Abstract
Background:
DNA barcoding is a widely used tool that enables rapid and accurate identification of species based on standardized DNA regions.
Materials and Methods:
In this study, potential DNA barcodes, namely three plastid regions (rbcL, trnH-psbA and matK) and one nuclear ribosomal internal transcribed spacer (ITS) were adopted for species identification of original plants of Fritillariae Cirrhosae Bulbus.
Results:
The rbcL and trnH-psbA regions showed better success rate of PCR amplification and DNA sequencing, as well as superior discriminatory ability. On the contrary, ITS region did not possess effective genetic variation and matK was faced with low success rate of sequencing. Combination of multi-loci sequences could improve identification ability of DNA barcoding. The trnH-psbA + rbcL could discriminate 25% - 100% species based on the Blast, Tree-Building and Distance methods.
Conclusion:
The potential DNA barcodes could not completely solving species identification of botanic origins of Fritillariae Cirrhosae Bulbus. In future, we should pay more attention to super-barcoding or specific barcode that enhance ability to discriminate the closely related plants.
Keywords: Fritillariae Cirrhosae Bulbus, species identification, DNA barcoding, internal transcribed spacer (ITS), traditional Chinese medicine (TCM)
Introduction
DNA barcoding is currently in use and has proved to be an effective tool that enables rapid and accurate identification of plant species using short, standardized DNA markers (Chen et al., 2014; China Plant BOL Group 2011; Hebert et al., 2003; Li et al., 2015). It has become an efficient supplement to traditional morphology-based taxonomy and enhanced relative studies, such as detecting undescribed/cryptic species, identifying medicinal plant material, food traceability etc. (Hajibabaei et al., 2007; Kress et al., 2005; Li et al., 2015). Compared with perfect performance of CO1 (cytochrome c oxidase subunit 1) in animals, the marker was not adopted to identify plants due to its low substitution rates in plant mitochondrial genome (Fazekas et al., 2008, China Plant BOL Group, 2011). In recent years, plenty of experts paid attention to screening and testing on potential plant DNA barcodes (CBOL Plant Working Group, 2009; Chase et al., 2007; China Plant BOL Group, 2011; Dong et al., 2012; Hollingsworth et al., 2011).
However, none of the available loci could work perfectly across all species. As a result, combinations of multi-locus became unavailable choice to obtain adequate discriminatory power. Specific combinations of potential DNA barcodes were debated and controversial all long. The CBOL Plant Working Group (2009) recommended a two-marker combination of plastid rbcL and matK as the core plant barcode, to be supplemented with additional markers such as plastid trnH-psbA and nuclear ribosomal internal transcribed spacer (ITS). However, universality and discriminatory ability of the core barcodes were extremely limited in many taxa, so other researchers suggested that ITS/ITS2 should be listed as one of the core DNA barcodes based on the analysis of massive data from much more species range (China Plant BOL Group, 2011; Yao et al., 2010). Moreover, super-barcodes using the complete chloroplast genome and specific barcode for closely relative plants were proposed to solve the current predicament of plant DNA barcoding in recent years (Li et al., 2015).
Fritillariae Cirrhosae Bulbus, namely “Chuanbeimu” or “Chuanbei” is an important traditional Chinese medicine that is mainly used to relieve cough and eliminate phlegm. According to the Chinese Pharmacopoeia (2010), original plants of the medicine are composed of five species, namely Fritillaria cirrhosa D. Don, F. przewalskii Maxim., F. unibracteata Hsiao et K.C.Hsia, F. delavayi Franch. and F. taipaiensis P. Y. Li. In morphology, F. delavayi is obviously different from other species; whereas, other four species are similar and share a series of morphological characters (Chen, 2000). Due to difference on botanical origins and producing areas, Fritillariae Cirrhosae Bulbus is divided into different types which have different efficacy and prices. “Songbei” is the most famous kind of “Chuanbeimu” which is mainly produced in Songpan county and adjacent areas in Sichuan province. It is dry bulbs of F. unibracteata (Liu et al., 2008).
However, “Songbei” and its botanical origin are not easy to be distinguished from other types using traditional methods. So it is necessary to develop rapid and accurate identification on Fritillariae Cirrhosae Bulbus and their original plants. Until now, many researchers carried out massive studies on molecular identification of original plants of Fritillariae Cirrhosae Bulbus and relative species (Li et al., 2003; Li et al., 2009; Luo et al., 2012; Wang et al., 2005). However, these studies did not adopt the potential DNA barcodes to identify original plants of Fritillariae Cirrhosae Bulbus.
In this study, we use potential DNA barcodes to identify the confused botanic origins of Fritillariae Cirrhosae Bulbus and explore possibility of molecular identification based on plant DNA barcoding.
Material and methods
Material sampling
In this study, 21 individuals belonging to 4 botanic origins of Fritillariae Cirrhosae Bulbus were adopted (Table 1). Fresh and clear leaves of 3 to 6 individuals for each species were sampled from wild habitats and then dried by silica gel as soon. In order to avoid sampling individuals from the same maternal, geographical distance among individuals was kept above 20m basically. All voucher specimens were identified by author and deposited in the Herbarium of Medicinal Plants and Crude Drugs of the College of Pharmacy and Chemistry, Dali University (Table 1).
Table 1.
Collecting information of all samples in this study.
| Species | Locality | Latitude/Longitude | Altitude (m) | Voucher specimen | DNA Accessions |
|---|---|---|---|---|---|
| Fritillaria przewalskii Maxim. | Ganzi, Sichuan, China | N31°33’/ E100°01’ | 3682 | ZDQ130018 | BM10-BM12 |
| Fritillaria przewalskii Maxim. | Luhuo, Sichuan, China | N31°46’/ E100°46’ | 4047 | ZDQ130029 | BM13-BM15 |
| Fritillaria unibracteata Hsiao et K.C.Hsia | Hongyuan, Sichuan, China | N32°11’/ E102°31’ | 3621 | ZDQ130030 | BM16-BM18 |
| Fritillaria unibracteata Hsiao et K.C.Hsia | Songpan, Sichuan, China | N32°53’/ E103°30’ | 3199 | ZDQ130032 | BM19-BM21 |
| Fritillaria cirrhosa D. Don | Xiangcheng, Sichuan, China | N29°06’/ E99°39’ | 3425 | ZDQ130040 | BM22-BM24 |
| Fritillaria cirrhosa D. Don | Xianggelila, Yunnan, China | N28°07’/ E99°51’ | 4258 | ZDQ130053 | BM25-BM27 |
| Fritillaria taipaiensis P. Y. Li | Wuxi, Chongqing, China | N31°38’/ E108°50’ | 2274 | ZDQZNTB | BM28-BM30 |
DNA extraction, amplification and sequencing
Total DNA was extracted from dried leaves using modified CTAB method (Doyle, 1987). Primers of DNA barcodes and reaction system were performed according to technique requirement of the China Plant BOL Group (2011). Bidirectional DNA sequencing was conducted on the ABI 3730 DNA Sequencer (Applied Biosystems, Foster City, CA, USA). Sequences that only met quality requirement of DNA barcodes were adopted in final analysis.
DNA barcode analysis
DNA sequences were assembled using the program SeqMan Pro (DNASTAR, Lasergene) and then aligned by MEGA 5.0 (Tamura et al., 2011). The alignments were adjusted manually in BioEdit version 7.1.3.0 (Hall, 1999). Gaps were treated using Simple Indel Coding (SIC) method. Three different methods (Distance, Blast and Tree-Building) were adopted to assess the ability of species identification for single-locus and their combinations (China Plant BOL Group, 2011). Due to low success rate of DNA sequencing, matK region was not used for final analysis. P-distances intra - and inter-species for all barcodes and combinations were calculated using MEGA 5.0.
For the Blast method, all sequences of the three markers and their combinations were used as query sequences, and BioEdit 7.2.5 was used to query the reference database with each sample in turn to establish whether the closest hit was the conspecific species (Hall, 1999). Moreover, Neighbor-Joining (NJ) trees were constructed using MEGA 5.0 with the recommended settings. Judged standards of successful species identification were identical to relative requirements of the China Plant BOL Group (2011). Moreover, all DNA sequences have been submitted to the GenBank database with the following accession numbers (The numbers will be added in the later).
Results
PCR and sequencing success
Primers for the four DNA regions were universally applicable to the studied individuals. All samples could be successfully PCR amplified for the DNA barcodes. The success rate of bidirectional sequencing was 100% for all markers except matK (62%) in the present study.
Alignment and variability
Length of the aligned ITS sequences was 614 bp which had 4 variable sites. The aligned trnH-psbA matrix was 372 bp long with 2 variable sites. For the rbcL matrix, the aligned sequences are 731 bp long with 2 variable sites. Due to low generality of DNA sequencing (62%), matK was not adopted for final analysis of species identification.
Species identification
To assess discriminatory ability of the DNA barcodes, three methods (Distance, Blast and Tree-Building) were performed in this study (Table 2 & Figure 2). In these methods, Tree-Building method showed the lowest success rate, especially using single DNA barcode. On the contrary, Distance method possessed relatively superior identified ability. For combination of trnH-psbA and rbcL regions, all species could be discriminated successfully by Distance method. Finally, Blast method showed relatively stable success rate for different region combinations.
Table 2.
Properties of the four potential DNA barcodes evaluated in this study.
| DNA regions | ITS | trnH-psbA | matK | rbcL |
|---|---|---|---|---|
| Universal primer | Yes | Yes | Yes | Yes |
| PCR success (%) | 100% | 100% | 100% | 100% |
| Sequencing success (%) | 100% | 100% | 62% | 100% |
| Aligned sequence length (bp) | 614 | 372 | - | 731 |
| Indel number (length in bp) | 0 | 0 | - | 0 |
| No. informative sites/variable sites | 3/4 | 2 | - | 2/2 |
| No. samples (individuals) | 21 | 21 | - | 21 |
| Mean inter-specific distance (range) | 0.0000 | 0.0024 (0.0000-0.0040) | - | 0.0010 (0.0000-0.0021) |
| Mean intra-specific distance (range) | 0.0000 | 0.0006 (0.0000-0.0016) | - | 0.0002 (0.0000-0.0008) |
Figure 1.

Original plants of Fritillariae Cirrhosae Bulbus in the present study.
(A) Fritillaria przewalskii Maxim., (B) F. unibracteata Hsiao et K.C.Hsia, (C) F. taipaiensis P. Y. Li, (D) and (E) F. cirrhosa D. Don, (F) F. delavayi Franch.
Figure 2.
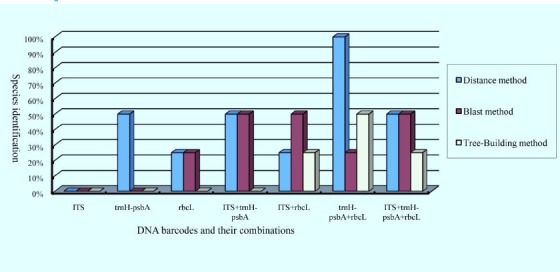
Species identification each DNA barcode and their combinations based on Distance method, Blast method and Tree-Building method.
Figure 3.
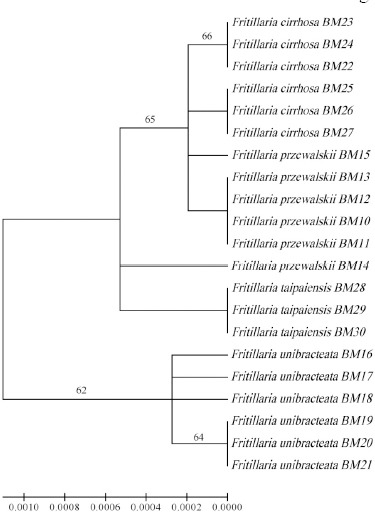
Neighbor-Joining (NJ) tree of the studied species based on combination of trnH-psbA + rbcL. Bootstrap value (>50%) were shown above the relevant branches.
For single DNA barcode, rbcL was probably the best barcode with stable success rate (25%) using Distance and the Blast method. However, ITS region could not discriminate any species though it had 4 variable sites in this study. For combinations of 2-barcodes and 3-barcodes, trnH-psbA + rbcL was the most powerful to identify these Fritillaria L. species. In the combination, 25%-100% species could be identified successfully based the three methods. ITS region did not improve discriminatory ability of trnH-psbA + rbcL, so 3-barcodes was not the best choice for identification of botanic origins of Fritillariae Cirrhosae Bulbus.
Discussion
As a famous traditional Chinese medicine, Fritillariae Cirrhosae Bulbus or “Chuanbeimu” attracts a lot of attention and it has been studied from many areas, including physiology, resources, molecular identification, chemical compositions and pharmacology etc. (Konchar et al. 2011; Li et al., 2003; Li et al., 2012; Liu et al., 2008; Luo et al., 2012; Zhang et al., 2010). The medicine includes multiple botanic origins belonging to Fritillaria L. These species are easily confused due to their similarity and complex variation in morphology (Fig.1). Meanwhile, Fritillariae Cirrhosae Bulbus, coming from different species and producing areas, are obviously different in efficacy (Yan, 2009). But it is not easy to distinguish them from each other based on morphology. As a result, difficulty of identify various Fritillariae Cirrhosae Bulbus and their original plants becomes current question waiting to be solved urgently. It is necessary to develop a quick and accurate method for identifying them.
DNA barcoding, an approach to identify species based on sequences from a short, standardized DNA region, opens up a unique avenue for the identification of organisms (China Plant BOL Group, 2011). For traditional Chinese medicines, ITS2 region is regarded as core barcodes, to be supplemented with additional markers trnH-psbA considering to probably DNA damage (Chen et al., 2010; Chen et al., 2014; Yao et al., 2010). There were a lot of studies on species identification to these medicines using DNA barcodes (Chen, 2012; Pang et al., 2013; Sun and Chen, 2013; Zhang et al., 2014). However, ITS2 marker is too short to provide enough variation so the barcode might be powerless to identify medicines with complex botanic origins. Luo et al. (2012) identified original plants of Fritillariae Cirrhosae Bulbus and its adulterants using ITS2 region. The results showed that the barcode could distinguish original species of the genuine from that of the adulterants but it did not discriminate these original plants of Fritillariae Cirrhosae Bulbus. Meanwhile, other researchers also identified certified products of “Chuanbeimu” from adulterants and spurious breeds using different molecular methods (Li et al., 2003; Li et al, 2009; Su et al., 2014). However, these molecular tools could not efficiently identify botanic origins of Fritillariae Cirrhosae Bulbus.
In the present study, the ability of potential DNA barcodes was still limited to discriminate the original plants, especially single marker (Fig.2). In most of plant taxa, ITS and ITS2 showed strong ability of species identification (China Plant BOL Group, 2011; Yao et al., 2010), but it was useless in Fritillaria L. species. On the contrary, chloroplast regions were much better in success rate of species identification for original plants of Fritillariae Cirrhosae Bulbus, except matK region. Although matK was proposed as one of the core barcodes, it was not an ideal region in many group due to low success rate of PCR amplifying and DNA sequencing (Zhang et al., 2014). RbcL was useful barcode across most of taxa due to good universality in despite of limited variation (Chase et al., 2007; China Plant BOL Group, 2011). Compared with rbcL, trnH-psbA was probably more effective to identify species. In this study, combination of trnH-psbA + rbcL possessed the best discriminatory ability for original plants of Fritillariae Cirrhosae Bulbus (25%-50% for the three methods). However, these potential DNA barcodes and their combinations could not discriminate all original plants of Fritillariae Cirrhosae Bulbus.
In my opinion, there might be two main reasons on difficulty of identifying these species. On the one hand, SW China is the main center of distribution and diversification of Fritillaria L. in China, and the taxa forms a complex group including a series of species with complicated morphological variation, namely complex group of F. cirrhosa D. Don (Luo and Chen, 1996; Xiao et al., 2007; Zhang et al., 2010). On the other hand, Fritillaria L. species have the largest genome sizes known in diploid plants that vary between 30.15 and 85.38 GB (Kelly et al., 2015; Leitch et al., 2007). The giant genomes in Fritillaria might be an important reason for awful species identification of DNA barcodes in this study. The reliable evidence was that identified ability of DNA barcodes was not ideal in most of taxa belonging to Liliaceae in which genome sizes are generally huge (Leitch et al., 2007).
For such a complex but important taxa, super-barcodes using complete chloroplast genome and specific barcodes for Fritillaria L. species might be new choices that improve identified ability on closely related plants at the species level (Li et al., 2015).
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant no. 31200180) and the General Program of Yunnan Provincial Department of Education (Grant no. 2014Z134).We also thank Junbo Yang and Jing Yang in Kunming Institution of Botany (CAS) for their help in molecular experiments.
References
- 1.CBOL Plant Working Group. A DNA barcode for land plants. PNAS. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chase M.W, Cowan R.S, Hollingsworth P.M. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007;56:295–299. [Google Scholar]
- 3.Chen S.C, Mordak H.V. Fritillaria L. In: Wu Z.Y, Raven P.H, editors. Flora of China. Vol. 24. Science Press/ Missouri Botanical Garden Press; Beijing/St. Louis: 2000. pp. 127–133. [Google Scholar]
- 4.Chen S.L. DNA barcoding of traditional Chinese medicine: molecular identification. Beijing: People’s Health Publishing House; 2012. [Google Scholar]
- 5.Chen S.L, Pang X.H, Song J.Y, Shi L.C, Yao H, Han J.P. A renaissance in herbal medicine identification: from morphology to DNA. Biotech. Advan. 2014;32:1237–1244. doi: 10.1016/j.biotechadv.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Chen S.L, Yao H, Han J.P, Liu C, Song J.Y, Shi L.C. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.China Pllant BOL Group. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. PNAS. 2011;108:19641–19646. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinse Pharmacopoeia Committee. Chinese Pharmacopoeia. 1st part. Beijing: China Medical Science and Technology Press; 2010. [Google Scholar]
- 9.Dong W.P, Liu Yu J, Wang J. L, Zhou S.L. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PloS ONE 7. 2012:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyle J.J, Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry. 1987:11–15. [Google Scholar]
- 11.Fazekas A.J, Burgess K.S, Kesanakurti P.R, Graham S.W, Newmaster S.G, Husband B.C. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE. 2008;3:e2802. doi: 10.1371/journal.pone.0002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajibabaei M, Singer G.A.C, Hebert P.D.N, Hickey D.A. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007;23:167–172. doi: 10.1016/j.tig.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 14.Hebert P.D, Ratnasingham S, de Waard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. PNAS. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollingsworth P.M, Graham S.W, Little D.P. Choosing and using a plant DNA barcode. PloS ONE. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly L.J, Renny N, Byfield S, Pellicer J, Macas J, Novák P, Neumann P. Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytologist (Online version) 2015 doi: 10.1111/nph.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konchar K, Li X.L, Yang Y.P, Emshwiller E. Phytochemical variation in Fritillaria cirrhosa D. Don (Chuan Bei Mu) in relation to plant reproductive stage and timing of harvest. Econ. Bot. 2011;65:283–294. [Google Scholar]
- 18.Kress W.J, Wurdack K.J, Zimmer E.A, Weigh L.A, Janzen D.H. Use of DNA barcodes to identify flowering plants. PNAS. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leitch I.J, Beaulieu J.M, Cheung K, Hanson L, Lysak M.A, Fay M.F. Punctuated genome size evolution in Liliaceae. J Evol. Biol. 2007;20:2296–2308. doi: 10.1111/j.1420-9101.2007.01416.x. [DOI] [PubMed] [Google Scholar]
- 20.Li K.Q, Wu W, Zheng Y.L, Dai Y, Xiang L, Liao K. Genetic diversity of Fritillaria from Sichuan province based on ISSR. China J. Chin. Mat. Med. 2009;34:2149–2154. [PubMed] [Google Scholar]
- 21.Li X.W, Song J.Y, Wei J.H, Hu Z.G, Xie C.X, Luo G.A. Natural fostering in Fritillaria cirrhosa integrating herbal medicine production with biodiversity conservation. Acta Pharmcol. Sin. B. 2012;2:77–82. [Google Scholar]
- 22.Li X.W, Yang Y, Henry R.J, Rosetto M, Wang Y.T, Chen S.L. Plant DNA barcoding: from gene to genome. Biol. Rev. 2015;90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 23.Li YF, Li Y.X, Lin J, Xu Y, Yan F, Tang L. Identification of bulb from Fritillaria cirrhosa by PCR with specific primers. Planta Med. 2003;69:186–188. doi: 10.1055/s-2003-37699. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Chen S.L, Yao H, Li X.W, Zhang Y. Research progress on resources in Bulbus Fritillariae Cirrhosae. China J. Chin. Mat. Med. 2008;33:1645–1648. [PubMed] [Google Scholar]
- 25.Luo K, Ma P, Yao P. H, Song J.Y, Chen K.L, Liu YM. Molecular identification of Frtillariae Cirrhosae Bulbus and its adulterants. World Sci. Technol.: Moderniz. Tradit. Chin. Medic. Mat. Med. 2012;14:1153–1158. [Google Scholar]
- 26.Luo Y.B, Chen S.C. A revision of Fritillaria L. (Liliaceae) in the Hengduan mountais and adjacent regions, China (I) - A study of Frtillaria cirrhosa D.Don and its related species. Acta Phytotax. Sin. 1996;34:304–312. [Google Scholar]
- 27.Pang X.H, Shi L.C, Song J.Y, Chen XC, Chen S.L. Use of the potential DNA barcode ITS2 to identify herbal materials. J. Nat. Med. 2013;67:571–575. doi: 10.1007/s11418-012-0715-2. [DOI] [PubMed] [Google Scholar]
- 28.Su P, Hu L, Dong P.L. Identification of eight varieties of bulbus Fritillaria cirrhosa. Southwest China J. Agricult. Sci. 2014;27(6):2559–2563. [Google Scholar]
- 29.Sun Z, Chen S. Identification of cortex herbs using the DNA barcode nrITS2. J. Nat. Med. 2013;67:296–302. doi: 10.1007/s11418-012-0681-8. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, P. D., Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C.Z, Li P, Ding J. Y, Jin G. Q, Yuan C.S. Identification of Fritillariapallidiflora using diagnostic PCR and PCR-RFLP based on nuclear ribosomal DNA internal transcribed spacer sequences. Planta Med. 2005;71:384–386. doi: 10.1055/s-2005-864112. [DOI] [PubMed] [Google Scholar]
- 32.Xiao P.G, Jiang Y, Li P, Luo YB, Liu Y. The botanical origin and pharmacophylogentic treatment of Chinese materia medica Beimu. Acta Phytotax. Sin. 2007;45:473–487. [Google Scholar]
- 33.Yan B.H. The research on the functional difference among different species of Chuan Fritillaria in relieving cough and resolving phlegm. Chengdu: Chengdu University of Traditional Chinese Medicine; 2009. [Google Scholar]
- 34.Yao H, Song J.Y, Liu C, Luo K, Han J.P, Li Y. Use of ITS2 region as the universal DNA barcode for plants and animals. PloS ONE. 2010;5:e13102. doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D. Q, Gao L.M, Yang Y.P. Genetic diversity and structure of a traditional Chinese medicinal plant species Fritillaria cirrhosa (Liliaceae) in southwest China and implications for its conservation. Bioch. Syst. Ecol. 2010;38:236–242. [Google Scholar]
- 36.Zhang D.Q, Duan L.Z, Zhou N. Application of DNA barcoding in Roscoea (Zingiberaceae) and a primary discussion on taxonomic status of Roscoea cautleoides var pubescens. Bioch. Syst. Ecol. 2014;52:14–19. [Google Scholar]
- 37.Zhang D.Q, Yang Y.S, Jiang B, Duan L.Z, Zhou N. How to correctly identify herbal materials in market: a case study based on DNA barcodes. Afr. J. Tradit. Complement. Altern. Med. 2014;11:66–76. [Google Scholar]


