Secretory preproteins contain a mature domain fused to a signal peptide that targets the protein to the translocase, which mediates secretion. In this study, the authors show that the mature domains bear independent targeting signals (MTS) that consist of multiple, degenerate, interchangeable, linear or 3D hydrophobic stretches that are essential for proper secretion.
Abstract
Secretory proteins are only temporary cytoplasmic residents. They are typically synthesized as preproteins, carrying signal peptides N-terminally fused to their mature domains. In bacteria secretion largely occurs posttranslationally through the membrane-embedded SecA-SecYEG translocase. Upon crossing the plasma membrane, signal peptides are cleaved off and mature domains reach their destinations and fold. Targeting to the translocase is mediated by signal peptides. The role of mature domains in targeting and secretion is unclear. We now reveal that mature domains harbor their own independent targeting signals (mature domain targeting signals [MTSs]). These are multiple, degenerate, interchangeable, linear or 3D hydrophobic stretches that become available because of the unstructured states of targeting-competent preproteins. Their receptor site on the cytoplasmic face of the SecYEG-bound SecA is also of hydrophobic nature and is located adjacent to the signal peptide cleft. Both the preprotein MTSs and their receptor site on SecA are essential for protein secretion. Evidently, mature domains have their own previously unsuspected distinct roles in preprotein targeting and secretion.
Introduction
The Sec system is essential for cellular viability and mediates the export of most membrane and secretory proteins (Beckwith, 2013; Chatzi et al., 2013; De Geyter et al., 2016; Tsirigotaki et al., 2017). Some of them, mainly membrane proteins, are targeted cotranslationally, via the signal recognition particle (Grudnik et al., 2009). In contrast, most bacterial secretory proteins bind posttranslationally to the translocase, comprising the SecYEG protein-conducting channel and the peripheral ATPase SecA (Park and Rapoport, 2012; Chatzi et al., 2014; Tsirigotaki et al., 2017). They are synthesized as preproteins with N-terminal signal peptides fused to their mature domains (Martoglio and Dobberstein, 1998) and are maintained soluble, in poorly understood nonnative states, during their ribosome to membrane trafficking.
Signal peptides target preproteins to the translocase (Blobel and Dobberstein, 1975; Hegde and Bernstein, 2006). They bind with micromolar affinities to SecA (Roos et al., 2001; Papanikou et al., 2005; Gouridis et al., 2009) and allosterically activate the translocase (Gouridis et al., 2009). It has been shown that signal peptides can even target some cytoplasmic proteins artificially fused to them (Le Loir et al., 2001; Schierle et al., 2003).
In contrast to signal peptides, little is known about the role of mature domains in secretion. They are commonly considered as passive passengers. This view, however, cannot explain why signal peptides cannot export just any polypeptide fused to them (Le Loir et al., 2001), how at least some mature domains can be self-targeted to the translocase with nanomolar affinities (Gouridis et al., 2009), or the pioneering demonstration of in vivo secretion of signal peptide-less mature domains in prl strains (Bieker et al., 1990; Derman et al., 1993). In vitro, mature domains not only stimulated the SecA lipid (Lill et al., 1990) and translocation (Lill et al., 1990; Gouridis et al., 2009) ATPase activities but also could be fully translocated by in trans addition of signal peptide (Gouridis et al., 2009). Each of the two preprotein components binds on distinct SecA sites. Although the binding site of the mature domain remains unknown, that of the signal peptide is an elongated groove on the SecA preprotein-binding domain (PBD; Gelis et al., 2007; Gouridis et al., 2009).
We now demonstrate that mature domains contain their own targeting signals (mature domain targeting signals [MTSs]) that can act independently of or in addition to those of signal peptides. MTSs are universal, multiple, distributed, linear or 3D hydrophobic patches that become available for translocase binding because of the extensive loss of structure in targeting-competent preproteins. We also identified and mapped the previously elusive mature domain–binding site of SecA on a flat cytoplasmic platform. Multiple hydrophobic patches on SecA are available to bind the accessible MTSs. Some of them are substrate-binding motifs common to helicases. The main one is proximal to and forms an L-shaped groove with the signal peptide–binding cleft. Preproteins with mutationally impaired MTSs can still be targeted to the translocase thanks to their signal peptides, albeit with lower affinities. Nevertheless, they fail to become translocated. Both the preprotein MTSs as well as their receptor site on SecA are essential for preprotein secretion. We conclude that preprotein mature domains have their own, previously unsuspected, significant roles in preprotein targeting and secretion.
Results
Mature domains bear signal peptide–independent targeting signals
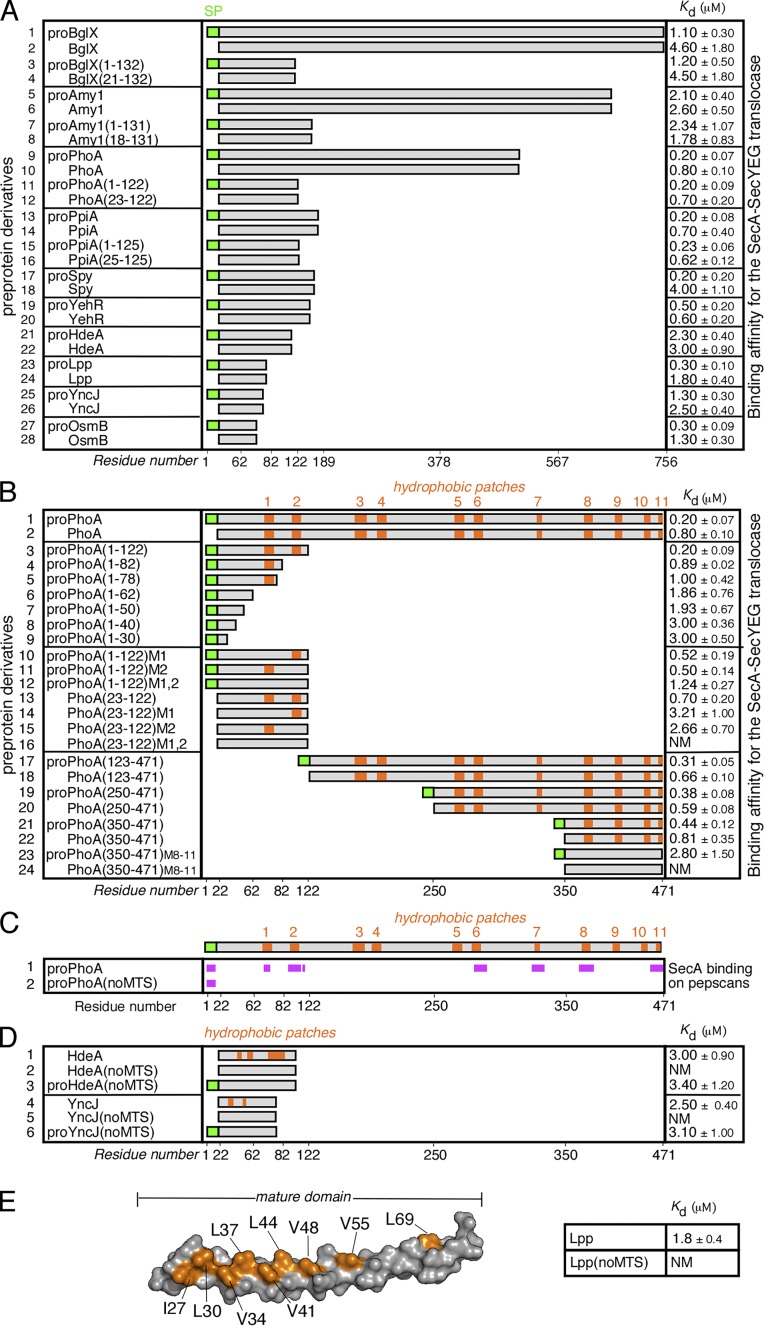
The mature domains from 11 preproteins were targeted to the SecA receptor of the membrane-embedded translocase in the absence of signal peptides with affinities that ranged from 0.6 to 4.6 µM (Fig. 1 A, even numbers; Gouridis et al., 2009). This is best rationalized by assuming that mature domains contain MTSs. To identify them, we first determined the minimal preprotein length that is required for high-affinity binding to SecA/SecYEG. Judging by the length of several native Escherichia coli preproteins (Orfanoudaki and Economou, 2014), ∼100 residues are sufficient for high-affinity targeting of preproteins or mature domains alone to the translocase (Fig. 1 A, lanes 17–28). In agreement to this, when we truncated four different long preproteins down to ∼100 aa, both the preproteins and the corresponding mature domains retained the affinities of their full-length parent molecules (lanes 1–16). The presence of signal peptides improved binding by ∼1.2- to 4-fold and, in the case of proSpy, by 20-fold (lanes 17 and 18; Gouridis et al., 2009).
Figure 1.
MTSs in preproteins. (A) Equilibrium dissociation constants (Kd; micromoles; right) of the indicated preproteins, their mature domains and truncated analogues for the wild-type SecA-SecYEG translocase. No detectable binding of proPhoA or derivatives occurs to the SecYEG-inverted membrane vesicles in the absence of SecA (for detailed analysis, see Gouridis et al., 2009). n = 3–9. x axis indicates preprotein length in residues. SP, signal peptide. (B) Hydrophobic patches (HPs; orange) in the mature domain of PhoA (see also Fig. S1 A) and their contribution to targeting. The Kd of the indicated protein derivatives for the translocase were determined; n = 3–9. M1,2 and M8–11, hydrophobicity-reducing mutations in the indicated HPs of PhoA; the mutated residues are detailed in Fig. S1 A. x axis: proPhoA residues. (C) Binding experiments of soluble SecA onto proPhoA or proPhoA(noMTS) peptide arrays are summarized; n = 6 (see also Fig. S2 A). HPs are indicated; x axis: proPhoA residues. (D) HPs (orange) in the secretory proteins HdeA and YncJ (see also Fig. S1 A) and their contribution to targeting. The Kd of the indicated protein derivatives for the translocase were determined; n = 3–6. noMTS, hydrophobicity-reducing mutations in all HPs; the mutated residues are detailed in Fig. S1 A. (E) Left: 3D surface representation of the Lpp structure (PDB: 1EQ7; a single protomer is shown). The residues that were mutated in Lpp(noMTS), shown in orange, are detailed in Fig. S1 B. The Kd of Lpp(noMTS) for the translocase was determined (right); n = 3. NM in C–E, nonmeasurable binding for the translocase (i.e., >20 µM). Affinity values in A, B, D, and E represent means ± SEM.
Preproteins and derivatives thereof were purified in a chaotrope as nonstructured, denatured polypeptides and then transferred to an aqueous environment to determine binding to the translocase or/and translocation (see the following paragraph and Materials and methods). In all steps, the conditions used were those of established, well-characterized protocols widely used for the past several years (Lecker et al., 1990; Gouridis et al., 2010). Solubility and circular dichroism measurements (see Materials and methods) performed under the same conditions verified that all preproteins and their mutated or truncated derivatives remained soluble and nonfolded for at least 1 h (see Materials and methods section Solubility/aggregation testing of preproteins).
MTSs are hydrophobic patches in the protein primary sequence
To determine the elements that are important for high-affinity translocase targeting, we focused on proPhoA(1–122) that retained the affinity of the full-length proPhoA for the translocase (Fig. 1 B, compare lane 3 with lane 1) and performed further truncation analysis. Additional deletions gave rise to stable polypeptides that could be purified by affinity chromatography. However, C-terminal truncations of increasing length progressively lost translocase affinity (Fig. 1 B, lanes 4–9) down to that of the signal peptide (dissociation constant [Kd] ∼3 µM; lane 9). Full-length proPhoA has 11 hydrophobic patches (HPs) with no sequence conservation, spread throughout its sequence (Figs. 1 B, lane 1, orange; and Fig. S1 A, top; see also Materials and methods for a definition and description of HPs). The minimal proPhoA(1–122) retained two of them (Fig. 1 B, lane 3). The loss of targeting efficiency that was observed in the soluble C-terminal truncations of proPhoA(1–122) coincided with deletion of one (Fig. 1 B, lanes 4 and 5) or both (lanes 6–9) of these hydrophobic patches. A similar decrease in affinity was seen by introducing mutations that reduced the hydrophobicity of those hydrophobic patches in an otherwise intact proPhoA(1–122) (i.e., M1, M2, or M1,2; Fig. 1 B, compare lanes 10–12 with lane 3). The effect became pronounced in the absence of the signal peptide. Targeting of PhoA(23–122) was either substantially affected when one hydrophobic patch was mutated (Fig. 1 B, compare lanes 14 and 15 with lane 13) or rendered nondetectable when both were (lane 16). Because all PhoA derivatives retained a nonfolded state and the solubility properties of the parent molecules, their impaired affinity for the translocase can only reflect a direct association defect, verifying the significance of MTSs in mature domain targeting. By comparing PhoA(23–122)M1,2 with proPhoA(1–122)M1,2, and taking into account that the signal peptide alone was targeted to the translocase with a Kd = 3 µM (Fig. 1 B, lane 9), it became evident that targeting of proPhoA(1–122)M1,2 could occur exclusively because of its signal peptide (compare lanes 16 and 12).
MTSs are multiple, distributed, and interchangeable
To test whether hydrophobic patches 1 and 2 act as the sole MTSs of PhoA, we deleted the 100 aa that contained them. The derivative PhoA(123–471) (Fig. 1 B, lane 18) had the same affinity for SecY-bound SecA as full-length PhoA (lane 2), indicating that the remaining hydrophobic patches acted as targeting signals of equal strength to HP1 and HP2. So did all PhoA derivatives with additional consecutive deletions (Fig. 1 B, lanes 20 and 22), unless their hydrophobic patches were mutated (lane 24). Clearly, one mature domain has multiple, distributed, and mutually interchangeable MTSs with degenerate sequences.
A minimum of one MTS allowed signal peptide–independent mature domain targeting (Fig. 1 B, lanes 14 and 15), and two MTSs optimized it (lane 13). This can complement and enhance the alternative targeting solution, represented by a single signal peptide on each preprotein (Fig. 1 B, lanes 6–9, 12, and 23).
Physical association of SecA with MTSs of PhoA on peptide arrays
To further interrogate our findings, we used arrays of immobilized proPhoA peptides and probed the ability of SecA to bind to them. We observed direct SecA binding to the signal peptide and on six linear HPs of PhoA but no binding to any of the dozens of flanking peptides that included various polar stretches (Figs. 1 C, lane 1; and Fig. S2 A, left). Because SecA recognized the PhoA HPs even as 13-mer peptides, we presume that the surrounding mature domain regions play negligible role. We also probed SecA binding using “mutated” proPhoA peptide arrays, in which all of the hydrophobic patches present in the mature domain region were mutated with the purpose of reducing their hydrophobicity. This time, we observed that SecA binding on mature domain regions of proPhoA was practically eliminated, whereas in the same experiment, binding to peptides containing signal peptide segments was retained (Fig. 1 C, compare lane 2 with 1; and Fig. S2 A, compare right with left).
We concluded that SecA binds specifically to hydrophobic patches on PhoA.
MTSs are universal preprotein elements
All of the mature domains we tested (Fig. 1 A), with one exception (Lpp; see the following paragraph), were predicted to have two to three hydrophobic patches per 100 aa (unpublished data). To test whether hydrophobic patches are universal MTSs, crucial for translocase targeting, we mutated the hydrophobic patches of two more preproteins, HdeA and YncJ (Fig. 1 D, lanes 1 and 4, HdeA(noMTS) and YncJ(noMTS); and Fig. S1 A). In both cases the mutant mature domains lost their targeting efficiency completely (Fig. 1 D, compare lane 2 with lane 1 and lane 5 with lane 4). Similarly to our previous observation with proPhoA(1–122), the mutant preproteins could still be targeted to the translocase because of their signal peptides (Fig. 1 D, lanes 3 and 6).
MTSs can be 3D
Are all hydrophobic patches linear stretches? We have observed that 20 E. coli mature domains lack any detectable continuous hydrophobic segments in their primary sequence (Table S1). The major outer membrane lipoprotein Lpp (Shu et al., 2000) belongs to this group (Fig. S1 A) and may represent a one-helix minimal exportable structure that displays continuous hydrophobicity only on one face (Fig. 1 E, left, orange). Once in the periplasm, native Lpp trimerizes through the same hydrophobic face of its single helix (Fig. S1 B; Shu et al., 2000). To test whether this 3D hydrophobic patch is functional as an MTS, we mutated it (Lpp(noMTS)) and examined the effect on Lpp targeting. Although Lpp bound to the translocase, its reduced hydrophobicity derivative Lpp(noMTS) did not (Fig. 1 E, right). We concluded that MTSs can also be 3D.
MTSs are essential for secretion in vitro
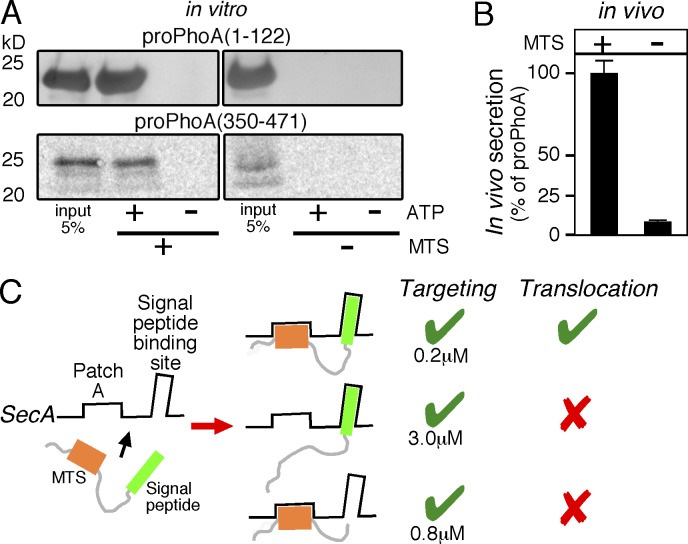
So far, we have demonstrated that mature domains carry MTSs that can target them to the translocase in a signal peptide–independent way. However, is this preprotein feature important for the translocation process per se? Would a preprotein targeted to the translocase only by its signal peptide get subsequently translocated, or is binding of both mature domain and signal peptide on SecA needed downstream of targeting? To address this, we compared the secretion efficiency of preproteins with wild-type mature domains (proPhoA(1–122); proPhoA(350–471)) to those with mature domains that lost their MTSs (proPhoA(1–122)M1,2; proPhoA(350–471)M8-11; Fig. 2 A) in vitro. We observed that both proPhoA(1–122) and proPhoA(350–471) are secreted in vitro unless their mature domains lost their MTSs (Fig. 2 A, compare left and right lanes). Because in the in vitro translocation assay all preproteins were used at 8 µM, thus securing association to the translocase even for those with a Kd = 2.5 µM, the inability to get translocated can only reflect an after-targeting defect.
Figure 2.
MTSs are essential for protein secretion. (A) Representative, in vitro, SecA-dependent translocation assays of proPhoA(1–122)M1,2 compared with proPhoA(1–122) (top) and proPhoA(350–471)M8-11 compared with proPhoA(350–471) (bottom) into the lumen of SecYEG containing inverted membrane vesicles; n = 3. 5% of the input is indicated. (B) In vivo secretion of proYncJ-PhoA (left) and proYncJ(noMTS)-PhoA (right) was compared with that of proPhoA (considered as 100%) under identical conditions (MC4100 cells; OD600 = 0.2; 0.002% wt/vol arabinose; 30min; 30°C). In all cases, the measured PhoA enzymatic activity (Gouridis et al., 2009) was normalized to the amount of PhoA or fusion-PhoA protein produced. This provided a means to quantitate the in vivo secretion of all three proteins. The alkaline phosphatase units per microgram PhoA for cells expressing proPhoA was considered 100%; proYncJ-PhoA and proYncJ(noMTS)-PhoA values were expressed as a percentage of this value. n = 5. Values are expressed as means ± SEM. (C) Schematic summary of requirements for preprotein targeting and translocation (as indicated). Preproteins are bivalent ligands with distinct binding sites on SecA. Targeting to the translocase is efficiently achieved by either targeting element, the signal peptide (middle) or one/more MTSs (bottom), independently. However, preprotein translocation requires binding of both (top).
Clearly, the MTSs are essential for preprotein translocation in vitro. This requirement cannot be bypassed by the presence of a functional signal peptide.
MTSs are essential for secretion in vivo
To confirm that the MTSs are essential for protein translocation in vivo as well, we selected one example from the ones used in the in vitro system. We chose YncJ; its MTSs set the low threshold in hydrophobicity that defined a functional MTS (Figs. 1 D and S1 A and Materials and methods). proYncJ and proYncJ(noMTS) were fused in front of PhoA, acting as a sensitive secretion reporter, because it only becomes active in the periplasm (Michaelis et al., 1983). These plasmids, as well as one carrying only the proPhoA gene acting as a control, were transformed in MC4100 cells. The expression of either gene was induced (OD600 = 0.2; 0.002% wt/vol arabinose; 30 min; 30°C). Half of the collected samples were used to measure the enzymatic activity of PhoA in the periplasm, as described previously (Gouridis et al., 2009), and the other half to analyze the total protein content on SDS-PAGE. PhoA or PhoA-fusion proteins were visualized after Western blot analysis and immunodetection using α-PhoA antibody and then quantified. The enzymatic activity was normalized per microgram PhoA or PhoA fusion protein produced. This provided us with the means of quantitating protein translocation of the fusion proteins relative to proPhoA (Fig. 2 B).
In agreement with the in vitro results, the ability of YncJ to become translocated in vivo was drastically affected by mutating its MTSs. A signal peptide alone was not sufficient for protein secretion. We anticipate that the HPs of other proteins, more hydrophobic than those of YncJ, would have the same or even more exaggerated effects.
In summary (Fig. 2 C), a preprotein can become targeted to the translocase by either its signal peptide or by its MTSs or by both. However, only the preprotein that gets both its signal peptide and mature domain bound on SecA can proceed and become translocated.
Lack of native structure in translocation-competent preproteins makes the MTSs available
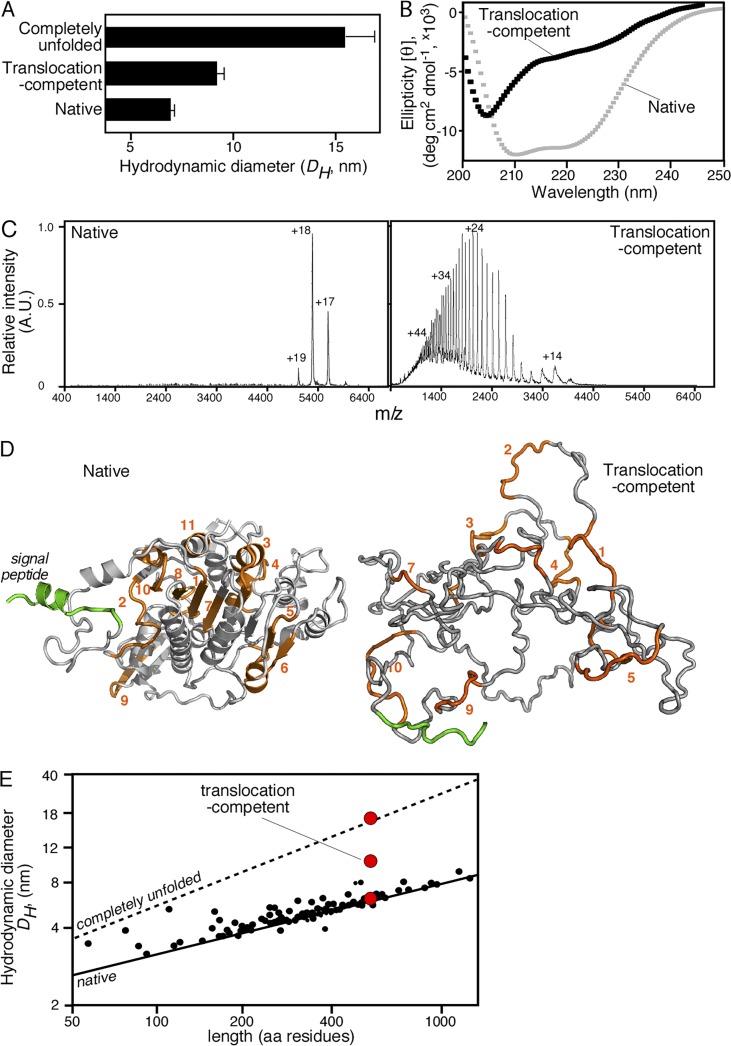
How do MTSs become available for SecA binding in translocation-competent preproteins? Natively folded proPhoA is an alkaline phosphatase with no affinity for the translocase (Gouridis et al., 2009). On the other hand, translocation-competent proPhoA has no phosphatase activity but binds with high affinity to the translocase (Gouridis et al., 2009; Fig. 1 A, lane 10). We probed the structural features of the translocation-competent proPhoA using various biophysical techniques (Figs. 3 and S3).
Figure 3.
Biophysical characterization of translocation-competent proPhoA. (A) Hydrodynamic diameter (DH, nanometers; x axis) of native (no urea; no DTT), translocation-competent (no urea; 1 mM DTT) and completely unfolded proPhoA (8M urea; 1 mM DTT) as determined by quasielastic laser light scattering that was performed online after gel permeation chromatography on a Superdex HR200 (see also Fig. S3 A); n = 6–15. Values represent means ± SD. For the native species, natively purified proPhoA was diluted and chromatographed in buffer L. For the translocation-competent and the completely unfolded species, urea-purified proPhoA (0.5 mM; 6 M urea), preincubated with DTT (10mM; 30min; ice), was diluted and chromatographed in buffer L supplemented with the indicated urea and DTT concentration. (B) Representative circular dichroism spectra of natively folded (no DTT) and translocation-competent (1 mM DTT) proPhoA; n > 3. x axis: wavelength (nanometers); y axis: mean residue molar ellipticity ([θ]MRW). For the translocation-competent species, urea-purified proPhoA was preincubated with 10 mM DTT (30 min; ice) and dialyzed in buffer U supplemented with 8M urea and 1 mM DTT. The natively purified proPhoA was dialyzed in 5 liters buffer U (15 h; 4°C). Spectra for both were recorded in buffer U supplemented with 1 mM EDTA, 0.2 M urea, and DTT (as indicated). Natively folded proPhoA exhibits two minima (208 and 222 nm) typical of folded, predominantly α-helical proteins, whereas the translocation-competent proPhoA does not. The urea-purified proPhoA, dialyzed in buffer U in the absence of 5 liters DTT (15 h), folds and gives spectra similar to those of the natively purified proPhoA (not depicted). (C) Representative native nano–electrospray ionization mass spectrometry spectra of native PhoA and translocation-competent proPhoA; n = 3. Translocation-competent proPhoA acquires many charges with broad distribution, typical of unfolded proteins with increased solvent-accessible surface area (Testa et al., 2013), whereas native PhoA acquires few charges with narrow distribution, typical of well-folded, compact proteins, and is a dimer. (D) Ribbon 3D model of folded E. coli PhoA (PDB: 1KHN; a single protomer is shown; left). A signal peptide (green) was modeled. Model of disordered proPhoA derived from the trigger factor–bound structure solved by nuclear magnetic resonance (Saio et al., 2014), with a DH in accordance with the quasielastic laser light scattering measurements of disordered PhoA (right). (E) Predicted and measured hydrodynamic diameters (DH) of SecA-dependent preproteins. Lines show the predicted DH of either folded (solid) or completely unfolded (dotted) preproteins as a function of their length (Wilkins et al., 1999b). Small black dots represent the calculated DH for 40 mature domains with solved structures (Table S8), using Hydropro (García De La Torre et al., 2000). Red circles represent the experimental DH measurements for proPhoA (as indicated; see also A and Fig. S3 B).
Gel permeation chromatography coupled online to multiangle and quasielastic laser light scattering allowed us to measure the hydrodynamic diameter (DH) of proPhoA under different regimes. The DH of the natively folded, monomeric protein (6.9 [±0.47] nm) became 15.4 (±1.95) nm in 8 M urea (Fig. 3 A and Fig. S3, A and B), suggestive of a fully extended molecule. However, the translocation-competent proPhoA in an aqueous environment had a DH of 9.09 (±0.65) nm. In summary, the translocation-competent proPhoA was measured to be ∼30% more expanded than the native form yet significantly more compacted than a random coil.
Moreover, far-UV circular dichroism and native mass spectrometry demonstrated that the translocation-competent proPhoA had significantly less secondary (Fig. 3 B) and tertiary (Fig. 3 C) structure than the natively folded form under the same conditions. Because the targeting-competent, signal-less PhoA demonstrated a similarly loose structure under the same conditions (Fig. S3, C–E), these structural properties are inherent to the mature domain and independent of any contribution from the signal peptide. We have observed that the translocation-competent states of other secretory proteins have similarly loose structures (unpublished data).
We concluded that translocation-competent preproteins are looser than their native states but are not random coils. Some or all of the multiple, linear (Fig. 1, B–D), or discontinuous, 3D (Fig. 1 E) MTSs might be buried in the natively folded Lpp (Fig. S1 B) or PhoA (e.g., MTS 1, 7, and 8; Fig. 3 D, left) but would be exposed in the nonfolded/disordered states that are competent for targeting (Fig. 1 E and Fig. 3 D, right). Other MTSs are more peripheral (e.g., MTS 2, 3, and 5; Fig. 3 D, left). All MTSs could be occasionally exposed for translocase recognition by the expanded, nonfolded mature domains (Fig. 3 D, right).
SecA handles a wide spectrum of preprotein dimensions
In our effort to understand how hundreds of nonfolded preprotein structures are recognized by SecA, we calculated the DH of the folded and unfolded states of secretory proteins using bioinformatic prediction for the hydrodynamic dimensions of folded secretory proteins for which structures exist (García De La Torre et al., 2000) and for those of the nonfolded states (Wilkins et al., 1999a; García De La Torre et al., 2000; Uversky, 2002; Materials and methods). Folded preprotein mature domains span a wide range of diameters (∼2.8–9.1 nm; Fig. 3 E, solid line; and Table S8). Complete unfolding is predicted to increase their diameters (random coil upper limits ∼3.6–37.4 nm; Fig. 3 E, dashed line; and Table S8). Based on our experimental measurements on proPhoA (Fig. 3 A), it is anticipated that translocation-competent, nonfolded preproteins are particles of significant dimensions, with their diameters ranging between those of the folded and unfolded states. Many of them would be as large as their receptor, the SecA dimer (maximal dimension of 10.5 nm; Gouridis et al., 2013). We examined how the SecA receptor recognizes these large nonfolded polypeptides.
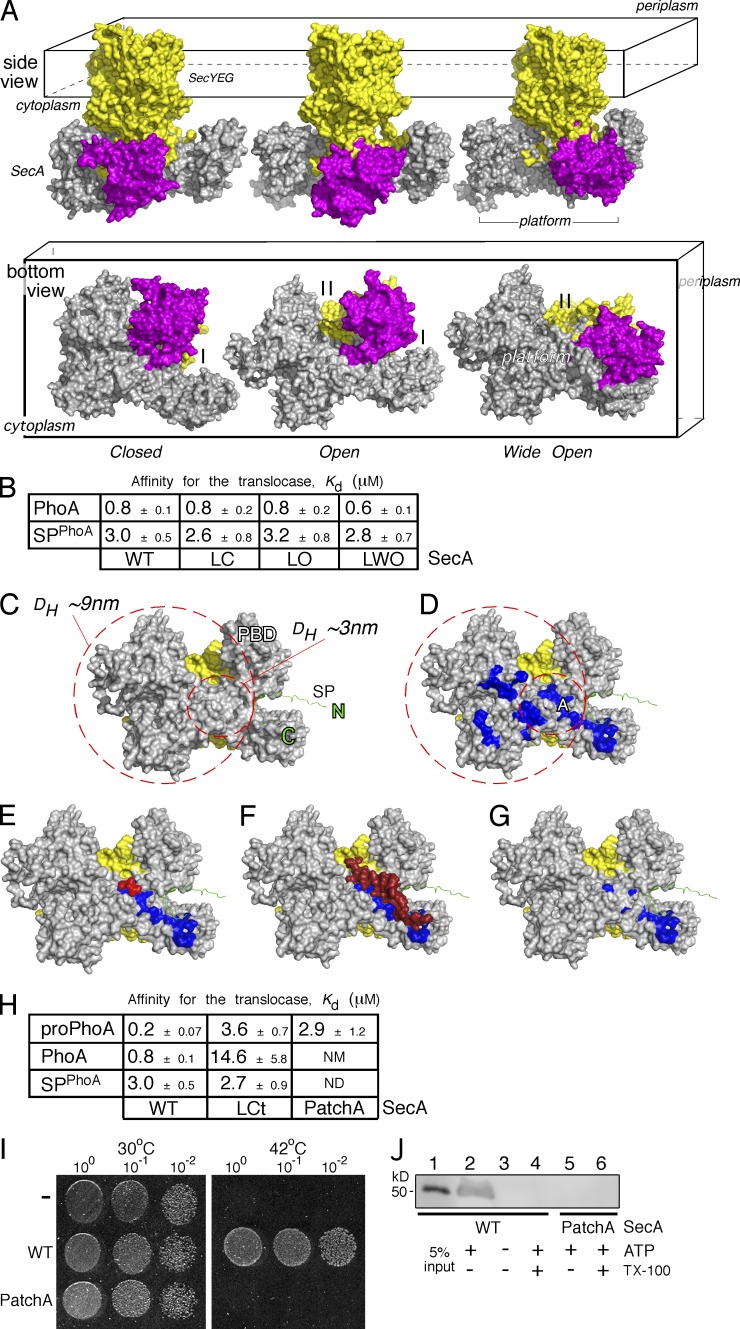
SecA clamps I and II are not used for mature domain docking
PBD swiveling around its stem forms two apparent “clamps” (I and II; Fig. 4 A, bottom; and Fig. S4, A and B). Clamp II was proposed to “grab” preproteins in advanced stages of translocation (Zimmer et al., 2008; Bauer and Rapoport, 2009; Chen et al., 2015). To test whether SecA clamps I and II play a role as mature domain docking sites, we “locked” PBD in its three discernible states using engineered disulfides (Fig. S4 C and Materials and methods). Locking PBD in the wide open conformation (locked wide open [LWO]) blocks clamp I, closed conformation (locked closed [LC]) blocks clamp II, and open conformation (locked open [LO]) blocks neither clamp for preprotein binding (Fig. 4 A, bottom view). After purification, all three mutants were shown to have formed quantitatively intraprotomeric disulfide bonds, representative of each locked state (Fig. S4 C). Next, we determined the Kd of the derivative translocases for PhoA or for its signal peptide and observed that blocking either clamp does not alter the affinity of either element for the translocase (Fig. 4 B). Although clamp II is part of the preprotein route toward the SecYEG channel during active translocation (Bauer and Rapoport, 2009) our results clearly suggest that at least the initial mature domain docking occurs on more peripheral SecA regions, outside the two clamps.
Figure 4.
The mature domain–binding site onto SecA. (A) The E. coli SecA (gray)–SeYEG (yellow) was modeled after the Thermotoga maritima translocase in three conformational states, based on PBD (purple) positioning: closed (left), open (middle), and wide open (right). Side and bottom views are shown (as indicated). I, II: SecA clamps. (B) Kd measurements of PhoA and its signal peptide (SPPhoA) for the wild-type (WT), locked closed (LC), locked open (LO), and locked wide open (LWO) SecA bound to SecYEG-inverted membrane vesicles. proPhoA(1–30) was used as SPPhoA. Affinity values represent means ± SEM; n = 3. (C) Potential space occupied by an incoming preprotein onto the cytoplasmic side (platform) of a SecA(gray)–SecYEG(yellow) translocase; signal peptide is in green. The inner circle represents the minimum area a translocation-competent preprotein would occupy, depicted here by the predicted DH of the smallest known preprotein (proEcnA; ∼3 nm; Table S8). The bigger circle represents the area that the expanded, translocation-competent proPhoA would occupy, based on our experimental measurements (∼7 nm; Figs. 3 A and S3 A). (D) Hydrophobic patches (blue; PatchA is indicated) on SecA’s cytoplasmic platform (the rest as in C). (E–G) Structural models of (E) a tripeptide (red; Zimmer and Rapoport, 2009) bound on PatchA (blue) of SecA (detailed interactions in Fig. S5 B) and (F) the C-tail of SecA (dark red) shielding PatchA (blue; Hunt et al., 2002). The C-tail directly interacts with or shields PatchA residues (detailed interactions in Fig. S5 B) but only partially occludes the signal peptide–binding site on SecA (Gelis et al., 2007; see also Fig. S6 E). Signal peptide–binding-determinant residues (i.e., L306; Fig. S6 E) remain exposed and available for interaction at this state. (G) SecA(PatchA) mutant. Four conserved PatchA amino acids (see also Fig. S5 C) were substituted with alanyl residues (M191A/F193A/I224A/I225A) to disrupt the continuum of its hydrophobic surface. (H) Kd measurements of PhoA, proPhoA, and its signal peptide (SPPhoA) for the wild-type, locked C-tail (LCt), and PatchA SecA bound to SecYEG-inverted membrane vesicles; Affinity values represent means ± SEM; n = 3–12. proPhoA(1–30) was used as SPPhoA. NM: nonmeasurable binding (i.e., >20 µM). (I) Representative, in vivo, genetic complementation assay of the E. coli BL21.19(secAts) strain by either an empty vector (−) or wild-type secA or secA(PatchA) mutant; n = 3. Only wild-type secA allows cell growth at 42°C. An identical plate, grown in parallel, at 30°C is shown; the dilutions of cells that were used are indicated. (J) Representative, in vitro SecA-dependent translocation of proPhoA into wt SecYEG-inverted membrane vesicles using wild-type SecA or SecA(PatchA) mutant under the same conditions; n = 3. 5% of the proPhoA input is indicated. Migration of ovalbumin (prestained protein molecular mass marker; Thermo Fisher Scientific).
Mature domains might dock onto the flat cytoplasmic platform of SecA
The signal peptide binds onto the PBD of SecA with its C terminus oriented toward the helicase motor, facing the cytoplasm (Fig. 4 C, light green; Gelis et al., 2007). Given (a) this orientation, (b) mature domain docking outside clamps I and II (see previous paragraph), and (c) the diameters of the expanded, translocation-competent preproteins (Fig. 3 E and Table S8), mature domains may bind to the flat cytoplasmic face of SecA (hereafter “platform”; Fig. 4, A [right] and C), that is formed at an interface of all four SecA domains (Fig. S4 A).
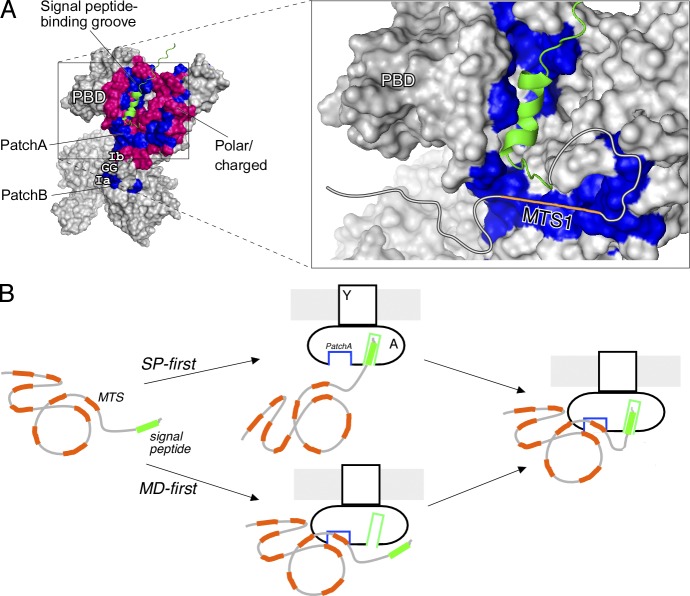
Mature domain docking on SecA relies on hydrophobic MTSs (Fig. 1). Analysis of the surface of the SecA platform revealed at least four hydrophobic patches (Fig. 4 D, blue) that remain generally exposed irrespective of PBD swiveling (Fig. S5A, blue). Among them, the largest one, PatchA, is formed by residues from three SecA domains (Fig. S5B), is the most proximal to the signal peptide cleft (Fig. 4 D, “A”) and is highly conserved (Fig. S5C, blue). This proximity might secure interaction even with the smallest mature domain (Fig. 4, C and D, small circle). A tripeptide bound on one end of PatchA has been cocrystallized with SecA (Zimmer and Rapoport, 2009; Fig. 4 E, red). Moreover, the flexible C-tail of SecA that shields PatchA (Fig. 4 F, dark red; Hunt et al., 2002) might act as an auto-inhibitory element mimicking a preprotein (Gelis et al., 2007). The extensive hydrophobic islands that are characteristic of the cytoplasmic platform are not common on the SecYEG-binding face of SecA (Fig. S6, A and B).
The PatchA of SecA is a binding determinant for mature domains
To test the role of PatchA in mature domain binding we locked on it the C-tail of SecA (hereafter LCt), quantitatively, via engineered disulfides (Fig. S4 D) and determined the Kd of SecA(LCt) for PhoA. Clearly, shielding PatchA severely reduced the affinity of PhoA for the translocase (∼18-fold; Fig. 4 H). In contrast, proPhoA exhibited the same affinity as the SPPhoA, presumably because it could still bind to the unobstructed signal peptide groove of SecA via its signal peptide (Fig. 4 H). DTT addition restored mature domain binding (not depicted).
To further examine the importance of PatchA we substituted four of its main conserved hydrophobic residues with alanines (M191A/F193A/I224A/I225A; hereafter SecA(PatchA)), with the purpose of reducing its hydrophobicity (Fig. 4 G). SecA(PatchA) retained wild-type affinity for SecYEG (SecA(PatchA) Kd = 100 nM [±15]; SecA Kd = 62 nM [±10]; Karamanou et al., 2008; Gouridis et al., 2013), indicating that the mutated SecA is structurally intact. Nevertheless, its binding affinity for PhoA was eliminated (Fig. 4 H), demonstrating that PatchA is a major mature domain–binding determinant. In agreement with the results seen with SecA(LCt), proPhoA could still be targeted to the translocase, with a Kd = 2.9 µM characteristic of the signal peptide alone binding (Fig. 4 H).
PatchA is essential for protein secretion
If mature domain targeting to SecA was an important feature of protein secretion, it is anticipated that the PatchA mutant derivative (secA(PatchA)) would be defective in protein translocation. secA(PatchA) was tested for its ability to complement a thermo-sensitive BL21secAts strain at 42°C (Mitchell and Oliver, 1993), in vivo. We observed that growth of the BL21secAts was fully restored by the wild-type SecA, but not by secA(PatchA) (Fig. 4 I).
We next examined the ability of purified SecA(PatchA) to support ATP-driven protein translocation in vitro (Fig. 4 J). In contrast to SecA (Fig. 4 J, lane 2), SecA(PatchA) (lane 5) was unable to catalyze the translocation of proPhoA into SecYEG-containing inverted inner membrane vesicles.
We concluded that PatchA is an essential element of SecA-mediated protein translocation.
Discussion
We have identified novel, universal, targeting signals that lie within the mature domain of preproteins and are essential for secretion, as well as their receptor sites on SecA. MTSs are either linear or 3D hydrophobic patches that are multiple, distributed, degenerate, and mutually interchangeable. Additional electrostatic or polar interactions, not detected here, might contribute in preprotein targeting to the translocase. MTSs increase a preprotein’s targeting information repertoire and ensure high-affinity posttranslational targeting to the translocase. MTSs and signal peptides are autonomous preprotein targeting elements, as they bind independently to distinct but adjacent SecA sites, forming a characteristic L-shaped receptor (Figs. 1 and 5 A; Gouridis et al., 2009). Although the signal peptide binds to its groove on PBD (Gelis et al., 2007; Figs. 5 A and S6 D), the MTSs bind on hydrophobic patches on the flat cytoplasmic platform of SecYEG-bound SecA (Figs. 4 and 5 A). Some of these patches include motifs known to bind RNA helicase substrates such as DNA or RNA molecules (Fig. S5 B). Three of these motifs (Ia, Ib, and GG) assemble close together in 3D space, with motifs Ib and GG forming an elevated hydrophobic ledge (PatchB) overlooking PatchA (Fig. 5 A, left). Preproteins trapped on these motifs would be on pathway to enter the SecY channel (Fig. 5 A, right). Our findings expand and update current views of posttranslational protein targeting.
Figure 5.
Preprotein docking on SecA and targeting to the translocase. (A) Structural model for preprotein docking on SecA. Complete view of the SecA features surrounding PatchA and the signal peptide binding groove (left; blue, nonpolar SecA residues; pink, polar/charged SecA residues; green, signal peptide). Zoom-in view of PatchA and the signal peptide–binding groove (right). The signal peptide–binding site on SecA lies within a hydrophobic groove between the PBD and IRA1 (Gelis et al., 2007; for details, see also Fig. S6, C and D). PatchA is a shallow furrow that converges at a 90° angle, forming an “L” shape with the signal peptide groove, and is surrounded by motives (Ia, Ib, and GG) on which helicases bind their nucleic acid substrates (e.g., RNA helicases; Sengoku et al., 2006; Papanikolau et al., 2007). A hypothetical mature domain is drawn as an extension of the signal peptide, bound on PatchA with its MTS1 (orange), based on the mainly hydrophobic nature of the interaction (Fig. 1 and Materials and methods, Bioinformatics approach to define hydrophobic patches on proteins). Additional minor electrostatic or polar contacts might contribute in the association of some mature domains with SecA. (B) Model of preprotein targeting/docking to the Sec translocase initiated by stochastic binding of either the signal peptide (SP-first) or the mature domain (MD-first; see text for details). A, SecA; Y, SecY. Green, signal peptide; orange, mature domain. Only one SecA protomer is shown (the one activated for high-affinity preprotein binding; Gouridis et al., 2013). The second regulatory protomer is omitted for simplicity.
Outfitting mature domains with hydrophobic targeting signals is somewhat unexpected. As signal peptides are add-on, temporary appendages, absent from the final protein structure, they are an obvious, simple choice as targeting tags. Evolution can tinker with signal peptides, until they are “custom-optimized” for targeting every single preprotein of the secretome, without affecting the exported protein’s structure/function that resides in its mature domain. In contrast, MTSs are both targeting and structural elements of the final natively folded structure of any secretory protein. As such, their targeting potential can be optimized only up to the point that it does not interfere with the subsequent structure/function of the exported protein. This pressure may explain why even within a single preprotein the MTSs appear degenerate and of weak hydrophobicity and far outnumber the single signal peptide tag. Multiple, degenerate MTSs endorse the hundreds of different mature domains with nanomolar affinities for the single SecA receptor.
How do MTSs become exposed for chaperone/translocase recognition? We presume that, as seen for PhoA (Fig. 3 D) or Lpp (Fig. S1 B), at least some of the MTSs are buried/hidden in the natively folded structure. Exposing hydrophobic MTSs while remaining aggregation resistant during translocase targeting, in the absence of chaperones (Fig. 1), is a conundrum. Whether this is achieved through cycles of fleeting MTS solvent exposure/retraction, or/and MTSs being flanked by aggregation-avoiding gatekeeper residues (Beerten et al., 2012), or/and other unique structural features of secretory mature domains is currently unknown and a future challenge for structural studies.
Either one of the two preprotein targeting elements suffices for nanomolar to low-micromolar targeting to the translocase: MTSs (Fig. 1; Papanikou et al., 2005; Gouridis et al., 2009) or signal peptides (Roos et al., 2001; Gelis et al., 2007; Gouridis et al., 2009). However, in a posttargeting step, both elements become essential for secretion. The role of the signal peptide in translocase triggering cannot be bypassed, except partially by conformational mutant derivatives of the translocase (known as prl; Bieker et al., 1990; Gouridis et al., 2009). Why MTSs are essential for preprotein secretion (Fig. 2) is currently unknown. We anticipate that their efficient targeting revealed here secures the engagement of mature domains in the translocase, perhaps in the “trapping” step that is a prerequisite for stimulation of the SecA ATPase activity (Gouridis et al., 2009).
Recognition of MTSs by SecA is significantly more complex than a simple, nonspecific, hydrophobic interaction: (a) SecA binds to both linear and 3D MTSs (Fig. 1, B, D, and E). (b) Apart from the signal peptide, soluble dimeric SecA binds well only to 6 of the 11 PhoA MTSs on peptide arrays (Fig. 1 C). This suggests that MTSs may have varying affinities for SecA; some may only associate with SecYEG-bound SecA, justifying the measured 10-fold-higher affinity (Gouridis et al., 2009). (c) Despite apparent similarities, the SecA-binding sites on the preprotein are distinct from those of chaperones (trigger factor and SecB; Fig. S2 B; Saio et al., 2014; Huang et al., 2016). (d) MTSs might be dynamically recognized in a 3D search space. Although preproteins are commonly depicted as extended coils in their targeting-competent state, our data imply that these polymers are significantly collapsed and yet expanded and clearly nonnative (Fig. 3, A and C), with little (Fig. 3 B) or some (Lecker et al., 1990) secondary structure. In these states, some of the available MTSs will spend a lot of time internally in the polypeptide structure, possibly making near-native but ephemeral hydrophobic contacts, whereas only some of the others will be solvent-exposed. Polymer “breathing” might expose/present different combinations of MTSs for SecA recognition.
How has SecA evolved to recognize preproteins with high affinity? PatchA, lying adjacent to the signal peptide–binding site (Fig. 5 A; Gelis et al., 2007), is a major, conserved (Fig. S5 C) mature domain receptor site (Fig. 4). The proximity of the two sites suggests that the MTSs of short preproteins or the most N-terminal MTSs of longer ones may interact preferentially with PatchA. In parallel, interaction of additional Patches (Figs. 4 D; Fig. S5, A and B [e.g., PatchB located near PatchA]; and Fig. S6) with more MTSs (i.e., in larger preproteins with multiple MTSs) might enhance the affinity of a mature domain. To avoid illicit or premature interactions, SecA covers PatchA with its mature domain–mimicking C-tail. The C-tail physically occupies PatchA but only partially occludes the signal peptide–binding groove by sterically decreasing accessibility to it (Figs. S5 B and S6 E), which can lead to reduced binding of some signal peptides to C-tail–containing SecA in solution (Gelis et al., 2007), but not in the SecYEG-bound SecA state (Fig. 4 H). PatchA formed by residues that come together in space from three different SecA domains, and its proximity to PBD and overlap or/and proximity with known substrate binding motifs in helicase motors (Figs. 5 A and S5 B) suggest a simple mechanism for mature domain dissociation through nucleotide-driven allosteric changes during translocase cycles (Chatzi et al., 2014).
Our data lead to the following hypothesis of posttranslational targeting (Fig. 5 B). Ribosome-released preproteins, alone or assisted by chaperones, diffuse and bind stochastically to single protomers of SecY-bound dimeric SecA (Gouridis et al., 2013). This SecA protomer is the activated as a high-affinity two-site preprotein receptor (Fig. 5 B). Preproteins may dock first either their signal peptide (Fig. 5 B, SP-first) or their mature domain (MD-first), but both elements eventually bind to the same SecA protomer. Evidently, large mature domains with multiple MTSs and/or mature domains fused to lower-affinity signal peptides have a higher probability of binding first than signal peptides. In such cases, the docked mature domains will bias the subsequent positioning of signal peptides (Gouridis et al., 2009) along their cleft (Gelis et al., 2007) on the same SecA protomer. Signal peptide docking on SecA will initiate triggering, a prerequisite for protein translocation (Gouridis et al., 2009, 2013; Fig. 5 B, right). Even if cytoplasmic proteins with exposed hydrophobic patches dock by chance on the translocase, their lack of a signal peptide ensures that they cannot initiate triggering (Gouridis et al., 2009) and will thus be rejected.
Materials and methods
Antisera used in this study
Polyclonal rabbit antisera were raised against purified proPhoA or SecA (Davids Biotechnologie). For His-tagged proteins, mouse anti–His antibodies (MCA 1396; Serotec) were used. HRP-AP goat anti–rabbit IgG (111-035-0030) or HRP AP goat anti–mouse IgG (115-035-1460) secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc.
Protein purification
E. coli BL21.19 cells transformed with pET22b plasmids carrying the indicated preprotein derivative gene or pET5 plasmids carrying secA derivative genes were grown (LB; 30°C; OD600 0.5–0.6), and gene expression was induced (0.2 mM IPTG; 3 h; 30°C). Cells were collected (13,000 g; 4°C; 15 min; Avanti J-26S XPI; Beckman Coulter), solubilized in buffer A for preproteins or buffer G for SecA derivatives, and lysed by using a French press (8,000 psi; three to five passes; precooled cylinder at 4°C; Thermo Fisher Scientific).
For preproteins and their derivatives, lysed samples were centrifuged (13,000 g; 4°C; Optima XPN-80; Beckman Coulter) to remove soluble proteins. The pellet was solubilized in buffer B, using a Dounce homogenizer and centrifuged (13,000 g, Optima XPN-80; Beckman Coulter). The new supernatant was diluted with buffer A (to 6 M urea) before applying it on a Ni+2-NTA agarose (QIAGEN) column preequilibrated with buffer C (gravity flow; 1 ml/min). The column was washed sequentially with buffers C and D (10 and 15 column volumes, respectively). Preproteins were eluted in buffer E, collected, incubated with 10 mM EDTA (30 min; 4°C), dialyzed (12–14,000 D molecular mass cutoff, buffer F, overnight; 4°C; Medicell Membranes Ltd.), aliquoted, and stored at −80°C.
For SecA and derivatives, lysed samples were centrifuged (13,000 g; 4°C, Optima XPN-80; Beckman Coulter) to remove membrane insoluble material. The supernatant was subjected to metal affinity chromatography, performed as described in the previous paragraph but using buffer G for column equilibration and washing, followed by washing in buffer H and elution in buffer I. The eluted material was incubated with 10 mM EDTA (30 min, ice) and dialyzed first in buffer J (2 h; 4°C) and then in buffer K (overnight; 4°C) before storage at −80°C. Further purification was followed for SecA(LC) and SecA(LCt); their eluate from Ni+2-NTA chromatography was concentrated and loaded on a preparative, preequilibrated with buffer L, Hi-Load Superdex 200 26/60 (GE Healthcare), under a flow rate of 1 ml/min at 4°C, to remove SecAs that had not formed intraprotomeric disulfide bonds. For SecA(LC), gel filtration was in buffer L. For SecA (LCt), gel filtration was in buffer M. 2-ml fractions were collected and analyzed by SDS-PAGE. Those containing intraprotomerically oxidized SecAs were pooled together. Elution samples were dialyzed first in buffer J supplemented with 2 mM EDTA and then in buffer K. SecA(LO) and SecA(LWO) oxidized spontaneously to >95% of the total protein; therefore, for these two proteins, there was no need to enrich the oxidized species by further purification.
Determination of protein concentration
Protein concentration was measured spectroscopically, in the range of 0.5–5 mg/ml, which is within the linear range of the Nanodrop instrument used (Thermo Fisher Scientific 2000 series; 280 nm; buffer L, 8 M urea; Stoscheck, 1990) or/and by Bradford protein assay (Bio-Rad Laboratories; Bradford, 1976) using BSA to generate a linear control standard curve (0.5–20 µg). The Expasy server (http://web.expasy.org/protparam/) was used to determine the molecular extinction coefficients and molecular masses of the different proteins needed for A280 analysis. Protein samples were concentrated using an ultrafiltration device (Amicon Ultra-15, 3K cutoff; EMD Millipore; 3,000 g; Sigma-Aldrich 3-16KL; 4°C).
Solubility/aggregation testing of preproteins
All proteins and their derivatives used in this study were tested for their solubility in aqueous buffer, as a function of time. Proteins in buffer F were diluted >30 times with buffer L supplemented with 1 mM DTT, 1 mM EDTA to 1–30 µM and incubated (at 4°C for 0, 10, and 60 min and 24 h). Samples were centrifuged (20,000 g; 10 min; 4°C), and protein concentration in the supernatant was monitored spectroscopically and compared with that of the input (0 time; 100%). Only proteins that remained soluble (≥90% for at least 1 h) were used in targeting or/and secretion experiments in vitro. proPhoA, as an example, remains soluble and competent for translocation ATPase activity for more than 24 h and does not bind to membranes nonspecifically (Gouridis et al., 2009). Moreover, these soluble forms elute in single, sharp peaks, far away from the exclusion volume in size exclusion chromatography (Fig. S3, A and B). These properties of proPhoA make it a preprotein of choice for biophysical/structural studies over the previously predominant, more established proOmpA that is aggregation prone and once aggregated cannot interact with the translocase or stimulate the SecA ATPase.
Preparation of inverted inner membrane vesicles
BL31(DE3) cells, transformed with a pET610 carrying the his-secYEG operon and grown (LB; overnight; 37°C; 500 ml) were used to inoculate a fermentor (30 liters LB; 100 mg/ml ampicillin; agitation). Protein expression was induced (OD600 = 0.6; 0.2 mM IPTG; 3 h). Cells were collected (5,000 g; 20 min; 4°C; JLA 8.1000 rotor; Beckman Coulter), resuspended in buffer N, and lysed using a French press (8,000 psi; three to five times; Chang et al., 1978). Unbroken cells were removed first (4,000 g; 10 min; Sigma-Aldrich 3-16KL; rotor 11180) and then the supernatant was ultra-centrifuged (95,000 g; 90 min; 4°C; fixed angle 45 Ti rotor; Optima XPN-80; Beckman Coulter). The membrane pellet was resuspended in buffer O using a Dounce-homogenizer, loaded (2 ml) on top of a five-step sucrose gradient (1.9, 1.7, 1.5, 1.3, and 1.1 M sucrose in 50 mM Tris, pH 8.0; 6 ml each layer) and centrifuged (84,000 g; 16 h, 4°C; swinging bucket SW 32 Ti rotor, Optima XPN-80; Beckman Coulter; equilibrium centrifugation). Inverted inner membrane vesicles were collected from gradient fraction 2 (Nikaido, 1994), resuspended in buffer P, and recentrifuged (95,000 g; 90 min; 4°C; fixed angle T647.5 rotor; Sorvall). The membrane pellet was homogenized in 6 M urea, 50 mM Tris-Cl, pH 8.0 (20 min; ice), loaded on top of an equal volume of buffer Q, and centrifuged (95,000 g; T647.5 rotor; 90 min; 4°C; Sorvall). Inverted inner membrane vesicles (Chang et al., 1978; Rhoads et al., 1984; Cunningham and Wickner, 1989; Lill et al., 1989, 1990) were collected and homogenized in buffer R through an Avestin LiposoFast-Basic system (100-nm pore size filter; 15–21 passes), and stored in aliquots at −80°C.
[35S]-labeling of preproteins and derivatives
Preproteins were labeled with 1,000 Ci/mM [35S]-methionine (PerkinElmer) during their in vitro synthesis using TNT Quick coupled Transcription/Translation system (Promega; according to the manufacturer’s instructions). Labeled proteins were separated from the free unincorporated radiolabeled amino acids by centrifugal gel filtration (1 ml homemade G-50 columns, packed in an insulin syringe, and preequilibrated with buffer S) and supplemented with 1 mM DTT.
Preprotein binding to the translocase
0.2 µM SecA derivatives and wild-type SecYEG-inverted inner membrane vesicles (0.2 µM SecY) were mixed and preincubated on ice (10 min; in 20 µl buffer R). Non-labeled preproteins and their derivatives were diluted into these reactions to achieve a range of 0.05 to 10 µM (final urea concentration <0.2 M). [35S]-preproteins or their derivatives were added as a tracer (2 µl) to all reactions, as indicated. Samples were incubated on ice, for 20 min, overlaid to an equal volume of a sucrose cushion (6.85% wt/vol sucrose dissolved in buffer R), and ultracentrifuged (320,000 g; 30 min; 4°C). The membrane-bound material was resuspended in buffer R and immobilized on a nitrocellulose membrane using a vacuum manifold (Bio-Rad Laboratories). [35S]-signals were visualized on a phosphorimager system (Typhoon 9500; GE Healthcare) using a high-resolution phosphor storage screen, quantified using ImageJ 1.48v (Schneider et al., 2012), and then extrapolated to the amount of preprotein derivative bound to the translocase taking into account that, each signal represented x concentration of nonlabeled preprotein (0.05–10 µM) + 2 µl [35S]-preprotein. Bound preprotein to the translocase (y axis) was plotted as a function of preprotein concentration (x axis). Nonlinear regression fitting for one binding site was used. Kd values were determined using Prism 5.0c (GraphPad). No detectable binding of preproteins or derivatives occurs to the SecYEG-inverted inner membrane vesicles in the absence of SecA.
In vitro secretion
100 µl translocation reactions (buffer T), with 0.4 µM SecA or derivatives, SecYEG-IMVs (1 µM SecY), 8 µM preproteins (or as indicated), and 2 mM ATP, was incubated at 37°C (12 min) and then transferred on ice. Disulfide-locked SecA derivatives were tested in the absence of any reducing agent. Nontranslocated preprotein molecules were digested by addition of 1 mg/ml proteinase K (20 min; 4°C). Proteins were precipitated with 15% wt/vol TCA (30 min; 4°C), analyzed by 12% SDS-PAGE (or as indicated) and visualized by immunostaining with α-proPhoA or α-His antisera using the appropriate horseradish peroxidase–coupled secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.). Chemiluminescent signals were derived using the Supersignal West Pico kit (Thermo Fisher Scientific), visualized on a CCD-camera system (LAS-4000; Fujifilm), and quantified with ImageJ 1.48v (Schneider et al., 2012) using a standard curve made from control samples with known amounts of purified protein concentration.
In vivo secretion
MC4100 cells were transformed with pBAD33 derivatives carrying either the proPhoA or proYncJ-PhoA or proYncJ(noMTS)-PhoA genes. The expression of these genes was induced under identical conditions (30°C, OD600 = 0.2; 0.002% wt/vol arabinose; 30 min). Samples were collected, and the PhoA enzymatic activity in the periplasm was measured as described previously (Gouridis et al., 2009). Total protein content was analyzed on SDS-PAGE, the PhoA or PhoA-fusion proteins were immunodetected using α-PhoA antibody, and their amounts were quantified using ImageJ 1.48v. In all cases, the enzymatic activity was normalized per microgram PhoA or fusion-PhoA protein produced. This provided us with the means to quantitate secretion of each protein compared with the wild-type proPhoA.
In vivo genetic complementation
BL21.19 cells (secAts; Mitchell and Oliver, 1993) transformed with either an empty vector or a plasmid carrying wild-type secA or the indicated mutant derivative were grown in LB, at 30°C, until OD600 = 0.5–0.6. Cells were diluted (as indicated), spotted on two identical LB plates that were air-dried for 15 min, and then incubated, one at 30°C and the other at 42°C. Only cells that had a functional secA gene could grow at 42°C.
Binding of SecA to proPhoA peptide arrays
Peptide arrays for the E. coli protein proPhoA and of one derivative missing all hydrophobic segments except that of the signal peptide (Figs. S1 A and S2 B) were prepared by automated spot synthesis (Jerini Peptide Technologies GmbH; Reineke et al., 2001) as 13-mers with 10 residue overlaps, which were C-terminally attached to PEG cellulose membrane via a b-Ala-spacer. Each spot carries ∼5 nmol peptide. Before screening, the dry membrane was immersed in methanol (1 min), then in high-salt buffer (50 mM Trizma-base, pH 8, 6.4 mM KCl, and 170 mM NaCl; RT; 3 × 5 min), next in in equilibration buffer (50 mM Trizma-base, pH 8.0, 50 mM KCl, and 5 mM MgCl2; RT; 5 × 5 min), and finally in 50 ml equilibration buffer (supplemented with 5 g sucrose; 100 mg/ml BSA; 2 mM DTT; 1 mM ADP; 0.01% Tween-20; RT; 15 min). 150–200 nM SecA was allowed to react with the peptide array membrane for 1 h at 30°C before removing the excess with equilibration buffer rinses (3 × 5 min). SecA bound to peptides was electrotransferred to polyvinylidene fluoride (EMD Millipore) using cathode buffer (25 mM Trizma-base, 40 mM aminocaproic, 20% methanol, and 0.01% SDS, pH 9.2) and anode buffers ABI and ABII (ABI = 30 mM Trizma-base, 20% methanol; ABII = 300 mM Trizma-base, 20% vol/vol methanol) using a Trans-Blot SD Semi dry electrophoretic transfer cell (Bio-Rad Laboratories). Bound SecA was detected by α-SecA–specific polyclonal rabbit sera and the Supersignal West Pico kit (Thermo Fisher Scientific) using an imaging CCD system (LAS 4000; GE Healthcare).
Bioinformatics approach to define hydrophobic patches on proteins
The hydrophobicity profile of each protein, based on the Kyte–Doolittle scale, was calculated for a window size of 9, and a linear weight variation model was applied (Wilkins et al., 1999b). Each profile was normalized against the maximum of the scale (i.e., the hydrophobicity value of an isoleucyl-nonapeptide) to reflect the differences in the levels of hydrophobicity for various HPs in each protein. Hydrophobic peaks were then identified within the hydrophobic profiles and selected by maximum hydrophobicity and length. As potential MTSs, we considered those HPs in secretory proteins with a maximum (normalized) hydrophobicity equal or greater than that of the first HP of YncJ (equal to 0.14288) and with a length of at least 4 aa.
Hydrophobicity was sufficient for the initial identification of HPs in all the protein examples tested here (PhoA, HdeA, and YncJ). Bioinformatics analysis failed to detect any sequence conservation among them. For an exhaustive characterization of MTS features in the whole secretome, detailed bioinformatics efforts will be required. The 3D HPs of Lpp (Fig. 1 E) were determined by visual inspection of the helical structure. In all cases, the functional importance of HPs and their role as MTSs were experimentally proven by alanyl or threonyl substitutions causing reduced hydrophobicity at the specified regions. YciG (Uniprot entry P21361), a hydrophilic, polar, and nonfolded polypeptide (negative absorbance band at 200 nm in circular dichroism, characteristic of unstructured proteins), has no measurable affinity for the translocase in the in vitro binding assay.
Choice of cysteine mutants to lock PBD in three different states and the C-tail of SecA on its PatchA
The cysteine pairs that were introduced on SecA with the purpose of locking, through disulfide bridges, PBD at the Closed, Open and Wide open states and the C-tail on PatchA were decided using the following criteria: (a) the mutated residue pairs should be surface exposed, preferentially serines and proximal, within 6Å distance at the relevant, representative state; and (b) the mutations should not affect protein stability or/and SecA enzymatic function under reducing conditions. Proximity of residues was determined by the analysis of available SecA crystal structures at the corresponding states (Protein Data Bank [PDB] accession numbers 3DIN for LC, 2FSF for LO, and 1M6N for LWO and LCt). The cysteine pairs were introduced in an otherwise cysteine-free protein background, and therefore, the observed spontaneous (no addition of oxidizing agent) intraprotomeric disulfide cross-links presented in Fig. S4 are unique to, and demonstrative of, the locking of the specified states. These disulfides are in complete agreement with the available crystal structures. Formation of the disulfide bridges is quantitative. This is verified by the complete conversion of SecA to the slow-migrating oxidized state, which is reversed by DTT addition (Fig. S4 C). As the locked SecA mutants have downstream functional defects, such as compromised translocation ATPase activity that is quantitatively reversed by DTT addition as observed by in vitro ATPase activity assays, they were only used to determine whether the surface each one hindered affects mature domain binding.
Gel-permeation chromatography and multiangle and quasielastic light scattering analysis
Multiangle and quasielastic light scattering experiments were performed online after gel-permeation chromatography on a Superdex HR200 10/300GL that was mounted on an HPLC system (LC10A-VP; Shimadzu) coupled to a photodiode-array detector (SPD-M10AVP; Shimadzu), a multiangle light scattering detector (DAWN-EOS; Wyatt), a quasielastic light scattering detector (Wyatt), and a refractive index detector (RID10A; Shimadzu). Data collection, analysis, and plotting were performed using Astra v.5.0 software from Wyatt. 50–100 µM protein was loaded using a 100-µl injection loop, in buffer L, and chromatographed at 22°C at 0.8 ml/min. Where indicated, urea and/or DTT was included (concentration 0–8 M, as indicated).
Circular dichroism analysis
Experiments were carried on a Jasco J-1500 spectrometer equipped with a Peltier-cooled, six-position cuvette holder (0.3–10 μΜ protein; 20°C; buffer U or as indicated; data pitch, 1 nm; slit, 1 nm; signal averaging time, 1 s; 0.1 mm [Hellma] or 10 mm [Jasco] quartz cuvettes). Urea-purified proteins were incubated with 10 mM DTT (30 min; ice), dialyzed in buffer U supplemented with 5 liters 8M urea (15 h; 4°C). Natively purified proteins were dialyzed in 5 liters buffer U (15 h; 4°C). Samples were centrifuged (20,000 g; 20 min; 4°C) before determining protein concentration. Spectra were recorded (190–260 nm) in buffer U supplemented with 1 mM EDTA, 0.2 M urea, and DTT as indicated at 5 min, 1 h, or 24 h after the chaotrope dilution (n = 5). Data analysis was performed using the Spectra Manager v.2 software (Jasco).
Native nano–electrospray ionization mass spectrometric analysis
Native PhoA in a stock solution of buffer L and translocation-competent PhoA in a stock solution with buffer F (without glycerol) were exchanged in 100 mM ammonium acetate, pH 6.9, on a spin column (Bio-Spin Columns P-6; Bio-Rad Laboratories). The samples were further diluted in 100 mM ammonium acetate, pH 6.9, to a final protein concentration of 5 µM and introduced into the gas phase using nano–electrospray ionization with in-house–prepared gold-coated borosilicate glass capillaries at an ionization voltage of +1.6 kV. All spectra were recorded on a traveling-wave quadruple-time-of-flight instrument (Synapt G2; Waters). Critical voltages were a sampling cone of 25 V, extraction cone of 0 V, trap collision energy 4 V, transfer collision energy 0 V, and bias 45 V. Gas pressures throughout the instrument were 2.6, 1.77 × 10–2, and 1.82 × 10–2 mbar for the source region, trap, and transfer cell, respectively. Spectra, recorded for either protein instantly upon buffer exchange and 3 h after buffer exchange were identical. For data analysis the MassLynX interface (version 4.1 SCN870; Waters) was used.
Statistical analysis
In all cases, values of independent experimental replicates were analyzed by Prism 5.0c (GraphPad) for the determination of mean values and error, expressed as SEM or SD as indicated.
Calculation of hydrodynamic radii based on solved structures
160 out of the 505 secretory proteins with known structures (via crystallization or nuclear magnetic resonance) were selected via the corresponding information downloaded from UniProt (Dimmer et al., 2012). PDB files were downloaded from the RCSB database from which 82 are solved as homomultimers or in complex with other proteins (e.g., PhoA is solved as a homodimer). PyMol was used to remove redundant polypeptide chains. For each protein, the radius of gyration was calculated using WinHydroPRO v1.0 (Table S8; Ortega et al., 2011). For each protein, the molecular mass was specified, “radius of atomic elements” and temperature set to default (2.84 Å and 20°C, respectively), and specific volume set to 1.
Analysis of protein structures
Protein structures were modeled with Swiss PDB viewer and PyMol/MacPyMol software. Structures with PDB codes were downloaded from RSCB, PDB (http://www.rcsb.org/). Models were generated with maximum quality, anti-alias set at 2, ray trace mode 1 and ray-traced at 1,500, 1,500. Adobe Photoshop was used to add structural elements for visualization purposes. FASTA protein sequences were downloaded from http://www.ncbi.nlm.nih.gov/protein. ClustalW (Larkin et al., 2007) was used for sequence alignments (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Structural alignment of proteins was performed with the Swiss PDB viewer (Guex and Peitsch, 1997).
Online supplemental material
Fig. S1 shows hydrophobicity plots for the detection of linear MTSs in preproteins and the structure of Lpp with 3D MTSs. Fig. S2 details the SecA-binding sites on proPhoA and proPhoA(noMTS) using peptide arrays and shows a collective map of the proPhoA recognition by SecA and chaperones. Fig. S3 shows the hydrodynamic properties of proPhoA and PhoA at the targeting-competent and native states, using SEC-MALS/QELS, circular dichroism and native electrospray ionization mass spectrometry. Fig. S4 describes the domains of SecA, the motions of the SecA PDB domain, and the experimental proof of the intraprotomeric SecA locked mutants. Fig. S5 details the hydrophobic patches on the SecA cytoplasmic platform and the conservation of PatchA among species and summarizes known interactions of and allosteric effects on PatchA. Fig. S5 shows the properties of the SecA surface and details the interaction of SecA with signal peptides and the SecA C-tail. The vicinity but independence of the SecA signal peptide–binding cleft and PatchA is explained with structural insights. Table S1 summarizes preproteins with weak or no detectable linear hydrophobic patches. Buffers, host strains, cloning vectors, synthetic genes, genetic constructs, and primers used in this study are listed in Tables S2, S3, S4, S5, S6, and S7, respectively. Table S8 provides the predicted hydrodynamic radii of secretory proteins that use the Sec secretion system. Table S9 provides the sequences of proPhoA peptides used in the peptide arrays shown in Figs. 1 C and S2 A.
Supplementary Material
Acknowledgments
We thank G. Gouridis for useful discussions, T. Saio for the unfolded PhoA model, and G. Orfanoudaki for help with the hydrodynamic diameters and definition of hydrophobic patches.
Our research was funded by KUL-Spa (Onderzoekstoelagen 2013; Bijzonder Onderzoeksfonds; Katholieke Universiteit Leuven), RiMembR (Vlaanderen Onderzoeksprojecten; Fonds Wetenschappelijk Onderzoek grant G0C6814N), and StrepSynth (Seventh Framwork Programme FP7 KBBE.2013.3.6-02: Synthetic Biology toward applications; grant 613877; to A. Economou) and Fonds Wetenschappelijk Onderzoek grant G0B4915N (to S. Karamanou).
The authors declare no competing financial interests.
Author contributions: K.E. Chatzi and M.F. Sardis engineered preprotein derivatives, measured their affinity for the wild-type translocase, and performed in vitro and in vivo secretion assays. M. Koukaki prepared genetic constructs and set up the disulfide locking assay for PBD. M.F. Sardis engineered and isolated the SecA mutant derivatives, monitored their functionality for preprotein binding and secretion, and performed in silico calculations of preprotein hydrodynamic diameters. N. Šoštarić contributed to in vivo genetic complementation. S. Karamanou performed binding assays on peptide arrays and biophysical analysis of proPhoA and PhoA and supervised biochemical and biophysical assays. A. Tsirigotaki contributed to the biophysical and structural analysis. A. Konijnenberg performed native nano–electrospray ionization mass spectrometry with contributions from A. Tsirigotaki and F. Sobott. C.G. Kalodimos contributed the unfolded PhoA model. A. Tsirigotaki, S. Karamanou, and A. Economou wrote the paper with contributions from K.E. Chatzi and M.F. Sardis. S. Karamanou and A. Economou conceived and managed the project. All authors reviewed the manuscript.
Footnotes
Abbreviations used:
- DH
- hydrodynamic diameter
- HP
- hydrophobic patch
- Kd
- dissociation constant
- LC
- locked closed
- LO
- locked open
- LWO
- locked wide open
- MTS
- mature domain targeting signal
- PBD
- preprotein-binding domain
References
- Bauer B.W., and Rapoport T.A.. 2009. Mapping polypeptide interactions of the SecA ATPase during translocation. Proc. Natl. Acad. Sci. USA. 106:20800–20805. 10.1073/pnas.0910550106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith J. 2013. The Sec-dependent pathway. Res. Microbiol. 164:497–504. 10.1016/j.resmic.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerten J., Jonckheere W., Rudyak S., Xu J., Wilkinson H., De Smet F., Schymkowitz J., and Rousseau F.. 2012. Aggregation gatekeepers modulate protein homeostasis of aggregating sequences and affect bacterial fitness. Protein Eng. Des. Sel. 25:357–366. 10.1093/protein/gzs031 [DOI] [PubMed] [Google Scholar]
- Bieker K.L., Phillips G.J., and Silhavy T.J.. 1990. The sec and prl genes of Escherichia coli. J. Bioenerg. Biomembr. 22:291–310. 10.1007/BF00763169 [DOI] [PubMed] [Google Scholar]
- Blobel G., and Dobberstein B.. 1975. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67:835–851. 10.1083/jcb.67.3.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Chang C.N., Blobel G., and Model P.. 1978. Detection of prokaryotic signal peptidase in an Escherichia coli membrane fraction: Endoproteolytic cleavage of nascent f1 pre-coat protein. Proc. Natl. Acad. Sci. USA. 75:361–365. 10.1073/pnas.75.1.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzi K.E., Sardis M.F., Karamanou S., and Economou A.. 2013. Breaking on through to the other side: Protein export through the bacterial Sec system. Biochem. J. 449:25–37. 10.1042/BJ20121227 [DOI] [PubMed] [Google Scholar]
- Chatzi K.E., Sardis M.F., Economou A., and Karamanou S.. 2014. SecA-mediated targeting and translocation of secretory proteins. Biochim. Biophys. Acta. 1843:1466–1474. 10.1016/j.bbamcr.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Chen Y., Bauer B.W., Rapoport T.A., and Gumbart J.C.. 2015. Conformational changes of the clamp of the protein translocation ATPase SecA. J. Mol. Biol. 427:2348–2359. 10.1016/j.jmb.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K., and Wickner W.T.. 1989. Detergent disruption of bacterial inner membranes and recovery of protein translocation activity. Proc. Natl. Acad. Sci. USA. 86:8673–8677. 10.1073/pnas.86.22.8673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter J., Tsirigotaki A., Orfanoudaki G., Zorzini V., Economou A., and Karamanou S.. 2016. Protein folding in the cell envelope of Escherichia coli. Nat. Microbiol. 1:16107 10.1038/nmicrobiol.2016.107 [DOI] [PubMed] [Google Scholar]
- Derman A.I., Puziss J.W., Bassford P.J. Jr., and Beckwith J.. 1993. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 12:879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer E.C., Huntley R.P., Alam-Faruque Y., Sawford T., O’Donovan C., Martin M.J., Bely B., Browne P., Mun Chan W., Eberhardt R., et al. . 2012. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 40(D1):D565–D570. 10.1093/nar/gkr1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García De La Torre J., Huertas M.L., and Carrasco B.. 2000. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 78:719–730. 10.1016/S0006-3495(00)76630-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelis I., Bonvin A.M., Keramisanou D., Koukaki M., Gouridis G., Karamanou S., Economou A., and Kalodimos C.G.. 2007. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 131:756–769. 10.1016/j.cell.2007.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouridis G., Karamanou S., Gelis I., Kalodimos C.G., and Economou A.. 2009. Signal peptides are allosteric activators of the protein translocase. Nature. 462:363–367. 10.1038/nature08559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouridis G., Karamanou S., Koukaki M., and Economou A.. 2010. In vitro assays to analyze translocation of the model secretory preprotein alkaline phosphatase. Methods Mol. Biol. 619:157–172. 10.1007/978-1-60327-412-8_10 [DOI] [PubMed] [Google Scholar]
- Gouridis G., Karamanou S., Sardis M.F., Schärer M.A., Capitani G., and Economou A.. 2013. Quaternary dynamics of the SecA motor drive translocase catalysis. Mol. Cell. 52:655–666. 10.1016/j.molcel.2013.10.036 [DOI] [PubMed] [Google Scholar]
- Grudnik P., Bange G., and Sinning I.. 2009. Protein targeting by the signal recognition particle. Biol. Chem. 390:775–782. 10.1515/BC.2009.102 [DOI] [PubMed] [Google Scholar]
- Guex N., and Peitsch M.C.. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 18:2714–2723. 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Hegde R.S., and Bernstein H.D.. 2006. The surprising complexity of signal sequences. Trends Biochem. Sci. 31:563–571. 10.1016/j.tibs.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Huang C., Rossi P., Saio T., and Kalodimos C.G.. 2016. Structural basis for the antifolding activity of a molecular chaperone. Nature. 537:202–206. 10.1038/nature18965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt J.F., Weinkauf S., Henry L., Fak J.J., McNicholas P., Oliver D.B., and Deisenhofer J.. 2002. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 297:2018–2026. 10.1126/science.1074424 [DOI] [PubMed] [Google Scholar]
- Karamanou S., Bariami V., Papanikou E., Kalodimos C.G., and Economou A.. 2008. Assembly of the translocase motor onto the preprotein-conducting channel. Mol. Microbiol. 70:311–322. 10.1111/j.1365-2958.2008.06402.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. . 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lecker S.H., Driessen A.J., and Wickner W.. 1990. ProOmpA contains secondary and tertiary structure prior to translocation and is shielded from aggregation by association with SecB protein. EMBO J. 9:2309–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Loir Y., Nouaille S., Commissaire J., Brétigny L., Gruss A., and Langella P.. 2001. Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 67:4119–4127. 10.1128/AEM.67.9.4119-4127.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Cunningham K., Brundage L.A., Ito K., Oliver D., and Wickner W.. 1989. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 8:961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Dowhan W., and Wickner W.. 1990. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 60:271–280. 10.1016/0092-8674(90)90742-W [DOI] [PubMed] [Google Scholar]
- Martoglio B., and Dobberstein B.. 1998. Signal sequences: More than just greasy peptides. Trends Cell Biol. 8:410–415. 10.1016/S0962-8924(98)01360-9 [DOI] [PubMed] [Google Scholar]
- Michaelis S., Inouye H., Oliver D., and Beckwith J.. 1983. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J. Bacteriol. 154:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C., and Oliver D.. 1993. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol. Microbiol. 10:483–497. 10.1111/j.1365-2958.1993.tb00921.x [DOI] [PubMed] [Google Scholar]
- Nikaido H. 1994. Isolation of outer membranes. Methods Enzymol. 235:225–234. 10.1016/0076-6879(94)35143-0 [DOI] [PubMed] [Google Scholar]
- Orfanoudaki G., and Economou A.. 2014. Proteome-wide subcellular topologies of E. coli polypeptides database (STEPdb). Mol. Cell. Proteomics. 13:3674–3687. 10.1074/mcp.O114.041137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega A., Amorós D., and García de la Torre J.. 2011. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 101:892–898. 10.1016/j.bpj.2011.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolau Y., Papadovasilaki M., Ravelli R.B., McCarthy A.A., Cusack S., Economou A., and Petratos K.. 2007. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J. Mol. Biol. 366:1545–1557. 10.1016/j.jmb.2006.12.049 [DOI] [PubMed] [Google Scholar]
- Papanikou E., Karamanou S., Baud C., Frank M., Sianidis G., Keramisanou D., Kalodimos C.G., Kuhn A., and Economou A.. 2005. Identification of the preprotein binding domain of SecA. J. Biol. Chem. 280:43209–43217. 10.1074/jbc.M509990200 [DOI] [PubMed] [Google Scholar]
- Park E., and Rapoport T.A.. 2012. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41:21–40. 10.1146/annurev-biophys-050511-102312 [DOI] [PubMed] [Google Scholar]
- Reineke U., Volkmer-Engert R., and Schneider-Mergener J.. 2001. Applications of peptide arrays prepared by the SPOT-technology. Curr. Opin. Biotechnol. 12:59–64. 10.1016/S0958-1669(00)00178-6 [DOI] [PubMed] [Google Scholar]
- Rhoads D.B., Tai P.C., and Davis B.D.. 1984. Energy-requiring translocation of the OmpA protein and alkaline phosphatase of Escherichia coli into inner membrane vesicles. J. Bacteriol. 159:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos T., Kiefer D., Hugenschmidt S., Economou A., and Kuhn A.. 2001. Indecisive M13 procoat protein mutants bind to SecA but do not activate the translocation ATPase. J. Biol. Chem. 276:37909–37915. [DOI] [PubMed] [Google Scholar]
- Saio T., Guan X., Rossi P., Economou A., and Kalodimos C.G.. 2014. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science. 344:1250494 10.1126/science.1250494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierle C.F., Berkmen M., Huber D., Kumamoto C., Boyd D., and Beckwith J.. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185:5706–5713. 10.1128/JB.185.19.5706-5713.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T., Nureki O., Nakamura A., Kobayashi S., and Yokoyama S.. 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 125:287–300. 10.1016/j.cell.2006.01.054 [DOI] [PubMed] [Google Scholar]
- Shu W., Liu J., Ji H., and Lu M.. 2000. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 A resolution. J. Mol. Biol. 299:1101–1112. 10.1006/jmbi.2000.3776 [DOI] [PubMed] [Google Scholar]
- Stoscheck C.M. 1990. Quantitation of protein. Methods Enzymol. 182:50–68. 10.1016/0076-6879(90)82008-P [DOI] [PubMed] [Google Scholar]
- Testa L., Brocca S., Santambrogio C., D’Urzo A., Habchi J., Longhi S., Uversky V.N., and Grandori R.. 2013. Extracting structural information from charge-state distributions of intrinsically disordered proteins by non-denaturing electrospray-ionization mass spectrometry. Intrinsically Disord. Proteins. 1:e25068 10.4161/idp.25068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigotaki A., De Geyter J., Šoštaric N., Economou A., and Karamanou S.. 2017. Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 15:21–36. 10.1038/nrmicro.2016.161 [DOI] [PubMed] [Google Scholar]
- Uversky V.N. 2002. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 11:739–756. 10.1110/ps.4210102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D.K., Grimshaw S.B., Receveur V., Dobson C.M., Jones J.A., and Smith L.J.. 1999a Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 38:16424–16431. 10.1021/bi991765q [DOI] [PubMed] [Google Scholar]
- Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., and Hochstrasser D.F.. 1999b Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112:531–552. [DOI] [PubMed] [Google Scholar]
- Zimmer J., and Rapoport T.A.. 2009. Conformational flexibility and peptide interaction of the translocation ATPase SecA. J. Mol. Biol. 394:606–612. 10.1016/j.jmb.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J., Nam Y., and Rapoport T.A.. 2008. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 455:936–943. 10.1038/nature07335 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.