Abstract
Camel milk is consumed in the Middle East because of its high nutritional value. Traditional heating methods and the duration of heating affect the protein content and nutritional quality of the milk. We examined the denaturation of whey proteins in camel milk by assessing the effects of temperature on the whey protein profile at room temperature (RT), moderate heating at 63 °C, and at 98 °C, for 1 h. The qualitative and quantitative variations in the whey proteins before and after heat treatments were determined using quantitative 2D-difference in gel electrophoresis (DIGE)-mass spectrometry. Qualitative gel image analysis revealed a similar spot distribution between samples at RT and those heated at 63 °C, while the spot distribution between RT and samples heated at 98 °C differed. One hundred sixteen protein spots were determined to be significantly different (p < 0.05 and a fold change of ≥1.2) between the non-heated and heated milk samples. Eighty protein spots were decreased in common in both the heat-treated samples and an additional 25 spots were further decreased in the 98 °C sample. The proteins with decreased abundance included serum albumin, lactadherin, fibrinogen β and γ chain, lactotransferrin, active receptor type-2A, arginase-1, glutathione peroxidase-1 and, thiopurine S, etc. Eight protein spots were increased in common to both the samples when compared to RT and included α-lactalbumin, a glycosylation-dependent cell adhesion molecule. Whey proteins present in camel milk were less affected by heating at 63 °C than at 98 °C. This experimental study showed that denaturation increased significantly as the temperature increased from 63 to 98 °C.
Keywords: camel milk, whey protein, heat treatment, 2D-DIGE, proteomics, matrix-assisted laser desorption/ionization-Time of Flight (MALDI-TOF)
1. Introduction
The one-humped camel (Camelus dromedarius) lives in the arid and semi-arid desert regions of the Middle East and produces a considerable amount of milk despite this hot and dry environment. It is thus a valuable source of nutrition, providing both milk and meat in these areas of the world [1,2].
Camel milk is popular locally and is mainly consumed as fresh raw milk or as soured milk [1,2]. Its characteristics and composition are attributed to many factors such as the age and breed of the camel, health of the animal, feeding conditions, stage of lactation, calving number, season, and geographical location; the latter two are the most important factors [1,3,4]. High amounts of active compounds, critical for the nutritional requirements of human neonates and adults, have been identified in camel milk [5].
Camel milk is mainly composed of lipids, proteins, lactose (as the major oligosaccharide), ash or minerals, numerous vitamins, essential amino acids, nucleotides, and other metabolites [6,7]. An extensive review by Konuspayeva (1993–2006) provides the mean values of the components as: fat matter 3.78 ± 1.31, total protein 3.19 ± 0.60, dry matter 11.98 ± 1.78, lactose 4.34 ± 0.54, and ash 0.81 ± 0.08 [8]. More recently, it was also reported to possess various health and therapeutic benefits, including antibacterial, immunological, anti-carcinogenic, anti-diabetic, and anti-hypertensive properties. Because of its low allergenic properties, camel milk is recommended for consumption for neonates and children allergic to bovine milk [1].
The protein in the milk is classified into three broad classes: colloidal casein, soluble whey, and milk fat globule membrane proteins (MFGMP) [9,10]. Proteins are highly heat labile and easily undergo conformational changes with increasing temperatures [11]. Heat treatment of milk is an essential step, performed domestically and industrially, to render milk safe for human consumption and improve its shelf life [12]. Heat treatment methods include thermization, extended pasteurization at low temperature, rapid pasteurization at high temperature, and sterilization at an ultra-high temperature [13,14]; pasteurization is the method of choice. However, in many rural areas, people consume milk after boiling it using charcoal or gas [15]. This practice can cause inefficient killing of microorganisms or deterioration in the quality of milk when excessive heat is applied.
As thermal treatment is the major step involved in the processing of milk and milk products, its effects on total milk and on whey proteins has been extensively studied. Several immune-active compounds identified in bovine, camel, caprine, and human milk are reduced after pasteurization [16,17,18,19].
Thus far only a few studies have investigated the effect of heat treatment on camel milk whey protein and a comprehensive proteome of camel milk and of proteins secreted in the lactating mammary gland of camels is not yet available [10]. There are limited studies that have examined the sameusing proteomic approach. Felfoul et al. have recently used a semi-quantitative-based one dimensional-gel electrophoresis (1D-GEL) and LC-MS/MS approach, at 80 °C for 1 h to study these changes [20].
We have thus carried out this study to determine quantitatively the changes in dromedary camel whey protein with relation to temperature using two different heating strategies, namely heating at 63 and 98 °C in comparison to room temperature, in order to identify proteins affected or stable by heating using a complementary proteomics and 2D-DIGE mass spectrometry approach.
2. Results and Discussion
The effects of heat on the whey proteins present in camel milk were examined using two-dimensional differential gel electrophoresis and matrix-assisted laser desorption ionization-time of flight mass spectrometry (2D-DIGE-MALDI/TOF). We used quantitative proteomics to obtain an overall representation of the changes in the profile of acid whey proteins present in camel milk after heating. Earlier studies on camel milk employed a targeted approach, studying a fraction, or a specific protein; no comprehensive profiling is currently available for the entire proteome. In this study, we found that a number of whey proteins were differentially affected by heating, with distinct changes noted at the different selected temperatures.
2.1. Overall Changes in Whey Proteins, Caused by Heating, Shown by 2D-DIGE
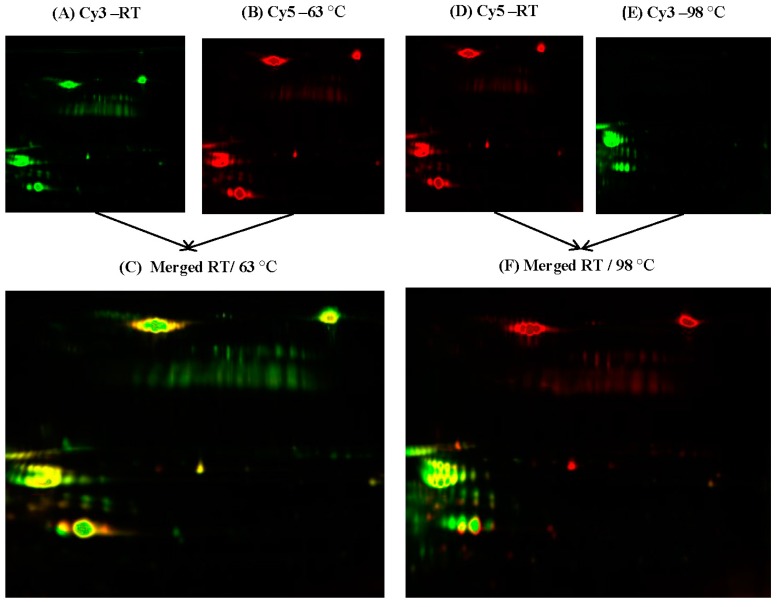
The 2D-DIGE experiments were carried out in triplicate and yielded reproducible spot patterns for all the milk samples, i.e., those at—RT, 63 and 98 °C. Approximately 1300 spots were mapped to the gels. The protein spots for the samples at room temperature (red/green), those at 63 °C (red), and those at 98 °C (green) were compared (Figure 1A,B,D,E). The gel images showed a high degree of protein similarity between the RT and those heated at 63 °C, as shown by the number of yellow spots (Figure 1C); protein similarity was minimal between the samples maintained at room temperature and those heated at 98 °C (Figure 1F); yellow spots represent proteins with the same isoelectric point, molecular weight, and nearly equal protein fluorescence intensity. Statistical analysis of the gels was conducted using the Progenesis statistical software v.3.3 (Nonlinear Dynamics, Newcastle, UK). One hundred and ninety protein spots, exhibiting a fold change of ≥1.2, and ANOVA p-value ≤ 0.05, were chosen for identification via mass spectrometry.
Figure 1.
Two-dimensional fluorescence difference gel electrophoresis (2D-DIGE) image showing heat-treated and non-heat-treated samples of camel milk. Whey proteins were focused on linear IPG (immobilized pH gradient) strips (pH 3–11, 24 cm) and then separated using 12.5% polyacrylamide gels. Individual 2D-DIGE gel images of camel milk samples: (A) Cy3 and (D) Cy5 represent non-heated samples (at room temperature); gel images (B) Cy5 and (E) Cy3 represent samples heated at 63 and 98 °C, respectively; images (C,F) represent a channel overlap image between Cy3 and Cy5.
2.2. PCA Analysis
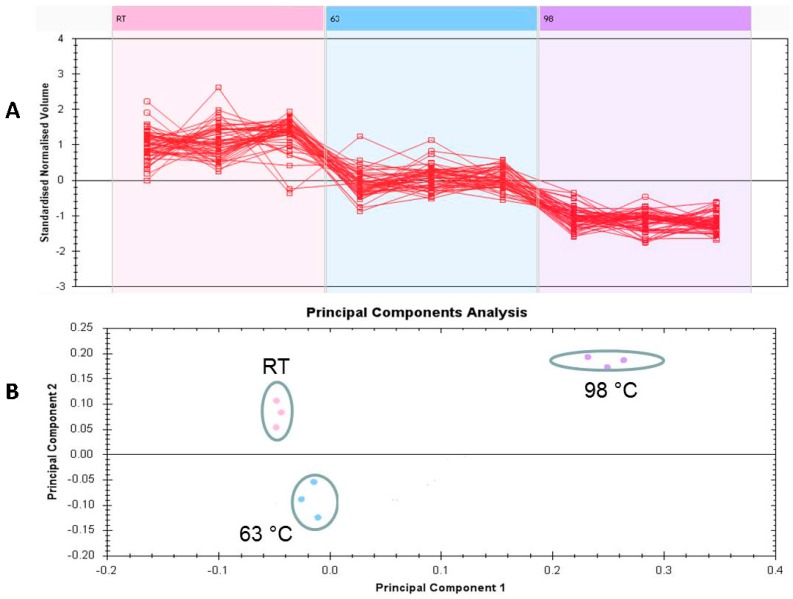
The unsupervised principal component analysis (PCA) bi-plot of gels and spots (Figure 2B) shows distinct gel grouping that agrees with the experimental groups. PCA plot of the two first principal components explained 85.15% of the selected spot’s variability within the three groups. The differentially abundant spots showed expression pattern clusters according to their abundant patterns based on a hierarchical clustering analysis (Figure 2A). The clustering pattern showed minimum amount of change in the protein intensities for the selected spots between RT and 63 °C while the difference for the same spots between RT and 98 °C was higher and significantly different.
Figure 2.
(A) Expression profiles, separated into clusters of expression patterns, indicating the number of spots for each cluster. Each line represents the standardized abundance of a spot across all gels and belongs to one of the clusters generated by hierarchical cluster analysis (Progenesis Same Spots); and (B) PCA plot of the first two principal components showing 85.15% variability of the selected spots. Colored dots and numbers represent gels and spots, respectively.
2.3. MALDI-TOF-MS
The 190 protein spots of interest were excised from the preparative 2-D gels. After trypsin digestion, the digested spots were analyzed by MALDI-TOF-MS; then, 116 protein spots, isolated using MALDI-TOF-MS analysis, were successfully identified using the MASCOT Swiss Prot database. The database search results, with the coverage and score, are listed in Table S1. Thirty-one proteins were successfully matched with high certainty with entries in the C. dromedarius database, and an additional 85 assignments were made by matching to known homologous peptides identified in other mammalian databases (Table 1). Not all spots of interest could be identified because some proteins were low in abundance and did not yield sufficiently intense mass of fingerprints; other spots were mixtures of multiple proteins.
Table 1.
Identified proteins, with changes in abundance, after application of heat treatment at 63 and 98 °C, compared with RT (room temperature). Table shows average ratio values for 63 °C/room temperature and 98 °C/room temperature, with their corresponding levels of fold changes and one-way ANOVA (p-value ≤ 0.05).
| Spot Number | Accession Number | Protein Name | Function of Protein | ANOVA (p) | Fold Change | |||
|---|---|---|---|---|---|---|---|---|
| 63 °C/RT | Expression | 98 °C /RT | Expression | |||||
| 219 | Q9TUM0 | Lactotransferrin | Enzyme | 0.001 | −1.251 | Down | −13.731 | Down |
| 242 | P31428 | Dipeptidase 1 | Enzyme | 0.005 | −1.752 | Down | −4.361 | Down |
| 247 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | −1.505 | Down | −5.241 | Down |
| 253 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | −1.348 | Down | −4.250 | Down |
| 255 | P01009 | α-1-Antitrypsin | Enzyme | 0.001 | −1.187 | Down | −4.648 | Down |
| 256 | P02647 | Apolipoprotein A-I | Binding protein | 0.001 | 1.003 | Non-significant | −2.337 | Down |
| 270 | P02675 | Fibrinogen β chain | Enzyme | 0.001 | 2.152 | Down | −6.851 | Down |
| 276 | Q8CG71 | Prolyl 3-hydroxylase 2 | Enzyme | 0.030 | −1.690 | Down | −3.460 | Down |
| 280 | P02679 | Fibrinogen γ chain | Enzyme | 0.003 | −2.042 | Down | −4.523 | Down |
| 285 | P02679 | Fibrinogen γ chain | Enzyme | 0.004 | −1.451 | Down | −2.023 | Down |
| 288 | Q32KP7 | Uncharacterized protein C17orf64 homolog | Others | 0.000 | −1.264 | Down | −2.313 | Down |
| 309 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | −1.053 | Down | −2.309 | Down |
| 316 | Q9TUM0 | Lactotransferrin | Enzyme | 0.004 | −1.187 | Down | −1.709 | Down |
| 318 | P79385 | Lactadherin | Cell adhesion | 0.000 | −1.103 | Down | −2.633 | Down |
| 333 | P02768 | Serum albumin | Enzyme | 0.003 | −1.466 | Down | −4.597 | Down |
| 340 | Q9TUM0 | Lactotransferrin | Enzyme | 0.001 | −1.421 | Down | −5.242 | Down |
| 338 | Q9TUM0 | Lactotransferrin | Enzyme | 0.001 | −1.421 | Down | −5.242 | Down |
| 339 | Q9TUM0 | Lactotransferrin | Enzyme | 0.013 | −1.483 | Down | −4.500 | Down |
| 343 | Q9TUM0 | Lactotransferrin | Enzyme | 0.033 | −1.512 | Down | −4.958 | Down |
| 348 | Q9TUM0 | Lactotransferrin | Enzyme | 0.014 | −1.417 | Down | −8.383 | Down |
| 353 | Q9TUM0 | Lactotransferrin | Enzyme | 0.003 | −1.297 | Down | −10.836 | Down |
| 357 | Q9TUM0 | Lactotransferrin | Enzyme | 0.001 | −1.291 | Down | −11.745 | Down |
| 358 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | 1.148 | Non-significant | −22.961 | Down |
| 359 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | 1.031 | Non-significant | −44.852 | Down |
| 360 | P15539 | Cytochrome P450 11B2, mitochondrial | Enzyme | 0.004 | −1.243 | Down | −9.594 | Down |
| 361 | Q6IML7 | DnaJ homolog subfamily C member 27 | Binding protein | 0.007 | −1.222 | Down | −6.650 | Down |
| 368 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | −1.060 | Down | −67.105 | Down |
| 373 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | 1.042 | Non-significant | −45.000 | Down |
| 384 | P02750 | Leucine-rich α-2-glycoprotein | Binding protein | 0.001 | −1.138 | Down | −27.366 | Down |
| 396 | Q9GK12 | Peptidoglycan recognition protein 1 | Binding protein | 0.001 | 1.044 | Non-significant | −26.553 | Down |
| 398 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | 1.095 | Non-significant | −36.065 | Down |
| 400 | Q7Z713 | Ankyrin repeat domain-containing protein 37 | Binding protein | 0.007 | 1.295 | Non-significant | −11.880 | Down |
| 407 | Q9H3Z7 | Protein ABHD16B | Binding protein | 0.000 | 1.307 | Non-significant | −15.143 | Down |
| 409 | Q8MJ14 | Glutathione peroxidase 1 | Enzyme | 0.000 | 1.231 | Non-significant | −13.938 | Down |
| 413 | P02790 | Hemopexin | Transport protein | 0.003 | 1.258 | Up | −16.162 | Down |
| 433 | Q9TUM0 | Lactotransferrin | Enzyme | 0.008 | 1.039 | Non-significant | −6.116 | Down |
| 438 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | 1.115 | Non-significant | −10.873 | Down |
| 440 | Q8R0F8 | Acylpyruvase FAHD1, mitochondrial | Enzyme | 0.000 | −1.419 | Down | −1.708 | Down |
| 454 | Q5E9H8 | Probable G-protein coupled receptor 173 | Enzyme | 0.001 | −1.213 | Down | −29.425 | Down |
| 459 | Q9TUM0 | Lactotransferrin | Enzyme | 0.002 | −1.257 | Down | −15.872 | Down |
| 462 | P01876 | Ig α-1 chain C region | Immune response protein | 0.001 | 1.117 | Non-significant | −34.941 | Down |
| 472 | Q96SZ5 | 2-Aminoethanethiol dioxygenase | Enzyme | 0.010 | −1.358 | Down | −2.038 | Down |
| 474 | Q9TUM0 | Lactotransferrin | Enzyme | 0.002 | −1.363 | Down | −15.500 | Down |
| 492 | Q95KI3 | Endothelial cell-selective adhesion molecule | Cell adhesion | 0.011 | −1.374 | Down | −3.065 | Down |
| 500 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | 1.156 | Non-significant | −7.593 | Down |
| 501 | P00435 | Glutathione peroxidase 1 | Enzyme | 0.035 | −1.218 | Down | −3.432 | Down |
| 516 | Q6GPH4 | XIAP-associated factor 1 | Binding protein | 0.006 | −1.068 | Down | −5.456 | Down |
| 517 | Q8NBT0 | POC1 centriolar protein homolog A | Binding protein | 0.023 | −1.460 | Down | −2.404 | Down |
| 539 | P70097 | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial | Enzyme | 0.002 | 1.140 | Non-significant | −1.760 | Down |
| 287 | P02679 | Fibrinogen γ chain | Enzyme | 0.003 | −2.042 | Down | −4.523 | Down |
| 543 | Q68G74 | LIM/homeobox protein Lhx8 | Binding protein | 0.009 | −1.284 | Down | −3.709 | Down |
| 540 | Q68G74 | LIM/homeobox protein Lhx8 | Binding protein | 0.009 | −1.284 | Down | −4.609 | Down |
| 546 | Q6AYB4 | Heat shock 70 kDa protein 14 | Immune response protein | 0.008 | −1.993 | Down | −2.279 | Down |
| 548 | Q6AYB4 | Heat shock 70 kDa protein 14 | Immune response protein | 0.007 | −1.793 | Down | −3.35 | Down |
| 553 | Q2KJ64 | Arginase-1 | Enzyme | 0.009 | −2.488 | Down | −2.541 | Down |
| 555 | Q2KJ64 | Arginase-1 | Enzyme | 0.087 | −2.47 | Down | −2.81 | Down |
| 561 | P79385 | Lactadherin | Cell adhesion | 0.013 | −1.910 | Down | −6.104 | Down |
| 566 | P79385 | Lactadherin | Cell adhesion | 0.002 | −1.510 | Down | −8.104 | Down |
| 562 | P79385 | Lactadherin | Cell adhesion | 0.000 | −1.480 | Down | −11.000 | Down |
| 563 | P79385 | Lactadherin | Cell adhesion | 0.001 | −1.580 | Down | −6.000 | Down |
| 564 | P79385 | Lactadherin | Cell adhesion | 0.000 | −2.652 | Down | −14.107 | Down |
| 560 | P79385 | Lactadherin | Cell adhesion | 0.000 | −2.022 | Down | −12.007 | Down |
| 568 | P46065 | Guanylyl cyclase-activating protein 1 | Enzyme | 0.000 | −1.742 | Down | −8.144 | Down |
| 570 | P46065 | Guanylyl cyclase-activating protein 1 | Enzyme | 0.000 | −1.82 | Down | −9.100 | Down |
| 573 | P06323 | T-cell receptor α chain V region CTL-F3 | Immune response protein | 0.05 | −1.417 | Down | −14.836 | Down |
| 576 | P06323 | T-cell receptor α chain V region CTL-F3 | Immune response protein | 0.002 | −1.357 | Down | −10.35 | Down |
| 572 | P06323 | T-cell receptor α chain V region CTL-F3 | Immune response protein | 0.004 | −2.17 | Down | −8.572 | Down |
| 574 | P79385 | Lactadherin | Cell adhesion | 0.000 | −2.304 | Down | −18.431 | Down |
| 579 | P79385 | Lactadherin | Cell adhesion | 0.000 | −2.65 | Down | −13.251 | Down |
| 578 | Q9TUM0 | Lactotransferrin | Enzyme | 0.003 | −1.357 | Down | −17.075 | Down |
| 580 | Q9TUM0 | Lactotransferrin | Enzyme | 0.000 | −1.564 | Down | −15.005 | Down |
| 582 | P04217 | α-1B-glycoprotein | Enzyme | 0.002 | −1.572 | Down | −2.461 | Down |
| 584 | P04217 | α-1B-glycoprotein | Enzyme | 0.015 | −1.980 | Down | −3.542 | Down |
| 585 | Q9QYE2 | Solute carrier organic anion transporter family member 1A6 | Binding protein | 0.000 | 1.036 | Non-significant | −9.092 | Down |
| 586 | Q9QYE2 | Solute carrier organic anion transporter family member 1A6 | Binding protein | 0.003 | 1.259 | Non-significant | −7.058 | Down |
| 587 | Q6AYB4 | Heat shock 70 kDa protein 14 | Immune response protein | 0.000 | −1.365 | Down | −16.099 | Down |
| 590 | Q6AYB4 | Heat shock 70 kDa protein 14 | Immune response protein | 0.000 | −1.572 | Down | −12.557 | Down |
| 588 | Q28560 | Activin receptor type-2A | Binding protein | 0.007 | −2.822 | Down | −16.337 | Down |
| 592 | Q28560 | Activin receptor type-2A | Binding protein | 0.000 | −2.092 | Down | −11.777 | Down |
| 594 | Q2KJ64 | Arginase-1 | Enzyme | 0.000 | −1.917 | Down | −14.343 | Down |
| 593 | P34896 | Serine hydroxymethyltransferase, cytosolic | Enzyme | 0.000 | −1.717 | Down | −16.343 | Down |
| 595 | Q13595 | Transformer-2 protein homolog α | Binding protein | 0.002 | −1.295 | Down | −8.503 | Down |
| 600 | Q13595 | Transformer-2 protein homolog α | Binding protein | 0.01 | −2.3 | Down | −6.3 | Down |
| 596 | P02675 | Fibrinogen β chain | Enzyme | 0.002 | −1.154 | Down | −7.207 | Down |
| 599 | P02675 | Fibrinogen β chain | Enzyme | 0.005 | −2.12 | Down | −5.02 | Down |
| 597 | P70097 | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial | Enzyme | 0.000 | −2.043 | Down | −15.508 | Down |
| 604 | Q68SB1 | Double-stranded RNA-binding protein Staufen homolog 2 | Binding protein | 0.001 | −1.002 | Down | −1.651 | Down |
| 615 | P00738 | Haptoglobin | Transport protein | 0.001 | 1.011 | Non-significant | −3.377 | Down |
| 616 | P00747 | Plasminogen | Enzyme | 0.001 | 1.008 | Non-significant | −3.657 | Down |
| 621 | P32261 | Antithrombin-III | Enzyme | 0.000 | 1.030 | Non-significant | −5.271 | Down |
| 629 | Q62803 | Hyaluronidase PH-20 | Enzyme | 0.000 | −1.025 | Down | −8.901 | Down |
| 644 | Q91Z85 | Steroid 17-α-hydroxylase/17,20 lyase | Enzyme | 0.000 | 1.079 | Non-significant | −10.522 | Down |
| 662 | P16261 | Graves disease carrier protein/Mitochondrial solute carrier protein homolog | Enzyme | 0.000 | 1.013 | Non-significant | −14.205 | Down |
| 664 | Q8HX86 | Thiopurine S-methyltransferase | Enzyme | 0.000 | 1.020 | Non-significant | −15.146 | Down |
| 723 | P10909 | Clusterin | Binding protein | 0.001 | −1.132 | Down | −2.792 | Down |
| 769 | Q9NSQ0 | Putative ribosomal RNA-processing protein 7 homolog B | Binding protein | 0.000 | −1.024 | Down | −2.642 | Down |
| 793 | O14791 | Apolipoprotein L1 | Binding protein | 0.001 | 1.371 | Non-significant | −2.684 | Down |
| 796 | Q3MID3 | ADP-ribosylation factor GTPase-activating protein 2 | Binding protein | 0.002 | 1.280 | Up | −2.509 | Down |
| 803 | P15169 | Carboxypeptidase N catalytic chain | Enzyme | 0.003 | 1.213 | Non-significant | −4.831 | Down |
| 819 | Q62803 | Hyaluronidase PH-20 | Enzyme | 0.000 | −1.264 | Down | −3.213 | Down |
| 827 | Q9UHR4 | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 | Binding protein | 0.000 | −1.161 | Down | −3.327 | Down |
| 828 | Q9CTN5 | Protein SIX6OS1 | Binding protein | 0.000 | −1.327 | Down | −4.083 | Down |
| 832 | Q3UR70 | Transforming growth factor-β receptor-associated protein 1 | Binding protein | 0.000 | −1.417 | Down | −3.559 | Down |
| 847 | P24310 | Cytochrome c oxidase subunit 7A1, mitochondrial | Enzyme | 0.001 | −1.507 | Down | −3.142 | Down |
| 849 | Q8NBP0 | Tetratricopeptide repeat protein 13 | Enzyme | 0.000 | −1.565 | Down | −4.503 | Down |
| 856 | Q29563 | l-lactate dehydrogenase C chain | Enzyme | 0.036 | 1.923 | Up | −1.445 | Down |
| 917 | P15522 | Glycosylation-dependent cell adhesion molecule 1 | Enzyme | 0.000 | 1.249 | Up | 2.805 | Up |
| 985 | P15522 | Glycosylation-dependent cell adhesion molecule 1 | Enzyme | 0.000 | 1.283 | Up | 3.925 | Up |
| 1013 | P15522 | Glycosylation-dependent cell adhesion molecule 1 | Enzyme | 0.000 | 1.345 | Up | 2.301 | Up |
| 1041 | O14618 | Copper chaperone for superoxide dismutase | Others | 0.060 | 1.535 | Up | 1.597 | Up |
| 1069 | P00710 | α-Lactalbumin | Enzyme | 0.000 | 1.391 | Up | 11.925 | Up |
| 1090 | P00710 | α-Lactalbumin | Enzyme | 0.001 | 1.321 | Up | 4.790 | Up |
| 1105 | P00710 | α-Lactalbumin | Enzyme | 0.014 | 1.831 | Up | 2.547 | Up |
| 1147 | Q8NFU3 | Thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 | Enzyme | 0.029 | −1.661 | Down | −2.326 | Down |
| 1155 | Q86X95 | Corepressor interacting with RBPJ 1 | Enzyme | 0.002 | −1.389 | Down | −3.541 | Down |
| 1167 | A8K979 | ERI1 exoribonuclease 2 | Enzyme | 0.000 | 1.196 | Up | 15.624 | Up |
2.4. Changes in Abundance of Proteins after Application of Different Heating Strategies
Heat treatment of milk subjects it to the highly complex Maillard reaction, which greatly affects the structure and properties of its constituents including the whey proteins [21]. The reaction commonly occurs between the milk sugar, lactose, and the lysine residues of the milk proteins, leading to formation of large high molecular weight aggregates. This reaction is mostly seen to take place between the different casein fractions and lactoglobulin. β-Lactoglobulin is present in the milk of other dairy animals, such as the cow and the buffalo, but is characteristically absent from camel milk; this renders the composition of camel milk similar to that of human milk and accounts for the decreased allergenic property of camel milk. Camel milk is also known to have a lower amount of lactose in comparison to bovine milk. The major whey proteins in camel milk are α-lactoglobulin, lactoferrin, lactoperoxidase, serum albumin, immunoglobulin G, and secretory immunoglobulin A. Thermal denaturing, and aggregation or gelation, in the bovine milk has been extensively studied, while only a few studies have examined it in camel milk [16,22]. We found that the major fraction of proteins affected by heat treatment included 61% enzymes, 20% binding proteins, 10% cell adhesion proteins, 5% proteins involved in the immune response, 2% transport proteins, and 2% others.
2.5. Proteins That Showed Decreased Abundance after Heating at 63 or 98 °C, Compared to Room Temperature
We found that a total of 80 protein spots, corresponding to samples heated at 63 and 98 °C, were decreased, with an appreciable difference in fold change. Statistical analysis revealed a significant decrease in the levels of these spots, which related to proteins including: lactotransferrin, spot #348 (fold change of −1.417/−8.383 at 63/98 °C, respectively); lactadherin, spot #564 (fold change of −2.652/−14.107 at 63/98 °C, respectively); serum albumin, spot #333 (fold change of −1.466/−4.597 at 63/98 °C, respectively); cytochrome P450 11B2 mitochondrial, spot # 360 (fold change of −1.243/−9.594 at 63/98 °C respectively); arginase-1, spot #553 (fold change of −2.488/−2.541 at 63/98 °C, respectively); heat shock 70 kDa protein 14, spot #587 (fold change of −1.365/−16.099 at 63/98 °C, respectively); succinate dehydrogenase cytochrome b560 subunit, spot #597 (fold change of −2.043/−15.508 at 63/98 °C, respectively); Ig α-1 chain C region #462 (fold change of −1.117/−34.941 at 63/98 °C, respectively) (Table 1). Intensive heat treatment affected the functional properties [15] and solubility of milk serum proteins [23,24]. Caseins and whey proteins are engaged in protein aggregates, found in heat-treated milk; the formation of intermolecular disulfide bonds is mostly responsible for this heat-induced protein association. Proteins, especially enzymes, are heat-sensitive, and are denatured at higher temperatures. Therefore, moderate and high heating temperatures eradicated 68% of the protein spots in our dataset, in which enzymes accounted for 61% of the spots. Spots relating to mitochondrial enzymes, such as cytochrome P450 11B2 mitochondrial spot and succinate dehydrogenase cytochrome b560, were completely absent at 98 °C. Spots relating to lactoferrin, a protein involved in iron metabolism and having antioxidant activity, were found to have variable intensities, this was likely caused by the different temperatures, because some of the spots representing the isoforms of lactoferrin were absent at 63 °C, while others were present.
2.6. Proteins Stable at 63 °C with Decrease at 98 °C Compared to RT
Camel milk whey has been shown to be more heat stable than bovine or buffalo whey. A number of studies reported that the denaturation of camel milk was reported to be lower (32%–35%) than that reported for bovine whey proteins (70%–75%) at 80 °C for 30 min [16,25]. We identified 25 protein spots stable and showed no significant change when heated at 63 °C; the same proteins were found to be significantly decreased, with a greater fold change, when heated at 98 °C (Table 1). This group of proteins included apolipoprotein A-I, spot #256 (fold change of 1.003/−2.337 at 63/98 °C, respectively); carboxypeptidase N catalytic chain, spot # 803 (fold change of 1.213/−4.831 at 63/98 °C, respectively); fibrinogen γ chain, spot #539 (fold change of 1.140/−1.760 at 63/98 °C, respectively); glutathione peroxidase 1, spot #409 (fold change of 1.231/−13.938 at 63/98 °C, respectively); haptoglobin, spot #615 (fold change of 1.011/−3.377 at 63/98 °C, respectively); Ig α-1 chain C region, spot #462 (fold change of 1.117/−34 at 63/98 °C, respectively); lactotransferrin, spot #500 (fold change of 1.156/−7.593 at 63/98 °C, respectively), peptidoglycan recognition protein 1, spot #396 (fold change of 1.044/−26.553 at 63/98 °C respectively); plasminogen, spot #616 (fold change of 1.008/3.657 at 63/98 °C, respectively); steroid 17-α-hydroxylase/17,20 lyase, spot #644 (fold change of 1.079/−10.522 at 63/98 °C, respectively); and succinate dehydrogenase cytochrome b560 subunit, spot #539 (fold change of 1.140/−1.760 at 63/98 °C, respectively). The heating of milk causes the denaturing of the globular whey protein, which can result in the exposure of reactive amino acid side groups, normally buried within the native conformation. The denaturation process is either reversible, where partial unfolding of the whey proteins with a loss of helical structure, or irreversible where an aggregation process occurs involving sulfhydryl (–SH)/disulfide (S–S) interchange reactions and other intermolecular interactions, such as hydrophobic and electrostatic interactions [26,27,28]. Protective proteins, and enzymes possessing antibacterial, antimicrobial, and antioxidant properties, such as lactoferrin, lactoperoxidase, and peptidoglycan recognition protein (PGRP), were destroyed after heating at 98 °C but remained unchanged after heating at 63 °C. PGRP, which possesses anti-carcinogenic properties, controls metastasis, stimulates the immune system, and exerts antimicrobial activity, is particularly important in this group of proteins [29].
2.7. Proteins That Increased in Abundance after Heating at 63 or 98 °C, Compared to Room Temperature
We found that eight protein spots had increase in abundance after heating at 63 and 98 °C; the fold change was greater at the latter temperature. These spots related to: α-lactalbumin, spot #1069 (fold change of 1.4/11.9 at 63/98 °C, respectively); copper chaperone for superoxide dismutase, spot #1041 (fold change of 1.5/1.6 at 63/98 °C, respectively); ERI1 exoribonuclease 2, spot #1167 (fold change of 1.2/15.6 at 63/98 °C, respectively); and glycosylation-dependent cell adhesion molecule 1, spot #985 (fold change of 1.3/3.9 at 63/98 °C, respectively). Using SDS-PAGE, Farah showed that the heat denaturation of the individual camel whey proteins is not as pronounced as that in bovine whey proteins; more intensive heat treatment of camel milk is necessary to obtain the same degree of denaturation as that observed in bovine milk [22]. Heat treatment at more than 63 °C results in unfolding of the globular structure of whey proteins and they denature [30,31]. These non-native structures can then form aggregates with other unfolded monomers or aggregate with other types of protein molecules [32,33], forming co-aggregates [34], which is a characteristic feature of the Maillard reaction.
2.8. Proteins with an Increase in Abundance at 63 °C but a Decrease at 98 °C and RT
We found that three protein spots, relating to hemopexin spot, #413 (fold change of 1.258/−16.162 at 63/98 °C, respectively); lactate dehydrogenase, spot #856 (fold change of 1.923/−1.445 at 63/98 °C, respectively); and ADP-ribosylation factor GTPase-activating, spot #796 (fold change of 1.280/−2.509 at 63/98 °C, respectively), were found to be differentially regulated between the two heating temperatures. These spots increased in abundance at 63 °C but decreased significantly, or were undetectable at, 98 °C. When subjected to different temperatures at low pH, milk forms different types of gels, causing the whey proteins to partially unfold and adopt an intermediate state, depending on the degree of heating [35]. Hattem et al. showed that the highest level of denaturation occurs after heating 90 °C for 30 min, and the lowest at 63 °C for 30 min; the rate of protein denaturation is proportional to increases in temperature [36]. Moderate thermal treatment (60–70 °C) induces structural unfolding in milk proteins, whereas at higher temperatures, protein aggregation occurs [37]. Lactate dehydrogenase is part of the glycolytic pathway mediating the oxidative/reductive connection between pyruvate and lactic acid. Avallone et al. found that lactate dehydrogenase is heat stable and retains most of its activity up to 70 °C; it, then, loses its activity upon further heating. This agrees with the presence of the spots indicating lactate dehydrogenase, where we observed a band at 63 °C, which was absent at 98 °C [38]. The enzyme levels in buffalo milk are used as markers for pasteurization because of their sensitivity to heat inactivation [39]. Hemopexin is a plasma glycoprotein present in mature camel milk; it acts as a binding protein for iron and possesses antioxidant activity. In their proteomic study, Le et al. used ion exchange fractionation to identify hemopexin in the colostrum and mature bovine milk, with a higher abundance in the colostrum. The presence of this protein in camel milk signifies the important properties of camel milk [40].
3. Materials and Methods
3.1. Animals and Sample Collection
Camel milk was obtained from three different healthy camels from Saudi Arabia (cream, tan, and black subtypes of Camelus dromedarius), fed annually by grazing, from a local farm in the city of Al Majmaah, Saudi Arabia. Approximately 600 mL of milk, from each subtype of camel, was collected at 5:00 p.m. by milking into sterile cans and transported to the ORC (Obesity Research Center), College of Medicine, King Saud University proteomic lab in an ice box at 4 °C within 4 h of collection. Upon reaching the laboratory, the pH of the milk samples was determined to be 6.6 with a pH meter (PB11, Sartorius, Gottingen, Germany).
3.2. Sample Preparation
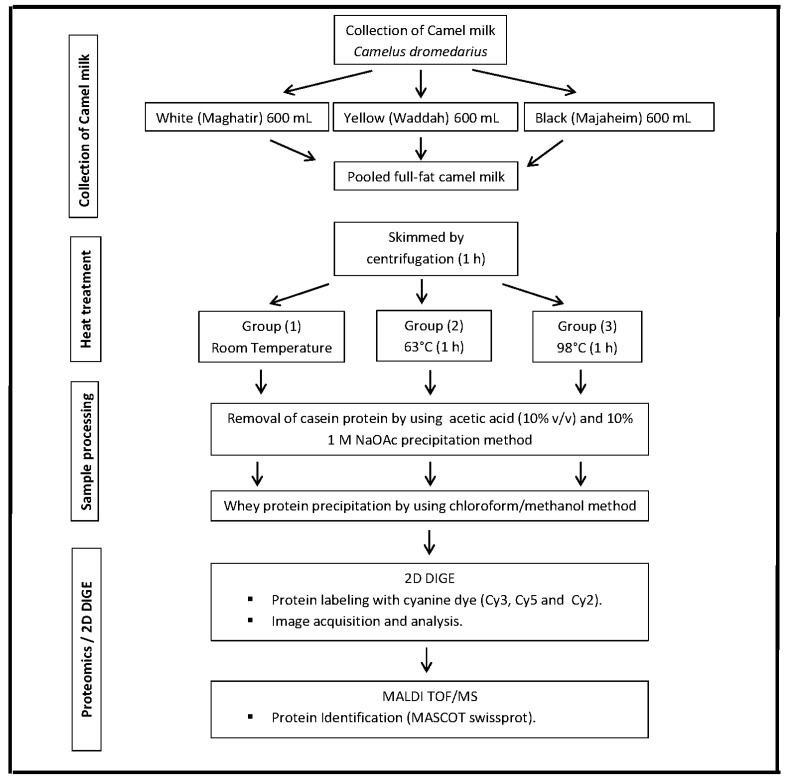
The milk samples from the three different subtypes of the same breed of camel were pooled together to produce a homogenous sample. The pooled sample was then triplicated to represent three independent biological samples for the application of heat treatment. The initial step, conducted on the samples, involved the removal of fat by centrifugation at 400× g and 4 °C for 30 min to obtain skim milk (Figure 3).
Figure 3.
Schema of the proteomics workflow, showing the key experimental steps in this study.
3.3. Heat Treatment
From the previously skimmed milk samples, one group of the samples, kept at room temperature, served as the control. The other two groups served as thermally treated samples that were heated at 63 and 98 °C for 1 h each, using a thermostatically controlled water bath (Shaking Water Bath SHWB10, Cole Parmer, San Diego, CA, USA) with rapidly circulated water. The treated and untreated samples were again triplicated for assessing reproducibility.
3.4. Whey Protein Extraction
Following the individual heat treatments, casein was precipitated out, as described previously, by adding 10% acetic acid (10% v/v), followed by 10% 1 M NaOAc, with gentle shaking; the samples were allowed to rest for 30 min at 35 °C between each addition. After acidification, the samples were centrifuged at 20,000× g for 30 min at 5 °C and the supernatants were collected [41]. The acidified whey proteins were then precipitated out using the methanol/chloroform (4:4:1; v:v:v) precipitation method at room temperature, and centrifuged at 5000× g for 5 min to remove the upper phase. Another three volumes of methanol were added to the supernatant, and the samples were again mixed and centrifuged at 16,000× g for 5 min to obtain the protein pellets, which were dried at room temperature in a vacuum concentrator (Concentrator Plus, Eppendorf, Hamburg, Germany) [42].
3.5. Protein Labeling with Cyanine Dyes
The protein pellets were solubilized in a labeling buffer (7 M urea, 2 M thiourea, 30 mM Tris–HCl, 4% CHAPS, pH 8.5). The insoluble material was pelleted by centrifugation at 12,000× g at room temperature for 5 min. Protein concentrations were determined in triplicate using the 2D-Quantkit (GE Healthcare, Piscataway, NJ, USA), and the pH of the samples was adjusted to 8.5 using NaOH (100 mM). The proteins were labeled using 400 pmol CyDye™ DIGE Fluor dyes (GE Healthcare, Buckinghamshire, UK) in 1 μL of dimethylformamide (DMF) and then mixed with a sample containing 50 μg of protein. Samples were incubated on ice for 30 min in the dark. The labeling reaction was terminated by adding 1 μL of 10 mM lysine. Each sample was covalently labeled with a Cy3 or Cy5 fluorophore. A mixture of equal amounts of protein, isolated from each sample, was labeled with Cy2 and used as an internal standard (see Table S2).
3.6. Two-Dimensional Electrophoresis, Image Scanning, and Preparative Gel
First dimension analytical gel electrophoresis was performed as follows. Five Immobiline Dry Strips (24 cm, pH 3–11; GE Healthcare, Uppsala, Sweden) were passively re-hydrated at 30 V for 12 h. This was followed by isoelectric focusing using an Ettan IPGphor IEF unit (GE Healthcare, Sweden). Focusing was performed at 20 °C, at 50 μA per strip, according to the following hold sequence: (1) 500 V for 3 h, (2) 1000 V for 3 h, (3) 8000 V for 4 h, and (4) 8000 V for 6.25 h totalling up to 50,000 Vhrs. IPG (immobilized pH gradient) strips were then stored at −80 °C until second-dimension separation. Before the second-dimension separation, the IPG strips were first equilibrated with dithiothreitol for 15 min at room temperature with gentle stirring, then with 5 mM Tris–HCl (pH 8.8), 6 M urea, 30% glycerol, 2% SDS, and 65 mM DTT. The strips were then equilibrated for an additional 15 min in the same solution containing 250 mM iodoacetamide. Polyacrylamide fixed gels (12.5%) were prepared on low fluorescence glass using a 2-D Optimizer (NextGen Sciences, London, UK). Next, we performed second-dimension separation using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; Ettan DALT six vertical units (GE Healthcare, Uppsala, Sweden), at 15 °C, 1 W per gel, for 1 h, and then at 2 W per gel until the bromophenol blue dye front reached the bottom of the gel. Then, the gels were scanned at the appropriate individual excitation and emission wavelengths using the Typhoon Trio Imager fluorescence gel scanner (GE Healthcare) with the values of filters and photomultiplier optimized for Cy3, Cy5, and Cy2.
3.7. Colloidal Coomassie Blue Staining of the Preparative Gel
Total protein (1 mg) was obtained from a pool of equal protein amounts of the nine camel milk samples. This was denatured in a lysis buffer, then mixed in a rehydration buffer. Gels were fixed in 40% (v/v) ethanol with 10% (v/v) acetic acid (overnight) and then washed (3×, 30 min each, ddH2O). The gels were incubated (1 h, 34% (v/v) methanol containing 17% (w/v) ammonium sulphate and 3% (v/v) phosphoric acid) prior to the addition of 0.5 g/L Coomassie G-250. After five days the stained gels were briefly rinsed with Milli-Q water and stored until the spots could be picked and identified by MS.
3.8. Protein Identification by MALDI-TOF MS
The spots from Coomassie-stained gels were excised manually, washed, and digested according to a previously published protocol [43]. The mixture of tryptic peptides (1 µL), derived from each protein, was spotted onto a MALDI (Matrix-assisted laser desorption/ionization) target (384 anchorchip MTP 800 µm Anchorchip; Bruker Daltonics, Bremen, Germany) together with 0.8 μL of matrix (10 mg α-cyano-4-hydroxycinnamic acid (CHCA) in 1 μL of 30% CH3CN and 0.1% aqueous CF3COOH) and then left to dry (RT) before MS analysis. Spectra were acquired using a MALDI-TOF MS (UltraFlexTrem, Bruker Daltonics, Bremen, Germany) in the positive mode with target voltage of 25 kV and pulsed ion extraction voltage of 20 kV. The reflector voltage was set to 21 kV and detector voltage to 17 kV. Peptide mass fingerprints (PMF) were calibrated against a standard (Peptide Calibration Standard II, Bruker Daltonics). The PMF were processed using the Flex AnalysisTM software (version 2.4, Bruker Daltonics). The MS data were interpreted using BioTools v3.2 (Bruker Daltonics), together with the MASCOT search algorithm (version 2.0.04 updated 09/05/2015; Matrix Science Ltd., London, UK). MASCOT search parameters were set as follows: fixed propionamide modification of cysteine, oxidation of methionine as variable modification, one missed cleavage site (such as in the case of incomplete trypsin hydrolysis), and a mass tolerance of 100 ppm. Identified proteins were accepted as correct if they showed a MASCOT score greater than 56 and p < 0.05. Not all spots of interest could be identified because some proteins were low in abundance and did not yield a sufficiently intense mass of fingerprints; other spots were mixtures of multiple proteins.
3.9. Image Acquisition
DIGE images were analyzed using the Progenesis Same Spots v3.3 software (Nonlinear Dynamics Ltd., Newcastle, UK). First, images were aligned. Then an automatic vector tool, using prominent spots, was employed to detect 400 total vectors for warping and aligning the gel images with a reference image of one internal standard across and within each gel. Gel groups were designated according to the experimental design; normalized spot volume was used to select statistically significant spots. The Progenesis Same Spots v3.3 software was used to calculate the normalized volume (NV) of each spot, on each gel, for Cy3 and Cy5, to Cy2 spot volume ratio. The software performs log transformations of the spot volumes to generate normally distributed data. Log normalized volume (LNV) was used to quantify differential expression. Independent direct comparisons were made between differently heat-treated milk samples. Fold differences and p-values were calculated using one-way ANOVA. All spots were pre-filtered and manually examined before applying the statistical criteria (ANOVA test, p ≤ 0.05 and fold ≥1.2). Instead of spot intensities, normalized spot volumes were used for statistical analysis. Only those spots that fulfilled the abovementioned statistical criteria were submitted for MS analysis.
4. Conclusions
This study aimed to identify the total protein composition during the heat treatment of camel milk whey proteins at a medium temperature of 63 °C and a high temperature of 98 °C for 60 min. A total of 116 protein were detected as significantly changing using 2-DIGE and identified by MALDI-MS. The obtained results showed that camel whey proteins were significantly affected by heat treatment at 98 °C and several proteins disappeared completely from the gel patterns; however, the whey proteins remain slightly stable under heat treatment at 63 °C for 60 min. This experimental study showed that denaturation increased significantly as the temperature increased from 63 to 98 °C. The fold change in the abundance of proteins identified between RT and 63 °C ranged from 15%–61% and for RT and 98 °C from 79%–98%. Further studies are needed to elucidate the mechanism involved in the heat denaturation of camel milk whey proteins and study the mechanisms in further detail.
Acknowledgments
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding this research through the Research Group Project no. RGP-334. The authors would also like to acknowledge .Khalid Mohamed Alrabiah for providing samples of camel milk for the study.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/4/721/s1.
Author Contributions
Hicham Benabdelkamel, Afshan Masood, and Assim A. Alfadda conceived and designed the study. Hicham Benabdelkamel, Ibrahim O. Alanazi, Deema K. Alrabiah, Dunia A Alzahrani, and Sami A. AlYahya designed the experiments and workflow, and conducted the experiments. Hicham Benabdelkamel, Afshan Masood, Deema Al-Rabiah, and Dunia Al-Zahrani performed the data analysis. Hicham Benabdelkamel, Afshan Masood, Deema Al-Rabiah, Dunia Al-Zahrani, and Ibrahim O. Alanazi prepared the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Al haj O.A., Al Kanhal H.A. Compositional, technological and nutritional aspects of dromedary camel milk. Int. Dairy J. 2010;20:811–821. doi: 10.1016/j.idairyj.2010.04.003. [DOI] [Google Scholar]
- 2.Park Y.W., Haenlein G.F.W. Handbook of Milk of Non-Bovine Mammals. 1st ed. Blackwell Pub; Ames, IA, USA: 2006. p. 297. [Google Scholar]
- 3.Riyadh S., Aljumaah F.F.A., Ismail E., Alshaikh M.A., Sami A., Ayadi M. Effects of production system, breed, parity and stage of lactation on milk composition of dromedary camels in Saudi Arabia. J. Anim. Vet. Adv. 2012;11:141–147. [Google Scholar]
- 4.Yang Y., Bu D., Zhao X., Sun P., Wang J., Zhou L. Proteomic analysis of cow, yak, buffalo, goat and camel milk whey proteins: Quantitative differential expression patterns. J. Proteome Res. 2013;12:1660–1667. doi: 10.1021/pr301001m. [DOI] [PubMed] [Google Scholar]
- 5.McSweeney P.L.H. Advanced Dairy Chemistry: Volume 1a: Proteins: Basic Aspects. 4th ed. Springer; New York, NY, USA: 2012. [Google Scholar]
- 6.El-Agamy E.I. Bioactive Components in Milk and Dairy Products. Wiley-Blackwell; Ames, IA, USA: 2009. Bioactive components in camel milk; pp. 159–194. [Google Scholar]
- 7.Ahamad S.R., Raish M., Ahmad A., Shakeel F. Potential health benefits and metabolomics of camel milk by GC-MS and ICP-MS. Biol. Trace Elem. Res. 2017;175:322–330. doi: 10.1007/s12011-016-0771-7. [DOI] [PubMed] [Google Scholar]
- 8.Konuspayeva G., Faye B., Loiseau G. The composition of camel milk: A meta-analysis of the literature data. J. Food Compos. Anal. 2009;22:95–101. doi: 10.1016/j.jfca.2008.09.008. [DOI] [Google Scholar]
- 9.Zhang L., Boeren S., Smits M., van Hooijdonk T., Hettinga K. Proteomic study on the stability of proteins in bovine, camel, and caprine milk sera after processing. Food Res. Int. 2016;82:104–111. doi: 10.1016/j.foodres.2016.01.023. [DOI] [Google Scholar]
- 10.Hailu Y., Hansen E.B., Seifu E., Eshetu M., Ipsen R., Kappeler S. Functional and technological properties of camel milk proteins: A review. J. Dairy Res. 2016;83:422–429. doi: 10.1017/S0022029916000686. [DOI] [PubMed] [Google Scholar]
- 11.Wijayanti H.B., Bansal N., Deeth H.C. Stability of whey proteins during thermal processing: A review. Compr. Rev. Food Sci. Food Saf. 2014;13:1235–1251. doi: 10.1111/1541-4337.12105. [DOI] [Google Scholar]
- 12.Felfoul I., Lopez C., Gaucheron F., Attia H., Ayadi M.A. A laboratory investigation of cow and camel whey proteins deposition under different heat treatments. Food Bioprod. Process. 2015;96:256–263. doi: 10.1016/j.fbp.2015.09.002. [DOI] [Google Scholar]
- 13.De Jong P. Advanced Dairy Science and Technology. Blackwell Publishing Ltd.; Oxford, UK: 2008. Thermal processing of milk; pp. 1–34. [Google Scholar]
- 14.Gedam K., Prasad R., Vijay V. The study on UHT processing of milk: A versatile option for rural sector. World J. Dairy Food Sci. 2007;2:49–53. [Google Scholar]
- 15.Abdalla M.O.M., Daffalla M.S. Comparison of chemical and microbiological parameters of charcoal versus gas and solar energy treated milk. Adv. J. Food Sci. Technol. 2010;2:286–290. [Google Scholar]
- 16.Elagamy E.I. Effect of heat treatment on camel milk proteins with respect to antimicrobial factors: A comparison with cows’ and buffalo milk proteins. Food Chem. 2000;68:227–232. doi: 10.1016/S0308-8146(99)00199-5. [DOI] [Google Scholar]
- 17.Ewaschuk J.B., Unger S., O’Connor D.L., Stone D., Harvey S., Clandinin M.T., Field C.J. Effect of pasteurization on selected immune components of donated human breast milk. J. Perinatol. 2011;31:593–598. doi: 10.1038/jp.2010.209. [DOI] [PubMed] [Google Scholar]
- 18.Laleye L.C., Jobe B., Wasesa A.A. Comparative study on heat stability and functionality of camel and bovine milk whey proteins. J. Dairy Sci. 2008;91:4527–4534. doi: 10.3168/jds.2008-1446. [DOI] [PubMed] [Google Scholar]
- 19.Li-Chan E., Kummer A., Losso J.N., Kitts D.D., Nakai S. Stability of bovine immunoglobulins to thermal treatment and processing. Food Res. Int. 1995;28:9–16. doi: 10.1016/0963-9969(95)93325-O. [DOI] [Google Scholar]
- 20.Felfoul I., Jardin J., Gaucheron F., Attia H., Ayadi M.A. Proteomic profiling of camel and cow milk proteins under heat treatment. Food Chem. 2017;216:161–169. doi: 10.1016/j.foodchem.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Singh H., Havea P. Thermal denaturation, aggregation and gelation of whey proteins. In: Fox P.F., McSweeney P.L.H., editors. Advanced Dairy Chemistry—1 Proteins. Springer; Boston, MA, USA: 2003. pp. 1261–1287. [Google Scholar]
- 22.Farah Z. Effect of heat treatment on whey proteins of camel milk. Milchwissenschaft. 1986;41:763–765. [Google Scholar]
- 23.Miyamoto Y., Matsumiya K., Kubouchi H., Noda M., Nishimura K., Matsumura Y. Effects of heating conditions on physicochemical properties of skim milk powder during production process. Food Sci. Technol. Res. 2009;15:631–638. doi: 10.3136/fstr.15.631. [DOI] [Google Scholar]
- 24.Manzo C., Nicolai M.A., Pizzano R. Thermal markers arising from changes in the protein component of milk. Food Control. 2015;51:251–255. doi: 10.1016/j.foodcont.2014.11.029. [DOI] [Google Scholar]
- 25.Farah Z., Atkins D. Heat coagulation of camel milk. J. Dairy Res. 1992;59:229–231. [Google Scholar]
- 26.McKinnon I.R., Yap S.E., Augustin M.A., Hemar Y. Diffusing-wave spectroscopy investigation of heated reconstituted skim milks containing calcium chloride. Food Hydrocoll. 2009;23:1127–1133. doi: 10.1016/j.foodhyd.2008.08.009. [DOI] [Google Scholar]
- 27.Hoffmann M.A.M., van Mil P.J.J.M. Heat-induced aggregation of β-lactoglobulin: Role of the free thiol group and disulfide bonds. J. Agric. Food Chem. 1997;45:2942–2948. doi: 10.1021/jf960789q. [DOI] [Google Scholar]
- 28.Anema S.G., Li Y. Further studies on the heat-induced, pH-dependent dissociation of casein from the micelles in reconstituted skim milk. LWT Food Sci. Technol. 2000;33:335–343. doi: 10.1006/fstl.2000.0665. [DOI] [Google Scholar]
- 29.Kustikova O.S., Kiselev S.L., Borodulina O.R., Senin V.M., Afanas’eva A.V., Kabishev A.A. Cloning of the tag7 gene expressed in metastatic mouse tumors. Genetika. 1996;32:621–628. [PubMed] [Google Scholar]
- 30.Dannenberg F., Kessler H.-G. Reaction kinetics of the denaturation of whey proteins in milk. J. Food Sci. 1988;53:258–263. doi: 10.1111/j.1365-2621.1988.tb10223.x. [DOI] [Google Scholar]
- 31.Kontopidis G., Holt C., Sawyer L. Invited review: β-Lactoglobulin: Binding properties, structure, and function. J. Dairy Sci. 2004;87:785–796. doi: 10.3168/jds.S0022-0302(04)73222-1. [DOI] [PubMed] [Google Scholar]
- 32.Carbonaro M., Bonomi F., Iametti S., Cappelloni M., Carnovale E. Aggregation of proteins in whey from raw and heat-processed milk: Formation of soluble macroaggregates and nutritional consequences. LWT Food Sci. Technol. 1998;31:522–529. doi: 10.1006/fstl.1998.0408. [DOI] [Google Scholar]
- 33.Tolkach A., Kulozik U. Reaction kinetic pathway of reversible and irreversible thermal denaturation of β-lactoglobulin. Lait. 2007;87:301–315. doi: 10.1051/lait:2007012. [DOI] [Google Scholar]
- 34.Corredig M., Dalgleish D.G. Effect of temperature and pH on the interactions of whey proteins with casein micelles in skim milk. Food Res. Int. 1996;29:49–55. doi: 10.1016/0963-9969(95)00058-5. [DOI] [Google Scholar]
- 35.Livney Y.D., Verespej E., Dalgleish D.G. Steric effects governing disulfide bond interchange during thermal aggregation in solutions of β-lactoglobulin B and α-lactalbumin. J. Agric. Food Chem. 2003;51:8098–8106. doi: 10.1021/jf034582q. [DOI] [PubMed] [Google Scholar]
- 36.Hattem H.E., Sakr H.S., Abouel-Einin E.H. A study on the effect of heat treatments on composition and some properties of camel milk. J. Brew. Distill. 2011;2:51–55. doi: 10.5261/2011.GEN2.05. [DOI] [Google Scholar]
- 37.Schmidt R.H., Morris H.A. Gelation properties of milk proteins, soy proteins, and blended protein systems. Food Technol. 1984;5:85–94. [Google Scholar]
- 38.Avallone L., Lombardi P., Florio S., d’Angelo A., Bogin E. Age-dependent variations of lactate dehydrogenase and creatine kinase activities in water buffalo calf serum. Eur. J. Clin. Chem. Clin. Biochem. 1996;34:961–964. doi: 10.1515/cclm.1996.34.12.961. [DOI] [PubMed] [Google Scholar]
- 39.Lombardi P., Avallone L., d’Angelo A., Mor T., Bogin E. Buffalo-milk enzyme levels, their sensitivity to heat inactivation, and their possible use as markers for pasteurization. J. Food Prot. 2000;63:970–973. doi: 10.4315/0362-028X-63.7.970. [DOI] [PubMed] [Google Scholar]
- 40.Le A., Barton L.D., Sanders J.T., Zhang Q. Exploration of bovine milk proteome in colostral and mature whey using an ion-exchange approach. J. Proteome Res. 2010;10:692–704. doi: 10.1021/pr100884z. [DOI] [PubMed] [Google Scholar]
- 41.Wangoh J., Farah Z., Puhan Z. Iso-electric focusing of camel milk proteins. Int. Dairy J. 1998;8:617–621. doi: 10.1016/S0958-6946(98)00092-2. [DOI] [Google Scholar]
- 42.Alhaider A., Abdelgader A.G., Turjoman A.A., Newell K., Hunsucker S.W., Shan B., Ma B., Gibson D.S., Duncan M.W. Through the eye of an electrospray needle: Mass spectrometric identification of the major peptides and proteins in the milk of the one-humped camel (Camelus dromedarius) J. Mass Spectrom. 2013;48:779–794. doi: 10.1002/jms.3213. [DOI] [PubMed] [Google Scholar]
- 43.Alfadda A.A., Benabdelkamel H., Masood A., Moustafa A., Sallam R., Bassas A., Duncan M. Proteomic analysis of mature adipo cytes from obese patients in relation to aging. Exp. Gerontol. 2013;48:1196–1203. doi: 10.1016/j.exger.2013.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.