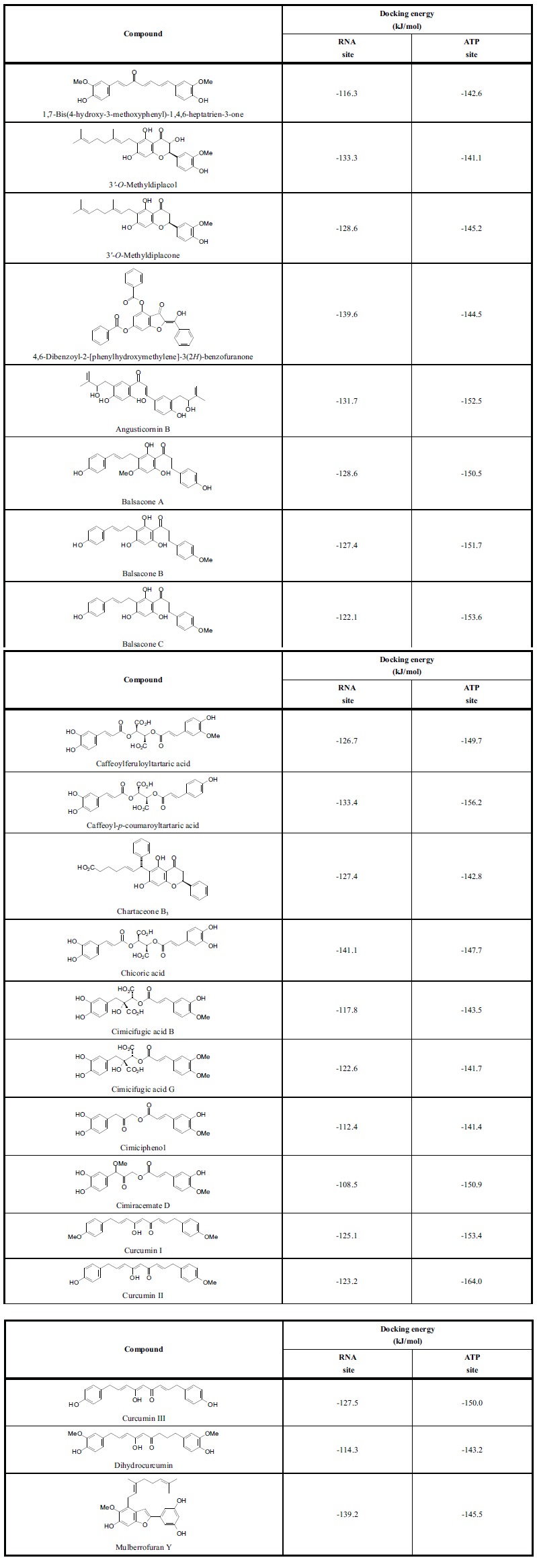
|
In addition to docking energies, it is useful to consider proximity of electrophilic ligands (e.g., epoxides or enones) to nucleophilic residues in the active site. A search of the docked ligands with DENV protease has revealed three sesquiterpenoid α-methylene lactones that dock in close proximity to the –OH group of Ser196. Ineupatorolide A, bigelovin, and 2-de-ethoxy-2-methoxyphantomolin (Fig. 2) dock with the methylene carbon within 3.6 Å of the serine –OH. These ligands can presumably undergo conjugate addition with the –OH group and form a covalent intermediate thereby inhibiting the protease.
Fig. (2).
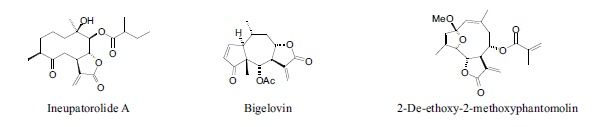
α-Methylenelactone ligands that can potentially alkylate DENV NW2B-NS3 protease.
3.2. Dengue Virus Helicase
The strongest docking (most exothermic) ligands that do not violate Lipinski’s rule of five are listed in Table 2 along with their docking energies to the RNA binding site and the ATP binding site. These ligands were selected based on their docking energies compared to the docking energies of the co-crystallized ligands in the ATP binding site, ANP (phosphoaminophosphonic acid adenylate ester, -156.9 kJ/mol) and ADP (adenosine 5’-diphosphate, -142.1 kJ/mol). Note that the phytochemical ligands showed a notable docking preference for the ATP site of DENV helicase over the RNA binding site. The ATP site of DENV helicase is dominated by protonated basic amino acids (Arg463, Lys199, Lys201, Arg460) that form ionic interactions and hydrogen bonds with the phosphate groups of ATP and the co-crystallized ligands ADP or ANP. Additional hydrogen-bonding amino acids include Gly196, Gly198, and Thr200. Lys201 and Arg418 form hydrogen-bonds with the adenine moiety.
As was the case with DENV protease, the strongest docking class of phytochemicals with DENV helicase ATP site were the chalcones (average Edock = -126.1 kJ/mol) and the stilbenoids (average Edock = -117.6 kJ/mol) while the worst were the terpenoids. The strongest docking ligands were curcumin II (Edock = -164.0 kJ/mol), caffeoyl-p-coumaroyltartaric acid (Edock = -156.2 kJ/mol), balsacone C (Edock = -153.6 kJ/mol), curcumin I (Edock = -153.4 kJ/mol), and angusticornin B (Edock = -152.5 kJ/mol). These ligands dock into the phosphate site of the ATP binding site and not in the adenine site of co-crystallized ADP or ANP. The common structural features of the best docking ligands are two phenolic groups with a flexible linker between them. One of the phenolic groups occupies the phosphate site while the other occupies a site flanked on either side by Arg463 and Arg418 (Figure 3). Several cinnamic acid derivatives from black cohosh (Cimicifuga racemosa) [56] have shown exceptional docking to the ATP site of DENV helicase, including cimicifugic acids B and G, cimiciphenol, and cimiracemate D (Table 2).
Fig. (3).
(A) Lowest-energy docking poses of angusticornin B (green), balsacone C (yellow), caffeoyl-p-coumaroyltartaric acid (aqua), and curcumin II (magenta) with DENV helicase (PDB 2JLX) [26]. (B) Curcumin II in the active site of DENV helicase (PDB 2JLX) showing key intermolecular contacts (hydrogen-bonds are shown as blue dashed lines). The ADP co-crystallized ligand is shown as a yellow wire figure.
3.3. Dengue Virus Methyltransferase
DENV MTase has a relatively large binding cavity (around 425-515 Å3) that accommodates the viral RNA cap (the GTP site) and SAM (the SAM site). The strongest docking phytochemical ligands all docked at the SAM site. In addition, it is beneficial for antiviral agents to show selectivity for DENV MTase over human RNA MTase. Table 3 lists those compounds that docked with DENV MTase with docking energies comparable or more exothermic than SAM and also with docking energies significantly more exothermic with DENV MTase than with Hs RNA MTase. Not surprisingly, the strongest docking ligands were relatively large molecules (MW around 680-802 g/mol), and therefore violate Lipinski’s rule of five. Based on this molecular docking analysis, the ligands in
Table 3.
MolDock docking energies (kJ/mol) of phytochemical ligands with dengue virus methyltransferase and human RNA methyltransferase.
|
a SAM and SFG are co-crystallized ligands.
Table 3 are predicted to show inhibition of DENV MTase by competition with SAM and to selectively inhibit viral MTase over human MTase. Additionally, there are several phytochemical ligands that showed more exothermic docking energies than SAM for the SAM site of DENV MTase, more exothermic docking with DENV MTase than with human RNA MTase, and no Lipinski’s rule violations: Isoborreverine, diplacone, styracifolin B, neosilyhermin A, and 8β-[4-hydroxy-5-(5-hydroxytigloyloxy)tigloyl]-sant-amarin.
3.4. Dengue Virus RNA-Dependent RNA Polymerase
Like DENV MTase, DENV RdRp has a very large binding cavity (ca. 3000 Å3), so there are several sites available for docking. Eight phytochemical ligands from our docking analysis showed excellent docking energies to DENV RdRp (Table 4). These ligands meet the rule-of-five criteria for drug likeness and have significantly lower (more exothermic) docking energies than the co-crystallized ligands guanosine 5ʹ-triphosphate (GTP) [29] or 5-{[(4-chlorophenyl)sulfonyl]amino}-2-methyl-1-benzofuran-3-carboxylic acid (VWS) [30]. Four of the strongly docking ligands docked preferentially at the GTP site of DENV RdRp, dimethylisoborreverine, drummondin D, flinderole B, and pungiolide A. The other four ligands docked preferentially at sites removed from either the GTP binding site or the VWS site. Flinderole A and 4,6-dibenzoyl-2-[phenylhydroxymethyl]-3(2H)-benzofuranone preferentially docked in a hydrophobic pocket surrounded by Trp302, Phe354, Val358, Val577, Val579, and Gly599. Neosilyhermin B and kanzonol Y docked preferentially in a hydrophobic pocked formed by Ala406, Ala407, Asn492, Glu507, Val603, Tyr606, and Ile797.
Table 4.
Lowest MolDock docking energies for phytochemical ligands with dengue virus RNA-dependent RNA polymerase.
|
Curcuma longa extracts have shown activity against hepatitis B virus replication [57] and dengue-2 virus protease [58]. In addition, curcumin, one of the major components of C. longa, has shown inhibition of HIV-1 and HIV-2 proteases [59] and in-vitro infectivity of dengue virus type 2 [60]. García Ariza and co-workers have suggested that dengue virus RNA-dependent RNA polymerase inhibition by curcumin, based on molecular docking studies, may be responsible for the anti-dengue activity of curcumin [61]. In this present docking study, several curcuminoids (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,4,6-heptatrien-3-one, curcu-min I, curcumin II, curcumin III, and dihydrocurcumin) showed notably strong docking energies to the ATP site of dengue virus helicase (Table 3). These ligands also showed docking energies somewhat stronger than those of the co-crystallized ligands for DENV RdRp (e.g., curcumin I, Edock = -119.3 kJ/mol). Curcumin I also showed excellent docking properties to the DENV envelope protein hydrophobic pore (see below, Table 5).
Table 5.
MolDock docking energies (kJ/mol) for phytochemical ligands with dengue virus envelope protein hydrophobic channel.

|
A number of alkylated flavanones, the chartaceones A-F, isolated from the bark of Cryptocarya chartacea, have shown potent activity against dengue virus NS5 RdRp [62]. The chartaceones C-F had shown the highest potency and these ligands also showed very exothermic docking energies (ranging from -125 to -139 kJ/mol). These compounds, however, all violate Lipinski’s rule (MW = 660.75 g/mol). The lower molecular weight chartaceones A-B, however, docked with DENV RdRp with docking energies (Edock = -101 to -117 kJ/mol) comparable to GTP (Edock = -107.7 kJ/mol). Chartaceone A2 showed selective docking to DENV protease (Table 1), chartaceone B3 docked strongly with the ATP site of DENV helicase (Table 2), and chartaceone B4 docked well with DENV MTase (Table 3).
3.5. Dengue Virus Envelope Protein
Several phytochemical compounds have been found that dock into the small hydrophobic channel of DENV envelope protein with docking energies significantly more exothermic than the co-crystallized inhibitor, n-octyl-β-d-glucoside [12] (Table 5). Additionally, these phytochemical ligands completely occupy the channel of the envelope protein (see Fig. 4). Thus, for example, the flavonoid canniflavin A docks inside the cavity pore, which is largely hydrophobic. There are, however, key hydrogen-bonding contacts between the flavonoid ligand and Thr48 and Thr280 (Fig. 4C). An obvious structural feature of strongly docking phytochemical ligands to the hydrophobic pore is the ability to form an extended structure, particularly involving hydrophobic groups such as prenyl and geranyl.
Fig. (4).
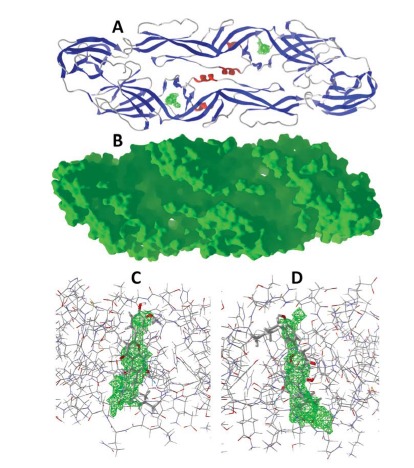
Dengue virus envelope protein. (A) Ribbon structure of DENV envelope protein (PDB 1OKE [31]). The cavity pores in each monomer are shown in green. (B) Solid structure of DENV envelope protein (PDB 1OKE). (C) Lowest-energy docked pose of canniflavin A with monomer A of the dimeric envelope protein (PDB 1OKE). The cavity pore is shown in green; hydrogen-bonds are shown as blue dashed lines. (D) Lowest-energy docked pose of paratocarpin L with monomer B of the dimeric envelope protein (PDB 1OKE).
A number of flavonoids from Silybum marianum have shown exceptional docking to the hydrophobic pore of DENV Ep (2,3-dehydroxilybin, cisilandrin, isosilybins A-D, silandrins A-B, and silybin (Table 5). Silybin (silibinin, silymarin) has shown antiviral activity against hepatitis C virus [63] and chikungunya virus [64]. C-geranylated flavanones from Paulownia tomentosa fruits have shown antibacterial [65, 66], cytotoxic [67], and anti-inflammatory [68] activities as well as SARS-CoV protease inhibitory activity [69]. Diplacone showed excellent docking properties with DENV MTase (Table 3) and both 3ʹ-O-methyldiplacone and 3ʹ-O-methyldiplacol docked well with the ATP site of DENV helicase (Table 2). Diplacone, bonannione A, 3ʹ-O-methyldiplacone and 4ʹ-O-methyldiplacone docked well with the hydrophobic channel of DENV Ep (Table 5). Likewise, the C-geranylated flavone cannflavin A, from Cannabis sativa [70], also docked well with DENV Ep (Table 5).
SUMMARY AND CONCLUSION
A molecular docking analysis of 2194 plant-derived secondary metabolites with dengue virus protein targets has been carried out. The analysis has revealed 24 compounds that docked strongly to dengue virus NS2B-NS3 protease (Edock < -130 kJ/mol) but not as strongly with mammalian trypsin, 21 compounds that docked strongly to the ATP binding site of DENV NS3 helicase (Edock < -140 kJ/mol), 13 compounds that docked strongly to DENV methyltransferase (Edock < -130 kJ/mol) but not as strongly with human RNA methyltransferase, 8 phytochemicals that showed notable docking to DENV RNA-dependent RNA polymerase, and 32 compounds that showed excellent docking properties with the hydrophobic pore of DENV envelope protein. The results of this study mirror previous docking studies that showed polyphenolic phytochemicals to be excellent docking ligands to dengue protein targets [42, 43, 46, 61]. These results and the current study underscore the importance of natural products from higher plants in drug discovery and may provide potential avenues for development of chemotherapeutic agents for the treatment of dengue fever.
ACKNOWLEDGEMENTS
Declare none.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., Myers M.F., George D.B., Jaenisch T., Wint G.R., Simmons C.P., Scott T.W., Farrar J.J., Hay S.I. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzmán M.G., Kourí G. Dengue: an update. Lancet Infect. Dis. 2002;2:33–42. doi: 10.1016/s1473-3099(01)00171-2. [DOI] [PubMed] [Google Scholar]
- 3.Halstead S.B. Dengue. Lancet. 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 4.De Paula S.O., da Fonseca B.A. Dengue: A review of the laboratory tests a clinician must know to achieve a correct diagnosis. Braz. J. Infect. Dis. 2004;8:390–398. doi: 10.1590/s1413-86702004000600002. [DOI] [PubMed] [Google Scholar]
- 5.Messina J.P., Brady O.J., Pigott D.M., Golding N., Kraemer M.U., Scott T.W., Wint G.R., Smith D.L., Hay S.I. Nat. Rev. Microbiol. 2015;13:230–239. doi: 10.1038/nrmicro3430. [DOI] [PubMed] [Google Scholar]
- 6.Vikram K., Nagpal B.N., Pande V., Srivastava A., Saxena R., Singh H., Anushrita Gupta S.K., Tuli N.R., Yadav N.K., Olivier T., Richard P., Velecha N. Detection of dengue virus in individual Aedes aegypti mosquitoes in Delhi, India. J. Vector Borne Dis. 2015;52:129–133. [PubMed] [Google Scholar]
- 7.Afreen N., Deeba F., Naqvi I., Shareef M., Ahmed A., Broor S., Parveen S. Molecular investigation of 2013 dengue fever outbreak from Delhi, India. . PLoS Currents, 2014. [DOI] [PMC free article] [PubMed]
- 8.Honório N.A., Nogueira R.M., Codeço C.T., Carvalho M.S., Cruz O.G., Magalhães M.A., de Araújo J.M., de Araújo E.S., Gomes M.Q., Pinheiro L.S., Pinel C.S., Lourenço-de-Oliveira R. Spatial evaluation and modeling of dengue seroprevalence and vector density in Rio de Janeiro, Brazil. PLoS Negl. Trop. Dis. 2009;3(11):e545. doi: 10.1371/journal.pntd.0000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heringer M., Nogueira R.M., de Filippis A.M., Lima M.R., Faria N.R., Nunes P.C., Nogueira F.B., dos Santos F.B. Impact of the emergence and re-emergence of different denque viruses’ serotypes in Rio de Janeiro, Brazil, 2010 to 2012. Trans. R. Soc. Trop. Med. Hyg. 2015;109:268–274. doi: 10.1093/trstmh/trv006. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Barraquer I., Cordeiro M.T., Braga C., de Souza W.V., Marques E.T., Cummings D.A. From re-emergence to hyperendemicity: The natural history of the dengue epidemic in Brazil. PLoS Negl. Trop. Dis. 2011;5(1):e935. doi: 10.1371/journal.pntd.0000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kikuti M., Cunha G.M., Paploski I.A., Kasper A.M., Silva M.M., Tavares A.S., Cruz J.S., Queiroz T.L., Rodrigues M.S., Santana P.M., Lima H.C., Calcagno J., Takahashi D., Gonçalves A.H., Araújo J.M., Gauthior K., Kiuk-Wasser M.A., Kitron U., Ko A.I., Reis M.G., Ribeiro G.S. Spatial distribution of dengue in a Brazilian urban slum setting: Role of socioeconomic gradient in disease risk. PLoS Negl. Trop. Dis. 2015;9(7):e0003937. doi: 10.1371/journal.pntd.0003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geiss B.J., Stahla H., Hannah A.M., Gari H.H., Keenan S.M. Focus on flaviviruses: current and future drug targets. Future Med. Chem. 2009;1(2):327. doi: 10.4155/fmc.09.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cragg G.M., Newman D.J. Natural products: A continuing source of novel drug leads. 2013. [DOI] [PMC free article] [PubMed]
- 14.Stevens A.J., Gahan M.E., Mahalingam S., Keller P.A. The medicinal chemistry of dengue fever. J. Med. Chem. 2009;52:7911–7926. doi: 10.1021/jm900652e. [DOI] [PubMed] [Google Scholar]
- 15.Tomlinson S.M., Malmstrom R.D., Watowich S.J. New approaches to structure-based discovery of dengue protease inhibitors. Infect. Disord. Drug Targets. 2009;9:327–343. doi: 10.2174/1871526510909030327. [DOI] [PubMed] [Google Scholar]
- 16.Schüller A., Yin Z., Chia C.S., Doan D.N., Kim H.K., Shang L., Loh T.P., Hill J., Vasudevan S.G. Tripeptide inhibitors of dengue and West Nile virus NS2B-NS3 protease. Antiviral Res. 2011;92:96–101. doi: 10.1016/j.antiviral.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Phong W.Y., Moreland N.J., Lim S.P., Wen D., Paradkar P.N., Vasudevan S.G. Dengue protease activity: the structural integrity and interaction of NS2B with NS3 protease and its potential as a drug target. Biosci. Rep. 2011;31:399–409. doi: 10.1042/BSR20100142. [DOI] [PubMed] [Google Scholar]
- 18.Luo D., Xu T., Watson R.P., Scherer-Becker D., Sampath A., Jahnke W., Yeong S.S., Wang C.H., Lim S.P., Strongin A., Vasudevan S.G., Lescar J. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble C.H., Chen Y.L., Dong H., Gu F., Lim S.P., Schul W., Wang Q.Y., Shi P.Y. Strategies for development of dengue virus inhibitors. Antiviral Res. 2010;85:450–462. doi: 10.1016/j.antiviral.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Noble C.G., Shi P.Y. Structural biology of dengue virus enzymes: Towards rational design of therapeutics. Antiviral Res. 2012;96:115–126. doi: 10.1016/j.antiviral.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Noble C.G., Li S.H., Dong H., Chew S.H., Shi P.Y. Crystal structure of dengue virus methyltransferase without S-adenosyl-L-methionine. Antiviral Res. 2014;111:78–81. doi: 10.1016/j.antiviral.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble C.G., She C.C., Chao A.T., Shi P.Y. Ligand-bound structures of the dengue virus protease reveal the active conformation. J. Virol. 2012;86:438–446. doi: 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yildiz M., Ghosh S., Bell J.A., Sherman W., Hardy J.A. Allosteric inhibition of the NS2B-SN3 protease from dengue virus. ACS Chem. Biol. 2013;8:2744–2752. doi: 10.1021/cb400612h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbs A., Tounge B., Steele R. NMR structure of an inhibitor bound dengue NS3 protease provides new insights into the NS2B NS3 ligand interactions. To be published.
- 25.Xu T., Sampath A., Chao A., Wen D., Nanao M., Chene P., Vasudevan S.G., Lescar J. Structure of the dengue virus helicase/nucleoside triphosphates catalytic domain at a resolution of 2.4 Å. J. Virol. 2005;79:10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo D., Xu T., Watson R.P., Scherer-Becker D., Sampath A., Jahnke W., Yeong S.S., Wang C.H., Lim S.P., Strongin A., Vasudevan S.G., Lescar J. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo D., Wei N., Doan D.N., Paradkar P.N., Chong Y., Davidson A.D., Kotaka M., Lescar J., Vasudevan S.G. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim S.P., Sonntag L.S., Noble C., Nilar S.H., Ng R.H., Zou G., Monaghan P., Chung K.Y., Dong H., Liu B., Bodenreider C., Lee G., Ding M., Vedananda T.R., Keller T.H., Shi P.Y. Small molecule inhibitors that selectively block dengue virus methyltransferase. J. Biol. Chem. 2011;286:6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap T.L., Xu T., Chen Y.L., Malet H., Egloff M.P., Canard B., Vasudeval S.G., Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-Angstrom resolution. J. Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noble C.G., Lim S.P., Chen Y.L., Liew C.W., Yap L., Lescar J., Shi P.Y. Conformational flexibility of the dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 2013;87:5291–5295. doi: 10.1128/JVI.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modis Y., Ogata S., Clements D., Harrison S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H., Lunin V.V., Zeng H., Antoshenko T., MacKenzie F., Weigelt J., Arrowsmith C.H., Edwards A.M., Bochkarev A., Min J., Plotnikov A.N. The crystal structure of human RNA (guanine-7-) methyltransferase in complex with SAH. To be published. DOI:10.2210/pdb3bgv/pdb. To be published.
- 33.Zeng H., Amaya M.F., Loppnau P., Bountra C., Weigelt J., Arrowsmith C.H., Edwards A.M., Botchkarev A., Min J., Plotnikov A.N., Wu H. Crystal structure of mRNA cap guanine- N7 methyltransferase (RNMT) in complex with sinefungin. To be published. DOI:10.2210/pdb3epp/pdb. To be published.
- 34.Smietanski M., Werner M., Purta E., Kaminska K.H., Stepinski J., Darzynkiewicz E., Nowotny M., Bujnicki J.M. Structural analysis of human 2ʹ-O-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun. 2014;5:3004. doi: 10.1038/ncomms4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perona J.J., Tsu C.A., Craik C.S., Fletterick R.J. Crystal structures of rat anionic trypsin complexed with the protein inhibitors APPI and BPTI. J. Mol. Biol. 1993;230:919–933. doi: 10.1006/jmbi.1993.1210. [DOI] [PubMed] [Google Scholar]
- 36.Brandt T., Holzmann N., Muley L., Khayat M., Wegscheid-Gerlach C., Baum B., Heine A., Hangauer D., Klebe G. Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties. J. Mol. Biol. 2011;405:1170–1187. doi: 10.1016/j.jmb.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 37.Wagner S., Heine A., Steinmetzer T. Bovine trypsin in complex with small molecule inhibitor. To be published.
- 38.Li de la Sierra I., Papamichael E., Sakarellos C., Dimicoli J.L., Prangé T. Interaction of the peptide CF3-Leu-Ala-NH-C6H4-CF3 (TFLA) with porcine pancreatic elastase. X-ray studies at 1.8 Å. J. Mol. Recognit. 1990;3:36–44. doi: 10.1002/jmr.300030104. [DOI] [PubMed] [Google Scholar]
- 39.Thomsen R., Christensen M.H. MolDock: a new technique for high-accuracy molecular docking. J. Med. Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 40.Halgren T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF 94. J. Comput. Chem. 1996;17:490–519. [Google Scholar]
- 41.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 42.de Sousa L.R., Wu H., Nebo L., Fernandes J.B., da Silva M.F., Kiefer W., Kanitz M., Bodem J., Diederich W.E., Schirmeister T., Vieira P.C. Flavonoids as noncompetitive inhibitors of Dengue virus NS2B-NS3 protease: Inhibition kinetics and docking studies. Bioorg. Med. Chem. 2015;23:466–470. doi: 10.1016/j.bmc.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 43.ul Qamar T., Mumtaz A., Ashfaz U.A., Azhar S., Fatima T., Hassan M., Hussain S.S., Akram W., Idrees S. Computer aided screening of phytochemicals from Garcinia against the dengue NS2B/NS3 protease. Bioinformation. 2014;10:115–118. doi: 10.6026/97320630010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yong K.S., Choi S.B., Wahab H.A. Docking of dengue NS2B-NS3 protease with Murraya koenigii.; 3rd International Conference on Computation for Science and Technology; 2015. pp. 82–84. [Google Scholar]
- 45.Kiat T.S., Pippen R., Yusof R., Ibrahim H., Khalid N., Rahman N.A. Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, Boesenbergia rotunda (L.), towards dengue-2-virus NS3 protease. Bioorg. Med. Chem. Lett. 2006;16:3337–3340. doi: 10.1016/j.bmcl.2005.12.075. [DOI] [PubMed] [Google Scholar]
- 46.Kee L.Y., Kiat T.S., Wahab H.A., Yusof R., Rahman N.A. Nonsubstrate based inhibitors of dengue virus serine protease: A molecular docking approach to study binding interactions between protease and inhibitors. Asia Pac. J. Mol. Biol. Biotechnol. 2007;15:53–59. [Google Scholar]
- 47.Hatano T., Yasuhara T., Miyamoto K., Okuda T. Anti-human immunodeficiency virus phenolics from licorice. Chem. Pharm. Bull. (Tokyo) 1988;36:2286–2288. doi: 10.1248/cpb.36.2286. [DOI] [PubMed] [Google Scholar]
- 48.Manfredi K.P., Vallurupalli V., Demidova M., Kindscher K., Pannell L.K. Isolation of an anti-HIV diprelylated bibenzyl from Glycyrrhiza lepidota. Phytochemistry. 2001;58:153–157. doi: 10.1016/s0031-9422(01)00177-7. [DOI] [PubMed] [Google Scholar]
- 49.Omer M.O., Al Malki W.H., Shakid I., Khuram S., Altaf I., Imran S. Comparative study to evaluate the anti-viral efficacy of Glycyrrhiza glabra extract and ribavirin against the Newcastle disease virus. Pharmacol. Res. 2014;6:6–11. doi: 10.4103/0974-8490.122911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeh C.F., Wang K.C., Chiang L.C., Shieh D.E., Yen M.H., Chang J.S. Water extract of licorice had anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J. Ethnopharmacol. 2013;148:466–473. doi: 10.1016/j.jep.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghannad M.S., Mohammadi A., Safiallahy S., Faradmal J., Azizi M., Ahmadvand Z. The effect of aqueous extract of Glycyrrhiza glabra on herpes simplex virus 1. Jundishapur J. Microbiol. 2014;7(7):e11616. doi: 10.5812/jjm.11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lavoie S., Legault J., Simard F., Chiasson E., Pichette A. New antibacterial dihydrochalcone derivatives from buds of Populus balsimifera. Tetrahedron Lett. 2013;54:1631–1633. [Google Scholar]
- 53.Sánchez I., Gómez-Garibay F., Taboada J., Ruiz B.H. Antiviral effect of flavonoids on the dengue virus. Phytother. Res. 2000;14:89–92. doi: 10.1002/(sici)1099-1573(200003)14:2<89::aid-ptr569>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 54.Schütz B.A., Wright A.D., Rali T., Sticher O. Prenylated flavanones from leaves of Macaranga pleiostemona. Phytochemistry. 1995;40:1273–1277. [Google Scholar]
- 55.Mitscher L.A., Gollapudi S.R., Khanna I.K., Drake S.D., Hanamaiah T., Ramaswamy T., Rao K.V. Antimicrobial agents from higher plants: Activity and structural revision of flemiflavanone-D from Flemingia stricta. Phytochemistry. 1985;24:2885–2887. [Google Scholar]
- 56.Nuntanakorn P., Jiang B., Einbond L.S., Yang H., Kronenberg F., Weinstein B., Kennelly E.J. Polyphenolic constituents of Actaea racemosa. J. Nat. Prod. 2006;69:314–318. doi: 10.1021/np0501031. [DOI] [PubMed] [Google Scholar]
- 57.Kim H.J., Yoo H.W., Kim J.C., Park C.S., Choi M.S., Kim M., Choi H., Min J.S., Kim Y.S., Yoon S.W., Ahn J.K. Antiviral effect of Curcuma longa Linn extract against hepatitis B virus replication. J. Ethnopharmacol. 2009;124:189–196. doi: 10.1016/j.jep.2009.04.046. [DOI] [PubMed] [Google Scholar]
- 58.Kiat T.S., Pippen R., Yusof R., Rahman N.A., Ibrahim H., Khalid N. Screening of selected Zingiberaceae extracts for dengue-2 virus protease inhibitory activities. Sunway Acad. J. 2003;3:1–7. [Google Scholar]
- 59.Sui Z., Salto R., Li J., Craik C., Ortiz de Montellano P.R. Inhibition of the HIV-1 and HIV-2 proteases by curcumin and curcumin boron complexes. Bioorg. Med. Chem. 1993;1:415–422. doi: 10.1016/s0968-0896(00)82152-5. [DOI] [PubMed] [Google Scholar]
- 60.Padilla-S L., Rodríguez A., Gonzales M.M., Gallego-G J.C., Castaño-O J.C. Inhibitory effects of curcumin on dengue virus type 2-infected cells in vitro. Arch. Virol. 2014;159:573–579. doi: 10.1007/s00705-013-1849-6. [DOI] [PubMed] [Google Scholar]
- 61.García Ariza L.L., Téllez Ramirez G.A., Cortes Hernández H.F., Padilla Sanabria L., Castaño Osorio J.C. Molecular cloning, modelling and docking with curcumin of the dengue virus 2 NS5 polymerase domain. In: Castillo L.F., Cristancho M., Isaza G., Pinzón A., Corchado Rodríguez J.M., editors. Advances in Computational Biology. Cham, Switzerland: Springer; 2014. pp. 273–278. [Google Scholar]
- 62.Allard P.M., Dau E.T., Eydoux C., Guillemot J.C., Dumontet V., Poullain C., Canard B., Guéritte F., Litaudon M. Alkylated flavanones from the bark of Cryptocarya chartacea as dengue virus NS5 polymerase inhibitors. J. Nat. Prod. 2011;74:2446–2453. doi: 10.1021/np200715v. [DOI] [PubMed] [Google Scholar]
- 63.Ferenci P., Scherzer T.M., Kerschner H., Rutter K., Beinhardt S., Hofer H., Schöniger-Hekele M., Holzmann H., Steindl-Munda P. Silibinin is a potent antiviral agent in patients with chronic hepatitis C not responding to pegylated interferon/ribavirin therapy. Gastroenterology. 2008;135:1561–1567. doi: 10.1053/j.gastro.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 64.Lani R., Hassandarvish P., Chiam C.W., Moghaddam E., Chu J.J., Rausalu K., Merits A., Higgs S., Vanlandingham D., Bakar S.A., Zandi K. Antiviral activity of silymarin against chikungunya virus. Nature Sci. Rep. 2015;5:11421. doi: 10.1038/srep11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Šmejkal K., Chudík S., Klouček P., Marek R., Cvačka J., Urbanová M., Julínek O., Kokoška L., Šlapetová T., Zima A., Dvorská M. Antibacterial C-geranylflavonoids from Paulownia tomentosa fruits. J. Nat. Prod. 2008;71:706–709. doi: 10.1021/np070446u. [DOI] [PubMed] [Google Scholar]
- 66.Navrátilová A., Schneiderová K., Veselá D., Hanáková Z., Fontana A., Dall’Acqua S., Cvačka J., Innocenti G., Novotná J., Urvanová M., Pelletier J., Čížek A., Žemličková H., Šmejkal K. Minor C-geranylated flavanones from Paulownia tomentosa fruits with MRSA antibacterial activity. Phytochemistry. 2013;89:104–113. doi: 10.1016/j.phytochem.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 67.Šmejkal K., Svačinová J., Šlapetová T., Schneiderová K., Dall’Acqua S., Innocenti G., Závalová V., Kollár P., Chudík S., Marek R., Julínek O., Urbanová M., Kartal M., Csöllei M., Doležal K. Cytotoxic activities of several geranyl-substituted flavanones. J. Nat. Prod. 2010;73:568–572. doi: 10.1021/np900681y. [DOI] [PubMed] [Google Scholar]
- 68.Hanáková Z., Hošek J., Babula P., Dall’Acqua S., Václavík J., Šmejkal K. C-geranylated flavanones from Paulownia tomentosa fruits as potential anti-inflammatory compounds acting via inhibition of TNF-α production. J. Nat. Prod. 2015;78:850–863. doi: 10.1021/acs.jnatprod.5b00005. [DOI] [PubMed] [Google Scholar]
- 69.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barrett M.L., Scutt A.M., Evans F.J. Cannflavin A and B, prenylated flavones from Cannabis sativa L. Experientia. 1986;42:452–453. doi: 10.1007/BF02118655. [DOI] [PubMed] [Google Scholar]


