Abstract
Cadmium is an environmental electrophile that modifies reactive thiols in proteins, indicating that this heavy metal may modulate redox-signal transduction pathways. The current consensus is that reactive persulfides and polysulfides produced by cystathionine γ-lyase (CSE) and cystathionine β-synthase are highly nucleophilic and thus cadmium may be captured by these reactive sulfur species. It has previously been found that electrophile-mediated covalent modifications of the heat shock protein (HSP) are involved in the activation of heat shock factor 1 (HSF1) pathway. The effects of cadmium on the activation of HSP/HSF1 pathway were investigated in this study. Exposure of bovine aortic endothelial cells to cadmium resulted in modification of HSP90 and HSF1 activation, thereby up-regulating the downstream protein HSP70. The siRNA-mediated knockdown of HSF1 enhanced the cytotoxicity induced by cadmium, suggesting that the HSP90/HSF1 pathway contributes to protection against cadmium toxicity. The knockdown of CSE and/or cystathionine β-synthase decreased the levels of reactive sulfur species in the cells and increased the degree of HSP70 induction and cytotoxicity caused by exposure to cadmium. Overexpression of CSE diminished cadmium-mediated up-regulation of HSP70 and cytotoxicity. These results suggest that cadmium activates HSF1 by modifying HSP90 and that reactive sulfur species regulate the redox signal transduction pathway presumably via capture of cadmium, resulting in protection against cadmium toxicity under toxic conditions.
Keywords: cadmium, heat shock protein, heat shock factor 1, cystathionine γ-lyase, persulfides/polysulfides.
Cadmium is a ubiquitous environmental pollutant that can affect the vascular endothelium (Prozialeck et al. , 2006, 2008). Cadmium is electrophilic, so it easily modifies protein thiols, causing the affected enzymes to become dysfunctional (Choong et al., 2014). However, it has been found that cells contain sensor proteins that act against electrophilic chemicals, activating redox signal transduction pathways in response to electrophilic chemical insults (Rudolph and Freeman, 2009). For example, Keap1, which is a typical sensor protein with low pKa thiols, acts as a negative regulator for the transcription factor Nrf2 (Taguchi et al., 2011). We have previously found that environmental electrophiles such as methylmercury (MeHg) and cadmium modify Keap1 thiols, activating Nrf2 to upregulate downstream proteins involved in protecting against electrophilic stress (Shinkai et al., 2016; Toyama et al., 2007). Another sensor protein with reactive thiols, PTEN, negatively regulates anti-apoptotic Akt signaling pathways (Stambolic et al., 1998; Worby and Dixon, 2014). We have found that MeHg activates the Akt/CREB/Bcl-2 signaling pathway by inactivating PTEN through S-mercuration, resulting in a cellular protection against MeHg (Unoki et al., 2016). These observations indicate that several defense systems protect cells from the stresses caused by electrophilic chemicals.
Heat shock proteins (HSPs) act as chaperones for protein folding, preventing the irreversible aggregation of non-native conformations, thereby protecting cells from injury (Saibil, 2013). HSPs are constitutively expressed and induced further in response to various stress conditions, including heat shock, oxidative stress, and the presence of certain chemicals (Lindquist and Craig, 1988). It has been found that 2 major HSP isoforms, HSP70 and HSP90, interact with heat shock factor 1 (HSF1) under basal conditions. However, these interactions become dissociated in response to stress, causing HSF1 to be translocated into the nucleus, where it activates genes that target heat shock elements (HSEs) (Akerfelt et al., 2010; Bjork and Sistonen, 2010; Voellmy, 2004). Interestingly, it has been found that electrophilic chemicals such as 4-hydroxynonenal and 6-methysulfinylhexyl isothiocyanate covalently modify HSP70 and HSP90, and therefore activate the HSF1/HSE pathway, inducing HSPs (Carbone et al., 2005; Jacobs and Marnett, 2007; Shibata et al., 2011). We therefore hypothesize that cadmium will also activate the HSF1/HSE signaling pathway via modification of HSP70/HSP90.
In addition to cellular response systems that use sensor proteins, there is a defense system in which electrophiles are made inactive through interactions with sulfur-containing nucleophiles, such as cysteine and glutathione (DeLeve and Kaplowitz, 1991). We recently found that cystathionine γ-lyase (CSE) and cystathionine β-synthase (CBS) produce reactive sulfur species (RSS) such as cysteine persulfides/polysulfides when cystine is used as a substrate and that these, in turn, produce glutathione persulfides/polysulfides (Ida et al., 2014). RSS have strongly nucleophilic properties compared with the parent nucleophiles, so it is likely that RSS will readily trap environmental electrophiles, forming sulfur adducts of the electrophiles. Consistent with this, MeHg reacts with RSS (eg, hydrogen sulfide, GSSH, GSSSG) forming bismethylmercury sulfide, which is not electrophilic and is not very toxic (Abiko et al., 2015; Yoshida et al., 2011). We therefore hypothesized that RSS will modulate the cadmium-mediated activation of redox signal transduction pathways such as the HSP/HSF1 pathway and modulate cadmium-induced cytotoxicity by capturing cadmium. The aims of the study presented here were to clarify the mechanism underlying the activation of the HSP/HSF1 pathway by cadmium and to investigate the functions of RSS in regulating the activation of signaling pathways and cytotoxicity under toxic conditions.
EXPERIMENTAL PROCEDURES
Materials
Cadmium chloride was obtained from Nacalai Tesque (Kyoto, Japan). Biotin-PEAC5-maleimide (BPM) and SSP4 were obtained from Dojindo (Kumamoto, Japan). Anti-HSP70 antibodies (ADI-SPA-810) and recombinant human HSP90β (ADI-SPP-777) were obtained from Enzo Life Sciences (Farmingdale, NY, USA). Anti-HSP90 antibodies (sc-7947), anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies (FL-335) and anti-Lamin B antibodies (M-20) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-glucose-6-phosphate dehydrogenase (G6PD) antibody was from Bethyl Laboratories (Montgomery, TX, USA). Anti-HSF1 antibodies (#4356), anti-rabbit IgG conjugated to horseradish peroxidase (HRP), and HRP-conjugated anti-mouse IgG were obtained from Cell Signaling Technology (Beverly, MA, USA). Anti-CBS antibodies (M01) were purchased from Abnova (Taipei, Taiwan). HSE-luciferase cDNA was purchased from Stratagene (La Jolla, CA, USA). The specific rabbit-derived polyclonal antibody against CSE was prepared by immunizing rabbits with recombinant human CSE protein kindly gifted from Prof. Y. Watanabe (Showa Pharmaceutical University). The IgG fraction of anti-CSE was purified by protein A-sepharose CL-4B column chromatography. All other reagents and chemicals were of the highest grade available.
Cell culture and treatment
The bovine aortic endothelial cells (BAECs) used were obtained from Cell Applications, Inc. (San Diego, CA, USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (Nissui, Tokyo, Japan) supplemented with 10% fetal bovine serum and antibiotics (100 units/ml penicillin and 100 µg/ml streptomycin) in a humidified atmosphere containing 5% CO2 at 37 °C. Confluent cultures were used in all of the experiments except transfection experiments. The culture medium was replaced with serum-free medium before BAECs were treated with chemicals.
Transfection
For transfection experiments, BAECs grown to subconfluence were transiently transfected by lipofection. Synthetic siRNAs were purchased from Bex (Tokyo, Japan) and the transfection of siRNAs was performed using RNAiMAX (Invitrogen, Carlsbad, CA, USA) following a protocol provided by the manufacturer. The sense and antisense siRNA strand sequences were shown in Table 1. A nonspecific sequence (Qiagen, Valencia, CA, USA) was used as the negative siRNA control. HA-tagged human cDNA of CSE in the pClneo vector was kindly gifted from Prof. Y. Watanabe (Showa Pharmaceutical University). The transfection of cDNA was performed using Lipofectamine LTX (Invitrogen) according to the manufacturer’s instructions.
TABLE 1.
Bovine Gene-Specific siRNA Oligo Sequences
| siRNA | Sense (5′–3′) | Antisense (5′–3′) |
|---|---|---|
| HSF1 siRNA-1 | GCGGCAGCUCAACAUGUAUdTdT | AUACAUGUUGAGCUGCCGCdTdT |
| HSF1 siRNA-2 | GUGCUGCCCAAGUACUUCAdTdT | UGAAGUACUUGGGCAGCACdTdT |
| CBS siRNA | CAUCUACAAGCAGUUCAAAdTdT | UUUGAACUGCUUGUAGAUGdTdT |
| CSE siRNA | GAGCAGUUCCAUCUCCUAUdTdT | AUAGGAGAUGGAACUGCUCdTdT |
Luciferase assay
The transfection of cDNA for luciferase assay was performed using FuGENE HD transfection reagent (Roche Applied Sciences, Indianapolis, IN, USA) following the manufacturer’s instructions. Briefly, cells were cultured in 24-well plates. HSE-luciferase cDNA (0.5 µg/well) and pRL-TK cDNA (0.05 µg/well) or transfection reagent (1 µl/well) were mixed with serum-free medium. The DNA solution and transfection reagent solution were mixed together and incubated at room temperature for 15 min to allow complexes to form before being added to the cells. The complexes were mixed with the culture medium containing the cells, and the mixture was incubated for 24 h. After the transfection process, the cells were treated with cadmium chloride and then the luciferase activity in the cellular extract was determined using the dual-luciferase reporter assay system (Promega) and a luminometer (Promega) following the manufacturer’s instructions.
Real-time polymerase chain reaction
Total RNA was extracted using an RNeasy Lipid Tissue Mini Kit (Qiagen), and cDNA was synthesized from the mRNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster, CA, USA). The real-time polymerase chain reaction (PCR) process was performed using a Power SYBR Green PCR Master Mix (Applied Biosystems) using 5 µg cDNA and 0.2 µM primers (Table 2) and a 7500 real-time PCR system (Applied Biosystems). The thermal treatment was 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Melting curve analysis and agarose gel electrophoresis with ethidium bromide staining were conducted to ensure a single PCR product with the correct amplicon length was obtained. The HSP70-1A, HSPA6, HSP90α, HSP90β, and β2-microglobulin (B2M) mRNA levels in each RNA sample were determined using the relative standard curve method. Fold-changes in each gene were assessed after the intensity value had been normalized to B2M.
TABLE 2.
Bovine Gene Primers Used in the Quantitative Real-Time Reverse Transcription PCR
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Product Size (bp) |
|---|---|---|---|
| HSP70-1A | ATGGGGGACAAGTCGGAGAAC | GTTGTCCGAGTAGGTGGTGAAGA | 152 |
| HSPA6 | TGTTCGACGTCAAGCGACTG | CCGTCTCCTTCATCTTGCTCA | 192 |
| HSP90α | AAGCAAGATCGAACCCTCACCA | CGGTCACTTTCTCAGCCACCAA | 199 |
| HSP90β | ACAGACCCTTCCAAATTGGACAG | AGACTTGGCAATGGTTCCCAAA | 147 |
| B2M | CCATCCAGCGTCCTCCAAAGA | TTCAATCTGGGGTGGATGGAA | 110 |
Western blotting analysis
After a treatment had been performed, the cells were washed twice with ice-cold phosphate-buffered saline (PBS). The total cell proteins were extracted by lysing the cells in SDS sample buffer (50 mM Tris–HCl, 2% SDS, 10% glycerol, at pH 6.8), then incubating the mixture at 95 °C for 10 min. The nuclear fraction was prepared using a nuclear extraction reagent kit (Pierce, Rockford, IL, USA) following a protocol provided by the manufacturer. The protein concentration was determined using a bicinchoninic acid protein assay reagent kit (Nacalai Tesque), then 2-mercaptoethanol was added. The proteins in a sample were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidene difluoride membrane at 2 mA/cm2 for 1 h following a previously published method (Kyhse-Andersen, 1984). The membrane was blocked using 5% skim milk in 20 mM Tris–HCl, 150 mM NaCl, and 0.1% Tween 20 at pH 7.5, then the membrane was incubated with primary antibodies at room temperature for 1 h. The membrane was washed, then incubated with HRP-conjugated secondary antibodies at room temperature for 1 h. The immunoreactive bands were visualized using enhanced chemiluminescence and scanned using an LAS 3000 instrument (Fujifilm, Tokyo, Japan).
Cytotoxicity assay
The 3-(4,5-dimethylthiazol-2-yl)-2,5-triphenyl tetrazolium bromide (MTT) assay, described previously (Denizot and Lang, 1986), was used to estimate cell viability. Briefly, cells were exposed to cadmium chloride and then treated with 5 mg/ml MTT (1/20 volume of medium), and the mixture was incubated at 37 °C for 2 h. The medium was removed, then dimethyl sulfoxide was added to dissolve the MTT formazan. The absorbance at a wavelength of 540 nm was measured using a plate reader.
Immunoprecipitation assay
BAECs were exposed to cadmium chloride (2 or 10 µM) for 3 h and then washed twice with Dulbecco’s PBS. Cell lysates were prepared by suspending the cells in radioimmunoprecipitation assay (RIPA) buffer supplemented with a protease inhibitor cocktail and keeping the mixture on ice for 20 min. Anti-rabbit IgG conjugated magnetic beads (Dynabeads M-280 sheep anti-rabbit IgG; Thermo Fisher Scientific, Waltham, MA, USA) were washed 3 times with Tris-buffered saline and Tween 20, then incubated with HSF1 antibodies at 4 °C for 2 h. The unbound antibodies were then removed and the beads were resuspended in the cell lysate prepared as described above. The mixture was then incubated at 4 °C overnight. The beads were then washed 4 times with lysis buffer, then protein complexes were eluted by adding 30 µl of SDS-PAGE loading buffer. The eluted proteins were incubated at 95 °C for 5 min, then subjected to Western blotting analysis.
BPM assay
The BPM-labeling assay and BPM-precipitation assay were performed as previously described (Toyama et al., 2013). Briefly, recombinant human HSP90 (1 µM) was reacted with cadmium chloride (1, 5, 25, or 100 µM) in 50 mM Tris–HCl (pH 7.5) at 25 °C for 30 min, then the mixture was incubated with BPM (25 µM) at 25 °C for 30 min. A 12 µl aliquot of a sample was mixed with 6 µl of SDS-PAGE loading buffer containing 50 mM tris(2-carboxyethyl)phosphine, then the mixture was subjected to Western blotting analysis using anti-biotin HRP-conjugated antibodies. BAECs were seeded on a 60 mm dish, exposed to cadmium chloride (2 or 10 µM) for 3 h, then washed twice with Dulbecco’s PBS. Cell lysates were prepared by suspending the cells in RIPA buffer supplemented with protease inhibitor cocktail and keeping the mixture on ice for 20 min. The mixture was then centrifuged, and the supernatant was incubated with BPM (20 µM) at 37 °C for 30 min. The BPM-modified proteins were then separated by adding Dynabeads M-280 streptavidin (40 µl/sample), and the mixture was incubated at 4 °C overnight. The mixture was then centrifuged, and the precipitated Dynabeads were collected and washed with 1 ml of RIPA buffer twice. The beads were then added to 20 µl of SDS-PAGE loading buffer containing 2-mercaptoethanol, and the mixture was heated to 95 °C for 20 min. The mixture was then centrifuged, and the supernatant was subjected to Western blotting analysis using antibodies against HSP90 and GAPDH. Although these BPM-using assays would detect not only covalent modification but also other Cys modification such as oxidation, they can easily assess the modification of protein thiols by electrophiles.
Liquid chromatography-mass spectrometry
Recombinant human HSP90 (2.5 µM) in 50 mM Tris–HCl (pH 7.5) was incubated with or without 10 µM cadmium chloride at 25 °C for 30 min. The native or cadmium-modified human HSP90 was then incubated with 2 mM tris(carboxyethyl)phosphine at 25 °C for 10 min, then the mixture was alkylated by adding 5 mM 2-iodoacetamide in 50 mM ammonium bicarbonate buffer and incubating the mixture at 25 °C in the dark for 20 min. The resulting protein was digested using mass spectrometry grade modified trypsin (6.7 ng/µl; Promega, Madison, WI, USA) at 37 °C overnight. The resulting peptides were analyzed using a nanoAcquity ultrahigh performance liquid chromatography system (nanoUPLC; Waters, Milford, MA, USA) equipped with a BEH130 nanoAcquity C18 column (100 mm long, 75 µm i.d., 1.7 µm particle diameter), which was kept at 35 °C. Mobile phase A (water containing 0.1% (v/v) formic acid) and mobile phase B (acetonitrile containing 0.1% (v/v) formic acid) were mixed in a linear fashion using a gradient program. The instrument was calibrated immediately before each series of experiments. The flow rate was 0.3 µl/min, and the mobile phase composition started at 3% B, which was held for 1 min, then linearly increased over 74 min to 40% B, which was maintained for 4 min, then linearly increased over 1 min to 95% B, which was maintained for 5 min, then linearly decreased over 1 min to 3% B. The total runtime, including conditioning the column at the initial conditions, was 100 min. The eluted peptides were transferred to the nano-electrospray source of a Synapt high definition Q-TOF mass spectrometer (Waters) through a Teflon capillary union and a precut PicoTip (Waters). The system was controlled and the mass spectral data analyzed using MassLynx version 4.1 software (Waters). The mass spectrometer used electrospray ionization with a capillary voltage of 3.5 kV and a sampling cone voltage of 40 V. A low collision energy (6 eV) was used to generate intact peptide precursor ions, and an elevated collision energy (stepped from 25 to 40 eV) was used to generate peptide product ions. The source temperature was 100 °C, and the detector was operated in positive ion mode. Data were collected in centroid mode, and the m/z range was 50–1990. All analyses were acquired using an independent reference. Glu-1-fibrinopeptide B (m/z 785.8426), used as an external mass calibrant, was infused through the nanoLockSpray ion source and sampled every 10 s. Biopharmlynx version 1.2 software (Waters) was used to perform baseline subtraction, smoothing, and deisotoping, to identify de novo peptide sequences, and to perform database searches.
Reactive sulfur species measurement
Cellular RSS imaging was performed using the specific probe SSP4. BAECs were seeded onto glass chamber slides. The cultured semi-confluent cells were washed with serum-free Dulbecco’s modified Eagle’s medium, and then incubated with 50 µM SSP4 in serum-free Dulbecco’s modified Eagle’s medium containing 200 µM cetyltrimethylammonium bromide at 37 °C for 20 min. The medium was removed, then the cells were washed with PBS and incubated in PBS at 37 °C for 20 min. Fluorescence images of the cells were acquired using a Nikon C2 confocal laser microscope (Nikon, Tokyo, Japan) using an excitation wavelength of 488 nm and an emission wavelength of 521 nm. BAECs were seeded into a 96-well black plate to allow the fluorescence intensity to be measured. The cultured semi-confluent cells were treated in the same way as described above, and the fluorescence (λex 500 nm, λem 521 nm) was monitored using a Varioskan plate reader (Thermo Fisher Scientific).
Statistical analysis
The statistical significances of the differences between results were assessed by performing Student’s t tests, and P < .05 was considered to indicate a significant difference.
RESULTS
Activation of the HSF1/HSE Pathway by Cadmium
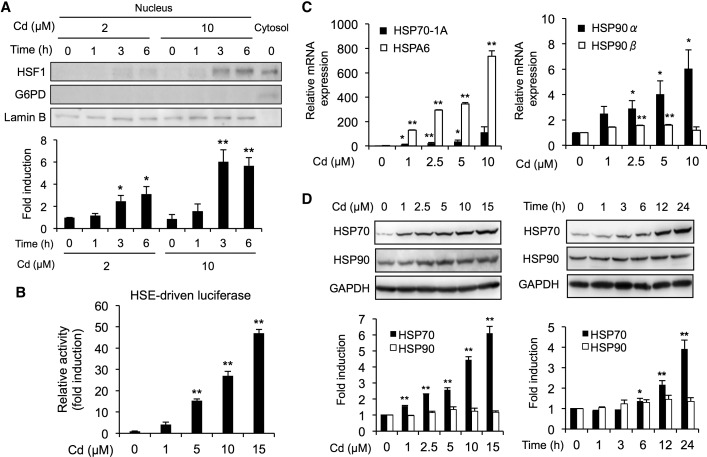
It has been reported that cadmium activates HSF1/HSE pathway in several cell lines (Galan et al., 2001; Holmberg et al., 2001). Consistent with these reports, cadmium caused a significant accumulation of HSF1 in the nucleus of BAECs (Figure 1A). The molecular weight of HSF1 in the nucleus was slightly higher than that in the cytosol (Figure 1A). Such observations are presumably due to the phosphorylation of HSF1 because cadmium was reported to induce phosphorylation of HSF1 (Holmberg et al., 2001). Also, cadmium significantly increased HSE-driven luciferase activity (Figure 1B). To examine the alteration of HSF1-target genes by cadmium, we determined the levels of HSP genes such as HSP70-1A, HSPA6, HSP90α, and HSP90β. Cadmium markedly up-regulated gene expressions of HSP70-1A and HSPA6 (Figure 1C), whereas a slight increase in gene expressions of HSP90α and HSP90β was detected in BAECs (Figure 1C). Similar phenomena were also observed in the protein expression levels of HSP70 and HSP90 during exposure to cadmium (Figure 1D).
FIG. 1.
Activation of the HSF1/HSE pathway by cadmium in BAECs. A, Translocation of HSF1 into the nucleus by cadmium. Cells were exposed to cadmium chloride (2 or 10 µM) for 1, 3, or 6 h, then the nucleus and cytosol fractions were subjected to Western blotting analysis using the antibodies indicated. G6PD and Lamin B were used as cytosolic maker and nucleus marker, respectively. B, Activation of HSE-driven transcriptional luciferase activity by cadmium. The HSE-driven luciferase activity was measured using cells that had been exposed to cadmium chloride (1, 5, 10, or 15 µM) for 12 h. C, Cadmium-mediated upregulation of heat shock protein (HSP) mRNA expression. Cells were exposed to cadmium chloride (1, 2.5, 5, or 10 µM) for 12 h, then real-time PCR analyses was performed for the HSP70-A1, HSPA6, HSP90α, and HSP90β genes. D, Increase in HSP proteins caused by exposure to cadmium. Cells were exposed to cadmium chloride (1, 2.5, 5, or 10 µM) for 24 h, then the total cell lysates were subjected to Western blotting analysis using the antibodies indicated. Each value is the mean ± SE of 3 independent experiments. * P < .05 and ** P < .01 compared with the controls.
Protective Role of HSF1 in Cadmium Toxicity
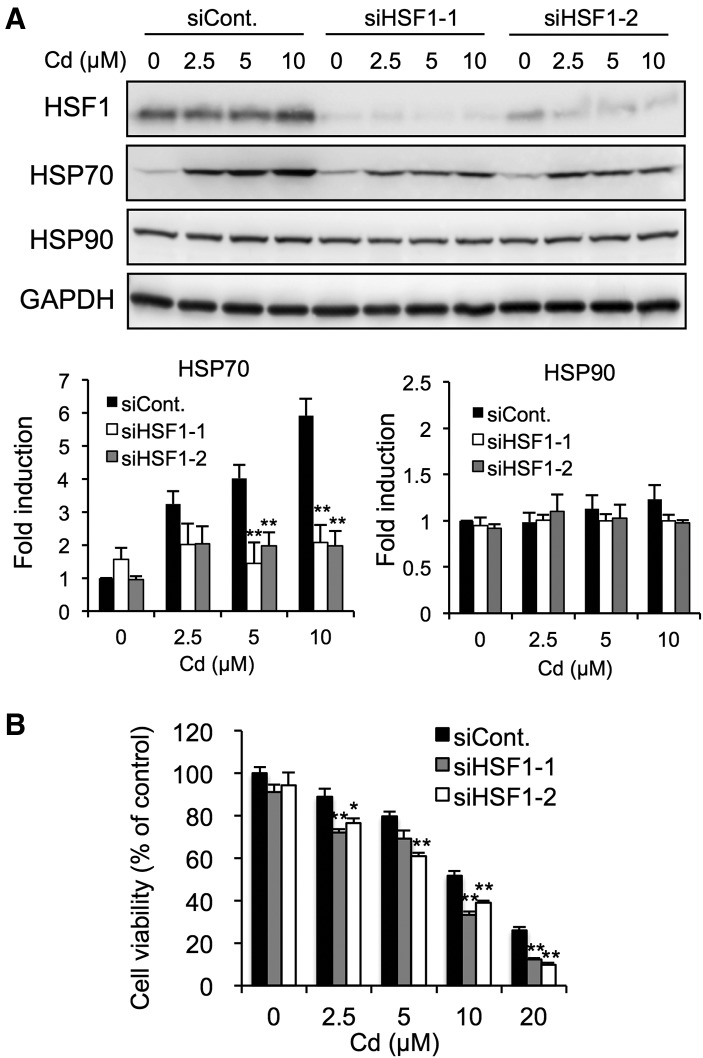
We performed siRNA-mediated HSF1 knockdown to confirm that HSF1 was involved in the induction of HSPs in BEACs. The transfection of HSF1 siRNA1 or HSF1 siRNA2 caused HSF1 protein expression to decrease markedly (Figure 2A). Under these conditions, the cadmium-mediated induction of HSP70 was significantly blocked by HSF1 knockdown but the basal expression of HSP70 as well as HSP90 was not affected (Figure 2A). This indicated that HSF1 predominantly regulates the induction of HSP70 but not HSP90 in BAECs. HSP70 protects against proteotoxic damage in cells, so we examined the protective role of HSF1 in cadmium toxicity. As is shown in Figure 2B, HSF1 knockdown enhanced the concentration-dependent cytotoxicity of cadmium, suggesting that HSF1 plays an important role in protection against cadmium toxicity.
FIG. 2.
Effect of HSF1 knockdown on HSP induction and cytotoxicity caused by the exposure of BAECs to cadmium. A, Cells were transfected with control siRNA, HSF1 siRNA1, or HSF1 siRNA2 for 48 h, then exposed to cadmium chloride (2.5, 5, or 10 µM) for 12 h. The total cell lysates were subjected to Western blotting analysis using the antibodies indicated. B, Cells were transfected with control siRNA, HSF1 siRNA1, or HSF1 siRNA2 for 48 h, then exposed to cadmium chloride (5, 10, 20, or 30 µM) for 24 h. Cell viability was measured using the MTT assay. Each value is the mean ± SE of 3 independent experiments. * P < .05 and ** P < .01 compared with the siControl results.
Modification of HSP90 by Cadmium
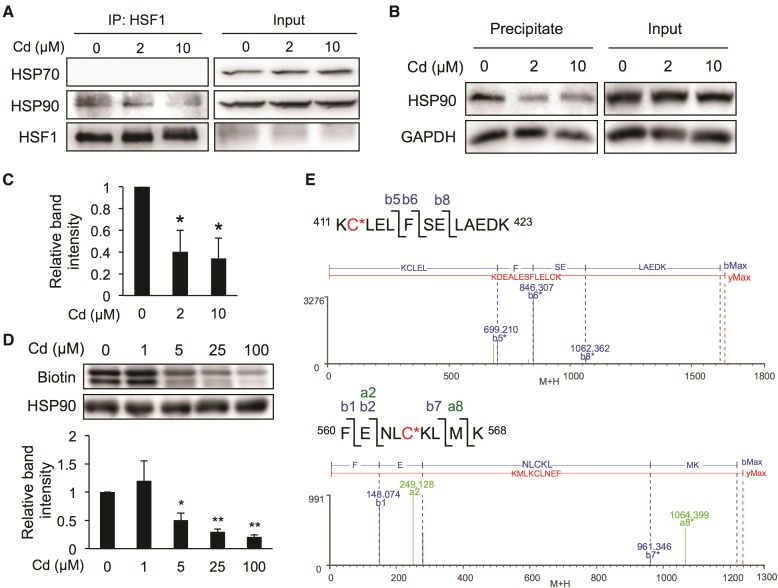
To clarify the mechanism underlying cadmium-mediated activation of HSF1, we examined the interaction between HSF1 and HSPs by performing immunoprecipitation assays using HSF1 antibodies in BAECs. Under basal conditions, HSF1 interacted with HSP90 but not HSP70 in BAECs, and such an interaction was decreased by exposure to cadmium (Figure 3A). Since HSF1 released from HSPs is reported to translocate into the nucleus to bind HSEs, thereby upregulating the downstream HSP genes (Akerfelt et al., 2010; Bjork and Sistonen, 2010; Voellmy, 2004), it seems likely that cadmium-mediated activation of HSF1 is attributable to disruption of the interaction between HSF1 and HSP90. In this context, the HSP90 modification caused by cadmium was evaluated using the BPM-precipitation assay that we previously developed (Toyama et al., 2013). Exposure of the cells to cadmium significantly decreased the HSP90 band intensity (Figs. 3B and C), suggesting that cadmium modified HSP90. We also determined whether cadmium could modify cysteine residues of HSP90 using a BPM-labeling assay (Toyama et al., 2013). Incubating human recombinant HSP90β with cadmium chloride decreased the BPM-bound HSP90β band intensity in a concentration-dependent manner (Figure 3D), suggesting that cadmium modified some of the cysteine residues of HSP90. We used trypsin to digest HSP90β, and the fragments produced were analyzed by nanoUPLC-mass spectrometry. Two peptides with masses increased by the mass of cadmium were detected only when HSP90β was incubated with cadmium chloride. The mass spectrometry and MSE fragmentation results (Figure 3E and Tables 3 and 4) suggested that cadmium modified, at least partly, human HSP90β at Cys412 and Cy564 under the conditions we used.
FIG. 3.
Effect of cadmium on interactions between HSF1 and HSPs, and the modification of HSP90 by cadmium. A, Dissociation of HSF1 from HSP90, caused by cadmium, in BAECs. Cells were exposed to cadmium chloride (2 or 10 µM) for 3 h, then the total cell lysates were subjected to immunoprecipitation assay using the antibodies indicated. B, Chemical modification of HSP90 by cadmium in BAECs. Cells were exposed to cadmium chloride (2 or 10 µM) for 3 h, then the total cell lysates were subjected to the BPM precipitation assay using the antibodies indicated. C, Quantitative band intensity results for the experiments described in part B. D, Chemical modification of recombinant HSP90 by cadmium. Recombinant human HSP90 (1 µM) was incubated with cadmium chloride (1, 5, 25, or 100 µM) in 50 mM Tris–HCl (pH 7.5) at 25 °C for 30 min. The resulting protein was subjected to the BPM-labeling assay. E, Recombinant human HSP90 (2.5 µM) was incubated with cadmium chloride (10 µM) in 50 mM Tris–HCl (pH 7.5) at 25 °C for 30 min. The resulting proteins were digested with trypsin and analyzed by nanoUPLC-MS system as described in the experimental procedures section. The mass spectrometry and MSE data are shown in Tables 3 and 4, respectively. Each value is the mean ± SE of 3 independent experiments. * P < .05 and ** P < .01 compared with the controls.
TABLE 3.
Cadmium-Modified Peptides in Human HSP90 Identified by NanoUPLC-MS
| Position | Peptide Sequence | Calculated Mass | Observed Mass | Cys | Increase in Mass |
|---|---|---|---|---|---|
| 411–423 | KCLELFSELAEDK + Cd | 1635.65 | 1635.61 | Cys412 | 111.90 |
| 560–568 | FENLCKLMK + Cd | 1237.47 | 1237.45 | Cys564 | 112.90 |
Recombinant human HSP90 beta (2.5 µM) was reacted with 10 µM cadmium chloride for 30 min. The native or cadmium-treated HSP90 was digested with trypsin and analyzed by nanoUPLC-MSE, as described in the experimental procedures section. A mass number of 111.90 or 112.90 was used to calculate the effect of the modification of a cysteine residue by cadmium. The MSE data are shown in Table 4.
TABLE 4.
MSE Data for the Cadmium-Modified Peptides Produced From Human HSP90
| Position | Assignment | Calculated Mass | Observed Mass | Analyte Modifier |
|---|---|---|---|---|
| 411–423 | b5* | 699.223 | 699.210 | + Cd (Cys) |
| b6* | 846.291 | 846.307 | + Cd (Cys) | |
| b8* | 1062.366 | 1062.362 | + Cd (Cys) | |
| b5 − H2O* | 681.212 | 681.217 | + Cd (Cys) | |
| b6 − H2O* | 828.281 | 828.289 | + Cd (Cys) | |
| 560–568 | b1 | 148.076 | 148.074 | |
| b2 | 277.119 | 277.123 | ||
| b7* | 961.334 | 961.346 | + Cd (Cys) | |
| a2 | 249.124 | 249.128 | ||
| a8* | 1064.380 | 1064.399 | + Cd (Cys) |
Regulation of Cadmium-Mediated HSP Induction and Toxicity by RSS
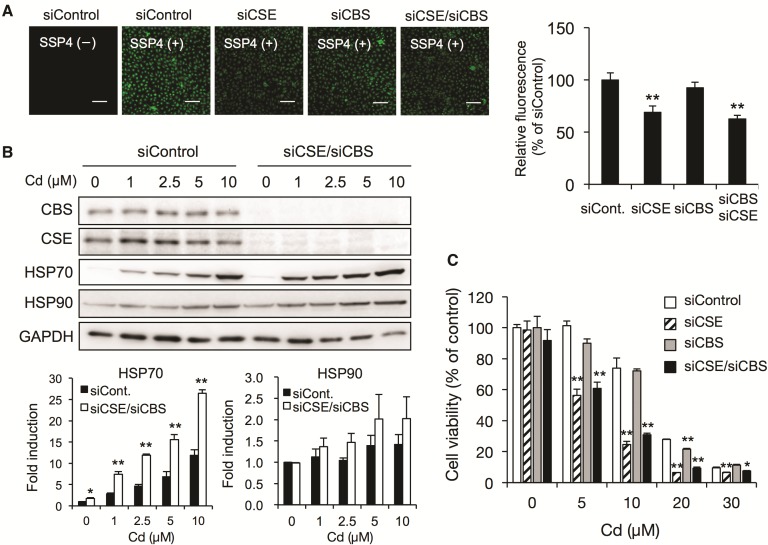
Several lines of evidence suggest that RSS derived from CSE and CBS are highly nucleophilic and inactivate electrophiles such as MeHg by forming sulfur adducts of the electrophiles (Abiko et al., 2015; Yoshida et al., 2011). We therefore expected that RSS derived from CSE and CBS will regulate the cadmium-mediated activation of the signal transduction pathway at low concentrations and regulate cadmium-mediated cytotoxicity at high concentrations by trapping cadmium via formation of sulfur adducts of the cadmium. We performed the siRNA-mediated knockdown of both enzymes and analyzed the RSS levels using the fluorescent probe SSP4 to determine the contributions of CSE and CBS to the production of RSS in BAECs. As is shown in Figure 4A, the knockdown of CSE but not CBS caused the RSS levels to decrease significantly, indicating that CSE rather than CBS is involved in RSS production in BAECs. This observation was in agreement with our previous findings that the enzymatic activity of recombinant CBS for generation of RSS such as cysteine persulfide from cystine was lower than that of CSE (Ida et al., 2014). The double knockdown of CSE and CBS caused a marked decrease in the RSS levels, and the cadmium-mediated induction of HSP70 was enhanced by the double knockdown of CSE and CBS (Figure 4B). Under the same conditions, the expression of HSP90 tended to be induced by exposure to cadmium. Consistent with the decrease in cellular RSS levels, cadmium-mediated cytotoxicity was significantly increased by the knockdown of CSE and/or CBS (Figure 4C).
FIG. 4.
Effect of siRNA-mediated knockdown of CSE and/or CBS on HSP induction and cytotoxicity in BAECs caused by exposure to cadmium. A, Effect of CSE and CBS knockdown on RSS levels. Cells were transfected with control siRNA, CSE siRNA, and/or CBS siRNA for 48 h, then the cellular RSS levels were determined using the fluorescent probe SSP4. The fluorescence images of the cells (left) and the relative intensity (right) were shown. The scale bar indicates 100 µm. B, Effect of CSE/CBS double knockdown on HSP induction caused by exposure to cadmium. Cells were transfected with control siRNA, CSE siRNA, or CBS siRNA for 48 h, then exposed to cadmium chloride (1, 2.5, 5, or 10 µM) for 24 h. The total cell lysates were subjected to Western blotting analysis using the antibodies indicated. C, Effect of CSE and CBS knockdown on cadmium-induced cytotoxicity. Cells were transfected with control siRNA, CSE siRNA, and/or CBS siRNA for 48 h, then exposed to cadmium chloride (5, 10, 20, or 30 µM) for 24 h. Cell viability was measured using the MTT assay. Each value is the mean ± SE of 3 independent experiments. * P < .05 and ** P < .01 compared with the siControl results.
Suppression of Cadmium-Mediated HSP Induction and Toxicity Through the Overexpression of CSE
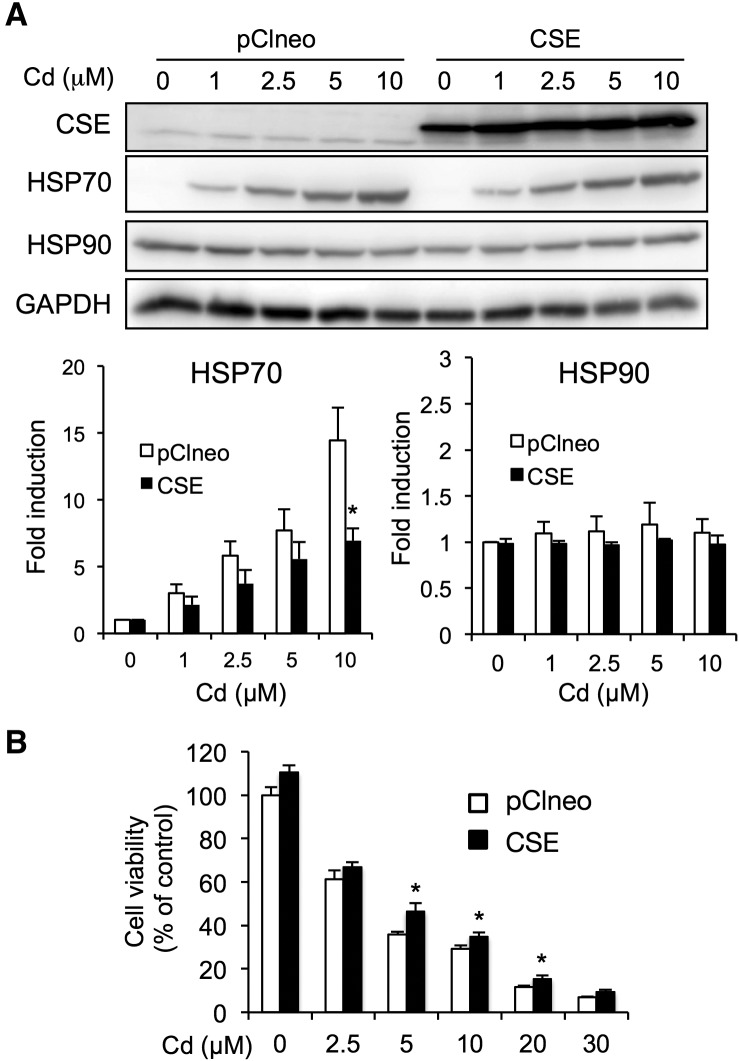
We performed CSE overexpression because CSE rather than CBS is involved in RSS production in BAECs (Figure 4). As is shown in Figure 5A, transfection of the pClneo-HA-CSE vector increased CSE expression. The cadmium-mediated induction of HSP70 was significantly suppressed under the conditions we used (Figure 5A). In contract with the results of the CSE knockdown, the overexpression of CSE significantly conferred protection against cadmium toxicity (Figure 5B).
FIG. 5.
Effect of CSE overexpression on HSP induction and cytotoxicity in BAECs caused by exposure to cadmium. A, Effect of CSE overexpression on HSP induction caused by exposure to cadmium. Cells were transfected with pClneo alone or pClneo-HA-CSE for 24 h, then exposed to cadmium chloride (1, 2.5, 5, or 10 µM) for 24 h. The total cell lysates were subjected to Western blotting analysis using the antibodies indicated. B, Effect of CSE overexpression on cadmium-induced cytotoxicity. Cells were transfected with pClneo alone or pClneo-HA-CSE for 24 h, then exposed to cadmium chloride (2.5, 5, 10, 20, or 30 µM) for 24 h. Cell viability was measured using the MTT assay. Each value is the mean ± SE of 3 independent experiments. * P < .05 and ** P < .01 compared with the siControl results.
DISCUSSION
The induction of HSPs by cadmium has been investigated in a number of studies (Beyersmann and Hechtenberg, 1997; Luparello et al., 2011), but the exact mechanisms underlying the activation of the signal transduction pathway have not been identified. Here, we propose that cadmium induces HSPs through the activation of HSF1, triggered by cadmium directly modifying HSP90 cysteine residues.
It has been found that both HSP70 and HSP90 interact with and act as repressors of HSF1 (Akerfelt et al., 2010; Bjork and Sistonen, 2010; Voellmy, 2004). In this study we showed that HSF1 interacts with HSP90 but not HSP70 in endothelial cells (Figure 3A), suggesting that HSP90 is the preferred stress condition sensor in endothelial cells. In fact, cadmium facilitated the dissociation of HSF1 and HSP90 coupled to the modification of HSP90 by cadmium (Figure 3). HSP90 is comprised of 3 domains: (1) an N-terminal domain containing nucleotide and co-chaperone binding site; (2) a middle domain containing binding sites for client proteins; (3) a C-terminal domain containing a dimerization motif (Mollapour and Neckers, 2012). HSP90 is highly conserved, and HSP90β in both humans and bovines has 6 cysteine residues (Cys366, Cys412, Cys521, Cys564, Cys589, and Cys590) (Mollapour and Neckers, 2012; Nardai et al., 2000). We found, for the first time, that cadmium can modify HSP90β through Cys412 located in the middle domain and Cys564 located in the C-terminal domain. Consistent with this, it has been found that sulfoxythiocarbamates modify Cys412 and Cys564 in HSP90β (Zhang et al., 2014) and that 4-hydroxynonenal modifies HSP90α at Cys572, corresponding to Cys564 in HSP90β (Carbone et al., 2005). We have also found that Cys412 and Cys564 in HSP90β are covalently modified by the atmospheric electrophile 1,4-naphthoquinone (Abiko Y et al. 2017). It is still unclear why environmental electrophiles such as cadmium and 1,4-naphthoquinone selectively target Cys412 and Cys564 in the 6 cysteine residues, but the proximal amino acids may be an explanation because Cys412 and Cys564 are adjacent to Lys411 and Lys565, respectively. Proximity to a basic amino acid decreases the pKa of the thiol group of a cysteine residue, increasing the degree of deprotonation (Snyder et al., 1981). However, Shibata et al. found that 6-methysulfinylhexyl isothiocyanate modifies Cys521 in HSP90β, and they suggested that a hydrophobic pocket adjacent to Cys521 may provide a suitable environment for the binding of 6-methysulfinylhexyl isothiocyanate (Shibata et al., 2011).
It has been suggested that cadmium forms complexes with amino acids not only via single sulfur donation but also via multiple sulfur donors and/or coordinating side chains, such as nitrogen and oxygen donors (Sovago and Varnagy, 2013). The peptide containing Cys412 increased in mass number by 111.90 but not 112.90, suggesting that cadmium may interact not only with Cys412 but also with another oxygen donor, such as Glu414. With regard to Cys564, the three-dimensional model showed that there are no proximal Cys residues (Shibata et al., 2011). The peptide containing Cys564 also contained Lys565, suggesting that cadmium may also interact with this basic amino acid through the formation of coordination bonds (Sovago and Varnagy, 2013).
Cadmium significantly increased the mRNA levels of both HSP70 and HSP90, but cadmium induced the protein expression of HSP70 but not HSP90 (Figure 1D). This may have been because cadmium strongly induced the HSPA6 gene. We speculate that such a strong induction of the HSPA6 gene is caused by multiple HSEs in the promoter region and other factors such as AP-1 (Ramirez et al., 2015) because cadmium is known to activate AP-1 (Thevenod and Lee, 2013). In our study, HSF1 knockdown suppressed the inducible HSP70 but not its basal expression (Figure 2A), suggesting that HSF1 is essential to the expression of the inducible HSP70. HSF1 knockdown increased cadmium-mediated cytotoxicity, so HSP70 induction via the activation of HSF1 is a cytoprotective response to cadmium. Consistent with this, HSF1/HSP70 has been found to have a protective function in in vivo experiments using HSF1 knockout mice (Wirth et al., 2003).
It has been found that 3 enzymes, CSE, CBS, and 3-mercaptopyruvate sulfurtransferase, responsible for the production of persulfides/polysulfides are expressed in endothelial cells (Mistry et al., 2016; Sen et al., 2012; Shibuya et al., 2009). We have previously found that the overexpression of CSE and CBS in A549 cells increased cellular persulfide/polysulfide levels (Ida et al., 2014). Similar to those results, the knockdown of CSE but not CBS decreased cellular persulfide/polysulfide concentrations in BAECs, suggesting that CSE rather than CBS contributes to the production of persulfides/polysulfides in endothelial cells. Interestingly, more than half of the RSS by concentration remained when CSE/CBS double-knockdown tests were performed (Figure 4A), indicating that other enzymes are also involved in the production of RSS. Nevertheless, the cadmium-mediated induction of HSP70 was enhanced by CSE/CBS double knockdown (Figure 4B), and cadmium-induced cytotoxicity was enhanced by the knockdown of these enzymes (Figure 4C). Such a regulative function of CSE was also confirmed in the overexpression experiments (Figure 5). Taken together, the results suggest that RSS derived particularly from CSE play critical roles in regulating activation of the cellular signaling pathway by cadmium and in protecting against cadmium toxicity, presumably via the inactivation of cadmium within the cells. We have previously found that MeHg reacts with RSS to form the detoxified metabolite (MeHg)2S (Abiko et al., 2015; Yoshida et al., 2011). Sulfur adducts formed with cadmium have yet to be identified, but we speculate that RSS can trap cadmium to form inert sulfur adducts, protecting cells from cadmium injury. Such a regulation process appears to be an initial defense system before sensor proteins such as HSP90 are captured because RSS can modulate the activation of electrophilic signaling.
In summary, we have identified Cys412 and Cys564 in HSP90β as being potential sites that are modified by cadmium. Cadmium activates HSF1 by dissociating it from HSP90, thereby upregulating downstream proteins such as HSP70 to protect cells from cadmium toxicity. In addition to this protective response to cadmium, we have found for the first time that RSS derived from CSE can modulate the activation of the HSF1/HSP70 signaling pathway by cadmium, presumably via inactivation of cadmium. This, in turn, protects cells from injury by cadmium.
CONFLICT OF INTEREST STATEMENT
These authors have no conflicts of interest.
FUNDING
Ministry of Education, Culture, Sports, Science, and Technology of Japan (#25220103 to Y. K. and 15K08042 to Y. S.); National Institutes of Health (R01HL116571 to M.X.).
REFERENCES
- Abiko Y., Sha L., Shinkai Y., Unoki T., Luong NC., Tsuchiya Y., Watanabe Y., Hirose R., Akaike T., Kumagai Y. (2017). 1,4-Naphthoquinone activates the HSP90/HSF1 pathway through the S-arylation of HSP90 in A431 cells: Negative regulation of the redox signal transduction pathway by persulfides/polysulfides. Free Radic. Biol. Med. 104, 118-128. [DOI] [PubMed] [Google Scholar]
- Abiko Y., Yoshida E., Ishii I., Fukuto J. M., Akaike T., Kumagai Y. (2015). Involvement of reactive persulfides in biological bismethylmercury sulfide formation. Chem. Res. Toxicol. 28, 1301–1306. [DOI] [PubMed] [Google Scholar]
- Akerfelt M., Morimoto R. I., Sistonen L. (2010). Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyersmann D., Hechtenberg S. (1997). Cadmium, gene regulation, and cellular signalling in mammalian cells. Toxicol. Appl. Pharmacol. 144, 247–261. [DOI] [PubMed] [Google Scholar]
- Bjork J. K., Sistonen L. (2010). Regulation of the members of the mammalian heat shock factor family. FEBS J. 277, 4126–4139. [DOI] [PubMed] [Google Scholar]
- Carbone D. L., Doorn J. A., Kiebler Z., Ickes B. R., Petersen D. R. (2005). Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J. Pharmacol. Exp. Ther. 315, 8–15. [DOI] [PubMed] [Google Scholar]
- Choong G., Liu Y., Templeton D. M. (2014). Interplay of calcium and cadmium in mediating cadmium toxicity. Chem. Biol. Interact. 211, 54–65. [DOI] [PubMed] [Google Scholar]
- DeLeve L. D., Kaplowitz N. (1991). Glutathione metabolism and its role in hepatotoxicity. Pharmacol. Ther. 52, 287–305. [DOI] [PubMed] [Google Scholar]
- Denizot F., Lang R. (1986). Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 89, 271–277. [DOI] [PubMed] [Google Scholar]
- Galan A., Troyano A., Vilaboa N. E., Fernandez C., de Blas E., Aller P. (2001). Modulation of the stress response during apoptosis and necrosis induction in cadmium-treated U-937 human promonocytic cells. Biochim. Biophys. Acta 1538, 38–46. [DOI] [PubMed] [Google Scholar]
- Holmberg C. I., Hietakangas V., Mikhailov A., Rantanen J. O., Kallio M., Meinander A., Hellman J., Morrice N., MacKintosh C., Morimoto R. I., et al. (2001). Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20, 3800–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida T., Sawa T., Ihara H., Tsuchiya Y., Watanabe Y., Kumagai Y., Suematsu M., Motohashi H., Fujii S., Matsunaga T., et al. (2014). Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U. S. A. 111, 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. T., Marnett L. J. (2007). Heat shock factor 1 attenuates 4-Hydroxynonenal-mediated apoptosis: Critical role for heat shock protein 70 induction and stabilization of Bcl-XL. J. Biol. Chem. 282, 33412–33420. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. (1984). Electroblotting of multiple gels: A simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10, 203–209. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Craig E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. [DOI] [PubMed] [Google Scholar]
- Luparello C., Sirchia R., Longo A. (2011). Cadmium as a transcriptional modulator in human cells. Crit. Rev. Toxicol. 41, 75–82. [DOI] [PubMed] [Google Scholar]
- Mistry R. K., Murray T. V., Prysyazhna O., Martin D., Burgoyne J. R., Santos C., Eaton P., Shah A. M., Brewer A. C. (2016). Transcriptional regulation of cystathionine-gamma-lyase in endothelial cells by NADPH oxidase 4-dependent signaling. J. Biol. Chem. 291, 1774–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M., Neckers L. (2012). Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim. Biophys. Acta 1823, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardai G., Sass B., Eber J., Orosz G., Csermely P. (2000). Reactive cysteines of the 90-kDa heat shock protein, Hsp90. Arch. Biochem. Biophys. 384, 59–67. [DOI] [PubMed] [Google Scholar]
- Prozialeck W. C., Edwards J. R., Nebert D. W., Woods J. M., Barchowsky A., Atchison W. D. (2008). The vascular system as a target of metal toxicity. Toxicol. Sci. 102, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prozialeck W. C., Edwards J. R., Woods J. M. (2006). The vascular endothelium as a target of cadmium toxicity. Life Sci. 79, 1493–1506. [DOI] [PubMed] [Google Scholar]
- Ramirez V. P., Stamatis M., Shmukler A., Aneskievich B. J. (2015). Basal and stress-inducible expression of HSPA6 in human keratinocytes is regulated by negative and positive promoter regions. Cell Stress Chaperones 20, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T. K., Freeman B. A. (2009). Transduction of redox signaling by electrophile–protein reactions. Sci. Signal 2, re7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H. (2013). Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 14, 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen U., Sathnur P. B., Kundu S., Givvimani S., Coley D. M., Mishra P. K., Qipshidze N., Tyagi N., Metreveli N., Tyagi S. C. (2012). Increased endogenous H2S generation by CBS, CSE, and 3MST gene therapy improves ex vivo renovascular relaxation in hyperhomocysteinemia. Am. J. Physiol. Cell Physiol. 303, C41–C51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Kimura Y., Mukai A., Mori H., Ito S., Asaka Y., Oe S., Tanaka H., Takahashi T., Uchida K. (2011). Transthiocarbamoylation of proteins by thiolated isothiocyanates. J. Biol. Chem. 286, 42150–42161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N., Mikami Y., Kimura Y., Nagahara N., Kimura H. (2009). Vascular endothelium expresses 3-mercaptopyruvate sulfurtransferase and produces hydrogen sulfide. J. Biochem. 146, 623–626. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Kimura T., Itagaki A., Yamamoto C., Taguchi K., Yamamoto M., Kumagai Y., Kaji T. (2016). Partial contribution of the Keap1-Nrf2 system to cadmium-mediated metallothionein expression in vascular endothelial cells. Toxicol. Appl. Pharmacol. 295, 37–46. [DOI] [PubMed] [Google Scholar]
- Snyder G. H., Cennerazzo M. J., Karalis A. J., Field D. (1981). Electrostatic influence of local cysteine environments on disulfide exchange kinetics. Biochemistry 20, 6509–6519. [DOI] [PubMed] [Google Scholar]
- Sovago I., Varnagy K. (2013). Cadmium(II) complexes of amino acids and peptides. Metal Ions Life Sci. 11, 275–302. [DOI] [PubMed] [Google Scholar]
- Stambolic V., Suzuki A., de la Pompa J. L., Brothers G. M., Mirtsos C., Sasaki T., Ruland J., Penninger J. M., Siderovski D. P., Mak T. W. (1998). Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95, 29–39. [DOI] [PubMed] [Google Scholar]
- Taguchi K., Motohashi H., Yamamoto M. (2011). Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16, 123–140. [DOI] [PubMed] [Google Scholar]
- Thevenod F., Lee W. K. (2013). Cadmium and cellular signaling cascades: Interactions between cell death and survival pathways. Arch. Toxicol. 87, 1743–1786. [DOI] [PubMed] [Google Scholar]
- Toyama T., Shinkai Y., Kaji T., Kumagai Y. (2013). A convenient method to assess chemical modification of protein thiols by electrophilic metals. J. Toxicol. Sci. 38, 477–484. [DOI] [PubMed] [Google Scholar]
- Toyama T., Sumi D., Shinkai Y., Yasutake A., Taguchi K., Tong K. I., Yamamoto M., Kumagai Y. (2007). Cytoprotective role of Nrf2/Keap1 system in methylmercury toxicity. Biochem. Biophys. Res. Commun. 363, 645–650. [DOI] [PubMed] [Google Scholar]
- Unoki T., Abiko Y., Toyama T., Uehara T., Tsuboi K., Nishida M., Kaji T., Kumagai Y. (2016). Methylmercury, an environmental electrophile capable of activation and disruption of the Akt/CREB/Bcl-2 signal transduction pathway in SH-SY5Y cells. Sci. Rep. 6, 28944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellmy R. (2004). On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9, 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth D., Christians E., Li X., Benjamin I. J., Gustin P. (2003). Use of Hsf1(-/-) mice reveals an essential role for HSF1 to protect lung against cadmium-induced injury. Toxicol. Appl. Pharmacol. 192, 12–20. [DOI] [PubMed] [Google Scholar]
- Worby C. A., Dixon J. E. (2014). Pten. Annu. Rev. Biochem. 83, 641–669. [DOI] [PubMed] [Google Scholar]
- Yoshida E., Toyama T., Shinkai Y., Sawa T., Akaike T., Kumagai Y. (2011). Detoxification of methylmercury by hydrogen sulfide-producing enzyme in Mammalian cells. Chem. Res. Toxicol. 24, 1633–1635. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Dayalan Naidu S., Samarasinghe K., Van Hecke G. C., Pheely A., Boronina T. N., Cole R. N., Benjamin I. J., Cole P. A., Ahn Y. H., et al. (2014). Sulphoxythiocarbamates modify cysteine residues in HSP90 causing degradation of client proteins and inhibition of cancer cell proliferation. Br. J. Cancer 110, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]