Drosophila neural stem cells, or neuroblasts, ingress from the neuroepithelium in an EMT-like process, during which the apical cell domain is lost. Apical constriction of neuroblasts and the serial loss of cell–cell contacts require periodic pulses of actomyosin that cause progressively stronger ratcheted contractions of the neuroblast apical cortex.
Abstract
Epithelial–mesenchymal transitions play key roles in development and cancer and entail the loss of epithelial polarity and cell adhesion. In this study, we use quantitative live imaging of ingressing neuroblasts (NBs) in Drosophila melanogaster embryos to assess apical domain loss and junctional disassembly. Ingression is independent of the Snail family of transcriptional repressors and down-regulation of Drosophila E-cadherin (DEcad) transcription. Instead, the posttranscriptionally regulated decrease in DEcad coincides with the reduction of cell contact length and depends on tension anisotropy between NBs and their neighbors. A major driver of apical constriction and junctional disassembly are periodic pulses of junctional and medial myosin II that result in progressively stronger cortical contractions during ingression. Effective contractions require the molecular coupling between myosin and junctions and apical relaxation of neighboring cells. Moreover, planar polarization of myosin leads to the loss of anterior–posterior junctions before the loss of dorsal–ventral junctions. We conclude that planar-polarized dynamic actomyosin networks drive apical constriction and the anisotropic loss of cell contacts during NB ingression.
Introduction
Epithelial–mesenchymal transitions (EMTs) are fundamental to animal development and the dissemination of epithelial tumor cells (Baum et al., 2008; Kalluri and Weinberg, 2009; Thiery et al., 2009; Lim and Thiery, 2012; Ye and Weinberg, 2015; Nieto et al., 2016). In a developmental context, many EMT-like processes are termed ingression and involve the loss of apical–basal polarity, including the disassembly of cell–cell junctions and the acquisition of stem cell and/or migratory capacity. When coupled to cell death, cells are often extruded from the epithelium through forces generated by neighboring cells in response, for example, to tissue overcrowding or mechanical tension (Marinari et al., 2012; Sokolow et al., 2012; Gudipaty and Rosenblatt, 2016; Levayer et al., 2016). In contrast, ingression events that generate novel cell types are promoted by cell shape change as a result of intrinsic cell specification programs (Wu et al., 2007; Hartenstein and Wodarz, 2013; Lamouille et al., 2014).
Cell–cell junctions organize epithelial tissues into cohesive polarized sheets. Adherens junctions (AJs) and their core component, E-cadherin, are linked to the cortical actomyosin cytoskeleton, allowing tension transmission across the tissue (Harris and Tepass, 2010; Yonemura et al., 2010; Desai et al., 2013; Lecuit and Yap, 2015). Loss of E-cadherin is widespread in epithelial tumors and is thought to be crucial in many cases for the escape of cells from their native epithelium (Jeanes et al., 2008; Thiery et al., 2009; Balzer and Konstantopoulos, 2012). A central notion has been that transcriptional repression of E-cadherin by factors that drive the EMT program, such as Snail, can initiate EMT (Batlle et al., 2000; Cano et al., 2000; Peinado et al., 2004). More recent work has also implicated posttranscriptional mechanisms in the disassembly of AJs, including cortical constriction driven by the nonmuscle myosin II motor (referred to as myosin in the following: Bertet et al., 2004; Fernandez-Gonzalez et al., 2009; Rauzi et al., 2010; Simões et al., 2010). However, the relative importance and level of cooperation of transcriptional and posttranscriptional mechanisms directing the loss of cell junctions during ingression/EMT remains unclear.
Several developmental models have been used to study cell ingression/EMT, including the primary mesenchymal cells of sea urchin embryos (Wu and McClay, 2007; Wu et al., 2007), formation of the inner cell mass in the early mouse embryo (Abell et al., 2011; Samarage et al., 2015), the neural crest cells in vertebrate embryos (Sauka-Spengler and Bronner-Fraser, 2008; Minoux and Rijli, 2010; Theveneau and Mayor, 2011), the internalization of endoderm cells in Caenorhabditis elegans (Pohl et al., 2012; Roh-Johnson et al., 2012), and cardiomyocytes in the developing hearts of zebrafish (von Gise and Pu, 2012). It can be challenging to track the molecular features of ingressing cells throughout the entire process at high temporal and spatial resolution because of tissue topography and temporal constraints or because ingression of single cells in epithelia can be a stochastic process (Marinari et al., 2012).
In trying to overcome some of these limitations, we examined ingressing neural stem cells or neuroblasts (NBs) in the Drosophila melanogaster embryo. NBs ingress as single cells, detaching from their neighbors and moving inside the embryo (Fig. 1 A), where they undergo asymmetric division to produce the neurons and glia cells of the central nervous system (Hartenstein and Wodarz, 2013). In this study, we analyzed the pattern and dynamics of apical constriction and junctional disassembly of NBs. In addition, we address questions about the dynamics and function of actomyosin and the role of neighboring noningressing cells (NICs) in controlling NB ingression.
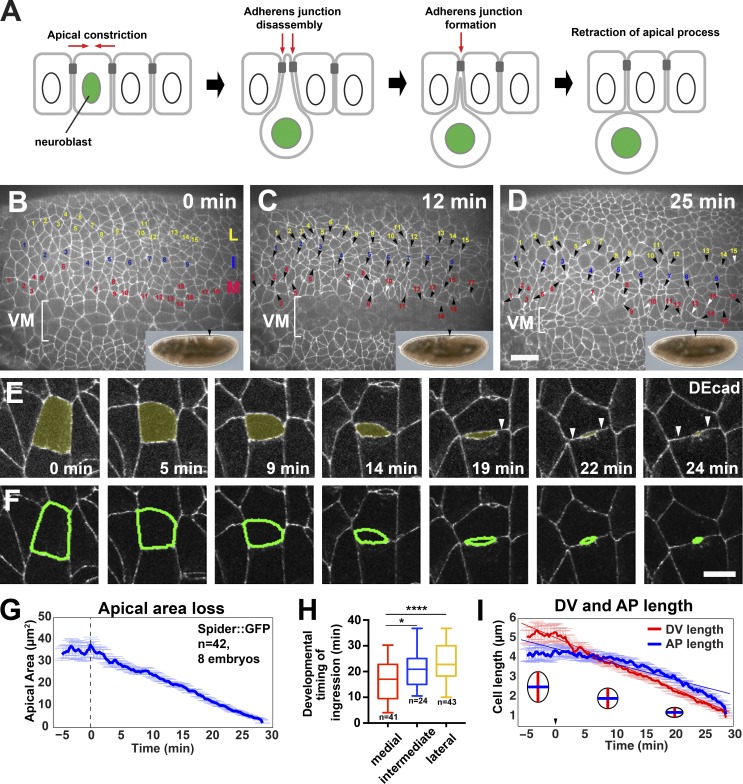
Figure 1.
NBs ingress via an anisotropic loss of apical cell–cell junctions. (A) An NB undergoes apical constriction and disassembles its AJs while moving out of the epithelium. (B–D) Stills of a time-lapse video of the ectoderm during stage 8 (membrane marker Spider::GFP). Numbered cells are medial (red, M), intermediate (blue, I), and lateral (yellow, L) NBs, located 1–3, 4, and 7–9 cell rows away from the VM, respectively. White arrowheads indicate the position of NBs that have lost their apical membrane. Insets show the corresponding stage of embryonic axis elongation (black arrowheads, posterior end). Anterior, left; dorsal, up. (E) Stills of a time-lapse video of an NB (apical domain, yellow; ubi-DEcad::GFP). Arrowheads point to new horizontal junctions forming between DV NICs. (F) Watershed segmentation (green outline) of the ingressing NB in E. Bars: (B–D) 15 µm; (E and F) 5 µm. (G) Mean loss of apical cell area during ingression. Time 0 (dashed line) indicates the onset of ingression. (H) Developmental timing of completion of ingression by NB row. Medial NBs complete ingression first, followed by the intermediate and lateral rows. Time 0 corresponds to the onset of cytokinesis in VM cells. Box plot with IQRs and minimum/maximums displayed. *, P = 0.037; ****, P = 0.0001 (two-tailed t test). (I) DV and AP apical cell length during ingression (Spider::GFP, n values as in G). The apical surface shrinks along the DV axis 1.6× faster than along the AP axis. Time 0 is the onset of ingression. Data presented are means ± SEM.
Results
NBs ingress through the anisotropic loss of cell–cell junctions
NBs ingress from the ventral ectoderm in multiple waves between stages 8 and 11 of embryogenesis (Hartenstein and Campos-Ortega, 1984; Hartenstein and Wodarz, 2013). NBs of the first wave (S1 NBs), which we have analyzed here, ingress as individual cells surrounded by NICs and are patterned as three longitudinal rows—medial, intermediate, and lateral—that run parallel to the ventral midline (VM; Fig. 1, B–D; and Video 1). Live imaging using the membrane marker Spider::GFP (Buszczak et al., 2007) revealed that 93% of the initial apical cell area of NBs (37.5 ± 3.6 µm2) was progressively lost within 29 min (Fig. 1, E–G). The total time required for cells to complete ingression was not statistically different among NBs of different rows (medial row, 31.4 ± 5.9 min; intermediate row, 36.2 ± 7.9 min; lateral row, 30.5 ± 5.3 min). However, we found that medial NBs completed ingression earlier than intermediate and lateral NBs (Fig. 1 H). Proneural clusters of medial NBs showed expression of the proneural transcription factor Achaete (Ac) during stage 7 (3 h and 10 min to 3 h and 20 min after egg laying), preceding lateral clusters, which were first detected at early stage 8 (3 h and 20 min after egg laying; Fig. 2, A and B). These findings suggest a ventral–dorsal specification and ingression gradient, with medial NBs specified and finishing ingression 4–7 min earlier than intermediate and lateral NBs.
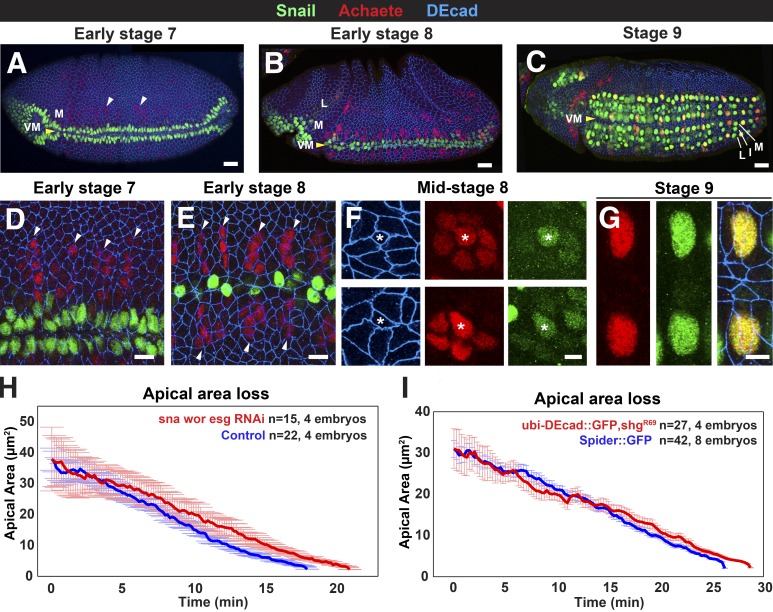
Figure 2.
Sna family proteins and DEcad transcriptional down-regulation are not essential for NB ingression. (A–C) Embryos expressing Sna::GFP (green) and stained for Ac (red) and DEcad (blue). I, intermediate row; L, lateral row; M, medial row. Medial proneural clusters express Ac at early stage 7 (A, arrowheads), preceding lateral expression seen at the onset of stage 8 (B); Sna is robustly expressed in ingressed NBs during stage 9 (C). (D and E) Sna (green) is expressed by VM cells but not in Ac-positive medial proneural clusters (arrowheads) at stage 7 (D) and early stage 8 (E). (F) Ingressing NBs (asterisks) express Sna and enrich Ac at mid–stage 8. (G) Two medial NBs flanking the midline expressing Ac and Sna at stage 9. Bars: (A–C) 50 µm; (D and E) 10 µm; (F and G) 5 µm. (H) Mean apical area loss during NB ingression in mock-injected controls (blue) and embryos depleted for Sna, Esg, and Wor (red) by RNAi. (I) Mean apical area loss during NB ingression in Spider::GFP controls (blue) and shgR69 mutants expressing ubi-DEcad::GFP (red). Data presented are means ± SEM.
Live imaging of NB apical junctions revealed that their loss was anisotropic: anterior–posterior (AP)/vertical junctions started shrinking 7 min earlier, on average, than dorsal–ventral (DV)/horizontal junctions, and AP junctions shrunk at a faster rate of 0.51 µm/min, versus 0.3 µm/min for DV junctions, resulting in the acquisition of an ellipsoid, lentil-like apical cell shape (Fig. 1, E, F, and I). Adjacent NICs that were separated by an NB formed new junctions concomitant with NB ingression (Fig. 1 E, arrowheads). Thus, ingression is achieved by a planar-polarized sequential loss of cell–cell junctions and is tightly coupled to the formation of new junctions to ensure epithelial continuity.
NB ingression does not require transcriptional repression of Drosophila E-cadherin (DEcad)
The expression of the Snail family of transcriptional repressors has been directly associated with EMT initiation (Peinado et al., 2007). One of the proposed mechanisms by which Snail induces EMT is through the transcriptional repression of E-cadherin (Batlle et al., 2000; Cano et al., 2000). Drosophila Snail (Sna) and the Sna paralogs Escargot (Esg) and Worniu (Wor) are known to be up-regulated in NBs (Ashraf et al., 1999; Ashraf and Ip, 2001). We examined the dynamics of Sna protein distribution with antibodies (Fig. S1 A) and a Sna::GFP reporter (Fig. 2, A–G). Sna was first detected in NBs during mid–stage 8, when ingression was under way. Ac- and Sna-positive cells showed a reduced apical cell surface (10.8 ± 5.7 µm2; n = 35) at mid–stage 8 (Fig. 2 F) compared with Ac-positive Sna-negative cells at early stage 8 (30.6 ± 8.6 µm2; n = 57). Upon completion of ingression, Sna was expressed in all NBs (Fig. 2, C and G).
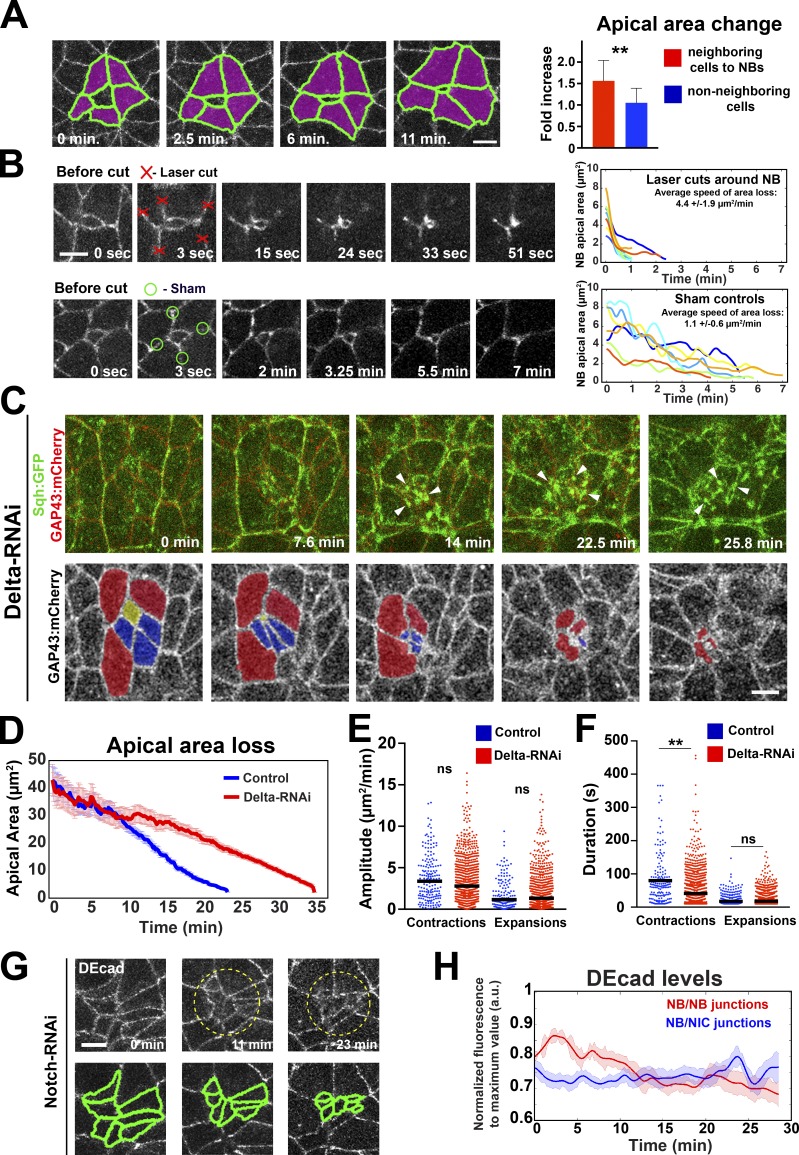
To assess the function of Sna and its paralogues, we simultaneously depleted Sna, Esg, and Wor and found no difference in ingression kinetics in these embryos compared with controls (Figs. 2 H and S1 B). NBs also ingressed in embryos homozygous for a deletion that uncovers sna, esg, and wor, similar to heterozygous controls (Fig. S1 C). We conclude that Snail family transcriptional repressors are not essential for NB ingression.
shotgun (shg), the gene that encodes DEcad, is down-regulated in NBs (Tepass et al., 1996). To directly test whether the transcriptional repression of shg by Snail family proteins or other transcription factors is required for ingression, we examined embryos that constitutively expressed DEcad under the control of the ubiquitin promoter (Oda and Tsukita, 2001) in an shg-null mutant background. The ingression dynamics in these embryos were indistinguishable from controls (Fig. 2 I). However, DEcad loss from NBs was progressive and correlated with the reduction of apical circumference so that DEcad concentration per surface unit remained constant during ingression (Figs. 1 E and S2). Collectively, these results suggest that posttranscriptional mechanisms, but not DEcad transcriptional down-regulation, cause the disassembly of AJs during NB ingression.
Apical domain loss in NBs results from progressively stronger contractions followed by short and weak expansions
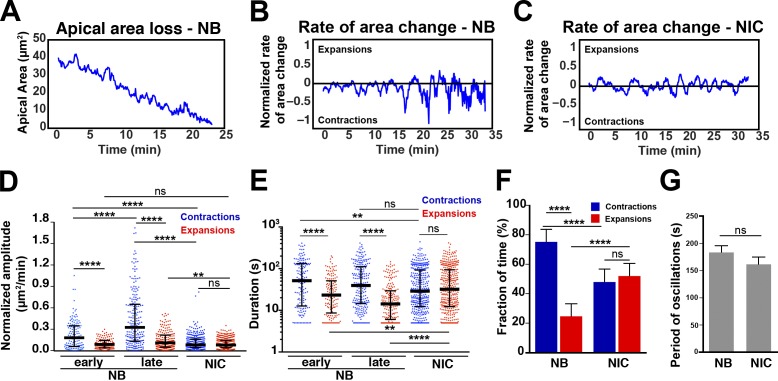
Actomyosin contractility is responsible for apical constriction and the loss of cell contacts in several tissues (Martin and Goldstein, 2014). To establish a framework for the interpretation of actomyosin function in NBs, we characterized the dynamics of apical membrane loss. Individual NBs underwent ∼12 cycles of apical contraction and expansion through which apical domain size was progressively reduced, pointing to a ratchet-like mechanism for apical constriction (Fig. 3, A and B). NICs displayed periodic fluctuations of their apical membrane with balanced rates and timing of expansions and contractions, thus maintaining their apical domains (Fig. 3 C). In contrast, NBs underwent consecutive cycles of strong and long contractions followed by weak and short expansions (Fig. 3 B). During the first two thirds of ingression, the amplitude (maximum relative change in apical area per cycle) and duration of contractions were 2.1-fold and 1.8-fold higher in NBs than in NICs, respectively (Fig. 3, D and E), whereas the duration of expansions was 28% lower in NBs compared with NICs (Fig. 3 E). Notably, the amplitude of contractions increased further by fourfold during the last 10 min of ingression compared with NICs, and expansions became 56% shorter (Fig. 3, D and E). Overall, NBs spent 25% longer contracting their apical surface during ingression than NICs (Fig. 3 F). We observed no significant difference in the periodicity of apical oscillations between NBs and temporally matched NICs (Fig. 3 G). Collectively, we found that the apical constriction of NBs results from a progressive increase in the strength of apical contractions and a corresponding decrease in the duration of apical expansions.
Figure 3.
Apical constriction of NBs is achieved by cycles of strong contractions followed by short and weak expansions. (A) Single profile of apical membrane area loss during ingression. (B and C) Rate of apical area change in an NB (B) and a NIC (C) normalized to the apical area size at each time point. Positive rates are expansions, negative rates are contractions, and time 0 depicts the onset of ingression. (D) Relative area change (area(t) − area(t − 1 min)/area(t)) during contractions and expansions in NBs and NICs. Onset to end minus 10 min was considered early ingression, and the last 10 min was considered late ingression. n = 123–328 contractions/expansions per condition. From left to right: ****, P = 2.4 × 10−9; 5.3 × 10−7; 9.6 × 10−12; 4.7 × 10−17; and 2.4 × 10−27; **, P = 0.005; ns (not significant), P = 0.97 and 0.33 (KS test). n = 19 NBs and 19 NICs (seven embryos). (E) Durations of contractions and expansions in seconds. Median values: 51.5, 39.4, and 28.8 s for contractions and 23.1, 14.2, and 32.2 s for expansions. From left to right: ****, P = 3.6 × 10−6; 6.6 × 10−12; and 1.7 × 10−10; **, P = 0.003; ns, P = 0.11 and 0.75 (KS test). Bars are IQRs. n values are as in D. (F) Fraction of time NBs and NICs spend contracting and expanding their apical membranes. From left to right: ****, P = 1.2 × 10−9; 1.4 × 10−11; and 1.1 × 10−11; ns, P = 0.155 (KS test). n values are as in D. (G) Mean period of apical oscillations in NBs and NICs; 183.6 ± 48.9 s for NBs and 161.7 ± 58.9 s for NICs. P = 0.25 (KS test). Data presented are means ± SEM. n values are as in D.
Junctional and medial myosin contributes to apical domain loss in NBs
The anisotropic and oscillatory patterns of apical domain loss during NB ingression is similar to the planar-polarized shrinkage of cell–cell junctions during epithelial cell intercalation (Bertet et al., 2004; Zallen and Wieschaus, 2004; Blankenship et al., 2006) and the apical constriction seen in Drosophila mesoderm cells (Martin et al., 2009; Xie and Martin, 2015). During intercalation, a junctional pool of myosin is thought to drive planar disassembly of AJs, whereas in invagination, a pulsatile medial pool of myosin pulls on AJs to promote apical contractions. NBs combine apical constriction with the concurrent planar loss of AJs. We hypothesized therefore that both junctional and medial pools of myosin could make important contributions to NB ingression.
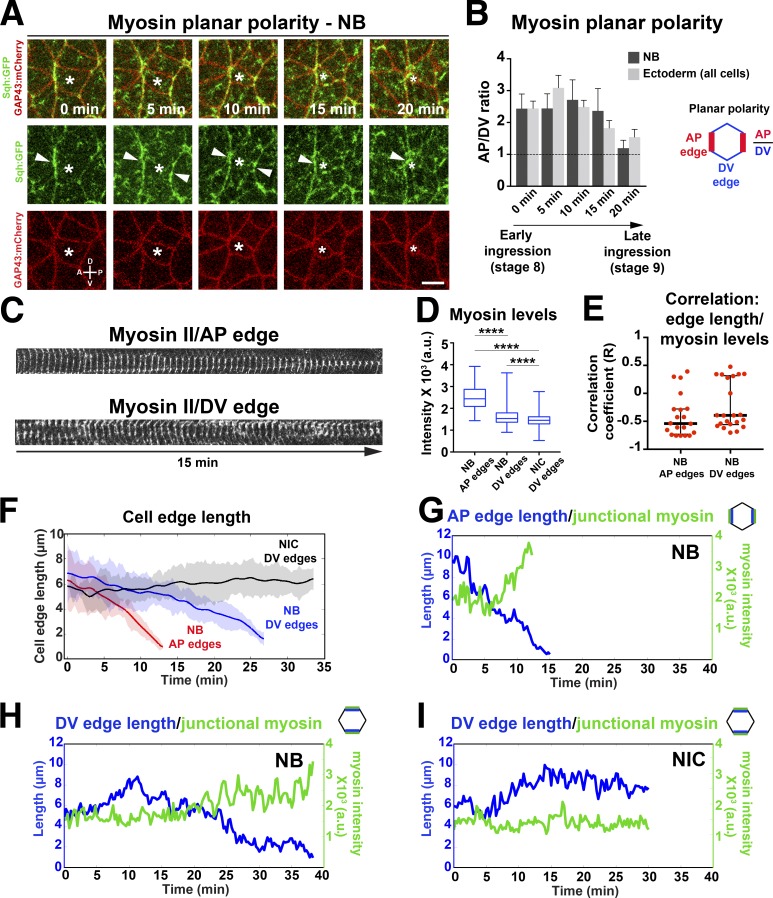
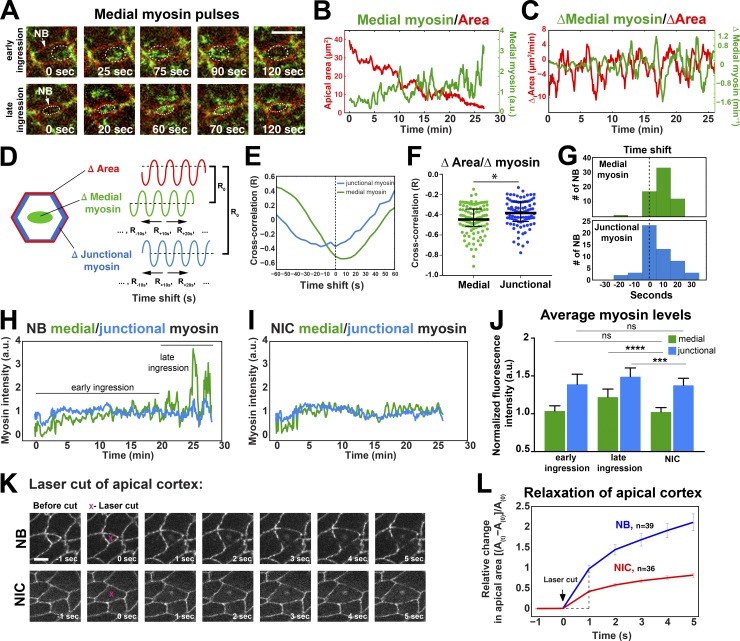
We examined myosin dynamics in relation to oriented AJs (edge) disassembly (Fig. 4) and apical membrane loss (Fig. 5). Junctional myosin was planar polarized and enriched at AP edges in NBs (Fig. 4, A–D). AP myosin enrichment correlated with faster AP edge contraction compared with DV edges (Figs. 1 I and 4 F). AP junctions displayed a pulsatile and cumulative pool of myosin as they contracted (period of pulses, 138 ± 7 s; Fig. 4, C and G), and increasing myosin levels correlated with reductions in edge length (Fig. 4 E). Myosin levels at disassembling DV junctions were also pulsatile (period of pulses, 150 ± 6 s; Fig. 4, C and H) and inversely correlated with edge length (Fig. 4 E). In contrast to AP edges, myosin was less persistent at DV edges (Fig. 4, C, G, and H) and showed levels intermediate between NB AP junctions and DV junctions of NICs (Fig. 4, D and G–I). Thus, NBs have two pulsatile junctional pools of myosin with distinct persistence: a cumulative pool at AP junctions associated with fast edge contraction and a weaker, less persistent pool at DV junctions associated with a slower rate of AJ disassembly.
Figure 4.
Planar-polarized myosin distribution correlates with planar disassembly of NB cell contacts. (A) Stills of a time-lapse video of an NB (asterisk) labeled with Sqh::GFP (myosin, green) and GAP43::mCherry (cell membrane, red). Myosin is enriched at AP junctions (arrowheads). Time point 20 min corresponds to a 2.5-µm2 apical area in NBs. Bar, 5 µm. P, posterior, A, anterior, D, dorsal, V, ventral. (B) Ratios between myosin intensity at AP (75–90°) and DV (0–15°) edges in NBs (n = 13 cells, seven embryos; data presented are means ± SEM) and in ectoderm cells during ingression. Time points are as in A. (C) Kymographs of myosin (Sqh::GFP) during the last 15 min of AP and DV edge disassembly in NBs. DV edges show more discontinuous myosin distribution than AP edges. Time intervals, 15 s. (D) Myosin levels (IQRs) in AP junctions (n = 12) and DV junctions (n = 11) in NBs versus DV junctions in NICs (n = 10). From left to right: ****, P = 2.5 × 10−173; 2.8 × 10−219; and P = 2.7 × 10−13 (KS test). (E) Correlation coefficients between AP/DV edge length and junctional myosin levels during ingression. Median correlation coefficient for AP edges was R = −0.54 and for DV edges was R = −0.39. Bars are IQRs. n = 20 junctions per condition, nine embryos. (F) AP and DV NB edge length during ingression. t = 0 min, onset of ingression. Speed of AP and DV edge contraction in NBs (means ± SD): −0.72 ± 0.3 µm/min and −0.33 ± 0.1 µm/min, respectively. P = 5 × 10−5 (two-tailed t test). Speed of DV edge length in NICs: 0.05 ± 0.06 µm/min. n = 11 junctions per condition, two embryos. (G–I) Representative plots of junctional myosin levels and AP or DV edge length in an NB and NIC. a.u., arbitrary units.
Figure 5.
Oscillations in medial and junctional myosin levels drive progressively stronger contractions in NBs. (A) Cycles of medial myosin assembly and disassembly during early and late NB ingression. The cell membrane is labeled in red (GAP43::mCherry) and myosin in green (Sqh::GFP). Dashed circles track a pulse of medial myosin II assembly and disassembly during early and late ingression. (B and C) Representative plots of medial myosin levels versus apical area (B) and rates of medial myosin and area change (C) during ingression. (D and E) Myosin data are shifted backward and forward in time to calculate cross-correlations between changes in medial or junctional myosin and cell area during ingression. R0 is the correlation coefficient with no time shift. A typical result is shown in E. (F and G) Maximum cross-correlation coefficients (F) and time shift distributions (G) as indicated in D and E. n = 63 NBs, 15 embryos. IQRs for medial myosin were −0.51, −0.45, and −0.34 and for junctional myosin were −0.47, −0.38, and −0.27. *, P = 0.01 (two-tailed t test). (H and I) Mean medial and junctional levels of myosin in an NB (H) and a temporally matched NIC (I). (J) Mean medial and junctional myosin levels during early ingression (up to last 10 min), late ingression (last 10 min), and in temporally matched NICs. n = 22 cells per condition. ***, P = 0.001; ****, P = 2 × 10−9; ns (not significant), P = 0.5 (left) and 0.7 (right; two-tailed t test). Data presented are means ± SD. a.u., arbitrary units. (K and L) Expansion of the apical cortex after laser ablation in NBs and NICs in endo-DEcad::GFP embryos. Maximum relative apical expansion (vertical dashed line in L): 97.0 ± 8.6% for NBs (n = 39) and 41.6 ± 3.3% for NICs (n = 36). P = 2.28 × 10−7 (two-tailed t test). Data presented are means ± SEM. (A and K) Bars, 5 µm.
Actomyosin also forms a pulsatile network at the medial free apical cortex during NB ingression, assembling and disassembling with a periodicity of 132 ± 29 s (Fig. 5, A–C; and Video 2). To assess which myosin pool could drive the ratchet-like contractions underlying apical domain loss, we calculated the correlation between medial or junctional myosin and apical area changes. Changes in apical cell area negatively correlated with changes in both myosin pools, with a slightly higher anticorrelation seen for medial myosin (Fig. 5 F), suggesting that both myosin pools contribute to apical contractions. Moreover, we found that medial myosin maximally accumulated within 20 s before the cell area was reduced in 71% of NBs, whereas in 27% of cells, maximal levels of medial myosin and cell area reduction coincided. In contrast, peak junctional myosin levels coincided with area change in 44% of NBs and preceded area change within 30 s in 40% of NBs (Fig. 5, D, E, and G). The remaining cells showed peak myosin accumulation—either medial or junctional—within seconds after apical area reduction. Assuming that the reduction of apical area in NBs is the immediate consequence of actomyosin contractility and stable linkage of actomyosin to the cell junctions, our findings suggest that assembly/activity of medial or junctional myosin may directly cause apical contraction.
Apical myosin levels of both junctional and medial pools were strongly enriched during the last 10 min of NB ingression compared with temporally matched NICs (Fig. 5, H–J). The striking increase in myosin during late ingression correlated with strong apical contractions (Fig. 3) and complete AP and DV edge disassembly. In contrast, during early ingression, when most of the myosin-rich AP edges disassemble, we found no significant differences in myosin level between ingressing cells and NICs (Fig. 5, H–J). To directly assess whether NBs during late ingression sustain increased apical tension, we used laser ablation and examined the retraction speed of the apical cortex. Late ingressing NBs displayed retraction velocities that were three times higher than those of NICs (Fig. 5, K and L). Collectively, these results suggest that ingression is not triggered by a detectable increase in apical myosin levels but that ingressing NBs progressively sustain stronger myosin pulses and, correspondingly, apical tension as they constrict their apical domain.
Medial myosin flows support AP and DV junctional myosin accumulation in NBs
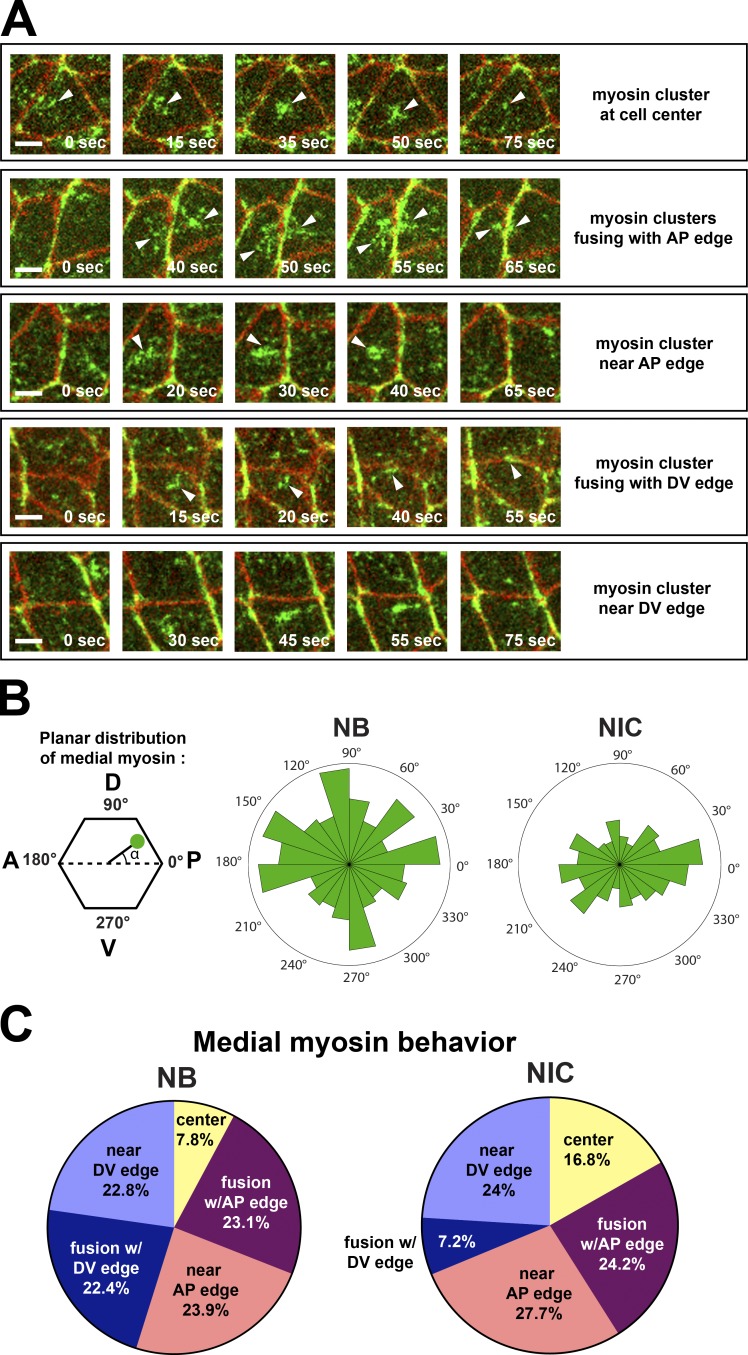
Medial myosin clusters move within the apical plane and may fuse with cell junctions (Rauzi et al., 2010; Weng and Wieschaus, 2016). To ask whether medial myosin could differentially contribute to increased myosin levels at AP and DV junctions in NBs, we compared the directionality of medial myosin and its frequency of fusion with cell junctions in NBs versus NICs. In NICs, medial myosin clusters were biased to flow toward AP edges (Fig. 6, A and B), fusing with them (24.2%) or disassembling in their vicinity (27.7%), but rarely fusing with nonconstricting DV edges (7.2%; Fig. 6 C). In contrast, medial myosin clusters in NBs flowed equally toward AP or DV edges (Fig. 6, A and B) and fused with constricting AP and DV edges at similar frequencies (23.1% and 22.4%, respectively; Fig. 6 C). Moreover, medial myosin fused 44% more frequently with junctions in NBs than in NICs (Fig. 6 C). Medial myosin pulses in NICs adjacent to NBs only correlated significantly with AP but not DV edge disassembly during ingression (Fig. S3). Collectively, these findings suggest that medial myosin flows to both AP and DV junctions to promote their disassembly in NBs. Medial myosin from both NBs and their NIC neighbors can contribute to junctional AP myosin pools, accounting at least in part for the enhanced myosin levels at AP edges and, consequently, the earlier and faster disassembly of these cell contacts.
Figure 6.
Medial myosin flows and fuses with AP and DV edges during NB ingression. (A) Classes of medial myosin behavior. Myosin clusters can assemble and coalesce at the cell center (>1 µm away from cell edges) or near (<1 µm) an AP or DV edge and either fuse (second and fourth rows) or not fuse (third and fifth rows) with that edge. Arrowheads track the movement of medial myosin clusters during NB ingression. Bars, 3 µm. (B) Planar distribution of medial myosin clusters. NBs show uniform planar distribution of myosin at the apical cortex, whereas medial myosin is more biased to flow parallel to the AP axis in NICs. P, posterior (0°), A, anterior (180°), D, dorsal (90°), V, ventral (270°). (C) Quantification of medial myosin behavior in NBs and NICs as described in A. n = 268 and 512 myosin clusters in NBs and NICs, respectively (19 cells per condition; seven embryos).
Myosin drives apical contractions and the anisotropic loss of cell junctions during ingression
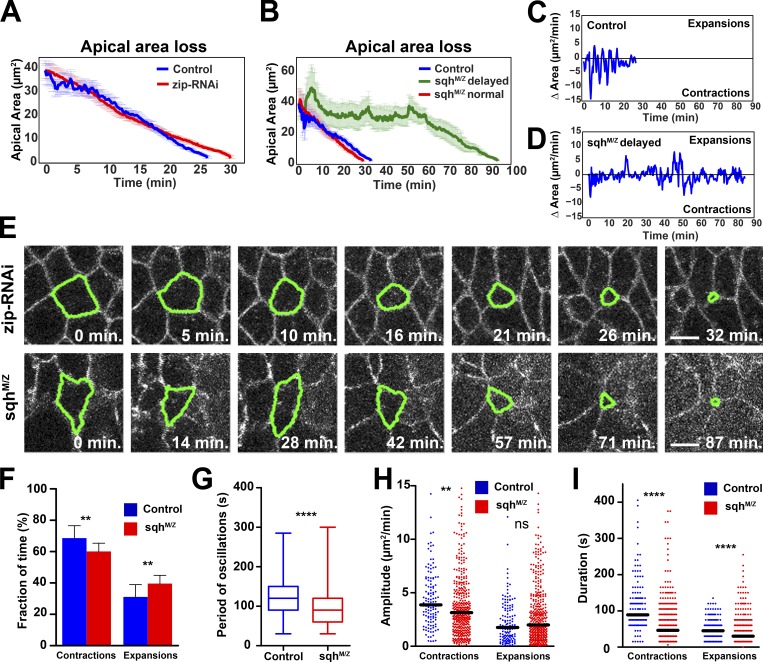
To directly assess the function of myosin during ingression, we examined embryos maternally depleted for the myosin heavy chain, encoded by zipper (zip) using RNAi, or the myosin regulatory light chain, encoded by spaghetti squash (sqh) through generating germline clones for the strong hypomorphic allele sqh1 (sqhM/Z; Karess et al., 1991). zip-RNAi embryos retained ∼40% of normal myosin levels during ingression (Fig. S4 B), whereas sqhM/Z embryos were reported to contain ∼10% of normal myosin levels (Karess et al., 1991; Vasquez et al., 2014). Complete removal of myosin function through genetic strategies is not possible, as myosin is required for oogenesis (Wheatley et al., 1995; Edwards and Kiehart, 1996). Acute myosin inhibition with the Rok inhibitor Y-27632 resulted in epithelial multilayering and precluded time-lapse analysis of ingression (unpublished data). Both zip-RNAi and sqhM/Z embryos showed severe disruptions in mesoderm invagination and germband extension, as previously reported (Fig. S4 A; Bertet et al., 2004; Simões et al., 2010; Vasquez et al., 2014). In contrast, 90.0 ± 0.08% and 87.5 ± 0.08% of NBs were found beneath the neuroepithelium in zip-RNAi or sqhM/Z embryos, respectively, at stage 9 compared with 97.6 ± 0.02% in controls (Fig. S4 A). These surprising findings suggest that NB ingression is much less sensitive to the loss of myosin function than other morphogenetic processes.
Analysis of ingression dynamics revealed a mild delay specifically during late ingression in zip-RNAi embryos, when myosin levels are normally elevated (Fig. 7, A and E). In the more severely myosin-depleted sqhM/Z embryos, we found a subpopulation of NBs that took three times longer to ingress than control NBs (Fig. 7, B and E; Fig. S4 C; and Video 3). Slowly ingressing mutant NBs showed abnormal anisotropy in their patterns of junction loss (Fig. S4 D). To address the concern that these late ingressing NBs belonged to the second wave (S2) of ingressing NBs, we examined the behavior of S2 NBs in normal embryos. All 38 S2 NBs (four embryos) that we traced underwent mitotic rounding, followed by cell division, before one daughter cell initiated ingression. This behavior was not observed in the delayed NBs in sqhM/Z embryos. This, together with the fact that these cells lost their apical surface gradually over time (Fig. S4 C), suggests that the slowly ingressing cells are S1 NBs. These data show that myosin contributes to the anisotropic loss of AJs and apical constriction during NB ingression.
Figure 7.
Myosin controls the ratcheting behavior during apical constriction of NBs. (A) Apical area loss during ingression in videos of Spider::GFP-labeled control embryos (overexpressing myristoylated GFP; 28 NBs, four embryos) and zip-RNAi embryos (26 NBs, three embryos). (B) Apical area loss during ingression in ubi-DEcad::GFP-labeled wild-type (control, 26 NBs, four embryos) and sqhM/Z mutants (17 NBs, three embryos). Two classes of mutant cells were observed, one with normal ingression speed (9/17 cells) and another significantly delayed group (8/17 cells). Data presented are means ± SEM. (C and D) Rate of apical area change during ingression of a control (C) and a delayed sqhM/Z mutant NB (D). (E) Stills from time-lapse videos of NBs (segmented in green) in zip-RNAi and sqhM/Z delayed NBs labeled with Spider::GFP and ubi-DEcad::GFP, respectively. See Fig. 1 F for control NB. Bars, 5 µm. (F) Fractions of time NBs spend contracting and expanding apically during ingression in control and sqhM/Z class 2 NBs. **, P = 0.0078 (two-tailed t test). Data presented are means ± SD. (G) Period of apical oscillations in control (median, 120 s; n = 152 periods, 26 NBs) and sqhM/Z delayed cells (median, 90 s; n = 433 periods, eight NBs). Box plot with IQRs and minimums/maximums displayed. ****, P = 3.2 × 10−9 (KS test). (H and I) Amplitude and duration of contractions and expansions in control and sqhM/Z delayed NBs. Median amplitudes (bars; squared micrometers/minute): contractions, 3.88/3.14; **, P = 0.005; expansions, 1.75/1.99; ns (not significant), P = 0.11 (H). Median durations (s): contractions, 90/45; ****, P = 5 × 10−19; expansions, 45/30; ****, P = 1 × 10−7 (KS test in H and I). 126–393 events per condition.
Next, we assessed myosin function in directing the ratcheting behavior driving apical membrane loss. Delayed NBs in sqhM/Z embryos spent 13% less time contracting their apical membrane than control NBs (Fig. 7 F), and their decrease in apical surface was frequently interrupted by apical expansions with greater variance compared with normal NBs (Fig. 7, C, D, H, and I; and Fig. S4 C). The amplitude and duration of apical contractions were reduced by 19% and 50%, respectively, compared with normal NBs (Fig. 7, H and I). Thus, myosin function is required for the sustained contractions that occur during apical membrane loss. In contrast, we found that the mean amplitude of apical expansions remained unchanged in sqhM/Z embryos despite higher variability, whereas the average duration of expansions was reduced by 33% (Fig. 7, H and I). Consequently, myosin-depleted NBs had shorter and more frequent apical oscillations than control cells (Fig. 7 G). Our findings indicate that myosin levels regulate the periodicity of apical ratcheting and that myosin promotes ingression by driving and stabilizing apical contractions.
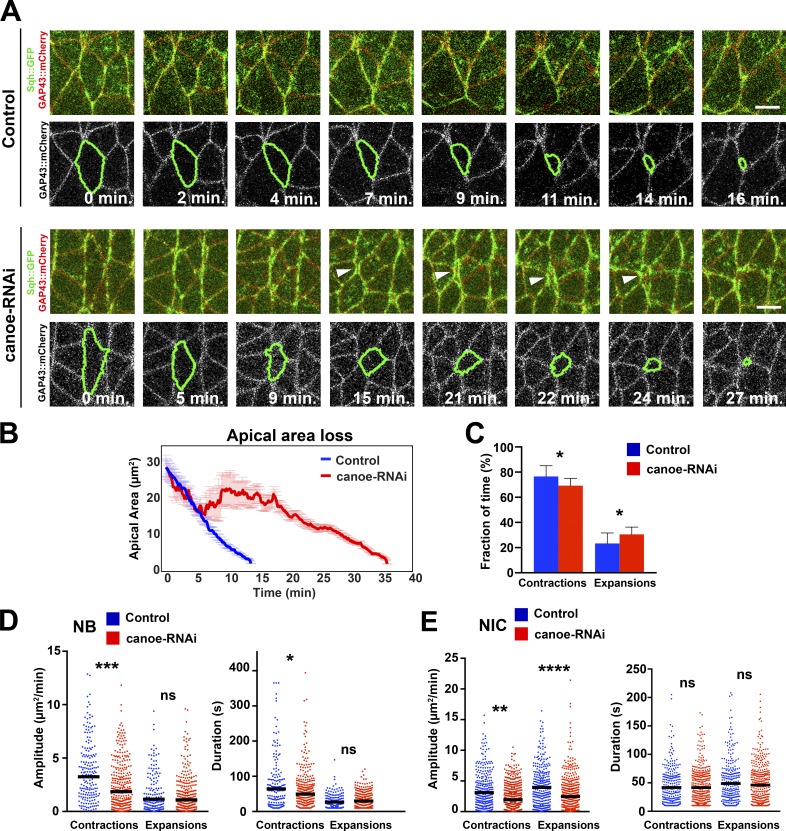
Myosin coupling to junctions regulates apical contractions during ingression
Multiple adapter proteins including Canoe link actomyosin to cell junctions. The regulation of this linkage can trigger apical constriction (Sawyer et al., 2009, 2011; Roh-Johnson et al., 2012). We examined canoe-RNAi embryos to assess whether the connection between myosin and AJs was required during ingression. Depletion of Canoe led to a detachment of junctional myosin from vertical junctions in the ectoderm (Sawyer et al., 2011), including ingressing NBs (Fig. 8 A), but did not impact on medial myosin levels or its pulsatility (not depicted). NBs took 2.5× longer to complete ingression in canoe-RNAi embryos (Fig. 8 B), a delay that was linked to reductions in the amplitude and duration of apical contractions by 37% and 17.6%, respectively (Fig. 8, C and D). Interestingly, the amplitude and duration of expansions remained unchanged compared with controls, suggesting that myosin engagement with junctions may be dispensable during apical expansions when myosin levels are reduced in normal NBs.
Figure 8.
Canoe is required for NB ingression. (A) Stills from time-lapse videos of ingressing NBs in water-injected Sqh::GFP GAP43:mCherry embryos (control) and canoe-RNAi embryos. Arrowheads indicate the detachment of junctional myosin from an NB edge during ingression. Bars, 5 µm. (B) Apical area loss during ingression in control (22 NBs, four embryos) and canoe-RNAi embryos (24 NBs, three embryos). Data presented are means ± SEM. (C) Fractions of time ingressing NBs spend contracting and expanding in control and canoe-RNAi. *, P = 0.037 (KS test). Data presented are means ± SD. (D) Amplitude and duration of individual contractions/expansions during ingression in control and canoe-RNAi. Median amplitudes for control/RNAi (bars; squared micrometers/minute): contractions, 3.38/2.12; ***, P = 3 × 10−4; expansions, 1.15/1.08; ns (not significant), P = 0.599. Median duration for control/RNAi (seconds): contractions, 61.57/50.71, *, P = 0.012; expansions, 17.17/19.04, ns, P = 0.385 (KS test); n = 154–297 events per condition. (E) Amplitude and duration of apical contractions and expansions in NICs in control and canoe-RNAi embryos. Median amplitudes for control/RNAi (bars; squared micrometers/minute): contractions, 3.09/2.38; **, P = 0.005; expansions, 3.94/2.44; ****, P = 7.2 × 10−7. Median duration for control/RNAi (seconds): contractions, 33.35/33.73; ns, P = 0.983; expansions, 38.12/35.45; ns, P = 0.3613 (KS test). n = 275–401 events per condition.
We also asked whether myosin attachment or force transmission to junctions was specifically increased in ingressing NBs relative to NICs. We tested this by comparing the change in contraction amplitude between NBs and NICs of Canoe-depleted embryos. Whereas ingressing NBs in canoe-RNAi embryos showed 37% reduction in the amplitude of contractions compared with controls, contraction amplitudes were reduced by only 22% in NICs (Fig. 8, D and E). As a second approach to disrupt the interactions between myosin and junctions, we reduced the levels of junctional myosin by depleting Smog, a G protein–coupled receptor that controls junctional, but not medial, myosin levels (Kerridge et al., 2016). Smog depletion reduced apical contractions in NBs but had no significant effect on apical area oscillations of NICs (Fig. S5). Collectively, our results argue that linkage between myosin and junctions is not only critical for apical contractions but also is enhanced in NBs during ingression.
Normal ingression requires adhesion to neighboring cells and their apical expansion
NB ingression occurs in response to the activation of specification factors such as Ac (Hartenstein and Wodarz, 2013). Ingression may therefore largely or exclusively be a cell-autonomous process. Nevertheless, NBs ingress from an epithelial layer in which cells are linked through AJs. Therefore, we wondered whether normal ingression depends on the biophysical properties of adjacent NICs, which expand their apical domains while NBs constrict (Fig. 9 A).
Figure 9.
Cell contact and apical expansion of NICs adjacent to NBs are required for normal ingression. (A) NICs (purple) next to NBs expand apically 1.56 ± 0.47–fold during the last 25 min of ingression, in contrast to temporally matched NICs not in contact with NBs (1.05 ± 0.34). n = 25 cells per condition. **, P = 9.8 × 10−3 (KS test). (B) Mechanical uncoupling of NBs from neighbors by laser ablation increases ingression speed (top panels and plot) compared with sham-irradiated controls (bottom panels and plot). Each line represents the apical area of one NB after laser/sham cut. Time 0 depicts the apical area before cut. n = 8–9 cells per condition. (C) Cluster of NBs in Delta-RNAi embryos expressing Sqh::GFP (green) and GAP43::mCherry (red). Bottom panels show the same cluster with pseudocolored cells (yellow, blue, and red) ingressing sequentially. Myosin coalesces into foci (arrowheads) during late ingression. (D) Mean apical area loss in control (water-injected Sqh::GFP GAP43::mCherry; 22 NBs, four embryos) and Delta-RNAi embryos (58 NBs, three embryos). (E and F) Amplitude and duration of apical contractions and expansions during ingression in control and Delta-RNAi embryos. Median amplitudes for control/RNAi (E, squared micrometers/minute): contractions, 3.37/2.78; ns (not significant), P = 0.08; expansions, 1.15/1.31; ns, P = 0.509. Median durations for control/RNAi (F, seconds): contractions, 61.57/41.24; **, P = 0.001; expansions, 17.7/17.8; ns, P = 0.6 (KS test). 154–874 events per condition. (G) Cluster of ingressing NBs in Notch-RNAi. Cells are outlined by ubi-DEcad::GFP. DEcad is down-regulated between neighboring NBs (segmented in bottom panels). Bars, 5 µm. (H) DEcad::GFP levels at NB–NB junctions (Notch-RNAi) decrease during ingression, in contrast to NB–NIC junctions in controls. n = 15–16 junctions per condition. Data presented are means ± SEM. a.u., arbitrary units.
To assess whether neighboring cells provide mechanical resistance to NB apical constriction, we released tissue tension around NBs through laser ablation of apical junctions in neighboring cells (Fig. 9 B). Strikingly, ingression proceeded four times faster when NBs were mechanically uncoupled from their neighbors than in controls (Fig. 9 B). This suggests that neighboring cells exert dragging forces on NBs that counteract NB apical constriction. In addition, NBs showed a rapid linear decrease in apical surface area without the oscillatory behavior seen normally (Fig. 9 B), suggesting that cycles of apical contraction and expansion depend on forces exerted by neighboring cells.
Conversely, we expected that increasing mechanical tension in neighboring cells would slow down ingression. We tested this prediction by converting neighbors into NBs through blocking Notch signaling, which compromises lateral inhibition within proneural clusters so that all cells within a cluster are specified as NBs (Hartenstein and Wodarz, 2013). Ectopic NBs in Delta-RNAi or Notch-RNAi embryos ingressed as clusters of two to eight cells (Fig. 9 C). Rather than ingressing simultaneously, cells in the larger clusters constricted sequentially in subgroups of two to three cells at a speed that was on average 46% slower than control cells (Fig. 9 D). The slower speed of ingression in Delta-RNAi was caused by a reduction in the duration, but not amplitude, of apical contractions (Fig. 9, E and F). However, Delta-RNAi NBs had identical mean levels of medial and junctional myosin compared with controls, and changes in area and myosin levels were similarly anticorrelated (not depicted). We conclude that expansion of neighboring cells is required for the normal duration of apical contractions during ingression.
We also examined DEcad in Notch-RNAi embryos and found that DEcad was lost more rapidly at contacts between NBs than between NBs and NICs (Fig. 9, G and H; and Video 5). The down-regulation of DEcad was posttranscriptional, as it was observed even when DEcad was expressed under a constitutive promoter. One consequence of DEcad loss was the appearance of persistent myosin foci during the late stages of ingression (Fig. 9 C, 14–25.8 min; and Video 4), as was previously observed in mesoderm cells depleted of AJs (Martin et al., 2010). In addition, NB clusters depleted of DEcad often lost adhesion between cells (Video 5) and showed NBs dividing on the surface of the embryo (not depicted). Our findings indicate that NBs rely on cell contacts with NICs to stabilize their junctions and the associated myosin for completion of a normal ingression process.
Discussion
We found that Drosophila NBs are a genetically and optically approachable model for cell ingression. AJs and DEcad are lost from ingressing NBs progressively, as NBs constrict their apical membrane via a sequential disassembly of cell–cell contacts. Transcriptional repression of E-cadherin is thought to play a core role in EMT (Lamouille et al., 2014; Nieto et al., 2016). However, the transcriptional regulation of DEcad is not essential to NB ingression. Also, the ingression of primary mesenchymal cells in sea urchins was found to be independent of the transcriptional regulation of E-cadherin (Wu et al., 2007), and E-cadherin retention was reported in newly formed mesodermal cells during chick gastrulation (Nakaya et al., 2008) and in Twist-induced migration of mammary epithelial cells (Shamir et al., 2014). These results together with our findings suggest that transcriptional repression of E-cadherin may not be as widespread a trigger for EMT-like processes as previously thought, and they point to the importance of alternative mechanisms of junctional disassembly.
Although the mechanism that leads to the removal of DEcad from NBs is unknown, one contributing process controlling the disassembly of AJs in NBs is apparently the persistent contact of NBs to NICs. This became evident in Notch signaling–compromised embryos that formed NB clusters instead of a single NB. The premature loss of DEcad from cell contacts between NBs in these clusters points to an NB’s intrinsic mechanism to remove DEcad from the cell surface that is counteracted by contacts to NICs.
Actomyosin drives NB ingression
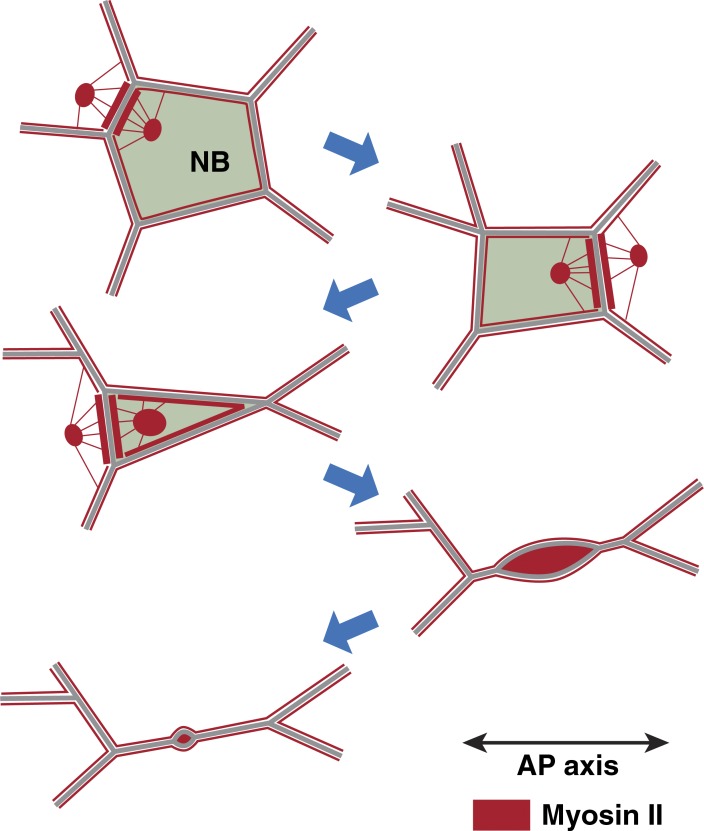
NBs contract their vertical edges earlier and faster than their horizontal edges, giving the apical domain a typical “lentil” shape during late ingression (Fig. 10). This anisotropic pattern correlated with an enrichment of myosin at vertical edges similar to that observed during cell intercalation in the same tissue (Bertet et al., 2004; Zallen and Wieschaus, 2004). Compromising myosin function often changed the anisotropy of edge contraction and significantly delayed ingression. Thus, global polarity cues guiding actomyosin planar polarity in the germband directly influence the spatial and temporal patterns of apical constriction in NBs.
Figure 10.
Model of NB ingression. A typical example of the organization of junctional and medial myosin (red) during five stages of progressive apical constriction in an NB is illustrated. During early ingression (stages 1–3), medial myosin in the NB and neighboring cells flows toward AP NB edges, which become myosin enriched and disassemble. During late ingression (stages 4–5), progressively stronger pulses of medial myosin fuse with DV NB edges, which subsequently disassemble.
In addition to the junctional myosin pool, we also detected rapid pulses of myosin assembly and disassembly at the free apical cortex of NBs. Medial myosin flows toward AP and DV junctions and may fuse with them, correlating with contractions of both AP and DV edges. Peak myosin concentrations of either junctional or medial myosin often coincided with apical area reduction, suggesting that the actomyosin activity of either pool can be the immediate cause for apical contraction. In contrast to NICs, medial myosin clusters more frequently fused with DV junctions in NBs. We therefore propose that the coupling of actomyosin to DV junctions is specifically enhanced in ingressing NBs.
Myosin pulses were previously reported in the context of AP edge disassembly during cell intercalation (Rauzi et al., 2010; Fernandez-Gonzalez and Zallen, 2011) and during cell shape stabilization in apically constricting cells, including the Drosophila mesoderm (Martin et al., 2009, 2010; Jodoin et al., 2015; Xie and Martin, 2015), amnioserosa cells during dorsal closure (Blanchard et al., 2010; David et al., 2010; Sokolow et al., 2012), and the C. elegans endoderm (Roh-Johnson et al., 2012). Medial myosin pulses increased in amplitude throughout NB ingression, correlating with stronger contractions of the apical cortex. Junctional myosin levels were also progressively enhanced and temporally matched with changes in cell edge length. These data suggest that transmission of tension from the medial cortex to the junctions is a conserved feature of apical constriction (Martin and Goldstein, 2014). Currently, there is no clear evidence from any system as to whether medial or junctional myosin alone can effectively promote apical constriction.
The regulation of the linkage between a pulsatile actomyosin network and apical junctions is thought to be crucial for the ratcheting that stabilizes apical contractions (Roh-Johnson et al., 2012; Xie and Martin, 2015). The loss of Canoe, a linker between actomyosin and apical junctions (Sawyer et al., 2009, 2011), and Smog, a G protein–coupled receptor that controls the levels of junctional myosin (Kerridge et al., 2016), significantly delayed NB ingression. Notably, apical contractions in Canoe- and Smog-compromised embryos were more strongly reduced in NBs than in NICs, suggesting that cytoskeletal coupling to the junctions might be differentially regulated during ingression. Stronger connections between actomyosin and the junctions could therefore enhance cortical contractions in NBs and further promote myosin recruitment via biomechanical feedbacks (Fernandez-Gonzalez et al., 2009).
Ingression of individual cells is a community effort
NICs adjacent to an NB expanded their apical domain up to 50% as NBs ingressed. These cells also displayed lower levels of myosin at late stages of ingression in contrast with NBs, in which myosin is elevated, suggesting tension anisotropy between NBs and their neighbors. Indeed, reducing local tension around NBs by laser ablation increased ingression speed fourfold, whereas converting neighbors into NBs that also constrict slowed down ingression. Thus, tension anisotropy between NBs and neighbors regulates normal ingression dynamics. Such a balance of forces model should also shape the interpretation of tissue-wide manipulations of myosin activity or changes in actomyosin–AJ coupling. For example, the normal ingression speed seen in embryos with ∼40% of normal myosin levels (zip-RNAi) may be the result of a readjusted force balance where actomyosin contractility is reduced in both NBs and neighbors. Nevertheless, it was surprising to see that even strong reduction of myosin to ∼10% of normal allowed many NBs to ingress, albeit with irregular apical shape and contractile activity. This finding points to other mechanisms that make major contributions to apical domain loss in NBs. A likely candidate is the endocytosis of apical and junctional proteins, which is currently being investigated.
In addition to ingression, cells can segregate from an epithelium in response to cell death or tissue crowding via cell extrusion, a process that plays a role in epithelial homeostasis (Marinari et al., 2012; Gudipaty and Rosenblatt, 2016). Contrary to cell ingression, the extruded cell is either dying or targeted to die, and it is apically squeezed out of the epithelium by a basal supracellular actomyosin cable that is assembled by neighboring cells (Rosenblatt et al., 2001). In contrast, NBs follow a cell-autonomous transcriptional program downstream of the basic Helix–Loop–Helix transcriptional factors of the Ac–Scute complex that results in NB specification and cell ingression (Hartenstein and Wodarz, 2013). How this program is linked to apical actomyosin, its enhanced coupling to the junctions, junctional disassembly, and other mechanisms of apical domain removal remain to be explored.
Materials and methods
Markers and mutants
The following fly markers and mutants were used: Spider::GFP (a gift from J. Zallen, Memorial Sloan Kettering Cancer Center, New York, NY), yw; ubi-DEcad::GFP (Oda and Tsukita, 2001), endoDEcad::GFP (a gift from Y. Hong, University of Pittsburgh, Pittsburgh, PA), f_TRG71 Sna::GFP (TransgeneOme; Ejsmont et al., 2009), sqh-GAP43::mCherry (Martin et al., 2010), sqh-Sqh::GFP (Royou et al., 2004), sqh-Sqh::mCherry (Martin et al., 2009), UAS-myristoylated::GFP (Bloomington Drosophila Stock Center), matαtub67-Gal4; matαtub15-Gal4 (a gift from D. St. Johnston, University of Cambridge, Cambridge, England, UK), UAS-zip-RNAi (Trip line HMS01618; Drosophila RNAi Screening Center), UAS-Notch-RNAi (Trip line HMS00009; Harvard Medical School), UAS-mCherry-RNAi control Trip line (Bloomington Drosophila Stock Center), yw P[FRT(whs)]101 sqh1/FM7 (Karess et al., 1991), w; shgR69/CyO (Godt and Tepass, 1998), and Df(2L)osp29/CyO (Ashraf et al., 1999).
To knock down myosin heavy chain during ingression, we analyzed the F2 progeny of matαtub67-Gal4; matαtub15-Gal4 Spider::GFP females or matαtub67-Gal4; matαtub15-Gal4 Sna::GFP females crossed to UAS-zip-RNAi males. Control embryos were generated similarly by using UAS-mCherry-RNAi or UAS-myristoylated::GFP. To knock down Notch, we analyzed the F2 progeny of UAS-Notch-RNAi females crossed to matαtub67-Gal4 ubi-DEcad::GFP; matαtub15-Gal4 males. Females carrying sqh1 germline clones expressing ubi-DEcad::GFP or Sna::GFP were generated using the flipase site-specific recombinase–dominant female sterile (FLP-DFS) system (Chou and Perrimon, 1996).
Immunohistochemistry
Embryos were fixed in a 1:1 mixture of 3.7% formaldehyde in phosphate buffer, pH 7.4, and heptane for 20 min under agitation and then devitellinized in a methanol/heptane mixture (Figs. 2 and S1 C) or a 1:1 mixture of 37% formaldehyde and heptane for 5 min followed by hand devitalization (Figs. S1 B and S4 A). The following antibodies were used: 1:150 rabbit anti-GFP (Torrey Pines), 1:100 guinea pig anti-Sna (a gift from E. Wieschaus, Princeton University, Princeton, NJ), 1:5 mouse anti-Ac (Developmental Studies Hybridoma Bank), 1:25 rat anti-DEcad2 (Developmental Studies Hybridoma Bank), 1:50 mouse anti-Dlg (Developmental Studies Hybridoma Bank), and 1:100 rat anti-Crumbs (Pellikka et al., 2002). Secondary antibodies conjugated to Alexa Fluor 488, 568, or 647 (Molecular Probes) were used at 1:400. Embryos were mounted in ProLong Gold (Molecular Probes) and imaged with zoom factors ranging from 0.75 to 2× on a confocal microscope (TCS SP8) with an HCX Plan-Apochromat 63×/1.4 NA CS2 objective (Leica Microsystems) via sequential scanning between channels. 1-µm z slices were acquired at 0.5-µm steps. Maximum projections of 2–3 µm including the apical domain or the cell nuclei were analyzed. Cross-sectional views of embryos were obtained by cutting embryos manually with a 27-gauge syringe.
Time-lapse imaging
Embryos were dechorionated for 2 min in 50% bleach, transferred to a drop of halocarbon oil 27 (Sigma-Aldrich) on a coverslip, and mounted on an oxygen-permeable membrane (YSI; Xylem Inc.). GFP was excited with an optically pumped semiconductor laser (488 nm; 2–2.5%), and mCherry was excited with an optically pumped semiconductor laser (514 nm; 3–5%). An HCX Plan-Apochromat 63×/1.4 NA CS2 objective on a resonant scanning confocal microscope (TCS SP8) was used for imaging at 20–22°C. 12-bit images of one- or two-color z stacks (five to six planes) were acquired at 0.5-µm steps in 5-, 10-, or 15-s intervals using Application Suite X software (Leica Microsystems) and maximally projected for analysis in ImageJ (National Institutes of Health; Schneider et al., 2012). Pixel dimensions ranged from 144 to 180 nm/pixel.
Laser ablation
Ablations were done using a pulsed micropoint N2 laser (Andor Technology) tuned to 365 nm in endo-DEcad::GFP embryos (Yu and Fernandez-Gonzalez, 2016). Embryos were mounted as described in the previous section for live imaging and then imaged using a Revolution XD spinning-disk confocal microscope equipped with an iXon Ultra 897 camera (Andor Technology) and a 1.5× coupling lens. All embryos were imaged with a 60×/1.35 NA oil immersion lens (Olympus) at 20–22°C. Images were acquired using Microscopy Automation and Image Analysis software (MetaMorph). For experiments involving ablation at the apical domain, five consecutive laser pulses of 60 µJ with a duration of 67 ms each were delivered to a single spot at the apical domain of late ingressing NBs or NICs. For ablation of cell boundaries around NBs, one pulse of 42 µJ and 67-ms duration was applied simultaneously at each junction. Two z stacks of 5–20 planes (0.35 µm each) were acquired before ablation, and after ablation every 1–3 s. In sham-irradiated controls, junctions were targeted with the laser completely attenuated, using the same number of pulses, to mimic the ablations performed in the corresponding experiments.
Double-stranded RNA (dsRNA) injections
Templates to produce dsRNA against sna, wor, esg, canoe, smog, and Delta were generated by PCR from genomic DNA using the following pairs of primers containing the T7 promoter sequence 5′-TAATACGACTCACTATAGGGAGACCAC-3′ at the 5′ end: sna forward, 5′-CCAGGATCAGCCGCAGGATCTATCC-3′, and sna reverse, 5′-CTCGCCGACGAGGCACTCATGGCCG-3′; wor forward, 5′-GGACCAGTGGACTTGAGTGTGGCCTC-3′, and wor reverse, 5′-CGCTCAGCACTCTCCTCTGGGTTAC-3′; esg forward, 5′-CAGATGATGAACACATCGAGGAGGAG-3′, and esg reverse, 5′-CTGCTGCTTGCTCTCCCGGCTGGCTAG-3′; canoe forward, 5′-CCCATCGAGCAGACAGATCTAAAC-3′, and canoe reverse, 5′-CTGAACCAGGCAATAGTGCAGGGG-3′; smog forward, 5′-ATGGAACTGTGCATAGCAACAAAG-3′, and smog reverse, 5′-CGGCTCATCTGGCAGCGTTTCGAG-3′; and Delta forward, 5′-GGAGCCTTGTGCAACGAGTGCGTTC-3′, and Delta reverse, 5′-CGCACGACAGGTGCACTGGTAATCG-3′.
PCR products were used as templates for T7 transcription reactions with the 5× MEGAscript T7 kit (Ambion). dsRNA was injected ventrally (for immunostaining) or dorsally (for live imaging). 0–1-h-old embryos from the stocks w; GAP43::mCherry; Sqh::GFP or fTRG_71 Sna::GFP were dechorionated, glued to a coverslip, dehydrated for 5 min, covered in 1:1 halocarbon oil 27/700 (Sigma-Aldrich), and injected with 100–200 pL of 1–2.5 µg/µl dsRNA each. Control embryos were injected with water. Embryos were incubated in a humidified chamber at 25°C and imaged at stages 7–9. For immunostaining, embryos were washed off the coverslip with heptane at stage 9, fixed for 5 min in 37% formaldehyde in PBS/heptane, and manually devitellinized.
Planar polarity quantifications
For myosin planar polarity measurements, we used Scientific Image Segmentation and Analysis (SIESTA; Fernandez-Gonzalez and Zallen, 2011; Leung and Fernandez-Gonzalez, 2015) to manually draw 3-pixel-wide lines (180 nm/pixel) for all cell edges to obtain the mean pixel intensity and orientation for each edge. Intensities were averaged for all edges in each angular range, and the mode (cytoplasmic) intensity was subtracted for background correction. The mean background-subtracted edge intensity for all edges in the 75–90° angular range was normalized to the mean background-subtracted intensity in the 0–15° bin. A single value was obtained for each NB at different time points of ingression (20 min in Fig. 4 A, the time point at which cells reached an apical surface of 2.5 µm2) and in temporally matched regions of the whole tissue (2 × 103 µm2).
To quantify the planar distribution of medial myosin foci relative to the AP axis, we used ImageJ to manually draw straight lines between the cell centroid and each myosin focus observed at the medial cortex. Angles were determined in the 0–360° range using the publicly available ImageJ macro Measure angle and length (Schneider et al., 2012). 0° and 180° corresponded to the posterior and anterior sides of the embryo, respectively. Because myosin foci can move as they coalesce, angles were measured when foci were fully coalesced and immediately before (5 s; one time frame) their disassembly or fusion with a cell edge.
Cell segmentation, tracking, and quantification
We used SIESTA to automatically identify cell outlines in time-lapse videos with a watershed algorithm as described previously (Fernandez-Gonzalez and Zallen, 2011; Leung and Fernandez-Gonzalez, 2015). To segment a single NB or NIC, we manually assigned one seed per NB or NIC and to all of its surrounding neighbors at the earliest time point that the cell of interest could be tracked in time-lapse videos (stage 7 or early stage 8) using the annotation tool Set fiducial markers, available in SIESTA. Seeds were propagated and expanded between consecutive time points using the tool Expand and propagate seeds, resulting in automatic cell segmentation. When manual correction was necessary, seeds were either manually assigned to single cells or a semiautomated method of manual tracing of cell interfaces included in SIESTA, the LiveWire, was used (Fernandez-Gonzalez and Zallen, 2013). Image analysis was performed using algorithms developed with Matlab (MathWorks) and DIPImage (Delf University of Technology; Fernandez-Gonzalez and Zallen, 2011; Leung and Fernandez-Gonzalez, 2015).
The cell length of NBs along the AP and DV axes was calculated by superimposing on the cell outline an array of lines 1 pixel apart (180 nm) parallel to the axis in question and measuring the mean length of the intersecting segments (Fernandez-Gonzalez and Zallen, 2011). To measure the mean apical surface area during ingression, 20–58 NBs from three to seven embryos of each genotype were temporally aligned (registered) based on the time when they reached an apical area of 2.5 µm2. We did not consider segmentation results below this threshold. Time 0 (onset of ingression) was defined as the time point at which the initial mean area (generally between 30 and 40 µm2) of the registered cells started declining persistently. To compare rates of ingression between genotypes, we matched the highest initial mean areas in control and mutant/RNAi embryos and determined the curve slope (an approximation to mean ingression speed) by linear fitting using the Matlab function polyfit. When mean fluorescence results from multiple cells were analyzed, cells were temporally registered using 2.5 µm2 as the area threshold. The mean rate of apical area loss in control embryos was reproducible for each fluorescent marker used, but we found consistent differences among different fluorescent markers: Spider::GFP (Fig. 1, B–D, G, and H; and Fig. 2 I), −1.3 ± 0.5 µm2/min; w; GAP43::mCherry; Sqh::GFP (Figs. 2 H, 3, 4, 5, 8, 9, C–F; and Figs. S3 and S5), −1.7 ± 0.8 µm2/min; ubi-DEcad::GFP, shgR69 (Figs. 2 I, 7 B, 9 G, and S4 C), −1.0 ± 0.5 µm2/min; endo-DEcad::GFP (Fig. 1, E and F; and Figs. 6, 9 B, and S2), −2.3 ± 1.2 µm2/min; and Mat67;15 Spider::GFP UAS-myrGFP (Fig.7 A), −1.6 ± 0.8 µm2/min (means ± SD). The speed of ingression was reproducible for each fluorescent marker when comparing videos made in three consecutive years (not depicted), suggesting that the marker itself or differences in the genetic background (and not temporal changes or sampling effects) are the cause for the observed differences in speed among markers. For all experiments, we compared controls to mutant or RNAi embryos carrying the same fluorescent marker or markers and imaged with the same settings and environmental conditions.
To quantify the total ingression time of medial, intermediate, and lateral NBs, individual cells were segmented and their apical areas were plotted against time. The onset of ingression was considered the first time point when apical area started decreasing after a plateau; the last time point of ingression was visually assigned when cells completely lost their apical domain.
To quantify junctional and medial mean protein levels, each cell was divided into two compartments (Fernandez-Gonzalez and Zallen, 2011). The junctional compartment was determined by a 3-pixel-wide (0.54 µm) dilation of the cell outline identified using watershed or LiveWire segmentation. The medial compartment was obtained by inverting a binary image representing the junctional compartment. Protein concentrations were quantified as the mean pixel intensity in each compartment considering either all ingression time points, up to the last 10 min of the process (early ingression), or the last 10 min (late ingression), as indicated. Total DEcad intensity was defined as the mean junctional intensity per pixel in the junctional compartment multiplied by the length in pixels of that compartment (180 nm/pixel). Changes in cell area and protein concentration were defined as described previously (Fernandez-Gonzalez and Zallen, 2011): Δarea(t) = area(t) − area(t − 60 s); and Δprotein(t) = protein(t) − protein(t − 60 s).
To combine results from different embryos, we normalized protein concentration at each time point by subtracting the fluorescence mode for the entire image (background) and dividing by the mean pixel value of each frame.
The duration of apical contractions and expansions was defined as the number of consecutive time points multiplied by time resolution, during which Δarea(t) was <0 or >0, respectively. The amplitude of contractions and expansions was the maximum value of area change within each event. Periods were defined as the total time between consecutive troughs or peaks after smoothening Δarea(t) using a Gaussian kernel of width of two times the temporal resolution in seconds. Because NBs reduce their apical surface during ingression, we normalized area change to the cell area at each time point (Δarea(t)/area(t)) when comparing amplitudes of contractions and expansions with those of NICs (Fig. 3, B–E).
Cross-correlation coefficients (R) between variables were determined using the Matlab function corrcoef at different time shifts. Maximum (significant, P < 0.05) correlation values and their corresponding time shifts were plotted (Figs. 4 E, 5 F, and S3 B). For cross-correlations between cell edge length and myosin levels at cell edges or in their vicinity (<1.8 µm), we considered 30–60 consecutive time points (5–10 s apart) per edge analyzed. For cross-correlations between cell areas and myosin levels at the free apical cell domain or the junctional perimeter (medial and junctional pools, respectively), we used 60–120 consecutive time points per cell during early ingression and late ingression (as defined in this section) and pooled the results.
To quantify myosin levels and edge length in AP and DV edges during ingression with respect to DV edges in NICs, we imaged embryos expressing GAP43::mCherry and Sqh::GFP and used SIESTA to measure fluorescence and edge length in manually traced cell interfaces. We subtracted the image mode from the myosin fluorescence measurements as an estimate of the background. Periods of myosin II pulses in cell edges were determined after detrending myosin fluorescence levels using the Matlab function detrend. Constricting edges were traced until their lengths reached 0.7–1.3 µm and aligned in time to their shortest lengths in this range. Nonconstricting DV cell edges were traced in temporally matched NICs. To measure medial myosin levels in the vicinity of shrinking junctions, we manually drew polygons in SIESTA on both sides of each junction covering a maximum distance of 1.8 µm away from the junction. Myosin levels inside these polygons were normalized by subtracting the image mode and dividing by the image mean fluorescence at each time point.
Statistics
Mean or median values were determined based on the n value of embryos/cells/cell edges/contractions/expansions, as indicated in each figure legend. Error bars are SD, SEM (which is SD/√n), or interquartile ranges (IQRs) as indicated. We used Prism 7 (GraphPad Software) to test whether the n values of each sample followed a normal distribution by performing a D’Agostino and Pearson normality test. We used unpaired two-tailed t tests to determine p-values when samples passed the normality test, and we used nonparametric two-sample Kolmogorov–Smirnov (KS) or Mann–Whitney tests otherwise.
Online supplemental material
Fig. S1 shows how NB ingression is independent of the Sna family of transcriptional repressors. Fig. S2 shows how DEcad concentration per surface unit is kept constant while its total levels decrease during ingression. Fig. S3 shows how AP edge disassembly correlates with medial myosin levels in NBs and neighboring NICs, whereas DV edge disassembly only correlates with medial myosin levels in NBs. Fig. S4 shows how myosin controls the anisotropic disassembly of cell junctions and depicts the ratchet mechanism of apical membrane loss during ingression. Fig. S5 shows how a partial reduction in junctional myosin by Smog knockdown alters ratcheted contractions of ingressing NBs. Video 1 shows NB ingression. Video 2 shows how myosin is planar polarized and pulsatile in ingressing NBs. Video 3 shows NB ingression in a sqhM/Z-maternal mutant. Video 4 shows myosin dynamics in NBs ingressing as clusters of cells in a Delta-RNAi embryo. Video 5 shows DEcad dynamics in NBs ingressing as clusters of cells in a Notch-RNAi embryo.
Supplementary Material
Acknowledgments
We would like to acknowledge Teresa Zulueta-Coarasa and Jessica Yu for help with Matlab scripts, Adam Martin and Jennifer Zallen for fly stocks, Eric Wieschaus for the Snail antibody, the Imaging Facility at the Department of Cell and Systems Biology for technical support, and Milena Pellikka for technical support and helpful discussions. We thank Dorothea Godt for critical reading of the manuscript.
This work was supported by a Canadian Cancer Society Innovation Grant to U. Tepass and R. Fernandez-Gonzalez.
The authors declare no competing financial interests.
Author contributions: The project was conceived and experiments were designed by U. Tepass and S. Simões. Experiments were conducted by S. Simões. Experimental data were analyzed by S. Simões, Y. Oh, and M.F.Z. Wang. Algorithms used to perform quantitative analysis and training were provided by R. Fernandez-Gonzalez, who also helped with data interpretation. The paper was written by S. Simões and U. Tepass.
Footnotes
Abbreviations used:
- AJ
- adherens junction
- AP
- anterior–posterior
- DEcad
- Drosophila E-cadherin
- DV
- dorsal–ventral
- EMT
- epithelial–mesenchymal transition
- IQR
- interquartile range
- KS
- Kolmogorov–Smirnov
- NB
- neuroblast
- NIC
- noningressing cell
- SIESTA
- Scientific Image Segmentation and Analysis
- VM
- ventral midline
- dsRNA
- double-stranded RNA
References
- Abell A.N., Jordan N.V., Huang W., Prat A., Midland A.A., Johnson N.L., Granger D.A., Mieczkowski P.A., Perou C.M., Gomez S.M., et al. 2011. MAP3K4/CBP-regulated H2B acetylation controls epithelial-mesenchymal transition in trophoblast stem cells. Cell Stem Cell. 8:525–537. 10.1016/j.stem.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf S.I., and Ip Y.T.. 2001. The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila. Development. 128:4757–4767. [DOI] [PubMed] [Google Scholar]
- Ashraf S.I., Hu X., Roote J., and Ip Y.T.. 1999. The mesoderm determinant snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. EMBO J. 18:6426–6438. 10.1093/emboj/18.22.6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E.M., and Konstantopoulos K.. 2012. Intercellular adhesion: mechanisms for growth and metastasis of epithelial cancers. Wiley Interdiscip. Rev. Syst. Biol. Med. 4:171–181. 10.1002/wsbm.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., and García De Herreros A.. 2000. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2:84–89. 10.1038/35000034 [DOI] [PubMed] [Google Scholar]
- Baum B., Settleman J., and Quinlan M.P.. 2008. Transitions between epithelial and mesenchymal states in development and disease. Semin. Cell Dev. Biol. 19:294–308. 10.1016/j.semcdb.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Bertet C., Sulak L., and Lecuit T.. 2004. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 429:667–671. 10.1038/nature02590 [DOI] [PubMed] [Google Scholar]
- Blanchard G.B., Murugesu S., Adams R.J., Martinez-Arias A., and Gorfinkiel N.. 2010. Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development. 137:2743–2752. 10.1242/dev.045872 [DOI] [PubMed] [Google Scholar]
- Blankenship J.T., Backovic S.T., Sanny J.S., Weitz O., and Zallen J.A.. 2006. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell. 11:459–470. 10.1016/j.devcel.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A.D., Nystul T.G., Ohlstein B., Allen A., et al. 2007. The Carnegie Protein Trap Library: A versatile tool for Drosophila developmental studies. Genetics. 175:1505–1531. 10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A., Pérez-Moreno M.A., Rodrigo I., Locascio A., Blanco M.J., del Barrio M.G., Portillo F., and Nieto M.A.. 2000. The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2:76–83. 10.1038/35000025 [DOI] [PubMed] [Google Scholar]
- Chou T.B., and Perrimon N.. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 144:1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David D.J., Tishkina A., and Harris T.J.. 2010. The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development. 137:1645–1655. 10.1242/dev.044107 [DOI] [PubMed] [Google Scholar]
- Desai R., Sarpal R., Ishiyama N., Pellikka M., Ikura M., and Tepass U.. 2013. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat. Cell Biol. 15:261–273. 10.1038/ncb2685 [DOI] [PubMed] [Google Scholar]
- Edwards K.A., and Kiehart D.P.. 1996. Drosophila nonmuscle myosin II has multiple essential roles in imaginal disc and egg chamber morphogenesis. Development. 122:1499–1511. [DOI] [PubMed] [Google Scholar]
- Ejsmont R.K., Sarov M., Winkler S., Lipinski K.A., and Tomancak P.. 2009. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods. 6:435–437. 10.1038/nmeth.1334 [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., and Zallen J.A.. 2011. Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys. Biol. 8:045005 10.1088/1478-3975/8/4/045005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., and Zallen J.A.. 2013. Wounded cells drive rapid epidermal repair in the early Drosophila embryo. Mol. Biol. Cell. 24:3227–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R., Simoes S.M., Röper J.C., Eaton S., and Zallen J.A.. 2009. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 17:736–743. 10.1016/j.devcel.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt D., and Tepass U.. 1998. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature. 395:387–391. 10.1038/26493 [DOI] [PubMed] [Google Scholar]
- Gudipaty S.A., and Rosenblatt J.. 2016. Epithelial cell extrusion: Pathways and pathologies. Semin. Cell Dev. Biol. In press. 10.1016/j.semcdb.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J., and Tepass U.. 2010. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11:502–514. 10.1038/nrm2927 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., and Campos-Ortega J.. 1984. Early neurogenesis in wild-type Drosophila melanogaster. Rouxs Arch. Dev. Biol. 193:308–325. 10.1007/BF00848159 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., and Wodarz A.. 2013. Initial neurogenesis in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2:701–721. 10.1002/wdev.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanes A., Gottardi C.J., and Yap A.S.. 2008. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 27:6920–6929. 10.1038/onc.2008.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodoin J.N., Coravos J.S., Chanet S., Vasquez C.G., Tworoger M., Kingston E.R., Perkins L.A., Perrimon N., and Martin A.C.. 2015. Stable force balance between epithelial cells arises from F-actin turnover. Dev. Cell. 35:685–697. 10.1016/j.devcel.2015.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., and Weinberg R.A.. 2009. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119:1420–1428. 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R.E., Chang X.J., Edwards K.A., Kulkarni S., Aguilera I., and Kiehart D.P.. 1991. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 65:1177–1189. 10.1016/0092-8674(91)90013-O [DOI] [PubMed] [Google Scholar]
- Kerridge S., Munjal A., Philippe J.M., Jha A., de las Bayonas A.G., Saurin A.J., and Lecuit T.. 2016. Modular activation of Rho1 by GPCR signalling imparts polarized myosin II activation during morphogenesis. Nat. Cell Biol. 18:261–270. 10.1038/ncb3302 [DOI] [PubMed] [Google Scholar]
- Lamouille S., Xu J., and Derynck R.. 2014. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15:178–196. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., and Yap A.S.. 2015. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 17:533–539. 10.1038/ncb3136 [DOI] [PubMed] [Google Scholar]
- Leung C.Y., and Fernandez-Gonzalez R.. 2015. Quantitative image analysis of cell behavior and molecular dynamics during tissue morphogenesis. Methods Mol. Biol. 1189:99–113. 10.1007/978-1-4939-1164-6_7 [DOI] [PubMed] [Google Scholar]
- Levayer R., Dupont C., and Moreno E.. 2016. Tissue crowding induces caspase-dependent competition for space. Curr. Biol. 26:670–677. 10.1016/j.cub.2015.12.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., and Thiery J.P.. 2012. Epithelial-mesenchymal transitions: insights from development. Development. 139:3471–3486. 10.1242/dev.071209 [DOI] [PubMed] [Google Scholar]
- Marinari E., Mehonic A., Curran S., Gale J., Duke T., and Baum B.. 2012. Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature. 484:542–545. 10.1038/nature10984 [DOI] [PubMed] [Google Scholar]
- Martin A.C., and Goldstein B.. 2014. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development. 141:1987–1998. 10.1242/dev.102228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., Kaschube M., and Wieschaus E.F.. 2009. Pulsed contractions of an actin–myosin network drive apical constriction. Nature. 457:495–499. 10.1038/nature07522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., Gelbart M., Fernandez-Gonzalez R., Kaschube M., and Wieschaus E.F.. 2010. Integration of contractile forces during tissue invagination. J. Cell Biol. 188:735–749. 10.1083/jcb.200910099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M., and Rijli F.M.. 2010. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 137:2605–2621. 10.1242/dev.040048 [DOI] [PubMed] [Google Scholar]
- Nakaya Y., Sukowati E.W., Wu Y., and Sheng G.. 2008. RhoA and microtubule dynamics control cell–basement membrane interaction in EMT during gastrulation. Nat. Cell Biol. 10:765–775. 10.1038/ncb1739 [DOI] [PubMed] [Google Scholar]
- Nieto M.A., Huang R.Y., Jackson R.A., and Thiery J.P.. 2016. EMT: 2016. Cell. 166:21–45. 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Oda H., and Tsukita S.. 2001. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J. Cell Sci. 114:493–501. [DOI] [PubMed] [Google Scholar]
- Peinado H., Marin F., Cubillo E., Stark H.J., Fusenig N., Nieto M.A., and Cano A.. 2004. Snail and E47 repressors of E-cadherin induce distinct invasive and angiogenic properties in vivo. J. Cell Sci. 117:2827–2839. 10.1242/jcs.01145 [DOI] [PubMed] [Google Scholar]
- Peinado H., Olmeda D., and Cano A.. 2007. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 7:415–428. 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- Pellikka M., Tanentzapf G., Pinto M., Smith C., McGlade C.J., Ready D.F., and Tepass U.. 2002. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature. 416:143–149. 10.1038/nature721 [DOI] [PubMed] [Google Scholar]
- Pohl C., Tiongson M., Moore J.L., Santella A., and Bao Z.. 2012. Actomyosin-based self-organization of cell internalization during C. elegans gastrulation. BMC Biol. 10:94 10.1186/1741-7007-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauzi M., Lenne P.F., and Lecuit T.. 2010. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 468:1110–1114. 10.1038/nature09566 [DOI] [PubMed] [Google Scholar]
- Roh-Johnson M., Shemer G., Higgins C.D., McClellan J.H., Werts A.D., Tulu U.S., Gao L., Betzig E., Kiehart D.P., and Goldstein B.. 2012. Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science. 335:1232–1235. 10.1126/science.1217869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J., Raff M.C., and Cramer L.P.. 2001. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 11:1847–1857. 10.1016/S0960-9822(01)00587-5 [DOI] [PubMed] [Google Scholar]
- Royou A., Field C., Sisson J.C., Sullivan W., and Karess R.. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell. 15:838–850. 10.1091/mbc.E03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarage C.R., White M.D., Álvarez Y.D., Fierro-González J.C., Henon Y., Jesudason E.C., Bissiere S., Fouras A., and Plachta N.. 2015. Cortical tension allocates the first inner cells of the mammalian embryo. Dev. Cell. 34:435–447. 10.1016/j.devcel.2015.07.004 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., and Bronner-Fraser M.. 2008. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9:557–568. 10.1038/nrm2428 [DOI] [PubMed] [Google Scholar]
- Sawyer J.K., Harris N.J., Slep K.C., Gaul U., and Peifer M.. 2009. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186:57–73. 10.1083/jcb.200904001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J.K., Choi W., Jung K.C., He L., Harris N.J., and Peifer M.. 2011. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol. Biol. Cell. 22:2491–2508. 10.1091/mbc.E11-05-0411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir E.R., Pappalardo E., Jorgens D.M., Coutinho K., Tsai W.T., Aziz K., Auer M., Tran P.T., Bader J.S., and Ewald A.J.. 2014. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol. 204:839–856. 10.1083/jcb.201306088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões S.M., Blankenship J.T., Weitz O., Farrell D.L., Tamada M., Fernandez-Gonzalez R., and Zallen J.A.. 2010. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell. 19:377–388. 10.1016/j.devcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolow A., Toyama Y., Kiehart D.P., and Edwards G.S.. 2012. Cell ingression and apical shape oscillations during dorsal closure in Drosophila. Biophys. J. 102:969–979. 10.1016/j.bpj.2012.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U., Gruszynski-DeFeo E., Haag T.A., Omatyar L., Torok T., and Hartenstein V.. 1996. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 10:672–685. [DOI] [PubMed] [Google Scholar]
- Theveneau E., and Mayor R.. 2011. Collective cell migration of the cephalic neural crest: the art of integrating information. Genesis. 49:164–176. 10.1002/dvg.20700 [DOI] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., and Nieto M.A.. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Vasquez C.G., Tworoger M., and Martin A.C.. 2014. Dynamic myosin phosphorylation regulates contractile pulses and tissue integrity during epithelial morphogenesis. J. Cell Biol. 206:435–450. 10.1083/jcb.201402004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A., and Pu W.T.. 2012. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ. Res. 110:1628–1645. 10.1161/CIRCRESAHA.111.259960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., and Wieschaus E.. 2016. Myosin-dependent remodeling of adherens junctions protects junctions from Snail-dependent disassembly. J. Cell Biol. 212:219–229. 10.1083/jcb.201508056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley S., Kulkarni S., and Karess R.. 1995. Drosophila nonmuscle myosin II is required for rapid cytoplasmic transport during oogenesis and for axial nuclear migration in early embryos. Development. 121:1937–1946. [DOI] [PubMed] [Google Scholar]
- Wu S.Y., and McClay D.R.. 2007. The Snail repressor is required for PMC ingression in the sea urchin embryo. Development. 134:1061–1070. 10.1242/dev.02805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.Y., Ferkowicz M., and McClay D.R.. 2007. Ingression of primary mesenchyme cells of the sea urchin embryo: a precisely timed epithelial mesenchymal transition. Birth Defects Res. C Embryo Today. 81:241–252. 10.1002/bdrc.20113 [DOI] [PubMed] [Google Scholar]
- Xie S., and Martin A.C.. 2015. Intracellular signalling and intercellular coupling coordinate heterogeneous contractile events to facilitate tissue folding. Nat. Commun. 6:7161 10.1038/ncomms8161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., and Weinberg R.A.. 2015. Epithelial–mesenchymal plasticity: A central regulator of cancer progression. Trends Cell Biol. 25:675–686. 10.1016/j.tcb.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M.. 2010. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12:533–542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Yu J.C., and Fernandez-Gonzalez R.. 2016. Local mechanical forces promote polarized junctional assembly and axis elongation in Drosophila. eLife. 5:e10757 10.7554/eLife.10757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J.A., and Wieschaus E.. 2004. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell. 6:343–355. 10.1016/S1534-5807(04)00060-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.