Abstract
A patterned gold nanoparticle microarray, functionalized with a nanoscale silicate coating, has been developed for on-chip, high-throughput mass spectrometric analyses of biomolecules with minimal sample preparation and reagent costs. Fabrication was realized by the combination of layer-by-layer functionalization of the nanoparticles with suitable polyelectrolytes, followed by fluidic patterning of the glass microarray support and calcination for permanent fixation of the nano-coating. Performance of the microarray was evaluated for surface-assisted laser-desorption/ionization mass spectrometry (SALDI-MS), where the nano-silicate coating was found to enhance SALDI efficiency, resulting in comparable performance to some common organic matrices for small and medium sized molecules. Performance contributing factors of this material have been discussed; heat confinement and interband transition/plasmonic resonance may play important roles. Taking the accessibility of fabrication, performance, and reusability of this substrate together, the material developed here provides a new tool for multiplexed and chip-based mass spectrometric analysis.
Introduction
The characterization of proteome, genome, and metabolome wide patterns represents a difficult and complex task in the face of a myriad of toxicants and stressors that affect living systems.1 While biosensors have served as effective tools for the detection of biomolecules and characterization of biophysical processes,2 higher throughput in the performance is constantly demanded. The combination of biosensing and microarray technologies offers robust, multiplex detection strategies to meet this demand for a variety of target analytes.3–5 Different from optical detection, various mass spectrometric techniques, capable of sophisticated tasks such as DNA and protein sequencing,6,7 are often difficult to integrate with chip-based samples, with the exception of matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) which has shown great promise as a powerful surface-based method.8 MALDI arrays utilize an organic matrix to assist in the ionization of target analytes, and with appropriate functionalization of the array chip, may be used for profiling global cellular activities.9 However, due to the presence of matrix related ions, MALDI-MS is rarely applied to low molecular weight samples, hindering its application in clinical and industry settings where small molecule detection and identification is greatly needed.
To circumvent this issue, the field of surface-assisted laser desorption/ionization mass spectrometry (SALDI-MS) has undergone rigorous development and is gaining considerable attention for low molecular weight analytes.10 SALDI-MS relies on the properties of the underlying material, rather than those of a separate organic matrix, to assist in analyte desorption and ionization from the substrate surface. Naturally, this results in minimized sample preparation and little to no interference in the low mass region, as an external matrix no longer needs to be applied or incorporated into the samples. Porous silicon (also known as desorption/ionization on silicon, or DIOS) is one of the most established SALDI materials, and its ionization performance benefits from strong UV absorption and heat transfer properties.11–13 Gold nanomaterials share many of the same performance benefits as DIOS for SALDI-MS, both in colloid14–17 and thin film18,19 formats. This class of materials has the benefit of being relatively inert compared to DIOS, which is prone to oxidation, leading to a decline or even loss of performance. While effective, gold nanoparticles for SALDI have certain weaknesses; gold clusters may ionize with the target analyte and introduce contaminating peaks into mass spectra.14,16 Furthermore, little attempt has been made to create patterned microarrays of these materials, likely due to the high expense of sophisticated cleanroom and lithography techniques, or to coffee ring formation effects in arrayed, drop-deposited colloids.20
Herein we demonstrate the layer-by-layer modification of gold nanoparticles, coupled with an inexpensive photocatalytic patterning process for the development of a highly effective, inexpensive, and reusable microarray substrate that is optimized for SALDI-MS. Gold nanoparticle modification is realized by a method our group developed for localized surface plasmon resonance (LSPR) and SALDI.17 Using photocatalytic patterning to create hydrophilic deposition areas with hydrophobic surroundings, we are able to alleviate the inhomogeneous sample spreading issue during formation of our AuNP array spots, and during deposition of target analytes. Calcination of the substrate before sample deposition permanently fixes these features in place, leaving robust films of AuNPs protected by ultrathin layers of silicate. In this work, performance-contributing factors of this material will be discussed. The calcinated AuNP microarray is found to be more efficient than many developed SALDI materials, while exhibiting comparable performance to other MS methods using organic matrices for small to medium sized molecules. In addition, the microarray can be regenerated under mild conditions with no deterioration of performance over many cycles. The sensitive and multiplexed measurements achieved here with minimized sample preparation and in a reusable format may lead to a large reduction in material and reagent costs for chip-based detection and screening.
Experimental section
Materials and reagents
Gold(III) chloride trihydrate, trisodium citrate dihydrate, poly-(allylamine hydrochloride) (PAH, MW ~56 kDa), n-octadecyltrichlorosilane (OTS, 90+%), α-cyano-4-hydroxycinnamic acid (CHCA), 2,5-dihydroxybenzoic acid (DHB), trifluoroacetic acid (TFA), titanium(IV) chloride, [Sar1, Thr8]-angiotensin II, and neurotensin were from Sigma-Aldrich (St. Louis, MO). Citric acid, sodium silicate (37%), ethanol, toluene, and chloroform were from Fisher Scientific (Pittsburgh, PA). Acetyl-tetradecapeptide renin substrate was from AnaSpec (San Jose, CA). BK-7 glass microscope slides were from Corning (Painted Post, NY).
Instrumentation
All extinction spectra were obtained using a Cary 50 UV-Vis spectrophotometer (Agilent Technologies, Santa Clara, CA). Scanning electron microscopy (SEM) was conducted with a Hitachi TM-1000 Tabletop SEM (Tokyo, Japan). Transmission electron microscopy (TEM) was conducted with a Philips FEI Tecnai 12 TEM (Andover, MA) in CFAMM at UC Riverside.
Mass spectra were collected using a Voyager-DE STR MALDI-TOF mass spectrometer (Applied Biosystems, Framingham, MA) set in positive reflector mode at an accelerating voltage of 20 kV. The spectrometer is equipped with a pulsed nitrogen laser operating at 337 nm, with each spectrum acquired as an average of 60 laser shots.
Sample preparation
The peptides, [Sar1, Thr8]-angiotensin II, neurotensin, and acetyl-tetradecapeptide renin substrate, were prepared in stock solutions of 50% acetonitrile containing 0.1% TFA and 10 mM citric acid to a final concentration of 20 μM. Cholesterol was dissolved in chloroform to a concentration of 2.59 mM (1 mg mL−1). These solutions were diluted to desired experimental concentrations, and aliquots of 1 μL were spotted onto the AuNP array, followed by drying under vacuum. The array was then attached to a custom stainless steel MALDI plate using polyimide tape. When matrices were utilized in ionization, the analyte and matrix were set to a volume ratio of 1: 10, respectively, and 1 μL aliquots were spotted onto a commercial stainless steel MALDI plate.
Synthesis and modification of gold nanoparticles
Gold nanoparticles (AuNPs), ca. 13 nm in diameter, were prepared using previously established methods.21 In short, a round bottom flask containing 500 mL of 1 mM gold(III) chloride trihydrate was heated until boiling, after which, 50 mL of 38.8 mM trisodium citrate dihydrate was added. The solution was continuously boiled and stirred until a deep red color was obtained. Nanoparticle formation and size distribution were verified using the LSPR band measured by UV-Vis absorption, and transmission electron microscopy.
AuNPs were modified using layer-by-layer immobilization of polyelectrolytes at the nanoparticle surface.22 To modify AuNPs for immobilization and calcination on glass slides, an aliquot (1.5 mL) of as-prepared citrate-capped AuNP solution was first centrifuged at 10 000 rpm for 15 minutes, and the supernatant was discarded. Thereafter, an aqueous solution of 1 mg per mL PAH (1.5 mL, pH 8.0) was added to the AuNP pellet. The mixture was bath sonicated for 5 minutes to disperse the AuNPs and immobilize PAH at the AuNP surface. The AuNP–PAH solution was centrifuged again followed by removal of the supernatant. The addition of an aqueous solution of 22 mg per mL sodium silicate (1.5 mL, pH 9.5) to the AuNP–PAH pellet was followed by sonication for 5 minutes to immobilize silicate ions and yield AuNP–(PAH-silicate)1. This process was repeated to yield the desired number of (PAH-silicate) layers on the AuNPs. The resulting solution was concentrated 15-fold before arraying.
Microarray patterning of glass substrates
A photocatalytic method to prepare alternating hydrophobic and hydrophilic areas on glass microscope slides was used with modification.23 BK-7 glass microscope slides were cleaned using a boiling piranha solution (3 : 1 v/v H2SO4 and 30% H2O2) for 30 minutes, followed by rinsing with DI water and drying under a stream of nitrogen gas. Glass slides were then immersed in 10 mM titanium(IV) chloride in toluene with 0.1% (v/v) water for 15 minutes, in order to hydrolyze TiCl4 and obtain a photocatalytic nanofilm of TiO2 on the glass surface.24 The TiO2 coated microscope slides were then rinsed alternately with toluene and ethanol three times. Thereafter, the slides were immersed in 10 mM OTS in toluene to generate a hydrophobic C18 layer, then rinsed alternately with toluene and ethanol three more times before drying under a stream of nitrogen gas. The entire glass/TiO2/C18 substrate was covered by a photomask and irradiated under a 450 mW cm−2, λ = 254 nm, light source (CL-1000, UVP Inc., Upland, CA) for one hour to cleave C18 from the exposed areas, and obtain a pattern of hydrophilic spots with hydrophobic surroundings. The photomask was perforated with holes representing the final array, separated into areas of 2 mm dia. spots with 3 mm pitch, 1 mm dia. spots with 3 mm pitch, and 800 μm dia. spots with 1.5 mm pitch. After UV irradiation, 1 μL of prepared AuNP–(PAH-silicate)n solution was deposited on each hydrophilic array spot and dried. The number of deposited and dried droplets per spot was varied to determine optimal performance. Finally, all slides were heated in a furnace to 450 °C for 4 hours at a rate of 17 °C per minute. Each slide was thoroughly rinsed with ethanol and DI water before use.
Results and discussion
Photocatalytic patterning of arrayed calcinated AuNPs
The layer-by-layer (LbL) assembly technique is a versatile and cost-effective strategy for creating nanostructures possessing specifically tailored compositions and morphologies.25 In this work, we applied the LbL technique to gold nanoparticles in solution, followed by high temperature calcination, to generate dense coatings of silicate with tunable, nanoscale thicknesses around individual nanoparticles (scheme in Fig. 1). The calcination step took place on BK-7 glass microscope slides, patterned with hydrophilic spots and hydrophobic surroundings, to assist in generating a well-defined microarray of the silicate-coated product (Fig. 1b). Each step of this fabrication was optimized for high SALDI-MS performance.
Fig. 1.
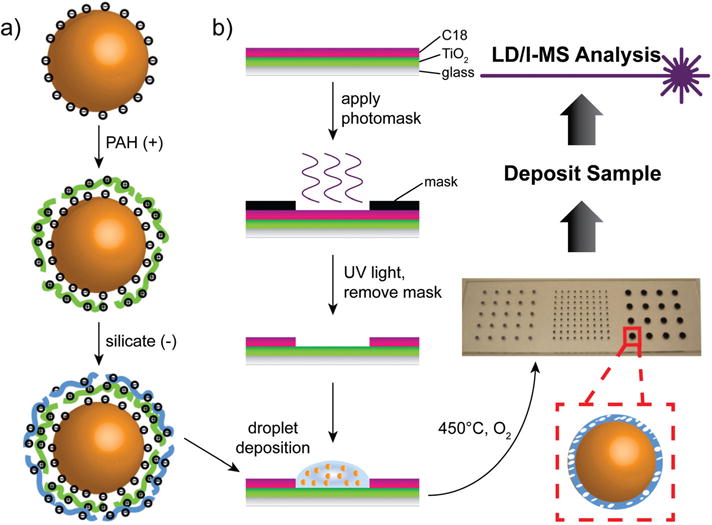
Fabrication of the calcinated AuNP array and analytical scheme. (a) Layer-by-layer modification of gold nanoparticle surface in solution. (b) Photocatalytic method for creating a hydrophilic spot pattern with hydrophobic surroundings on glass microscope slides.
The deposition of alternately charged polyelectrolytes at the nanoparticle surface was monitored using changes in the localized surface plasmon resonance (LSPR) band of the colloidal nanoparticle solutions, with transmission electron microscopy (TEM) providing additional verification of the results (Fig. 2). Due to the LSPR phenomenon, gold nanoparticles may function as optical nanoantennae, capable of absorbing light at discrete wavelengths and generating localized, oscillating electromagnetic fields from the delocalized electrons of the bulk material.26 In response to changes in the local dielectric environment, the wavelength at which these nanoparticles absorb light shifts as a function of the surrounding refractive index, allowing for a unique sensing modality that has been broadly exploited.17,27–33 Here, as each layer of polyelectrolyte is added to the gold nanoparticle surface, the absorbance peak shifts to a higher wavelength (Fig. 2a), in agreement with the underlying mechanism of LSPR.27 Starting with the citrate capped and negatively charged AuNP surface (AuNP, λpeak =520 nm), the first layer of positively charged polyallylamine hydrochloride (PAH) results in a red shift of the absorbance band (AuNP–PAH, λpeak = 535 nm), followed by a further red shift once a layer of negatively charged silicate is added to the surface (AuNP–(PAH-silicate)1, λpeak = 545 nm). Further increases in the number of (PAH-silicate)n layers result in additional red shifts of the absorbance band (λpeak = 550 nm). The decrease in LSPR sensitivity in the higher layer number samples is due to the short decay length of the plasmonic sensing field, which only extends ca. 10 nm and decreases with distance from the nanoparticle surface. TEM analysis agrees with the LSPR results, showing increasing shell thicknesses around the AuNPs as more layers are added (Fig. 2b).
Fig. 2.
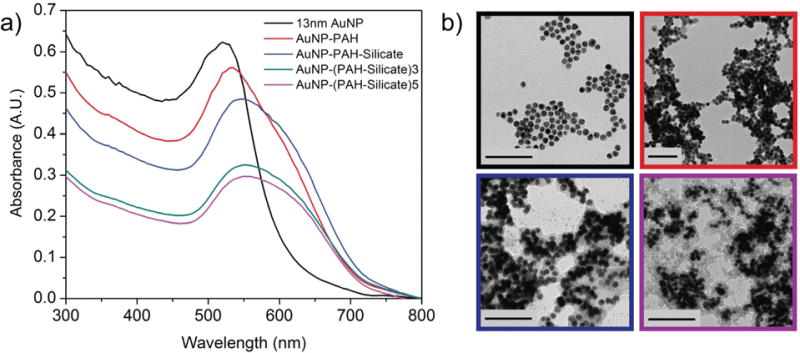
Layer-by-layer modification of gold nanoparticles. (a) Monitoring of polymer and silicate depositions using localized surface plasmon resonance absorbance bands. (b) Corresponding transmission electron micrographs. Scale bars represent 100 nm.
Fabricating a precisely defined microarray from a colloidal suspension presents a challenge, as liquid droplets deposited on solid surfaces often do not dry uniformly, which can adversely affect microarray patterning and performance.34 Capillary flow from the center of a liquid drop with a pinned contact line (e.g. a deposited nanoparticle suspension) results in dissolved particles migrating outwardly from the center of the droplet toward the contact line in order to replenish evaporating solvent at this location.20,35 The formation of a ringlike deposit and heterogeneous sample spreading is often unavoidable, but properties of this deposit may be controlled by altering the deposition environment.36 Lubrication theory shows that if the evaporative flux of solvent can be driven away from the contact line, a uniform deposition can be achieved,37 and this was successfully realized for fluorescence based proteomic arrays in which fixed, hydrophobic patterns were used to confine aqueous solutions and create uniform drying profiles.38 We used a modification of previously established methods23,24 to photocatalytically pattern hydrophilic deposition areas with hydrophobic surroundings on BK-7 glass microscope slides and achieve even drying profiles of deposited AuNPs within each array spot (Fig. S1†). While hydrophobic C18 layers may be applied directly to glass, the thin film of TiO2 utilized in this and prior work is applied for its photocatalytic acceleration of C18 breakdown during UV treatment, decreasing fabrication time for this material.23,24 The OTS monolayer, with a thickness of 2–3 nm,39–41 functions as a hydrophobic corral for the nanoparticle suspensions, and is removed during the calcination step (Fig. S2†). After deposition of the modified AuNP solutions, followed by heating of the entire microarray substrate in a furnace set at 450 °C, the physical properties were examined using scanning electron microscopy (SEM).
SEM images in Fig. 3 reveal that the nanoparticles are remarkably evenly distributed throughout the entire array spot (dia. 800 μm shown), and no ring-like deposit is observed along the defined spot edge. Increasing magnification of the arrayed AuNPs reveals a regular pattern of porosity, with a relatively high roughness across the surface of the material (Fig. S3†). These features are beneficial; high surface area increases sample-loading capacity within smaller areas for MS analysis, and may confine heat to promote higher laser desorption/ionization performance.11,15,19,23,42 For this microarray, we accomplished this through the addition of a high molecular weight (56 kDa) polyelectrolyte during AuNP modification, and by utilizing multiple (n = 9), consecutive droplet depositions on the patterned glass slides. The high molecular weight PAH induces bridging flocculation of neighboring AuNPs, producing a regular pattern of AuNP clusters, or “flocs”, when deposited on the solid surface.22 The resulting roughness and porosity are controllable, which is advantageous for high shot-to-shot reproducibility in mass spectra. Calcination permanently fixes these nanoscale (floc) and microscale (array spot) patterns in place on the glass slides, as the high temperature combusts the sacrificial polymer layers used for immobilization (PAH) and forms a dense network of silicate glass covering the AuNPs. Each (PAH-silicate)n layer results in ca. 2 nm of glass coating43 and protects the nano-particle morphologies during the calcination process, with individual diameters of ca. 13 nm retained after calcination and with varying layer numbers (Fig. S4†). In addition to increasing the structural robustness of the nanoparticles, this nanoscale coating also confines heat at the AuNP surface for highly effective SALDI-MS performance.17
Fig. 3.
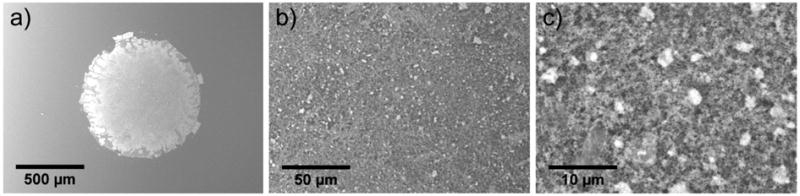
Scanning electron micrographs of arrayed and calcinated gold nanoparticles. (a) 100× magnification. (b) 1000× magnification. (c) 5000× magnification.
Characterization of arrayed AuNPs by SALDI-MS
Ionization of two peptides, [Sar1, Thr8]-angiotensin II (M1 = 956.1 Da) and neurotensin (M2 = 1672 Da) was carried out to demonstrate how the calcinated AuNP array performs for matrix-free laser desorption/ionization (LD/I) mass spectrometry. These peptides were applied as a mixture in the amount of 20 pmol and ionization was carried out at the same arbitrary laser fluence (1900 a.u.). The solutions are deposited on the calcinated AuNP array, along with several surfaces for comparison that include a bare AuNP array (bearing no silicate), an established SALDI material (46 nm evaporated Au, 20 nm silicate, see ESI† for fabrication),19 and a commercial, stainless steel MALDI plate. The mass spectra are shown in Fig. 4. Only the peptides deposited on the commercial MALDI plate required an organic matrix for ionization, and in this study, CHCA proved suitable. While the peptide solution was diluted in CHCA matrix for optimal signal, the amount of analyte applied to the plate remained constant (fixed 20 pmol peptides per 1 μL spot). In the other substrates, which did not utilize a matrix, 10 mM citric acid was incorporated into the experimental solutions as an external proton donor to promote the formation of protonated ions over alkali-adducted ions.44 Three peaks are prominent in each spectrum: the signals at m/z 957, 978, and 1673 correspond to [M1 + H]+, [M1 + Na]+, and [M2 + H]+, respectively, and all exhibit signal intensities that are within the same order of magnitude. The sodium adducted peak, [M1 + Na]+, appears in the spectra from the calcinated substrates (Fig. 4a and c), which we have attributed to the sodium silicate precursor used during the array fabrication process. However, these were largely suppressed through competition of protons donated from citric acid, resulting in an ion intensity ratio of protonated ions to single sodium adducted ions greater than 7.5. Furthermore, all substrates tested showed little fragmentation of target analytes, indicating that the calcinated AuNP substrate functions effectively as a soft ionization source, with mass spectra that are simplified and easy to interpret.
Fig. 4.
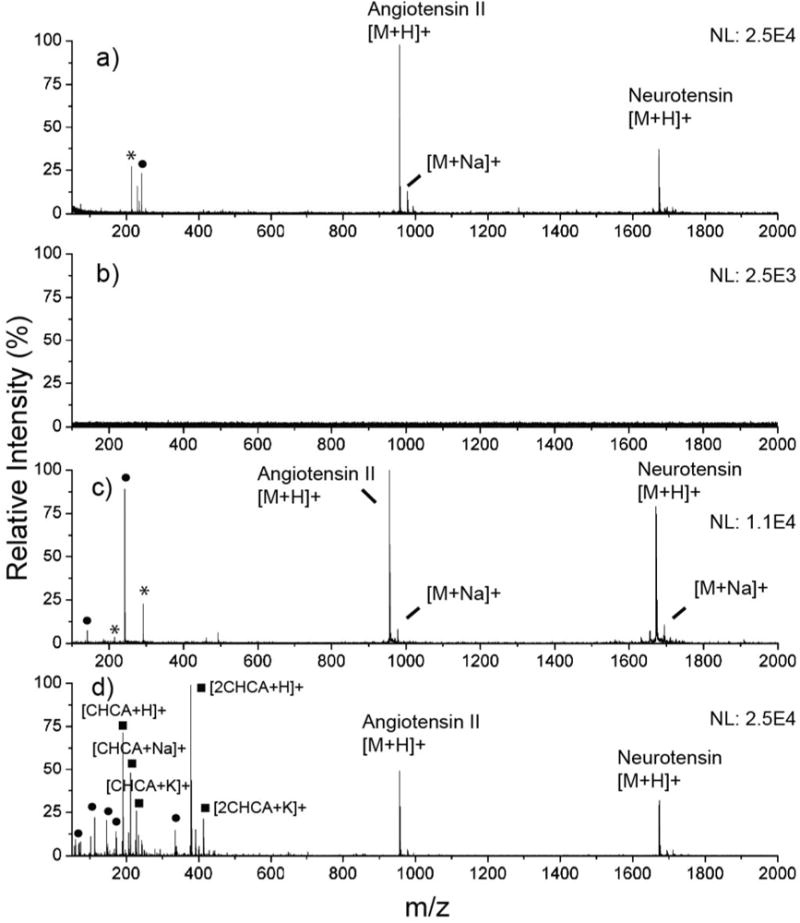
Ionization of [Sar1, Thr8]-angiotensin II and neurotensin from various laser desorption/ionization substrates at laser fluence of 1900 a.u. S/N ratios are given for the angiotensin II [M + H]+ peak. (a) Calcinated AuNP microarray (S/N = 72.0). (b) Bare AuNPs, with no silicate protection or calcination (no analyte signal). (c) Evaporated gold substrate with ca. 20 nm of silicate (S/N = 74.0). (d) CHCA matrix (S/N = 37.0).
AuNPs alone have served as an effective matrix for the desorption and ionization of biomolecules.14 However, the nanoglassification process used in this work further improves the efficiency of LD/I and renders the material superior for chip-based, mass spectrometric characterizations. This is clearly demonstrated in comparing Fig. 4a and b, in which the ultrathin silicate layer is present, and not present, respectively. In the absence of silicate (Fig. 4b), no effective ionization of either peptide was achieved at the laser fluence used (1900 a.u.). While ionization is still possible on the bare AuNP substrate, higher laser fluences are required. For all SALDI experiments involving the AuNPs, we used only one layer of silicate, as this demonstrated the best ionization performance compared to other silicate thicknesses. Further increases in thickness attenuated signal intensity, suggesting that heat generated at the nanoparticle surface is gradually subjected to insulation with thicker silicate layers. These findings were also supported by investigating the ionization of N-acetyl-tetradecapeptide renin substrate from AuNP–(PAH-silicate)1 constructs before (Fig. S5a†) and after calcination (Fig. S5b†), where the calcinated substrate displayed a much higher ionization intensity and signal-to-noise (S/N) ratio (S/N of 5.5 and 60 in the uncalcinated and calcinated substrates, respectively). PAH has been shown to potentially act as an insulator during the LD/I process,15 and will therefore further confine heat when left deposited with silicate. As PAH is removed during the calcination step, the overall insulation thickness is decreased by several nanometers, and tuned to optimal experimental conditions.
Performance toward small biomolecules and reusability
Cholesterol and its oxidation products are important small molecules of broad interest, mainly due to their roles in atherosclerosis,45 participation in a number of biosynthetic pathways,46 and unique biophysical trafficking mechanisms.47 Traditionally, one major drawback of MALDI-MS has been the presence of matrix-related ions at m/z <600, which convolute the spectra and make it difficult to discern meaningful data when ionizing small molecules in this mass range (e.g. sterols, hormones, amino acids, nucleic acids, and short chain carbohydrates). While sputtered silver films have successfully been used in MALDI imaging for high resolution spatial mapping of cholesterol in tissues,48 little success has been seen in ionizing cholesterol with gold nanomaterials, and with no matrix present. We have tested the ionization of cholesterol using the calcinated AuNP array and compared the results to those obtained from two common organic matrices to demonstrate the versatility of the calcinated film for small molecule detection and identification (Fig. 5). Cholesterol is known to oxidize under LD/I mass spectrometric methods, generating unique fragments and providing additional information for structural analysis and elucidation of various oxidation mechanisms.49–51 The laser-induced oxidation products for cholesterol show distinct peaks in each spectrum at m/z 288 and 316. The peaks in Fig. 5a, corresponding to products ionized from the calcinated AuNP film, have a very clean background compared to those obtained from CHCA (Fig. 5c) and DHB (Fig. 5d) matrices. The uncalcinated AuNP–(PAH-silicate)1 construct is also capable of generating a clean spectrum, albeit with slightly more background noise and lower absolute signal intensity (Fig. 5b). It is clear that in the matrix-assisted ionization analyte detection is overwhelmed by CHCA fragments and clusters (Fig. 5c), and while DHB seems to fare better for detection below m/z 600, many interfering ions still remain which may prohibit a comprehensive analysis (Fig. 5d).
Fig. 5.
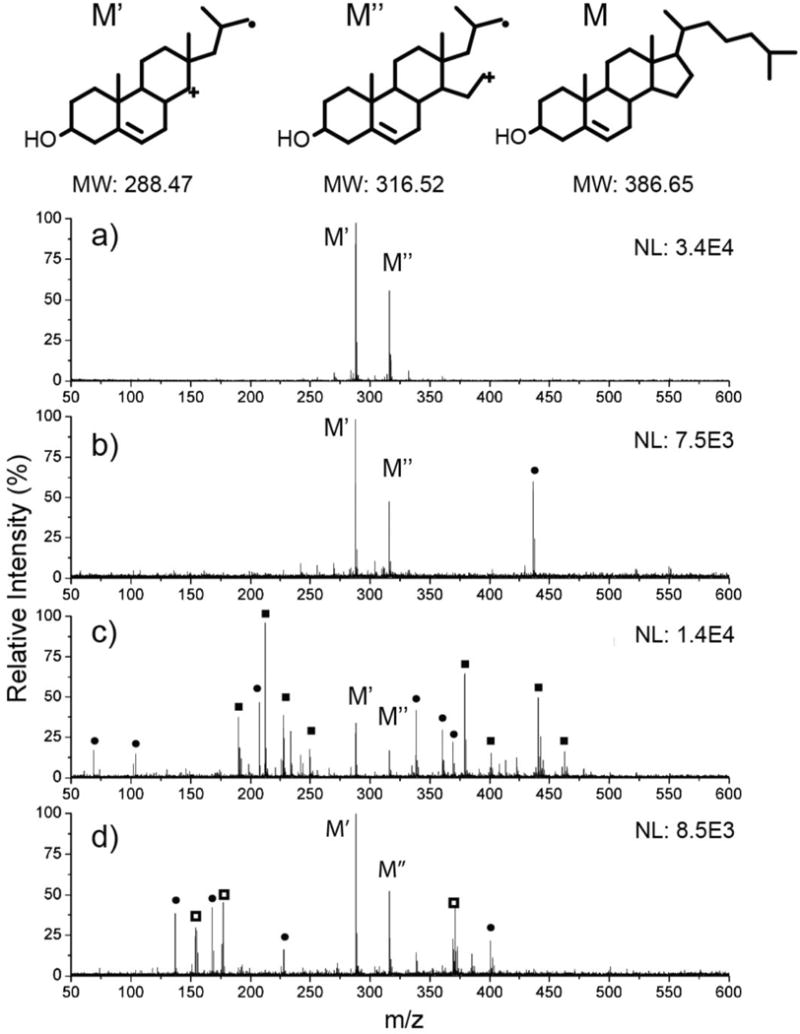
Ionization of cholesterol from various surfaces and matrices. S/N ratios are given for the M′ peak. (a) Calcinated gold nanoparticle array (S/N = 70.0). (b) Uncalcinated AuNP–(PAH-silicate)1 construct (S/N = 35.5). (c) CHCA matrix (S/N = 12.5). (d) DHB matrix (S/N = 36.0).
Aside from the lack of matrix interference, one of the advantages of using chip-assisted ionization is the ability to reuse the substrate for repeated measurements, saving costs in materials. The microarray presented in this work is no exception, and was tested for reusability. We applied 2 pmol of [Sar1, Thr8]-angiotensin II to each array spot, and after a mass spectrum was obtained, the chip was rinsed with 0.1 M ammonia and DI water (Fig. 6). Each time the surface is rinsed, no trace of the analyte remains, yet the signal can be regenerated within the same order of magnitude for subsequent measurements. The resulting signal abundance exhibits only an 8.4% relative standard deviation (n = 3 surface regenerations), with no observable damage to the substrate induced by the source laser. This is vastly different from many SALDI structures that are often subjected to surface damage over repeated use, resulting in a decline of ionization performance.52,53 The improved reusability is likely due to the fact that we require a much lower laser power for ionization compared to other SALDI materials, and due to the additional durability appended by the nano-silicate coating. The robust silicate coatings installed through the calcination process can in fact with stand significantly higher laser fluences than used here (up to 2500 a.u.), as we demonstrated in the previous work.18 Furthermore, the durability can be further increased through the application of long-chain monolayer coatings.18
Fig. 6.
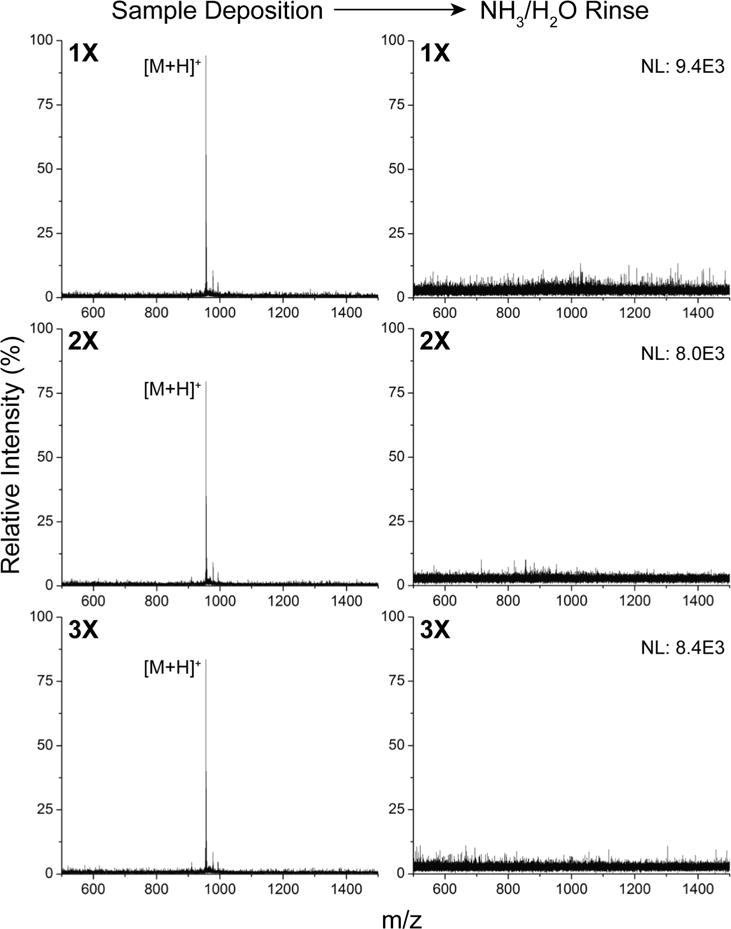
Reusability of the microarray. Ionization and rinsing of [Sar1, Thr8]-angiotensin II was repeated three times on the substrate, and the mass spectra for each deposition and rinse step is shown above. S/N is >35 for each deposition and regeneration.
The effect of laser fluence on the analyte signal abundance has previously been used to investigate LD/I efficiency from various SALDI materials,19,54 and we have applied this method to evaluate the performance of the calcinated AuNP array against CHCA. The laser fluence threshold for detecting measurable peptide signals on the calcinated AuNP array was determined to be 1450 a.u. (Fig. S6†), which is 150 units lower than analytes ionized from CHCA, and 470 units lower than those ionized from our previously developed evaporated gold SALDI material.19 In contrast, other SALDI structures, including porous silicon (DIOS) and porous alumina, possess much higher threshold laser fluences than CHCA.54 This shows that the calcinated AuNP array developed here has a clear advantage compared to other nano- and micro-sized structures. In addition, this array exhibits comparable performance to common organic matrices for small and medium sized molecules. This is reflected in its sensitivity for [Sar1, Thr8]-angiotensin II, which exhibits a limit of detection in the low femtomole range on a Voyager-DE STR MALDI-TOF mass spectrometer. Ionization of this analyte at 20 fmol gives a S/N ratio of 12.8, and a discernable signal for 2 fmol of peptide can still be achieved without any pre-enrichment or concentration steps (Fig. S7†).
Mechanistic investigations and simulations
We have demonstrated that the calcinated AuNP array is capable of achieving sensitive mass spectrometric measurements for small molecular mass compounds. While the mechanism of LD/I on the calcinated AuNP microarray could be very complex, here we discuss some of the factors that may contribute to the high LD/I efficiency observed. DIOS remains one of the most well studied systems for SALDI-MS, and it has been shown that DIOS performance benefits from the following features: confinement of heat within submicron sized gaps, crevices, and pores; effective heat transfer to analytes for desorption/ionization; and strong UV laser light absorption.12,13,54–56 Many of these properties align well with those of gold nanoparticle films, and Kawasaki et al. propose that during UV illumination, energy is absorbed by the interband transition of gold and converted to heat confined to the nanoparticle surface, causing analytes to thermally desorb from the material.15 Here, due to polymer induced bridging flocculation of neighboring AuNPs, a high number of nanoscale gaps and crevices have been created to further confine heat generated during the LD/I process (Fig. S3†), similar to other AuNP based SALDI materials.15–17 The ultrathin silicate layer also provides heat confinement, mostly to individual AuNP clusters, so that this energy is not dissipated throughout the entire gold substrate but instead turned into localized “hot spots” for analytes to desorb from.19 A fine balance appears to exist in heat confinement at the AuNP surface and total insulation, however. Increasing the thickness of this silicate layer beyond optimized conditions leads to lower LD/I efficiency, possibly due to isolation of heat transfer between the gold surface and the surrounding analyte medium, which has been seen in this and other work.19
In regards to absorption of UV light to produce heat, gold nanoparticles exhibit a reasonably high absorption coefficient in the UV range (Fig. 2a) and it has been shown this is mainly due to interband transition of d-orbital electrons.57 Following their photoexcitation, these electrons undergo quick (picoseconds) relaxation back to ground state through electron–electron and electron–phonon collisions, generating heat at the nanoparticle surface.15,57 This photoexcitation/relaxation mechanism is not only applicable to gold nanoparticles, but also to gold thin films, which function as effective SALDI substrates when properly insulated.19 Recently, the field of plasmon-induced hot carriers and engineered heating from metallic nanostructures has gained attention.58,59 Following the excitation of a localized surface plasmon, resulting in a nanoparticle-localized, oscillating electromagnetic field, this resonance is non-radiatively damped by the creation of hot electron–hole pairs via a process termed Landau damping.60 After carrier relaxation, in which the hot electrons redistribute energy via electron–electron interactions within the metal, heat is generated and transferred from the nanoparticle to the surroundings, resulting in an overall process that ranges from 100 ps to 10 ns.58 Given that the laser pulse width from our instrument is 3 ns, plasmon-induced excitation and heating may be allowed for within the provided timeframe, and in conjunction with the interband transition process. To test whether any plasmonic field exists for calcinated AuNP array structure at λ = 337 nm (operating wavelength of the instrument’s pulsed nitrogen laser), finite-difference time-domain (FDTD) simulations were conducted for various nanoparticle geometries consistent with the material developed here (Fig. 7). While the electromagnetic fields are in general weaker than those excited at λ = 520 nm (Fig. S8†), there appear to be localized surface plasmons at λ = 337 nm (Fig. 7a–c) and the intensities vary according to cluster geometry. It is interesting to note that plasmonic fields at λ = 337 nm are absent for the planar gold film (Fig. 7d). These results suggest that for both isolated gold nanoparticles and their aggregated states, plasmonic fields are generated under UV illumination, which may subsequently undergo Landau damping and produce heat. While the interband transition proposed by Kawasaki et al.15 for SALDI is likely the main source for localized heating, we speculate that the LSPR character of the AuNPs may also contribute, thereby enhancing ionization performance in comparison to planar thin film structures (as seen in Fig. 4a and c).
Fig. 7.
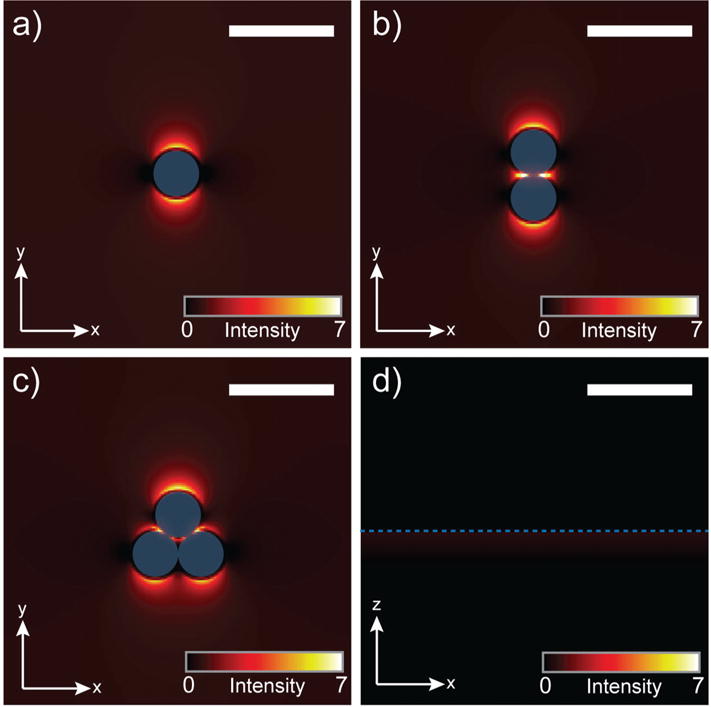
Numerical simulations of plasmonic fields induced by λ = 337 nm light source for different nanoscale geometries. Scale bars represent 30 nm and blue represents the gold surface. (a) Single gold nanoparticle. (b) Gold nanoparticle dimer. (c) Gold nanoparticle trimer. (d) Planar gold film.
Conclusions
The calcinated AuNP microarray represents an attractive tool for multiplexed analysis of biomolecules by SALDI-MS. The fabrication method is simple, and thus accessible to a wide audience, as no sophisticated cleanroom equipment is required. Layer-by-layer modification of the nanoparticles allows for nanoscale features that are easily tailored in a consistent fashion. This includes the formation of small AuNP clusters through selection of a suitable polyelectrolyte, and formation of a nanoscale layer of glass on the material surface. Performance was found to be better than many existing SALDI structures, and comparable to that of commonly used organic matrices. We attribute this to the rational design in the nanoscale morphology of the material, which has resulted in efficient heat transfer from the surface to the analyte. The more significant benefits this material has are less sample preparation and reusability of the substrate. As the accessibility of silane chemistry to the glassy surface allows for easy derivitization, the material may be further modified for on-chip enrichment, purification, and detection,18 providing a multifaceted and multifunctional substrate. The high performance reported here should inspire more widespread use of SALDI microarrays for higher throughput detection and characterization.
Supplementary Material
Acknowledgments
This research was supported by the National Science Foundation (CHE-1413449). S.S.H. was supported by an NIEHS T32 training grant (T32 ES018827). SEM images were generated at the Microscopy Core/Center for Plant Cell Biology at the Institute for Integrative Genome Biology at the University of California, Riverside. The authors thank Ronald B. New from the UCR Analytical Chemistry Instrumentation Facility for training and assistance in the MS measurements.
Footnotes
Electronic supplementary information (ESI) available: Additional methods and figures. See DOI: 10.1039/c5nr06635b
Notes and references
- 1.National Research Council (U.S.). Committee on Toxicity Testing and Assessment of Environmental Agents. Toxicity testing in the 21st century: a vision and a strategy. National Academies Press; Washington, DC: 2007. [Google Scholar]
- 2.Turner APF. Chem Soc Rev. 2013;42:3184–3196. doi: 10.1039/c3cs35528d. [DOI] [PubMed] [Google Scholar]
- 3.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 4.Hoheisel JD. Nat Rev Genet. 2006;7:200–210. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 5.Miller MB, Tang YW. Clin Microbiol Rev. 2009;22:611–633. doi: 10.1128/CMR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aebersold R, Goodlett DR. Chem Rev. 2001;101:269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 7.Edwards JR, Ruparel H, Ju JY. Mutat Res, Fundam Mol Mech Mutagen. 2005;573:3–12. doi: 10.1016/j.mrfmmm.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Clark AE, Kaleta EJ, Arora A, Wolk DM. Clin Microbiol Rev. 2013;26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo HY, DeLuca TA, Miller WM, Mrksich M. Anal Chem. 2013;85:10635–10642. doi: 10.1021/ac402614x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen LH, Gusev AI. Anal Bioanal Chem. 2002;373:571–586. doi: 10.1007/s00216-002-1321-z. [DOI] [PubMed] [Google Scholar]
- 11.Buriak JM, Wei J, Siuzdak G. Nature. 1999;399:243–246. doi: 10.1038/20400. [DOI] [PubMed] [Google Scholar]
- 12.Shen ZX, Thomas JJ, Averbuj C, Broo KM, Engelhard M, Crowell JE, Finn MG, Siuzdak G. Anal Chem. 2001;73:612–619. doi: 10.1021/ac000746f. [DOI] [PubMed] [Google Scholar]
- 13.Xiao YS, Retterer ST, Thomas DK, Tao JY, He L. J Phys Chem C. 2009;113:3076–3083. [Google Scholar]
- 14.McLean JA, Stumpo KA, Russell DH. J Am Chem Soc. 2005;127:5304–5305. doi: 10.1021/ja043907w. [DOI] [PubMed] [Google Scholar]
- 15.Kawasaki H, Sugitani T, Watanabe T, Yonezawa T, Moriwaki H, Arakawa R. Anal Chem. 2008;80:7524–7533. doi: 10.1021/ac800789t. [DOI] [PubMed] [Google Scholar]
- 16.Pilolli R, Ditaranto N, Di Franco C, Palmisano F, Cioffi N. Anal Bioanal Chem. 2012;404:1703–1711. doi: 10.1007/s00216-012-6243-9. [DOI] [PubMed] [Google Scholar]
- 17.Chen CY, Hinman SS, Duan J, Cheng Q. Anal Chem. 2014;86:11942–11945. doi: 10.1021/ac503808r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan JC, Wang H, Cheng Q. Anal Chem. 2010;82:9211–9220. doi: 10.1021/ac102262m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan J, Linman MJ, Cheng Q. Anal Chem. 2010;82:5088–5094. doi: 10.1021/ac100132x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deegan RD, Bakajin O, Dupont TF, Huber G, Nagel SR, Witten TA. Nature. 1997;389:827–829. doi: 10.1103/physreve.62.756. [DOI] [PubMed] [Google Scholar]
- 21.Frens G. Nature. 1973;241:20–22. [Google Scholar]
- 22.Schneider G, Decher G. Langmuir. 2008;24:1778–1789. doi: 10.1021/la7021837. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Duan JC, Cheng Q. Anal Chem. 2011;83:1624–1631. doi: 10.1021/ac1024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Addamo M, Augugliaro V, Di Paola A, Garcia-Lopez E, Loddo V, Marci G, Palmisano L. Colloids Surf, A. 2005;265:23–31. [Google Scholar]
- 25.Decher G. Science. 1997;277:1232–1237. [Google Scholar]
- 26.Willets KA, Van Duyne RP. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 27.Jensen TR, Duval ML, Kelly KL, Lazarides AA, Schatz GC, Van Duyne RP. J Phys Chem B. 1999;103:9846–9853. [Google Scholar]
- 28.Haes AJ, Van Duyne RP. J Am Chem Soc. 2002;124:10596–10604. doi: 10.1021/ja020393x. [DOI] [PubMed] [Google Scholar]
- 29.Raschke G, Kowarik S, Franzl T, Sönnichsen C, Klar TA, Feldmann J, Nichtl A, Kürzinger K. Nano Lett. 2003;3:935–938. [Google Scholar]
- 30.Dahlin AB, Jonsson MP, Hook F. Adv Mater. 2008;20:1436–1442. [Google Scholar]
- 31.Zhao SS, Bichelberger MA, Colin DY, Robitaille R, Pelletier JN, Masson JF. Analyst. 2012;137:4742–4750. doi: 10.1039/c2an35839e. [DOI] [PubMed] [Google Scholar]
- 32.Jackman JA, Zhdanov VP, Cho NJ. Langmuir. 2014;30:9494–9503. doi: 10.1021/la502431x. [DOI] [PubMed] [Google Scholar]
- 33.Yockell-Lelievre H, Bukar N, McKeating KS, Arnaud M, Cosin P, Guo Y, Dupret-Carruel J, Mougin B, Masson JF. Analyst. 2015;140:5105–5111. doi: 10.1039/c5an00694e. [DOI] [PubMed] [Google Scholar]
- 34.Hu JB, Chen YC, Urban PL. Anal Chim Acta. 2013;766:77–82. doi: 10.1016/j.aca.2012.12.044. [DOI] [PubMed] [Google Scholar]
- 35.Deegan RD. Phys Rev E: Stat Phys, Plasmas, Fluids, Relat Interdiscip Top. 2000;61:475–485. doi: 10.1103/physreve.61.475. [DOI] [PubMed] [Google Scholar]
- 36.Deegan RD, Bakajin O, Dupont TF, Huber G, Nagel SR, Witten TA. Phys Rev E: Stat Phys, Plasmas, Fluids, Relat Interdiscip Top. 2000;62:756–765. doi: 10.1103/physreve.62.756. [DOI] [PubMed] [Google Scholar]
- 37.Fischer BJ. Langmuir. 2002;18:60–67. [Google Scholar]
- 38.Li Y, McKenna EO, Parkes W, Pitt AR, Walton AJ. Appl Phys Lett. 2011;99:073703. [Google Scholar]
- 39.Tillman N, Ulman A, Schildkraut JS, Penner TL. J Am Chem Soc. 1988;110:6136–6144. doi: 10.1021/ja00226a031. [DOI] [PubMed] [Google Scholar]
- 40.Wasserman SR, Tao YT, Whitesides GM. Lang-muir. 1989;5:1074–1087. [Google Scholar]
- 41.Vallant T, Brunner H, Mayer U, Hoffmann H, Leitner T, Resch R, Friedbacher G. J Phys Chem B. 1998;102:7190–7197. [Google Scholar]
- 42.Nayak R, Knapp DR. Anal Chem. 2010;82:7772–7778. doi: 10.1021/ac1017277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips KS, Han JH, Martinez M, Wang ZZ, Carter D, Cheng Q. Anal Chem. 2006;78:596–603. doi: 10.1021/ac051644y. [DOI] [PubMed] [Google Scholar]
- 44.Chen CT, Chen YC. Rapid Commun Mass Spectrom. 2004;18:1956–1964. doi: 10.1002/rcm.1572. [DOI] [PubMed] [Google Scholar]
- 45.Kumar N, Singhal OP. J Sci Food Agric. 1991;55:497–510. [Google Scholar]
- 46.Rao KN. Anticancer Res. 1995;15:309–314. [PubMed] [Google Scholar]
- 47.Ikonen E. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 48.Dufresne M, Thomas A, Breault-Turcot J, Masson JF, Chaurand P. Anal Chem. 2013;85:3318–3324. doi: 10.1021/ac3037415. [DOI] [PubMed] [Google Scholar]
- 49.Andreu I, Bosca F, Sanchez L, Morera IM, Camps P, Miranda MA. Org Lett. 2006;8:4596–4600. doi: 10.1021/ol061854c. [DOI] [PubMed] [Google Scholar]
- 50.van den Brink OF, Ferreira ESB, van der Horst J, Boon JJ. Int J Mass Spectrom. 2009;284:12–21. [Google Scholar]
- 51.McAvey KM, Guan B, Fortier CA, Tarr MA, Cole RB. J Am Soc Mass Spectrom. 2011;22:659–669. doi: 10.1007/s13361-011-0074-3. [DOI] [PubMed] [Google Scholar]
- 52.Wada Y, Yanagishita T, Masudata H. Anal Chem. 2007;79:9122–9127. doi: 10.1021/ac071414e. [DOI] [PubMed] [Google Scholar]
- 53.Alimpiev S, Grechnikov A, Sunner J, Karavanskii V, Simanovsky Y, Zhabin S, Nikiforov S. J Chem Phys. 2008;128:014711. doi: 10.1063/1.2802304. [DOI] [PubMed] [Google Scholar]
- 54.Okuno S, Arakawa R, Okamoto K, Matsui Y, Seki S, Kozawa T, Tagawa S, Wada Y. Anal Chem. 2005;77:5364–5369. doi: 10.1021/ac050504l. [DOI] [PubMed] [Google Scholar]
- 55.Li J, Lipson RH. J Phys Chem C. 2013;117:27114–27119. [Google Scholar]
- 56.Alhmoud HZ, Guinan TM, Elnathan R, Kobus H, Voelcker NH. Analyst. 2014;139:5999–6009. doi: 10.1039/c4an01391c. [DOI] [PubMed] [Google Scholar]
- 57.Shoji M, Miyajima K, Mafune F. J Phys Chem C. 2008;112:1929–1932. doi: 10.1021/jp803247h. [DOI] [PubMed] [Google Scholar]
- 58.Brongersma ML, Halas NJ, Nordlander P. Nat Nanotechnol. 2015;10:25–34. doi: 10.1038/nnano.2014.311. [DOI] [PubMed] [Google Scholar]
- 59.Govorov AO, Richardson HH. Nano Today. 2007;2:30–38. [Google Scholar]
- 60.Li XG, Xiao D, Zhang ZY. New J Phys. 2013;15:023011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


