Abstract
Background:
DNA barcoding is a technique used to identify species based on species-specific differences in short regions of their DNA. It is widely used in species discrimination of medicinal plants and traditional medicines.
Materials and Methods:
In the present study, four potential DNA barcodes, namely rbcL, matK, trnH-psbA and ITS (nuclear ribosomal internal transcribed spacer) were adopted for species discrimination in Crawfurdia Wall (Genetiaceae). Identification ability of these DNA barcodes and combinations were evaluated using three classic methods (Distance, Blast and Tree-Building).
Results:
As a result, ITS, trnH-psbA and rbcL regions showed great universality for a success rate of 100%; whereas matK was disappointing for which only 65% samples gained useful DNA sequences. ITS region, which could clearly and effectively identify the five species in Crawfurdia, performed very well in this study. On the contrary, trnH-psbA and rbcL performed poorly in discrimination among these species.
Conclusion:
ITS marker was an ideal DNA barcode in Crawfurdia and it should be incorporated into one of the core barcodes for seed plants.
Keywords: Crawfurdia Wall, DNA barcode, species discrimination, internal transcribed spacer (ITS), DNA sequence
Introduction
DNA barcoding is a useful molecular tool that could rapidly and accurately identify species using standardized DNA markers. The approach has become an efficient supplement to traditional taxonomy based on morphology (Hebert et al., 2003; Li et al., 2011; Packer et al., 2009). Moreover, it is also used for detecting cryptic species, identifying medicinal plant material, food traceability etc. (Chen et al., 2014; Galimberti et al., 2013; Shokralla et al., 2014; Techen et al., 2014). CO1 (cytochrome c oxidase subunit 1), as an universal barcode, performs well in animals. However, it is not appreciate to plants due to low substitution rates in plant mitochondrial genome (Fazekaset al., 2008; China Plant BOL Group, 2011). An perfect DNA barcode should be reliable, powerful and universal in indentifying species, as well as cost-effective (CBOL Plant Working Group, 2009). In recent years, many experts carried out a great deal of researches on screening and testing for potential DNA barcodes (Kress and Erickson, 2007; CBOL Plant Working Group, 2009; China Plant BOL Group, 2011; Hollingsworth et al., 2011; Dong et al., 2012). But variable sites of single gene are relatively limited, and thus none of the potential barcodes could meet the afore mentioned criteria well. As a result, multi-locus method became a smart choice to gain enough discriminatory ability for plant DNA barcoding. Specific combinations of potential regions were debated and controversial all long (Hollingsworth et al., 2011). The CBOL Plant Working Group (2009) recommended that rbcL and matK (cpDNA) were listed as core plant barcode; meanwhile, trnH-psbA (cpDNA) and ITS (nuclear ribosomal internal transcribed spacer, ncDNA) were also added as supplement. However, this recommendation was concluded based on limited studies and plant taxa. What’s more, rbcL and matK performed not so well as it was declared in universality and identified ability. And thus China scientists recommended ITS/ITS2 as one of the core DNA barcodes due to good ability of identifying species based on analysis of massive data from more representative species (China Plant BOL Group, 2011; Yao et al., 2010).
Crawfurdia is a small genus (Gentianaceae) that is mainly distributed in China and adjacent regions (Ho and Pringle, 1995). It is extremely similar to Tripterospermum Bl. in morphology with minor difference in flower. The genus includes about 16 species. Most of the species have voluble stems except C. semialata (Marq.) H. Smith. Furthermore, the genus is close to Tripterospermum, Metagentiana and Genetiana in phylogenetic relationship (Chen et al., 2005). Moreover, there were also some studies on surface feature of seeds, embryology and floral anatomy etc. (Chen et al., 2005; Chen and He, 2002; Chen et al., 2000). However, no genetic study using molecular methods has been reported until now.
Gentianaceae is an extremely important family with many medicinal plants that is widely distributed across the world (Ho and Pringle, 1995). However, there was only a few DNA barcoding researches on species of the family (China Plant BOL Group, 2011; Xue and Li, 2011). In the present study, four potential barcodes (rbcL, trnH-psbA, matK and ITS) were tested using five Crawfurdia species. This study would be beneficial to know discriminatory ability of these potential DNA barcodes in species of Gentianaceae.
Material and Methods
Material sampling
In the present study, 26 individuals belonging to five species in Crawfurdia were collected in fieldwork, namely 4 to 6 individuals per species (Table 1). Clear and young leaves were sampled from wild habitats and then dried using silica gel as soon. Geographical distance among individuals was generally more than 20m. All voucher specimens of Crawfurdia species were identified by authors and they were deposited in Herbarium of Kunming Institute of Botany, Chinese Academy of Science (KUN).
Table 1.
Collecting information of 5 species in Crawfurdia in the present study.
| Species | Locality | Latitude/Longitude | Altitude (m) | Voucher specimen | DNA Accessions |
|---|---|---|---|---|---|
| Crawfurdia maculaticaulis C.J.Wu | Delong, Napo, Guangxi, China | N23°19’/ E105°48’ | 1128 | DLUZDQ031 | MLD02-MLD05 |
| C. maculaticaulis C.J.Wu | Lida, Funing, Yunnan, China | N23°29’/ E105°36’ | 1365 | DLUZDQ032 | MLD10A, MLD10B |
| C. angustata C.B.Clarke | Shiyueliang, Fugong, Yunnan, China | N27°10’/ E98°53’ | 1219 | DLUZDQ024 | MLD11-MLD13 |
| C. angustata C.B.Clarke | Puladi, Fugong, Yunnan, China | N27°32’/ E98°50’ | 1341 | DLUZDQ025 | MLD14-MLD16 |
| C. price i (Marq.) H. Smith | Maoershan, Xingan, Guangxi, China | N25°54’/ E110°27’ | 886 | DLUZDQ026 | MLD17-MLD22 |
| C. campanulacea Wall. et Griff. ex C.B. Clarke | Zhenan, Longling, Yunnan, China | N24°40’/ E98°49’ | 1817 | DLUZDQ028 | MLD27-MLD30 |
| C. campanulacea Wall. et Griff. ex C.B. Clarke | Shiyueliang, Fugong, Yunnan, China | N27°10’/ E98°53’ | 1350 | DLUZDQ029 | MLD33 |
| C. delavayi Franch. | Cangshan, Dali, Yunnan, China | N25°45/ E100°06’ | 2900 | DLUZDQ030 | MLD34-MLD37 |
DNA extraction, amplification and sequencing
Modified CTAB method was used for extracted total DNA from dried leaves using (Doyle and Doyle, 1987). Primers for the four DNA markers, as well as their reaction systems were in accordance with requirements of the China Plant BOL Group (2011). DNA sequencing was performed on the ABI 3730 DNA Sequencer (Applied Biosystems, USA). DNA Sequences would be sequenced again or discarded in final analysis if they could not meet with quality requirement of plant DNA barcodes (China Plant BOL Group, 2011).
DNA barcode analysis
Lasergene SeqMan Pro 7.1.0 (DNASTAR) was used to assemble DNA sequences. And then assembled sequences were aligned using MEGA 5.0 (Tamura et al., 2011) and adjusted manually in BioEdit version 7.1.3.0 (Hall, 1999). For all sequences, gaps were treated according to Simple Indel Coding (SIC) method (Liu et al., 2012). Three methods (Distance, Blast and Tree-Building) were used to evaluated species discrimination success (China Plant BOL Group, 2011). Due to terrible sequencing success of matK region, it was excluded from final analysis. The P-distances intra- and inter-species for all barcodes and combinations were calculated using MEGA 5.0. For the Blast method, all sequences and possible combinations were used as query sequences, and BioEdit 7.2.5 was used to query the reference database with each DNA sequence successively to establish if the closest hit was a conspecific species and to provide statistics for identifying species (Hall, 1999). Neighbor-Joining (NJ) trees were also constructed by MEGA 5.0 according to the recommended settings. Judged standards of successful species identification were identical to relative requirements of the China Plant BOL Group (2011).
Results
Primers for the DNA barcodes were universally applicable to the studied individuals. All samples could be successfully PCR amplified for the four DNA barcodes. The success rate of bidirectional sequencing was 100% for all markers except matK 65% in the present study. In total, 26 rbcL sequences, 26 trnH-psbA sequences, 17 matK sequences and 26 ITS sequences were generated (Table 2).
Table 2.
Properties of the four DNA barcodes evaluated in Crawfurdia.
| DNA regions | ITS | trnH-psbA | matK | rbcL |
|---|---|---|---|---|
| Universal primer | Yes | Yes | Yes | Yes |
| PCR success (%) | 100% | 100% | 100% | 100% |
| Sequencing success (%) | 100% | 100% | 65% | 100% |
| Aligned sequence length (bp) | 684 | 323 | 638 | 650 |
| Indel number (length in bp) | 1 (1) | 7 (1-12) | 0 | 0 |
| No. informative sites/variable sites | 77/78 | 5/6 | 1/8 | 3/3 |
| No. samples (individuals) | 26 | 26 | 17 | 26 |
| Mean inter-specific distance (range) | 0.0002 (0.0000-0.0006) | 0.0009 (0.0000-0.0019) | - | 0.0000 |
| Mean intra-specific distance (range) | 0.0490 (0.0254-0.0626) | 0.0129 (0.0005-0.0209) | - | 0.0020 (0.0000-0.0049) |
The length of the aligned ITS sequences was 684bp which had 78 variable sites, including 1 indels. The aligned trnH-psbA matrix was 323bp long with 6 variable sites. In the matK matrix, the sequences were 638bp in length with 8 variable sites. For the rbcL matrix, the aligned sequences were 650bp long with 3 variable sites.
To evaluate ability of species discrimination for these DNA barcodes and combinations, three methods (Distance, Blast and Tree-Building) were performed in this study (Table 2 & Fig. 1). For matK region, only 65% (17/26) samples gained available sequence. DNA sequences of matK were gained only in one individual for C. pricei and C. delavayi respectively, so the marker was excluded in final analysis. Analyzed results were similar to species identification based on the three methods. ITS region could perfectly identify all species in Crawfurdia but trnH-psbA and rbcL performed much poorly in identifying species (Fig. 2). Due to perfect discriminatory power of ITS, combinations of DNA barcodes were seemly redundant in this study.
Figure 1.
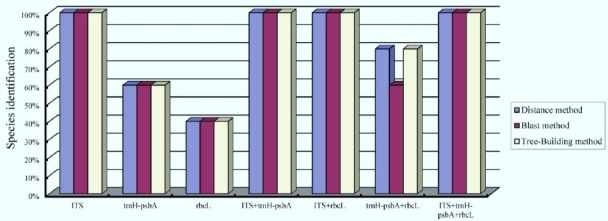
Species discrimination of each DNA barcode and their combination based on methods of Distance, Blast and Tree-Building.
Figure 2.
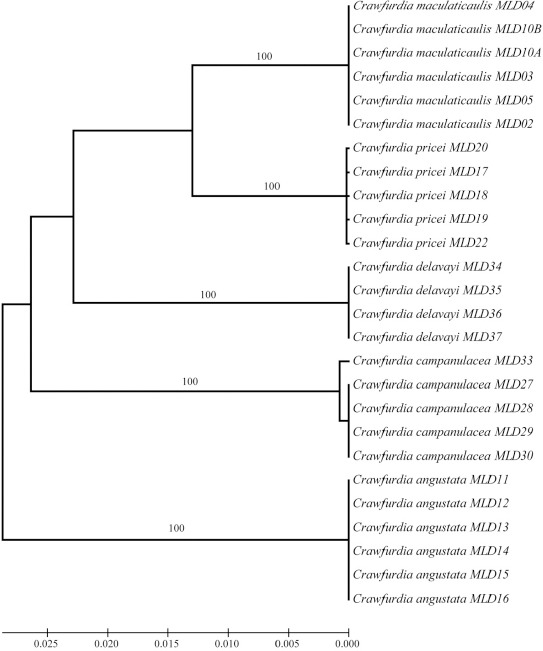
Neighbor-Joining (NJ) tree of the Crawfurdia species based on ITS marker. Bootstrap values (>50%) were shown above the relevant branches
Discussion
Primer universality and species discrimination are important criterion for an ideal DNA barcode (China Plant BOL Group, 2011). In the four potential DNA barcodes, ITS, trnH-psbA and rbcL showed well success rates of PCR amplification and DNA sequencing, as well as obviously different ability of species identification. On the contrary, matK was difficult to sequence using currently available primer sets though it possessed high evolutionary rate, suitable length. In this study, 9 individuals in 26 samples of Crawfurdia species could not generated available matK sequences, thus it was discarded in final analysis. As a plastid marker, rbcL is widely used DNA marker in phylogenetic studies and it is very useful at the family and genus levels with well universality cross different taxon (China Plant BOL Group, 2011). However, rbcL evolves slowly so it is not suitable to lower taxonomic level and complex group. Although the barcode was listed as core DNA barcode in the previous researches, it is not an ideal marker for Crawfurdia obviously. Moreover, trnH-psbA is the most widely used cpDNA barcode at present (China Plant BOL Group, 2011; Li et al., 2014). The barcode shows rich sequence variation and possesses high rates of insertion/deletion (Kress and Erickson, 2007). Thus Kress et al. (2005) and Chase et al. (2007) suggested that trnH-psbA marker could be used in two-locus or three-locus barcode systems to afford better discriminatory ability. However, this region was relatively short in Crawfurdia species and its ability of species identification was also limited in the present study.
The ITS region, as a powerful DNA marker at the species level, shows high levels of inter-specific divergence (Álvarez and Wendel, 2003). But CBOL Plant Working Group only regarded ITS as a supplementary barcode due to its limitations, such as difficulty of amplification and sequencing, fungal contamination and incomplete concerted evolution etc. (CBOL Plant Working Group, 2009). However, China Plant BOL Group recommended that when direct sequencing was possible, the ITS marker should be listed as the core barcodes due to its better discriminatory ability than cpDNA barcode (China Plant BOL Group, 2011). In this study, ITS region showed perfect universality and species discrimination for Crawfurdia species. All species was obviously identified by using Distance, Blast and Tree-Building methods. And this result further proved that ITS was a powerful barcode in majority group and should be listed as one of the core barcodes (China Plant BOL Group, 2011; Yao et al., 2010).
Moreover, multi-locus DNA barcodes have been regarded as a possible resolution because of inadequate variation of single loci (Chase et al., 2007; China Plant BOL Group, 2011; Kress et al., 2005). According to suggestion of The CBOL Plant Working Group, rbcL+matK was regarded as the universal barcode combination (CBOL Plant Working Group, 2009). Then many experts gave large amount of suggestions on DNA barcodes and combinations (Chase et al., 2007; China Plant BOL Group, 2011; Hollingsworth et al., 2011; Kress and Erickson, 2007). However, multi-locus DNA barcodes did not solve with the current difficulty of identifying species completely. Data analysis also faced with more difficulties for combined barcodes than single-locus maker, especially when one of the target loci fails to amplify. Moreover, CBOL revealed that using seven candidate loci did not obviously enhance discriminatory ability at species level compared to rbcL+matK (Hollingsworth et al., 2011). In the present study, multi-locus combinations could not improve the ability of species discrimination due to good performance of ITS region.
In conclusion, ITS marker was undoubtedly an ideal DNA barcode in Crawfurdia, and it was also proved to be powerful in most of seed plants by previous studies. So it should be incorporated into one of the core barcodes for seed plants.
Acknowledgments
This study was supported by Key Project of Yunnan Provincial Department of Education (2014Z134) and National Natural Science Foundation of China (Grant no. 31200180). The authors thank Junbo Yang, Jing Yang and Wuxiang Fu in Kunming Institution of Botany (CAS) for their help in molecular experiments. Moreover, we would like to thank Ting Zhang in Kunming Institution of Botany (CAS) for providing molecular materials of part species.
References
- 1.Alvarez I, Wendel J.F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. E. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 2.CBOL Plant Working group. A DNA barcode for land plants. Proc. Nat. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chase M.W, Cowan R.S, Hollingsworth P.M. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007;56:295–299. [Google Scholar]
- 4.Chen S.Y, Xia T, Wang Y.J, Liu J.Q, Chen S. Molecular systematics and biogeography of rawfurdia, Metagentiana and Tripterospermum (Gentianaceae) based on nuclear ribosomal and plastid DNA sequences. Ann. Bot. 2005;96:413–424. doi: 10.1093/aob/mci188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S.L, He T.N, Liu J.Q, Hong D.Y. Embryology of rawfurdia delavayi (Gentianaceae) and its systematic value. Israel J. Plant Sci. 2000;48:113–119. [Google Scholar]
- 6.Chen S.L, He T.N. Surface features of seeds in Tripterospermum and rawfurdia (Gentianaceae) Acta Bot. Bor-Occid. Sin. 2002;22:37–42. [Google Scholar]
- 7.Chen S.L, Pang X.H, Song J.Y, Shi L.C, Yao H, Han J.P, Leon C. A renaissance in herbal medicine identification: from morphology to DNA. Biotech. Advances. 2014;32:1237–1244. doi: 10.1016/j.biotechadv.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 8.China Pant BOL Group. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Nat. Acad. Sci. USA. 2011;108:19641–19646. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong W.P, Liu J, Yu J, Wang L, Zhou S.L. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyle J.J, Doyle J.L. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 11.Fazekas A.J, Burgess K.S, Kesanakurti P.R, Graham S.W, Newmaster S.G, Husband B.C, Percy D.M, Hajibabaei M, Barrett S.C. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One. 2008;3:e2802. doi: 10.1371/journal.pone.0002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galimberti A, Mattia F.D, Losa A, Bruni I, Federici S, Casiraghi M, Martellos S, Labra M. DNA barcoding as a new tool for food traceability. Food Res. Intern. 2013;50:55–63. [Google Scholar]
- 13.Hall T.A. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Series. 1999;41:95–98. [Google Scholar]
- 14.Hebert D, Ratnasingham S, de Waard J.R. Barcoding animal life, cytochrome c oxidase subunit 1 divergences among closely related species. Proceed. Roy. Soc. B. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho T.N, Pringle J.S. Gentianaceae. In: Wu C.Y, Raven P.H, editors. Flora of China. Vol. 16. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press; 1995. pp. 1–139. [Google Scholar]
- 16.Hollingsworth P.M, Graham S.W, Little D.P. Choosing and using a plant DNA barcode. PLoS One. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kress W.J, Wurdack K.J, Zimmer E.A, Weigh L.A, Janzen D.H. Use of DNA barcodes to identify flowering plants. Proc. Nat. Acad. Sci. USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kress W.J, Erickson D.L. A two-locus global DNA barcode for land plants, the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2007;6:e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D.Z, Liu J.Q, Chen Z.D, Wang H, Ge X.J, Zhou S.L, Gao L.M, Fu C.X, Chen S.L. Plant DNA barcoding in China. J. Syst. E. 2011;49:165–168. [Google Scholar]
- 20.Li X.W, Yang Y, Henry R.J, Rosetto M, Wang YT, Chen S.L. Plant DNA barcoding, from gene to genome. Biological Reviews. Biol. Reviews. 2014;90(1):157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Möller M, Gao L.M, Zhang D.Q, Li D.Z. DNA barcoding for the discrimination of Eurasian yews Taxus L, Taxaceae and the discovery of cryptic species. Mol. Ecol. Resour. 2011;111:89–100. doi: 10.1111/j.1755-0998.2010.02907.x. [DOI] [PubMed] [Google Scholar]
- 22.Packer L, Gibbs J, Sheffield C, Hanner R. DNA barcoding and the mediocrity of morphology. Mol. Ecol. Resour. 2009;9:42–50. doi: 10.1111/j.1755-0998.2009.02631.x. [DOI] [PubMed] [Google Scholar]
- 23.Shokralla S, Gibbson J.F, Nikbakht H, Janzen D.H, Hallwachs W, Hajibabaei M. Next-generation DNA barcoding, using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Mol. Ecol. Resour. 2014;14:892–901. doi: 10.1111/1755-0998.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5, molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. E. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Techen N, Parveen I, Pan Z, Khan I.A. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotechnol. 2014;25:103–110. doi: 10.1016/j.copbio.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Xue C.Y, Li D.Z. Use of DNA barcode sensu lato to identify traditional Tibetan medicinal plant Gentianopsis paludosa (Gentianaceae) J. Syst. Evvol. 2011;49:267–270. [Google Scholar]
- 27.Yao H, Song J.Y, Liu C, Luo K, Han J.P, Li Y, Pang X.H, Xu H.X, Zhu YJ, Xiao PG, Chen S.L. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 2010;5:e13102. doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]


